Abstract
The human MC1R gene is highly polymorphic among lightly pigmented populations, and several variants in the MC1R gene have been associated with increased risk of both melanoma and non-melanoma skin cancers. The functional consequences of MC1R gene variants have been studied in vitro and in vivo in postulated causal pathways, such as G-protein-coupled signaling transduction, pigmentation, immune response, inflammatory response, cell proliferation, and extracellular matrix adhesion. In a case-control study nested within the Nurses’ Health Study, we utilized hierarchical modeling approaches, incorporating the quantitative information from these functional studies, to examine the association between particular MC1R alleles and the risk of skin cancers. Different prior matrices were constructed according to the phenotypic associations in controls, cell surface expression, and enzymatic kinetics. Our results showed the parameter variance estimates of each SNP were smaller when using a hierarchical modeling approach compared to standard multivariable regression. Estimates of second-level parameters gave information about the relative importance of MC1R effects on different pathways, and odds ratio estimates changed depending on prior models (for example, the change ranged from −21% to 7% for melanoma risk assessment). In addition, the estimates of prior model hyper-parameters in the hierarchical modeling approach allow us to determine the relevance of individual pathways on the risk of each of the skin cancer types. In conclusion, hierarchical modeling provides a useful analytic approach in addition to the widely used conventional models in genetic association studies that can incorporate measures of allelic function.
Keywords: hierarchical modeling, MC1R, functional variants, pigmentation, skin cancer
Introduction
Skin cancer is the most common form of cancer in the United States and accounts for more than 1 million new cases per year, including approximately 87,000 cases of cutaneous malignant melanoma (hereafter referred to simply as melanoma) [2017; Howe, et al. 2001; Siegel, et al. 2017]. There are three major types of skin cancer, with melanoma being the most fatal; the most common types of non-melanoma skin cancer are basal cell carcinoma (BCC) followed by squamous cell carcinoma (SCC). Ultra-violet (UV) radiation exposure is an important risk factor for skin cancer, and dark pigmentation is an important inherited factor protecting against UV-induced skin cancer. Pigmentary melanin is synthesized in melanocytes and secreted into keratinocytes. Brown/black eumelanin absorbs UV and neutralizes free radicals to protect skin from UV damage, whereas yellow/red phaeomelanin generates free radicals in response to UV. Human pigmentation is an inherited trait partially determined by the melanocortin 1 receptor (MC1R) gene, located at 16q24.3 [Gantz, et al. 1994]. It encodes a 317-amino acid seven-pass-transmembrane G protein-coupled receptor. When activated by α-melanocyte-stimulating hormone (α-MSH) or adrenocorticotrophic hormone (ACTH), MC1R activates adenylate cyclase, thereby elevating intracellular cyclic adenosine monophosphate (cAMP). MC1R was shown to be the limiting factor controlling the output of the cAMP signaling pathway in heterologous cells expressing the wild-type MC1R gene [Mas, et al. 2003]. This signaling induces the maturation of the phenomelanosome to eumelanosome, resulting in darker pigmentation [Rees 2003; Sturm, et al. 2001].
The human MC1R gene is highly polymorphic among lightly pigmented populations [Harding, et al. 2000; Sturm 2002]. Among more than 80 nonsynonymous variants identified to date, Arg151Cys, Arg160Trp, and Asp294His were associated with red-hair phenotype [Beaumont, et al. 2005; Box, et al. 1997; Valverde, et al. 1995] and are known as “red hair color” (RHC) variants; other variants with weaker association are referred to as “non-red hair color” (NRHC) variants. Several variants in the MC1R gene have been associated with increased risks for melanoma and non-melanoma skin cancers after pigmentation phenotype was taken into account [Bastiaens, et al. 2001; Box, et al. 2001; Kennedy, et al. 2001; Landi, et al. 2005; Matichard, et al. 2004; Palmer, et al. 2000]. Previous studies grouped RHC alleles and NRHC alleles in tests of genetic associations between the MC1R alleles and risk of skin cancer [Han, et al. 2006; Landi, et al. 2005; Matichard, et al. 2004]. Distinct functions of specific MC1R variants in relation to pigmentation, immune response, inflammatory response, cell proliferation, and extracellular matrix adhesion pathways have been examined using data on pigmentation phenotypes or quantitative in vitro assays.
In this study, we utilized this knowledge regarding the role of MC1R alleles in biological processes potentially relevant to skin cancers to improve the accuracy of the estimated effects of MC1R variants on predisposition to skin cancer. Using a nested case-control design within the Nurses’ Health Study, we explored hierarchical modeling analyses, incorporating quantitative information on the function of particular MC1R alleles [Aragaki, et al. 1997; Brenner, et al. 2013; Greenland 2000; Hung, et al. 2004; Witte 1997]. We evaluated the associations of seven common MC1R variants (Val60Leu, Val92Met, Arg151Cys, Ile155Thr, Arg160Trp, Arg163Gln, and Asp294His) with three major types of skin cancer (melanoma, SCC, and BCC) in women of European ancestry. Hierarchical regression approach allows the integration of biological data, reduces the overall estimation error by shrinking the estimated coefficients to their mean, and at the same time protects against over-parameterization of the model.
Methods
Study Population
The Nurses’ Health Study was established in 1976, when 121,700 female registered nurses between the ages of 30 and 55 completed a self-administered questionnaire on their medical histories and baseline health-related exposures. Updated information has been obtained by questionnaires every 2 years since then. Between 1989 and 1990, blood samples were collected from 32,826 of the cohort members. Detailed methods of this nested case-control study were described previously [Han, et al. 2006]. The distributions of risk factors for skin cancer in the subcohort of those who donated blood samples were very similar to those in the overall cohort. Briefly, eligible cases in this study consisted of women with incident skin cancer from the subcohort who had given a blood specimen, including cases of SCC and BCC with a diagnosis any time after blood collection up to June 1, 1998, and cases of melanoma up to June 1, 2000, with no previously diagnosed skin cancer. A common control series was randomly selected from participants who gave a blood sample and were free of diagnosed skin cancer up to and including the questionnaire cycle in which the case was diagnosed. One or two controls were matched to each case by year of birth (±1 year) and self-reported race (Caucasian/missing or others). Fewer than 5% of cases and controls had missing or other race/ethnicity. The nested case-control study consisted of 219 melanoma cases, 286 SCC cases, 300 BCC cases, and 874 matched controls. We restricted analysis to 197 melanoma cases, 264 SCC cases, 263 BCC cases, and 791 controls with complete genotype information. The study protocol was approved by the Committee on Use of Human Subjects of Brigham and Women’s Hospital, Boston, MA.
Information regarding skin cancer risk factors was obtained from the prospective biennial questionnaires and the retrospective supplementary questionnaire. Information on natural hair color and childhood and adolescent tendency to tan was solicited in the 1982 prospective questionnaire. To avoid potential population stratification, we excluded one Asian melanoma case and one control, one Hispanic SCC case and two controls.
Single Nucleotide Polymorphism (SNP) Identification
The distribution and frequency of MC1R variants in 179 Caucasian controls from the US were determined by Kanetsky et al. using a direct-sequencing method [Kanetsky, et al. 2004]. Seven non-synonymous polymorphisms with allele frequency >1% were identified (Val60Leu, Val92Met, Arg151Cys, Ile155Thr, Arg160Trp, Arg163Gln, and Asp294His) in the coding region. We genotyped these variants in our case-control study. There are comprehensive and quantitative in vitro assays on the function of these particular variants, which we incorporated into hierarchical modeling analyses. We did not genotype the three variants (86insA, Asp84Glu, and Arg142His) with allele frequency ≤ 1% [Han, et al. 2004].
Laboratory Assays
The Arg160Trp polymorphism was genotyped by restriction fragment length polymorphism (RFLP), and the other polymorphisms were genotyped by the 5’-nuclease assay (TaqMan®) in 384-well format, using the ABI PRISM 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA). TaqMan® primers and probes were designed using the Primer Express® Oligo Design software v2.0 (ABI PRISM). Laboratory personnel were blinded to case-control status, and blinded quality-control samples were inserted to validate genotyping procedures; concordance for the blinded samples was 100%. Primers, probes, and conditions for genotyping assays are available upon request.
Statistical Analyses
Hierarchical modeling outline
We utilized a two-level hierarchical modeling approach that incorporates functional information in a second-level (prior) model for regression parameters. Specifically, it assumes the log-odds ratio parameters β from the first-level logistic regression of disease risk on MC1R variants are themselves independent random variables with means Z π and variance τ2. The first level of the regression models the log odds of disease on the seven MC1R SNPs, X, and the vector of covariates, W.
The second level is a linear model on which the first-level regression parameter β is regressed on a matrix obtained using functional information, Z, and this helps improve the estimation of β.
