Abstract
Catalytic modules of assembly-line polyketide synthases (PKSs) have previously been observed in two very different conformations—an “extended” architecture and an “arch-shaped” architecture—although the catalytic relevance of neither has been directly established. By the use of a fully human naïve antigen-binding fragmenẗ (Fab) library, a high-affinity antibody was identified that bound to the extended conformation of a PKS module, as verified by X-ray crystallography and tandem size-exclusion chromatography–small-angle X-ray scattering (SEC–SAXS). Kinetic analysis proved that this antibody-stabilized module conformation was fully competent for catalysis of intermodular polyketide chain translocation as well as intramodular polyketide chain elongation and functional group modification of a growing polyketide chain. Thus, the extended conformation of a PKS module is fully competent for all of its essential catalytic functions.
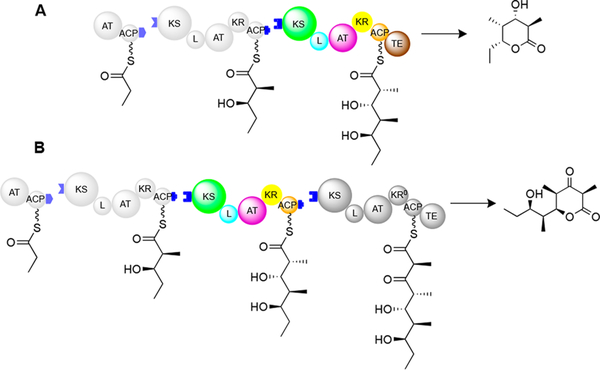
Assembly-line polyketide synthases (PKSs), such as 6-deoxyerythronolide B synthase (DEBS), are multifunctional, multimodular enzymes that synthesize structurally diverse natural products.1–3 Two truncated derivatives of DEBS are shown in Figure 1.
Figure 1.
(A) Bimodular and (B) trimodular derivatives of DEBS. Domains are shown as spheres. KS = ketosynthase; AT = acyl transferase; ACP = acyl carrier protein; L = KS-AT linker domain; KR = ketoreductase; KR0 = redox-inactive, epimerase-active KR homologue; TE = thioesterase. Blue tabs depict docking domains that facilitate non-covalent association of successive modules. Module 2 domains are colored for ease of comparison with Figure 2.
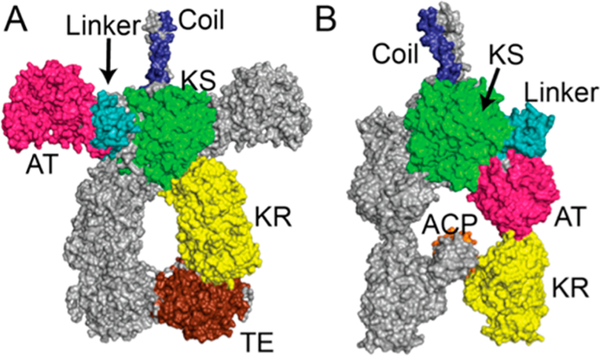
Although high-resolution structures of individual domains and multidomain fragments from several PKS modules have been solved, no X-ray structures of intact modules are available.4 Complete understanding of PKS structure–function relationships is further challenged by the possibility of large, catalytically relevant conformational changes within a module. Indeed, two very different conformations have been proposed. Small-angle X-ray scattering (SAXS) analysis of DEBS module 3 supported a conformation with the ketosynthase–acyltransferase (KS-AT) core in the same extended conformation as that previously observed in the crystal structures of two stand-alone KS-AT fragments5 (Figure 2A). In contrast, cryo-electron microscopy (cryo-EM) analysis of module 5 of the related pikromycin synthase revealed an arch-shaped conformation with the homodimeric KS as the capstone and each AT domain oriented downward to form an extensive interface with the KS of the opposite dimer6 (Figure 2B). Hypothetically, these apparent differences might be reconciled if one conformation corresponds to the state of the module during intermodular translocation of the polyketide chain (step I, Figure 3) while the other reflects the chain elongation state (step III, Figure 3). To address this question, we have now trapped individual PKS modules in defined conformational states and analyzed their catalytic properties.
