Abstract
Characterizing the reciprocal interactions between toxicants, the gut microbiota, and the host, holds great promise for improving our mechanistic understanding of toxic endpoints. Advances in culture-independent sequencing analysis (e.g., 16S rRNA gene amplicon sequencing) combined with quantitative metabolite profiling (i.e., metabolomics) have provided new ways of studying the gut microbiome and have begun to illuminate how toxicants influence the structure and function of the gut microbiome. Developing a standardized protocol is important for establishing robust, reproducible, and importantly, comparative data. This protocol can be used as a foundation for examining the gut microbiome via sequencing-based analysis and metabolomics. Two main units follow: (1) analysis of the gut microbiome via sequencing-based approaches; and (2) functional analysis of the gut microbiome via metabolomics.
Keywords: bioinformatics, microbiome, metabolomics, toxicology
INTRODUCTION
Alterations of the gut microbiome can occur following exposure to xenobiotics (Spanogiannopoulos, Bess, Carmody, & Turnbaugh, 2016). Reports indicate the lung microbiome is altered following exposure to aerosolized polycyclic aromatic hydrocarbons (Hosgood et al., 2015). Additional data supports that the skin microbiome can be altered by xenobiotics (Lee et al., 2017). Despite the importance of the microbiome in toxicology, investigation into the impact xenobiotics can have on the microbiome, or the potential influence the microbiome can have on toxicologic endpoints remains limited. Below is a step by step, protocol created to start the process of incorporating gut microbiome analyses into a toxicologic study. While this protocol focuses on the gut microbiome, it can provide a foundation for other investigations of other microbiomes.
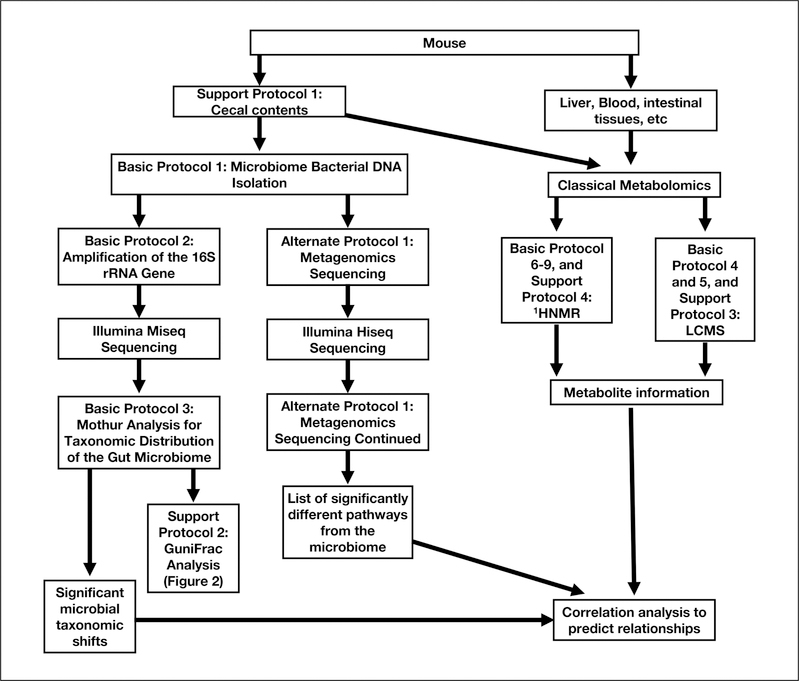
The following unit describes the process of bacterial DNA extraction from mouse cecal contents. A flow chart describing the following units can be seen in Figure 1. The bacterial DNA isolation kit used in this protocol can also be used for bacterial DNA isolation of rodent fecal pellets or human stool samples and the data generation and analysis protocols also apply to human fecal bacterial DNA.
Figure 1.
Analysis flow chart using sequence- and metabolomics-based analysis to uncover structural and functional changes in the gut microbiome.
Integration of microbiome analyses with toxicology studies can provide insights into cryptic or previously uncharacterized toxic endpoints. Comprehensive microbiome analysis requires basic terminal commands and basic skills in R. There are numerous online resources available. For example, the R cookbook, a comprehensive manual for R programming, is a freely available, (https://www.cookbook-r.com/) and there are many Websites for terminal-based coding. This unit covers 16S rRNA gene analysis using the mothur software package (Kozich, Westcott, Baxter, Highlander, & Schloss, 2013) and metagenomic sequence analysis using the HUMAnN2 (Human microbiome project Unified Metabolic Analysis Network) software package (Abubucker et al., 2012). The resulting files from 16S rRNA gene analysis are a taxonomic distribution that can be used to create illustrations of the significant changes. The resulting files from the metagenomic analysis represent pathways that are present in the gut microbiome and demonstrate if the relative abundance of these pathways have increased or decreased in response to a specific treatment. Importantly, sequence analyses revealing the presence of a given panel of genes associated with specific metabolic pathways does not imply phenotypic expression of the pathway, additional functional assessment is required. Functional verification using metabolomics is covered in Basic Protocols 4 to 9. Overall this unit provides a comprehensive and easy to follow method for gut microbiome analysis. Readers are encouraged to visit the various wiki resources as software and databases are routinely updated. Further, kits and other reagents may also change.
Sample data, a script for the mothur analysis, and a R mark down file for GUnifrac analysis are included to accompany this protocol in a zip file (test_data.zip; see Supporting Information). Other sample data can be found on the mothur wiki site (https://www.mothur.org/wiki/MiSeq_SOP) and sample data for HUMAnN2 can be found on the HUMAnN2 bitbucket page (https://bitbucket.org/biobakery/humann2/wiki/Home). To run the included sample script, simply unzip the sample folder, open terminal, and navigate to the test_data directory. Type ./mothur/mothur.Mothur.test.batch.txt. This script should take roughly 15 min (script specifies two processors, it can be edited for more processors if more are available) and will result in a summary table, which includes the taxonomic distribution for the test data. It should be noted that the commands to make a phylogenic tree with mothur are included but will not run without removing the hashtags before the commands. These commands are not run because these commands will add an extra 30 min to the run time of this script and the resulting .tre and count files (see Supporting Information) WILL NOT work for GUnifrac analysis due to the small size of the subset. Instead both a separate .tre file and a count file are provided to illustrate the GUnifrac analysis with the R mark down file in a folder called GUnifrac_data. Also, the included mothur script is a guide and each user should modify the file names and parameters as necessary.
BASIC PROTOCOL 1
BACTERIAL DNA EXTRACTION
This protocol explains the process of bacterial DNA isolation from mouse cecal contents. For information on how to extract cecal contents, see Support Protocol 1. This protocol is adopted from the Omega-BioTek E.Z.N.A stool isolation kit and has been used extensively within our laboratory (Hubbard et al., 2017; Li et al., 2017; Murray, Nichols, Zhang, Patterson, & Perdew, 2016; Zhang et al., 2016). The PowerSoil DNA Isolation kit (moBio) has also been used and can be implemented instead of the Omega-BioTek E.Z.N.A kit. A recent study has shown that the use of different bacterial DNA isolation kits leads to less variation than the use of different 16S rRNA gene primers (V3-V4 yields different results than V4-V5) (Rintala et al., 2017). Listed below is a modified version of the protocol provided by Omega-BioTek.
Materials
Omega-BioTek E.Z.N.A Stool DNA kit (200 preps) containing:
DNA wash buffer
VHB buffer HTR reagent
SLX-Mlus buffer
DS buffer
Proteinase K solution
SP2 buffer
Elution buffer
BL buffer
HiBind DNA mini column
2-ml collection tubes
100% Ethanol (Any brand as long as it meets USP specifications)
Cecal contents (see Support Protocol 1)
Zirconia/silica 1.0-mm diameter homogenization beads (BioSpec Products)
Benchmark Multi-Therm Shaker with Heating
Sterile 10- to 200-μl pipette (Denville)
Incubators
Ice bath
Set of sterile sample labeled 1.5-ml screw-cap homogenizer tubes (VWR)
Precellys 24 lysis and homogenization (Bertin Technologies) (Optional)Vortex mixer (any brand)
Centrifuge (Eppendorf 5409 R)
Sterile sample labeled 2-ml nuclease-free Eppendorf tubes (Eppendorf)
Sterile 1000-μl pipette (Denville)
Dilute the DNA wash buffer from the E.Z.N.A kit with 80 ml of 100% Ethanol (only if the 200-prep kit is purchased). If this was previously done go to step three.
Dilute the VHB buffer from the E.Z.N.A kit with 84 ml of 100% Ethanol (only if the 200-prep kit is purchased). If this was previously done go to step three.
Set one incubator to 70°C.
If a second incubator is available, set it to 95°C.
Place the HTR reagent from the E.Z.N.A kit into the ice bath.
Take between 50 and 100 mg of cecal contents (can be as high as 200 mg) and deposit it into the labeled screw-cap tubes.
Add 10 to 30 Zirconia/Silica beads to each tube and place the tube into the ice bath.
Add 540 μl of the SLX-Mlus buffer from the E.Z.N.A kit to each tube.
If homogenizer is available, homogenize samples at 6,500 rpm for 15 sec, pause for 30 sec, then homogenize for another 15 sec. Samples will look foamy. Go to step 11.
If homogenizer is not available, vortex each sample for at least 10 min or until each sample is thoroughly homogenized.
Add 60 μl of the DS buffer and 20 μl of the Proteinase K solution from the E.Z.N.A kit. Vortex for 30 sec to mix.
Place samples in the incubator (70°C) for 10 min. Vortex each sample twice for 15 sec during the incubation, once at minute 2 and once at minute 7.
Immediately after incubation place the samples in the 95 °C incubator for 5 min. This step is optional but improves DNA isolation from Gram-positive bacteria.
Add 200 μl SP2 buffer from the E.Z.N.A kit and vortex for 30 sec to mix. Place samples for 5 min in an ice bath.
Centrifuge for 5 min at maximum speed (at least 13,000 × g), room temperature.
-
While the samples are spinning, transfer 5 ml of the provided elution buffer to separate 2-ml Eppendorf tubes and incubate them at 65 °C until needed.
Each sample requires 150 μl of elution buffer at the end of this protocol, so adjust the total amount of elution buffer accordingly.
Remove 400 μl of the supernatant from step 15 and transfer it to the first set of labeled nuclease-free Eppendorf tubes. Be careful when transferring to not disturb the pellet.
Make sure the cap is secure on the HTR reagent and shake it vigorously to completely mix the reagent. Cut the tip off of a 1000-μl pipette tip (this helps pipetting the HTR reagent) and transfer 200 μl of the HTR reagent to each sample.
Incubate for 2 min at room temperature and then centrifuge for 2 min at maximum speed, room temperature.
Remove 250 μl of the supernatant and place it in the second set of labeled Eppendorf tubes.
Add 250 μl of the BL buffer from the E.Z.N.A kit and 250 μl of 100% ethanol to each sample and vortex for 10 sec to mix.
Place one HiBind DNA Mini Column into a 2-ml collection tube, both provided in the E.Z.N.A kit. Label each column appropriately.
Transfer the entire sample from step 21 into each respective column (including any precipitates). Centrifuge for 1 min at maximum speed, room temperature.
Discard the filtrate and collection tube. Transfer the column into a new collection tube and add 500 μl of VHB buffer from the E.Z.N.A kit.
Centrifuge for 30 sec at maximum speed, room temperature. Discard the filtrate but reuse the collection tube.
Add 700 μl of the DNA wash buffer to each sample. Centrifuge for 1 min at maximum speed, room temperature. Discard the filtrate but reuse the collection tube.
Repeat step 26 to wash the DNA once again.
Centrifuge for 2 min at maximum speed, room temperature, to dry out the column and remove any excess wash buffer.
Transfer the column to the third set of labeled Eppendorf tubes.
Add 150 μl of the heated elution buffer to the middle of each column and incubate them for 2 min at room temperature.
-
Centrifuge for 1 min at maximum speed, room temperature.
NOTE: Do not be alarmed if some of the Eppendorf caps come off during the centrifugation. Since the caps of the Eppendorf tubes cannot be closed during the centrifugation, the g-force will sometimes rip them off.
Store the samples up to 1 year at −20°C.
BASIC PROTOCOL 2
V4-V4 AMPLIFICATION FOR 16S RRNA GENE SEQUENCING
After DNA isolation, samples can either be directly submitted for bacterial metagenomic shotgun sequencing (see Alternate Protocol for metagenomic analysis) or they can be further modified for 16S rRNA gene sequencing. Here the process for PCR amplification of the fourth variable region of the 16S rRNA gene is described. The V4 region of the 16S rRNA gene has been reported to provide the most taxonomic information of the 8 variable regions present in the 16S rRNA gene, but other variable regions like V5 and V6 can provide comparable results (Yang, Wang, & Qian, 2016). Also, if there is access to a long-read sequencer like the Pacbio Sequel II system, the entire variable region can be amplified and sequenced. Sequencing the entire variable region is one way to get reliable species level taxonomy assignment (Martinez, Muller, & Walter, 2013). Using V4-V4 16S rRNA gene sequencing provides reliable genus level sequencing (Kozich et al., 2013). This protocol will describe how to amplify the V4 region of the 16S rRNA gene by PCR and sequence it.
Materials
Isolated DNA (see Basic Protocol 1)
Nuclease-free water (Any Brand)
V4-V4 primer set (515F and 806R) (10 μM concentration)
Invitrogen Platinum SuperFi Enzyme Kit (ThermoFisher Scientific)
1 × TAE (Tris base, acetic acid and EDTA) buffer
Omnipur agarose (Calbiochem)
GelRed dye (Biotium)
6× Gel loading dye, no SDS (Biolabs)
100-bp DNA ladder (Omega)
Ice bath
NanoDrop UV-Vis Spectrophotometer Lite (Thermo-Scientific)
Sterile 0.2-ml thin-wall PCR Tubes, strips of 8 tubes (Denville)
Sterile 0.5- to 10-μl pipettes (Denville)
Sterile 10- to 200-μl pipettes (Denville)
Sterile 1000-μl pipettes (Denville)
T100 Thermal cycler (Bio rad)
Gel electrophoresis box (Labnet)
ChemiDoc XRS+ (BioRad)
Prepare DNA for amplification
-
1
Thaw the isolated bacterial DNA from Basic Protocol 1.
