Abstract
Low molecular weight polycyclic aromatic hydrocarbons (LMW PAHs; < 206.3 g/mol) are under regulated environmental contaminants (eg, secondhand smoke) that lead to gap junction dysregulation, p38 MAPK activation, and increased mRNA production of inflammatory mediators, such as cytokines and cyclooxygenase (COX2), in lung epithelial cells. However, the early mechanisms involving lipid signaling through the arachidonic acid pathway and subsequent eicosanoid production leading to these downstream events are not known. Common human exposures are to mixtures of LMW PAHs, thus C10 cells (a mouse lung epithelial cell line) were exposed to a representative binary PAH mixture, 1-methylanthracene (1-MeA) and fluoranthene (Flthn), for 30 min–24 h with and without p38 and cytosolic phospholipase A2 (cPLA2) inhibitors. Cytosolic phospholipase A2 inhibition reversed PAH-induced phospho-p38 MAPK activation and gap junction dysregulation at 30 min. A significant biphasic increase in cPLA2 protein was observed at 30 min, 2, and 4 h, as well as COX2 protein at 2 and 8 h. Untargeted metabolomics demonstrated a similar trend with significantly changing metabolites at 30 min and 4 h of exposure relative to 1 h; a “cPLA2-like” subset of metabolites within the biphasic response were predominately phospholipids. Targeted metabolomics showed several eicosanoids (eg, prostaglandin D2 (PGD2), PGE2α) were significantly increased at 4, 8, and 12 h following exposure to the binary PAH mixture and this effect was p38-dependent. Finally, PAH metabolism was not observed until after 8 h. These results indicate an early lipid signaling mechanism of LMW PAH toxicity in lung epithelial cells due to parent PAH compounds.
Keywords: lung, p38 MAPK/cPLA2, eicosanoids, metabolomics, polycyclic aromatic hydrocarbons, secondhand smoke
Polycyclic aromatic hydrocarbons (PAHs) are ubiquitous environmental and occupational pollutants (ATSDR, 2005) and abundant in secondhand smoke (SHS; cigarette and marijuana) (Lee et al., 2010; Moir et al., 2008); thus, the risk to human health is evident. Toxicological research in this field has almost exclusively focused on the high molecular weight (HMW; ≥ 5 rings) PAHs and their carcinogenic potential (ATSDR, 2005; IARC, 2010; U.S.E.P.A., 2002). However, humans are exposed to non-genotoxic low molecular weight (LMW, 2–4 rings) PAHs at much greater concentrations. Nevertheless, low molecular weight polycyclic aromatic hydrocarbons (LMW PAHs) are understudied and under regulated (eg, lack of IARC classifications). Our previous studies provide evidence that exposure to LMW PAHs leads to gap junction dysregulation, activation of mitogen-activated protein kinase (MAPK) pathways (eg, p38, ERK1/2), and inflammatory mediator production in lung epithelial cells, critical cell signaling events that contribute to pulmonary disease (Osgood et al., 2013, 2017). Therefore, further investigation of LMW PAH toxicity in lung cells is imperative.
Gap junction intercellular communication (GJIC) is necessary for tissue homeostasis and, when dysregulated, can lead to alterations in cell survival, growth, calcium signaling, ciliary beating, and signaling (Cesen-Cummings et al., 1998; Chaudhuri et al., 1993; Freund-Michel et al., 2016; Upham, 2011). Therefore, dysregulated GJIC is considered an emerging hallmark of cancer and an established biomarker of PAH toxicity (Bauer et al., 2017; Montaner et al., 1997; Nahta et al., 2015; Osgood et al., 2013, 2017; Upham et al., 1998, 2008). Gap junction intercellular communication was decreased in a time- and dose-dependent manner in C10 cells (a mouse alveolar type II epithelial cell line) exposed to 1-methylcholanthrene (1-MeA), a LMW PAH, for 15 min–24 h. p38 MAPK has vital roles in the signal transduction pathways leading to inflammation involved in pulmonary diseases, such as chronic obstructive pulmonary disease (Renda et al., 2008) and cancer (Zarubin and Han, 2005), including activation of inflammatory lipid mediator pathways, such as eicosanoids (Chandrasekharan and Sharma-Walia, 2015; Yun et al., 2016). Significant p38 MAPK activation occurred concurrently with the GJIC dysregulation in these cells as early as 15 min after exposure to 1-MeA. Hence, these data support our working hypothesis; p38-MAPK is involved in regulating GJIC inhibition and subsequent early cell signaling events prior to 24 h in lung epithelial cells, and that GJIC and the p38 MAPK pathway are likely involved in events leading to inflammatory mediator production and PAH-induced toxicities, all at noncytotoxic doses (Osgood et al., 2017; Upham et al., 2008).
Single LMW PAH exposures are rare; therefore, we chose to investigate an environmentally relevant 1:1 binary PAH mixture of 1-MeA and fluoranthene (Flthn) to provide a more accurate assessment of LMW PAH toxicity (IARC, 2010; NTP Board of Scientific Counselors Meeting, 2012). In C10 cells, exposure to the binary PAH mixture for 4–8 h significantly elevated cyclooxygenase (Cox2) mRNA expression far greater than either compound alone (Osgood et al., 2017). COX2 is an enzyme, along with lipoxygenase and cytochrome P450 enzymes, that leads to the production of eicosanoids that primarily originate from arachidonic acid (AA) release from membrane phospholipids via cytosolic phospholipase A2 (cPLA2) and subsequent enzymatic processing (Leslie, 2015; Ricciotti and FitzGerald, 2011). Similar to the 1-MeA studies, the binary mixture also dysregulated GJIC, activated p38-MAPK, and induced inflammatory cytokines (Il6, Kc, Mcp1), metalloproteinases, and pathways associated with steroid synthesis, metabolism, and oxidative responses in both a time- and dose-dependent manner (primarily prior to 6 h) that was additive and/or potentially synergistic compared with single compound effects (Osgood et al., 2013, 2017). Given that Cox2 expression and p38-MAPK signaling were implicated in LMW PAH exposures prior to 24 h, we were driven to investigate the early mechanistic lipid signaling events of this binary PAH mixture leading to inflammatory lipid mediator production in lung cells.
Therefore, we hypothesized that a binary PAH mixture (1-MeA and Flthn) activates mechanistic events prior to 24 h leading to upregulation of eicosanoid signaling via lipid-derived activation of the AA pathway in a mouse lung epithelial cell model. To elucidate the temporal relationship between these inflammatory pathways and exposure to LMW PAHs, we used several methodologies including pharmaceutic inhibitors, measurement of p38 activation, GJIC activity, and protein expression for cPLA2, COX1, and COX2. In addition, the amount of PAH metabolism was also measured via gas chromatography/mass spectrometry (GC/MS) analysis of PAH concentration in the extracts. Finally, we used mass spectrometry-based targeted lipidomics methodologies to quantify intracellular eicosanoid profiles as well as untargeted lipidomics and corresponding pathway enrichment analyses. This study will provide the much-needed toxicity data on environmentally relevant PAHs that can be used to develop preventative and therapeutic outcomes pertaining to lung disease from exposure to SHS and environmental and occupational pollutants.
MATERIALS AND METHODS
Chemicals and Reagents
Fluoranthene (Flthn; purity 97.2%) was purchased from AccuStandard (New Haven, Connecticut), and 1-methylanthracene (1-MeA; purity 99.5%) from Crescent Chemical (Islandia, New York). Dimethyl sulfoxide (DMSO) and Lucifer Yellow was purchased from Sigma-Aldrich. The cPLA2 inhibitor, CAY10502, was purchased from Cayman Chemicals (Ann Arbor, Michigan) and p38 inhibitor, SB203580, was purchased from Tocris Bioscience (Bristol, UK). All PAH stock solutions and inhibitors were prepared in DMSO.
All internal standards used for liquid chromatography (LC)/mass spectrometry (MS)/MS analysis of AA-derived lipid mediators were purchased from Cayman Chemical. All HPLC solvents and extraction solvents (Sigma) for the metabolomics studies were LC-MS grade or better. Unless otherwise noted, chemicals were purchased from Sigma.
Cell Culture and Binary PAH Mixture Treatment
The C10 cell line was obtained from Dr Lori Nield (University of Colorado). These cells are an immortalized, non-transformed alveolar type II cell line originally derived from a BALB mouse (Bentel et al., 1989; Malkinson et al., 1997) that exhibits normal gap junctional communication, expression of surfactant proteins B and C (Tong et al., 2006), and were used previously for acute responses to LMW PAH exposure (Osgood et al., 2013, 2017). Because type II cells are a progenitor cell for type I cells, self-renewing, and known to initiate non-small cell lung carcinomas (eg, adenocarcinomas) (Desai et al., 2014; Lin et al., 2012), the C10 cells are one of the best cell models for these studies. This cell line was also extensively reviewed (Malkinson et al., 1997).
Cells (passage < 20) were maintained in CMRL 1066 media (Gibco, Thermo Fisher Scientific) containing 10% FBS and 1% glutamate in a humidified atmosphere at 37°C, 5% CO2, and 95% air (Osgood et al., 2013). Once cells were grown to confluence (2–3 days), they were serum-deprived for 24 h prior to treatment with the 1:1 PAH mixture (binary PAH mixture) of 1-MeA and Flthn at a final dose of 40 μM for time points 30 min–12 h and 15 µM for 24 h. This mixture has been used previously in our lab (Osgood et al., 2013, 2017) to represent 2 LMW PAHs common in SHS (Lee et al., 2010; Moir et al., 2008) and environmental exposures (Vondracek et al., 2007). The 40 μM dose was selected based on the IC50 for the dysregulation of GJIC at 30 min of exposure in the C10 and is not cytotoxic up to 48 h (Osgood et al., 2017). DMSO concentrations (< 0.01%) did not elicit cytotoxicity to the cells; no differences between the DMSO and media control were observed. For all inhibitor studies, inhibitors were applied to the cells 30 min–1 h prior to binary PAH mixture treatment. The p38 inhibitor concentration (5 μM) and incubation time was based on previous studies (Osgood et al., 2017), whereas the 10 μM cPLA2 inhibitor concentration was based on a dose-response conducted in the C10 cells (data not shown). Cytotoxicity measurements for the cPLA2 inhibitor resulted in no observable cytotoxicity (data not shown).
