Abstract
This study examined hypermethylated and downregulated genes specific to carbon tetrachloride (CCl4) by Methyl-Seq analysis combined with expression microarray analysis in the liver of rats treated with CCl4 or N-nitrosodiethylamine (DEN) for 28 days, by excluding those with DEN. Among 52 genes, Ldlrad4, Proc, Cdh17, and Nfia were confirmed to show promoter-region hypermethylation by methylation-specific quantitative PCR analysis on day 28. The transcript levels of these 4 genes decreased by real-time reverse transcription-PCR analysis in the livers of rats treated with nongenotoxic hepatocarcinogens for up to 90 days compared with untreated controls and genotoxic hepatocarcinogens. Immunohistochemically, LDLRAD4 and PROC showed decreased immunoreactivity, forming negative foci, in glutathione S-transferase placental form (GST-P)+ foci, and incidences of LDLRAD4− and PROC− foci in GST-P+ foci induced by treatment with nongenotoxic hepatocarcinogens for 84 or 90 days were increased compared with those with genotoxic hepatocarcinogens. In contrast, CDH17 and NFIA responded to hepatocarcinogens without any relation to the genotoxic potential of carcinogens. All 4 genes did not respond to renal carcinogens after treatment for 28 days. Considering that Ldlrad4 is a negative regulator of transforming growth factor-β signaling, Proc participating in p21WAF1/CIP1 upregulation by activation, Cdh17 inducing cell cycle arrest by gene knockdown, and Nfia playing a role in a tumor-suppressor, all these genes may be potential in vivo epigenetic markers of nongenotoxic hepatocarcinogens from the early stages of treatment in terms of gene expression changes. LDLRAD4 and PROC may have a role in the development of preneoplastic lesions produced by nongenotoxic hepatocarcinogens.
Keywords: hepatocarcinogenesis, nongenotoxic hepatocarcinogen, hypermethylation, rat, liver, glutathione S-transferase placental form (GST-P)
Evaluation of carcinogenicity is crucial for the assessment of chemical safety. However, standard carcinogenicity bioassays in which hundreds of rodent animals are administered test compounds over a prolonged period are time-consuming and costly. To identify early prediction marker molecules of hepatocarcinogenesis in rats, we previously reported that administration of carcinogens for 28 days induces expression changes in cell cycle-related molecules resulting in cell cycle arrest in many target organs (Kimura et al., 2016; Taniai et al., 2012; Yafune et al., 2013). Considering that cell cycle arrest is a typical feature of cellular senescence (Campisi, 2013), these study results suggest that chronic exposure to carcinogens may cause an increased number of liver cells to undergo cellular senescence.
Epigenetic alterations have been known to contribute to various biological phenomena. Without altering the DNA sequence itself, epigenetic alterations in the form of DNA methylation in concordance with chromatin-based modifications affect only the expression of targeted genes (Weinhold, 2006). DNA methylation is one of the major epigenetic alterations and has been investigated extensively in normal development and in association with key processes of many diseases (Brandeis et al., 1993; Robertson, 2005). CpG dinucleotides at high densities located primarily upstream of the coding region of genes are termed CpG islands (CGIs). CGI methylation directly prevents transcription factor binding and leads to changes in the chromatin structure, which limits promoter accessibility and causes gene silencing (Palacios et al., 2010). Importantly, the genomic methylation status can be inherited by daughter cells (Holliday, 1989). Furthermore, altered DNA methylation in the genome is found in almost all types of cancers and can lead to changes in gene expression, such as oncogene overexpression and silencing of tumor-suppressor genes to acquire further growth advantages in cancer development (Jones and Baylin, 2007). In our previous study, transmembrane protein 70, a mitochondrial protein involved in the biogenesis of mitochondrial adenosine triphosphate synthase, and ubiquitin-conjugating enzyme E2E 2, which plays a role in the ubiquitin proteasome pathway, were hypermethylated and downregulated after treatment for 28 days with the hepatocarcinogen thioacetamide (TAA) in the rat liver using CGI microarrays (Mizukami et al., 2017). We clarified that downregulation of these molecules promotes rat hepatocarcinogenesis from the early stages, suggesting that hypermethylated and downregulated genes from the early stages of carcinogen treatment may become early detection markers of carcinogens.
We have previously reported that TAA and methapyrilene hydrochloride (MP), nongenotoxic hepatocarcinogens that facilitate cellular proliferation of liver cells by repeated administration in rats for up to 90 days, clearly facilitate cell cycle arrest during both the G1/S and G2/M phases and accompany the upregulation of Tp53 and p21WAF1/CIP1 activation in liver cells (Kimura et al., 2015). In contrast, carbadox (CRB), a genotoxic hepatocarcinogen, slightly induced p21WAF1/CIP1 activation alone even after administration for up to 90 days (Kimura et al., 2015). These results indicate that the responding molecules related to cellular senescence may differ between genotoxic and nongenotoxic hepatocarcinogens.
This study was performed to search for early in vivo epigenetic markers for detection of nongenotoxic hepatocarcinogens, and we examined hypermethylated and downregulated genes specifically in the liver of rats treated with a nongenotoxic hepatocarcinogen for 28 days. For this purpose, we used carbon tetrachloride (CCl4) as a nongenotoxic hepatocarcinogen (Barber et al., 1981; Weisburger, 1977), and N-nitrosodiethylamine (DEN) as a genotoxic carcinogen (IARC Monographs on the Evaluation of Carcinogenic Risk of Chemicals to Humans, 1978; Verna et al., 1996). We first applied genome-wide Methyl-Seq analysis combined with gene expression microarray analysis. To identify CCl4-specific genes, we excluded those responding to DEN from those responding to CCl4. Obtained genes were selected in terms of methylation status and transcript levels in response to genotoxic or nongenotoxic hepatocarcinogen exposure for up to 90 days, as well as immunohistochemical cellular distribution of the corresponding proteins in relation to hepatocellular proliferative lesions induced by these hepatocarcinogens. Finally, we examined the transcript levels of the candidate genes in the kidneys of rats treated with genotoxic or nongenotoxic renal carcinogens for 28 days.
MATERIALS AND METHODS
Chemicals and Animals
CCl4 (CAS No. 56-23-5, purity: ≥ 99.5%), TAA (CAS No. 62-55-5, purity: ≥ 98%), CRB (CAS No. 6804-07-5, purity: ≥ 99%), 1-amino-2, 4-dibromoantraquinone (ADAQ; CAS No. 81-49-2, purity:≥ 97%), 1, 2, 3-trichloropropane (TCP; CAS No. 96-18-4, purity: ≥ 99%), ochratoxin A (OTA; purity: ≥ 98%), dimethyl sulfoxide (DMSO; purity: ≥ 99.5%) and corn oil were purchased from Fujifilm Wako Pure Chemical Corporation (Osaka, Japan). Aflatoxin B1 (AFB1; CAS No. 1162-65-8) was extracted from medial and mycelial fractions of cultivated A. flavus in M1 medium and purified by high-performance liquid chromatography (HPLC) as previously described (De Jesus et al., 1988). According to the AOAC official method 970.44, the purity of aflatoxin B1 was calculated to be approximately 90% according to absorption peak ratios of ultraviolet measurements on methanol (AOAC, 2005). DEN (CAS No. 55-18-5, purity: ≥ 99%) and nitrofurantoin (NFT; CAS No. 67-20-9, purity: > 98.0%) were obtained from Tokyo Chemical Industry Co. Ltd. (Tokyo, Japan), and N-nitrosopyrrolidine (NPYR; CAS No. 930-55-2, purity: ≥ 99%) and MP (CAS No. 135-23-9, purity: ≥ 98%) from Sigma-Aldrich Japan K.K. (Tokyo, Japan).
In all experiments, 5-week-old male F344/NSlc rats were purchased from Japan SLC, Inc. (Hamamatsu, Japan) and acclimatized to a basal diet (CRF-1; Oriental Yeast Co., Ltd., Tokyo, Japan) and tap water ad libitum for 1 week. They were housed in plastic cages with paper chip bedding in a barrier-maintained animal room on a 12-h light-dark cycle and conditioned at 23°C ± 2°C with a relative humidity of 55% ± 15%.
Experimental Design
There were 3 animal experiments.
Experiment 1
In Experiment 1, animals were randomized into 3 groups of 10 animals per group and were untreated (untreated controls) or treated with DEN (4 mg/5 ml/kg body weight, dissolved in saline) or CCl4 (100 mg/5 ml/kg body weight, dissolved in corn oil) daily by gavage for 28 or 90 days. In CCl4 group, the initial dose was set at 100 mg/kg body weight daily by gavage. To examine whether the dose level of CCl4 at 100 mg/kg body weight is appropriate for 90-day repeated oral dose study, we conducted a preliminary 28-day repeated oral dose study using 5-week-old male rats (n = 5/group; data not shown). As a result, the CCl4-treated rats only showed transient decreases in food consumption and body weight at day 3 of treatment, and therefore, we judged that the dose level of CCl4 at 100 mg/kg body weight is appropriate for 90-day study. However, as 2 animals died and the general condition of the remaining animals worsened at day 80, the dose was reduced to 50 mg/kg body weight after 80 days from starting administration with CCl4. At day 84, 1 animal further died and the general condition of other animals worsened in CCl4 group, and therefore, it was judged to terminate the experiment at this time point. Animals of all groups were euthanized by exsanguination from the posterior vena cava and abdominal aorta under CO2/O2 anesthesia at the end of the 28- or 84-day treatment. The dose level of DEN and CCl4, even after the dose change of the latter compound, has shown to induce liver tumors in rats (Suzuki et al., 2009; Weisburger, 1977). Livers were removed, weighed, and cut into small pieces (approximately 30 mg/sample). All samples were immediately frozen in liquid nitrogen and stored at −80°C until analysis. In addition, liver slices (2 slices per animal, one from median lobe and another from left lateral lobe) were fixed in 4% (w/v) paraformaldehyde (PFA) in 0.1 M phosphate buffer (pH 7.4) overnight and processed for histopathological examinations.
Experiment 2
In Experiment 2, to verify gene regulation alteration and cellular distribution of molecules in preneoplastic lesions induced by other hepatocarcinogens, animals were repeatedly administrated with genotoxic hepatocarcinogens (AFB1, NPYR, or CRB) or nongenotoxic hepatocarcinogens (TAA or MP) for up to 90 days. Animals were randomized into 6 groups of 10 animals per group and were untreated (untreated controls) or treated with AFB1 (15 µg/0.5 ml/kg body weight, dissolved in DMSO) daily by gavage, NPYR (13 mg/5 ml/kg body weight, dissolved in saline) daily by gavage, CRB (300 ppm) in diet, TAA (400 ppm) in diet, MP (1000 ppm) in diet for 28 or 90 days. AFB1, NPYR, and CRB were selected as genotoxic hepatocarcinogens in rats and the dose level of each compound has shown to induce preneoplastic liver lesions after 5 or 13 weeks of treatment or neoplastic lesions after 10 months of treatment, respectively (Kanki et al., 2005; King, 1976; Qian et al., 2013). TAA and MP were selected as nongenotoxic hepatocarcinogens, and the dose level of each compound has shown to induce neoplastic liver lesions in the carcinogenicity bioassay (Becker, 1983; NTP, 2000).