-
2
Measure DNA concentration on the NanoDrop
This requires only 1 μl of isolated bacterial DNA. Concentration values typically range from 100 ng/μl to 400 ng/μl. In addition, the NanoDrop gives only an estimate of the total bacterial DNA concentration. For a more accurate result, submit samples for quantification on a Bioanalyzer.
-
3
Create a 100 μl aliquot at 10 ng/μl concentration.
The easiest way to complete this is to first figure out how much original DNA to add and then subtract that from 100 to figure out how much nuclease-free water to add. To find out how much original bacterial DNA to add simply divide 1000 by the average concentration. For example, if the average concentration was 254 ng/μl, take 1000/254 = 3.94. Add 3.94 μl of original bacterial DNA sample to (100–3.94 = 96.06) 96.06 pl of nuclease-free water.
-
4
Place aliquots on ice and create 10 μM solutions of forward (515F) and reverse primers (806R).
Amplify master mix and perform PCR
-
5
Place 10 μl of the Platinum Superfi Enzyme mix, 0.4 μl of the forward primer (10 μM), 0.4 μl of the reverse primer (10 μM), and 8.2 μl of nuclease-free water to each PCR tube.
It is important to prepare a master mix. As an example, a master mix for 20 samples can be prepared as follows: The 20-sample master mix should be prepared for 23 samples (for blanks as well as to account for imprecise pipetting) samples and would contain 230 μl (10 × 23) of Platinum Superfi enzyme mix, 9.2 μl (0.4 × 23) of forward primer, 9.2 μl (0.4 × 23) of reverse primer, and200.1 μl (8.7 × 23) of nuclease-free water. Then 19.5 μl of the master mix is placed in each of the 21 PCR tubes (20 samples + 1 blank)
-
6
Add 1.0 μl of the 10 ng/μl aliquot of bacterial DNA and pipette to mix.
-
7Place the caps on the PCR tubes and place the sealed tubes into the PCR machine. Run the PCR machine at these settings:
1 cycle: 2 min 98°C (initial denaturation) 25 cycles: 10 sec 98°C (denaturation) 20 sec 56.6°C (annealing) 15 sec 72°C (extension) 1 cycle: 5 min 72°C (final extension) Final step: Indefinite 4°C (hold). Note that over amplification can affect the results. The more cycles of initial amplification completed, the more populated the abundant species become and it makes it much more difficult to observe the rare species. In addition, as the number of cycles increases, there is a greater chance of contamination amplification.
Gel creation and gel electrophoresis
-
8
While the PCR is running, create a 1 × agarose gel by mixing 1 g of Omnipur agarose in 100 ml of 1 × TAE buffer and microwaving for 2 min.
-
9
Before the gel sets, add 10 μl (10 μl per 100 ml of gel) of GelRed dye to the liquid gel.
GelRed can be used instead of ethidium bromide for several reasons: First, it is safer to use in the laboratory. Second, there is no need to add extra dye to the running buffer, so the buffer can be reused multiple times. Third, and most importantly, the gels are visibly clearer and there is no ethidium bromide band in the gel.
-
10
Once PCR is finished, add 5 μl of the PCR sample to 2 μl of 6× loading dye (BioLabs) and 4 μl of nuclease-free water in a separate tube.
-
11
Fill the gel electrophoresis box with 1 × TAE buffer and add 5 μl of 100-bp DNA ladder to the edges of the gel. Add the entire sample from step 10 to the empty wells. Run at 80 V for 50 min to an hour. The gel run will be complete when the purple band is ¾ of the way down the gel. The gel can also be placed back into the gel box for further running if the bands have not separated enough.
-
12
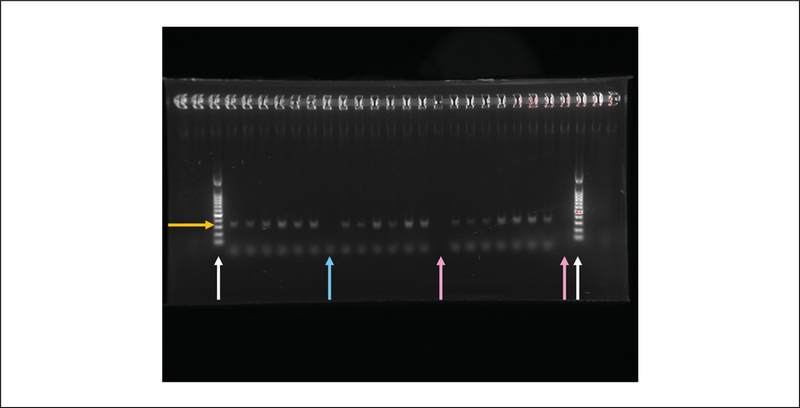
When the bands are at least ¾ the way down the gel, remove the gel and analyze it with the ChemiDoc. The correct band length should be 350 bp.
Do not be alarmed if the bands are not very bright (Fig. 2). Duller bands are preferred because another round of PCR will be completed before sequencing.
-
13
Submit samples to a sequencing core or a sequencing company and request 250 × 250 paired end sequencing on the Illumina Miseq.
IMPORTANT NOTE: Each sequencing core or sequencing company is different and may require a different end product for sample submission. Most will take the sample after the first round of PCR because this generates amplicons of the 16S rRNA gene variable region of the users choosing. If they require more PCR follow the detailed instructions provided by the sequencing core or company of the users choosing.
Depth is also an important specification to decide prior to sequencing. Typically, the Illumina Miseq will provide 10 million reads split across each of the user’s samples. This means if the user has 50 samples in one run on the Illumina Miseq the user will get roughly 200,000 reads per sample. Depth preference is generally between 50,000 and 100,000 reads per sample (Jovel et al., 2016)
When the data is returned, it should be demultiplexed, generating two files for each sample in FASTQ format.
Figure 2.
An example of the 1 x gel used to check the size of the amplified 16S V4V4 region.
BASIC PROTOCOL 3
16S rRNA GENE AMPLICON DATA ANALYSIS
The following protocol is directly based on the mothur miseq SOP created by Dr. Patrick Schloss. The Web site can be found here, https://www.mothur.org/wiki/MiSeq_SOP, and if this method is used, the 2013 paper by Kozich et al. must be cited (Kozich et al., 2013). The following command progression is exactly how it appears in the Schloss SOP, but the file names, values and explanations are different. For a more detailed explanation, please see the above website and consult the Wiki. This protocol covers the basic mothur analysis, normalization, identification of significantly different bacterial taxa, and Generalized unifrac analysis. If one chooses, QIIME is an alternative 16S rRNA gene sequence analysis pipeline, and more information can be found at https://qiime2.org/ (Caporaso et al., 2010).
This protocol requires that the analysis be performed within a Mac or PC Linux environment through the application terminal. It is also recommended that at least 8 processors with at least 100 Gb of memory be used. This analysis can be done on a personal laptop, but it is extremely time consuming; therefore, the use of an external server or a computing cluster is highly recommended. Since mothur is terminal-based, basic command line knowledge is required for this analysis. Also, all graphing and some statistical analysis can be done with R studio, thus basic R knowledge or an alternative statistical/graphing software is required.
The mothur github site and SOP describes how to download and install this software on a personal computer (https://github.eom/mothur/mothur/releases/tag/v1.39.5 and https://mothur.org/wiki/MiSeq_SOP). If one is using an external server or a computing cluster, the download is a little more complicated because the user does not have administrative privileges. The easiest way to “install” mothur on an external server is first by downloading the most recent version on the mothur github site. There are multiple options of how to download mothur, and the one used for this procedure is mothur.linux_64.zip. This file can be copied over to the external server or a cloud cluster and unzipped there. Then simply add the mother folder to the user’s path with the command export PATH= “$PATH: ~ /mothur”. To run mothur, simply type mothur in the command line.
In addition, on the mothur miseq SOP, there are several files that are required for the analysis. The first is the SILVA alignment file, which can be found under the Logistics section of the mothur miseq SOP. This provides a zip file, and only the silva.bacteria.fasta file is needed for this analysis. The SILVA alignment file is regularly updated, and new versions of this file can be downloaded from the Silva database Website (https://www.arb-silva.de/). The next two necessary files can be found directly below the SILVA link, in a link titled mothur-formated version of RDP training set. This will provide a second zip file that contains only two files; both are needed for this analysis. Like the SILVA alignment file, the RDP trainsets are also updated regularly and can be found at the RDP website (https://rdp.eme.msu.edu/mise/resourees.jsp#aligns). Once all three files are obtained (silva.bacteria.fasta, trainset9_032012.pds.fasta, and trainset9_032012.pds.tax), create a work folder on the external server or computing cluster for the mother analysis and move these files into it. For this analysis the provided RDP trainsets and the provided SILVA alignment files from the mothur miseq SOP (version 9) will be used. For future use, RDP and SILVA regularly puts out new trainsets and alignment files, as mentioned above.
Materials
Mac computer (or Windows with Linux environment)
External server or computing cluster with an allocation of at least 100 GB and 8 processors (can use personal computer but will drastically increase computational time)
Sequenced data (see Basic Protocol 2)
16S analysis set up and contig creation
-
1
Before the analysis, be sure to read the above information and have mothur installed and acquire all the necessary files. Check to see that you are using the most current version of mothur.
-
2
With the raw data make a stability file. This is a file that will help mothur know what two paired end files to combine and name it according to the user created sample names.
-
This file can be made with a text editor and will look like the example below (and an example stability file can be found in the provided sample data).
501 501_S21_L001_R1_001.fastq 501_S21_L001_R2_001.fastq
502 502_S22_L001_R1_001.fastq 502_S22_L001_R2_001.fastq
503 503_S23_L001_R1_001.fastq 503_S23_L001_R2_001.fastq
504 504_S24_L001_R1_001.fastq 504_S24_L001_R2_001.fastq
505 505_S25_L001_R1_001.fastq 505_S25_L001_R2_001.fastq
The first column contains the sample names; in this case they are 501, 502, 503, 504, and 505. After each sample name, it is important to tab, not space, to the next column. The second column contains the first file name for each pair. In this case, 501_S21_L001_R1_001.fastq is the name of the first file of the 501 pair. Again tab to create the third column, the second file name for each pair. In this case 501_S21_L001_R2_001.fastq is the second file name for the 501 pair. Continue this for each sample in the run.
This file should be named after the user’s project. In this example this file will be named Test.stab.txt. This file should then be sent to the mothur work folder along with all the FASTQ data and the required files mentioned above.
-
-
3
Execute mothur and run make.contigs (file=Test.stab.txt, processors=8).
Notice how the stability file created in the previous step is directly used and how mother needs to be told to use 8 processors. If mothur is not instructed how many processors to use, the default is 1.
This process will take about 1 min per sample and will result in six files. The only two required for this analysis are Test.stab.trim.contigs.fasta and Test.stab.contigs.groups.
Notice how the first part of these file names is the name of the stability file. This is why it is important to name the stability file something related to the experiment.
-
4
With the output files, run a summary with the command summary.seqs(fasta=Test.stab.trim.contigs.fasta).
The result will be a table that breaks down the fasta file from step 3. An example of this can be seen in Table 1.
The rows break down the data into various segments defined by the different columns. For example, the 25%-tile row says that 25% of the data has a start site at 1, an end site at 292, they are all at least 292 bases long with 0 ambiguous sites, an average of three polymers and has 670164 sequences in this group. This is typical, and the only column that is important from this specific summary file is the NBases column. Since the above protocol resulted in a 350 bp insert of the V4 region in Basic Protocol 2, the user would expect the average base length of the sequences to be around 320 base pairs long.
Table 1.
Example of Output from the Summary.seqs Command Described in Basic Protocol 3, Step 4
| Start | End | NBases | Ambigs | Polymer | NumSeqs | |
|---|---|---|---|---|---|---|
| Minimum | 1 | 90 | 90 | 0 | 3 | 1 |
| 2.5%-tile | 1 | 292 | 292 | 0 | 3 | 67017 |
| 25%-tile | 1 | 300 | 300 | 0 | 4 | 670164 |
| Median | 1 | 301 | 301 | 0 | 4 | 1340328 |
| 75%-tile | 1 | 307 | 307 | 1 | 5 | 2010492 |
| 97.5%-tile | 1 | 311 | 311 | 13 | 6 | 2613639 |
| Maximum | 1 | 602 | 602 | 128 | 300 | 2680655 |
| # of Seqs | 2680655 |
Trimming off large reads, condensing for unique reads, and preparing for alignment
-
5
Screen the sequences with the command screen.seqs(fasta= Test.stab.trim.contigs. fasta, group=Test.stab.contigs.groups, maxambig=0, maxlength=320).
This command screens the data and trims off any bad reads. The maxambig=0 part of the command indicates that this command will cut any sequence with ambiguous bases. Referring back to the above table the user can see that 128 sequences have ambiguous bases. In addition, this command cuts anything larger than 320 bases (maxlength = 320). 320 was picked because according to the above table, 97.5% of the data is 311 base pairs long or smaller and it is recommended on the mothur miseq wiki to go a few base pairs higher than the number at the 97.5% mark.
The screen.seqs command specifications is very dependent on the data, so the max length will change depending on which variable region is used and the type of Illumina miseq run is completed (150 × 150 or 250 × 250). As a general rule, the user wants the max length to be at least the nBases number for the 97.5-tile group.
-
6
Remove duplicate sequences by running unique.seqs (fasta= Test.stab.trim.contigs. good.fasta).
This step is included to save computational time by condensing the data. The resulting files represent a fasta file with only unique sequences and a name file that includes how many times each sequence occurred. This way when aligning and cleaning the data, each sequence is only seen once.
-
7
Combine the resulting name file from step 6 and the group file from step 3 to form a count table with the command count.seqs(name= Test.stab.trim.contigs.good. names, group=Test.stab.contigs.good.groups)
This command will now create a count table that will have the names for every unique sequence and how many times they occur in each sample.
-
8
Optional: To save on computational time, the silva.bacteria.fasta file can be modified to only include alignment for the V4-V4 region of the 16S rRNA gene with the command pcr.seqs(fasta=silva.bacteria.fasta, start=11894, end=25319, keepdots=F, processors=8).