Z is a user-specified prior design matrix, and the dimension of the second-level parameter vector, , can be much smaller than the dimension of β. For example, if β is a vector of seven SNP log odds ratios, Z might be a vector of seven ones, indicating that the average effect for each of the modeled SNPs is the same (namely ). By treating β as a random variable, this approach incorporates the uncertainty in this prior structure (e.g., prediction error in the second-level covariates in Z). The residuals in the second-level regression, δ, are assumed to have a normal distribution, with a mean 0 and variance τ2.
We coded each variant using a dominant model and fit all variants simultaneously. We used dominant models because the MC1R variants being studied follow a dominant inheritance pattern. Hierarchical analyses can be implemented using the %glimmix SAS macro [Witte, et al. 1998; Witte, et al. 2000], in which the two levels of the hierarchical regression are combined into a mixed model with fixed coefficients (π, and γ) and random coefficient (δ). By substituting the value of β from the second-level model into the first-level model, we get the following mixed-model equation:
Therefore, in addition to the SNPs X and the covariates W, we also included the product of X and Z matrices (XZ) in the mixed model, and their fixed effect coefficients can be interpreted as π.
In this analysis, we used a Semi-Bayes approach and pre-specified the common variance of the random coefficient vector δ, for effect sizes to be consistent with those found in previous studies of genetic variants. Rather than allowing τ to be estimated empirically, we fixed the value of τ2 at 0.1 [Greenland 1994; Witte, et al. 1994]. At this value of τ, we assume with 95% confidence that the variation in the true odds ratios for SNPs, after adjusting for the effects of the attributes considered in the Z matrix, will fall within a 1.5-fold range, i.e., (exp(0.1×3.92) ≈ 1.5).
Construction of prior design matrix
The prior design matrix Z is a critical component of the hierarchical approach. We considered several prior design matrices corresponding to different working hypotheses about the causal role of different MC1R variants. First, we simply assumed that all variants were exchangeable, i.e., they have the same average effect. This corresponds to the Z matrix described in the previous section. We then defined several prior matrices according to the functional parameters generated from phenotypic correlations and in vitro assays (Table 1). The functional parameters for model 4–9 in Table 2 were obtained from published literature [Beaumont, et al. 2005; Sanchez-Mas, et al. 2004] and not based on our samples in the NHS.
Table 1.
Hierarchical modeling of MC1R variants
| Model | Wildtype | Val60Leu | Val92Met | Arg151Cys | Ile155Thr | Arg160Trp | Arg163Gln | Asp294His | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Assumes that all variants have the same effects | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 2 | Regression coefficient with hair color in controls | 0 | −0.020 | −0.092 | 0.368 | 0.195 | 0.539 | 0.028 | 0.605 |
| 3 | Regression coefficient with skin color in controls | 0 | 0.149 | 0.083 | 0.310 | 0.140 | 0.250 | 0.026 | 0.428 |
| 4 | Regression coefficient with tanning ability in controls | 0 | 0.317 | 0.259 | 0.625 | 0.477 | 0.453 | 0.136 | 0.899 |
| 5 | Cell surface MC1R antibody binding | 0 | 0.772 | 0.004 | 1.505 | 1.911 | 1.537 | 0.411 | −0.378 |
| 6 | Cell surface MSH ligand binding | 0 | 1.234 | 0.126 | 1.966 | 3.808 | 2.137 | 0.648 | −0.466 |
| 7 | Kinetics, α-MSH Ki | 0 | 0.276 | 3.191 | −0.034 | 1,878 | 0.259 | 1.563 | 1.887 |
| 8 | Kinetics, NDP Ki | 0 | −0.084 | 0.069 | 0.808 | 0.313 | 0.425 | 0.468 | −0.166 |
| 9 | Maximum cAMP generation in response to MSH | 0 | 1.006 | 0.211 | 1.634 | 0.483 | 1.516 | −0.401 | 2.088 |
Parameters for models 5–9 were log transformed.
Table 2.
Estimates of prior-model hyper-parameters: univariate second-level models
| Model | Melanoma | SCC | BCC | ||||
|---|---|---|---|---|---|---|---|
| π0 est (SD) | π1 est (SD) | π0 est (SD) | π1 est (SD) | π0 est (SD) | π1 est (SD) | ||
| 1 | Fixed mean | 0.63 (0.18) | − | 0.46 (0.17) | − | 0.38 (0.17) | − |
| 2 | Hair color | 0.52 (0.22) | 0.56 (0.58) | 0.33 (0.21) | 0.61 (0.58) | 0.30 (0.21) | 0.38 (0.58) |
| 3 | Skin color | 0.37 (0.30) | 1.34 (1.24) | 0.22 (0.29) | 1.23 (1.25) | 0.26 (0.29) | 0.61 (1.22) |
| 4 | Tanning ability | 0.29 (0.35) | 0.79 (0.69) | 0.20 (0.34) | 0.59 (0.70) | 0.34 (0.33) | 0.10 (0.68) |
| 5 | Cell surface MC1R antibody binding | 0.62 (0.24) | 0.01 (0.21) | 0.29 (0.24) | 0.21 (0.20) | 0.36 (0.23) | 0.02 (0.20) |
| 6 | Cell surface MSH ligand binding | 0.64 (0.24) | −0.003 (0.13) | 0.32 (0.24) | 0.11 (0.13) | 0.45 (0.24) | −0.06 (0.14) |
| 7 | Kinetics, α-MSH Ki | 0.67 (0.24) | −0.03 (0.13) | 0.61 (0.23) | −0.13 (0.13) | 0.57 (0.23) | −0.16 (0.13) |
| 8 | Kinetics, NDP Ki | 0.58 (0.22) | 0.22 (0.47) | 0.34 (0.21) | 0.41 (0.46) | 0.23 (0.21) | 0.53 (0.46) |
| 9 | Maximum cAMP generation | 0.48 (0.26) | 0.16 (0.19) | 0.27 (0.25) | 0.20 (0.19) | 0.31 (0.24) | 0.08 (0.19) |
We performed multiple linear regression analyses among controls to evaluate the associations between the phenotypes and the MC1R variants (Table 1, models 2–4). We treated hair color (red, blond, light brown, dark brown or black), skin color (fair, medium, olive), and tanning ability (deep tan, average tan, light tan, practically none) as ordinal outcome variables. The three RHC alleles (Arg151Cys, Arg160Trp, and Asp294His) had a stronger association with skin color and hair color compared with other variant alleles. For tanning ability, the three RHC alleles and 155Thr allele showed strong associations. The variant-specific estimated regression coefficients zi from these analyses were then used in the second-level model, so that the mean log-odds ratio for variant i was π0 + π1 zi.
MC1R polymorphisms are also associated with a change in receptor localization. cAMP production depends on the number of surface MC1R sites per cell, and a lower receptor number would result in decreased production of cAMP and downstream signaling [Sanchez-Mas, et al. 2004]. Beaumont et al. [Beaumont, et al. 2005] quantified cell-surface MC1R antibody binding detected by immuno-fluorescence in transient cell expression of variant alleles. We used the log-transformed fold decrease relative to the wildtype allele in construction of Z matrix (Table 1, model 5). Cell-surface expression of MC1R variant alleles in relation to the wildtype was also quantified by binding of the radio-labeled MC1R ligand 125I-NDP-MSH [Beaumont, et al. 2005]. We used the log-transformed fold decrease in maximum binding values of the variant alleles relative to the wildtype allele (Table 1, model 6).
Reduced MC1R coupling activity is also an important factor contributing to the genetic association between the MC1R variants and the RHC phenotype. Pharmacological characterization of common MC1R variants was conducted previously [Ringholm, et al. 2004; Schioth, et al. 1999]. The log-transformed dissociation constants from competition studies (Ki) using α-MSH and NDP-MSH are listed in Table 1 (models 7–8). The Ki is the concentration of the competing ligand that binds to half of the binding sites at equilibrium in the absence of radioligand or other competitors. If the Ki is low, the affinity of the receptor for the inhibitor is high. The differential binding to MC1R between the α-MSH and NDP-MSH is related to the general binding characteristics of these peptides. The ability of the variant alleles to increase the production of cAMP after stimulation with α-MSH was measured [Ringholm, et al. 2004; Schioth, et al. 1999]. We listed the log-transformed fold decrease in the maximum production by the MC1R variant alleles relative to that of the wildtype in transfection assays (Table 1, model 9). In this signaling transduction process, the downstream signaling from cAMP production influences the ratio of phenomelanosome to eumelanosome. The cAMP production is most reduced for three RHC alleles, Arg151Cys, Arg160Trp, and Asp294His, which is in good agreement with their association with lighter skin color and hair color.
Finally, we considered a multivariable second-level model. We selected three representative measures, one for each “pathway”: hair color as pigmentary phenotype, cell surface MSH ligand binding as cell surface expression, and kinetics (Ki) using α-MSH as MC1R coupling activity. In this case, the mean log odds ratio for variant i is π1 + π2 ∑ z2i + π3 ∑ z3i + π4 ∑ z4i, where zhi are the variant-specific coefficients for functional measure h.