All animals were euthanized in the same way to Experiment 1 at the end of 28- and 90-day treatment. The livers of all animals were immediately removed and weighed, and small portions of the liver tissue were cut into small pieces and frozen in liquid nitrogen and stored at –80°C until analysis. Additional liver slices (2 slices per animal) were fixed in 4% PFA in 0.1 M phosphate buffer (pH 7.4) in the same way to the Experiment 1.
Experiment 3
In Experiment 3, to verify gene regulation alteration by genotoxic or nongenotoxic renal carcinogens, animals were repeatedly administered test compounds for 28 days. After administration, the candidate genes selected from Experiment 1 were examined for transcript levels using real-time reverse-transcription (RT) PCR. As genotoxic renal carcinogens, NFT, ADAQ, and TCP were selected (NTP, 1989a, 1993, 1996), and OTA was selected as a nongenotoxic renal carcinogen (FSCJ Food Safety Commission of Japan, 2015; NTP, 1989b). Animals were divided into 5 groups based on initial body weights of 10 animals per group and were untreated (untreated controls) or administered with NFT in diet, ADAQ (25 000 ppm) in diet, TCP (125 mg/5 ml/kg body weight, dissolved in corn oil) daily by gavage, OTA (210 µg/5 ml/kg body weight) daily by gavage for 28 days. OTA was dissolved in an appropriate volume of 0.1 M NaHCO3 solution (pH 8.22; Muto PureChemicals Co., Ltd., Tokyo, Japan) as a solvent by vigorous vortexing. In NFT group, the initial dose was set at 5000 ppm in the diet, because this dose level was twice higher than the renal carcinogenic dose (2500 ppm) and was not lethal in a 13-week repeated oral dose study (NTP, 1989a). However, as the general condition of the animals worsened, the dose was reduced to 4000 ppm after 9 days from starting administration and 3000 ppm after 14 days. The dose levels of NFT, ADAQ, TCP, and OTA, even after the dose change in case of NFT, have been shown to induce renal tumors in rats (NTP, 1989a, 1993, 1996, 1989b). The animals in each group were euthanized by exsanguination from the abdominal aorta under deep anesthesia with CO2/O2 and kidneys were removed. Half of the left and right kidneys were vertically sliced along the length of the center. Portions of the outer stripe of the outer medulla were collected using a biopsy punch (Ф 1.5 mm, Kai Industries Co., Ltd., Seki, Japan) and stored at −80°C until extraction.
All procedures of these animal experiments were conducted in compliance with the Guidelines for Proper Conduct of Animal Experiments (Science Council of Japan, 1 June 2006) and according to the protocol approved by the Animal Care and Use Committee of Tokyo University of Agriculture and Technology. All efforts were made to minimize animal suffering.
DNA and RNA Extraction
For methylation analysis, genomic DNA (gDNA) was extracted from liver tissue of untreated controls, DEN and CCl4 group on day 28 in Experiment 1 with an Allprep DNA/RNA Mini Kit (Qiagen, Hilden, Germany). Extracted gDNA was used for Methyl-Seq analysis (n = 5/group, pooled as one sample) and quantitative methylation-specific PCR analysis (n = 5/group). For gene expression microarray analysis, total RNA from liver tissue samples of untreated controls, DEN and CCl4 group (n = 3/group) on days 28 and 84 in Experiment 1 was extracted using QIAzol (Qiagen) together with the RNeasy Mini Kit (Qiagen) according to the manufacturer’s protocol. For transcript expression analysis of candidate genes, total RNA was extracted from tissue samples in Experiments 1–3 with an Allprep DNA/RNA Mini Kit. Extracted total RNA was used for real-time RT-PCR analysis (n = 6/group).
Methyl-Seq Analysis
To identify DEN or CCl4-induced epigenetic changes on day 28 in Experiment 1, SureSelect Target Enrichment System (Rat Methyl-Seq; Agilent Technologies, Santa Clara, California) was implemented according to the manufacturer’s protocol (SureSelectXT Methyl-Seq Target Enrichment System, version B.3, June 2015). Using the publicly available databases from the University of California Santa Cruz (UCSC) Genome Browser (http://genome.ucsc.edu), genomic coordinates for all known CGIs, shores and shelves in the rat genome were obtained. Briefly, 2 µg of gDNA from each animal was pooled from 5 animals of each group to prepare one sample. Each pooled gDNA sample was fragmented using a Covaris sonicator (Covaris, Woburn, Massachusetts). These fragments were end-repaired, 3’-adenylated, and further ligated with methylated primers. Following hybridization to biotinylated, plus-strand DNA-complementary RNA library “baits,” precipitation from the solution using streptavidin-coated magnetic beads, and RNase-digestion of the “baits,” captured DNA was bisulfite-converted using the EZ-DNA Methylation-Gold Kit (Zymo Research, Irvine, California). Subsequently, DNA samples were PCR-amplified using sample-specific indexed (“barcoding”) primers to allow for multiplexing and sequenced by 100 bp paired-end sequencing by Illumina HiSeq2500 as described in the manufacturer’s protocol. Three samples were loaded on each lane.
The adapter sequences and low-quality regions were trimmed by Cutadapt software version 1.1 (GitHub Inc., San Francisco, California) and Trimmomatic software version 0.32 (RWTH Aachen University, Aachen, Germany), respectively. Remaining reads after trimming were mapped to the Rattus norvegicus.rn6.fa (with GA, CT converted Index), which was produced by the Rat Genome Reference Consortium, using Bismark version 0.18.2 (Babraham Institute, Cambridgeshire, UK) with Bowtie 2 version 2.3.2 (Johns Hopkins University, Baltimore, Maryland). After running Bismark, PCR duplicates were removed from the mapped reads using the “deduplicate bismark” routine. Postalignment quality control was performed using Samtools version 1.3.1 (GitHub Inc.). The Default Bismark methylation extractor routine was used with the exception of paired-end, no-overlap, and minimum coverage of at least 1 read to extract all CpGs in individual samples. For the calculation of methylation ratio, detection of methylation candidate sites on the reference genome sequence and methylation ratio were performed using Bismark. Only cytosines with a minimal coverage of 10× and phred quality score of Q30 in the CpG context were analyzed. The metrics for the high-throughput sequencing of liver samples are shown in Supplementary Table 1.
Genes showing CpG site hypermethylation up to 1000 bp upstream from the transcription start site were selected with the criterion of the methylation ratio of ≥ 30% in CCl4 sample as compared with untreated control, excluding the genes showing methylation ratio of > 0% in DEN sample compared with untreated control.
Gene Expression Microarray Analysis in Experiment 1
Gene expression analysis was conducted using Agilent Rat Oligo arrays with approximately 60 000 probes for known genes and expressed sequence tags (Agilent Technologies). For sample preparation and array processing, the Agilent protocol “One-Color Microarray-Based Gene Expression Analysis” was used. Briefly, the recommended volume of control RNAs (Agilent One-Color RNA Spike-In Kit) was added to 100 ng of total RNA from the each liver of untreated controls, DEN and CCl4 groups on day 28 and 84 (n = 3/group) in Experiment 1. Thereafter, Cy3-labeled cRNA was produced using the Agilent Low Input Quick Amp Labeling (1-color), purified with the RNeasy Mini Kit, fragmented using the In Situ Hybridization Kit (Agilent Technologies), and subjected to hybridization by incubation in a hybridization oven (Agilent Technologies). Hybridized slides were scanned (G2565CA scanner, Agilent Technologies), and data were obtained using Agilent Feature Extraction software version 11.7.1.1 with defaults for all parameters.
Microarray data analyses were performed using GeneSpring GX software version 14.5 (Agilent Technologies). Expression values of <1 were substituted by 1, and 75th percentile normalization was performed using GeneSpring normalization algorithms. Reliability of each expression value was represented by a flag based on the default setting of GeneSpring (Detected, Compromised and Not Detected).
The entire data set has been deposited in National Center for Biotechnology Information Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) and is accessible through GEO Series accession number GSE123408.
Methylation-Specific Quantitative PCR in Experiment 1
Thirty-eight genes that were selected in terms of the presence of commercially available antibodies for immunohistochemical analysis among those showing hypermethylation and downregulation specific to CCl4 group were subjected to methylation-specific quantitative PCR. Isolated gDNA from untreated controls, DEN and CCl4 groups on day 28 in Experiment 1 (n = 5/group) was sonicated using a Bioruptor (UCD-250; Cosmo Bio Co., Ltd., Tokyo, Japan). A part of the fragmented DNA was stored as an input (control) DNA at −20°C until use. The remaining mixture was incubated with MBD2/magnetic bead complexes and then eluted. The methylation-enriched DNA was purified using an EpiXplore Methylated DNA Enrichment kit (Clontech Laboratories, Mountain View, California). Input and methylation-enriched DNA samples were used as templates for quantitative measurement of methylation at target CpG sites by real-time PCR using Power SYBR Green PCR Master Mix (Thermo Fisher Scientific, Waltham, Massachusetts) and a StepOnePlus Real-time PCR System (Thermo Fisher Scientific). The PCR primers for the target gene CpG sites were designed using Methyl Primer Express software version 1.0 (Thermo Fisher Scientific) and Primer Express software version 3.0 (Supplementary Table 2; Thermo Fisher Scientific). The Quantification was based on the comparative CT method and involved a comparison of the CT values of the IP DNA to the CT values of the input DNA.
Real-time RT-PCR Analysis in Experiments 1–3
Four genes, Ldlrad4, Proc, Cdh17, and Nfia, that were selected by confirming promoter-region hypermethylation for confirming transcript downregulation by real-time RT-PCR using the RNA samples (n = 6/group) isolated from the untreated controls and each treatment groups in Experiments 1–3. First-strand complementary DNA was synthesized from 2 µg of total RNA using SuperScript III Reverse Transcriptase (Thermo Fisher Scientific). Real-time PCR was performed using Power SYBR Green PCR Master Mix and StepOnePlus Real-Time PCR System (Thermo Fisher Scientific). PCR primers listed in Supplementary Table 3 for target genes were designed using Primer Express version 3.0 (Thermo Fisher Scientific). Using the threshold cycle (CT) values of actin, beta (Actb) or hypoxanthine phosphoribosyltransferase 1 (Hprt1) in the same sample as the endogenous control, the relative differences in gene expression were calculated using the 2−ΔΔCT method (Livak and Schmittgen, 2001).