Figure 2.
Alternative conformational models of homodimeric PKS modules: (A) SAXS model of DEBS module 3 + TE;5 (B) cryo-EM model of module 5 of the pikromycin synthase.6 Docking domain (“coil”), blue; KS, green; KS-AT linker, cyan; AT, pink; KR, yellow; ACP, orange; TE, brown. The collinear domain organization is as shown in Figure 1. The two conformations differ primarily in the orientation of the AT and L domains relative to the KS.
Figure 3.
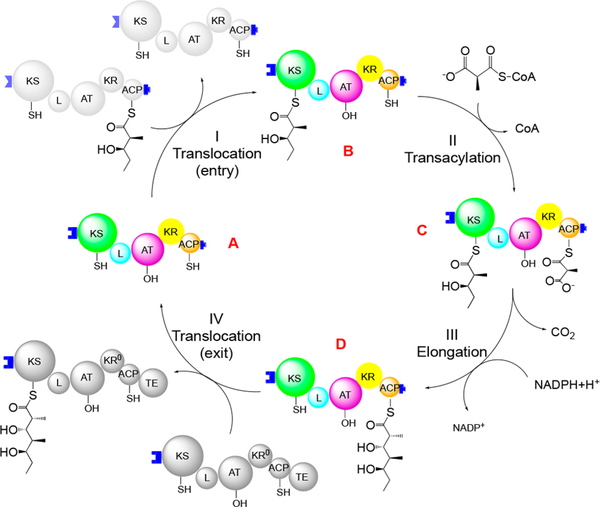
Catalytic cycle of DEBS module 2. Domains are colored as in Figure 1. The cycle includes four reactions: translocation of the growing polyketide chain (I) into and (IV) out of the module, (II) transacylation of an extender unit from methylmalonyl-CoA to its ACP domain, and (III) chain elongation via decarboxylative C–C bond formation. Module 2 also reduces the β-keto thioester product of chain elongation; this reaction is lumped into step III.
In this study, we chose DEBS modules 2 and 3 as representative proteins, principally because of (i) their shared organization and stereochemical control with PikAIII, (ii) the fact that activity assays and reference kinetic data for these modules are well-established, (iii) our early success in isolating antibodies against them, and (iv) the solved X-ray structure of the KS/AT fragment from module 3, which enabled structural analysis of its antibody-bound complex.
Monoclonal antibodies are powerful reagents for stabilizing specific conformations of individual proteins.7–10 In particular, antigen-binding fragments (Fabs) form rigid complexes with their protein antigens. We therefore sought to identify Fabs that would recognize DEBS module 3 + thioesterase (TE) (Figure 1) in a conformation consistent with retention of the enzymatic activity of this PKS.
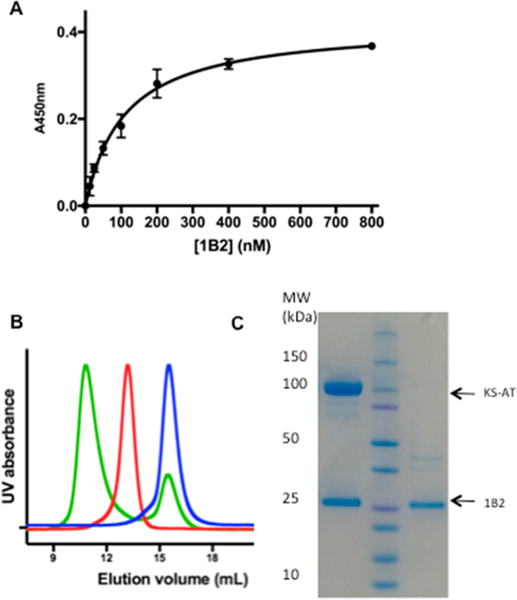
A large phage-displayed library of Fabs (3.7 × 1010) from the naive human antibody repertoirë 11 was used to identify Fabs against DEBS module 3 + TE.12 Briefly, module 3 + TE was partially biotinylated and immobilized onto streptavidin magnetic beads. These beads were incubated with the phage-displayed Fab library. After repeated washing to remove nonspecifically bound phage, the tightly bound phage particles were eluted and reinfected in Escherichia coli for further propagation. This panning process was repeated for several rounds and yielded multiple high-affinity clones against the target protein. One Fab, designated 1B2 (Figure S1A), bound module 3 + TE with high affinity (KD = 110 ± 10 nM, as measured by enzyme-linked immunosorbent assay (ELISA); Figure 4A). The stability of the antigen–Fab complex was verified by size-exclusion chromatography (SEC) (Figure 4B,C). Binding analysis with individual domains and multi-domains of module 3 + TE revealed that 1B2 recognized the KS-AT fragment of this PKS module (KD = 145 ± 25 nM, as measured by ELISA; Figure S1B).