GJIC Activity via the Scalpel-loaded Dye-transfer Assay
Dysregulation of GJIC was measured following the method by Upham (2011) and our laboratory (Osgood et al., 2017). Briefly, cells were grown as noted above prior to treatment and immediately following the allotted treatment time, the scalpel-loaded dye-transfer (SL/DT) assay performed as in Osgood et al. (2017). GJIC was observed as area of dye spread with an Eclipse Ti-S microscope at 100X, captured with a DS-QiMc camera (Nikon Instruments, Melville, New York), and quantified using ImageJ software (http://imagej.nih.gov/ij/; Accessed November 21, 2018). Area of dye spread was quantified by comparing the binary PAH mixture, with and without cPLA2 inhibitor, and the cPLA2 inhibitor alone to DMSO control for the final fraction of control (FOC) percentages. Three cut lines were analyzed per dish, 3 dishes were used per treatment, and the experiment was repeated 3 times, for a total n = 9.
Protein Extraction and Immunoblots
C10 cells were maintained and treated as stated above. Immediately following treatment, cells were harvested and resuspended in homogenization buffer [20 mM HEPES, pH 7.5, 2 mM ethylenediaminetetraacetic acid, 2 mM ethylene glycol-bis(2-aminoethyl)-N,N,N′,N′-tetraacetic acid, 1 mM dithiothreitol, 10% glycerol] with protease inhibitor (Protease Inhibitor Cocktail 100×, Sigma-Aldrich, St Louis, Missouri) and phosphatase inhibitor (Halt Phosphatase Inhibitor Cocktail 100×, Thermo Fisher Scientific, Waltham, Massachusetts), disrupted by sonication, and centrifuged at 16 000 × g for 30 min. The resulting soluble fractions were used for cPLA2 immunoblotting. The particulate fractions were resuspended in the above buffer with the addition of 0.1% NP-40 and used for COX1 and COX2 immunoblotting. For p38 immunoblots (phospho- and total), we followed our previous method, described in Osgood et al. (2017). For all immunoblots, 15 µg of protein was separated on 10% SDS page gels and transferred to a polyvinylidene fluoride membrane (Millipore). Anti-rabbit phosphorylated cPLA2 (Santa Cruz, Cat. No. 34391, 1:1000), anti-mouse cPLA2 (Santa Cruz, Cat. No. 454), anti-rabbit pp38 (Cell Signaling, Cat. No. 9215 S, 1:500), anti-rabbit total p38 (Cell Signaling, Cat. No. 9212 S, 1:1000), anti-goat COX1 (Santa Cruz, Cat. No. 1754, 1:500), anti-goat COX2 (Santa Cruz, Cat. No. 1747, 1:500), and anti-mouse β-Actin (Sigma-Aldrich, Cat. No. A1978; 1:1000) primary antibodies were used. Secondary antibodies used were mouse anti-goat-HRP (Santa Cruz, Cat. No. 2354, 1:10 000), mouse anti-rabbit-HRP (Santa Cruz, Cat. No. 2357, 1:7500), and goat anti-mouse-HRP (Santa Cruz, Cat. No. 2005, 1:100 000). Supersignal West Dura chemiluminescent (Thermo Scientific) were used to detect proteins of interest. Immunoblots were quantified by densitometry using the Image Studio Lite V5 software (Licor, Lincoln, Nebraska).
Untargeted Metabolomics
Cell lysate sample preparation
The C10 cells were maintained and treated as stated above. Immediately following treatment, cell monolayers were washed 3 times with cold PBS, scraped into 500 μl 70% MeOH, transferred into 1.5 ml Eppendorf tubes (Hamburg, Germany), and immediately stored at −80°C until further processing. The hydrophobic fraction was then extracted from the samples using an organic liquid-liquid extraction technique with methyl tert-butyl ether and water (Cruickshank-Quinn et al., 2014; Yang et al., 2013). The upper hydrophobic layer was transferred to a new tube, dried under nitrogen, reconstituted in 200 µl of methanol, and stored at −80°C until LCMS analysis. Each exposure was replicated 4 times and the experiment was not repeated, for a total n = 4.
Liquid chromatography mass spectrometry
The hydrophobic fractions were analyzed using reverse phase chromatography on an Agilent Technologies (Santa Clara, California) 1290 ultra-high precision liquid chromatography system on an Agilent Zorbax Rapid Resolution HD SB-C18, 1.8 µm (2.1 × 100 mm) analytical column with an Agilent Zorbax SB-C18, 1.8 µm (2.1 × 5 mm) guard column. An Agilent Technologies 6210 time-of-flight mass spectrometer (Agilent Technologies) in positive electrospray ionization modes was used for compound detection for the hydrophobic fractions as previously described in Heischmann et al. (2016).
Data processing
Compound data was extracted using Agilent Technologies Mass Hunter Profinder Version B.08.00 (Profinder) software in combination with Agilent Technologies Mass Profiler Professional Version 14 (MPP) as previously described in Heischmann et al. (2016). Briefly, a naive feature-finding algorithm “Find By Molecular Feature” was used in Profinder to extract compound data from all samples and sample preparation blanks. Extracted compounds were imported into MPP and compounds present in blanks were removed from further analysis. To reduce the presence of missing values, a theoretical mass and retention time database was then generated for compounds present in at least 2 samples. This database was then used to re-search the raw sample data in Profinder using the “Find By Ion” algorithm.
Compound identification
An in-house database containing METLIN (PMID: 16404815), Lipid Maps (PMC1669719), Kyoto Encyclopedia of Genes and Genomes (KEGG) (PMC102409), and Human Metabolomics Database (HMDB) (PMC1899095) spectral data was used to annotate metabolites based on exact mass, isotope ratios, and isotopic distribution with a mass error cutoff of 10 ppm. This corresponds to a Metabolomics Standards Initiative metabolite identification level 3 (Sumner et al., 2007). The data obtained in this study will be accessible at the NIH Common Fund’s Data Repository and Coordinating Center (supported by NIH grant, U01-DK097430) website, the Metabolomics Workbench,http://www.metabolomicsworkbench.org. It is under Study number ST001094.
Targeted Lipidomics
Treatment and sample collection
The C10 cells were maintained and treated as stated above. Each exposure was replicated 4 times and the experiment was not repeated, for total n = 4. Immediately following treatment, cell monolayers were washed 3 times with cold PBS, scraped into 500 μl 70% MeOH, transferred into 1.5 ml Eppendorf tubes (Hamburg, Germany), and immediately stored at −80°C until pretreatment for solid phase extraction (SPE), as follows.
Sample preparation
Briefly, before SPE cleanup, the frozen samples were removed from the freezer and thawed on ice, and centrifuged at 14 000 rpm at 4°C for 10 min. The supernatant was removed, transferred to a new microcentrifuge tube, and spiked with 10 μl of 10 pg/μl internal standard solution (100 pg of each: 5(S)-HETE-d8, 8-iso-PGF2a-d4, 9(S)-HODE-d4, LTB4-d4, LTD4-d5, LTE4-d5, PGE2-d4, PGF2a-d9, and RvD2-d5 in ethanol). The supernatant was dried in a vacuum centrifuge and reconstituted with 1.0 ml of 10% methanol. The reconstituted extracts were loaded on a Strata-X 33 μm 30 mg/1mal SPE column (Phenomenex, Torrance, California) preconditioned with 1.0 ml of methanol followed by 1.0 ml of water. The SPE column was washed with 10% methanol and then eluted directly into a reduced surface activity/maximum recovery glass autosampler vial with 1.0 ml of methyl formate. The methyl formate was evaporated completely from the vial with a stream of nitrogen and the SPE cartridge was then eluted with 1.0 ml of methanol directly into the same autosampler vial. The methanol was evaporated to dryness with a stream of nitrogen, and the sample was reconstituted with 20 µl of ethanol. The samples were analyzed immediately or frozen at −70°C until analysis. Liquid chromatography-mass spectrometry of Scanning probe microscopes was performed as previously described in Kosaraju et al. (2017). The data obtained in this study will be accessible at the NIH Common Fund’s Data Repository and Coordinating Center (supported by NIH grant, U01-DK097430) website, the Metabolomics Workbench, http://www.metabolomicsworkbench.org. It is under study number ST001124.
Statistical Analysis
All analyses (SL/DT assays, immunoblots, targeted metabolomics, and PAH metabolism) were performed using SAS/STAT (Version 9.4) for Windows. A mixed method ANOVA model was used to compare binary PAH mixture treatment groups to DMSO control with experimental repeats as the random effect and the toxicity end point of interest as the fixed effect. All differences were considered statistically significant at p < .05. However, the untargeted metabolomics analysis required more extensive statistical methods.
Untargeted metabolomics
Data were summarized across technical replicates by sample for each metabolite using updated MSPrep package (Hughes et al., 2014). Sample replicates were summarized with their mean if they had a coefficient of variation (CV) less than or equal to 50%. If their CV was greater than 50% and there was data for all 3 replicates they were summarized using their median. If their CV was greater than 50% and there was missing data for at least 1 replicate, that metabolite was assigned a missing value. We filtered out metabolites that had more than 1 missing value per group (ie, DMSO or binary PAH mixture) per time point. These data were then log2 transformed, median normalized using the full dataset, and quantile normalized within each unique treatment and time. Quantile normalization was done using the EDASeq package (v. 2.10.0) (Bullard et al., 2010). Quantile normalization caused some metabolites to have zero variance at a given time point (1 per time point); these metabolites were removed. Bayesian principle component analysis method (Nyamundanda et al., 2010) was used to impute data.
To determine whether either exposure, DMSO control or binary PAH mixture, produced a temporal effect, we used separate 1-way ANOVA models that included only metabolites present at 2 or 3 time points for each treatment group. p Values were adjusted to control false discovery rate (FDR) using the Benjamini and Hochberg method (Benjamini and Hochberg, 1995) using the function p.adjust in the stats package of R (R Core Team, 2017). Adjustment was done on all ANOVA tests within a specific treatment group. To determine temporal effects, t tests between each pairwise combination of times were performed. This was performed instead of an ANOVA due to various missing data between the different types of metabolites.
To examine the differential abundance for each metabolite detected, we performed 2-sample t tests to generate a fold change (FC) difference in abundance compared with DMSO control. Again, p values were adjusted to control FDR using the same method aforementioned. Metabolites that were present only in DMSO control samples were filtered out. Significance was determined as p < .05 after controlling for FDR. Those metabolites annotated with either KEGGID or HMDB-ID were included in the heat map. Biological relevance was determined to be a FC less than 0.6 or greater than 1.5.
