Abstract
Background
Methicillin resistance in staphylococci is conferred by an alternative PBP (PBP2a/2′) with low affinity for most β-lactam antibiotics. PBP2a is encoded by mecA, which is carried on a mobile genetic element known as SCCmec. A variant of mecA, mecC, was described in 2011 and has been found in Staphylococcus aureus from humans and a wide range of animal species as well as a small number of other staphylococcal species from animals.
Objectives
We characterized a novel mecC allotype, mecC3, encoded by an environmental isolate of Staphylococcus caeli cultured from air sampling of a commercial rabbit holding.
Methods
The S. caeli isolate 82BT was collected in Italy in 2013 and genome sequenced using MiSeq technology. This allowed the assembly and comparative genomic study of the novel SCCmec region encoding mecC3.
Results
The study isolate encodes a novel mecA allotype, mecC3, with 92% nucleotide identity to mecC. mecC3 is encoded within a novel SCCmec element distinct from those previously associated with mecC, including a ccrAB pairing (ccrA5B3) not previously linked to mecC.
Conclusions
This is the first description of the novel mecC allotype mecC3, the first isolation of a mecC-positive Staphylococcus in Italy and the first report of mecC in S. caeli. Furthermore, the SCCmec element described here is highly dissimilar to the archetypal SCCmec XI encoding mecC in S. aureus and to elements encoding mecC in other staphylococci. Our report highlights the diversity of mecC allotypes and the diverse staphylococcal species, ecological settings and genomic context in which mecC may be found.
Introduction
Methicillin resistance in Staphylococcus aureus is typically conferred by the gene mecA along with two variants, mecB and mecC.1–4mec gene resistance is mediated by an alternative PBP with reduced affinity for almost all β-lactam antimicrobials.5,6 Since its first discovery in bulk tank milk on an English dairy farm,1mecC has been found in S. aureus isolates from a wide range of host species, including human carriage and infection and various wildlife, companion and livestock species7,8 with genomic analysis indicating zoonotic transmission from livestock to humans.9,10mecC-MRSA have been reported from a range of countries and while this geographical distribution has centred on Europe,8mecC-MRSA have also been reported in Australia.11 In addition to S. aureus, mecC has been described in other species of staphylococci, albeit in only a limited number of species and a small number of isolates, which have all come from animals: Staphylococcus xylosus,12Staphylococcus sciuri,13Staphylococcus stepanovicci14 and Staphylococcus saprophyticus15,16 or in the case of Staphylococcus edaphicus from environmental sampling.17mecC in MRSA is found within a staphylococcal cassette chromosome mec (SCCmec) type XI while in other staphylococci it is found in a range of genomic contexts, although always within the orfX region and with features common to SCCmec elements. Divergent mecC allotypes mecC1 and mecC2 have been described in S. xylosus12 and S. saprophyticus,15 respectively, but have not been reported in S. aureus, suggesting a greater diversity and ancestral association of mecC with non-aureus staphylococci.
Here we describe a novel mecC allotype, mecC3, encoded within a distinct and novel SCCmec element in a newly described species, Staphylococcus caeli,18 which furthers our understanding of the diversity of mecC genes and the diverse species and genetic elements that carry it.
Materials and methods
Isolate collection and WGS
The S. caeli isolate 82BT (=NCTC 14063T=CCUG 71912T) was collected from air sampling in a commercial rabbit holding in Italy in 2013 as part of a previous study19 and has been described elsewhere, including its genome sequencing.18 MRSA was also present on the sampled farm.19 Six months prior to the isolation of 82BT, ST398 MRSA belonging to t034 and t5210 were isolated from farm workers, their relatives and rabbits on the farms.19 At the time of 82T isolation MRSA belonging to t034, t5210, t1190 and t2970 were isolated from rabbits and humans with non-typed MRSA isolates found in air samples and surface swabs.19
Sequence assembly and identification of SCCmec
Sequencing reads were de novo assembled using Velvet20 and annotated using the NCBI Prokaryotic Genome Annotation Pipeline.21 Contiguous sequences (contigs) containing the orfX and mecC genes were identified using BLAST, using the respective genes from S. aureus LGA251 (accession number FR821779) as query sequences. Contigs NZ_FMPG01000005.1 and NZ_FMPG01000008.1 were identified as containing the orfX and mecC genes, respectively. Primers were designed for the end of contig NZ_FMPG01000005.1 and start of contig NZ_FMPG01000008.1, followed by PCR, with the resulting amplicon being ABI sequenced (Source BioScience, Cambridge, UK), to close the gap between the contigs. Primers were designed using Primer3 (http://primer3.ut.ee/).