Figure 4.
Binding of 1B2 to the KS-AT fragment of DEBS module 3. (A) ELISA of binding of 1B2 with module 3 + TE. (B) SEC traces of 1B2 (blue), the KS-AT fragment (red), and the 1B2/KS-AT complex (green). (C) SDS-PAGE: (left lane) the major SEC peak is a stoichiometric mixture of 1B2 and KS-AT; (right lane) 1B2 alone.
Taking advantage of the high affinity of 1B2 for the KS-AT fragment of module 3 + TE, we grew crystals of this complex. Crystals belonged to the monoclinic space group C2 and diffracted to 2.1 Å resolution (Table S1). The asymmetric unit contained one molecule each of KS-AT and 1B2 and 680 waters; the homodimeric structure was obtained through symmetry expansion. The structure was refined to Rwork and Rfree values of 0.167 and 0.210, respectively.
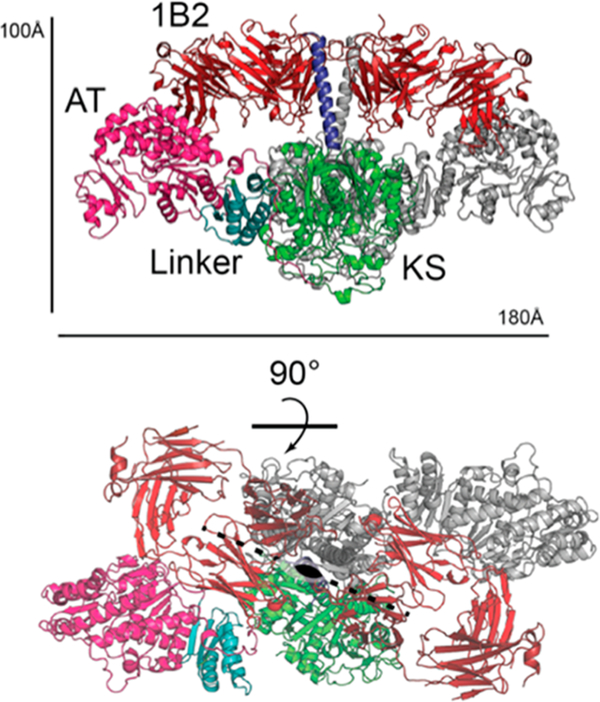
As can be seen in Figure 5, the structure of the complex established that the KS-AT portion assumed an extended dimeric architecture analogous to that shown in Figure 2A for the uncomplexed KS-AT didomain. The complementarity-determining regions (CDRs) of two 1B2 equivalents face each other on opposite sides of the N-terminal coiled-coil region (Figure S2A,B). Each Fab also contacts both the KS domain (Figure S2C) and the AT domain (Figure S2D). In particular, hydrogen bonds were observed between Arg98 of the light chain and the conserved Asp331 of the KS domain and between Asn211 of the heavy chain and the carbonyl oxygen of Pro775 of the AT domain. The extensive overall footprint of 1B2 across the KS-AT fragment is readily visualized in the space-filling model shown in Figure S2E.