RESULTS
P38 Pathway Activation and Dysregulation of GJIC: A Role for cPLA2
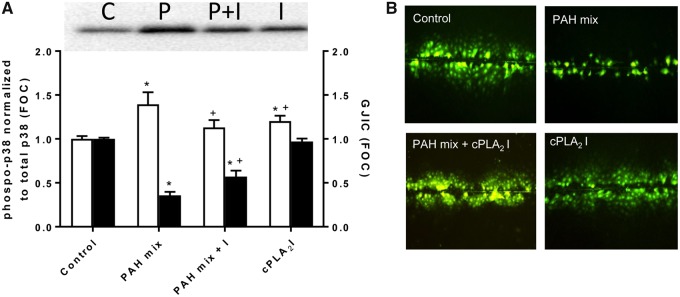
We previously demonstrated activation of p38 MAPK in response to exposure a 1:1 binary PAH mixture, 1-methylanthracene and fluoranthene, for 30 min–8 h which coincided with dysregulated GJIC (Osgood et al., 2017). In this study, exposure to the same binary PAH mixture for 30 min in the C10 cells significantly reduced p38 activity when cells were first exposed to CAY10502, a cPLA2 inhibitor (Figure 1A). In addition, GJIC was significantly reduced following exposure to the binary PAH mixture for 30 min; however, GJIC inhibition was significantly, albeit partially, reversed in the presence of CAY10502, indicating a role for cPLA2 in the early events involved in PAH-induced GJIC dysregulation (Figs. 1A and B).
Figure 1.
Cytosolic phospholipase A2 inhibition reverses phospho-p38 MAPK activation and gap junction intercellular communication (GJIC) dysregulation from 30 min exposure to binary polycyclic aromatic hydrocarbon (PAH) mixture in C10 cells. A, Representative pp38 immunoblot and graph (white bars) of cells exposed to DMSO control (C), binary PAH mix (P), binary PAH mix plus cPLA2 inhibitor (P + I), or cPLA2 inhibitor (I) (PAH mix of 40 μM, 1:1 ratio of 1-methylanthracene and fluoranthene; 10 μM of cPLA2 inhibitor, CAY10502). pp38 MAPK is normalized to total p38 and expressed as fraction of control (FOC). Black bars represent GJIC presented as FOC for the same 4 treatments using a SL/DT assay. Mean ± SE presented; n = 3 per treatment; repeated 3 times. *p < .05, significantly different from control; +p < .05, significantly different from PAH mix alone. B, Representative images of GJIC dysfunction in response to the binary PAH mixture and reversal by cPLA2 inhibition. Area of dye spread is used for GJIC quantification. The color version of this figure is available online.
Changes in Lipid Mediators: Eicosanoid Involvement
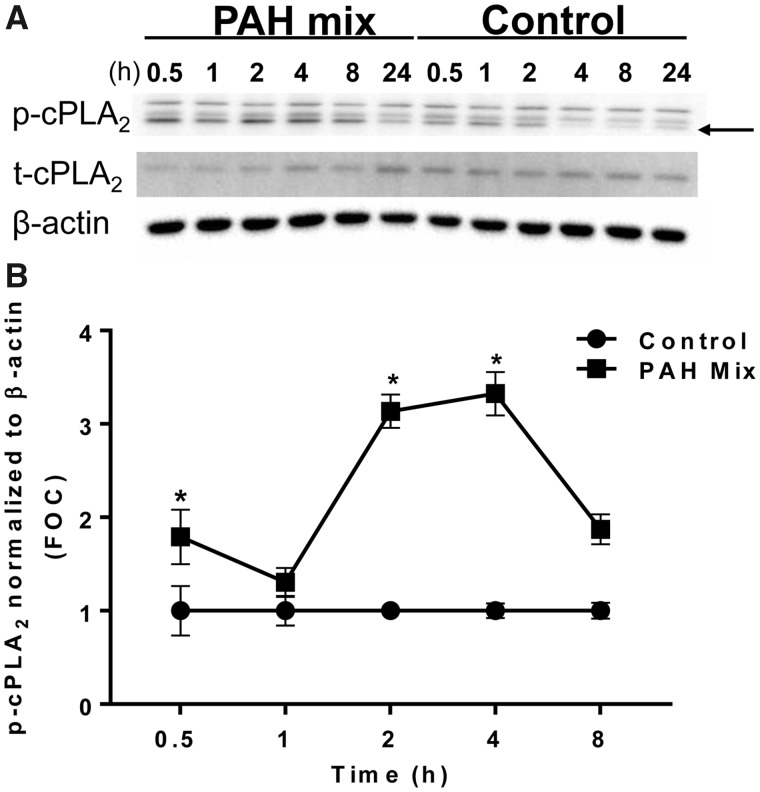
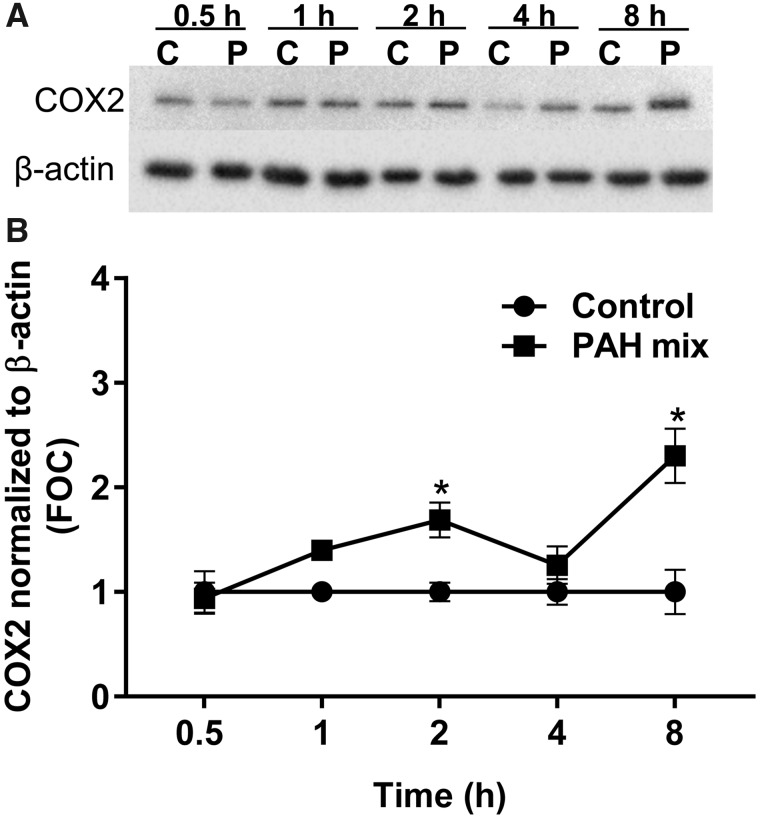
Due to the early involvement of cPLA2, a key enzyme in eicosanoid biosynthesis, in response to the binary PAH mixture, we investigated the changes in several enzymes and lipid mediators along this pathway. C10 cells were exposed to 40 μM of binary PAH mixture over time (0.5, 1, 2, 4, 8 h). Phospho-cPLA2 was significantly elevated after 0.5, 2, and 4 h compared with control (Figs. 2A and B). Following cPLA2 activation, the downstream enzyme COX2 was significantly elevated at 2 and 8 h after exposure, COX1 was unchanged (Figs. 3A and B, Supplementary Figs. 1A and B). Interestingly, both cPLA2 and COX2 enzymes demonstrated a biphasic response from exposure to the binary PAH mixture at these early time points.
Figure 2.
Cytosolic phospholipase A2 activation in response to the binary PAH mixture in C10 cells. A, Immunoblots of phosphorylated cPLA2 (p-cPLA2), total cPLA2 (t-cPLA2), and β-actin protein in C10 exposed to binary PAH mixture (40 µM for 0.5–8 h; 15 µM for 24 h) or DMSO control for 0.5–24 h. Arrow points to phosphorylated band, bottom. B, Densitometric analysis of p-cPLA2 protein activity normalized to β-actin and then presented as a ratio compared with t-cPLA2 shows a biphasic increase at 0.5, 2, and 4 h following exposure to binary PAH mixture (40 µM; 1:1 ratio of 1-methylanthracene and fluoranthene). Mean ± SE; n = 3, repeated twice. *p < .05, significantly different from control.
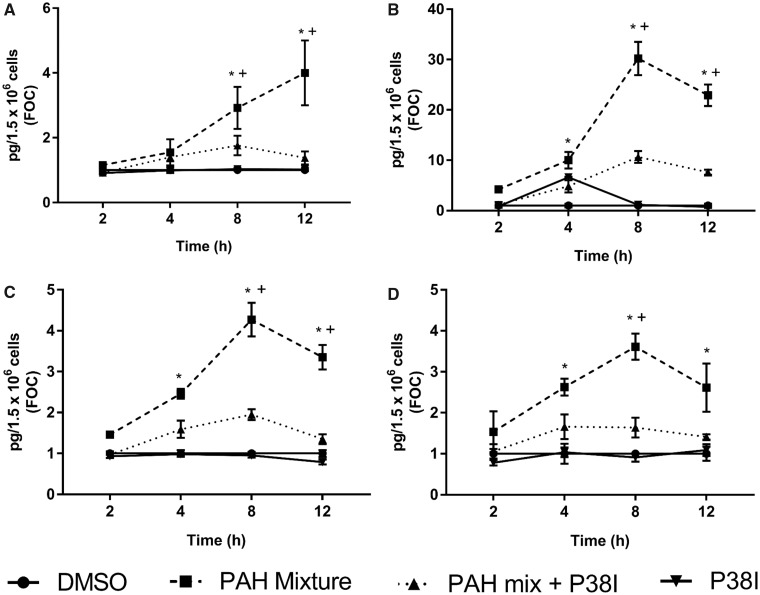
Intracellular targeted lipidomics analysis via LC/MS demonstrated significantly increased expression of several downstream prostaglandins, namely, PGD2, PGE2, PGF2α, and 15 R-PGF2α (a PGF2α metabolite, data not shown) after exposure to 4, 8, and 12 h of the binary PAH mixture (Figs. 4A–C). When cells were treated with p38 inhibitor, SB203580, 30 min prior to PAHs, the expression was reduced significantly at 8 and 12 h. Similarly, 8-iso-PGF2α, an isoprostane associated with oxidative stress, was significantly increased after 4, 8, and 12 h of exposure (Figure 4D), and was significantly reduced in the presence of p38 inhibitor at 8 and 12 h. In addition to the prostaglandins, other pro-inflammatory eicosanoids were significantly elevated following 4 h exposure to binary PAH mixture such as 14(15)-EET, and 8S-HETE (Supplementary Figure 2A), as well as some anti-inflammatory mediators, such as 15S-HETE and LXA4 (Supplementary Figure 2B); none of these lipid mediators were reduced with p38 inhibitor. Several other eicosanoid mediators were measured but did not change in response to the binary PAH mixture, such as 8-iso-15R-PGF2α and 8, 9-EET (Supplementary Figure 2A), as well as PGI2, 11B-PGF2α, TXB2, LTB4, LTD4, and LTE4 (data not shown). Figure 5 demonstrates the temporality of this lipid signaling pathway observed in the C10 cells for cPLA2, COX2, and the prostaglandins that were induced by this binary PAH mixture. Finally, we did this same targeted lipidomics analysis at the 24 h time point (Supplementary Table 1) and only isoprostane (8-iso-PGF2α) demonstrated a significant difference compared with control.