Phylogenetic analysis of mecA homologues
Phylogenetic analyses were carried out in MEGA7.22 All nucleotide sequences were obtained from NCBI databases, using the following accession numbers: S. aureus LGA251, FR821779, mecC; S. xylosus S04009, HE993884, mecC1; S. saprophyticus 210, KF955540, mecC2; S. caeli 82B, mecC3, FMPG01000008; S. sciuri K11, Y13094, mecA1; S. aureus N315, NC_002745, mecA; Staphylococcus vitulinus CSBO8, AM048810, mecA2; Macrococcus caseolyticus IMD0819, KY013611, mecD; and M. caseolyticus JCSC5402, NC_011996, mecB. The sequences were aligned using MUSCLE and a maximum likelihood tree was generated, using a general time reversible model. Site substitution rates were calculated using a discrete gamma distribution model.
Nucleotide accession numbers
The whole genome nucleotide sequences for S. caeli 82BT have been deposited previously18 in the NCBI database under accession numbers NZ_FMPG00000000 (assembly) and ERR473447 (sequence reads). The assembled SCCmec region of 82BT generated in this work has been deposited under accession number MH155596.
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was performed by Vitek2 using AST P260 cards following the manufacturer’s instructions. Disc diffusion was performed following the EUCAST disc diffusion method Version 6.0 January 2017. Growth on MRSA Brilliance (Oxoid, Basingstoke, UK) was tested by spreading a 1 μL loop taken from a 0.5 McFarland suspension (∼1.5 × 105 cfu) on to the Brilliance plate and incubating at 30°C, 35°C and 37°C for 24 h. Interpretation was done based on the criteria for CoNS with S. aureus NCTC 12497 and NCTC 12493 used as control strains for susceptibility testing. PBP2a detection was performed using the PBP2′ Latex Agglutination Test Kit (Oxoid) following the manufacturer’s instructions.
Results and discussion
S. caeli 82BT contains a novel mecC allotype, mecC3
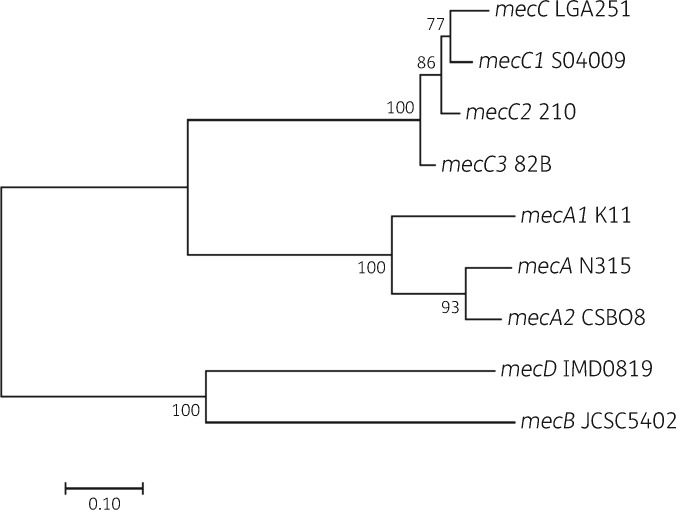
Isolate 82BT has been described as the type strain of a novel staphylococcal species, S. caeli, most closely related to S. xylosus, S. saprophyticus and S. edaphicus.18 As part of the characterization of 82BT it underwent WGS. Further investigation of the 82BT genome by BLAST analysis using the mecC gene from S. aureus LGA251 as the query sequence revealed that 82BT encodes a mecC homologue on contig NZ_FMPG01000008.1. Alignments of the 82BTmecC gene with known mecC allotypes showed that it shares 92% nucleotide identity with mecC from LGA251, 93% with mecC1 from S. xylosus S04009 and 94% with mecC2 from S. saprophyticus 210. Based on these results and in line with guidelines for the classification for mecA homologues,23 the mecC gene for S. caeli 82BT is a new allotype of mecC and therefore designated mecC3. The phylogenetic relationship of mecC3 to the other mecA homologues is illustrated in Figure 1; this suggests that the more common mecC has evolved from the more ancestral forms (mecC1, mecC2, mecC3) and that, of the three, mecC3 is closest to the ancestral form. The mecC complex of S. caeli 82BT shares 93% and 92% nucleotide identity with the complete mecC complex of LGA251 and S04009, respectively, with the partial mecC complex of S. saprophyticus 210 sharing 94%. 82BT gave a negative reaction using the commercial PBP2a latex agglutination test kit from Oxoid. This reaction was not altered after cefoxitin induction. mecC3 in 82BT was, however, detected by the published mecC primer pairs 1A and 1B24 and mecC-Uni-F and mecC-Uni-R.13
Figure 1.