Figure 5.
Crystal structure of 1B2 in complex with the KS-AT fragment of DEBS module 3 + TE. Two views of the complex (PDB entry 6C9U) are shown. The coloring scheme is identical to that in Figures 1–3 (docking domain, blue; KS, green; KS-AT linker, cyan; AT, pink). This homodimer binds to two copies of the Fab, shown in red. The axis and plane of symmetry are shown in the lower view.
With the exception of the N-terminal coiled coil, which is absent in the previously reported structure of the KS-AT fragment of DEBS module 3,13 no significant conformational changes were observed when the 1B2/KS-AT structure was compared to that of its antibody-free counterpart (Figure S3A). Excellent backbone overlap was also observed between the 1B2/KS-AT structure and the homologous KS-AT fragment of DEBS module 5, which also included an N-terminal coiled coil14 (Figure S3B). Notably, the extended KS-AT conformation in the 1B2/KS-AT complex was very different from the reported cryo-EM-deduced arched conformation attributed to the homologous KS-AT portion of module 5 of the pikromycin synthase (Figure S4).
The structure of 1B2 in complex with the KS-AT fragment of module 3 + TE led us to speculate that the same Fab would also recognize the module 2 + TE construct shown schematically in Figure 1A, as this hybrid PKS module includes the N-terminal docking domain from module 3 and the hydrogen-bonding functional groups highlighted above are conserved between the two DEBS modules. Indeed, native module 2 + TE bound just as tightly to 1B2 as did the KS-AT didomain of module 3 (KD = 100 ± 10 nM, as measured by ELISA; Figure S5). We therefore utilized previously developed UV-spectrophotometric kinetic assays with module 2 + TE15 to analyze the influence of the Fab on the individual rates of both intermodular chain translocation and intramodular chain elongation catalyzed by this module. Our expectation was that if access to the arch-shaped KS-AT conformation (Figure 2B) were essential for either reaction, then 1B2 would prove inhibitory in the corresponding assay.
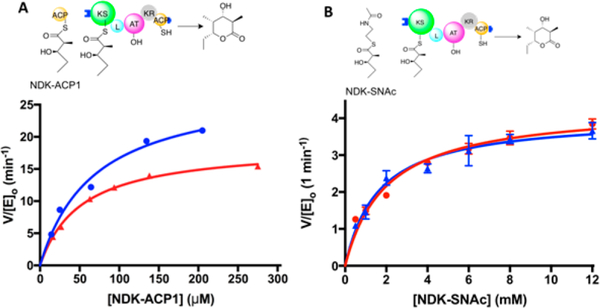
Chain translocation kinetics were measured for module 2 + TE with its natural diketide (NDK) substrate, (2S,3R)-2-methyl-3-hydroxypentanoyl-ACP1.16 As shown in Figure 6A, both the kcat and KM values were comparable in the presence and absence of 1B2 (kcat = 19 ± 1 and 28 ± 3 min−1 in the presence and absence of 1B2, respectively; KM = 52 ± 4 and 69 ± 16 μM in the presence and absence of 1B2, respectively). To quantify the chain elongation kinetics for the same module, an alternative assay was used involving a diffusible mimic of the same substrate.17 Again, as shown in Figure 6B, both the kcat and KM values were comparable in the presence and absence of 1B2 (kcat = 4.1 ± 0.2 and 4.3 ± 0.2 min−1 in the presence and absence of 1B2, respectively; KM = 1.6 ± 0.2 and 2.0 ± 0.3 mM in the presence and absence of 1B2, respectively). Trapping module 2 + TE in its extended conformation by addition of excess 1B2 had no significant adverse effect on the ability of this PKS module to catalyze either chain elongation or chain translocation. We therefore conclude that the extended KS-AT conformation (Figure 2A) is fully competent for both chain translocation and chain elongation, while the alternative arch-shaped conformation is not required for either activity. The data in Figure 6 also suggest that neither AT-catalyzed extender unit transacylation (step II, Figure 3) nor KR-catalyzed β-ketoreduction (lumped into step III, Figure 3) requires access to the arch-shaped conformation, although neither transacylation nor ketoreduction is rate-limiting in either assay.