Histopathology and Immunohistochemistry in Experiments 1 and 2
Liver slices in Experiments 1 and 2 (n = 10/group) were processed using a standard protocol for paraffin embedding and were serially sectioned to a thickness of 3 µm. Immunohistochemistry was performed by incubating liver tissue sections overnight at 4°C with primary antibodies against glutathione S-transferase placental form (GST-P), a preneoplastic liver cell lesion marker in rats (Ito et al., 1998; Shirai, 1997; rabbit polyclonal antibody, 1:1000; Medical & Biological Laboratories, Nagoya, Japan), low-density lipoprotein receptor class A domain-containing protein 4 (LDLRAD4), also known as C18ORF1, a negative regulator of transforming growth factor (TGF)-β signaling (Nakano et al., 2014; rabbit polyclonal antibody, 1:100; Bioss Antibodies, Woburn, Massachusetts), vitamin K-dependent protein C precursor (PROC), a liver cell-derived regulator of blood coagulation (Kovács et al., 2015; rabbit polyclonal antibody, 1:100; Bioss Antibodies), cadherin-17 precursor (CDH17), a member of nonclassical cadherin family (Jung et al., 2004; rabbit polyclonal antibody, 1:200; Bioss Antibodies), and nuclear factor 1 A-type (NFIA), a member of the nuclear factor I transcription factors (Chen et al., 2017; rabbit polyclonal antibody, 1:2000; Novus Biologicals, LLC, Littleton, Colorado). To inhibit endogenous peroxidase, deparaffinized sections were incubated in 0.3% (v/v) hydrogen peroxide solution in absolute methanol for 30 min. Immunodetection was performed using a Vectastain Elite ABC Kit (Vector Laboratories Inc., Burlingame, California) with the primary antibodies and 3, 3′,diaminobenzidine/H2O2 as the chromogen. All immunostained slides were counterstained with hematoxylin and coverslipped for microscopic examination.
Analysis of Immunolocalization in the Livers in Experiments 1 and 2
The number and area of GST-P+ liver cell foci >200 μm in diameter in liver sections from Experiments 1 and 2 (n = 10/group) were measured according to the method and equipment as described previously (Taniai et al., 2009). In DEN and CCl4 groups on day 84 of Experiment 1, and AFB1, NPYR, TAA, and MP groups on day 90 of Experiment 2 (n = 10/group), the immunoreactivity of LDLRAD4, PROC, and CDH17 was classified as increased (+) or decreased (−) in the GST-P+ foci compared with the surrounding hepatocytes, and the incidence of LDLRAD4−, PROC−, and CDH17− expression in total GST-P+ foci appeared in liver sections per animal was estimated. In DEN and CCl4 groups on day 84 of Experiment 1, and NPYR and TAA groups on day 90 of Experiment 2 (n = 10/group), ratio of nuclear NFIA+ cells to total liver cells was calculated for each of the inside and outside regions of GST-P+ foci in 10 randomly selected areas at a magnification of 400×.
Statistical Analysis
In microarray analysis, statistically significant differences in gene expression between 2 groups were determined using Welch’s t test. The expression levels of each gene were computed by the ratio of gene expression in DEN or CCl4 group to untreated controls. Significantly expressed genes were selected by following criterion: (1) significance level is 0.05 (p < .05). Gene expression ratio is > 1.50 or < 0.67. Samples flagged with “Not Detected” from the 2 groups were not included. (2a) Significance level is 0.01. Gene expression ratio is > 4. Samples in the exposure group flagged with “Not Detected” were not included, and one or more samples of untreated controls flagged with “Not Detected” flag of untreated controls were included. (2b) Significance level is 0.01. Gene expression ratio is < 0.25. Samples in untreated controls flagged with “Not Detected” flag were not included, and one or more samples of the exposure group flagged with “Not Detected” flag were included.
Numerical data are presented as mean ± SD. For comparison of the numerical data between multiple groups, values were analyzed by the Bartlett’s test for the homogeneity of variance. If there was no significant difference in variance, Dunnett’s test was performed for comparison between the groups. If a significant difference was found in variance, Steel’s test was performed. In case of the data that compare all pairs, such as the data of real-time RT-PCR and immunoreactive NFIA+ cells number in the inside and outside regions of GST-P+ foci, values were analyzed by the Bartlett’s test for the homogeneity of variance. If there was no significant difference in variance, Tukey’s test was performed for comparison among the groups. If a significant difference was found in variance, Steel-Dwass test was performed. In the case of numerical data consisting of 2 sample groups, data were assessed using F-test for homogeneity of variance. If the variance was homogenous between the groups, a Student’s t test was applied for comparison, and if it was heterogeneous, the Aspin-Welch’s t test was performed. With regard to categorical data, such as the incidence data of LDLRAD4−, PROC−, and CDH17− foci in population of GST-P+ foci, Fisher’s exact test was performed. All analyses were conducted using an Excel Statistics 2013 software package version 2.02 (Social Survey Research Information Co. Ltd., Tokyo, Japan), and p < .05 was considered statistically significant.
RESULTS
Hypermethylated and Downregulated Genes Specific to CCl4 in Experiment 1
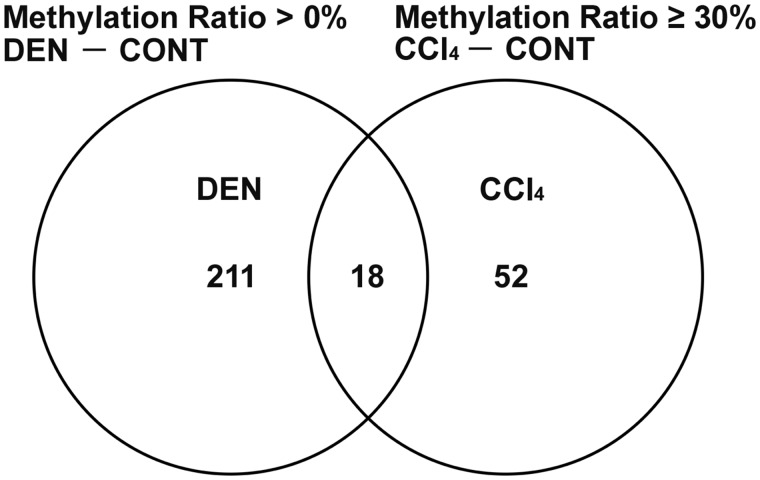
As a result of verifying the data of SureSelect Methyl-Seq and microarray analyses in Experiment 1, 52 genes showed promoter-region hypermethylation (difference in the methylation ratio ≥ 30%) on day 28 and statistically significant transcript downregulation on both days 28 and 84 specific to CCl4 group in comparison with untreated controls, after excluding hypermethylated (difference in the methylation ratio > 0%) and downregulated genes in DEN group compared with untreated controls (Figure 1, Tables 1 and 2). Among selected 52 genes, 18 genes were significantly downregulated and 1 gene was significantly upregulated on day 28, and 44 genes were significantly downregulated on day 84 in DEN group in comparison with untreated controls (Table 2).
Figure 1.
Venn diagram denoting 52 genes located at promoter region of gene sequence showing hypermethylation (methylation ratio ≥ 30%) and transcript downregulation specific to CCl4 group in comparison with untreated controls, after excluding hypermethylated (methylation ratio > 0%) and downregulated genes in DEN group compared with untreated controls in Experiment 1.
Table 1.
List of Hypermethylated and Downregulated Genes Specific to CCl4 After Excluding the Genes Hypermethylated and Downregulated With DEN (52 genes, ≥ 30% Methylation Ratio: CCl4–CONT)
| % Methylation |
|||||
|---|---|---|---|---|---|
| Location | Accession No. | Gene Symbol | Gene Description | CONT | CCl4 |
| Chr18: 63843080 | NM_001271365 | Ldlrad4 | Low density lipoprotein receptor class A domain containing 4 | 22.0 | 65.9 |
| Chr18: 63843085 | 21.8 | 57.0 | |||
| Chr18: 63843106 | 19.4 | 54.2 | |||
| Chr18: 24928994 | NM_012803 | Proc | Protein C, inactivator of coagulation factors Va and VIIIa | 12.2 | 48.6 |
| Chr5: 25390796 | NM_053977 | Cdh17 | Cadherin 17 | 50.0 | 100.0 |
| Chr5: 116421515 | NM_012988 | Nfia | Nuclear factor I/A | 16.7 | 48.6 |
| Chr8: 48582433 | NM_001191996 | Pdzd3 | PDZ domain containing 3 | 25.0 | 64.3 |
| Chr8: 77398743 | NM_012597 | Lipc | Lipase C, hepatic type | 9.3 | 46.3 |
| Chr8: 77398998 | 33.3 | 68.2 | |||
| Chr13: 50499698 | NM_001108343 | Etnk2 | Ethanolamine kinase 2 | 31.4 | 61.8 |
| Chr13: 75174405 | NM_053571 | Sec16b | SEC16 homolog B, endoplasmic reticulum export factor | 15.4 | 63.9 |
| Chr13: 75174440 | 11.8 | 42.1 | |||
| Chr2: 188449177 | NM_012624 | Pklr | Pyruvate kinase L/R | 0.0 | 33.3 |
| Chr2: 188449199 | 8.7 | 40.7 | |||
| Chr14: 19132230 | NM_172320 | Afm | Afamin | 7.1 | 55.0 |
| Chr3: 101466148 | NM_022629 | Bbox1 | Gamma-butyrobetaine hydroxylase 1 | 9.4 | 53.6 |
| Chr3: 101466153 | 17.2 | 58.6 | |||
| Chr3: 101466160 | 17.2 | 51.9 | |||
| Chr7: 70661881 | NM_022614 | Inhbc | Inhibin subunit beta C | 17.2 | 61.5 |
| Chr15: 18488366 | NM_001107253 | Kctd6 | Potassium channel tetramerization domain containing 6 | 50.0 | 92.3 |
| Chr15: 18488454 | 18.8 | 85.7 | |||
| Chr15: 18488459 | 18.8 | 54.8 | |||
| Chr11: 32551517 | NM_153724 | Rcan1 | Regulator of calcineurin 1 | 35.3 | 90.9 |
| Chr12: 45183219 | NM_001024254 | Taok3 | TAO kinase 3 | 9.1 | 53.8 |
| Chr8: 50558483 | NM_001277264 | Apoa5 | Apolipoprotein A5 | 15.8 | 62.5 |
| Chr2: 84644363 | NM_001008770 | Cmbl | Carboxymethylenebutenolidase homolog | 15.9 | 50.9 |
| Chr2: 84644555 | 6.7 | 47.1 | |||