This step will only work if the V4 region was sequenced but this step is not necessary for this analysis and will only save computational time.
Alignment and clean-up of the reads, preparing for classification
-
9
Align the raw reads to the SILVA database with the command align.seqs(fasta=Test. stab.trim.contigs.good.unique.fasta, reference= silva.bacteria.pcr.fasta, flip=t).
The reference file used in this example is the edited one from step 8. If step 8 is not completed the file for the reference option will simply be silva.bacter.fasta
With the optional step 8 the alignment time was about 9 min for 1618841 sequences.
Without the optional step 8 the alignment time was 30 min to align 1618841 sequences.
The flip=t option is included to attempt to align the reverse complement of sequences that do not align in the forward direction. This option will also produce more alignments and a more comprehensive look at the microbiome composition.
-
10
Investigate the alignment with another summary command, summary.seqs(fasta=Test. stab. trim. contigs. good. unique. align, count = Test. stab. trim. contigs. good. count_table).
The purpose of this step is to further clean the data by picking reads that start and end at particular values.
The summary table will be the same format as the one obtained in step 4 but the values will be different. Table 2 provides an example of this summary table.
When using the modified SILVA file, the start sequence will almost always be 1. The important variables to look at are the End and the NBases column. The Nbases column will show how large the sequences are and they should be similar to the cutoffs from step 4. In this case nothing should be larger than 320 and there should be no ambiguity. The end column will be used in the next step.
-
11
Screen the sequences again for poor alignment and any alignment errors with the command screen.seqs(fasta=Test.stab.trim.contigs.good.unique.align, count=Test.stab.trim.contigs.good.count_table, start=1, end= 13424, maxhomop=8).
The values for the start, end, and maxhomop options can be found in the summary file generated in step 10. The start option will select any sequence that starts at or before this value. The end value will select any sequence that ends at or after any value and the maxhomop removes any sequences that have more than 8 homopolymers. These details are important to know because occasionally the summary from step 10 will show that 50% of the values have an end site of 13424 and 50% will have an end site of 13425. Picking the higher value makes logical sense but this command actually wants the lower value because it selects any sequence that ends at or after the selected value. Deciding the threshold of homopolymers is completely arbitrary and 8 is used in this methods paper because 8 are used in the miseq SOP (Kozich et al., 2013).
-
12
Filter the raw data to remove any overhangs from the alignment with the command filter.seqs(fasta=Teststab.trim.contigs.good.unique.good.align, vertical=t).
The vertical option is used to ignore certain characters like the ‘-’and ‘.’ to prevent them from being removed.
-
13
Remove any duplicate sequences that resulted from the alignment with a second unique command, unique.seqs(fasta=Test.stab.trim.contigs.good.unique.good. filter.fasta, count=Test.stab.trim.contigs.good.good.count_table).
Like step 6, this step saves only the unique sequences and updates the count file with the number of times each sequence appears in each sample.
-
14
Further clean the data by addressing minor sequencing errors and combining sequences that are only different by 2 nucleotides with the command pre.cluster (fasta=Test.stab.trim.contigs.good.unique.good.filter.unique.fasta,count=Test.stab. trim.contigs.good.unique.good.filter.count_table, diffs=2).
The pre.cluster command is based off an algorithm developed for pyrosequencing by Sue Huse (Huse, Welch, Morrison, & Sogin, 2010).
-
15
Remove chimeras from the data with the command chimera.uchime(fasta=Test.stab.trim.contigs.good.unique.good.filter.unique.precluster.fasta, count=Test. stab.trim.contigs.good.unique.good.filter .unique.precluster.count_table,dereplicate=t).
Depending on the version ofmothur, this command may be called something else. Later versions ofmothur use the command chimera.vsearch, but the options within the command are exactly the same.
If this command discovers a chimera present in one sequence in one sample, the default option is to remove that sequence from every other sample in the data set, regardless of the presence of chimeras. To prevent this, the dereplicate=t option is implemented. This pulls out all identified sequences with chimeras and what sample they are present in. The next command will remove the chimeric sequences only from the samples where they were discovered.
-
16
Remove the chimeras from the FASTA file with the command remove.seqs (fasta=Test. stab. trim. contigs. good. unique. good. filter. unique. precluster. fasta, accnos=Test.stab.trim.contigs.good.unique.good.filter.unique.precluster.denovo.uc hime.accnos).
-
17
Optional: Change the file names to something smaller with the commands system (cp Test. stab. trim. contigs.good. unique. good. filter. unique. precluster. pick. fasta test.final.fasta) and system(cp Test.stab.trim.contigs.good.unique.good.filter.un ique.precluster.denovo.uchime.pick.count_table test.final.count).
This step is used to clean up the file names. Having long file names can lead to frustration and errors. At this point the data cleaning is completed and the file names can be shortened with the above commands if desired.
In addition, at any time, instead of typing in the entire FASTA or count name, one can use “current” to call the most recent FASTA or count file. For example, instead of typing summary.seqs(fasta=Test.stab.trim.contigs.good.unique.good.filter.fasta, count=Test.st ab.trim.contigs.good.good.count_table), one could type summary.seqs(fasta=current, count=current) to get the same output.
Table 2.
Example of Output from the Summary.seqs Command Described in Basic Protocol 3, step 10
| Start | End | NBases | Ambigs | Polymer | NumSeqs | |
|---|---|---|---|---|---|---|
| Minimum | 1 | 1984 | 25 | 0 | 3 | 1 |
| 2.5%-tile | 1 | 13424 | 290 | 0 | 3 | 46530 |
| 25%-tile | 1 | 13424 | 292 | 0 | 4 | 465299 |
| Median | 1 | 13424 | 292 | 0 | 4 | 930597 |
| 75%-tile | 1 | 13424 | 292 | 0 | 5 | 1395895 |
| 97.5%-tile | 1 | 13425 | 293 | 0 | 5 | 1814663 |
| Maximum | 10024 | 13425 | 312 | 0 | 10 | 1861192 |
| # of Seqs | 1618841 |
Read classification
-
18
Classify the sequences to the RDP trainsets with the command classify.seqs(fasta =Test.stab.trim.contigs.good.unique.good.filter.unique.precluster.pick.fasta, count =Test.stab.trim.contigs.good.unique.good.filter.unique.precluster.denovo.vsearch. pick.count_table, reference=trainset9_032012.pds.fasta, taxonomy=trainset9_03 2012.pds.tax, cutoff=75).
The FASTA and count names can vary depending on whether step 17 was completed.
As mentioned in the introduction of this protocol, the taxonomy and reference files can vary depending on which version is used.
The cutoff value is, again, arbitrary. This value provides a threshold of classification. As it is now, only 75% of the sequence has to align to the RDP trainset to be classified. This value can be higher leading to a more stringent analysis, or lower leading to a less stringent analysis.
-
19
Create a text file of the taxonomic summary obtained from step 18 with the command system(mv Test.stab.trim.contigs.good.unique.good.filter.unique.preclus ter.pick.pds.wang.tax.summary Test.summary.txt).
This command creates a text file that now can be opened on a personal computer.
This file can be copied and pasted into excel for normalization. To normalize this data, simply divide all taxonomic values in each sample by the root value in each respective sample. This normalization will show the percentage each taxa for each taxonomic level. This means that the members of each taxonomic level (phyla, class, order, family, genus) will add up to 100%.
Significance can be found with a students’ T-test.
-
20
Proceed to Support Protocol 2 for population-based gut microbiome analysis, if needed.
SUPPORT PROTOCOL 1
CECAL CONTENT EXTRACTION
The cecum is a rich source of intestinal microbiota in mice. However, other sources including feces or stool, intestinal tissues, or biopsies can be used.
The mice are transferred to a euthanasia chamber (Patterson Scientific) for CO2 asphyxiation, which is then followed by cervical dislocation. The cecal contents are obtained immediately following mouse dissection. The cecum is the intraperitoneal pouch connected to the junction of the small and large intestines. It is located at the beginning of the ascending colon of the large intestine. Once the cecum is identified, it will be resected with a surgical scissors from the rest of the digestive tract and placed on a piece of foil. Then, roll the cecum using a sterile pipette tip (1000-μl tip works the best), and the contents will easily come out. Use the same pipette tip to then scrape up the cecal contents and place them into a 1.5-ml screw-top vial. The foil allows for an easy collection of the cecal contents once they have been removed from the cecum. All procedures must be performed in accordance with the Institute of Laboratory Animal Resources guidelines and, in the case of this protocol, were approved by the Pennsylvania State University Institutional Animal Care and Use Committee.
COMMUNITY-BASED ANALYSIS OF THE GUT MICROBIOME
This protocol is intended to describe the steps for a Generalized Unifrac analysis with the R package GUniFrac. Generalized unifrac is a measure that combines weighted and unweighted unifrac (Chen et al., 2012). Weighted unifrac is a measure used to analyze differences in abundant species within several populations. Unweighted unifrac is a measure of the differences in rare species within several populations. Generalized unifrac combines these measures to look at both rare and abundant species between two or more populations. This algorithm works by aligning a table of raw sequence reads to a customized hierarchal phylogenic tree. The output is a plot showing the two populations (control and treatment) and how distinctly different, or similar they are. This protocol will only address using two populations (control and treatment) but GUniFrac analysis can be done with many groups as well. As mentioned above, an R markdown file and sample files have been included with this unit discussion.
Materials
Computer with R Studio installed
Analyzed raw sequence reads (see Basic Protocol 3)
External server or computing cluster with an allocation of at least 100 GB and 8 processors (can use personal computer but will drastically increase computational time)
GUnifrac file creation and Mothur software
-
1
Return to the folder with the mothur output files via terminal and create a new folder for GUnifrac analysis.
-
2
Move the Test.stab.trim.contigs.good.unique.good.filter.unique.precluster. pickfasta (or test.final. fasta) and the Test.trim.contigs.good.unique.good.filter.unique.precluster.denovo.vsearch.pick.count_table (or test.final.count) file to the new folder.
It is recommended to rename these files as described in Basic Protocol 3, step 17. Shortening the names of these files makes the downstream analysis much simpler.
-
3
Start mothur and create a distance table with the command dist.seqs(fasta=test.final.fasta, output=lt, processors=8).
-
This command will take several hours and may crash. If it does crash, the command line will say “killed”, and the mothur program will close. In the event that dist.seqs crashes, follow the following steps:
Take a subsample of the fasta and the count files with the command sub.sample(fasta=test.final.fasta, count=test.final.count).
This command will take a random 10% of test.final.fasta and the same random 10% from test.final.count and create files with the name test.final.subsample.fasta and test.final.subsample.count.
-
If a larger subsample is desired, run the command count.groups(count=test. final.count). This will show how many sequences are in each group and how many total sequences are present (shown below).
501 contains 20554.
502 contains 4474.
503 contains 19336.
504 contains 2101.
505 contains 11445.
601 contains 23595.
602 contains 22541.
603 contains 22195.
604 contains 12943.
605 contains 13733.
Total seqs: 152917.
Note that the above sequences are uneven for each group. This occurs because the Illumina miseq provides 10 million reads, randomly distributed between the samples of the run. The above example data comes from a 50-sample run, giving each sample about 150,000 to 200,000 reads. Concordantly step iii takes a 10% subsample of the data. This means that each sample should have between 15,000 and 20,000 reads. The above data table is variable, but most samples are around that range.
The total sequences are 152917. For a 50% subsample take 50% of 152917, which is 76459. Run the command sub.sample(fasta=test.final.fasta, count=test.final.count, size=76459).
The size option tells the command to take 76459 random sequences from both the files, thus resulting in a 50% random sampling.
Rerun dist.seqs(fasta=test.final.subsample.fasta, output=lt, processors=8).
If it crashes again, take a smaller subsample.
Note this protocol will use file names that have not been subsampled. If a subsample is needed, change the names accordingly.
-
-
4
Create a phylogenic tree with the command clearcut(phylip=test.final.phylip.dist).
When this command is running, it will appear that the command line is frozen, but that is completely normal. When it is complete, this command will result in test.final.phylip.tre.
-
5
Exit mothur and change directories on terminal to be in the directory with all the GUnifrac files. Convert the count file to a text file with the command cp test.final.count test.final.count.txt.
-
6
Create a meta file containing sequence names and group identification.
This can be done in excel where the first column, labeled “samples,” are the respective sample names and the second column, labeled “treatment,” are the treatment groups (control, treatment).
For this analysis, the metafile will be named test.meta.txt.
-
7
Open R studio and install the GUnifrac package.
Loading and formatting the files for GUnifrac analysis
-
8
Import the resulting tree file from step 4 with the command read.tree(file=“//Users/setup/Desktop/mothur_files/test.final.phylip.tre”)-> test.tre.
The file path is where the tree file is on the computer.
-
9
Search the imported tree for nodes by typing test.tre in the R command line.
This is very important because a tree with nodes will not work with the GUnifrac command.
If a tree has nodes, it will be only one sequence and can be found under ‘Node labels’ in the output.
If there are no “Node labels” or if “Node labels” does not show any sequence ids then proceed to step 11
-
10
Open test.final.count.txt and search (using command f) for the node label from step 9. When found, delete this sequence and the entire row associated with it, and save.
-
11
Import test.final.count.txt into R studio, making sure the first column is used as row names with the command read.delim(“~/Desktop/mothur_files/test.final.count.txt”, row.names=1)->test.count.
Again the file path will be different for everyone, adjust accordingly.
This command imports the count table into the variable test.count.
This file will also be referred to as the OTU table by the GUniFrac software.
-
12
Test the row names and column names with the commands head(row.names (test.count)) and colnames(test.count), respectively.
The head modifier is used with the row names because there will be over 100,000 rows
The row names should look like “M00946_96_000000000-AEE8U_1_1119_3781_11413”.
The column names should be the sample names. If the sample names are numbers, for example: 501, 502, 503 . . . 605. They will appear different after the colnames command. They will look like “X501, X502, X503 . . . X605”. This occurs because when importing, R puts an X in front of the column names to distinguish them from numbers. To fix this issue run the command colnames(test.count)=c(“501”, “502”, “503”, . . . ,”605).
-
13
Transpose the rows and columns with the command t(test.count)=test.transpose. count. Check the column names with the command head(colnames(test.trans pose.count)).