Results
The genotype distributions of the seven non-synonymous polymorphisms did not show departure from Hardy-Weinberg equilibrium among controls. The minor allele frequency of the seven polymorphisms among the controls was 12.7% for Val60Leu, 9.8% for Val92Met, 7.0% for Arg151Cys, 1.3% for Ile155Thr, 7.8% for Arg160Trp, 4.0% for Arg163Gln, and 1.6% for Asp294His, which were compatible with the previous report on 179 US Caucasian controls [Han, et al. 2004]. Overall, based upon the seven polymorphisms genotyped, 31% of the controls were homozygous for the consensus allele. Half of the controls carried one variant allele; 32% carried one NRHC allele and 18% carried one RHC allele. 20% of the controls carried two variant alleles; 9% carried two NRHC alleles, 8% carried one NRHC allele and one RHC allele, and 3% carried two RHC alleles.
The fixed-effect parameter (π0 and π1) estimates are listed in Table 2. The results of the genetic main effect of each variant on melanoma risk are presented in Table 3 and Figures 1–6, which compare the conventional and hierarchical analyses. In the conventional analysis, five of the seven common variants included in the analysis (except 155Thr and 163Gln) were associated with a significantly increased risk of melanoma. All genetic factors were first assumed to be exchangeable in the hierarchical model (model 1). We then pre-specified the prior second-level covariates according to regression coefficients with phenotypes (models 2–4), MC1R cell-surface expression (models 5–6), and parameters of in vitro pharmacologic assays (models 7–9). We observed that the CIs from hierarchical modeling were narrower than those from conventional estimates, i.e., the precision of the risk estimates was enhanced. The change of ORs ranged from −21% to 7%. As was observed by Hung et al. [Hung, et al. 2004], the shrinkage of estimates was not always towards the null, but toward the prior means.
Table 3.
Hierarchical modeling of associations between MC1R variants and skin cancer risk
| Val60Leu | Val92Met | Arg151Cys | Ile155Thr | Arg160Trp | Arg163Gln | Asp294His | ||
|---|---|---|---|---|---|---|---|---|
| Melanoma | Conventional model | 1.52 (1.04 – 2.24) |
1.64 (1.10 – 2.45) |
2.47 (1.63 – 3.75) |
1.87 (0.80 – 4.40) |
1.79 (1.15 – 2.80) |
1.71 (0.96 −3.04) |
2.67 (1.35 – 5.31) |
| Model 1 | 1.60 (1.13 – 2.28) |
1.69 (1.18 – 2.44) |
2.30 (1.58 – 3.36) |
1.88 (1.07 – 3.31) |
1.82 (1.22 – 2.70) |
1.78 (1.12 – 2.84) |
2.22 (1.32 – 3.72) |
|
| Model 2 | 1.55 (1.09 – 2.23) |
1.63 (1.12 – 2.37) |
2.35 (1.61 – 3.44) |
1.87 (1.06 – 3.44) |
1.92 (1.27 – 2.88) |
1.71 (1.06 – 2.75) |
2.50 (1.42 – 4.41) |
|
| Model 3 | 1.58 (1.11 – 2.25) |
1.63 (1.13 – 2.37) |
2.39 (1.63 – 3.51) |
1.79 (1.01 – 3.18) |
1.86 (1.25 – 2.76) |
1.62 (0.98 – 2.68) |
2.63 (1.45 – 4.77) |
|
| Model 4 | 1.57 (1.10 – 2.23) |
1.64 (1.13 – 2.37) |
2.39 (1.63 – 3.50) |
1.92 (1.09 – 3.38) |
1.82 (1.23 – 2.71) |
1.61 (0.97 – 2.66) |
2.71 (1.47 – 4.98) |
|
| Model 5 | 1.60 (1.13 – 2.28) |
1.69 (1.17 – 2.45) |
2.31 (1.57 – 3.40) |
1.90 (1.01 – 3.58) |
1.82 (1.22 – 2.73) |
1.78 (1.12 – 2.85) |
2.20 (1.24 – 3.92) |
|
| Model 6 | 1.60 (1.13 – 2.28) |
1.70 (1.17 – 2.45) |
2.30 (1.57 – 3.37) |
1.87 (0.92 – 3.80) |
1.81 (1.21 – 2.71) |
1.79 (1.12 – 2.85) |
2.22 (1.26 – 3.93) |
|
| Model 7 | 1.61 (1.13 – 2.30) |
1.67 (1.14 – 2.45) |
2.33 (1.58 – 3.43) |
1.86 (1.05 – 3.30) |
1.83 (1.23 – 2.74) |
1.78 (1.12 – 2.84) |
2.19 (1.30 – 3.71) |
|
| Model 8 | 1.58 (1.10 – 2.26) |
1.68 (1.16 – 2.42) |
2.38 (1.59 – 3.56) |
1.90 (1.08 – 3.33) |
1.84 (1.23 – 2.73) |
1.82 (1.14 – 2.92) |
2.10 (1.20 – 3.70) |
|
| Model 9 | 1.60 (1.13 – 2.28) |
1.65 (1.14 – 2.39) |
2.37 (1.61 – 3.47) |
1.79 (1.00 – 3.19) |
1.86 (1.25 – 2.78) |
1.62 (0.96 – 2.73) |
2.46 (1.40 – 4.32) |
|
| SCC | Conventional model | 1.43 (1.02 – 2.01) |
1.25 (0.86 – 1.81) |
2.13 (1.47 – 3.09) |
1.61 (0.72 – 3.59) |
1.97 (1.34 – 2.89) |
1.27 (0.74 – 2.17) |
1.55 (0.75 – 3.23) |
| Model 1 | 1.46 (1.06 – 2.00) |
1.32 (0.94 – 1.85) |
1.98 (1.41 – 2.79) |
1.59 (0.92 – 2.73) |