Figure 6.
Effect of 1B2 binding on chain translocation and chain elongation by DEBS module 2 + TE. 1B2 binds tightly to DEBS module 2 + TE. The initial slope of the graph in (A) reflects conditions where chain translocation is rate-limiting, whereas the maximum velocities in (A) and (B) reflect conditions where elongation is rate-limiting. Blue curves show the activity without 1B2; red curves show the activity in the presence of 1.2 equiv of 1B2.
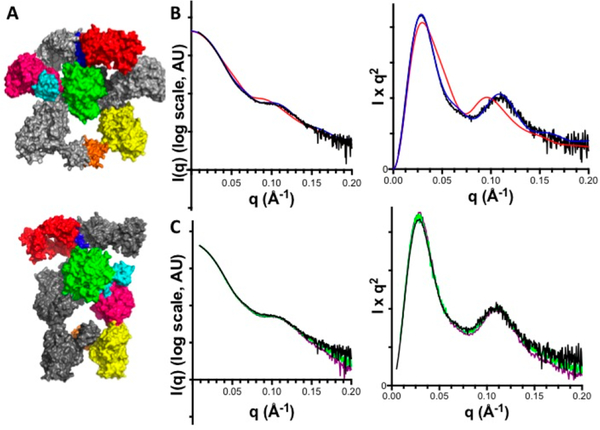
To directly correlate the above findings from crystallography and activity assays of 1B2-bound PKS modules, we performed SEC in tandem with SAXS on the 1B2/module 2 complex under solution conditions corresponding to high specific activity of this PKS module.18 Samples of the complex were freshly prepared in three of the four states shown in Figure 3 (states A, B, and D) and immediately subjected to SEC−SAXS analysis using methods very similar to those reported for DEBS module 3.5 The data from each SEC peak fraction were compared to theoretical scattering curves generated by CRYSOL19 based on the two alternative docking models shown in Figure 7A. In every case, the experimental data correlated much more strongly to the theoretically predicted curve from the extended model (χ2 = 0.55) than to the curve from the arched model (χ2 = 7.74) (Figure 7B,C). These findings strongly reinforce the conclusion that module 2 predominantly assumes the extended conformation in the preacylation state (state A, Figure 3), the acylated state (state B, Figure 3), and the postelongation state (state D, Figure 3), establishing that conformational reorganization of the KS, AT, and KR domains is not required during catalysis.
Figure 7.
Tandem (SEC−SAXS) analysis of the 1B2/module 2 complex. (A) Docking models of 1B2 bound to each conformation from Figure 2 were generated by aligning the X-ray structure of the 1B2/KSAT complex (Figure 5) with each module conformation. The Fab complexed to the extended conformation is shown at the top, whereas the arched conformation is shown below. Domain coloring is the same as in Figures 2 and 5. (B) Comparisons of (left) log(I) vs q and (right) Kratky plots generated by SEC−SAXS analysis of the 1B2/module 2 complex (black curve) and CRYSOL analysis of the extended (blue curve) and arched (red curve) models from (A). (C) Comparison of (left) log(I) vs q and (right) Kratky plots from SEC−SAXS analysis of 1B2/module 2 trapped in states A (black curve), B (green curve), and D (purple curve). (cf. Figure 3).