A second check is required because occasionally the row names do not get transposed to the column names.
If they did not get transferred, use the command colnames(test.transpose.count) = row.names(test.count).
Running GUnifrac analysis and creating the representative figure
-
14
Run GUnifrac with the command GUniFrac(test.transpose.count, test.tre, alpha=c(0,.5,1))$unifracs -> TestUni
This command will take roughly half an hour to run, and will most likely end in an error. If it immediately ends with the error “Warning message: In GUniFrac(test.transpose.count, test.tre, alpha=c(0,.5,1)): The tree has more OTUthan the OTU table!” there is a problem, please see the above troubleshooting, or the Troubleshooting section at the end of this protocol.
If the above error is seen at the end of 10 to 30 min, then the command worked. This is because the command will work if there are less sequences in the count table then are represented on the tree, but it will not work if there are more sequences in the count table than are represented on the tree. When deleting the node label from the count table, the user is reducing the count table by 1. The count table is now one less than the mapped tree, thus this error will be reported at the end of the analysis.
The alpha value is used to tell how much weight to put on abundance species, so in this example alphas of 0, 0.5, and 1 are being used. An alpha of 0 will put no weight on abundance species, an alpha of 0.5 will put half the weight on abundant species, and an alpha value of 1 will put all of the weight on abundant species. For this analysis the most important alpha value is 0.5 because this corresponds to a generalized unifrac measure.
The resulting data frames will be in the variable TestUni.
-
15
Extract the generalized unifrac data frame with the command TestUni[,, “d_0.5”]->TestGU.
If interested, the weighted and unweighted unifrac analysis can be extracted with the command TestUni[,, “d_1”]->TestWandTestUni[,, “d_UW”]->TestUW, respectively.
-
16
Import the meta file and create a meta variable with the command read.delim (“ ~/Desktop/mothur_files/test.meta.txt”)$treatment->meta.
This command will import the treatment groups into a variable called meta.
-
17
Create a color and a shape variable with the commands coul= coul<-c(“red”, “blue”) and shape= c(15, 15, 15, 15, 15, 16, 16, 16, 16,16), respectively.
The colors can be changed to any color desired.
The shape codes come from the PCH table (https://www.endmemo.com/program/R/pchsymbols.php), which has numerical values for different shapes. In this case they are squares (15) and circles (16).
-
18
Plot the results with the command s.class(cmdscale(TestGU, k=2), fac=meta, cpoint=1, pch=shape, col=coul).
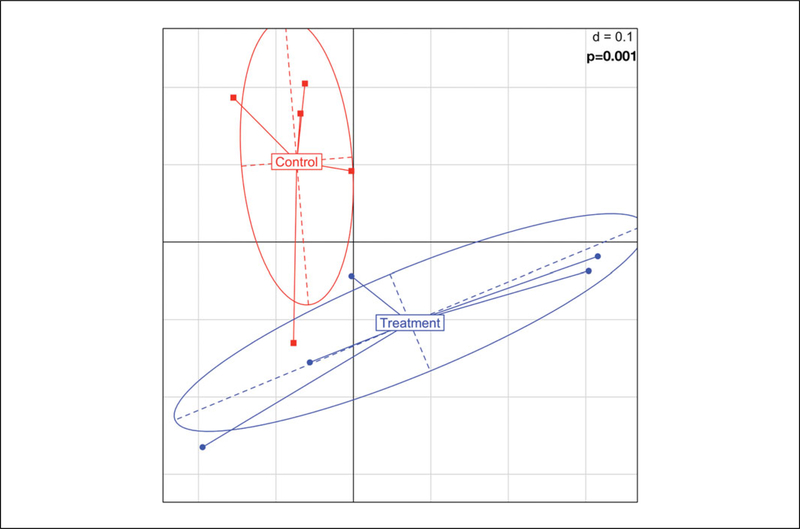
An example of a GUniFrac graph can be seen in Figure 3.
Since there are no axes measurements, the “d=0.1” measurement represents the length of each axis in the graph space.
-
19
Check for statistical significance with the command Adonis(as.dist(TestGU) ~ meta).
The Adonis command computes a multivariate analysis of variance using distance matrices. Since GUnifrac is a measurement of phylogenic distance, the Adonis command is the logical choice for statistical significance. Adonis is also recommended for use in the GUnifrac package details and will result in a p-value (Chen et al., 2012).
Figure 3.
An example of GUniFrac Output. The use of different colors and shapes make the population level differences clear and easy to see. The p-value must be manually added to the graph after running ADONIS.
ALTERNATE PROTOCOL
METAGENOMIC ANALYSIS OF THE GUT MICROBIOME
This protocol describes the process of metagenomic analysis with the HUMAnN2 software from the Huttenhower laboratory (Abubucker et al., 2012). HUMAnN2 is a powerful pipeline combining a taxonomic analysis through the software Metaphlan2 (Truong et al., 2015), alignment of raw sequences to a bacterial reference genome with Bowtie2 (Fonslow et al., 2013), and a secondary alignment to a protein database for unmapped reads with DIAMOND (Buchfink, Xie, & Huson, 2015). Together these programs work together to produce a comprehensive list of metabolic pathways present in the gut microbiome. This information can be used to help predict and validate metabolic changes seen in the host. Significantly different pathways will be discovered with the use of LEfSe (Linear discriminant analysis Effect Size) which combines statistical significance and biological relevance with the Wilcoxon and the Kruskal-Wallace statistical tests, respectively (Segata et al., 2011). This protocol uses the bacterial DNA isolated in Basic Protocol 1. There is also an online manual for HUMAnN2 on bitbucket (https://bitbucket.org/biobakery/humann2/wiki/Home).
Materials
Bacterial DNA (see Basic Protocol 1)
NanoDrop UV-Vis Spectrophotometer Lite (Thermo-Scientific)
Sequencing core facility or an Illumina Hiseq 2500
External server or computing cluster with an allocation of at least 100 GB and 8 processors (can use personal computer but will drastically increase computational time)
HUMAnN2 installed with all required dependencies (can be found at https://bitbucket.Org/biobakery/humann2/wiki/Home#markdown-header-requirements)
Internet connection and access to the Huttenhower galaxy site
Excel or Numbers
Preparing and submitting raw DNA for metagenomic analysis
-
Measure DNA concentration on the NanoDrop.
-
This requires only 1 μl of isolated bacterial DNA. Concentration values should range from 100 ng/μl to 400 ng/pl.
-
i
If values exceed 400 ng/μl, this is not an issue and less input bacterial DNA will be used. In addition, most sequencing cores will test the quality of DNA before sequencing
-
ii
If values are lower than 100 ng/μl, then PCR may be required to increase the input material before sequencing. This is not an issue but can introduce PCR bias into the results. PCR bias occurs when abundant species are amplified and end up masking rarer species. PCR can also amplify contaminants which can skew results
-
i
With metagenomic shotgun sequencing, no PCR is needed before submission as long as there is at least 1 to 2 μg of DNA.
-
Submit the DNA isolates to a sequencing core or an independent company for Illumina hiseq 150 × 150 sequencing with the PCR-free library construction kit.
Please note that HUMAnN2 cannot run both partners of a paired-end read simultaneously. This protocol will go through using only one partner from each pair. Due to this, single-end sequencing can be completed instead of paired-end sequencing if HUMAnN2 is the only analytical pipeline being used. If, however, further analysis is required, it is recommended to use the paired end sequencing because most analytical pipelines require paired end sequencing.
Install HUMAnN2 and all relevant dependencies
-
3
Install HUMAnN2 according to instructions, making sure that all dependencies are installed.
The dependencies include: MetaPhlAn2, Bowtie2, Diamond, and python (at least version 2.7). They should be automatically installed when installing HUMAnN2.
-
This can be difficult without administrative permissions. This will be the case if an external server or a computing cluster is being used for analysis.
-
i
To get around this, import the latest humann2.tar.gz file on to the server.
-
ii
Decompress the file, enter the resulting directory and run python setup.py install –user.
-
iii
This will put all the dependencies in a /.local directory, bypassing the need for administrative permissions.
-
iv
The user must also run export PATH= “$PATH:~/.local/bin”.
-
i
-
4
Install the chocophlan and uniref databases using the commands humann2_databases -download chocophlan full $Path_to_install and humann2_databases -download uniref uniref90_diamond $Path_to_install.
The $Path_to_install will be modified to the path of the desired location of the database. This is important because the configuration file that is used to run HUMAnN2 will be updated with this command, so do not move the databases once installed.
In addition, together both databases are about 20 GB.
It is also important to make sure that the version of HUMAnN2 being used is v 0.11.1 or higher. This command will not work with earlier versions of HUMAnN2.
-
5
Import the raw sequence file from the Illumina Hiseq to the server or computer cluster being used.
-
6
Create a directory for the output.
Run HUMAnN2 on the metagenomic reads
-
5
Run HUMAnN2 with the command humann2 -input./Raw_sequence_files/Test1. Rl.fastq -output./output_files -metaphlan./metaphlan2/-threads 8.
The paths to the input and the output depends on the environment being used and will be different for everyone.
It is important to tell HUMAnN2 where to find metaphlan2, because when running on the external server adding the location of metaphlan2 to the /.local directory does not work. Thankfully, the HUMAnN2 command allows the user to specify where the metaphaln2 dependencies are.
-
At any point HUMAnN2 crashes and has an error describing that bowtie2 or diamond cannot be found, they can also be added to the above HUMAnN2 command.
-
i
The resulting command could potentially read, humann2 –input. /Raw_sequence_files/Test.R1.fastq -output ./output_files-metaphlan ./metaphlan2/-bowtie2 ~/bowtie2–2.2.5 -diamond ~/diamond-0.7.9/bin -threads 8.
-
ii
This will work as long as the bowtie and diamond dependencies are at the ~ (home) location.
-
iii
With 8 processors, this process will take 12 hr to run. The run time can be shortened with more available processors.
-
i
-
The resulting files will be a pathway abundance file, a pathway coverage file and a gene families file. The pathway abundance file has abundance values for all HUMAnN2 identified pathways. The pathway coverage file contains the percentage of each pathway present. This is represented with a value from 0–1, with 1 being 100% covered and every gene family present in the pathway. The gene families file contains all the gene families identified with HUMAnN2.
This analysis will not utilize the gene families file, but the gene families file could be used for de novo pathway creation.
-
4
Transport all pathway abundance files and all coverage files off of the external server and onto the desktop.
-
5
Install the latest version of HUMAnN2 onto the computer in use, but do not install the databases.
Gene table editing and final figure creation
-
6
Combine the pathway abundance and pathway coverage file for each sample with the command humann2_join_tabels -input./test.1 -output./test.1.combo.txt.
Each sample should get its own directory and each respective pathway abundance and pathway coverage file will be placed into that directory. In this case the directory is called test.1. This directory contains the files test1.pathwayabundance.txt and test.Lpathwaycoverage.txt.
When combined, the resulting file will be called test.l.combo.txt and will contain the pathway abundances and respective coverages for all pathways discovered in sample test.l.
-
7
11. Copy and paste the contents of the combo files into Excel or Numbers. Sort the table by coverage (high to low), remove all pathways below a 0.3 (30%) coverage and create a new text file with the trimmed data.
The 0.30 (30%) cut off is completely arbitrary and can be higher or lower depending on the needs of the experiment.
The reason a cutoff value is needed is because multiple gene families can belong to multiple pathways, so the less the coverage is, the less likely the pathway is to be actually present.
The edited combo file should be placed in a directory called test.edits.
-
8
Combine all edited combo files into one file which contains all the pathway abundances with at least a 30% coverage with the command humann2Join_tabels - input./test.edits -output./Test.whole.txt.
Test.whole.txt contains all the pathway abundances with at least a 30% coverage for the experiment.
Depending on the version of HUMAnN2, the coverages may or may not be combined with the pathway abundances. If this is the case just delete the coverage columns, leaving only the pathway abundances.
-
9
Open Test.whole.txt and clean up the labels by replacing the column names with the sample names (testl, test2, test3 . . . testn). Also add a new row directly below the column names and title the row ‘Treatment’ and add the appropriate treatments to each sample.
The Test.whole.txt file example can be seen in Table 3.
A cleaned version can be seen in Table 4.
-
10
Export the cleaned version of Test.whole.txt as Test.whole.clean.txt and import it to the Huttenhower galaxy page (https://huttenhower.sph.harvard.edu/galaxy/).
-
11
Go to LeFSe tab A) and select the uploaded file. Make sure that rows are selected as the vector option and select ‘Treatment’ for the class option and “Sample” as the subject option. Click execute.
-
12
Move to LeFSe tab B) and select the file created from the previous step. Adjust the alpha values if needed (default is p = 0.05). Click execute.
-
13
Move to LeFSe tab C) select the resulting file from B and adjust the DPI if necessary and click execute.
The resulting file will show the significantly different and biologically relevant pathways from the gut microbiome.
Table 3.
Example of the First Four Rows of Test.whole.txt
| Test1_R1_ abundance | Test2_R1_ abundance | Test3_R1_ abundance | Test4_R1_ abundance | |
|---|---|---|---|---|
| PWY-6531: mannitol cycle | 1795.578222 5069 | 1725.750470 4103 | 2029.961280 5819 | 2440.945348 1149 |
| PWY-5097: lysine biosynthesis VI | 2323.337957 2841 | 2022.847331 4703 | 1611.732967 2909 | 1280.102152 8578 |
| PWY-5100: pyruvate fermentation to acetate and lactate II | 1675.222610 1548 | 1792.684429 8156 | 2250.172944 7694 | 2139.362683 4727 |
| VALSYN-PWY: valine biosynthesis | 1577.517860 9018 | 1469.875122 2891 | 1902.632068 1466 | 2261.711518 9351 |
Table 4.