| ChrX: 156318979 | NM_001109324 | Fam3a | Family with sequence similarity 3, member A | 40.0 | 82.4 |
| Chr6: 8218710 | NM_001270620 | Ppm1b | Protein phosphatase, Mg2+/Mn2+ dependent, 1B | 11.1 | 51.4 |
| Chr1: 65575816 | NM_024143 | Slc27a5 | Solute carrier family 27 member 5 | 22.2 | 63.6 |
| Chr18: 48200786 | NM_001008364 | Snx24 | Sorting nexin 24 | 9.5 | 50.0 |
| Chr10: 59172270 | NM_022690 | Ube2g1 | Ubiquitin-conjugating enzyme E2G 1 | 34.6 | 81.3 |
| Chr1: 176854020 | NM_001107542 | Usp47 | Ubiquitin specific peptidase 47 | 14.7 | 64.0 |
| Chr1: 176854025 | 22.2 | 54.2 | |||
| Chr4: 82703106 | NM_022243 | Hibadh | 3-hydroxyisobutyrate dehydrogenase | 43.9 | 82.6 |
| Chr3: 80013365 | NM_031627 | Nr1h3 | Nuclear receptor subfamily 1, group H, member 3 | 14.3 | 50.0 |
| Chr4: 84741495 | NM_001025063 | Scrn1 | Secernin 1 | 48.1 | 83.3 |
| ChrX: 111888669 | NM_031345 | Tsc22d3 | TSC22 domain family, member 3 | 13.9 | 48.7 |
| Chr2: 188707433 | NM_001106446 | Zbtb7b | Zinc finger and BTB domain containing 7B | 61.5 | 94.7 |
| Chr5: 56577130 | NM_001106646 | Ndufb6 | NADH: ubiquinone oxidoreductase subunit B6 | 12.3 | 45.5 |
| Chr6: 127656730 | NM_001166352 | Serpina11 | Serpin family A member 11 | 13.8 | 46.8 |
| Chr3: 150886421 | NM_181637 | Pigu | Phosphatidylinositol glycan anchor biosynthesis, class U | 30.0 | 61.3 |
| Chr3: 138708890 | NM_019219 | Rbbp9 | RB-binding protein 9, serine hydrolase | 50.0 | 80.0 |
| Chr7: 71035686 | NM_001130537 | Zbtb39 | Zinc finger and BTB domain containing 39 | 0.0 | 36.8 |
| Chr9: 16924520 | NM_053537 | Slc22a7 | Solute carrier family 22 member 7 | 3.7 | 37.9 |
| Chr9: 16924551 | 13.2 | 43.2 | |||
| Chr9: 16924762 | 35.7 | 66.7 | |||
| Chr15: 36917924 | NM_001017375 | Mphosph8 | M-phase phosphoprotein 8 | 9.5 | 43.1 |
| Chr1: 264741143 | NM_001108526 | Sema4g | Semaphorin 4G | 10.0 | 41.0 |
| Chr1: 264741769 | 7.7 | 41.2 | |||
| Chr17: 43661010 | NM_001173434 | Hfe | Homeostatic iron regulator | 9.4 | 40.0 |
| Chr17: 43661014 | 4.0 | 36.8 | |||
| Chr7: 11490192 | NM_001012115 | Creb3l3 | cAMP responsive element binding protein 3-like 3 | 6.1 | 38.5 |
| Chr6: 132761344 | NM_001001509 | Slc25a47 | Solute carrier family 25, member 47 | 18.4 | 50.0 |
| Chr12: 25092252 | NM_001006957 | Eif4h | Eukaryotic translation initiation factor 4H | 60.0 | 90.9 |
| Chr4: 154997088 | NM_001271376 | Cpamd8 | C3 and PZP-like, alpha-2-macroglobulin domain containing 8 | 0.0 | 33.3 |
| Chr4: 154997750 | 24.4 | 60.0 | |||
| Chr4: 154997751 | 20.0 | 80.0 | |||
| Chr20: 27308979 | NM_001100860 | Slc25a16 | Solute carrier family 25 member 16 | 50.0 | 92.9 |
| Chr9: 95501133 | NM_053577 | Spp2 | Secreted phosphoprotein 2 | 10.0 | 52.2 |
| Chr9: 95501143 | 12.5 | 65.0 | |||
| Chr9: 95501151 | 20.0 | 61.1 | |||
| Chr15: 42693578 | NM_022220 | Gulo | Gulonolactone (L-) oxidase | 13.6 | 45.8 |
| Chr15: 42693586 | 7.0 | 44.9 | |||
| Chr9: 65308552 | NM_001106913 | Clk1 | CDC-like kinase 1 | 18.2 | 56.0 |
| Chr9: 65308567 | 47.1 | 78.9 | |||
| Chr1: 199336659 | NM_001109156 | Prss53 | Serine protease 53 | 30.4 | 66.7 |
| Chr16: 21099849 | NM_001127654 | Tm6sf2 | Transmembrane 6 superfamily member 2 | 7.7 | 40.0 |
| Chr16: 21100135 | 0.0 | 31.3 | |||
| Chr1: 81047457 | NM_133323 | Zfp111 | Zinc finger protein 111 | 50.0 | 81.8 |
| Chr3: 94687482 | NM_001029916 | Depdc7 | DEP domain containing 7 | 2.6 | 33.3 |
| Chr20: 7761965 | NM_001134755 | Zfp523 | Zinc finger protein 523 | 25.0 | 61.5 |
| Chr3: 81282906 | NM_001012088 | Pex16 | Peroxisomal biogenesis factor 16 | 0.0 | 30.3 |
| Chr3: 81282923 | 4.2 | 40.0 | |||
| Chr3: 72329199 | NM_001107743 | Slc43a3 | Solute carrier family 43, member 3 | 5.9 | 38.3 |
Abbreviations: CCl4, carbon tetrachloride; CONT, untreated controls; DEN, N-nitrosodiethylamine.
Table 2.
List of Selected 52 Genes and Their Transcript Levels in the Liver of Rats After Day 28 and 84 of Treatment With DEN or CCl4 in Experiment 1 as Determined by Gene Expression Microarray Analysis
| DEN |
CCl4 |
|||
|---|---|---|---|---|
| Gene Symbol | Day 28 | Day 84 | Day 28 | Day 84 |
| Ldlrad4 | 0.73 | 0.71** | 0.47** | 0.62** |
| Proc | 0.88 | 0.61* | 0.38* | 0.44** |
| Cdh17 | 0.79 | 0.34 | 0.48** | 0.14* |
| Nfia | 0.79* | 0.48* | 0.32** | 0.50** |
| Pdzd3 | 1.01 | 0.86 | 0.48* | 0.56** |
| Lipc | 0.83 | 0.41* | 0.24* | 0.19** |
| Etnk2 | 0.90 | 0.58* | 0.49* | 0.48** |
| Sec16b | 1.04 | 0.72** | 0.59* | 0.53* |
| Pklr | 0.63 | 0.50** | 0.24* | 0.24** |
| Afm | 0.95 | 0.57 | 0.43* | 0.38** |
| Bbox1 | 0.79* | 0.65** | 0.41* | 0.59** |
| Inhbc | 0.74* | 0.28** | 0.26* | 0.23** |
| Kctd6 | 0.80 | 0.55* | 0.53* | 0.43** |
| Rcan1 | 0.95 | 0.86 | 0.63* | 0.63* |
| Taok3 | 0.76* | 0.45** | 0.66* | 0.53* |
| Apoa5 | 0.87 | 0.51* | 0.52** | 0.50** |
| Cmbl | 0.96 | 0.82* | 0.58* | 0.64** |
| Fam3a | 0.92 | 0.71** | 0.60* | 0.61* |
| Ppm1b | 0.81* | 0.73* | 0.62* | 0.68** |
| Slc27a5 | 0.76* | 0.33* | 0.30* | 0.33** |
| Snx24 | 0.83 | 0.49** | 0.49* | 0.36** |
| Ube2g1 | 0.90 | 0.70* | 0.70** | 0.68** |
| Usp47 | 0.86* | 0.83* | 0.74** | 0.68** |
| Hibadh | 0.83* | 0.75** | 0.49* | 0.51** |
| Nr1h3 | 0.99 | 0.73* | 0.70** | 0.64** |
| Scrn1 | 1.04 | 0.28* | 0.31* | 0.19** |
| Tsc22d3 | 0.45** | 0.62 | 0.19** | 0.43* |
| Zbtb7b | 0.94 | 0.61** | 0.69** | 0.63** |
| Ndufb6 | 0.95 | 0.79** | 0.66* | 0.62** |
| Serpina11 | 0.85* | 0.55** | 0.47** | 0.54** |
| Pigu | 1.25* | 0.57* | 0.63* | 0.47** |
| Rbbp9 | 0.81* | 0.73** | 0.60** | 0.60** |
| Zbtb39 | 0.70* | 0.54* | 0.44** | 0.37** |
| Slc22a7 | 0.88 | 0.49* | 0.64* | 0.52** |
| Mphosph8 | 1.07 | 1.11 | 0.60** | 0.68** |
| Sema4g | 0.83 | 0.51* | 0.58* | 0.51* |
| Hfe | 0.83 | 0.86 | 0.58* | 0.74** |
| Creb3l3 | 0.84* | 0.60 | 0.79** | 0.67** |
| Slc25a47 | 0.78 | 0.58** | 0.60** | 0.47** |
| Eif4h | 0.94* | 0.89** | 0.84** | 0.79** |
| Cpamd8 | 0.75* | 0.54** | 0.29* | 0.39** |
| Slc25a16 | 0.90 | 0.78* | 0.57* | 0.49** |
| Spp2 | 0.85* | 0.59* | 0.45* | 0.49* |
| Gulo | 0.74* | 0.39* | 0.44* | 0.51** |
| Clk1 | 0.96 | 0.72** | 0.65* | 0.72** |
| Prss53 | 0.80 | 0.63** | 0.38** | 0.49** |
| Tm6sf2 | 1.11 | 0.86* | 0.62* | 0.57** |
| Zfp111 | 0.90 | 0.73** | 0.75** | 0.81** |
| Depdc7 | 0.86* | 0.68** | 0.54* | 0.49** |
| Zfp523 | 0.94 | 0.85* | 0.79** | 0.73** |
| Pex16 | 0.97 | 0.68* | 0.64** | 0.60* |
| Slc43a3 | 0.90 | 0.38* | 0.51* | 0.43** |
Values are fold change with the expression level in untreated controls set as 1.
Abbreviations: Afm, Afamin; Apoa5, Apolipoprotein A5; Bbox1, Gamma-butyrobetaine hydroxylase 1; CCl4, carbon tetrachloride; Cdh17, cadherin 17; Clk1, CDC-like kinase 1; Cmbl, Carboxymethylenebutenolidase homolog; Cpamd8, C3 and PZP-like, alpha-2-macroglobulin domain containing 8; Creb3l3, cAMP responsive element binding protein 3-like 3; DEN, N-nitrosodiethylamine; Depdc7, DEP domain containing 7; Eif4h, eukaryotic translation initiation factor 4H; Etnk2, Ethanolamine kinase 2; Fam3a, Family with sequence similarity 3, member A; Gulo, Gulonolactone (L-) oxidase; Hfe, Homeostatic iron regulator; Hibadh, 3-hydroxyisobutyrate dehydrogenase; Inhbc, Inhibin subunit beta C; Kctd6, Potassium channel tetramerization domain containing 6; Ldlrad4, low-density lipoprotein receptor class A domain containing 4; Lipc, Lipase C, hepatic type; Mphosph8, M-phase phosphoprotein 8; Ndufb6, NADH: ubiquinone oxidoreductase subunit B6; Nfia, nuclear factor I/A; Nr1h3, Nuclear receptor subfamily 1, group H, member 3; Pdzd3, PDZ domain containing 3; Pex16, Peroxisomal biogenesis factor 16; Pigu, Phosphatidylinositol glycan anchor biosynthesis, class U; Pklr, Pyruvate kinase L/R; Ppm1b, Protein phosphatase, Mg2+/Mn2+ dependent, 1B; Proc, protein C, inactivator of coagulation factors Va and VIIIa; Prss53, Serine protease 53; Rbbp9, RB-binding protein 9, serine hydrolase; Rcan1, Regulator of calcineurin 1; Scrn1, Secernin 1; Sec16b, SEC16 homolog B, endoplasmic reticulum export factor; Sema4g, Semaphorin 4G; Serpina11, Serpin family A member 11; Slc22a7, Solute carrier family 22 member 7; Slc25a16, Solute carrier family 25 member 16; Slc25a47, Solute carrier family 25, member 47; Slc27a5, Solute carrier family 27 member 5; Slc43a3, Solute carrier family 43, member 3; Snx24, Sorting nexin 24; Spp2, Secreted phosphoprotein 2; Taok3, TAO kinase 3; Tm6sf2, Transmembrane 6 superfamily member 2; Tsc22d3, TSC22 domain family, member 3; Ube2g1, Ubiquitin-conjugating enzyme E2G 1; Usp47, Ubiquitin specific peptidase 47; Zbtb39, Zinc finger and BTB domain containing 39; Zbtb7b, Zinc finger and BTB domain containing 7B; Zfp111, Zinc finger protein 111; Zfp523, Zinc finger protein 523.
p < .05, **p < .01, significantly different from untreated controls by Welch’s t test.