1.87 (1.31 – 2.66) |
1.38 (0.89 – 2.13) |
1.56 (0.93 – 2.62) |
|
| Model 2 | 1.42 (1.03 – 1.96) |
1.26 (0.89 – 1.79) |
2.03 (1.43 – 2.86) |
1.58 (0.91 – 2.72) |
1.96 (1.37 – 2.82) |
1.32 (0.84 – 2.06) |
1.79 (1.01 – 3.18) |
|
| Model 3 | 1.44 (1.05 – 1.99) |
1.28 (0.90 – 1.81) |
2.05 (1.44 – 2.90) |
1.52 (0.88 – 2.65) |
1.90 (1.33 – 2.71) |
1.27 (0.79 – 2.03) |
1.84 (1.00 – 3.89) |
|
| Model 4 | 1.44 (1.05 – 1.98) |
1.29 (0.91 – 1.82) |
2.03 (1.44 – 2.88) |
1.61 (0.93 – 2.78) |
1.88 (1.32 – 2.67) |
1.28 (0.80 – 2.05) |
1.83 (0.98 – 3.43) |
|
| Model 5 | 1.46 (1.06 – 2.00) |
1.27 (0.90 – 1.80) |
2.06 (1.45 – 2.92) |
1.83 (1.00 – 3.33) |
1.94 (1.35 – 2.78) |
1.35 (0.87 – 2.09) |
1.36 (0.75 – 2.45) |
|
| Model 6 | 1.46 (1.06 – 2.00) |
1.28 (0.91 – 1.82) |
2.02 (1.43 – 2.86) |
1.87 (0.96 – 3.64) |
1.91 (1.34 – 2.73) |
1.35 (0.87 – 2.10) |
1.40 (0.79 – 2.51) |
|
| Model 7 | 1.49 (1.08 – 2.06) |
1.24 (0.86 – 1.78) |
2.06 (1.45 – 2.93) |
1.50 (0.86 – 1.78) |
1.92 (1.35 – 2.75) |
1.36 (0.88 – 2.11) |
1.48 (0.87 – 2.52) |
|
| Model 8 | 1.42 (1.03 – 1.96) |
1.29 (0.92 – 1.82) |
2.08 (1.45 – 2.99) |
1.61 (0.93 – 2.77) |
1.90 (1.33 – 2.70) |
1.43 (0.92 – 2.22) |
1.40 (0.79 – 2.49) |
|
| Model 9 | 1.46 (1.06 – 2.01) |
1.28 (0.90 – 1.80) |
2.05 (1.44 – 2.90) |
1.50 (0.86 – 2.62) |
1.92 (1.35 – 2.74) |
1.23 (0.76 – 2.01) |
1.79 (1.01 – 3.17) |
|
| BCC | Conventional model | 1.39 (0.99 – 1.95) |
1.02 (0.70 – 1.50) |
2.06 (1.41 – 2.99) |
0.51 (0.15 – 1.73) |
1.60 (1.07 – 2.37) |
2.10 (1.32 – 3.35) |
1.46 (0.71 – 3.04) |
| Model 1 | 1.40 (1.02 – 1.92) |
1.13 (0.80 – 1.59) |
1.90 (1.34 – 2.68) |
1.11 (0.62 – 1.98) |
1.57 (1.09 – 2.25) |
1.85 (1.23 – 2.78) |
1.47 (0.87 – 2.47) |
|
| Model 2 | 1.38 (1.00 – 1.90) |
1.10 (0.77 – 1.56) |
1.92 (1.36 – 2.72) |
1.11 (0.62 – 1.98) |
1.62 (1.12 – 2.34) |
1.81 (1.20 – 2.74) |
1.60 (0.90 – 2.85) |
|
| Model 3 | 1.40 (1.02 – 1.92) |
1.11 (0.78 – 1.57) |
1.93 (1.36 – 2.74) |
1.09 (0.60 – 1.96) |
1.58 (1.10 – 2.27) |
1.79 (1.17 – 2.75) |
1.60 (0.87 – 2.94) |
|
| Model 4 | 1.40 (1.02 – 1.92) |
1.12 (0.79 – 1.59) |
1.91 (1.34 – 2.70) |
1.11 (0.62 – 1.99) |
1.57 (1.10 – 2.25) |
1.84 (1.20 – 2.81) |
1.51 (0.80 – 2.85) |
|
| Model 5 | 1.40 (1.02 – 1.92) |
1.12 (0.79 – 1.60) |
1.90 (1.34 – 2.70) |
1.13 (0.58 – 2.17) |
1.57 (1.09 – 2.27) |
1.85 (1.23 – 2.78) |
1.45 (0.81 – 2.60) |
|
| Model 6 | 1.40 (1.02 – 1.92) |
1.14 (0.80 – 1.62) |
1.88 (1.32 – 2.66) |
1.00 (0.47 – 2.15) |
1.55 (1.07 – 2.23) |
1.87 (1.24 – 2.80) |
1.55 (0.87 – 2.74) |
|
| Model 7 | 1.45 (1.05 – 1.99) |
1.04 (0.72 – 1.51) |
1.99 (1.40 – 2.82) |
1.03 (0.56 – 1.87) |
1.62 (1.13 – 2.33) |
1.83 (1.22 – 2.75) |
1.38 (0.81 – 2.35) |
|
| Model 8 | 1.36 (0.98 – 1.87) |
1.09 (0.77 – 1.55) |
2.03 (1.41 – 2.91) |
1.13 (0.63 – 2.01) |
1.59 (1.11 – 2.89) |
1.93 (1.28 – 2.91) |
1.28 (0.72 – 2.28) |
|
| Model 9 | 1.40 (1.02 – 1.93) |
1.11 (0.78 – 1.58) |
1.92 (1.35 – 2.72) |
1.08 (0.60 – 1.96) |
1.58 (1.10 – 2.28) |
1.79 (1.15 – 2.78) |
1.55 (0.87 – 2.74) |
Each model includes seven variants (carrier versus non-carrier) simultaneously and adjusts for age.
Conventional model is the logistical regression model. Models 1–9 utilize hierarchical modeling approach.
Model 1 assumes that all variants have the same effects.
Model 2 is based on the associations with hair color among controls.
Model 3 is based on the associations with skin color among controls.
Model 4 is based on the associations with tanning ability among controls.
Model 5 is based on cell surface MC1R antibody binding.
Model 6 is based on cell surface MSH ligand binding.
Model 7 kinetics, Ki-a-MSH.
Model 8 kinetics, Ki-NDP.
Model 9 is based on maximum generation of cAMP in response to MSH.
Figure 1.
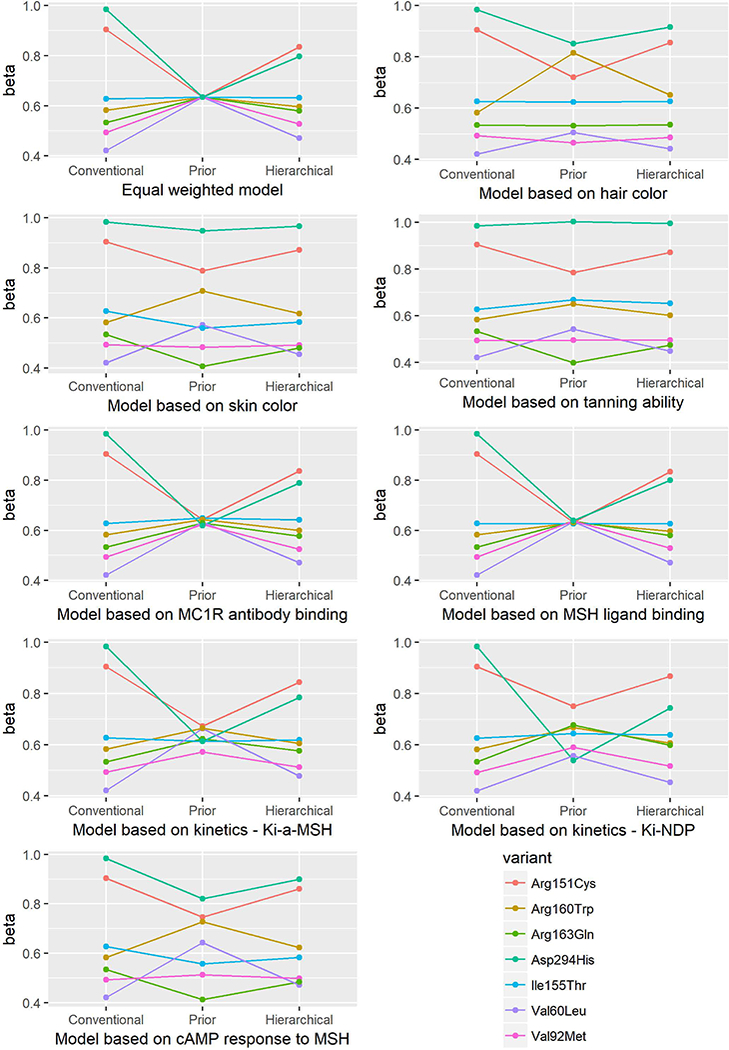
Beta estimates from the conventional, prior, and hierarchical models for association between MC1R variants and risk of melanoma.
Figure 6.
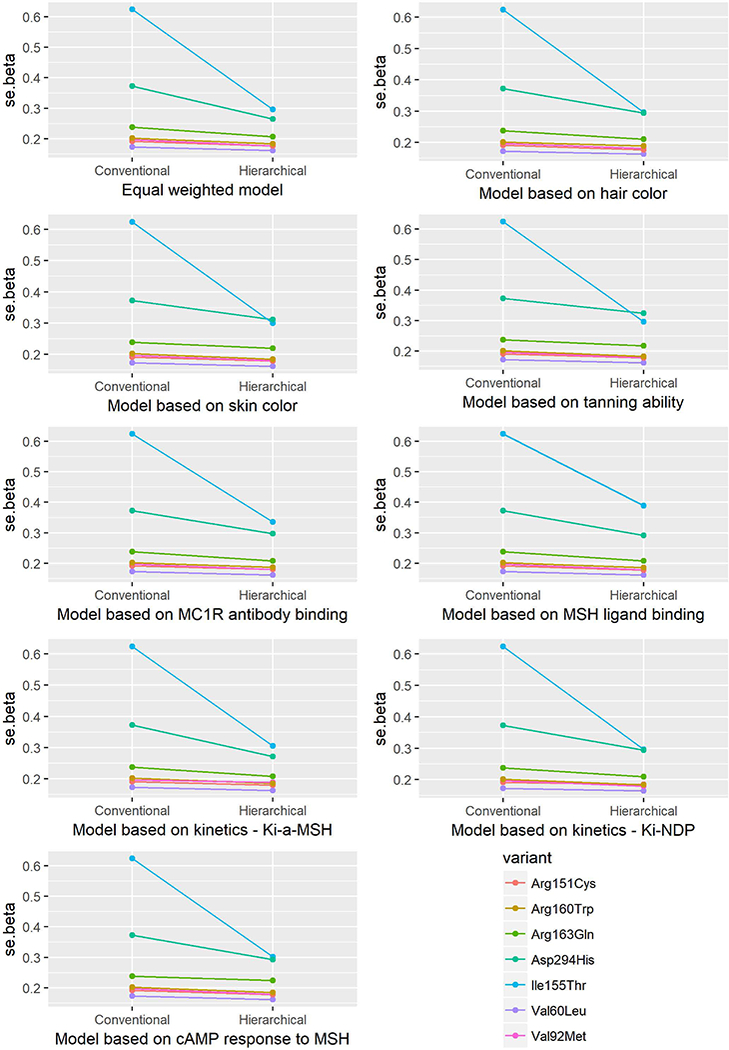
Standard error (se) of the beta estimates from the conventional and hierarchical models for the association between MC1R variants and risk of BCC.