Figure 4.
Prostaglandin production in C10 cells from exposure to the binary PAH mixture with and without p38 MAPK inhibition. Prostaglandin levels were determined using a LC/MS protocol. (A) prostaglandin D2 (PGD2), (B) prostaglandin E2 (PGE2α), (C) prostaglandin F2α (PGF2α), (D) 8-iso-prostaglandin F2α (8-iso-PGF2α). Mean ± SE presented; n = 4, repeated once. *p < .05, significantly different from control; +p < .05, significantly different from p38 MAPK inhibitor + binary PAH mixture treatment.
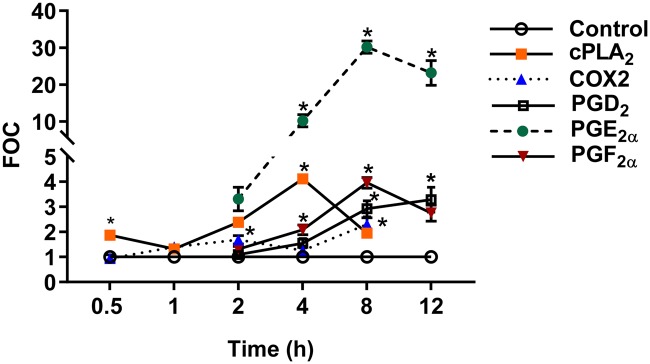
Figure 5.
Kinetics of lipid signaling events following C10 cell exposure to the binary PAH mixture. A time course of metabolic enzymes and downstream prostaglandins following exposure to binary PAH mixture. Cytosolic phospholipase A2 and COX2 protein expression were measured by immunoblotting and eicosanoids by LC/MS analysis. Phosphorylated cPLA2 densitometry was first normalized to β-actin and reported as a ratio compared with t-cPLA2; COX2 was normalized to β-actin as well. Each was then displayed as a fraction of control (FOC). The prostaglandins are reported as an FOC of the mean concentration across n−4. Error bars represent SE. *Significantly different from control, p < .05. The color version of this figure is available online.
Collectively, we provide sufficient evidence to support that the binary PAH mixture is influencing eicosanoid production that is likely initiated due to the release of phospholipids in the membrane. Figure 6 is a working hypothesis of the production of these lipid mediators in these C10 cells. Finally, we sought to evaluate the makeup of the cellular lipid metabolism at similar time points using an untargeted approach.
Figure 6.
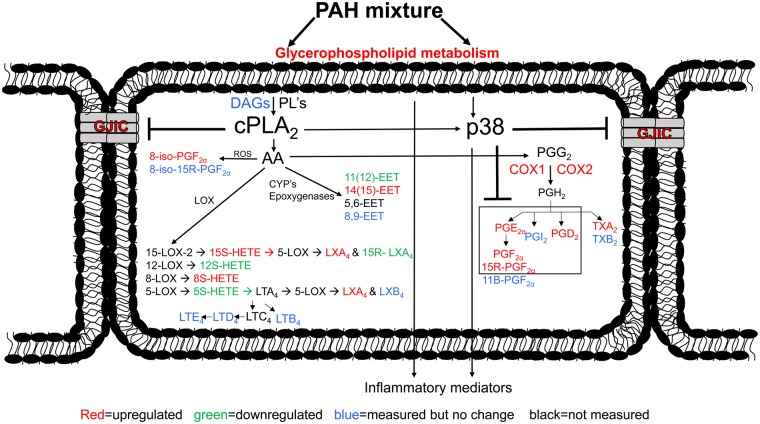
A schematic demonstrating the early mechanistic events leading to eicosanoid production from exposure to the binary PAH mixture prior to 24 h. Abbreviations: COX, cyclooxygenase; CYP, cytochrome p450 enzymes; DAGs, diacylglycerols; EET, epoxyeicosatrienoic acid; GJIC, gap junctional intercellular communication; HETE, hydroxyeicosatetraenoic acid; LT, leukotriene; LX, lipoxin; LOX, lipoxygenase; PG, prostaglandin; cPLA2, phospholipase A2; PL, phospholipids; ROS, reactive oxygen species; TX, thromboxane. The color version of this figure is available online.
Untargeted Metabolomics Demonstrates Involvement of Glycerophospholipid Metabolism
An untargeted metabolomics analysis was conducted to further establish the mechanism and potential pathological events associated with binary PAH mixture toxicity at 30 min, 1, and 4 h. Specifically, the early mechanistic events of PAH mixture toxicity leading to eicosanoid production via activation of cPLA2 could use several fatty acid substrates, from either endogenous (eg, phospholipids in the plasma membrane of the epithelial cell) or exogenous (eg, phospholipids contained in culture media containing serum) sources. To determine these substrates, untargeted metabolomics analysis via LC/MS were used to measure the abundance of intracellular metabolites associated with exposure to the binary PAH mixture or DMSO control over time (0.5, 1, and 4 h) and disease outcomes through pathway enrichment analyses.
First, to determine if the metabolomics assay was able to capture a temporal effect in response to treatment, an ANOVA was done for the 3489 metabolites present in at least 2 time points, regardless of exposure. After controlling for FDR, there were 232 and 1461 metabolites significantly changing over time from DMSO control and binary PAH mixture, respectively, for a total of 1693 metabolites. However, 151 of these metabolites were common between treatments and, subsequently, considered to be basal response metabolites. Therefore, a total of 1542 metabolites, 81 from DMSO and 1310 from binary PAH mixture, were significantly changing over time (Supplementary Figure 3). With a significant temporal relationship established between treatment and time, we then determined when these effects occurred, and which metabolites and/or disease outcomes are involved. Post hoc t tests were performed on the 1-way ANOVAs testing for a temporal relationship between time points. Of the 1693 metabolites that were detected in at least 2 time points, 395 and 72 were significantly different between 30 min and 1 h following binary PAH mixture and control exposure, respectively. Between 1 and 4 h, there were 284 and 25 significantly different metabolites from exposure to the binary PAH mixture and control, respectively (Supplementary Table 2). Finally, between 30 min and 4 h, there were 355 and 76 significantly different metabolites, respectively (Supplementary Table 2). The specific metabolites changing between each time point from each treatment can be found in Supplementary Tables 3 and 4.
To measure the differential abundance of significant metabolites from exposure to binary PAH mixture compared with DMSO control, a 2-sample t test was performed for each metabolite at each time point (Table 1). After removing those metabolites identified in DMSO only treated group, a total of 3093, 3332, and 3243 metabolites were used in the subsequent analysis. Of those metabolites detected in either 30 min treatment, only 224 (32%) were annotated with either KEGG or HMDB ID. Similarly, at 1 and 4 h of exposure, only 121 (45%) and 263 (36%) of those were annotated, respectively. All annotated and unannotated metabolites can be found in Supplementary Table 5 for all time points. One hundred and eighty-two, 77, and 181 metabolites were found to be biologically relevant (FC < 0.6 or > 1.5) with 125, 64, and 148 upregulated and 57, 13, and 33 downregulated, after 0.5, 1, and 4 h of exposure, respectively (Table 1). Each biologically relevant metabolite can be found grouped by compound class with corresponding FC at each time point in Figure 7.
Table 1.
Untargeted Metabolomics Analysis Determined Abundance of Significant Metabolites Over Time From Exposure to Binary PAH Mixture
| 0.5 h | 1 h | 4 h | |
|---|---|---|---|
| Detected metabolites | 3291 | 3561 | 3474 |
| Metabolites in DMSO only | 198 | 229 | 231 |
| Total metabolites analyzed (detected minus DMSO-only) | 3093 | 3332 | 3243 |
| Significant metabolites | 697 | 266 | 732 |
| Annotated metabolites | 224 | 121 | 263 |
| Biologically relevant FC | 182 | 77 | 181 |
| ↑ FC | 125 | 64 | 148 |
| ↓ FC | 57 | 13 | 33 |
| Significant w/o biologically relevant FC | 42 | 44 | 82 |
| Biologically relevant FC w/o significance | 110 | 72 | 63 |
C10 cells were exposed to binary PAH mixture (40 µM; 1:1 ratio of 1-methylanthracene and fluoranthene) or DMSO control for 0.5, 1, and 4 h and analyzed using untargeted metabolomics via LC/MS methods described above. This table displays the number of detected metabolites after exposure at each time point. Significance was determined via methods described above using p < .05. Metabolites were annotated with either KEGGID or HMDBID. Biologically relevant FC<0.6 or > 1.5.
Figure 7.