Example of the First Four Rows of Test.whole.clean.txt
| Sample | Test 1 | Test 2 | Test 3 | Test 4 |
|---|---|---|---|---|
| Treatment | Control | Control | Treatment | Treatment |
| PWY-6531: mannitol cycle | 1795.578222 5069 | 1725.750470 4103 | 2029.961280 5819 | 2440.945348 1149 |
| PWY-5097: lysine biosynthesis VI | 2323.337957 2841 | 2022.847331 4703 | 1611.732967 2909 | 1280.102152 8578 |
| PWY-5100: pyruvate fermentation to acetate and lactate II | 1675.222610 1548 | 1792.684429 8156 | 2250.172944 7694 | 2139.362683 4727 |
| VALSYNPWY: valine biosynthesis | 1577.517860 9018 | 1469.875122 2891 | 1902.632068 1466 | 2261.711518 9351 |
BASIC PROTOCOL 4
CECAL CONTENT EXTRACTION FOR LC-ORBITRAP-MS
LC-Orbitrap-MS offers high-throughput, high-resolution, accurate-mass (HRAM) performance, and has been extensively utilized as a powerful metabolomics tool to detect a wide range of compounds, especially small metabolites. Cecal contents are a rich source of microbiota, thus the metabolic profile of cecal content indicates bacterial and host metabolic activity. This protocol describes an untargeted hydrophilic phase extraction method of cecal content for LC-Orbitrap-MS (Thermo) analysis.
Materials
Cecal content (see Basic Protocol 1, step 6 and Support Protocol 1)
1-mm Silica homogenization beads (BioSpec)
HPLC graded methanol (Sigma-Aldrich)
HPLC graded water (Sigma-Aldrich)
Chlorpropamide (Sigma-Aldrich)
Liquid nitrogen
10–200 μl pipette (Denville)
1000 μl pipette (Denville)
Vortex mixer (Any Brand)
37°C water bath
Labeled 2-ml screw-cap homogenizer tubes (VWR)
Precellys 24 lysis and homogenization (Bertin Technologies)
Labeled 1.5-ml microcentrifuge tubes (Eppendorf)
Savant SPD121P SpeedVac Concentrator (Thermo Scientific)
250-μl autosampler vials (Thermo Fisher)
First cecal extraction
-
1
Inside a 2-ml screw-cap homogenizer tubes, mix cecal content (~50 mg) with 10 to 15 1-mm silica homogenization beads first, and then extract with 1 ml ice-cold methanol (50% v/v) containing 1 μM chlorpropamide.
-
2
Vortex the sample briefly, and then homogenize thoroughly (after homogenizing for 1 min, stop for 2 min to prevent overheating).
-
3
Freeze and thaw three times with liquid nitrogen and a 37°C water bath.
-
4
Centrifuge for 10 min at 12,000 × g, 4°C.
-
5
Transfer the supernatants into a new 1.5-ml tube.
Perform second cecal extraction
-
5
Re-extract cecal contents by adding an additional 500 μl of ice-cold methanol (50% v/v) containing 1 μM chlorpropamide; repeat step 2 to 4.
-
6
Combine the supernatants.
-
7
Dry down the samples in a SpeedVac (takes about 3 hr).
-
8
Resuspend the pellet in 200 μl of 3% methanol.
-
9
Centrifuge for 10 min, at 13000 × g, 4°C.
-
10
Transfer 150 μl of the supernatants into 250 μl autosampler vials and store up to 2 weeks at −20°C until ready to be run.
-
11
Pool 10 μl of each sample into a new tube for quality control. Pooled samples are prepared in triplicate.
-
12
See Support Protocol 3 for how to set up the LC-Orbitrap-MS.
SUPPORT PROTOCOL 3
LC-ORBITRAP-MS INSTRUMENTATION SETTINGS
The LC-MS system consists of a Dionex Ultimate 3000 quaternary HPLC pump, a Dionex 3000 column compartment, a Dionex 3000 autosampler, and an Exactive plus Orbitrap mass spectrometer controlled by Xcalibur 2.2 software (all from Thermo Fisher Scientific). Extracts are analyzed by LC-MS using a modified version of an ion pairing reversed-phase Negative-ion electrospray ionization method (Lu, Kimball, & Rabinowitz, 2006). A 10-μl sample is injected and separated on aPhenomenex (Torrance, CA) Hydro-RP C18 column (100 × 2.1-mm, 2.5-μm particle size) using a water/methanol gradient with tributylamine and acetic acid added to the aqueous mobile phase. The HPLC column is maintained at 30°C, and at flow rate of 200 μl/min. Solvent A is 3% aqueous methanol with 10 mM tributylamine and 15 mM acetic acid; solvent B is methanol. The gradient is 0 min, 0% B; 5 min, 20% B; 7.5 min, 20% B; 13 min, 55% B; 15.5 min, 95% B; 18.5 min, 95% B; 19 min, 0% B; and 25 min, 0% B. The Exactive plus is operated in negative ion mode at maximum resolution (140,000) and scanned from m/z 72 to m/z 1000 for the first 90 sec and then from m/z 85 to m/z 1000 for the remainder of the chromatographic run. The AGC target is 3 × 106 with a maximum injection time of 100 msec, the nitrogen sheath gas is set at 35, the auxiliary gas at 10 and the sweep gas at 1. The capillary voltage is 3.2 kV and both the capillary and heater set at 200°C, the S-lens was 55. To aid in the detection of metabolites, a database generated from pure metabolite standards (Table 5) using the same instrument and method to determine detection capability, mass/charge ratio (m/z), and retention time for each metabolite is used as a primary database for metabolite identification.
Table 5.
Orbitrap Metabolite Database
| m/z [M-H]- | Retention time (min) | Identity | Elemental composition |
|---|---|---|---|
| 168.0779 | 2 | 1-Methyl-histidine | C7H11N3O2 |
| 280.1051 | 3 | 1-Methyl-adenosine | C11H15N5O4 |
| 153.0193 | 13.58 | 2,Dihydroxybenzoic acid | C7H6O4 |
| 264.9520 | 14.68 | 2,3-Diphosphoglyceric acid | C3H8O10P2 |
| 158.1187 | 11.8 | 2-Aminooctanoic acid | C8H17NO2 |
| 834.1341 | 17.17 | 2-Butenoyl-CoA/Crotonoyl-CoA | C25H40N7O17P3S |
| 147.0338 | 12.06 | 2-Hydroxy-2-methylbutanedioic acid | C5H8O5 |
| 175.0612 | 13.87 | 2-Isopropylmalic acid | C7H12O5 |
| 193.0354 | 5 | 2-Keto-gluconate | C6H10O7 |
| 115.0401 | 13.06 | 2-Keto-isovalerate | C5H8O3 |
| 147.0121 | 13.75 | 2-Oxo-4-methylthiobutanoate | C5H8O3S |
| 101.0244 | 10.95 | 2-Oxobutanoate | C4H6O3 |
| 910.1502 | 15.94 | 3-Hydroxy-3-methylglutaryl-CoA | C27H44N7O20P3S |
| 103.0401 | 3.4 | 3-Hydroxybutyric acid | C4H8O3 |
| 852.1447 | 15.6 | 3-Hydroxybutyryl-CoA | C25H42N7O18P3S |
| 149.0608 | 15.41 | 3-Methylphenylacetic acid | C9H10O2 |
| 184.0017 | 8.2 | 3-Phosphoserine | C3H8NO6P |
| 184.9857 | 13.58 | 3-Phosphoglycerate | C3H7O7P |
| 119.0172 | 11.92 | 3-Methylthiopropionate | C4H8O2S |
| 102.0561 | 4 | 4-Aminobutyrate | C4H9NO2 |
| 181.0506 | 11.25 | 4-Hydroxyphenyllactate | C9H10O4 |
| 357.0891 | 12.14 | 4-Phosphopantetheine | C11H23N2O7PS |
| 298.0697 | 13.94 | 4-Phosphopantothenate | C9H18NO8P |
| 182.0459 | 14.14 | 4-Pyridoxic acid | C8H9NO4 |
| 116.0717 | 1.18 | 5-Aminopentanoic acid | C5H11NO2 |
| 233.0932 | 8.95 | 5-Methoxytryptophan | C12H14N2O3 |
| 458.1794 | 14.1 | 5-Methyl-THF | C20H25N7O6 |
| 388.9445 | 15.13 | 5-Phosphoribosyl-1-pyrophosphate | C5H13O14P3 |
| 275.0174 | 13.38 | 6-Phospho-gluconate | C6H13O10P |
| 442.1481 | 25.2 | 7,8-Dihydrofolate | C19H21N7O6 |
| 296.1000 | 12 | 7-Methylguanosine | C11H15N5O5 |
| 101.0244 | 7.74 | Acetoacetate | C4H6O3 |
| 850.1291 | 15.95 | Acetoacetyl-CoA | C25H40N7O18P3S |
| 174.0408 | 13 | Acetyl-aspartate | C6H9NO5 |
| 808.1184 | 16.18 | Acetyl-CoA | C23H38N7O17P3S |
| 116.0353 | 7.44 | Acetyl-glycine | CH3CONHCH2CO2H |
| 202.1085 | 2.4 | Acetylcarnitine | C9H17NO4 |
| 187.1088 | 1.1 | Acetyllysine | C8H16N2O3 |
| 138.9802 | 12.74 | Acetylphosphate | C2H5O5P |
| 173.0092 | 14 | Aconitate | C6H6O6 |
| 134.0472 | 3.5 | Adenine | C5H5N5 |
| 266.0895 | 1.27 | Adenosine | C10H13N5O4 |
| 426.0126 | 13.58 | Adenosine phosphosulfate | C10H14N5O10PS |
| 426.0221 | 14.16 | ADP | C10H15N5O10P2 |
| 588.0750 | 13.72 | ADP-glucose | C16H25N5O15P2 |
| 88.0404 | 1.15 | Alanine | C3H7NO2 |
| 175.0473 | 5.62 | Allantoate | C4H8N4O4 |
| 157.0367 | 0.82 | Allantoin | C4H6N4O3 |
| 145.0142 | 13.13 | Alpha-ketoglutarate | C5H6O5 |
| 160.0615 | 3.22 | Aminoadipic acid | C6H11NO4 |
| 130.0510 | 1.15 | Aminolevulinate | C5H9NO3 |
| 346.0558 | 11.3 | AMP | C10H14N5O7P |
| 136.0404 | 13.51 | Anthranilate | C7H7NO2 |
| 173.1044 | 0.87 | Arginine | C6H14N4O2 |
| 289.1154 | 7.7 | Arginino-succinate | C10H18N4O6 |
| 175.0248 | 6.59 | Ascorbic acid | C6H8O6 |
| 131.0462 | 1.15 | Asparagine | C4H8N2O3 |
| 132.0302 | 3.95 | Aspartate | C4H7NO4 |
| 505.9885 | 15.04 | ATP | C10H16N5O13P3 |
| 870.1342 | 17.78 | Benzoyl-CoA | C28H40N7O17P3S |
| 243.0809 | 12.92 | Biotin | C10H16N2O3S |
| 836.1498 | 17.5 | Butyryl/Isobutyryl-CoA | C25H42N7O17P3S |
| 139.9754 | 12.06 | Carbamoyl phosphate | CH4NO5P |
| 160.0979 | 1 | Carnitine | C7H15NO3 |
| 304.0340 | 4.91 | cCMP | C9H12N3O7P |
| 402.0109 | 13.53 | CDP | C9H15N3O11P2 |
| 487.1001 | 6.53 | CDP-choline | C14H26N4O11P2 |
| 445.0531 | 6.38 | CDP-ethanolamine | C11H2ON4O11P2 |
| 341.1089 | 3.86 | Cellobiose | C12H22O11 |
| 344.0402 | 8.2 | cGMP | C10H12N5O7P |
| 465.3044 | 17.25 | Cholesteryl sulfate | C27H46O4S |
| 407.2803 | 16.69 | Cholic acid | C24H40O5 |
| 129.0193 | 13.29 | Citraconic acid | C5H6O4 |
| 191.0197 | 13.6 | Citrate/Isocitrate | C6H8O7 |
| 174.0884 | 0.9 | Citrulline | C6H13N3O3 |
| 322.0446 | 8.61 | CMP | C9H14N3O8P |
| 766.1079 | 15.93 | Coenzyme A | C21H36N7O16P3S |
| 481.9772 | 14.75 | CTP | C9H16N3O14P3 |
| 689.0876 | 14.2 | Cyclic bis(3′->5′) dimeric GMP | C20H24N10O14P2 |
| 328.0452 | 12.97 | Cyclic-AMP | C10H12N5O6P |
| 221.0602 | 1.14 | Cystathionine | C7H14N2O4S |
| 120.0125 | 1.23 | Cysteine | C3H7NO2S |
| 242.0782 | 1.2 | Cytidine | C9H13N3O5 |
| 110.036 | 1.18 | Cytosine | C4H5N3O |
| 257.0068 | 6.17 | D-glucono-lactone-6-phosphate | C6H11O9P |
| 258.0384 | 1.9 | D-glucosamine-1-phosphate | C6H14NO8P |
| 258.0384 | 1.9 | D-glucosamine-6-phosphate | C6H14NO8P |