Validation of Hypermethylation Status of Candidate Genes in Experiment 1
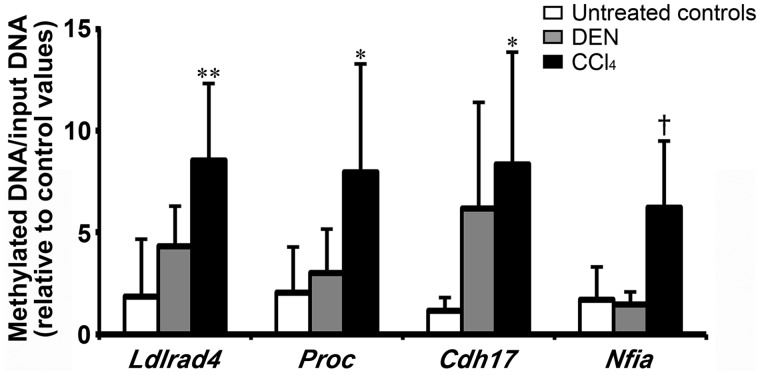
Among the 38 genes selected from the 52 genes in terms of the presence of commercially available antibodies for immunohistochemical analysis, methylation-specific quantitative PCR revealed that 4 genes, Ldlrad4, Proc, Cdh17, and Nfia, significantly increased in methylation status in CCl4 group compared with untreated controls on day 28, while these 4 genes displayed no significantly different changes in methylation status in DEN group on day 28 (Figure 2, Supplementary Table 4).
Figure 2.
Methylation-specific quantitative PCR data of selected 4 genes hypermethylated and downregulated specific to CCl4 group in the liver on day 28 in Experiment 1. Values represent mean + SD (n = 5). *p < .05, **p < .01, significantly different from untreated controls by Dunnett’s or Steel’s test. †p < .05, significantly different from untreated controls by Student’s or Aspin-Welch’s t test.
Transcript Expression Changes of Candidate Genes in Experiments 1 and 2
Experiment 1
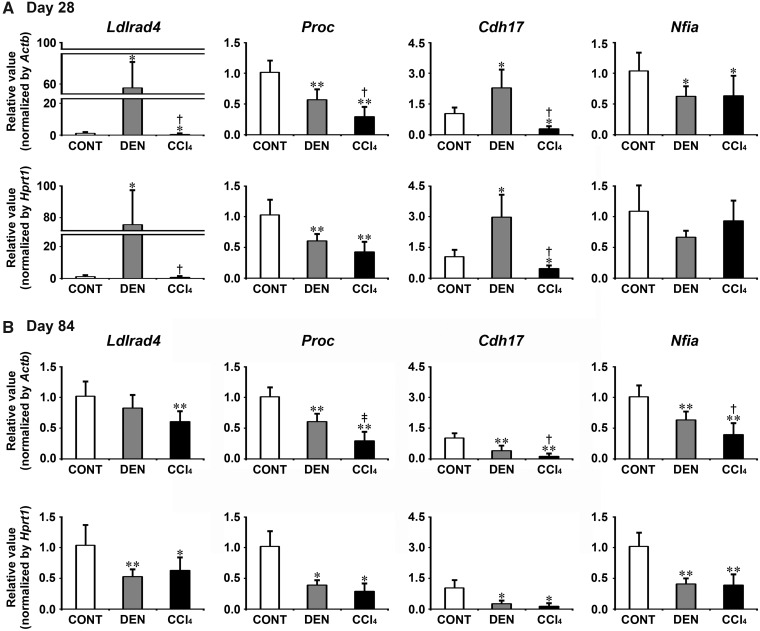
Transcript levels of the candidate genes, Ldlrad4, Proc, Cdh17, and Nfia were determined by real-time RT-PCR in DEN and CCl4 groups on day 28 and 84 (Figure 3). On day 28, DEN group showed significantly decreased transcript levels of Proc and Nfia after normalization with Actb and Hprt1 or Actb-alone compared with untreated controls. DEN group showed significantly increased transcript levels of Ldlrad4 and Cdh17 after normalization with Actb and Hprt1 compared with untreated controls. CCl4 group showed significantly decreased transcript levels of all genes (Ldlrad4, Proc, Cdh17, and Nfia) after normalization with Actb and Hprt1 or Actb-alone compared with untreated controls. When the transcript level was compared between DEN and CCl4 groups, CCl4 group significantly decreased transcript levels of Ldlrad4, Proc, and Cdh17 after normalization with Actb and Hprt1 or Actb-alone compared with DEN group. On day 84, DEN group showed significantly decreased transcript levels of all genes (Ldlrad4, Proc, Cdh17, and Nfia) after normalization with Actb and Hprt1 or Hprt1-alone compared with untreated controls. CCl4 group showed significantly decreased transcript levels of all genes after normalization with Actb and Hprt1 compared with untreated controls. When the transcript level was compared between DEN and CCl4 groups, CCl4 group significantly decreased transcript levels of Proc, Cdh17, and Nfia after normalization with Actb compared with DEN group.
Figure 3.
Data of real-time RT-PCR analysis of selected genes specifically responded to CCl4 group in the liver in Experiment 1. A, Day 28. B, Day 84. Values are normalized to Actb or Hprt1 and expressed as the mean + SD (n = 6). *p < .05, **p < .01, significantly different from untreated controls by Tukey’s or Steel-Dwass test. †p < .05, ‡p < .01, significantly different from DEN group by Tukey’s or Steel-Dwass test.
Experiment 2
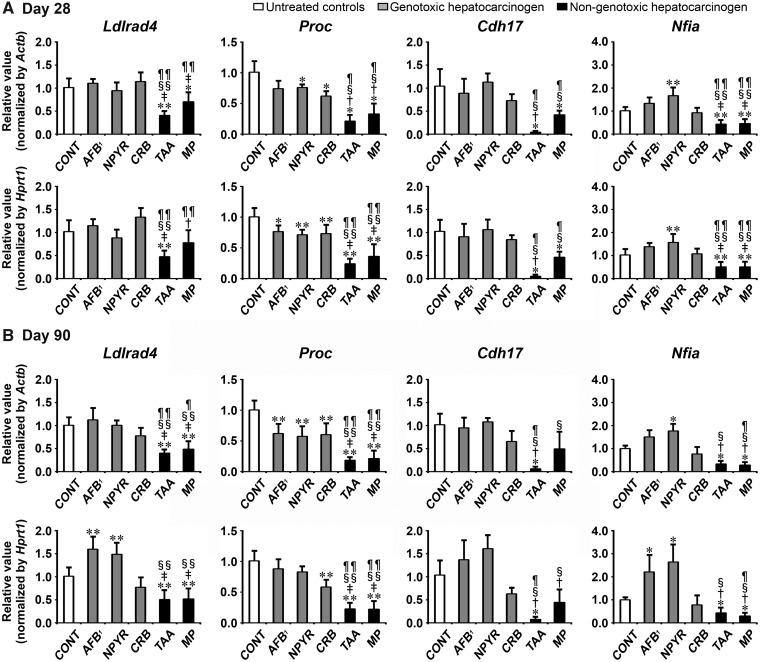
Transcript levels of the candidate genes, Ldlrad4, Proc, Cdh17, and Nfia were determined by real-time RT-PCR in groups of genotoxic hepatocarcinogens (AFB1, NPYR, and CRB) and nongenotoxic hepatocarcinogens (TAA and MP) on day 28 and 90 (Figure 4). On day 28, all genotoxic hepatocarcinogens showed significantly decreased transcript levels of Proc after normalization with Actb and Hprt1 or Hprt1-alone compared with untreated controls. NPYR group showed significantly increased transcript levels of Nfia after normalization with Actb and Hprt1 compared with untreated controls. Both of nongenotoxic hepatocarcinogens showed significantly decreased transcript levels of all genes (Ldlrad4, Proc, Cdh17, and Nfia) after normalization with Actb and Hprt1 or Actb-alone compared with untreated controls. When the transcript level was compared between genotoxic and nongenotoxic hepatocarcinogens, TAA group showed significantly decreased transcript levels of all genes after normalization with Actb and Hprt1 compared with all genotoxic hepatocarcinogens. MP group showed significantly decreased transcript levels of Proc and Nfia after normalization with Actb and Hprt1 compared with all genotoxic hepatocarcinogens. MP group showed significantly decreased transcript levels of Ldlrad4 after normalization with Actb and Hprt1 compared with AFB1 and CRB groups. MP group showed significantly decreased transcript levels of Cdh17 after normalization with Actb and Hprt1 compared with NPYR and CRB groups.
Figure 4.
Data of real-time RT-PCR analysis of selected genes to examine specificity to nongenotoxic hepatocarcinogens in Experiment 2. A, Day 28. B, Day 90. Values are normalized to Actb or Hprt1 and expressed as the mean + SD (n = 6). *p < .05, **p < .01, significantly different from untreated controls by Tukey’s or Steel-Dwass test. †p < .05, ‡p < .01, significantly different from AFB1 group by Tukey’s or Steel-Dwass test. §p < .05, §§p < .01, significantly different from NPYR group by Tukey’s or Steel-Dwass test. ¶p < .05, ¶¶p < .01, significantly different from CRB group by Tukey’s or Steel-Dwass test.
On day 90, all genotoxic hepatocarcinogens showed significantly decreased transcript levels of Proc after normalization with Actb and Hprt1 or Actb-alone compared with untreated controls. AFB1 and NPYR groups showed significantly increased transcript levels of Ldlrad4 and Nfia after normalization with Actb and Hprt1 or Hprt1-alone compared with untreated controls. Both of nongenotoxic hepatocarcinogens showed significantly decreased transcript levels of Ldlrad4, Proc, and Nfia after normalization with Actb and Hprt1 compared with untreated controls. TAA group showed significantly decreased transcript levels of Cdh17 after normalization with Actb and Hprt1 compared with untreated controls. When the transcript level was compared between genotoxic and nongenotoxic hepatocarcinogens, both of nongenotoxic hepatocarcinogens showed significantly decreased transcript levels of Ldlrad4 and Proc after normalization with Actb and Hprt1 or Actb-alone compared with all genotoxic hepatocarcinogens. TAA group showed significantly decreased transcript levels of Cdh17 after normalization with Actb and Hprt1 compared with all genotoxic hepatocarcinogens. TAA group showed significantly decreased transcript levels of Nfia after normalization with Actb and Hprt1 compared with AFB1 and NPYR groups. MP group showed significantly decreased transcript levels of Cdh17 after normalization with Actb and Hprt1 or Hprt1-alone compared with AFB1 and NPYR groups. MP group showed significantly decreased transcript levels of Nfia after normalization with Actb and Hprt1 compared with all genotoxic hepatocarcinogens.