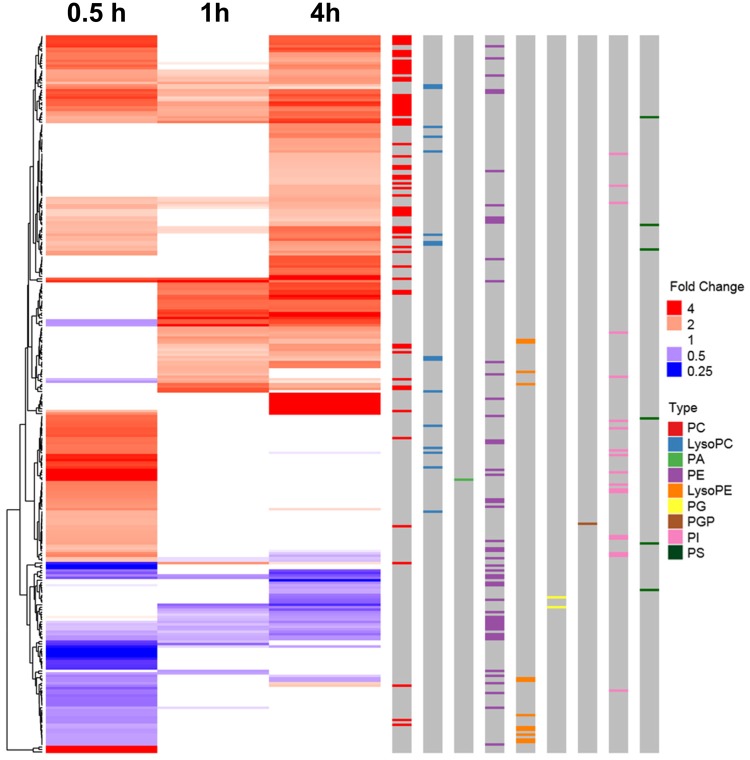
Heat map depicting the biologically relevant annotated metabolites that were significantly different in the C10 cells in response to the binary polycyclic aromatic hydrocarbon (PAH) mixture. Fold changes of the 293 biologically relevant metabolites (having KEGG annotation) differentially abundant between exposure groups, binary PAH mixture (40 µM) to DMSO control exposure, over time are shown here as a heatmap. Red indicates an increased fold change and blue indicates a decrease. If the fold change was not significant, the value for the heatmap was forced to 1 as it is not significantly different than 1 (white). The metabolites are clustered by fold change pattern over time (left-hand side). The colored bars on the right depict different metabolite types of interest: lysophosphatidylcholine (LysoPC; blue), lysophosphatidylethanolamine (LysoPE; orange), phosphatidic acid (PA; green), phosphatidylcholine (PC; red); phosphatidylethanolamine (PE; purple), phosphatidylglycerol (PG; yellow), phosphatidylglycerophosphate (PGP; brown), phosphatidylinositol (PI; pink), phosphatidylserine (PS; black).The color version of this figure is available online.
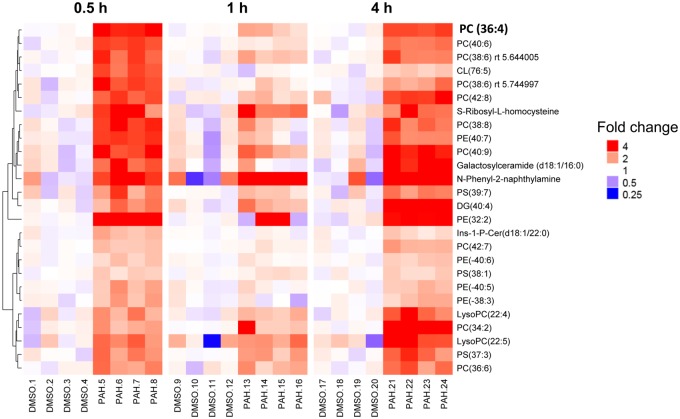
Metabolomics analyses enable to align significant metabolites with pathways associated with relevant pathological outcomes. Therefore, a list of annotated metabolites was combined, similar to genomics platform ones. In this study, those significant metabolites with biologically relevant FCs were included in pathway analyses via MetaboAnalyst at each time point based on their KEGGID. Significant pathways included glycerophospholipid metabolism, sphingolipid metabolism, phosphatidylinositol signaling system, glycerolipid metabolism, pathways in cancer, and metabolic pathways (Table 2). In addition, we have displayed how the glycerophospholipids are enriched in response to the binary PAH mixture on the heat map by color coding on the right side of Figure 7 to demonstrate the abundance of these metabolites. The most notable class of phospholipids was the phosphatidylcholines (PCs), for which many were upregulated in response to the binary PAH mixture (see red bar on the right side of Figure 7). Figure 8 depicts those significant metabolites that were identified at the time points that the cPLA2 protein was upregulated categorized by chain length of the phospholipids. Interestingly, one of the metabolites was a chain length for phosphatidylcholine, PC 36:4, that is known to be cleaved to AA (Banno et al., 2017).
Table 2.
Pathway Enrichment Analysis of the Biologically Relevant Metabolites at Each Time Point Annotated by KEGGID and Analyzed With MetaboAnalyst
| Pathway | Time (h) | No. of Matched Metabolites | Pathway p Value |
|---|---|---|---|
| Glycerophospholipid metabolism | 0.5 | 10 | <.0001 |
| 1 | 9 | <.0001 | |
| 4 | 9 | <.0001 | |
| Sphingolipid metabolism | 0.5 | 6 | <.0001 |
| 1 | 3 | .004 | |
| 4 | 3 | .004 | |
| Phosphatidylinositol signaling system | 0.5 | 4 | .0007 |
| 1 | 3 | .004 | |
| 4 | 3 | .004 | |
| Glycerolipid metabolism | 0.5 | 4 | .001 |
| 1 | 9 | .006 | |
| 4 | 9 | .006 | |
| Pathways in cancer | 0.5 | 3 | .003 |
| Metabolic pathways | 0.5 | 18 | .02 |
The biologically relevant metabolites detected at 0.5, 1, and 4 h (182, 77, and 181, respectively) were analyzed for possible pathological relevance through pathway enrichment using MetaboAnalyst. This table shows significant pathway with corresponding time point, number of matched metabolites, and p value determined through the software algorithm. There were no metabolites with KEGGID’s detected in “pathways in cancer” or “metabolic pathways” at the 1 and 4 h time points.
Figure 8.
A cPLA2-like subset of metabolites are predominately phospholipids. Sample-level ratio to time-specific DMSO abundance is shown for 26 metabolites displaying a cPLA2-like temporal pattern (upregulated in binary PAH mixture exposure at 0.5 and 4 h, but no statistical difference at 1 h). The metabolites are clustered by abundance at 30 min only (left-hand side) and labeled with corresponding chain length (right-hand side). The color version of this figure is available online.
PAH Metabolism Is Not a Factor Prior to 24 h
We observed that the parent compounds of the binary PAH mixture were not metabolized in these cells until 24 h (see Supplementary Figs. 4 and 5; Supplementary Methods). We analyzed the amount of each PAH metabolized (1-MeA and Flthn) via GC/MS at 0.5, 1, 4, 8, and 24 h time points. All times prior to 24 h observed approximately 100% recovery of the parent compounds, whereas at 24 h, only 50%–60% of parent compounds were recovered. This suggests that PAH parent compounds are active without metabolism prior to 24 h and that the early cell signaling events we have measured are elicited by these parent compounds rather than their metabolites.
DISCUSSION
We previously provided evidence that a binary mixture of 1-MeA and Flthn, both LMW PAHs, elicited GJIC-dysregulation and induced p38 MAPK activity leading to cytokine production at exposure times prior to 24 h indicating early mechanistic involvement of inflammatory mediators in C10 cells (Osgood et al., 2017). In a human bronchial epithelial cell line (HBE1) (Yankaskas et al., 1993), we also observed GJIC-dysregulation and p38 MAPK activation following exposure to the same LMW PAHs at 30 min (see Supplementary Figs. 6 and 7) supporting our previous findings. Because LMW PAHs are ubiquitous environmental contaminants (eg, SHS, air pollution) that humans are rarely exposed to as a single compound, we hypothesized that this binary PAH mixture activates mechanistic events prior to 24 h leading to upregulation of eicosanoid signaling via lipid-derived activation of the AA pathway in a mouse lung epithelial cell model. To elucidate the temporal relationship between these inflammatory pathways in response to exposure to LMW PAHs, we employed several methodologies, including specific signaling pathway inhibitors, measurement of p38 activation, GJIC activity, protein expression for enzymes involved in eicosanoid signaling, and PAH parent compound metabolism. Finally, we used targeted mass spectrometry-based lipidomics methodologies to quantify intracellular eicosanoid profiles as well as untargeted lipidomics and pathway enrichment analyses.
p38 MAPK has an important role in the signal transduction pathways leading to inflammation in lung diseases such as cancer (Zarubin and Han, 2005), including the production of eicosanoids (Chandrasekharan and Sharma-Walia, 2015; Yun et al., 2016). Eicosanoids are produced through the hydrolysis of AA, a fatty acid, via activated cPLA2 using components of the phospholipid membrane as substrates (Leslie, 2015; Ricciotti and FitzGerald, 2011). Arachidonic acid is further metabolized through 3 enzymatic pathways: cyclooxygenase (COX), lipoxygenase (LOX), and cytochrome P450 (CYP450). As a result, a variety of either pro- or anti-inflammatory eicosanoids are produced such as prostaglandins, leukotrienes, lipoxins, and resolvins (Dennis and Norris, 2015). Lipid signaling leading to eicosanoids through the COX pathway is further complicated due to different tissue-specific responses and bifunctional enzymatic activities; therefore, many therapeutics designed mainly for chronic inflammatory diseases that are based on this pathway fail (Dennis and Norris, 2015).
In our mouse lung epithelial model using C10 cells, exposure to the binary PAH mixture in the C10 cells for 30 min led to cPLA2-dependent p38 activation with corresponding partial cPLA2-dependent GJIC dysregulation. Because we did not have a positive control for the cPLA2 inhibitor, it is possible that the inhibitor was not fully active. However, we also know that p38 MAPK inhibition does not fully reverse the GJIC response, thus other unknown pathways are likely involved (Osgood et al., 2013, 2017), such as other phospholipases (Upham et al., 2008). Dysregulated GJIC can lead to disrupted communication between cells and, therefore, could potentially result in disrupted resolution of inflammation (Dennis and Norris, 2015; Islam et al., 2012). In addition, GJIC dysregulation has also been linked to early events in tumor promotion by known tumor promoters (eg, TPA) (Chaudhuri et al., 1993; Osgood et al., 2013; Upham et al., 2008) and GJIC inhibition is considered a component of the evasion of growth suppression Hallmark of Cancer (Nahta et al., 2015).