Alterations in the prior matrix influenced the risk estimates (Table 3). The results for the RHC variants -- Arg151Cys, Arg160Trp, and Asp294His -- were robust for melanoma risk; all three variants were significantly associated with melanoma risk in all the models. Similarly, the Val60Leu and Val92Met variants were significantly associated with melanoma risk in all models. The association of Ile155Thr variant with melanoma risk became non-significant in model 6 and borderline significant in model 9. In the models incorporating phenotypic parameters, the association between the Arg163Gln variant and melanoma risk was significant in some models, but not in others (model 3,4 and 9).
For SCC, the Val60Leu, Arg151Cys and Arg160Trp variants were significantly associated with risk in all models. The Asp294His variant was not associated with SCC risk in the conventional model but was significantly associated with SCC risk in three hierarchical models, incorporating hair color, skin color or generation of cAMP in response to MSH. The association of Ile155Thr variant with SCC was not significant in any model except in the hierarchical one based on cell surface MC1R antibody binding. The Val92Met and Arg163Gln variants were not associated with SCC risk in any model. (Table 3)
For BCC, Arg151Cys, Arg160Trp and Arg163Gln were significantly associated with increased risk in all models, whereas Asp294His, Val92Met and Ile155Thr were not associated with BCC risk in any model. The association of Val60Leu variant with BCC risk was not significant only in one of the hierarchical models (model 8).
We constructed a multivariable second-level model using three selected representative measures: hair color as pigmentary phenotype, cell surface MSH ligand binding as cell surface expression, and kinetics (Ki) using α-MSH as MC1R coupling activity. The parameter estimates are listed in Table 4. By comparing the π estimates, we found that cell-surface MSH ligand binding and Ki using α-MSH parameters showed relevance for BCC but not melanoma and SCC, while pigmentary hair color parameters did not strongly affect SNP parameters for any of the three outcomes. However the precision of these parameter estimates is low perhaps due to a small sample size. The results of the genetic main effect of each variant on each type of skin cancer are presented in Table 5. We found that the Val60Leu, Arg151Cys and Arg160Trp variants remained significant for the risk of each type of skin cancer. The Asp163Gln variant was associated with the risk of melanoma and BCC, but not SCC. The Asp294His and Val92Met variants were associated only with melanoma risk. The Ile155Thr variants were not associated with the risk of any type of skin cancer.
Table 4.
Estimates of prior-model hyperparameters: multivariate second-level model
| Melanoma | SCC | BCC | |
|---|---|---|---|
| Variable | π est (SD) | π est (SD) | π est (SD) |
| Mean | 0.52 (0.43) | 0.37 (0.41) | 0.97 (0.45) |
| Cell surface MSH ligand binding | −0.03 (0.15) | 0.05 (0.15) | −0.24 (0.18) |
| Kinetics, α-MSH Ki | 0.02 (0.16) | −0.06 (0.16) | −0.28 (0.18) |
| Hair color | 0.61 (0.65) | 0.44 (0.65) | 0.09 (0.64) |
Table 5.
Hierarchical modeling of MC1R variants using multivariate second-level model
| Val60Leu | Val92Met | Arg151Cys | Ile155Thr | Arg160Trp | Arg163Gln | Asp294His | |
|---|---|---|---|---|---|---|---|
| Melanoma | 1.55 (1.07–2.24) | 1.64 (1.11–2.42) |
2.3 (1.58–3.44) |
1.80 (0.80–4.05) | 1.91 (1.26–2.88) | 1.71 (1.06–2.77) | 2.60 (1.37–4.95) |
| SCC | 1.45 (1.04–2.02) | 1.23 (0.85–1.77) | 2.07 (1.46–2.94) | 1.67 (0.78–3.57) | 1.98 (1.38–2.86) | 1.31 (0.84–2.06) | 1.60 (0.82–3.14) |
| BCC | 1.46 (1.05–2.03) | 1.03 (0.71–1.50) | 1.97 (1.39–2.81) | 0.61 (0.22–1.97) | 1.59 (1.09–2.31) | 1.86 (1.23–2.81) | 1.66 (0.85–3.24) |
The model includes seven variants (carrier versus non-carrier) simultaneously and adjusts for age. We use multivariate second-level model incorporate three representative measures: cell surface MSH ligand binding, kinetics, α-MSH Ki, and hair color.
Discussion
Hierarchical modeling has previously been used in genetic association studies. Aragaki et al. [Aragaki, et al. 1997] evaluated the NAT2 genotype-specific dietary effects on adenomatous polyps by constructing a Z matrix of conversion rates calculated for NAT2 genotype-specific enzymatic activity and dietary item-heterocyclic amine combinations. Hung et al. [Hung, et al. 2004] evaluated multiple polymorphisms in different pathways in relation to bladder cancer risk by constructing a Z matrix according to the involvement of each polymorphism in specific pathways. The genetic variants in the MC1R gene have been evaluated in relation to pigmentary phenotypes and the risk of melanoma and non-melanoma skin cancer, and characterized quantitatively in in vitro phenotypic assays. To our knowledge, this is the first report incorporating biological functional relevance in evaluation of associations between MC1R genetic variants and skin cancer risk. We incorporated the quantitative phenotypic parameters into the Z matrix construction. Consistent with previous reports, our data indicate that hierarchical modeling increases the precision of the risk estimates by tightening the confidence intervals.
Defects in MC1R function are attributable to either decreased affinity for the ligand α-MSH or altered G-protein coupling. These alleles have important structural functions and in turn influence the binding to the ligand and/or signaling process. In this study, we used some phenotypic parameters of the MC1R variants in the construction of the Z matrix such as correlations with constitutional phenotypes (hair color, skin color, and tanning ability), cell surface expression, and enzymatic kinetic parameters (dissociation constant Ki and maximum cAMP generation in response to MSH). The MC1R variants were characterized quantitatively in other assays, such as measurements of hair color [Naysmith, et al. 2004] and experimentally induced erythemal response to UV [Flanagan, et al. 2001]. However, only some, but not all, of the seven common variants were assayed in these studies. Hence, we were not able to use these data to evaluate the seven variants simultaneously.
We emphasize that the construction of the Z matrix, i.e., the specification of the prior, is a key component of this approach. The MC1R gene is involved in G-protein-coupled signaling transduction, which in turn is thought to be involved in multiple pathways in addition to pigmentation, such as immune response, inflammatory response, cell proliferation, and extracellular matrix adhesion [Kalden, et al. 1999; Luger, et al. 2003; Murata, et al. 1997; Naysmith, et al. 2004; Robinson and Healy 2002; Smalley and Eisen 2000]. In the construction of the Z matrix in this study, we used three series of parameters: the phenotypic associations in controls (models 2–4), cell surface expression (models 5 and 6), and enzymatic kinetics (models 7–9). The first of these parameters pertained primarily to the pigmentation pathway; the second and third may represent signaling transduction in multiple pathways.
In the multivariable second-level model using three selected representative measures, the two RHC variants, Arg151Cys and Arg160Trp, showed robust association with the risk of each type of skin cancer. This was consistent with the results from the conventional multivariate model. The other RHC variant, Asp294His, was associated only with melanoma risk in the conventional model. After taking into account the three functional parameters in the hierarchical model, this variant became associated with the risk of SCC as well. This suggests that Asp294His may be involved in the etiology of SCC through these potential mechanisms. Neither the conventional model nor the hierarchical model indicated that Asp294His was associated with the risk of BCC. In addition, the Val60Leu variant was associated with the risk of BCC in most of the hierarchical models, but not in the conventional model. These results suggest that the hierarchical models may provide additional information on risk assessment by incorporating biological functional relevance.
We can also utilize the estimates of the second-level parameter vector, to make inferences about relevant mechanisms that may play a role in carcinogenesis for different skin cancer types. For example, altered affinity for the ligand α-MSH may play a role in basal cell carcinoma – as evidenced by higher estimates for cell-surface MSH ligand binding and α-MSH Ki parameters for BCC risk. In contrast, higher estimates of prior-model hyper-parameters suggest that variation in pathways relevant to hair color phenotype may play a bigger role in melanoma and SCC.
For this application several experimentally derived quantitative measures of variant effects on different etiologic pathways were available. However, this is not always the case. In the absence of such data, in silico estimates of variant effects could be used. For example, the effects of non-synonymous polymorphisms could be regressed on their SIFT score. The SIFT (Sorting Intolerant From Tolerant) algorithm predicts whether an amino acid substitution will have an impact on protein function based on the alignment of highly similar proteins by evaluating its identity and physicochemical characteristics [Ng and Henikoff 2001; Ng and Henikoff 2002; Ng and Henikoff 2003]. The predictions rely on the evolutionary conservation of amino acids among the protein’s family members, which can suggest their importance for the function/structure of the protein. For example, Kanetsky et al. [Kanetsky, et al. 2004] used SIFT analysis to identify MC1R positions that were predicted to be intolerant of amino acid substitutions; the predicted intolerant variants were Asp84Glu, Arg142His, Arg151Cys, Ile155Thr, Arg160Trp, and Asp294His, which is in good agreement with previous publications on their reduced function.