| 168.9908 | 7.35 | D-glyceraldehdye-3-phosphate | C3H7O6P |
| 289.0330 | 7.29 | D-sedoheptulose-1/7-phosphate | C7H15O10P |
| 330.0609 | 12 | dAMP | C10H14N5O6P |
| 489.9936 | 14.82 | dATP | C10H16N5O12P3 |
| 386.0160 | 13.58 | dCDP | C9H15N3O10P2 |
| 306.0497 | 9.8 | dCMP | C9H14N3O7P |
| 465.9823 | 14.8 | dCTP | C9H16N3O13P3 |
| 920.2436 | 19.35 | Decanoyl-CoA | C31H54N7O17P3S |
| 250.0946 | 11.99 | Deoxyadenosine | C10H13N5O3 |
| 391.2854 | 16.79 | Deoxycholate | C24H40O4 |
| 266.0895 | 4.8 | Deoxyguanosine | C10H13N5O4 |
| 251.0786 | 3.9 | Deoxyinosine | C10H12N4O4 |
| 213.0170 | 7.74 | Deoxyribose-phosphate | C5H11O7P |
| 227.0673 | 3.54 | Deoxyuridine | C9H12N2O5 |
| 686.1416 | 15.08 | Dephospho-CoA | C21H35N7O13P2S |
| 426.0221 | 14 | dGDP | C10H15N5O10P2 |
| 346.0558 | 11 | dGMP | C10H14N5O7P |
| 505.9885 | 14.77 | dGTP | C10H16N5O13P3 |
| 157.0255 | 6.9 | Dihydroorotate | C5H6N2O4 |
| 157.0255 | 6.9 | Dihydroorotate | C5H6N2O4 |
| 168.9908 | 9 | Dihydroxy-acetone-phosphate | C3H7O6P |
| 153.0049 | 4.37 | Dithioerythritol | C4H10O2S2 |
| 152.0717 | 2.1 | Dopamine | C8H11NO2 |
| 401.0157 | 14.41 | dTDP | C10H16N2O11P2 |
| 321.0493 | 11.27 | dTMP | C10H15N2O8P |
| 480.9820 | 14.75 | dTTP | C10H17N2O14P3 |
| 307.0337 | 10.28 | dUMP | C9H13N2O8P |
| 466.9663 | 14.81 | dUTP | C9H15N2O14P3 |
| 199.0013 | 7.45 | Erythrose-4-phosphate | C4H9O7P |
| 784.1499 | 15 | FAD | C27H33N9O15P2 |
| 221.0608 | 7.7 | Flavone | C15H10O2 |
| 455.0973 | 14.54 | FMN | C17H21N4O9P |
| 440.1324 | 13.99 | Folate | C19H19N7O6 |
| 338.9888 | 13.6 | Fructose-1,6-bisphosphate | C6H14O12P2 |
| 259.0224 | 7.98 | Fructose-6-phosphate | C6H13O9P |
| 115.0037 | 13.49 | Fumarate | C4H4O4 |
| 442.0170 | 13.9 | GDP | C10H15N5O11P2 |
| 313.0612 | 16.46 | Geranyl-PP | C10H20O7P2 |
| 177.0405 | 6.9 | Glucono-lactone | C6H10O6 |
| 178.0721 | 0.7 | Glucosamine | C6H13NO5 |
| 259.0224 | 7.98 | Glucose-1-phosphate | C6H13O9P |
| 259.0224 | 6.9 | Glucose-6-phosphate | C6H13O9P |
| 209.0303 | 13 | Glucarate | C6H10O8 |
| 195.0510 | 5 | Gluconate | C6H12O7 |
| 146.0459 | 3.52 | Glutamate | C5H9NO4 |
| 145.0619 | 1.17 | Glutamine | C5H10N2O3 |
| 306.0765 | 8.02 | Glutathione | C10H17N3O6S |
| 611.1447 | 12.94 | Glutathione disulfide | C20H32N6O12S2 |
| 105.0193 | 6.35 | Glycerate | C3H6O4 |
| 171.0064 | 7.3 | Glycerol-3-phosphate | C3H9O6P |
| 74.0248 | 1 | Glycine | C2H5NO2 |
| 448.3068 | 14.75 | Glycodeoxycholate | C26H43NO5 |
| 75.0088 | 6.95 | Glycolate | C2H4O3 |
| 72.9931 | 7.45 | Glyoxylate | C2H2O3 |
| 362.0507 | 10.5 | GMP | C10H14N5O8P |
| 521.9834 | 14.92 | GTP | C10H16N5O14P3 |
| 116.0466 | 1.89 | Guanidoacetic acid | C3H7N3O2 |
| 150.0421 | 3.4 | Guanine | C5H5N5O |
| 282.0844 | 4 | Guanosine | C10H13N5O5 |
| 601.9497 | 15.18 | Guanosine 5′-diphosphate-3′-diphosphate | C10H17N5O17P4 |
| 864.1811 | 18.15 | Hexanoyl-CoA | C27H46N7O17P3S |
| 259.0224 | 7 | Hexose-phosphate | C6H13O9P |
| 110.0724 | 1.92 | Histamine | C5H9N3 |
| 154.0622 | 0.87 | Histidine | C6H9N3O2 |
| 140.0829 | 0.8 | Histidinol | C6H11N3O |
| 182.0129 | 4.8 | Homocysteic acid | C4H9NO5S |
| 134.0281 | 1 | Homocysteine | C4H9NO2S |
| 118.0510 | 1 | Homoserine | C4H9NO3 |
| 131.0714 | 14.14 | Hydroxyisocaproic acid | C6H12O3 |
| 151.0401 | 14.68 | Hydroxyphenylacetic acid | C8H8O3 |
| 179.0350 | 13.58 | Hydroxyphenylpyruvate | C9H8O4 |
| 130.0510 | 1.18 | Hydroxyproline | C5H9NO3 |
| 135.0312 | 1.18 | Hypoxanthine | C5H4N4O |
| 427.0062 | 13.9 | IDP | C10H14N4O11P2 |
| 347.0398 | 10.5 | IMP | C10H13N4O8P |
| 116.0500 | 7.5 | Indole | C8H7N |
| 160.0404 | 14.31 | Indole-3-carboxylic acid | C9H7NO2 |
| 186.0561 | 14.94 | Indoleacrylic acid | C11H9NO2 |
| 267.0735 | 3.5 | Inosine | C10H12N4O5 |
| 179.0561 | 1.13 | Inositol | C6H12O6 |
| 317.0925 | 15.06 | Isopentyl-PP | C10H24O7P2 |
| 850.1655 | 17.82 | Isovaleryl/2-Methylbutyryl-CoA | C26H44N7O17P3S |
| 129.0557 | 14.3 | Ketoleucine | C6H10O3 |
| 188.0353 | 14.22 | Kynurenic acid | C10H7NO3 |
| 207.0775 | 3.6 | Kynurenine | C10H12N2O3 |
| 89.0244 | 7.28 | Lactate | C3H6O3 |
| 948.2750 | 19.99 | Lauroyl-CoA | C33H58N7O17P3S |
| 130.0874 | 2.22 | Leucine/isoleucine | C6H13NO2 |
| 205.0362 | 15.97 | Lipoate | C8H14O2S2 |
| 145.0983 | 0.85 | Lysine | C6H14N2O2 |
| 133.0142 | 12.7 | Malate | C4H6O5 |
| 852.1083 | 16.91 | Malonyl-CoA | C24H38N7O19P3S |
| 148.0438 | 1.77 | Methionine | C5H11NO2S |
| 134.0281 | 1 | Methylcysteine | C4H9NO2S |
| 117.0193 | 11.72 | Methylmalonic acid | C4H6O4 |
| 135.0564 | 3.5 | Methylnicotinamide | C7H8N2O |
| 179.0561 | 7.2 | Myo-inositol | C6H12O6 |
| 976.3063 | 21.8 | Myristoyl/tetradecanoyl-CoA | C35H62N7O17P3S |
| 300.0490 | 1.71 | N-Acetyl-glucosamine | C8H16NO9P |
| 300.0490 | 7.3 | N-Acetyl-glucosamine-1/6-phosphate | C8H16NO9P |
| 188.0564 | 13.37 | N-Acetyl-glutamate | C7H11NO5 |
| 187.0724 | 7 | N-Acetyl-glutamine | C7H12N2O4 |
| 130.0510 | 8.02 | N-Acetyl-L-alanine | C5H9NO3 |
| 173.0932 | 1.21 | N-Acetyl-L-ornithine | C7H14N2O3 |
| 129.1033 | 1 | N-Acetylputrescine | C6H14N2O |
| 175.0360 | 12.25 | N-Carbamoyl-aspartate | C5H8N2O5 |
| 662.1019 | 8.62 | NAD+ | C21H27N7O14P2 |
| 664.1175 | 14.14 | NADH | C21H29N7O14P2 |
| 742.0682 | 13.87 | NADP+ | C21H28N7O17P3 |
| 744.0838 | 14.87 | NADPH | C21H30N7O17P3 |
| 333.0493 | 1.71 | Nicotinamide mononucleotide | C11H15N2O8P |
| 122.0248 | 11.2 | Nicotinate | C6H5NO2 |
| 334.0333 | 11.44 | Nicotinic acid mononucleotide | C11H14NO9P |
| 146.0459 | 1.1 | O-acetyl-serine | C5H9NO4 |
| 892.2124 | 18.72 | Octanoyl-CoA | C29H50N7O17P3S |
| 319.0476 | 6 | Octoluse 8/1P | C8H17O11P |
| 399.0099 | 13.64 | Octoluse Bisphosphate | C8H18O14P2 |
| 131.0826 | 1 | Ornithine | C5H12N2O2 |
| 155.0098 | 8.1 | Orotate | C5H4N2O4 |
| 367.0184 | 13.58 | Orotidine-5-phosphate | C10H13N2O11P |
| 130.9986 | 13.64 | Oxaloacetate | C4H4O5 |
| 136.0404 | 8.7 | p-Aminobenzoate | C7H7NO2 |
| 137.0244 | 10.88 | p-Hydroxybenzoate | C7H6O3 |
| 277.1228 | 9.17 | Pantetheine | C11H22N2O4S |
| 218.1034 | 11.31 | Pantothenate | C9H17NO5 |
| 884.1498 | 17.78 | Phenylacetyl-CoA | C29H42N7O17P3S |
| 164.0717 | 4.37 | Phenylalanine | C9H11NO2 |
| 165.0557 | 14.68 | Phenyllactic acid | C9H10O3 |
| 145.0295 | 15.2 | Phenylpropiolic acid | C9H6O2 |
| 163.0401 | 15.1 | Phenylpyruvate | C9H8O3 |
| 166.9751 | 13.83 | Phosphoenolpyruvate | C3H5O6P |
| 128.0717 | 1.2 | Pipecolic acid | C6H11NO2 |
| 225.0405 | 13.87 | Prephenate | C10H10O6 |
| 114.0561 | 1.27 | Proline | C5H9NO2 |
| 822.1342 | 16.89 | Propionyl-CoA | C24H40N7O17P3S |
| 167.0826 | 7.6 | Pyridoxamine | C8H12N2O2 |
| 168.0666 | 1.8 | Pyridoxine | C8H11NO3 |
| 128.0353 | 7.69 | Pyroglutamic acid | C5H7NO3 |
| 176.9360 | 13.87 | Pyrophosphate | P2H4O7 |
| 87.0088 | 8.67 | Pyruvate | C3H4O3 |
| 166.0146 | 13.58 | Quinolinate | C7H5NO4 |
| 375.1310 | 12.56 | Rboflavin | C17H2ON4O6 |
| 229.0119 | 6.94 | Ribose-5-phosphate | C5H11O8P |
| 229.0119 | 6 | Ribose-phosphate | C5H11O8P |
| 229.0119 | 7.81 | Ribulose-5-phosphate | C5H11O8P |
| 383.1143 | 6.86 | S-adenosyl-L-homocysteine | C14H2ON6O5S |
| 353.1401 | 1 | S-adenosyl-L-methioninamine | C14H22N6O3S |
| 397.1300 | 7 | S-adenosyl-L-methionine | C15H22N6O5S |
| 296.0823 | 11.6 | S-methyl-5′-thioadenosine | C11H15N5O3S |
| 266.0704 | 0.68 | S-ribosyl-L-homocysteine | C9H17NO6S |
| 88.0404 | 1 | Sarcosine | C3H7NO2 |
| 368.9993 | 13.64 | Sedoheptoluse bisphosphate | C7H16O13P2 |
| 104.0353 | 1.14 | Serine | C3H7NO3 |
| 173.0455 | 10.92 | Shikimate | C7H10O5 |
| 253.0119 | 13.44 | Shikimate-3-phosphate | C7H11O8P |
| 117.0193 | 11.77 | Succinate | C4H6O4 |
| 866.1240 | 16.93 | Succinyl-CoA/Methylmalonyl-CoA | C25H40N7O19P3S |
| 124.0074 | 0.82 | Taurine | C2H7NO3S |
| 498.2895 | 16.23 | Taurodeoxycholic acid | C26H45NO6S |
| 240.1102 | 1.68 | Tetrahydrobiopterin | C9H15N5O3 |
| 263.0972 | 0.94 | Thiamine | C12H16N4OS |
| 423.0299 | 9.56 | Thiamine pyrophosphate | C12H18N4O7P2S |
| 343.0635 | 11.11 | Thiamine-phosphate | C12H17N4O4PS |
| 118.0510 | 1.19 | Threonine | C4H9NO3 |
| 241.0830 | 5.4 | Thymidine | C10H14N2O5 |
| 125.0357 | 2.6 | Thymine | C5H6N2O2 |
| 381.1238 | 16.74 | Trans_trans-farnesyl diphosphate | C15H28O7P2 |
| 421.0753 | 6.95 | Trehalose-6-Phosphate | C12H23O14P |
| 341.1089 | 1.18 | Trehalose/sucrose | C12H22O11 |
| 203.0826 | 7.5 | Tryptophan | C11H12N2O2 |
| 180.0666 | 2 | Tyrosine | C9H11NO3 |
| 402.9949 | 13.81 | UDP | C9H14N2O12P2 |
| 565.0478 | 13.44 | UDP-glucose | C15H24N2O17P2 |
| 579.0270 | 14.68 | UDP-glucuronate | C15H22N2O18P2 |
| 606.0743 | 13.37 | UDP-N-acetyl-glucosamine | C17H27N3O17P2 |
| 323.0286 | 10.32 | UMP | C9H13N2O9P |
| 111.0200 | 1.18 | Uracil | C4H4N2O2 |
| 167.0211 | 6.17 | Uric acid | C5H4N4O3 |
| 243.0623 | 1.7 | Uridine | C9H12N2O6 |
| 482.9613 | 14.81 | UTP | C9H15N2O15P3 |
| 116.0717 | 1.45 | Valine | C5H11NO2 |
| 151.0262 | 2.69 | Xanthine | C5H4N4O2 |
| 283.0684 | 7.77 | Xanthosine | C10H12N4O6 |
| 363.0347 | 12.7 | Xanthosine-5-phosphate | C10H13N4O9P |
| 204.0302 | 14.14 | Xanthurenic acid | C10H7NO4 |
| 149.0455 | 1.22 | Ribose | C5H10O5 |
BASIC PROTOCOL 5
LC-ORBITRAP-MS DATA ANALYSIS WITH MS-DIAL
MS-DIAL, an open-source software pipeline is used for untargeted metabolomics analysis (Tsugawa et al., 2015). Readers should note that other software tools are available including vendor-specific software. Additionally, readers are encouraged to check that they are using the most current version.