When C10 cells were exposed to this same binary PAH mixture for 0.5–12 h, a biphasic activation of cPLA2 protein was observed at 0.5, 2, and 4 h (Figure 2B). Although we have no definitive explanation for the observed biphasic response on PAH-induced activation of cPLA2, we hypothesize that there is an early transient response that is gap junction dependent, as gap junctions are not significantly closed under 10 min (Osgood et al., 2013), followed by a gap junction-independent activation of cPLA2, where gap junctions are significantly inhibited by the PAH mixture at 10 min and longer (Osgood et al., 2013, 2017). Also, a very similar biphasic activation of cPLA2 in response to estrogen in MCF7 cells (breast cancer cell line) was observed with nearly identical periods and methods of detection (Caiazza et al., 2010).
An increase in the downstream COX2 enzyme was observed at 2 and 8 h following exposure to the binary PAH mixture (Figure 3B). Targeted lipidomics provided evidence of increased production of eicosanoids that were p38-dependent, at the later time points between 4 and 12 h (Figs. 4A–C). The temporal relationship among cPLA2, COX2, and eicosanoids is a known inflammatory mechanistic pathway that has been implicated in lung disease and cancer (Heasley et al., 1997; Hida et al., 1998; Keith et al., 2006; Meyer et al., 2004) (Figure 5). In Figure 6, our working hypothesis, cPLA2 acts upon phospholipids in the cell membrane, leading to AA release and subsequent pro- and anti-inflammatory eicosanoid production (eg, prostaglandins, isoprostanes), but also activates p38 MAPK. p38 MAPK leads to additional inflammatory mediators known to be involved in lung inflammation (ie, KC, MCP1, IL6; Osgood et al., 2017) as well as prostaglandin production, supported by significant reductions in PGD2, PGE2, PGF2α, and 8-iso-PGF2α in response to the p38 MAPK inhibitor. Recently, a study using diesel exhaust (DE) known to contain both B[a]P and several LMW PAHs, observed changes in human bronchial epithelial cells in response to DE, such as increases in AA, PGE2, and isoprostanes, supporting our eicosanoid pathway observations (Rynning et al., 2018). In addition, in a human monocytic cell line (THP1), these same LMW PAHs also induced apoptosis, and more specifically, fluoranthene-induced phosphatidylinositol-phospholipase C and PGD2 synthase gene expression, further supporting a role for specific phospholipases and eicosanoids in the mechanism driving the effects we observed in the C10 lung cells (Wan et al., 2006, 2008).
Figure 3.
COX2 expression is upregulated in response to the binary PAH mixture in C10 cells. A, Immunoblots of COX2 and β-actin protein in C10 exposed to DMSO control (C) or binary PAH mixture (P; 40 µM) for 0.5–8 h. B, Densitometric analysis of COX2 protein expression normalized to β-actin shows a biphasic increase at 2 and 8 h following exposure to PAH mixture (40 µM; 1:1 ratio of 1-methylanthracene and fluoranthene). Mean ± SE; n = 3, repeated twice. *p < .05, significantly different from control.
Several studies provide additional support for our findings linking PAHs to eicosanoid signaling. Generation of AA in response to exposure to LMW PAHs has been measured in several other species (rat and human) and cell types (WB liver epithelial and endothelial cells) (Tithof et al., 2002; Upham et al., 2008). In addition, a mouse study using gavaged B[a]P demonstrated that COX2 was elevated in response to B[a]P (Barnwal et al., 2018). In a rat lung alveolar type II cell line (RLE-6TN), exposure to B[a]P with TNF-induced COX2 expression after 6 and 24 h with a subsequent increase in PGE2 after 24 h from exposure to B[a]P alone (Umannová et al., 2011). The “eicosanoid storm” can occur in conjunction with inflammasome activation and produce, not only inflammatory responses, but also resolving mediators (Dennis and Norris, 2015).
Although PGE2 and PGI2 are produced mostly by COX2 (Dennis and Norris, 2015), in the lung, there is a fine balance between PGE2:PGI2 levels that likely determines the inflammatory nature of this pathway, because in general, PGE2 is pro-inflammatory, promotes cell proliferation, and blocks apoptosis whereas PGI2 is anti-inflammatory in the lung (Keith et al., 2004; Leone et al., 2007; Sinicrope et al., 2004; Stolfi et al., 2008). If this balance is perturbed, the resulting response is either increased or decreased lung inflammation, or in this case, production of pro-inflammatory mediators (Bauer et al., 2001; Dwyer-Nield et al., 2005; Keith et al., 2004; Tennis et al., 2010). We did not observe any PGI2 nor other anti-inflammatory eicosanoids, from the C10 cells in this toxicant model, however, previous studies demonstrated that these cells can in fact produce PGI2 (Dwyer-Nield et al., 2005). Thus, this data supports that these PAHs are specifically eliciting a pro-inflammatory lipid mediator response.
Metabolomics analyses are more representative of phenotype compared with other “omics” because they reflect biochemical responses downstream of transcription, protein modification, and feedback mechanism (Bouhifd et al., 2013). Toxicometabolomics of in vitro studies is gaining more and more credibility within the toxicology community because the recent push to reduce the burdens associated with animal studies. Doing these analyses on both culture media and cell lysate samples can gather insight into biomarker discovery and, as in this study, mechanistic data on a cellular level (Bouhifd et al., 2013). This study used both targeted and untargeted metabolomics approaches to determine the cellular mechanism of how a binary PAH mixture induces toxicity through the p38-MAPK pathway and subsequent lipid-signaling involving eicosanoids. The untargeted analysis allowed us to cast a wide net to, not only measure significant cellular metabolites from exposures to a binary PAH mixture, but also to establish a temporal effect. Based on these results, we could then take a targeted approach, which focused on eicosanoids and other lipids involved in cell signaling.
An untargeted metabolomics analysis demonstrated the involvement of upstream glycerophospholipid metabolism, as early as 30 min, following exposure to the binary PAH mixture providing evidence that the plasma membrane is supplying substrates for the p38 MAPK/cPLA2 pathway (Figs. 7–9). The metabolites that were significantly different from DMSO control were grouped through hierarchical clustering and analyzed for compound class. The glycerophospholipid metabolism pathway following exposure to binary PAH mixture in the C10 cell is shown in Figure 9. Phosphatidylinositol, an early lipid signaling compound, was shown to be most active after 30 min exposure (Figs. 7 and 9). Phosphatidylcholine, perhaps the most well-characterized phospholipid, is hydrolyzed directly by cPLA2 to remove a fatty acid group to produce lysophosphatidylcholine, and was shown to be associated significantly with exposure to the binary PAH mixture (Figure 7) and increased primarily at 0.5 and 4 h after exposure (Figure 9). In addition, the top PC identified in Figure 8 (PC 36:4) has also been specifically linked to AA production (Banno et al., 2017), providing further evidence in support of our findings herein. Phospholipids, such as PC, LysoPC, PE, and LysoPE, were upregulated in the HepG2 cell line exposed to PAHs and SCCP and analyzed via metabolomics methods (Wang et al., 2018). Polycyclic aromatic hydrocarbon exposure can also activate PLA2 with fatty acid supplementation (Gdula-Argasińska et al., 2016; Tithof et al., 2002).
Figure 9.
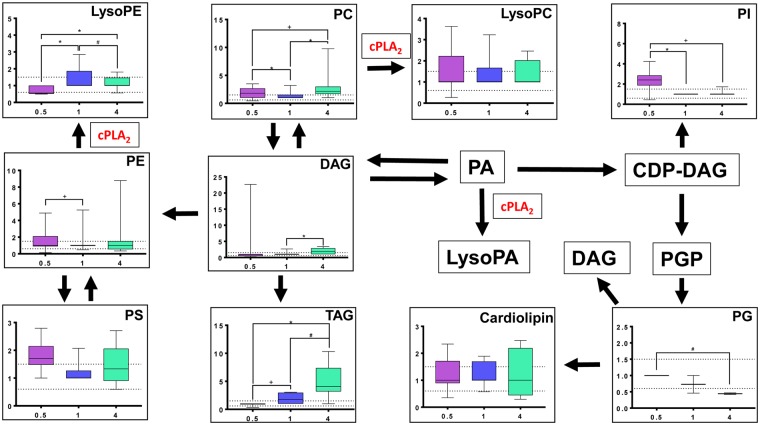
Glycerophospholipid metabolism pathway with corresponding temporal response from exposure to binary polycyclic aromatic hydrocarbon (PAH) mixture. The glycerophospholipid metabolism pathway is displayed through box and whisker plots and depicted with mean fold change (FC) of specific metabolites within glycerophopsholipid compound classes (ie, all metabolites within that specific class combined) changing from exposure to binary PAH mixture over time as measured through untargeted metabolomics analysis (n > or = 2). Graphs display variation in FC (y-axis) of compound class significantly changing compared with DMSO control at 0.5, 1, and 4 h (x-axis; purple, blue, and green, respectively). Dotted line represents biological relevance (0.6–1.5 FC) and error bars represent SE. Note: If no graph is displayed, n < 2. Abbreviations: CDP-DAG, CDP-diacylglycerol; LysoPA, lysophosphatidic acid; LysoPC, lysophosphatidylcholine; LysoPE, lysophosphatidylethanolamine; LysoPG, lysophosphatidylglycerol; PA, phosphatidic acid; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PGP, phosphatidylglycerophosphate; PI, phosphatidylinositol; PS, phosphatidylserine; DAG, sn-1, 2-diacylglycerol; TAG, triacylglycerol. *Significantly different between times, p < .001; +p < .01; #p < .05. The color version of this figure is available online.
With evidence of a biphasic response from cPLA2 protein, we chose a subset of 26 significant metabolites that reflected this cPLA2-trend, increasing in abundance only at 0.5 and 4 h (Figure 8). These metabolites were grouped by hierarchical clustering based on FC and displayed as a heat map of individual replicates for each treatment group. Through this display of the data, we show little variance between replicates for both treatment groups. In addition, this “cPLA2-like” subset showed an increased abundance with respect to a binary PAH mixture and the compounds were almost entirely phospholipids. We believe this strongly supports the proposed mechanism of toxicity from the binary PAH mixture involving the early activation of glycerophospholipids as substrates for cPLA2 enzyme activity leading to downstream responses such as increased COX2 and eicosanoids.