One limitation of the study is our use of self-reported phenotypes such as hair color, skin color, and tanning ability. Self-report has been shown to be an appropriate and widely used method of assessing risk factors for skin cancer. Test-retest reliability of collecting information on phenotypic risk factors, including skin color, tanning/burning tendency, and sunburn history, from questionnaires is moderate to substantial [Branstrom, et al. 2002; Glanz, et al. 2003; Westerdahl, et al. 1996]. Several previous studies have evaluated the validity of self-reported melanoma risk factors. Kang et al. reported 85% agreement between self-reported skin phototypes and those determined by a dermatologist, and these phototypes had a positive correlation with the minimal erythemal dose [Kang, et al. 1992]. Self-reported skin characteristics predict melanoma risk reasonably well. In our study, we observed that the OR of self-reported skin color (fair vs. medium/olive) for melanoma risk was 2.24 (95% CI, 1.59–3.16), which is in good agreement with two previous reports. In a case-control study of 511 cases and 511 age- and gender-matched controls in Western Australia, reflectance-measured skin color was associated with melanoma risk (fair vs. medium/olive: OR, 2.54; 95% CI, 1.94–3.31) [Holman and Armstrong 1984]. In a meta-analysis of 31 case-control studies, fair skin color was associated with a two-fold increased risk of melanoma (fair vs. medium/olive: OR, 2.06; 95% CI, 1.68–2.52) [Gandini, et al. 2005]. For other constitutional risk factors, such as hair color, sunburn history, and mole counts, the relative risks in our study were also similar to previous reports [Cho, et al. 2005].
In summary, we performed hierarchical modeling analyses, incorporating quantitative information on the function of particular MC1R alleles in an association study with skin cancer risk. Hierarchical modeling stabilizes estimates and provides an alternative analytic approach in genetic association studies.
Supplementary Material
Figure 2.
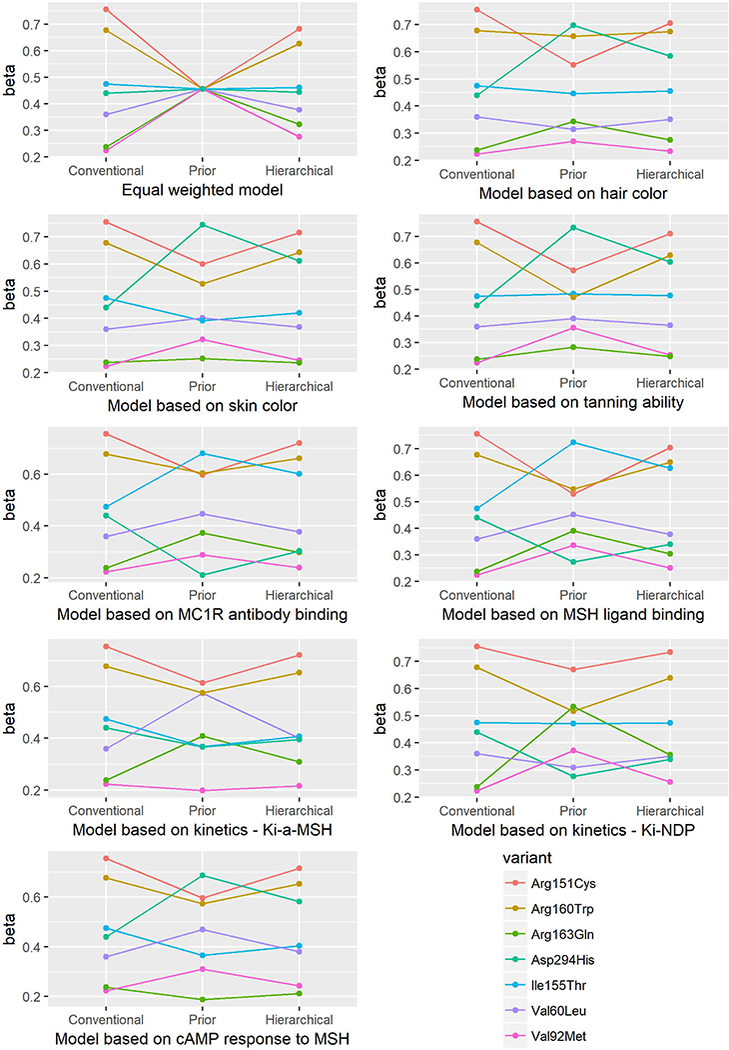
Beta estimates from the conventional, prior, and hierarchical models for association between MC1R variants and risk of SCC.
Figure 3.
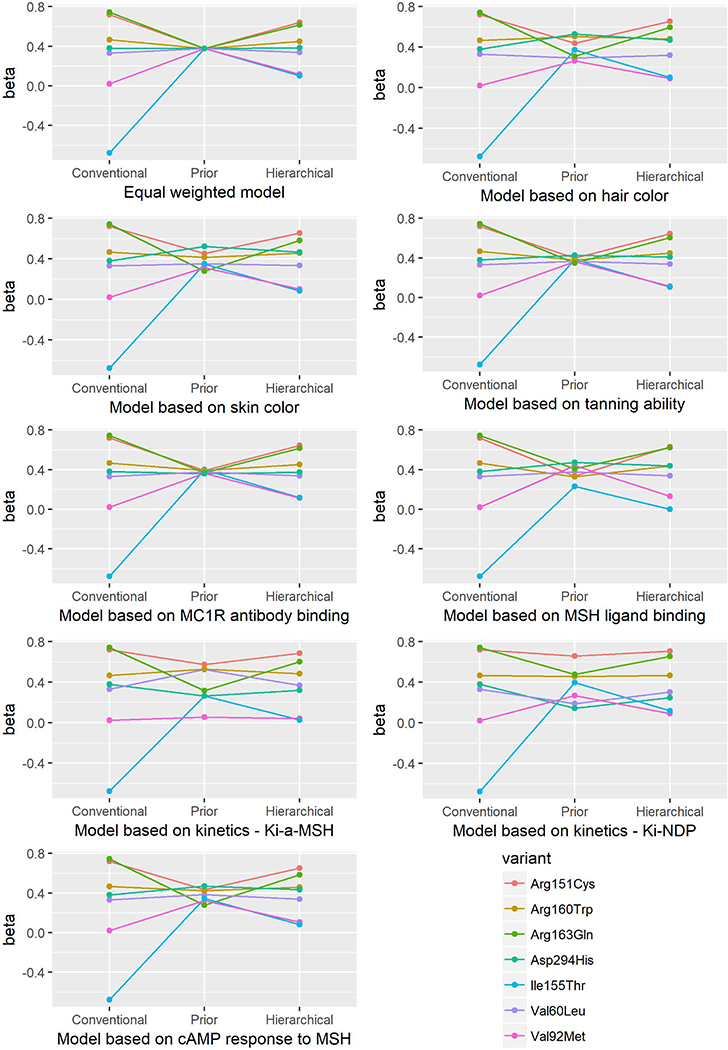
Beta estimates from the conventional, prior, and hierarchical models for association between MC1R variants and risk of BCC.
Figure 4.
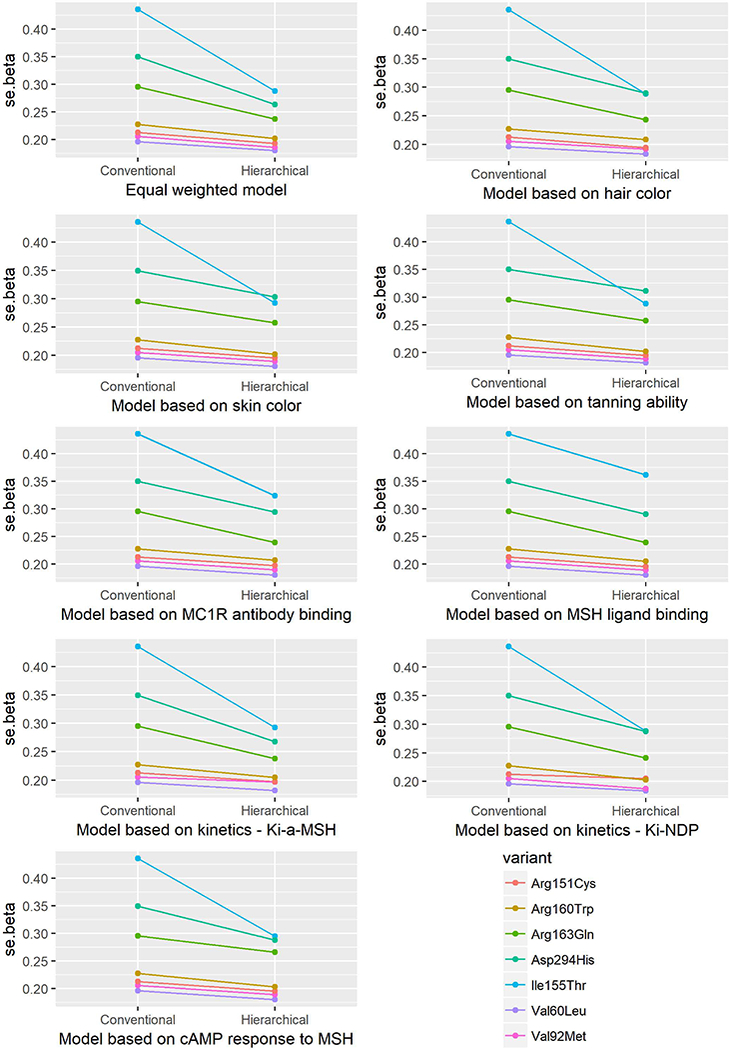
Standard error (se) of the beta estimates from the conventional and hierarchical models for the association between MC1R variants and risk of melanoma.