Materials
Proteowizard software, MS-DIAL software, LC-Orbitrap-MS Data (.raw)
-
1
Start up a project:
Before performing analysis, LC-Orbitrap-MS Data (.raw) needs to be converted to mzML format with open source software Proteowizard (Kessner, Chambers, Burke, Agus, & Mallick, 2008).
In the MS-DIAL interface, click “File-New project” and open “Start up a project” window. Select a directory that contains the converted mzML format MS files. Choose the “Soft Ionization” as ionization type; “Conventional LC/MS or data dependent MS/MS” as method type; “Profile data” as data type for MS1 and MS/MS; choose “Negative ion mode” as ion mode and “Metabolomics” as target omics, click next.
Browse the analysis file paths, change the file format to mzML file (*.mzml) and select all the mzML format files to be analyzed. Choose the correct type for each sample (sample, standard, quality control, or blank). Then based on the group information, add the corresponding Class ID for each sample (Control, low dose treatment, high dose treatment, etc.). Uncheck any samples under “Included” for exclusion if necessary, then click next.
-
2
Peak detection, identification, and alignment setting:
Under “Analysis parameter setting” window, click “Identification” tab, select the “MSMS-AllPubfic-Curated-Neg” MSP file from Public MSPs folder included in the software package.
Based on the accuracy of mass and retention time of the liquid chromatraphy and mass spectrometer platform, select the retention time tolerance within range of 0.2 to 0.5 min, accurate mass tolerance from 0.001 to 0.005 Da (2 to 10 ppm at 200 m/z).
If an in-house library generated from a list of pure metabolite standards using the same instrument and methods, and then select the text file (an accurate database) containing name, mass-to-charge ratio (m/z), and retention time for each metabolite. Select the stricter tolerance setting such as retention time tolerance ≤0.2 min and accurate mass tolerance ≤0.002 Da.
Click “Alignment” tab, choose a non-blank sample as a reference file for alignment. Recommended reference file: a pooled sample, or intermediate sample in injection sequence order. Recommended tolerance setting: RT tolerance of 0.2 to 0.5 min and MS1 tolerance of 0.0025 to 0.003 Da.
Click “Finish” and peak detection, identification, and alignment starts. Those processing steps take several hours based on sample number and computer capacity.
-
3
Browse the result window and export alignment results:
Double-click “Alignment navigator” at the bottom left of the result window.
Optional: Apply normalization method by clicking statistical analysis-normalization.
Optional: Perform PCA analysis by clicking statistical analysis-principal component analysis.
At the Peak spot navigator window, select “Identified display filter” (identified peaks with the database generated from a list of pure metabolite standards using the same instrument and method). Check the number of the alignment in “Peak spot navigator” with identified display filter. If the number is too low, check the “Annotated display filter” (identified the peaks with the public MSP file without MS/MS) or return to 2c and 2d to increase accurate mass and retention time tolerance.
Click individual spot in “Alignment spot viewer” window at the middle bottom, check the peak and compound information (right top window), bar chart of aligned spots (middle top window) and the MS1 spectrum (left bottom).
Click export-alignment result, select a folder for import, choose export format as “txt”, the most important files for import are “Raw data matrix (Area)”, “Parameters” and “normalized data matrix” (If normalization method is applied).
-
4
Post processing alignment result:
-
Check the data quality.
Check coefficient of variation value of the internal standard (chlorpropamide) and replicated pooled samples.
Check if the pooled samples are close to biological averaging.
Check the fill % (Percentage of samples having good shape, otherwise, apply “Gap Filling”).
-
Apply additional filter if necessary to clean the data.
Subtract blank values from averaged sample values and filter the compound only with positive values.
Filter the compound with fill % > 0.3–0.5 (good alignment).
-
BASIC PROTOCOL 6
CECAL CONTENT EXTRACTION FOR NMR
1H NMR is a reliable, stable, and cost-effective tool for global metabolomics analysis. The non-destructive, non-invasive, and instrument-independent nature of NMR techniques guarantees high reproducibility. The protocol below describes cecal content extraction, data processing, and statistical analysis for NMR spectroscopy.
Materials
Cecal contents (see Basic Protocol 1, step 6 and Support Protocol 1)
Potassium phosphate dibasic (K2HPO4; Sigma-Aldrich)
Sodium phosphate monobasic (NaH2PO4; Sigma-Aldrich)
3-(Trimethylsilyl)propionic-2,2,3,3-d4 acid sodium salt (TSP-d4; Sigma-Aldrich)
Distilled Water
Deuterium oxide (D2O; Cambridge Isotope Laboratories)
Liquid nitrogen
1-mm silica homogenization beads (BioSpec)
10- to 200-μl pipette (Denville)
1000-μl pipette (Denville)
Labeled 2-ml screw-cap homogenizer tubes (VWR)
Labeled 1.5-ml microcentrifuge tubes (Eppendorf)
Centrifuge (Eppendorf 5430 R)
Vortex mixer
Precellys 24 lysis and homogenization (Bertin Technologies)
5-mm NMR tube and lid (Norell)
Inside the 2-ml screw-cap homogenizer tubes, extract cecal content (~50 mg) with 1 ml phosphate buffer (K2HPO4/NaH2PO4, 0.1 M, pH 7.4,50% v/v D2O) containing 50 μg/ml (290 μM) TSP-d4 as a chemical shift reference (δ 0.00).
Freeze-thaw three times with liquid nitrogen.
Add five to six 1-mm silica homogenization beads to the screw-top tubes and homogenize for 1 min, 6500 rpm, 1 cycle and centrifuge for 10 min at 11,180 × g, 4°C.
Transfer the supernatants into a new 1.5-ml tube
Add another 600 μl PBS to the pellets, vortex for 1 min.
Centrifuge for 10 min at 11,180 × g, 4°C.
Transfer the additional supernatants into the 1.5-ml (combined supernatants are around 1.2 ml in total volume) tube.
Centrifuge the combined supernatants for 10 min at 11,180 × g, 4°C.
Transfer the supernatants into a 5-mm NMR tube and store up to 1 week at 4 °C until NMR spectroscopy analysis.
See Support Protocol 4 for information on acquisition settings.
SUPPORT PROTOCOL 4
NMR SPECTRA ACQUISITION SETTING
All 1H spectra are recorded at 298K on a Bruker NMR spectrometer (600 MHz for 1H) configured with a 5-mm inverse cryogenic probe. A standard one-dimensional pulse sequence noesyprld (recycle delay-90°-t1–90°-tm-90°-acquisition) is used with a 90-pulse length of approximately 10 μsec (−9.6 dbW) and 64 transients are recorded into 32k data points with a spectral width of 9.6 KHz. For quantitation purposes, arelaxation delay (5s) and a recycle delay (4s) are added to the cycle to ensure the total repetition time (relaxation time, recycle delay and acquisition time) is more than 5 times the longitudinal relaxation time (T1) of the compounds (Cai et al., 2017). Quantitation analysis is performed based on either TSP-d4 reference with known concentration (Dai, Xiao, Liu, & Tang, 2010) or calibration curve. To facilitate NMR resonance assignments, two-dimensional (2D) NMR spectra including 1H-1H total correlation spectroscopy (TOCSY), 1H-1H correlation spectroscopy (COSY), J-resolved (JRES), 1H-13C heteronuclear single quantum correlation (HSQC), and 1H-13 C heteronuclear multiple bond correlation (HMBC) are acquired. Chemical shifts are reported in ppm from TSP (δ = 0.00).
BASIC PROTOCOL 7
NMR SPECTRA PROCESSING
1H NMR spectra processing ensures quality of the data for integration and multivariate analysis. The protocol below describes the detailed NMR spectra processing steps including phase and baseline correction, calibration, residual water signal removal, normalization and binning using different software.
Materials
NMR spectra with Topsin 3.0 (Bruker Biospin)
AMIX software version 3.9.14 (Bruker Biospin)
1D NMR Spectra
-
NMR spectra processing with Topspin 3.0 (Bruker Biospin, Germany):
Before performing statistical analysis, an exponential window function (command code: efp) is applied with a line-broadening factor of 1 Hz (command code: lb 1) prior to Fourier Transformation.
Then all 1H NMR spectra qualities are improved by correcting the phase (command code: ph), baseline (command code: bas) and referencing to TSP (8 0.00) (command code: cal) automatically or manually (manually recommended).
-
Further process the spectra with AMIX software version: 3.9.14 (Brucker Biospin, Germany):
-
Import the data and check quality as follows:
Open Amix-File-Open TOPSPIN 1D file, choose the right directory where the 1H NMR spectra are stored and select all the spectra for analysis.
Check the layered spectra for proper overlay. If the spectra are not overlaid properly, repeat 1b to improve spectra phase, baseline, and calibration.
-
Bucketing:
Click “Amix-Tools-Bukcets,Statistics-Statistics-Bucket Table-New”.
Choose 1D NMR and simple rectangular buckets.
Change bucket width to 0.004 ppm (2.4 Hz).
Change the scaling mode to either scaling to total intensity (non-quantitative purpose) to compensate the overall concentration differences (Cai et al., 2016), or no scaling then normalize to the tissue weight later (for quantitative purpose) (Cai et al., 2017).
Select “edit exclusions” and remove the interference signals including the residual water signal (region 8 4.2–5.2), other contamination signal, like methanol (region δ 3.3–3.4), ethanol (region δ 1.1–1.2, 3.6–3.7) and Polyethylene glycol (8 3.6–3.8).
Select a directory to save the bucket table.
Select data source from TOPSPIN data tree and reselect all the spectra again.
Choose “no” for “select next” window, close the file display window.
Click “Statistic-Bucket table-Import”, rename the txt file, choose “table (spectrum per column)” as “output format”; Choose “blanks, commas, tables” as “delimiters used in table output”.
The imported txt file is under default path of Bruker-amix directory.
-
BASIC PROTOCOL 8
UNIVERSAL PROFILING OF NMR DATA
Multivariate data analysis is performed with SIMCA 13 (Umetrics, Sweden). Before performing statistical analysis, import the bucketed .txt file into Excel, delete the regions with a number 0 (the exclusion regions edited in Amix), save as “Excel 2003–2007 workbook” as other formats cannot be recognized by SIMCA.
Materials
SIMCA 13 (Umetrics)
-
Principal component analysis (PCA) as follows:
Open SIMCA, create a new regular project, select the saved Excel file as data source.
At import data wizard window, click “Edit-Transpose” to transpose the spread sheet. Now the first column is the data ID and first row is the ppm ID.
Click the arrows on the first column and row, choose primary observation ID and primary variable ID, respectively. Click “File-Save as-Finish import.”
Click “New model,” under “observations” tab, select all the samples from one treatment group, click “Set class,” and then select another group click “Set class,” until all samples are assigned. Under the “Scale” tab, select “Ctr” as scaling type, choose “PCA-X” as “Model type.”
Click “Two First” to calculate the first two components.
-
Click “Overview” to create summary plots.
Check Score Scatter Plot for outliers and other abnormalities.
If a sample is significantly away from the rest of the samples, check loading scatter plot for the contributed specific primary IDs (ppm IDs). Confirm this abnormality by checking the ppm regions from the original spectra with topspin. The data points could be removed if the signals are contamination or external signals.
Loading plot also reveals the significant contributors (metabolites) for the group separation. Future targeted analysis could be applied if necessary.
-
Orthogonal projection to latent structure-discriminant analysis (OPLS-DA):
Follow the same step described in 1a-c. Before import, insert one new column at position 2, select the column name as “Y variable.” This binary variable Y is created and assigned to defining a group (e.g., control group is 0, treatment group is 1).
Same as 1d, select “UV” instead of “Ctr” as scaling type for OPLS-DA analysis. A 7-fold cross validation method is employed to validate the OPLS-DA models. The quality of the model is indicated by the parameters R2X (predictive power) and Q2 (validity of the model). The validity of the OPLS-DA model is further assessed with CV-ANOVA tests by clicking “Analyze-CV-ANOVA” for significance with p < 0.05 (Eriksson, Trygg, & Wold, 2008).
BASIC PROTOCOL 9
QUANTITATIVE ANALYSIS OF NMR DATA
Quantitative analysis is performed with Chenomx NMR suite (Chenomx).
Materials
Chenomx NMR suite (Chenomx)
1D NMR Spectra
Converting and processing native spectra in a batch with Chenomx Processor
-
Convert native spectrum formats to Chenomx file format with the application named Chenomx processor within Chenomx NMR suite as follows:
Click “Tools-Batch Import” and select files or a fold that contains all native spectra to be processed. Click next.
Choose “Bruker 1r” as the type of data. Click next.
Select “TSP” as a Chemical Shape Indicator (CSI), the concentration is 0.29 mM. Click next.
Select “Automatic Phase Correction” and “Automatic Baseline Correction-Spline”. Click next.
Choose a folder to save the converted files.
-
Manually check the batch-processed spectra to ensure quality as follows:
-
a
Click “File-Open” and select the converted files generated from the last step.
-
b
Click “Processing history-Files”, a list of converted spectra shows at the left window. Go through those spectra to make sure the quality of the spectra is satisfied. If necessary, click the “Phase Correction” and “Baseline correction” below the spectrum window to adjust the processing parameters manually to improve the quality of the spectrum.
-
c
Click “File-Send to Profiler” for metabolite identification and quantitation.
-
a
Identifying and quantifying the metabolites in a batch with Chenomx Profiler
-
4
The converted NMR spectra are transferred to Chenomx Profiler, a function named “batch fit” allows to identify and quantify the metabolites across entire datasets with sophisticated computer-assisted fitting routines using Chenomx’s spectral library or a library with only targeted metabolites generated by the user.
Click “Tools-Batch Fit-Add Folder” and select a folder that contains all processed spectra. Click next.
-
Choose a list of interested compounds from:
A profiled spectrum (a library with only targeted metabolites generated by the user manually).
Chenomx Reference Compounds at 600 HZ (Default Chenomx spectral library). Click next (if choosing bi, jump to step 5 to learn how to generate a library with only targeted metabolites).
Refine the previous compound selection by moving the compounds of interest from the left to the right window. Click next and finish. It takes from several mins to several hours to process, depending on the number of compounds selected and the computer’s processing speed.