Phylogenetic relationship of representative mec allotypes. Nucleotide sequences were aligned using MUSCLE, with the maximum likelihood method, based on the general time reversible model, being used to build the tree. The highest log likelihood ( − 9785.4010) tree is shown. The percentage of trees in which the associated genes clustered together, based on bootstrapping with 500 replicates, is shown at the branches. A discrete gamma distribution was used to model site substitution rates, with branch lengths measured in the number of substitutions per site.
S. caeli 82BT mecC3 is encoded within a distinct SCCmec-like element
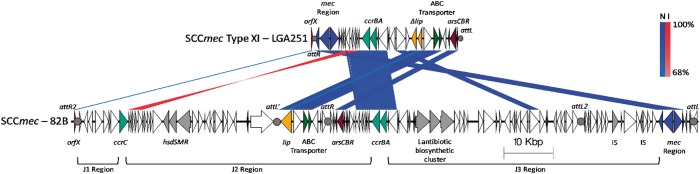
mecC genes are typically found within SCCmec elements,13,14 which insert into the genome at the 3′-end of a 23S rRNA methyltransferase gene, often referred to as orfX.25,26 BLASTn analysis using orfX from LGA251 as the query sequence identified the orfX region of 82BT within contig NZ_FMPG01000005.1. PCR and amplicon sequencing confirmed that the two contigs (NZ_FMPG01000005.1 containing orfX and NZ_FMPG01000008.1 containing mecC3) are contiguous in the 82BT genome with a gap of 140 bp between the ends of the two contigs. The assembled sequence generated in this study from these two contigs, encoding the orfX region of 82BT and mecC3 including a SCCmec-like element, has been deposited in NCBI with accession number MH155596.
The integration site on the genome for SCCmec elements within orfX, known as the attachment (attB) site, is identified by the recombinase CcrAB/CcrC via a 14 bp sequence.27 Insertion into the attB generally results in the SCCmec being flanked by direct repeats, attR and attL.13,27,28 We therefore searched for these repeat sequences within the orfX region of 82BT to define the limits of its putative SCCmec-like element. The search sequences; gc[ag]tatca[tc]aaatgatgcggttt and aacc[tg]catca[tc][tc][at][ac]c[tc]gataag[ct], produced from previously identified attR and attL sequences, respectively, identified an attR site 51 kbp downstream of orfX and an attL site 3.8 kbp downstream of mecC3 (Figure 2a). This indicates an SCCmec in 82BT, which is 127 kbp in size, with the orfX and mecC3 regions at opposite ends of the element (Figure 2a). However, it is not clear if this represents a single entity or a composite element generated by consecutive insertions. Within this SCCmec region, 82BT contains two ccr complexes and three joining (J) regions (Figure 2a). The ccrA and ccrB genes of 82BT share 90% and 91% nucleotide identity with ccrA5 and ccrB3, respectively, from Staphylococcus pseudintermedius KM241 (accession number AM904731). This represents a type 6 ccr allotype,25,28 which is novel for an SCCmec encoding a mecC. Indeed this pairing is rare among SCCmec in staphylococci, with a search of NCBI finding only two other staphylococcal isolates that contained this ccrAB pairing [Staphylococcus cohnii WC28 (accession number GU370073)29 and S. cohnii SNUDS-2 (CP019597; locus tags BZ166_04625 and BZ166_04620)]. In addition to the ccrAB genes described above, the SCCmec of 82BT also contains a ccrC gene, sharing 90% nucleotide identity with S. aureus PM1 (SCCmec VII, accession number AB462393, gene ccrC8). The attR site was identified 51 kbp away from the orfX gene (Figure 2a), which deviates from typical SCCmec elements and therefore the orfX region was examined to find any additional attR-like site.