In summary, we have investigated the structure and dynamics of modules from a prototypical assembly-line PKS using a combination of protein structural and functional approaches. The simplest model for polyketide biosynthesis, based on the well-established model for vertebrate fatty acid biosynthesis,20 posits that the catalytic domains of individual modules assemble into relatively rigid structures, with interdomain and intermodular motions being primarily restricted to acyl-ACP species. Alternatively, PKS modules might undergo larger conformational changes along their reaction coordinates, such as interconversions between the extended and the arched protein conformations shown in Figure 2. To test these alternative hypotheses, we identified a specific Fab that stabilizes a PKS module in an extended conformation. The targeted discovery of the specific Fab antibody 1B2 combined with crystallographic, SEC−SAXS, and kinetic analyses of 1B2/PKS complexes establish that the extended conformation of the KS-AT didomains of both DEBS modules 2 and 3 is catalytically competent for all of the essential PKS reactions shown in Figure 3. The stage is now set for a deeper mechanistic understanding of long-distance translocations of growing polyketide chains between successive PKS modules.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by grants from the National Institutes of Health (GM087934 to C.K., P41CA196276 and GM104659 to C.S.C., and GM022172 to D.E.C.). X-ray analysis was performed on beamlines 11-1 and 12-2 at the Stanford Synchrotron Radiation Lightsource (SSRL), SLAC National Accelerator Laboratory. Use of the SSRL, SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393).
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.8b02100.
Experimental methods and supporting figures and tables (PDF)
REFERENCES
- (1).Walsh CT Science 2004, 303, 1805. [DOI] [PubMed] [Google Scholar]
- (2).Cortes J; Haydock SF; Roberts GA; Bevitt DJ; Leadlay PF Nature 1990, 348, 176. [DOI] [PubMed] [Google Scholar]
- (3).Donadio S; Staver MJ; McAlpine JB; Swanson SJ; Katz L Science 1991, 252, 675. [DOI] [PubMed] [Google Scholar]
- (4).Robbins T; Liu YC; Cane DE; Khosla C Curr. Opin. Struct. Biol. 2016, 41, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Edwards AL; Matsui T; Weiss TM; Khosla C J. Mol. Biol 2014, 426, 2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Dutta S; Whicher JR; Hansen DA; Hale WA; Chemler JA; Congdon GR; Narayan ARH; Hakansson K; Sherman DH; Smith JL; Skiniotis G Nature 2014, 510, 512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Malia TJ; Obmolova G; Luo J; Teplyakov A; Sweet R; Gilliland GL Acta Crystallogr., Sect. F: Struct. Biol. Cryst. Commun 2011, 67, 1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Zhou Y; Morais-Cabral JH; Kaufman A; MacKinnon R Nature 2001, 414, 43. [DOI] [PubMed] [Google Scholar]
- (9).Uysal S; Vasquez V; Tereshko V; Esaki K; Fellouse FA; Sidhu SS; Koide S; Perozo E; Kossiakoff A Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Farady CJ; Egea PF; Schneider EL; Darragh MR; Craik CS J. Mol. Biol. 2008, 380, 351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Duriseti S; Goetz DH; Hostetter DR; LeBeau AM; Wei Y; Craik CS J. Biol. Chem. 2010, 285, 26878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Kim J; Wu S; Tomasiak TM; Mergel C; Winter MB; Stiller SB; Robles-Colmanares Y; Stroud RM; Tampe R; Craik CS; Cheng Y Nature 2015, 517, 396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Tang Y; Chen AY; Kim CY; Cane DE; Khosla C Chem. Biol. 2007, 14, 931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Tang Y; Kim CY; Mathews II; Cane DE; Khosla C Proc. Natl. Acad. Sci. U. S. A. 2006, 103, 11124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Lowry B; Robbins T; Weng CH; O’Brien RV; Cane DE; Khosla CJ Am. Chem. Soc. 2013, 135, 16809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Wu N; Cane DE; Khosla C Biochemistry 2002, 41, 5056. [DOI] [PubMed] [Google Scholar]
- (17).Chen AY; Schnarr NA; Kim CY; Cane DE; Khosla CJ Am. Chem. Soc. 2006, 128, 3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Lowry B; Li X; Robbins T; Cane DE; Khosla C ACS Cent. Sci. 2016, 2, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Svergun DI; Barberato C; Koch MHJ J. Appl. Crystallogr. 1995, 28, 768. [Google Scholar]
- (20).Maier T; Leibundgut M; Ban N Science 2008, 321, 1315. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.