Measurement of Proliferative Lesions
In Experiments 1 and 2, there were no significant changes in the number and area of GST-P+ foci of all treatment groups compared with untreated controls after 28 days (Supplementary Tables 5 and 6). There were significantly more and larger GST-P+ foci compared with untreated controls after DEN, CCl4, AFB1, NPYR, TAA, and MP treatment for 84 or 90 days.
Distribution of Immunolocalized Cells
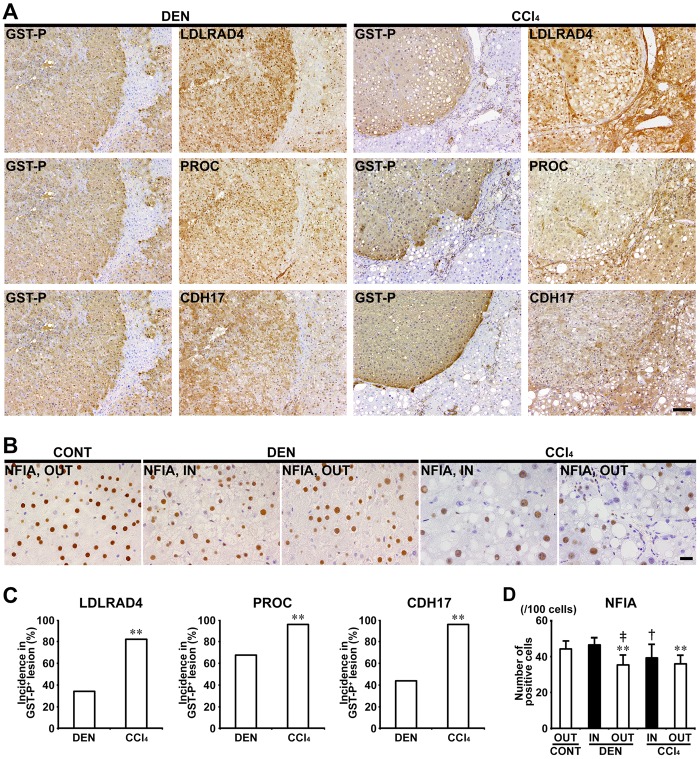
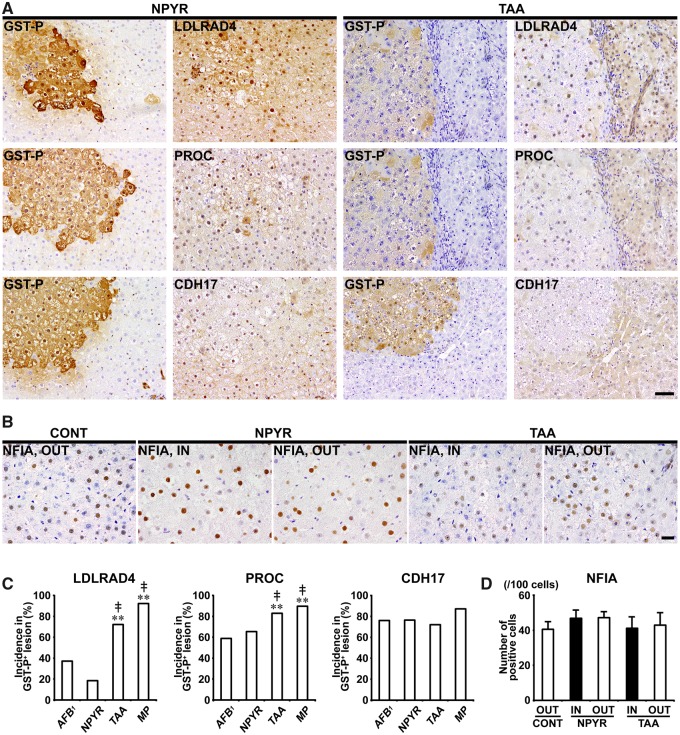
In Experiments 1 and 2, LDLRAD4, PROC, and CDH17 showed cytoplasmic expression in nonproliferative and proliferative liver cells. In both of DEN and CCl4 groups, GST-P+ foci showed either increased or decreased expression of these molecules. In CCl4 group, population of GST-P+ foci downregulating expression of LDLRAD4, PROC, and CDH17 was increased and incidences of LDLRAD4−, PROC−, and CDH17− foci in GST-P+ foci significantly increased compared with DEN group in Experiment 1 (Figs. 5A and 5C). In nongenotoxic hepatocarcinogens (TAA and MP), population of GST-P+ foci downregulating LDLRAD4 and PROC expression was increased and incidences of LDLRAD4− and PROC− foci in GST-P+ foci significantly increased compared with genotoxic hepatocarcinogens (AFB1 and NPYR) in Experiment 2 (Figs. 6A and 6C). On the other hand, population of GST-P+ foci downregulating CDH17 expression was observed in both genotoxic and nongenotoxic hepatocarcinogens, and there was no significant difference in incidence of CDH17− foci in GST-P+ foci between genotoxic and nongenotoxic hepatocarcinogens in Experiment 2 (Figs. 6A and 6C).
Figure 5.
Immunohistochemical cellular distribution of LDLRAD4, PROC, CDH17, and NFIA in association with GST-P+ liver cell foci after treatment with DEN or CCl4 for 84 days in Experiment 1. A, Expression of LDLRAD4, PROC and CDH17 in GST-P+ foci in DEN and CCl4 groups (×10 objective). Bar = 100 µm. B, Expression of NFIA of inside (IN) or outside (OUT) of GST-P+ foci (×40 objective). Bar = 20 µm. C, Incidences of LDLRAD4−, PROC−, and CDH17− foci in GST-P+ foci in DEN and CCl4 groups. Graphs show incidences (% value, mean + SD, n = 10) of negative foci of each molecule in GST-P+ foci in each group. **p < .01, significantly different from DEN group by Fisher’s exact test. D, Number of NFIA+ cells in IN or OUT of GST-P+ foci in DEN and CCl4 groups. Graph shows the number of NFIA+ cells (/100 cells) (value, mean + SD) IN or OUT of GST-P+ foci in each group. **p < .01, significantly different from OUT of untreated controls by Tukey’s or Steel-Dwass test. †p < .05, ‡p < .01, significantly different from IN of GST-P+ foci in DEN group by Tukey’s or Steel-Dwass test.
Figure 6.
Immunohistochemical cellular distribution of LDLRAD4, PROC, CDH17, and NFIA in association with GST-P+ liver cell foci after treatment with genotoxic (AFB1 or NPYR) or nongenotoxic hepatocarcinogens (TAA or MP) for 90 days in Experiment 2. A, Representative images in the expression of LDLRAD4, PROC, and CDH17 in GST-P+ foci in NPYR and TAA groups (×20 objective). Bar = 50 µm. B, Expression of NFIA of inside (IN) or outside (OUT) of GST-P+ foci in NPYR and TAA groups (×40 objective). Bar = 20 µm. C, Incidences of LDLRAD4−, PROC−, and CDH17− foci in GST-P+ foci in AFB1, NPYR, TAA and MP groups. Graphs show incidences (% value, mean + SD, n = 10) of negative foci of each molecule in GST-P+ foci in each group. **p < .01, significantly different from AFB1 group by Fisher’s exact test. ‡p < .01, significantly different from NPYR group by Fisher’s exact test. D, Number of NFIA+ cells in IN or OUT of GST-P+ foci in NPYR and TAA groups. Graphs show the number of NFIA+ cells (/100 cells) (value, mean + SD) IN or OUT of GST-P+ foci in each group.
In Experiments 1 and 2, NFIA showed immunolocalization in the nucleus of liver cells. In Experiment 1, the number of NFIA+ cells significantly decreased in liver cells distributed outside the GST-P+ foci in both DEN and CCl4 groups compared with untreated controls. Liver cells of the outside of GST-P+ foci in DEN group and inside of GST-P+ foci in CCl4 group, significantly decreased the number of nuclear NFIA+ expression compared with those distributed inside of GST-P+ foci in DEN group in Experiment 1 (Figs. 5B and 5D). In Experiment 2, no significant difference was observed in the number of NFIA+ liver cells between NPYR and TAA groups (Figs. 6B and 6D).
Transcript Expression Changes of Candidate Genes in Experiment 3
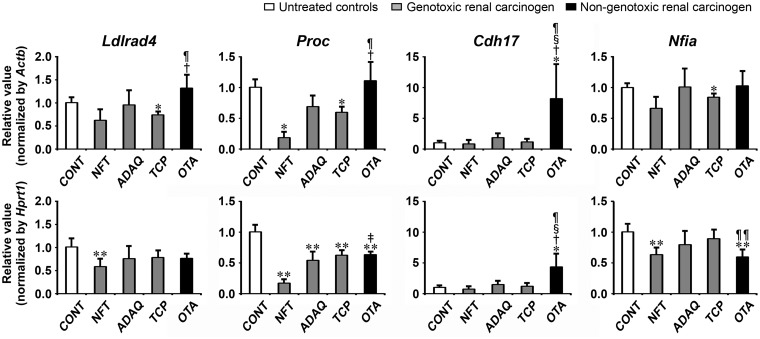
Transcript levels of the candidate genes, Ldlrad4, Proc, Cdh17, and Nfia, were determined by real-time RT-PCR in genotoxic renal carcinogens (NFT, ADAQ, and TCP) and nongenotoxic renal carcinogen (OTA) on day 28 of treatment (Figure 7). All genotoxic and nongenotoxic renal carcinogens showed significantly decreased transcript levels of Proc after normalization with Actb and Hprt1 or Hprt1-alone compared with untreated controls. NFT and TCP groups showed significantly decreased transcript levels of Ldlrad4 after normalization with Actb or Hprt1 compared with untreated controls. NFT, TCP, and OTA groups showed significantly decreased transcript levels of Nfia after normalization with Actb or Hprt1 compared with untreated controls. OTA group showed significantly increased transcript levels of Cdh17 after normalization with Actb and Hprt1 compared with untreated controls. When the transcript level was compared between genotoxic and nongenotoxic renal carcinogens, OTA group showed significantly decreased transcript levels of Nfia after normalization with Hprt1 compared with TCP group. OTA group showed significantly increased transcript levels of Ldlrad4 and Proc after normalization with Actb and Hprt1 or Actb-alone compared with NFT and TCP groups. OTA group showed significantly increased transcript levels of Cdh17 after normalization with Actb and Hprt1 compared with all genotoxic renal carcinogens.
Figure 7.