Figure 2.
Overview of S. caeli 82BT SCCmec. SCCmec Type XI–LGA251 corresponds to S. aureus LGA251, accession number FR821779, with SCCmec–82B corresponding to S. caeli 82BT. Regions of homology are represented by bands connecting the two sequences, with the percentage identity key shown on the right. Blue denotes normal sequence alignment (N); red denotes inverted sequence alignment (I). Key features associated with SCCmec elements are labelled. att sites are highlighted by filled circles and labelled above. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
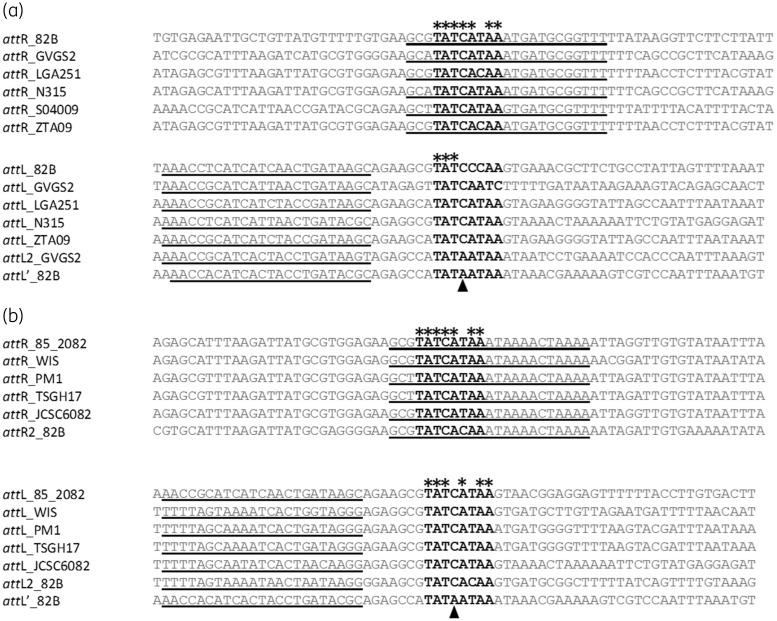
Within the 3′ region of orfX an att site with sequence homology to the attR SCCmec type VII of PM1 was identified. As there are two ccr regions within the 82BT SCCmec, it was proposed that the attR within the 3′-end of orfX (Figure 2, attR2) might be linked to the CcrC recombinase. Therefore, a search sequence: [at][at] [at][ct][ct][ga][cg][atc][ta][ca][at][ta][ct][ac]act[ga][ga][tc]a, based on the attL sequence of SCCmec elements containing only the ccrC gene, was used to identify the corresponding attL site linked to attR2, attL2. This revealed only one potential attL2 site, located 16.8 kbp upstream of the mec region (Figure 2, attL2). Sequence alignment of the attR/L sites of 82BT with attR/L sites of ccrAB-containing SCCmec, compared with the alignment of the attR/L2 sites of 82BT versus those attR/L sites of ccrC-containing SCCmec, suggest a varied att site for CcrAB/CcrC (Figure 3). Though it appears that the highly conserved central 8 bp sequence is consistent, there is notable variation between the ccrAB- and ccrC-associated att sites within those bases that flank the 8 bp region (Figure 3). Although the location of attL2 was identified with the predicted region, the only other potential attL site identified was attL′, recently described for a highly conserved type XI SCCmec found in S. xylosus 47–83.30 As with the attL′ site identified for S. xylosus 47–83, the attL′ of 82BT lies downstream of the lip gene; however, when compared with the attL of the ccrC-associated SCCmec attL sequences, it shares little similarity and lacks the central cysteine required for attB/attSCC recombination (Figure 3b).27 Indeed, the attL′ of 82BT shows more sequence similarity to that of attL2 from S. scuiri GVGS2 and the other attL sites related to CcrAB (Figure 3a). This suggests that attL2 is most likely linked with attR2, though its unusual genomic location within the SCCmec suggests various DNA recombination events may have occurred.