Interestingly, these adverse effects were observed at noncytotoxic doses (Osgood et al., 2017) challenging the traditional toxicological dogma requiring cytotoxicity from toxicants producing adverse health outcomes. In addition, these events were all measured prior to PAH metabolism (happening at 24 h, Supplementary Figure 4) indicating that these are parent compound effects. However, recovery yields are reported as being greater than 100%. Given the accepted error attributed from instrumentation and sample preparation, recovery yields ranging between 80% and 120% are regarded as complete recovery. In addition, matrix-induced signal enhancement (from the cell lysate) can occur in GC/MS when active surfaces in the system (eg, injector column, detector) cause retention and/or degradation of analytes. Conversely, standard analytes that do not contain cell matrix can fill active sites throughout the system which, in turn, reduces the percent recovery. Injected samples that contain cell matrix are biased to produce higher yields because cell matrix material will fill these active sites first allowing for increased efficiency of analyte transfer through the GC/MS system and then detector (Liu et al., 2012).
CONCLUSION
Collectively, the mechanism of toxicity from this environmentally relevant binary PAH mixture indicates the early involvement of fatty acid-derived eicosanoids. Therefore, this study will provide the much-needed toxicity data on environmentally relevant PAH mixtures that can be used to develop preventative and therapeutic interventions pertaining to lung disease from SHS, environmental, and occupational exposures (ATSDR, 2005; Ghoshal et al., 1999; Lee et al., 2010; Moir et al., 2008; U.S.E.P.A., 2002). We have tested our hypothesis using several established methodologies to determine a full spectrum of mechanisms of PAH toxicity at the cellular level; from transcription to protein concentration to cell signaling and, finally, cellular metabolomics. Because we have only used in vitro or cell lysate samples throughout the entire study, we were not able to capture extracellular events and possible biomarker identification. However, cell culture media samples were collected throughout these studies and will be evaluated for similar events in the future.
DECLARATION OF CONFLICTING INTERESTS
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
FUNDING
Flight Attendant Medical Research Institute (CIA130022 to A.K.B.); National Institute for Environmental Health Sciences at the National Institutes of Health (R15ES024893 to A.K.B.).
Supplementary Material
ACKNOWLEDGMENT
We would like to thank Dr Charmion Cruickshank-Quinn for her help uploading data to the metabolomics workbench and expert knowledge on metabolomics data graphics.
REFERENCES
- ATSDR. (2005). Toxicology Profile for Polyaromatic Hydrocarbons. CRC Press, Boca Raton, FL. [Google Scholar]
- Banno T., Omura T., Masaki N., Arima H., Xu D., Okamoto A., Costigan M., Latremoliere A., Matsuyama Y., Setou M. (2017). Arachidonic acid containing phosphatidylcholine increases due to microglial activation in ipsilateral spinal dorsal horn following spared sciatic nerve injury. PLoS One 12,e0177595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnwal P., Vafa A., Afzal S. M., Shahid A., Hasan S., Alpashree K., Sultana S. (2018). Benzo(a)pyrene induces lung toxicity and inflammation in mice: Prevention by carvacrol. Hum. Exp. Toxicol. 37,752–761. [DOI] [PubMed] [Google Scholar]
- Bauer A. K., Dwyer-Nield L. D., Keil K., Koski K., Malkinson A. M. (2001). Butylated hydroxytoluene (BHT) induction of pulmonary inflammation: A role in tumor promotion. Exp. Lung Res. 27,197–216. [DOI] [PubMed] [Google Scholar]
- Bauer A. K., Velmurugan K., Plottner S., Siegrist K. J., Romo D., Welge P., Bruning T., Xiong K. N., Kafferlein H. U (2017). Environmentally prevalent polycyclic aromatic hydrocarbons can elicit co-carcinogenic properties in an in vitro murine lung epithelial cell model. Arch. Toxicol. 92, 1311–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol.) 57,289–300. [Google Scholar]
- Bentel J. M., Lykke A. W., Smith G. J. (1989). Cloned murine non-malignant, spontaneously transformed and chemical tumour-derived cell lines related to the type 2 pneumocyte. Cell Biol. Int. Rep. 13,729–738. [DOI] [PubMed] [Google Scholar]
- Bouhifd M., Hartung T., Hogberg H. T., Kleensang A., Zhao L. (2013). Review: Toxicometabolomics. J. Appl. Toxicol. 33,1365–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullard J. H., Purdom E., Hansen K. D., Dudoit S. (2010). Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinform. 11,94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caiazza F., Harvey B. J., Thomas W. (2010). Cytosolic phospholipase A2 activation correlates with HER2 overexpression and mediates estrogen-dependent breast cancer cell growth. Mol. Endocrinol. 24,953–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesen-Cummings K., Fernstrom M. J., Malkinson A. M., Ruch R. J. (1998). Frequent reduction of gap junctional intercellular communication and connexin43 expression in human and mouse lung carcinoma cells. Carcinogenesis 19,61–67. [DOI] [PubMed] [Google Scholar]
- Chandrasekharan J. A., Sharma-Walia N. (2015). Lipoxins: Nature’s way to resolve inflammation. J. Inflamm. Res. 8,181–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri R., Sigler K., Dupont E., Trosko J. E., Malkinson A. M., Ruch R. J. (1993). Gap junctional intercellular communication in mouse lung epithelial cell lines: Effects of cell transformation and tumor promoters. Cancer Lett. 71,11–18. [DOI] [PubMed] [Google Scholar]
- Cruickshank-Quinn C., Quinn K. D., Powell R., Yang Y., Armstrong M., Mahaffey S., Reisdorph R., Reisdorph N (2014). Multi-step preparation technique to recover multiple metabolite compound classes for in-depth and informative metabolomic analysis. J. Visual. Exp. 89, e51670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis E. A., Norris P. C. (2015). Eicosanoid storm in infection and inflammation. Nat. Rev. Immunol. 15,511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai T. J., Brownfield D. G., Krasnow M. A. (2014). Alveolar progenitor and stem cells in lung development, renewal and cancer. Nature 507,190–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer-Nield L. D., Srebernak M. C., Barrett B. S., Ahn J., Cosper P., Meyer A. M., Kisley L. R., Bauer A. K., Thompson D. C., Malkinson A. M. (2005). Cytokines differentially regulate the synthesis of prostanoid and nitric oxide mediators in tumorigenic versus non-tumorigenic mouse lung epithelial cell lines. Carcinogenesis 26,1196–1206. [DOI] [PubMed] [Google Scholar]
- Freund-Michel V., Muller B., Marthan R., Savineau J. P., Guibert C. (2016). Expression and role of connexin-based gap junctions in pulmonary inflammatory diseases. Pharmacol. Ther. 164,105–119. [DOI] [PubMed] [Google Scholar]
- Gdula-Argasińska J., Czepiel J., Totoń-Żurańska J., Wołkow P., Librowski T., Czapkiewicz A., Perucki W., Woźniakiewicz M., Woźniakiewicz A. (2016). N-3 fatty acids regulate the inflammatory-state related genes in the lung epithelial cells exposed to polycyclic aromatic hydrocarbons. Pharmacol. Rep. 68,319–328. [DOI] [PubMed] [Google Scholar]
- Ghoshal B., Weber W. J., Rummel A. M., Trosko J. E., Upham B. L. (1999). Epigenetic toxicity of a mixture of polycyclic aromatic hydrocarbons on gap junctional intercellular communication before and after biodegradation. Environ. Sci. Technol. 33,1044–1050. [Google Scholar]
- Heasley L. E., Thaler S., Nicks M., Price B., Skorecki K., Nemenoff R. A. (1997). Induction of cytosolic phospholipase A2 by oncogenic Ras in human non-small cell lung cancer. J. Biol. Chem. 272,14501–14504. [DOI] [PubMed] [Google Scholar]
- Heischmann S., Quinn K., Cruickshank-Quinn C., Liang L. P., Reisdorph R., Reisdorph N., Patel M. (2016). Exploratory metabolomics profiling in the kainic acid rat model reveals depletion of 25-hydroxyvitamin D3 during epileptogenesis. Sci. Rep. 6,31424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hida T., Leyton J., Makheja A. N., Ben Av P., Hla T., Martinez A., Mulshine J., Malkani S., Chung P., Moody T. W. (1998). Non-small cell lung cancer cycloxygenase activity and proliferation are inhibited by non-steroidal antiinflammatory drugs. Anticancer Res. 18,775–782. [PubMed] [Google Scholar]
- Hughes G., Cruickshank-Quinn C., Reisdorph R., Lutz S., Petrache I., Reisdorph N., Bowler R., Kechris K. (2014). MSPrep—Summarization, normalization and diagnostics for processing of mass spectrometry-based metabolomic data. Bioinformatics 30,133–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IARC. (2010). Some Non-heterocyclic Polycyclic Aromatic Hydrocarbons and Some Related Exposures. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. World Health Organization. IARC, Lyon. [PMC free article] [PubMed] [Google Scholar]
- Islam M. N., Das S. R., Emin M. T., Wei M., Sun L., Westphalen K., Rowlands D. J., Quadri S. K., Bhattacharya S., Bhattacharya J. (2012). Mitochondrial transfer from bone-marrow-derived stromal cells to pulmonary alveoli protects against acute lung injury. Nat. Med. 18,759–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith R. L., Geraci M. W., Nana-Sinkam S. P., Breyer R. M., Hudish T. M., Meyer A. M., Malkinson A. M., Dwyer-Nield L. D. (2006). Prostaglandin E2 receptor subtype 2 (EP2) null mice are protected against murine lung tumorigenesis. Anticancer Res. 26,2857–2861. [PubMed] [Google Scholar]
- Keith R. L., Miller Y. E., Hudish T. M., Girod C. E., Sotto-Santiago S., Franklin W. A., Nemenoff R. A., March T. H., Nana-Sinkam S. P., Geraci M. W. (2004). Pulmonary prostacyclin synthase overexpression chemoprevents tobacco smoke lung carcinogenesis in mice. Cancer Res. 64,5897–5904. [DOI] [PubMed] [Google Scholar]