Data of real-time RT-PCR analysis of selected genes in the kidney in Experiment 3. Values are normalized to Actb or Hprt1 and expressed as the mean + SD (n = 6). *p < .05, **p < .01, significantly different from untreated controls by Tukey’s or Steel-Dwass test. †p < .05, ‡p < .01, significantly different from NFT group by Tukey’s or Steel-Dwass test. §p < .05, significantly different from ADAQ group by Tukey’s or Steel-Dwass test. ¶p < .05, ¶¶p < .01, significantly different from TCP group by Tukey’s or Steel-Dwass test.
DISCUSSION
In this study, 52 genes were found to be hypermethylated and downregulated specific to CCl4 by Methyl-Seq analysis combined with expression microarray analysis in the liver of rats treated with CCl4 or DEN for 28 days by excluding the genes hypermethylated and downregulated with DEN. Among them, 4 genes, Ldlrad4, Proc, Cdh17, and Nfia, were confirmed to be hypermethylated specific to CCl4 by methylation-specific PCR analysis on day 28. Furthermore, transcript downregulation of all of these genes was confirmed in many nongenotoxic hepatocarcinogens by real-time RT-PCR analysis at all time points examined. In contrast, genotoxic hepatocarcinogens did not consistently decrease transcript levels of these genes at any time point. In addition, nongenotoxic hepatocarcinogens showed lower transcript levels of these genes compared with genotoxic hepatocarcinogens. Immunohistochemically, LDLRAD4 and PROC showed decreased immunoreactivity, forming negative foci, in GST-P+ foci, and incidences of LDLRAD4− and PROC− foci in GST-P+ foci induced by treatment with nongenotoxic hepatocarcinogens for 84 or 90 days were increased compared with those with genotoxic hepatocarcinogens. CDH17 also showed a lack of expression in GST-P+ foci; however, the incidence of CDH17− foci in GST-P+ foci was high with both genotoxic and nongenotoxic hepatocarcinogens. With regard to NFIA, nuclear localization was found in the liver cells; however, temporal distribution of NFIA+ cells was not related to GST-P+ foci. None of the 4 genes responded to renal carcinogens after treatment for 28 days in rats. Thus, Ldlrad4, Proc, Cdh17, and Nfia may be potential in vivo epigenetic markers of nongenotoxic hepatocarcinogens from the early stages of treatment by means of transcript downregulation. LDLRAD4 and PROC may have a role in the development of preneoplastic lesions produced by nongenotoxic hepatocarcinogens.
Ldlrad4, a negative regulator of TGF-β signaling, plays a role in cell proliferation, differentiation, apoptosis, motility, extracellular matrix production, and immunosuppression (Nakano et al., 2014). It has been reported that LDLRAD4 promotes carcinogenesis of human hepatic cancer cells (Liu et al., 2017). However, TGF-β signaling regulated by Ldlrad4 has been considered as both a tumor-suppressor and a promoter of tumor progression (Derynck et al., 2001). In the process of liver cell carcinogenesis, overactivation of TGF-β signaling may contribute to tumor progression (Fabregat et al., 2016). We have previously reported the role of TGF-β signaling in rat hepatocarcinogenesis using an initiation promotion model and nongenotoxic hepatocarcinogens as tumor promoters (Ichimura et al., 2010). In that study, GST-P+ foci coexpressing TGF-β1, TGF-βR1, and phosphorylated and activated Smad3, a receptor-activated Smad (R-Smad), were increased by tumor promotion; however, Smad3 expression was localized only within the cytoplasm. Moreover, GST-P+ foci lacking Smad4 expression, a binding partner of R-Smad for nuclear translocation, were also increased by tumor promotion, suggesting an enhancement of disruptive TGF-β signaling during hepatocarcinogenesis. Therefore, our results may suggest that treatment with nongenotoxic hepatocarcinogens for 28 days induces epigenetic gene modification of Ldlrad4 in liver cells, which triggers the disruptive activation of TGF-β signaling for cancer development.
With regard to the possibility of the involvement of Ldlrad4 in cellular senescence, TGF-β induces the expression of p21WAF1/CIP1, a cyclin-dependent kinase inhibitor, by knockdown of Ldlrad4 (Nakano et al., 2014). It has been reported that p21WAF1/CIP1 plays an essential role in cell cycle arrest initiated by p53 activation in cellular senescence (Lujambio, 2016). Cellular senescent cells exist in premalignant tumors but not in malignant ones, suggesting that cellular senescence may help to restrict tumor progression (Collado et al., 2005). We previously found that liver cell tumor promotion using TAA as a nongenotoxic hepatocarcinogen for 4 weeks increased p21WAF1/CIP1+ liver cell foci in a 2-stage hepatocarcinogenesis model. However, this phenomenon was found to be transient and liver cell foci negative for p21WAF1/CIP1 were increased after 8 weeks of TAA promotion (Tsuchiya et al., 2012). This result may suggest disruption of cellular senescence during the early stage of hepatocarcinogenesis. Therefore, it may be possible that LDLRAD4 downregulation in nongenotoxic hepatocarcinogen-induced GST-P+ foci may play a role in the increase of cellular senescence-related molecules, such as p21WAF1/CIP1, during the early stage of hepatocarcinogenesis.
Proc, protein C encodes a vitamin K-dependent serine protease that regulates blood coagulation by inactivating factors Va and VIIIa in the presence of calcium ions and phospholipids (Kovács et al., 2015) and exerts a protective effect on the endothelial cell barrier function (Ding et al., 2015). The cytoprotective activities of activated protein C (APC) possess antiinflammatory, antiapoptotic, and cytoprotective activities mediated through engagement of its receptor, endothelial protein C receptor (EPCR; Bouwens et al., 2013; Esmon, 2006). In murine melanoma metastasis models, transgenic EPCR overexpressing mice exhibited marked reductions in liver and lung metastases compared with wild-type animals. These findings suggest a role of APC/EPCR pathway in reducing metastasis via inhibition of tumor cell adhesion and transmigration (Bezuhly et al., 2009). In addition, it is reported that APC induces enhanced expression of p21 WAF1/CIP1 and p27 KIP1 in rheumatoid synovial fibroblasts (Julovi et al., 2013). As aforementioned, p21WAF1/CIP1+ liver cell foci were increased after 4 weeks of TAA-promotion, and p21 WAF1/CIP1– liver cell foci were increased after 8 weeks of TAA-promotion (Tsuchiya et al., 2012), suggesting that APC plays a role in the facilitation of cellular senescence in liver cells, and its downregulation directs liver cells to begin the carcinogenic process. In addition, it is also reported that APC downregulates tumor necrosis factor α-stimulated cell proliferation and activation of p38 mitogen-activated protein kinase (MAPK), c-Jun NH2-terminal kinase and AKT in rheumatoid synovial fibroblasts (Julovi et al., 2013). We previously found an increase of liver cell foci expressing AKT-signaling molecules and activated p38 MAPK by phosphorylation in a population of GST-P+ foci produced by promotion with nongenotoxic hepatocarcinogens (Ichimura et al., 2010; Taniai et al., 2009). Therefore, our results may suggest that downregulation of Proc mRNA in liver cells by 28 days of nongenotoxic hepatocarcinogen treatment occurs through epigenetic gene modification that may inhibit the APC/EPCR pathway to downregulate p21 WAF1/CIP1 and upregulate AKT-signaling molecules and activated p38 MAPK for preneoplastic liver cell proliferation.
In this study, downregulation of mRNA levels was observed with Cdh17, a member of the nonclassical cadherin family (Jung et al., 2004) and Nfia, a member of the nuclear factor I transcription factors (Chen et al., 2017), in the liver of rats repeatedly treated with nongenotoxic hepatocarcinogens compared with untreated controls and genotoxic hepatocarcinogens. CDH17 is reportedly downregulated in human pancreatic carcinoma (Takamura et al., 2003) and knockdown of CDH17 in BGC823, a human gastric carcinoma cell line, induced cell cycle arrest (Liu et al., 2010). However, downregulation of CDH17 in GST-P+ foci was observed in all hepatocarcinogens without relation to genotoxic potential in this study. With regard to NFIA, copy number loss of Nfia has been reported in hepatocellular carcinoma, suggesting that NFIA has a tumor-suppressor role (Chen et al., 2017). However, NFIA+ cells were distributed without temporal relation to GST-P+ foci produced by treatment with hepatocarcinogens in this study. These findings may suggest that Cdh17 and Nfia play roles in creating a precarcinogenic environment in the early stages of treatment with nongenotoxic hepatocarcinogens. Of note, CDH17 may play another role in the hepatocarcinogenic process irrespective of the genotoxic potential of hepatocarcinogens.
In conclusion, we found 4 genes, ie, Ldlrad4, Proc, Cdh17, and Nfia, that show promoter-region hypermethylation and transcript downregulation in the liver after 28 days of CCl4 treatment in rats by excluding the genes hypermethylated and downregulated with DEN. Gene expression downregulation of Ldlrad4, Proc, Cdh17, and Nfia, and downregulation of LDLRAD4 and PROC in GST-P+ lesions were common among the nongenotoxic hepatocarcinogens examined. Thus, Ldlrad4, Proc, Cdh17, and Nfia may be potential in vivo epigenetic markers of nongenotoxic hepatocarcinogens from the early stages of treatment. LDLRAD4 and PROC may have a role in the development of preneoplastic lesions produced by nongenotoxic hepatocarcinogens.
DECLARATION OF CONFLICTING INTERESTS
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
FUNDING
This work was supported by a Research Fund from the Chemicals Evaluation and Research Institute, Japan, and by a Research Fund from Institute of Global Innovation Research, Tokyo University of Agriculture and Technology.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Shigeko Suzuki for her technical assistance in preparing the histological specimens.