Figure 3.
Comparison of attR/L sites from ccrAB- or ccrC-containing SCCmec. Conserved nucleotide bases within the core 8 bp region, represented in black, bold font, are indicated by an asterisk. The black triangle indicates the position of the central cytosine, thought to be essential for recombination between attB and attSCC.27 Inverted repeats are marked by the underlined bases. (a) Sequences of known attR and attL sites associated with ccrAB [from: S. aureus N315 (N315), NC_002745; S. aureus LGA251 (LGA251), FR821779; S. xylosus S04009 (S04009), HE993884; S. sciuri GVGS2 (GVGS2), HG515014; and S. aureus ZTA09 (ZTA09), LK024544] were aligned and compared with those identified in S. caeli 82BT. (b) Sequences of known attR and attL sites associated with ccrC [from: S. aureus 85/2082 (85_2082), AB037671; S. aureus WIS (WIS), AB121219; S. aureus PM1 (PM1), AB462393; S. aureus TSGH17 (TSGH17), AB512767; and S. aureus JCSC6082 (JCSC6082), AB373032] were aligned and compared with attR2 and attL2 from S. caeli 82BT.
SCCmec include J regions, defined by the areas between the orfX, ccr and mec genes, with the SCCmec of 82BT having three such J regions. The J1 region contains primarily hypothetical genes and the ccrC gene and shares 86% identity with the J1 region of S. aureus PM1 SCCmec. The J2 region upstream of attL2, contains genes similar to those of SCCmec type V, with the presence of type 1 restriction modification genes, although there is little similarity between the restriction modification genes of 82BT and those present on SCCmec type V.
The J2 region downstream of attL′ shows the greatest similarity to the SCCmec XI of LGA251, sharing 89% identity, with ABC transporters, genes associated with arsenic resistance and a lipase gene. Unlike the lipase gene in LGA251 the version in 82BT appears to be intact. The J3 region is divergent from the rest of the SCCmec of LGA251, with the exception of a putative membrane protein and a putative DNA helicase protein. A notable feature of the J3 region is the presence of a lantibiotic biosynthetic cluster that shares 90% nucleotide identity with one present on the plasmid pETB797 of S. aureus NRL 08/797 (accession number KY436025). The cluster encodes two peptide homologues of Lacticin 3147, produced by Lactococcus lactis, which has been shown to be active against Gram-positive bacteria.31 Within the cluster is also a gene encoding a homologue of LtnT, which is required for the transport of the Lacticin 3147 peptides as well as unrelated peptides.32 The J3 region also contains two ISL3-like transposases, with the majority of the other genes found within SCCmec being from different staphylococci.
Antimicrobial susceptibility of S. caeli 82BT
Using Vitek 2, isolate 82BT was found to be susceptible to ciprofloxacin, daptomycin, gentamicin, linezolid, mupirocin, rifampicin, teicoplanin, tigecycline and vancomycin. The isolate was resistant to clindamycin, erythromycin, fusidic acid, tetracycline and trimethoprim. Genomic analysis revealed that 82BT has the resistance genes erm(B), erm(C), fusD, tet(L) and dfrK, which is consistent with the resistance profile of the isolate. With regards to β-lactam antibiotics, it was resistant to benzylpenicillin (MIC ≥ 0.5 mg/L) and oxacillin (MIC 1–2 mg/L) but negative in the cefoxitin resistance screen. However, by disc diffusion it was resistant to cefoxitin and displayed an MIC of 3 mg/L when assessed by Etest. The isolate failed to grow on the MRSA screening agar MRSA Brilliance despite a high inoculum and the use of different incubation temperatures.