- Kosaraju R., Guesdon W., Crouch M. J., Teague H. L., Sullivan E. M., Karlsson E. A., Schultz-Cherry S., Gowdy K., Bridges L. C., Reese L. R. (2017). B cell activity is impaired in human and mouse obesity and is responsive to an essential fatty acid upon murine influenza infection. J. Immunol. 198,4738–4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. L., Hsieh D. P., Li L. A (2010). Polycyclic aromatic hydrocarbons in cigarette sidestream smoke particulates from a Taiwanese brand and their carcinogenic relevance. Chemosphere. 82, 477–482. [DOI] [PubMed] [Google Scholar]
- Leone V., di Palma A., Ricchi P., Acquaviva F., Giannouli M., Di Prisco A. M., Iuliano F., Acquaviva A. M. (2007). PGE(2) inhibits apoptosis in human adenocarcinoma Caco-2 cell line through Ras-PI3K association and cAMP-dependent kinase A activation. Am. J. Physiol. Gastrointest. Liver Physiol. 293,G673–G681. [DOI] [PubMed] [Google Scholar]
- Leslie C. C. (2015). Cytosolic phospholipase A(2): Physiological function and role in disease. J. Lipid Res. 56,1386–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C., Song H., Huang C., Yao E., Gacayan R., Xu S. M., Chuang P. T. (2012). Alveolar type II cells possess the capability of initiating lung tumor development. PLoS One 7,e53817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Shi X. W., Liu E. H., Sheng L. S., Qi L. W., Li P. (2012). More accurate matrix-matched quantification using standard superposition method for herbal medicines. J. Chromatogr. A 1254,43–50. [DOI] [PubMed] [Google Scholar]
- Malkinson A. M., Dwyer-Nield L. D., Rice P. L., Dinsdale D. (1997). Mouse lung epithelial cell lines—Tools for the study of differentiation and the neoplastic phenotype. Toxicology 123,53–100. [DOI] [PubMed] [Google Scholar]
- Meyer A. M., Dwyer-Nield L. D., Hurteau G. J., Keith R. L., O’Leary E., You M., Bonventre J. V., Nemenoff R. A., Malkinson A. M. (2004). Decreased lung tumorigenesis in mice genetically deficient in cytosolic phospholipase A2. Carcinogenesis 25,1517–1524. [DOI] [PubMed] [Google Scholar]
- Moir D., Rickert W. S., Levasseur G., Larose Y., Maertens R., White P., Desjardins S. (2008). A comparison of mainstream and sidestream marijuana and tobacco cigarette smoke produced under two machine smoking conditions. Chem. Res. Toxicol. 21,494–502. [DOI] [PubMed] [Google Scholar]
- Montaner L. J., Bailer R. T., Gordon S. (1997). IL-13 acts on macrophages to block the completion of reverse transcription, inhibit virus production, and reduce virus infectivity. J. Leukoc. Biol. 62,126–132. [DOI] [PubMed] [Google Scholar]
- Nahta R., Al-Mulla F., Al-Temaimi R., Amedei A., Andrade-Vieira R., Bay S. N., Brown D. G., Calaf G. M., Castellino R. C., Cohen-Solal K. A. et al. (2015). Mechanisms of environmental chemicals that enable the cancer hallmark of evasion of growth suppression. Carcinogenesis 36(Suppl. 1), S2–S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NTP Board of Scientific Counselors Meeting. (2012). NTP Research Concept: Polycyclic Aromatic Hydrocarbons (PAHs). National Toxicology Program. [Google Scholar]
- Nyamundanda G., Brennan L., Gormley I. C. (2010). Probabilistic principal component analysis for metabolomic data. BMC Bioinform. 11,571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osgood R. S., Upham B. L., Bushel P. R., Velmurugan K., Xiong K. N., Bauer A. K. (2017). Secondhand smoke-prevalent polycyclic aromatic hydrocarbon binary mixture-induced specific mitogenic and pro-inflammatory cell signaling events in lung epithelial cells. Toxicol. Sci. 157,156–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osgood R. S., Upham B. L., Hill T., 3rd, Helms K. L., Velmurugan K., Babica P., Bauer A. K. (2013). Polycyclic aromatic hydrocarbon-induced signaling events relevant to inflammation and tumorigenesis in lung cells are dependent on molecular structure. PLoS One 8,e65150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. (2017). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Vienna, Austria. [Google Scholar]
- Renda T., Baraldo S., Pelaia G., Bazzan E., Turato G., Papi A., Maestrelli P., Maselli R., Vatrella A., Fabbri L. M. et al. (2008). Increased activation of p38 MAPK in COPD. Eur. Respir. J. 31,62–69. [DOI] [PubMed] [Google Scholar]
- Ricciotti E., FitzGerald G. A. (2011). Prostaglandins and inflammation. Arterioscler. Thromb. Vasc. Biol. 31,986–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rynning I., Neca J., Vrbova K., Libalova H., Rossner P. Jr, Holme J. A., Gutzkow K. B., Afanou A. K. J., Arnoldussen Y. J., Hruba E. et al. (2018). In vitro transformation of human bronchial epithelial cells by diesel exhaust particles: Gene expression profiling and early toxic responses. Toxicol. Sci. 166,51–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinicrope F. A., Half E., Morris J. S., Lynch P. M., Morrow J. D., Levin B., Hawk E. T., Cohen D. S., Ayers G. D., Stephens L. C. (2004). Cell proliferation and apoptotic indices predict adenoma regression in a placebo-controlled trial of celecoxib in familial adenomatous polyposis patients. Cancer Epidemiol. Biomarkers Prev. 13,920–927. [PubMed] [Google Scholar]
- Stolfi C., Fina D., Caruso R., Caprioli F., Sarra M., Fantini M. C., Rizzo A., Pallone F., Monteleone G. (2008). Cyclooxygenase-2-dependent and -independent inhibition of proliferation of colon cancer cells by 5-aminosalicylic acid. Biochem. Pharmacol. 75,668–676. [DOI] [PubMed] [Google Scholar]
- Sumner L. W., Amberg A., Barrett D., Beale M. H., Beger R., Daykin C. A., Fan T. W., Fiehn O., Goodacre R., Griffin J. L. et al. (2007). Proposed minimum reporting standards for chemical analysis chemical analysis working group (CAWG) metabolomics standards initiative (MSI). Metabolomics 3,211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennis M. A., Vanscoyk M., Keith R. L., Winn R. A. (2010). The role of prostacyclin in lung cancer. Transl. Res. 155,57–61. [DOI] [PubMed] [Google Scholar]
- Tithof P. K., Elgayyar M., Cho Y., Guan W., Fisher A. B., Peters-Golden M. (2002). Polycyclic aromatic hydrocarbons present in cigarette smoke cause endothelial cell apoptosis by a phospholipase A2-dependent mechanism. FASEB J. 16,1463–1464. [DOI] [PubMed] [Google Scholar]
- Tong Q., Zheng L., Dodd-o J., Langer J., Wang D., Li D. (2006). Hypoxia-induced mitogenic factor modulates surfactant protein B and C expression in mouse lung. Am. J. Respir. Cell Mol. Biol. 34,28–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umannová L., Machala M., Topinka J., Schmuczerová J., Krčmář P., Neča J., Šujanová K., Kozubík A., Vondráček J. (2011). Benzo[a]pyrene and tumor necrosis factor-alpha coordinately increase genotoxic damage and the production of proinflammatory mediators in alveolar epithelial type II cells. Toxicol. Lett. 206,121–129. [DOI] [PubMed] [Google Scholar]
- Upham, B. L. (2011). “Role of integrative signaling through gap junctions in toxicology.” Curr Protoc Toxicol Chapter 2: Unit2 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upham B. L., Bláha L., Babica P., Park J.-S., Sovadinova I., Pudrith C., Rummel A. M., Weis L. M., Sai K., Tithof P. K. et al. (2008). Tumor promoting properties of a cigarette smoke prevalent polycyclic aromatic hydrocarbon as indicated by the inhibition of gap junctional intercellular communication via phosphatidylcholine-specific phospholipase C. Cancer Sci. 99, 696–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upham B. L., Weis L. M., Trosko J. E. (1998). Modulated gap junctional intercellular communication as a biomarker of PAH epigenetic toxicity: Structure-function relationship. Environ. Health Perspect. 106(Suppl. 4), 975–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S.E.P.A. (2002). Polycyclic aromatic hydrocarbons, 15 listings. Rep. Carcinog. 10,201–204. [PubMed] [Google Scholar]
- Vondracek J., Svihalkova-Sindlerova L., Pencikova K., Marvanova S., Krcmar P., Ciganek M., Neca J., Trosko J. E., Upham B., Kozubik A. et al. (2007). Concentrations of methylated naphthalenes, anthracenes, and phenanthrenes occurring in Czech river sediments and their effects on toxic events associated with carcinogenesis in rat liver cell lines. Environ. Toxicol. Chem. 26,2308–2316. [DOI] [PubMed] [Google Scholar]
- Wan B., Sayler G. S., Schultz T. W. (2006). Structure-activity relationships for flow cytometric data of smaller polycyclic aromatic hydrocarbons. SAR QSAR Environ. Res. 17,597–605. [DOI] [PubMed] [Google Scholar]
- Wan B., Yarbrough J. W., Schultz T. W. (2008). Structure-related clustering of gene expression fingerprints of THP-1 cells exposed to smaller polycyclic aromatic hydrocarbons. SAR QSAR Environ. Res. 19,351–373. [DOI] [PubMed] [Google Scholar]
- Wang F., Zhang H., Geng N., Ren X., Zhang B., Gong Y., Chen J. (2018). A metabolomics strategy to assess the combined toxicity of polycyclic aromatic hydrocarbons (PAHs) and short-chain chlorinated paraffins (SCCPs). Environ. Pollut. (Barking, Essex: 1987) 234, 572–580. [DOI] [PubMed] [Google Scholar]
- Yang Y., Cruickshank C., Armstrong M., Mahaffey S., Reisdorph R., Reisdorph N. (2013). New sample preparation approach for mass spectrometry-based profiling of plasma results in improved coverage of metabolome. J. Chromatogr. A 1300,217–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yankaskas J. R., Haizlip J. E., Conrad M., Koval D., Lazarowski E., Paradiso A. M., Rinehart C. A. Jr, Sarkadi B., Schlegel R., Boucher R. C. (1993). Papilloma virus immortalized tracheal epithelial cells retain a well-differentiated phenotype. Am. J. Physiol. 264(5 Pt 1), C1219–C1230. [DOI] [PubMed] [Google Scholar]
- Yun B., Lee H., Jayaraja S., Suram S., Murphy R. C., Leslie C. C. (2016). Prostaglandins from cytosolic phospholipase A2alpha/cyclooxygenase-1 pathway and mitogen-activated protein kinases regulate gene expression in Candida albicans-infected macrophages. J. Biol. Chem. 291,7070–7086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarubin T., Han J. (2005). Activation and signaling of the p38 map kinase pathway. Cell Res. 15,11–18. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.