Figure 5.
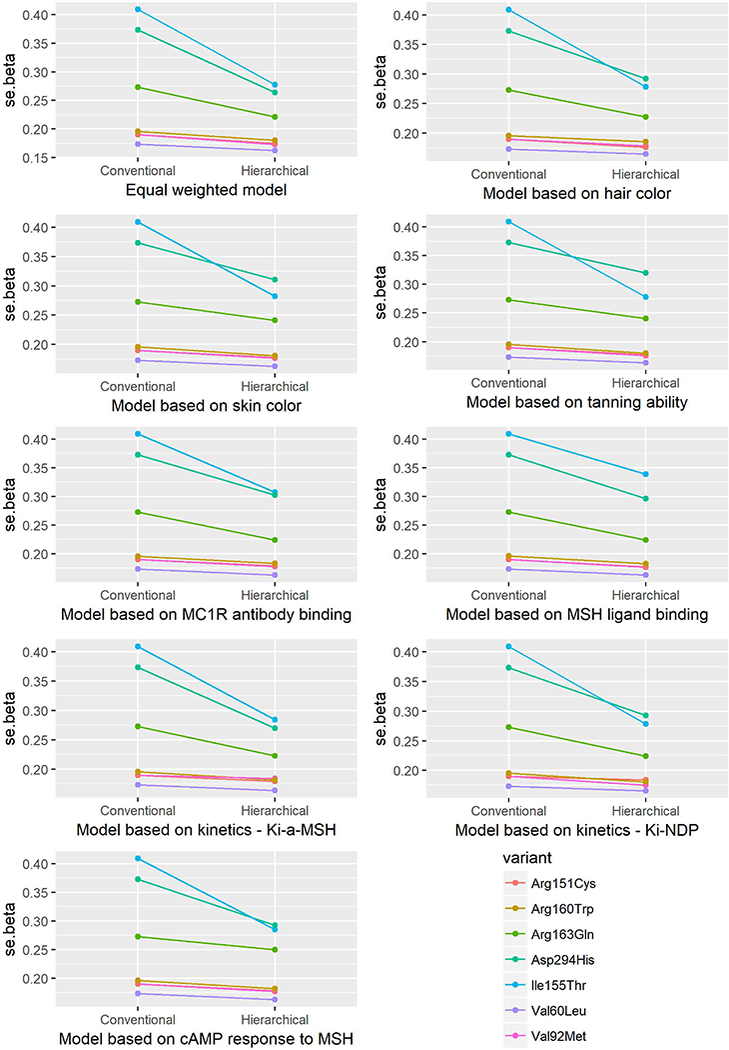
Standard error (se) of the beta estimates from the conventional and hierarchical models for the association between MC1R variants and risk of SCC.
Acknowledgements
We would like to thank Dr. Richard A. Sturm at University of Queensland for providing us with the absolute values corresponding to the bar graphs in his original publications. We also thank Dr. Helgi B. Schiöth at Uppsala University for his helpful discussions. We are grateful to the Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School for providing us the resources of the Nurses’ Health Study. We are also indebted to the participants in the Nurses’ Health Study for their dedication and commitment. We thank the following state cancer registries for their help: Alabama, Arizona, Arkansas, California, Colorado, Connecticut, Delaware, Florida, Georgia, Idaho, Illinois, Indiana, Iowa, Kentucky, Louisiana, Maine, Maryland, Massachusetts, Michigan, Nebraska, New Hampshire, New Jersey, New York, North Carolina, North Dakota, Ohio, Oklahoma, Oregon, Pennsylvania, Rhode Island, South Carolina, Tennessee, Texas, Virginia, Washington, and Wyoming. The authors assume full responsibility for analyses and interpretation of these data. This work is supported by NIH R01 CA49449, P01 CA87969, UM1 CA186107, UM1 CA167552 and K01DK110267 (ADJ).
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- 2017. Cancer facts and figures, American Cancer Society; Atlanta. [Google Scholar]
- Aragaki CC, Greenland S, Probst-Hensch N, Haile RW. 1997. Hierarchical modeling of gene-environment interactions: estimating NAT2 genotype-specific dietary effects on adenomatous polyps. Cancer Epidemiol Biomarkers Prev 6(5):307–14. [PubMed] [Google Scholar]
- Bastiaens MT, ter Huurne JA, Kielich C, Gruis NA, Westendorp RG, Vermeer BJ, Bavinck JN. 2001. Melanocortin-1 receptor gene variants determine the risk of nonmelanoma skin cancer independently of fair skin and red hair. Am J Hum Genet 68(4):884–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont KA, Newton RA, Smit DJ, Leonard JH, Stow JL, Sturm RA. 2005. Altered cell surface expression of human MC1R variant receptor alleles associated with red hair and skin cancer risk. Hum Mol Genet 14(15):2145–54. [DOI] [PubMed] [Google Scholar]
- Box NF, Duffy DL, Irving RE, Russell A, Chen W, Griffyths LR, Parsons PG, Green AC, Sturm RA. 2001. Melanocortin-1 receptor genotype is a risk factor for basal and squamous cell carcinoma. J Invest Dermatol 116(2):224–9. [DOI] [PubMed] [Google Scholar]
- Box NF, Wyeth JR, O’Gorman LE, Martin NG, Sturm RA. 1997. Characterization of melanocyte stimulating hormone receptor variant alleles in twins with red hair. Hum Mol Genet 6(11):1891–7. [DOI] [PubMed] [Google Scholar]
- Branstrom R, Kristjansson S, Ullen H, Brandberg Y. 2002. Stability of questionnaire items measuring behaviours, attitudes and stages of change related to sun exposure. Melanoma Res 12(5):513–9. [DOI] [PubMed] [Google Scholar]
- Brenner DR, Brennan P, Boffetta P, Amos CI, Spitz MR, Chen C, Goodman G, Heinrich J, Bickeboller H, Rosenberger A and others. 2013. Hierarchical modeling identifies novel lung cancer susceptibility variants in inflammation pathways among 10,140 cases and 11,012 controls. Hum Genet 132(5):579–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho E, Rosner BA, Feskanich D, Colditz GA. 2005. Risk factors and individual probabilities of melanoma for whites. J Clin Oncol 23(12):2669–75. [DOI] [PubMed] [Google Scholar]
- Flanagan N, Ray AJ, Todd C, Birch-Machin MA, Rees JL. 2001. The relation between melanocortin 1 receptor genotype and experimentally assessed ultraviolet radiation sensitivity. J Invest Dermatol 117(5):1314–7. [DOI] [PubMed] [Google Scholar]
- Gandini S, Sera F, Cattaruzza MS, Pasquini P, Zanetti R, Masini C, Boyle P, Melchi CF. 2005. Meta-analysis of risk factors for cutaneous melanoma: III. Family history, actinic damage and phenotypic factors. Eur J Cancer 41(14):2040–59. [DOI] [PubMed] [Google Scholar]
- Gantz I, Yamada T, Tashiro T, Konda Y, Shimoto Y, Miwa H, Trent JM. 1994. Mapping of the gene encoding the melanocortin-1 (alpha-melanocyte stimulating hormone) receptor (MC1R) to human chromosome 16q24.3 by Fluorescence in situ hybridization. Genomics 19(2):394–5. [DOI] [PubMed] [Google Scholar]
- Glanz K, Schoenfeld E, Weinstock MA, Layi G, Kidd J, Shigaki DM. 2003. Development and reliability of a brief skin cancer risk assessment tool. Cancer Detect Prev 27(4):311–5. [DOI] [PubMed] [Google Scholar]
- Greenland S 1994. Hierarchical regression for epidemiologic analyses of multiple exposures. Environ Health Perspect 102 Suppl 8:33–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenland S 2000. When should epidemiologic regressions use random coefficients? Biometrics 56(3):915–21. [DOI] [PubMed] [Google Scholar]
- Han J, Colditz GA, Samson LD, Hunter DJ. 2004. Polymorphisms in DNA double-strand break repair genes and skin cancer risk. Cancer Res 64(9):3009–13. [DOI] [PubMed] [Google Scholar]
- Han J, Kraft P, Colditz GA, Wong J, Hunter DJ. 2006. Melanocortin 1 receptor variants and skin cancer risk. Int J Cancer 119(8):1976–84. [DOI] [PubMed] [Google Scholar]
- Harding RM, Healy E, Ray AJ, Ellis NS, Flanagan N, Todd C, Dixon C, Sajantila A, Jackson IJ, Birch-Machin MA and others. 2000. Evidence for variable selective pressures at MC1R. Am J Hum Genet 66(4):1351–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holman CD, Armstrong BK. 1984. Pigmentary traits, ethnic origin, benign nevi, and family history as risk factors for cutaneous malignant melanoma. J Natl Cancer Inst 72(2):257–66. [PubMed] [Google Scholar]
- Howe HL, Wingo PA, Thun MJ, Ries LA, Rosenberg HM, Feigal EG, Edwards BK. 2001. Annual report to the nation on the status of cancer (1973 through 1998), featuring cancers with recent increasing trends. J Natl Cancer Inst 93(11):824–42. [DOI] [PubMed] [Google Scholar]
- Hung RJ, Brennan P, Malaveille C, Porru S, Donato F, Boffetta P, Witte JS. 2004. Using hierarchical modeling in genetic association studies with multiple markers: application to a case-control study of bladder cancer. Cancer Epidemiol Biomarkers Prev 13(6):1013–21. [PubMed] [Google Scholar]