-
5
Manually check the batch-fitted spectra for accurate quantitation.
Review the fitted spectra list on the left to make sure the peak fitting is correct. Adjust the fitting manually if necessary.
Click “File-Export-Compound Table” to export the quantified results.
Manually generate a profiled spectrum with only targeted metabolites (do this step first if using step 3bi)
-
6
Open a representative spectrum within the spectra batch with Chenomx Profiler.
-
7
Search for targeted compounds either by typing the metabolite name or the reference chemical shift at the “Find in Table” input box below the spectrum.
-
8
Select the targeted compound name in the candidate list below the spectrum. Once a compound name is selected, a corresponding reference peak in purple will appear in the spectrum window. Zoom in to adjust the purple arrow vertically and horizontally to fit the peak.
-
9
After fitting all targeted compounds, click “File-Save as”, name the file as “targeted library”.
-
10
Follow all of step 3, except in b, choose 3bi, use the profiled spectrum generated in this step as a reference.
REAGENTS AND SOLUTIONS
TAE, 50 x
Combine the following:
121 g Tris
28.55 ml acetic acid
50 ml of 0.5 M EDTA
500 ml distilled water
Mix and stir until fully dissolved (about 1 to 2 hr) using a magnetic stir bar
Dilute to 1 × by adding 20 ml of the 50 × solution to 980 ml distilled water Store up to 1 year at room temperature.
COMMENTARY
Background Information
There is a growing importance surrounding microbiome analyses and coupling them with toxicologic studies for novel pathway discovery, toxic endpoints, and risk factors. The above microbiome analysis is also not the only way to investigate the microbiome; QPCR methods have been established to analyze specific phyla or species of bacteria, and how they change with a given treatment. QPCR methods can and should be coupled with the above protocol because they can validate the sequence-based results. Deeper analysis of the microbiome via RNAseq can be used to investigate the metatranscriptome of the microbiome. RNAseq does involve an extensive isolation protocol and a 16S rRNA degradation or elimination step.
Critical Parameters
As previously discussed, some basic training in terminal-based coding and R programming is needed for microbiome analysis. For terminal coding, understanding how to connect to and move around an external server or computing cluster within terminal and how to move files to and from a server is required. R scripting may be used for graphing and statistics, but other graphing software can be used including Excel, Prism, or MATLAB. In addition, it is highly recommended that this analysis be done on a server or a computing cluster. If a computing cluster is not available, the Amazon cloud is affordable.
It is important to use blanks and method controls. The method controls will identify artifacts created during the extraction and amplification steps. Also, since 16S rRNA gene sequencing uses PCR, a small amount of contamination can have a dramatic influence on the data. To account for this, sequence the method blank to make sure the contamination seen in the blank is not in the samples. In addition, run a mock community with the samples. A mock community is a known quantity of bacteria, usually 12 to 15 different species. Mothur uses the mock community from BEI resources called HM-782D. Another mock community that can be used is from Zy- moBIOMICS called the Microbial Community DNA Standard #D6305. These mock communities can be used to validate other methods of 16S rRNA gene classification. Mothur has certain commands in the software that can be used to obtain an error rate based on the composition of mock communities. This can also help discover any sequence errors or any human errors in the analytic pipeline.
Troubleshooting
As with most computational workflows, errors may occur that are not mentioned in this protocol. If an error is not mentioned in this protocol, refer back to the mothur wiki (https://www.mothur.org/wiki/MiSeq_SOP), the HUMAnN2 bitbucket page (https://bitbucket.org/biobakery/humann2/wiki/Home), or the HUMAnN2 google group (https://groups.google.com/forum/#!forum/humann-users). This next section will discuss common errors that were not discussed in the above protocols.
“X. . . num.temp is blank. Please correct.” Error from mothur. This is a common error that results in a segmentation fault and an exit from the mothur program. When this error occurs, it is a notification that the server or the computer has run out of space. The best way to fix this is to delete all the files on the server that are not needed, especially dist files. Dist files can sometimes be over 100 GB, so deleting them will free up space.
After any command in mothur, there is an error of X sequence that is not present in Y file but is present in Z file. This could also have to do with memory but can usually be fixed by deleting all the files in the working directory except for the starting files (raw FASTQ files, stability file, silva.bacteria.fasta, and the two trainsets). When rerunning, this error should be eliminated. This also sometimes occurs after a subsample has been taken. If this is the case, delete the subsample files and take a new subsample. If this error is obtained multiple times, delete the files and start over from the beginning.
Negative sequence lengths after sum- mary.seqs command in mothur. This will not give a segmentation fault but the results will be corrupted. This is caused by a mistake in the first step (make.contigs) and is usually due to one of the FASTQ files missing or misspelling in the stability file. This is another important reason to do summary.seq commands with every run. To fix this, double check all the FASTQ files and make sure that both parts of the pair are present. Also check the stability file to make sure the format and the spelling are correct.
-
OTU’s do not match the tree error from GUnifrac. This was briefly mentioned in the above sections, but a different way to solve this can be done all in R. Use the following commands to fix the problem within R.
Test.count <- read.delim(“~/Desktop/
count_table.txt”, row.names=1)
row.names(Test.count)->A
Test.tre$tip.lable->B
setdiff(A,B)->C
Test.count.update<-Test.count [! row.
names(Test.count) %in% C,]
These commands will find all the different names between the count table and the tree file and remove them. This sometimes works better than the steps outlined in Support Protocol 2, because, occasionally, there are more sequence differences in the count_table than just the tree node. The above commands will find these differences and remove them.
Anticipated Results
As mentioned above, this protocol will result in a summary file that has the taxonomic distribution of the gut microbiome. This file can be used for statistical tests to illustrate the taxonomic shifts. If the alternative protocols are completed, a figure will be generated showing how different the two populations are based on distances mapped onto a phylogenic tree, as well as a list of pathways that are significantly different between a control and treatment group. With these outputs, many other applications can be done. A popular application is correlations between taxonomic changes and metabolomic changes. This can reveal potential relationships between bacterial genera and metabolites and can be used to validate the metagenomic results. Also this information can be used for modeling and predictive software.
Time Considerations
Basic Protocol 1 will take approximately 5 to 8 hr of bench work, depending on the number of samples. Basic Protocol 2 will take approximately 3 to 4 hr. The sequencing can take 1 to 4 weeks, depending on the queue or if samples are sent to private sequencing companies. Basic Protocol 3 will take between a day and a month depending on what is used for the analysis. If performed on an external server or computing cluster, Basic Protocol 3 will take approximately 24 hr but if the analysis is done on a personal laptop, then it could take up to a month to complete. Support Protocol 3 will take another day to a week, depending on the number of samples, size of the subsample, and what is being used to do the analysis. Dist.seqs is a command that can take a while and may be killed if the file being created is too large. Again this also depends on the computing power used for this analysis. Alternate Protocol will take 1 week to a month. The Illumina Hiseq takes considerably more time to sequence than the Illumina Miseq, one should factor in at least twice the Illumina Miseq sequencing run time for an Illumina Hiseq run. Also the actual analysis is easier but each HUMAnN2 command can take between 12 and 24 hr on an external server. A time estimate for a HUMAnN2 run on a personal computer cannot be provided. NMR sample preparation for 30 samples will take approximately 5 hr. NMR sample acquisition for 30 samples will take approximately 15 hr and the analysis of the 30 samples will take an additional 5 hr.
Supplementary Material
Acknowledgments
This material is based upon work that is supported by the National Institutes of Health (ES028244; ES028288; ES026684), the National Institute of Food and Agriculture, U.S. Department of Agriculture, under award number 2914-38420-21822 and the Big Data to Knowledge training grant number 1 T32 LM 12415-1.
Literature Cited
- Abubucker S, Segata N, Goll J, Schubert AM, Izard J, Cantarel BL, . . . Huttenhower C (2012). Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Computational Biology, 8, e1002358. doi: 10.1371/journal.pcbi.1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B, Xie C, & Huson DH (2015). Fast and sensitive protein alignment using DIAMOND. Nature Methods, 12, 59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- Cai J, Zhang J, Tian Y, Zhang L, Hatzakis E, Krausz KW, . . . Patterson AD (2017). Orthogonal comparison of GC-MS and 1H NMR spectroscopy for short chain fatty acid quantitation. Analytical Chemistry (Washington, DC, United States), 89, 7900–7906. doi: 10.1021/acs.analchem.7b00848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Zhang L, Jones RA, Correll JB, Hatzakis E, Smith PB, . . . Patterson AD (2016). Antioxidant drug tempol promotes functional metabolic changes in the gut microbiota. Journal of Proteome Research, 15, 563–571. doi: 10.1021/acs.jproteome.5b00957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, . . . Knight R, (2010). QIIME allows analysis of high-throughput community sequencing data. Nature Methods, 7, 335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Bittinger K, Charlson ES, Hoffmann C, Lewis J, Wu GD, . . . Li H (2012). Associating microbiome composition with environmental covariates using generalized UniFrac distances. Bioinformatics, 28, 2106–2113. doi: 10.1093/bioinformatics/bts342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai H, Xiao C, Liu H, & Tang H (2010). Combined NMR and LC-MS analysis reveals the metabonomic changes in Salvia miltiorrhiza Bunge induced by water depletion. Journal of Proteome Research, 9, 1460–1475. doi: 10.1021/pr900995m. [DOI] [PubMed] [Google Scholar]
- Eriksson L, Trygg J, & Wold S (2008). CVANOVA for significance testing of PLS and OPLS (R) models. Journal of Chemometrics, 22, 594–600. doi: 10.1002/cem.1187. [DOI] [Google Scholar]
- Fonslow BR, Stein BD, Webb KJ, Xu T, Choi J, Kyu S, & Iii JRY (2013). Fast gapped-read alignment with Bowtie 2 10, 54–56. [Google Scholar]
- Hosgood DH, Sapkota AR, Rothman N, Rohan T, Hu W, Xu J, . . . Lan Q (2015). The potential role of lung microbiota in lun cancer attributed to household coal burning exposures. Environmental Molecular Mutagenesis, 55, 643–651. doi: 10.1002/em.21878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard TD, Murray IA, Nichols RG, Cassel K, Podolsky M, Kuzu G, . . . Perdew GH (2017). Dietary broccoli impacts microbial community structure and attenuates chemically induced colitis in mice in an Ah receptor dependent manner. Journal of Functional Foods, 37, 685–698. doi: 10.1016/j.jff.2017.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse SM, Welch DM, Morrison HG, & Sogin ML (2010). Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environmental Microbiology, 12, 1889–1898. doi: 10.1111/j.1462-2920.2010.02193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovel J, Patterson J, Wang W, Hotte N, O’Keefe S, Mitchel T, . . . Wong GK-S (2016). Characterization of the gut mi- crobiome using 16S or shotgun metagenomics. Frontiers in Microbiology, 7, 1–17. doi: 10.3389/fmicb.2016.00459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessner D, Chambers M, Burke R, Agus D, & Mallick P (2008). ProteoWizard: Open source software for rapid proteomics tools development. Bioinformatics, 24, 2534–2536. doi: 10.1093/bioinformatics/btn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozich JJ, Westcott SL, Baxter NT, High-lander SK, & Schloss PD (2013). Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the miseq illumina sequencing platform. Applied and Environmental Microbiology, 79, 5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Jeong SE, Lee S, Kim S, Han H, & Jeon CO (2017). Effects of cosmetics on the skin microbiome of facial cheeks with different hydration levels. MicrobiologyOpen, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Xie C, Lu S, Nichols RG, Tian Y, Li L, . . . Gonzalez FJ (2017). Intermittent fasting promotes white adipose browning and decreases obesity by shaping the gut microbiota. Cell Metabolism, 26, 672–685.e674. doi: 10.1016/j.cmet.2017.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Kimball E, & Rabinowitz JD (2006). A high-performance liquid chromatographytandem mass spectrometry method for quantitation of nitrogen-containing intracellular metabolites. Journal of the American Society for Mass Spectrometry, 17, 37–50. doi: 10.1016/j.jasms.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Martínez I, Muller CE, & Walter J (2013). Long-term temporal analysis of the human fecal microbiota revealed a stable core of dominant bacterial species. PLoS One, 8, e69621. doi: 10.1371/journal.pone.0069621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray IA, Nichols RG, Zhang L, Patterson AD, & Perdew GH (2016). Expression of the aryl hydrocarbon receptor contributes to the establishment of intestinal microbial community structure in mice. Scientific Reports, 6, 33969. doi: 10.1038/srep33969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rintala A, Pietilä S, Munukka E, Eerola E, Pursiheimo JP, Laiho A, . . . Huovinen P (2017). Gut microbiota analysis results are highly dependent on the 16s rRNA gene target region, whereas the impact of DNA extraction is minor. Journal of Biomolecular Techniques, 28, 19–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, & Huttenhower C (2011). Metagenomic biomarker discovery and explanation. Genome Biology, 12, R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanogiannopoulos P, Bess EN, Carmody RN, & Turnbaugh PJ (2016). The microbial pharmacists within us: A metagenomic view of xenobiotic metabolism. Nature Reviews Microbiology, 14(5)273–287. doi: 10.1038/nrmicro.2016.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong DT, Franzosa EA, Tickle TL, Scholz M, Weingart G, Pasolli E, . . . Segata N (2015). MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nature Methods, 12, 902–903. doi: 10.1038/nmeth.3589. [DOI] [PubMed] [Google Scholar]
- Tsugawa H, Cajka T, Kind T, Ma Y, Higgins B, Ikeda K, . . . Arita M (2015). MS-DIAL: Data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nature Methods, 12, 523–526. doi: 10.1038/nmeth.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Wang Y, & Qian P-Y (2016). Sensitivity and correlation of hypervariable regions in 16S rRNA genes in phylogenetic analysis. BMC Bioinformatics, 17, 135. doi: 10.1186/s12859-016-0992-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Xie C, Nichols RG, Chan SHJ, Jiang C, Hao R, . . . Patterson AD (2016). Farnesoid X Receptor Signaling Shapes the Gut Microbiota and Controls Hepatic Lipid Metabolism. mSystems, 1, e00070–00016. doi: 10.1128/mSystems.00070-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.