This study extends the known distribution and diversity of mecC genes in terms of country of isolation, sample type, staphylococcal host species, genomic context and allotypes, all of which are novel to the best of our knowledge. These highlight the complex epidemiology of this resistance determinant, particularly among CoNS. It is interesting to note that the species most closely related to S. caeli (S. xylosus, S. saprophyticus and S. edaphicus) have all been reported to carry mecC or mecC allotypes, which may indicate a prominent role for this group in the origins, evolution and epidemiology of mecC and SCCmecC.
Acknowledgements
The help of the core sequencing and informatics teams and the Pathogens Informatics team at the Wellcome Trust Sanger Institute (WTSI) is gratefully acknowledged.
Funding
This project was supported by internal funding from the Royal (Dick) School of Veterinary Studies, University of Edinburgh, a Medical Research Council (MRC) partnership grant (G1001787/1) and the Wellcome Trust, grant number 098051. E. M. H. is supported by a UK Research and Innovation (UKRI) Fellowship: MR/S00291X/1. The funders had no role in the study design, data collection, analysis, decision to publish or preparation of the manuscript.
Transparency declarations
None to declare.
References
- 1. García-Álvarez L, Holden MTG, Lindsay H. et al. Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: a descriptive study. Lancet Infect Dis 2011; 11: 595–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shore AC, Deasy EC, Slickers P. et al. Detection of staphylococcal cassette chromosome mec type XI carrying highly divergent mecA, mecI, mecR1, blaZ, and ccr genes in human clinical isolates of clonal complex 130 methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 2011; 55: 3765–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Peacock SJ, Paterson GK.. Mechanisms of methicillin resistance in Staphylococcus aureus. Annu Rev Biochem 2015; 84: 577–601. [DOI] [PubMed] [Google Scholar]
- 4. Becker K, van Alen S, Idelevich EA. et al. Plasmid-encoded transferable mecB-mediated methicillin resistance in Staphylococcus aureus. Emerg Infect Dis 2018; 24: 242–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hartman BJ, Tomasz A.. Low-affinity penicillin-binding protein associated with β-lactam resistance in Staphylococcus aureus. J Bacteriol 1984; 158: 513–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kim C, Milheiriço C, Gardete S. et al. Properties of a novel PBP2A protein homolog from Staphylococcus aureus strain LGA251 and its contribution to the β-lactam-resistant phenotype. J Biol Chem 2012; 287: 36854–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Becker K, Ballhausen B, Köck R. et al. Methicillin resistance in Staphylococcus isolates: the “mec alphabet” with specific consideration of mecC, a mec homolog associated with zoonotic S. aureus lineages. Int J Med Microbiol 2014; 304: 794–804. [DOI] [PubMed] [Google Scholar]
- 8. Paterson GK, Harrison EM, Holmes MA.. The emergence of mecC methicillin-resistant Staphylococcus aureus. Trends Microbiol 2014; 22: 42–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Harrison EM, Coll F, Toleman MS. et al. Genomic surveillance reveals low prevalence of livestock-associated methicillin-resistant Staphylococcus aureus in the East of England. Sci Rep 2017; 7: 7406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Harrison EM, Paterson GK, Holden MTG. et al. Whole genome sequencing identifies zoonotic transmission of MRSA isolates with the novel mecA homologue mecC. EMBO Mol Med 2013; 5: 509–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Worthing KA, Coombs GW, Pang S. et al. Isolation of mecC MRSA in Australia. J Antimicrob Chemother 2016; 71: 2348–9. [DOI] [PubMed] [Google Scholar]
- 12. Harrison EM, Paterson GK, Holden MTG. et al. A Staphylococcus xylosus isolate with a new mecC allotype. Antimicrob Agents Chemother 2013; 57: 1524–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Harrison EM, Paterson GK, Holden MTG. et al. A novel hybrid SCCmec-mecC region in Staphylococcus sciuri. J Antimicrob Chemother 2014; 69: 911–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Semmler T, Harrison EM, Lübke-Becker A. et al. A look into the melting pot: the mecC-harboring region is a recombination hot spot in Staphylococcus stepanovicii. PLoS One 2016; 11: e0147150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Małyszko I, Schwarz S, Hauschild T.. Detection of a new mecC allotype, mecC2, in methicillin-resistant Staphylococcus saprophyticus. J Antimicrob Chemother 2014; 69: 2003–5. [DOI] [PubMed] [Google Scholar]