- Kalden DH, Scholzen T, Brzoska T, Luger TA. 1999. Mechanisms of the antiinflammatory effects of alpha-MSH. Role of transcription factor NF-kappa B and adhesion molecule expression. Ann N Y Acad Sci 885:254–61. [DOI] [PubMed] [Google Scholar]
- Kanetsky PA, Ge F, Najarian D, Swoyer J, Panossian S, Schuchter L, Holmes R, Guerry D, Rebbeck TR. 2004. Assessment of polymorphic variants in the melanocortin-1 receptor gene with cutaneous pigmentation using an evolutionary approach. Cancer Epidemiol Biomarkers Prev 13(5):808–19. [PubMed] [Google Scholar]
- Kang S, Fitzpatrick TB, Youn JI. 1992. Identification of population-at-risk for melanoma and epithelial skin cancer using skin phototyping. Book of Abstracts of 18th World Congress of Dermatology. [Google Scholar]
- Kennedy C, ter Huurne J, Berkhout M, Gruis N, Bastiaens M, Bergman W, Willemze R, Bavinck JN. 2001. Melanocortin 1 receptor (MC1R) gene variants are associated with an increased risk for cutaneous melanoma which is largely independent of skin type and hair color. J Invest Dermatol 117(2):294–300. [DOI] [PubMed] [Google Scholar]
- Landi MT, Kanetsky PA, Tsang S, Gold B, Munroe D, Rebbeck T, Swoyer J, Ter-Minassian M, Hedayati M, Grossman L and others. 2005. MC1R, ASIP, and DNA repair in sporadic and familial melanoma in a Mediterranean population. J Natl Cancer Inst 97(13):998–1007. [DOI] [PubMed] [Google Scholar]
- Luger TA, Scholzen TE, Brzoska T, Bohm M. 2003. New insights into the functions of alpha-MSH and related peptides in the immune system. Ann N Y Acad Sci 994:133–40. [DOI] [PubMed] [Google Scholar]
- Mas JS, Gerritsen I, Hahmann C, Jimenez-Cervantes C, Garcia-Borron JC. 2003. Rate limiting factors in melanocortin 1 receptor signalling through the cAMP pathway. Pigment Cell Res 16(5):540–7. [DOI] [PubMed] [Google Scholar]
- Matichard E, Verpillat P, Meziani R, Gerard B, Descamps V, Legroux E, Burnouf M, Bertrand G, Bouscarat F, Archimbaud A and others. 2004. Melanocortin 1 receptor (MC1R) gene variants may increase the risk of melanoma in France independently of clinical risk factors and UV exposure. J Med Genet 41(2):e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata J, Ayukawa K, Ogasawara M, Fujii H, Saiki I. 1997. Alpha-melanocyte-stimulating hormone blocks invasion of reconstituted basement membrane (Matrigel) by murine B16 melanoma cells. Invasion Metastasis 17(2):82–93. [PubMed] [Google Scholar]
- Naysmith L, Waterston K, Ha T, Flanagan N, Bisset Y, Ray A, Wakamatsu K, Ito S, Rees JL. 2004. Quantitative measures of the effect of the melanocortin 1 receptor on human pigmentary status. J Invest Dermatol 122(2):423–8. [DOI] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. 2001. Predicting deleterious amino acid substitutions. Genome Res 11(5):863–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. 2002. Accounting for human polymorphisms predicted to affect protein function. Genome Res 12(3):436–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. 2003. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res 31(13):3812–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer JS, Duffy DL, Box NF, Aitken JF, O’Gorman LE, Green AC, Hayward NK, Martin NG, Sturm RA. 2000. Melanocortin-1 receptor polymorphisms and risk of melanoma: is the association explained solely by pigmentation phenotype? Am J Hum Genet 66(1):176–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees JL. 2003. Genetics of hair and skin color. Annu Rev Genet 37:67–90. [DOI] [PubMed] [Google Scholar]
- Ringholm A, Klovins J, Rudzish R, Phillips S, Rees JL, Schioth HB. 2004. Pharmacological characterization of loss of function mutations of the human melanocortin 1 receptor that are associated with red hair. J Invest Dermatol 123(5):917–23. [DOI] [PubMed] [Google Scholar]
- Robinson SJ, Healy E. 2002. Human melanocortin 1 receptor (MC1R) gene variants alter melanoma cell growth and adhesion to extracellular matrix. Oncogene 21(52):8037–46. [DOI] [PubMed] [Google Scholar]
- Sanchez-Mas J, Hahmann C, Gerritsen I, Garcia-Borron JC, Jimenez-Cervantes C. 2004. Agonist-independent, high constitutive activity of the human melanocortin 1 receptor. Pigment Cell Res 17(4):386–95. [DOI] [PubMed] [Google Scholar]
- Schioth HB, Phillips SR, Rudzish R, Birch-Machin MA, Wikberg JE, Rees JL. 1999. Loss of function mutations of the human melanocortin 1 receptor are common and are associated with red hair. Biochem Biophys Res Commun 260(2):488–91. [DOI] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A. 2017. Cancer statistics, 2017. CA: A Cancer Journal for Clinicians 67(1):7–30. [DOI] [PubMed] [Google Scholar]
- Smalley K, Eisen T. 2000. The involvement of p38 mitogen-activated protein kinase in the alpha-melanocyte stimulating hormone (alpha-MSH)-induced melanogenic and anti-proliferative effects in B16 murine melanoma cells. FEBS Lett 476(3):198–202. [DOI] [PubMed] [Google Scholar]
- Sturm RA. 2002. Skin colour and skin cancer - MC1R, the genetic link. Melanoma Res 12(5):405–16. [DOI] [PubMed] [Google Scholar]
- Sturm RA, Teasdale RD, Box NF. 2001. Human pigmentation genes: identification, structure and consequences of polymorphic variation. Gene 277(1–2):49–62. [DOI] [PubMed] [Google Scholar]
- Valverde P, Healy E, Jackson I, Rees JL, Thody AJ. 1995. Variants of the melanocyte-stimulating hormone receptor gene are associated with red hair and fair skin in humans. Nat Genet 11(3):328–30. [DOI] [PubMed] [Google Scholar]
- Westerdahl J, Anderson H, Olsson H, Ingvar C. 1996. Reproducibility of a self-administered questionnaire for assessment of melanoma risk. Int J Epidemiol 25(2):245–51. [DOI] [PubMed] [Google Scholar]
- Witte JS. 1997. Genetic analysis with hierarchical models. Genet Epidemiol 14(6):1137–42. [DOI] [PubMed] [Google Scholar]
- Witte JS, Greenland S, Haile RW, Bird CL. 1994. Hierarchical regression analysis applied to a study of multiple dietary exposures and breast cancer. Epidemiology 5(6):612–21. [DOI] [PubMed] [Google Scholar]
- Witte JS, Greenland S, Kim LL. 1998. Software for hierarchical modeling of epidemiologic data. Epidemiology 9(5):563–6. [PubMed] [Google Scholar]
- Witte JS, Greenland S, Kim LL, Arab L. 2000. Multilevel modeling in epidemiology with GLIMMIX. Epidemiology 11(6):684–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