- 16. Loncaric I, Kubber-Heiss A, Posautz A. et al. Characterization of methicillin-resistant Staphylococcus spp. carrying the mecC gene, isolated from wildlife. J Antimicrob Chemother 2013; 14: 2222–5. [DOI] [PubMed] [Google Scholar]
- 17. Pantucek R, Sedlacek I, Indrakova A. et al. Staphylococcus edaphicus sp. nov., isolated in Antarctica, harbours mecC gene and genomic islands with suspected role in adaptation to extreme environment. Appl Environ Microbiol 2018; 84: e01746-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. MacFadyen AC, Drigo I, Harrison EM. et al. Staphylococcus caeli sp. nov., isolated from air sampling in an industrial rabbit holding. Int J Syst Evol Microbiol 2018; doi:10.1099/ijsem.0.003098. [DOI] [PubMed] [Google Scholar]
- 19. Agnoletti F, Mazzolini E, Bacchin C. et al. First reporting of methicillin-resistant Staphylococcus aureus (MRSA) ST398 in an industrial rabbit holding and in farm-related people. Vet Microbiol 2014; 170: 172–7. [DOI] [PubMed] [Google Scholar]
- 20. Zerbino DR, Birney E.. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 2008; 18: 821–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Angiuoli SV, Gussman A, Klimke W. et al. Toward an online repository of standard operating procedures (SOPs) for (meta)genomic annotation. OMICS 2008; 12: 137–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kumar S, Stecher G, Tamura K.. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 2016; 33: 1870–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ito T, Hiramatsu K, Tomasz A. et al. Guidelines for reporting novel mecA gene homologues. Antimicrob Agents Chemother 2012; 56: 4997–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Paterson GK, Larsen AR, Robb A. et al. The newly described mecA homologue, mecALGA251, is present in methicillin-resistant Staphylococcus aureus isolates from a diverse range of host species. J Antimicrob Chemother 2012; 67: 2809–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu J, Chen D, Peters BM. et al. Staphylococcal chromosomal cassettes mec (SCCmec): a mobile genetic element in methicillin-resistant Staphylococcus aureus. Microb Pathog 2016; 101: 56–67. [DOI] [PubMed] [Google Scholar]
- 26. Monecke S, Jatzwauk L, Müller E. et al. Diversity of SCCmec elements in Staphylococcus aureus as observed in south-eastern Germany. PLoS One 2016; 11: e0162654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang L, Safo M, Archer GL.. Characterization of DNA sequences required for the CcrAB-mediated integration of staphylococcal cassette chromosome mec, a Staphylococcus aureus genomic island. J Bacteriol 2012; 194: 486–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.International Working Group on the Classification of Staphylococcal Cassette Chromosome. Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob Agents Chemother 2009; 53: 4961–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zong Z, Lü X.. Characterization of a new SCCmec element in Staphylococcus cohnii. PLoS One 2010; 5: e14016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. MacFadyen AC, Harrison EM, Ellington MJ. et al. A highly conserved mecC-encoding SCCmec type XI in a bovine isolate of methicillin-resistant Staphylococcus xylosus. J Antimicrob Chemother 2018; 73: 3516–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Galvin M, Hill C, Ross RP.. Lacticin 3147 displays activity in buffer against Gram‐positive bacterial pathogens which appear insensitive in standard plate assays. Lett Appl Microbiol 1999; 28: 355–8. [DOI] [PubMed] [Google Scholar]
- 32. Kuipers A, Meijer-Wierenga J, Rink R. et al. Mechanistic dissection of the enzyme complexes involved in biosynthesis of lacticin 3147 and nisin. Appl Environ Microbiol 2008; 74: 6591–7. [DOI] [PMC free article] [PubMed] [Google Scholar]