REFERENCES
- AOAC. (2005). Preparation of standards for aflatoxins, Official Method 970.44 In Official Methods of Analysis of AOAC International (Horwitz W., G. W. Latimer Jr., Eds.), 18th ed, Chapter 49, pp. 47–53. AOAC International Press, Gaithersburg. [Google Scholar]
- Barber E. D., Donish W. H., Mueller K. R. (1981). A procedure for the quantitative measurement of the mutagenicity of volatile liquids in the Ames Salmonella/microsome assay. Mutat. Res. 90, 31–48. [DOI] [PubMed] [Google Scholar]
- Becker F. F. (1983). Thioacetamide hepatocarcinogenesis. J. Natl. Cancer Inst. 71, 553–558. [PubMed] [Google Scholar]
- Bezuhly M., Cullen R., Esmon C. T., Morris S. F., West K. A., Johnston B., Liwski R. S. (2009). Role of activated protein C and its receptor in inhibition of tumor metastasis. Blood 113, 3371–3374. [DOI] [PubMed] [Google Scholar]
- Bouwens E. A., Stavenuiter F., Mosnier L. O. (2013). Mechanisms of anticoagulant and cytoprotective actions of the protein C pathway. J. Thromb. Haemost. 11(Suppl. 1), 242–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandeis M., Ariel M., Cedar H. (1993). Dynamics of DNA methylation during development. Bioessays 15, 709–713. [DOI] [PubMed] [Google Scholar]
- Campisi J. (2013). Aging, cellular senescence, and cancer. Annu. Rev. Physiol. 75, 685–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K. S., Lim J. W. C., Richards L. J., Bunt J. (2017). The convergent roles of the nuclear factor I transcription factors in development and cancer. Cancer Lett. 410, 124–138. [DOI] [PubMed] [Google Scholar]
- Collado M., Gil J., Efeyan A., Guerra C., Schuhmacher A. J., Barradas M., Benguría A., Zaballos A., Flores J. M., Barbacid M., et al. (2005). Tumour biology: Senescence in premalignant tumours. Nature 436, 642.. [DOI] [PubMed] [Google Scholar]
- De Jesus A. E., Gorst-Allman C. P., Horak R. M., Vleggaar R. (1988). Large-scale purification of the mycotoxins aflatoxin B1, B2 and G1. J. Chromatogr. 450, 101–104. [DOI] [PubMed] [Google Scholar]
- Derynck R., Akhurst R. J., Balmain A. (2001). TGF-β signaling in tumor suppression and cancer progression. Nat. Genet. 29, 117.. [DOI] [PubMed] [Google Scholar]
- Ding Q., Yang L., Dinarvand P., Wang X., Rezaie A. R. (2015). Protein C Thr315Ala variant results in gain of function but manifests as type II deficiency in diagnostic assays. Blood 125, 2428–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esmon C. T. (2006). The endothelial protein C receptor. Curr. Opin. Hematol. 13, 382–385. [DOI] [PubMed] [Google Scholar]
- Fabregat I., Moreno-Càceres J., Sánchez A., Dooley S., Dewidar B., Giannelli G., Ten Dijke P; on behalf of IT-LIVER Consortium. (2016). TGF-β signalling and liver disease. FEBS J. 283, 2219–2232. [DOI] [PubMed] [Google Scholar]
- FSCJ Food Safety Commission of Japan. (2015). Ochratoxin A: Executive Summary. 3, 62–64. [Google Scholar]
- Holliday R. (1989). DNA methylation and epigenetic mechanisms. Cell Biophys. 15, 15–20. [DOI] [PubMed] [Google Scholar]
- IARC Monographs on the Evaluation of Carcinogenic Risk of Chemicals to Humans. (1978). N-Nitrosodiethylamine. In Some N-nitroso Compounds, Vol. 17, pp. 83–124. IARC, Lyon, France. [PubMed] [Google Scholar]
- Ichimura R., Mizukami S., Takahashi M., Taniai E., Kemmochi S., Mitsumori K., Shibutani M. (2010). Disruption of Smad-dependent signaling for growth of GST-P-positive lesions from the early stage in a rat two-stage hepatocarcinogenesis model. Toxicol. Appl. Pharmacol. 246, 128–140. [DOI] [PubMed] [Google Scholar]
- Ito N., Imaida K., Tamano S., Hagiwara A., Shirai T. (1998). Medium-term bioassays as alternative carcinogenicity test. J. Toxicol. Sci. 23(Suppl. 2), 103–106. [DOI] [PubMed] [Google Scholar]
- Jones P. A., Baylin S. B. (2007). The epigenomics of cancer. Cell 128, 683–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julovi S. M., Shen K., Mckelvey K., Minhas N., March L., Jackson C. J. (2013). Activated protein C inhibits proliferation and tumor necrosis factor α-stimulated activation of p38, c-Jun NH2-terminal kinase (JNK) and Akt in rheumatoid synovial fibroblasts. Mol. Med. 19, 324–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung R., Wendeler M. W., Danevad M., Himmelbauer H., Gessner R. (2004). Phylogenetic origin of LI-cadherin revealed by protein and gene structure analysis. Cell. Mol. Life Sci. 61, 1157–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki K., Nishikawa A., Masumura K., Umemura T., Imazawa T., Kitamura Y., Nohmi T., Hirose M. (2005). In vivo mutational analysis of liver DNA in gpt delta transgenic rats treated with the hepatocarcinogens N-nitrosopyrrolidine, 2-amino-3-methylimidazo [4, 5-f] quinoline, and di(2-ethylhexyl)phthalate. Mol. Carcinog. 42, 9–17. [DOI] [PubMed] [Google Scholar]
- Kimura M., Abe H., Mizukami S., Tanaka T., Itahashi M., Onda N., Yoshida T., Shibutani M. (2016). Onset of hepatocarcinogen-specific cell proliferation and cell cycle aberration during the early stage of repeated hepatocarcinogen administration in rats. J. Appl. Toxicol. 36, 223–237. [DOI] [PubMed] [Google Scholar]
- Kimura M., Mizukami S., Watanabe Y., Hasegawa-Baba Y., Onda N., Yoshida T., Shibutani M. (2015). Disruption of spindle checkpoint function ahead of facilitation of cell proliferation by repeated administration of hepatocarcinogens in rats. J. Toxicol. Sci. 40, 855–871. [DOI] [PubMed] [Google Scholar]
- King T. O. (1976). Target organ toxicity of GS-6244 (carbadox) and CP-17, 056 (desoxycarbadox) with chronic administration in rats. Submitted to WHO by Pfizer Central Research, Groton, CT. Unpublished.
- Kovács K. B., Pataki I., Bárdos H., Fekete A., Pfliegler G., Haramura G., Gindele R., Komáromi I., Balla G., Ádány R., et al. (2015). Molecular characterization of p.Asp77Gly and the novel p.Ala163Val and p.Ala163Glu mutations causing protein C deficiency. Thromb. Res. 135, 718–726. [DOI] [PubMed] [Google Scholar]
- Liu Q. S., Zhang J., Liu M., Dong W. G. (2010). Lentiviral-mediated miRNA against liver-intestine cadherin suppresses tumor growth and invasiveness of human gastric cancer. Cancer Sci. 101, 1807–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Huo X., Zhao S., Yang J., Shi W., Jing L., Li W., Li Y., Ma L., Gao Y., et al. (2017). Low density lipoprotein receptor class A domain containing 4 (LDLRAD4) promotes tumorigenesis of hepatic cancer cells. Exp. Cell Res. 360, 189–198. [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lujambio A. (2016). To clear, or not to clear (senescent cells)? That is the question. Bioessays 38 (Suppl. 1), S56–S64. [DOI] [PubMed] [Google Scholar]
- Mizukami S., Yafune A., Watanabe Y., Nakajima K., Jin M., Yoshida T., Shibutani M. (2017). Identification of epigenetically downregulated Tmem70 and Ube2e2 in rat liver after 28-day treatment with hepatocarcinogenic thioacetamide showing gene product downregulation in hepatocellular preneoplastic and neoplastic lesions produced by tumor promotion. Toxicol. Lett. 266, 13–22. [DOI] [PubMed] [Google Scholar]
- Nakano N., Maeyama K., Sakata N., Itoh F., Akatsu R., Nakata M., Katsu Y., Ikeno S., Togawa Y., Vo. Nguyen T. T., et al. (2014). C18ORF1, a novel negative regulator of transforming growth factor-β signaling. J. Biol. Chem. 289, 12680–12692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NTP (2000). NTP hepatotoxicity studies of the liver carcinogen methapyrilene hydrochloride (CAS no. 135-23-9) administered in feed to male F344/N rats. Toxic. Rep. Ser. 46, 1–C7. [PubMed] [Google Scholar]
- NTP (1989a). NTP Toxicology and carcinogenesis studies of nitrofurantoin (CAS No. 67-20-9) in F344/N rats and B6C3F1 mice (Feed Studies). Natl. Toxicol. Program Tech. Rep. Ser. 341, 1–218. [PubMed] [Google Scholar]
- NTP (1993). NTP Toxicology and carcinogenesis of 1, 2, 3-trichloropropane (CAS No. 96-18-4) in F344/N rats and B6C3F1 mice (Gavage studies). Natl. Toxicol. Program Tech. Rep. Ser. 384, 1–348. [PubMed] [Google Scholar]
- NTP (1996). NTP Toxicology and carcinogenesis of 1-amino-2, 4-dibromoantraquinone (CAS No. 81-49-2) in F344/N rats and B6C3F1 mice (Feed studies). Natl. Toxicol. Program Tech. Rep. Ser. 383, 1–370. [PubMed] [Google Scholar]
- NTP (1989b). NTP Toxicology and carcinogenesis studies of ochratoxin A (CAS No. 303-47-9) in F344/N rats (Gavage studies). Natl. Toxicol. Program Tech. Rep. Ser. 358, 1–142. [PubMed] [Google Scholar]
- Palacios D., Summerbell D., Rigby P. W., Boyes J. (2010). Interplay between DNA methylation and transcription factor availability: Implications for developmental activation of the mouse Myogenin gene. Mol. Cell. Biol. 30, 3805–3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian G., Wang F., Tang L., Massey M. E., Mitchell N. J., Su J., Williams J. H., Phillips T. D., Wang J. S. (2013). Integrative toxicopathological evaluation of aflatoxin B1 exposure in F344 rats. Toxicol. Pathol. 41, 1093–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson K. D. (2005). DNA methylation and human disease. Nat. Rev. Genet. 6, 597–610. [DOI] [PubMed] [Google Scholar]
- Shirai T. (1997). A medium-term rat liver bioassay as a rapid in vivo test for carcinogenic potential: A historical review of model development and summary of results from 291 tests. Toxicol. Pathol. 25, 453–460. [DOI] [PubMed] [Google Scholar]
- Suzuki S., Takeshita K., Asamoto M., Takahashi S., Kandori H., Tsujimura K., Saito F., Masuko K., Shirai T. (2009). High mobility group box associated with cell proliferation appears to play an important role in hepatocellular carcinogenesis in rats and humans. Toxicology 255, 160–170. [DOI] [PubMed] [Google Scholar]
- Takamura M., Sakamoto M., Ino Y., Shimamura T., Ichida T., Asakura H., Hirohashi S. (2003). Expression of liver-intestine cadherin and its possible interaction with galectin-3 in ductal adenocarcinoma of the pancreas. Cancer Sci. 94, 425–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniai E., Yafune A., Hayashi H., Itahashi M., Hara-Kudo Y., Suzuki K., Mitsumori K., Shibutani M. (2012). Aberrant activation of ubiquitin D at G2 phase and apoptosis by carcinogens that evoke cell proliferation after 28-day administration in rats. J. Toxicol. Sci. 37, 1093–1111. [DOI] [PubMed] [Google Scholar]
- Taniai E., Kawai M., Dewa Y., Nishimura J., Harada T., Saegusa Y., Matsumoto S., Takahashi M., Mitsumori K., Shibutani M. (2009). Crosstalk between PTEN/Akt2 and TGFβ signaling involving EGF receptor down-regulation during the tumor promotion process from the early stage in a rat two-stage hepatocarcinogenesis model. Cancer Sci. 100, 813–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya T., Wang L., Yafune A., Kimura M., Ohishi T., Suzuki K., Mitsumori K., Shibutani M. (2012). Disruptive cell cycle regulation involving epigenetic downregulation of Cdkn2a (p16Ink4a) in early-stage liver tumor-promotion facilitating liver cell regeneration in rats. Toxicology 299, 146–154. [DOI] [PubMed] [Google Scholar]
- Verna L., Whysner J., Williams G. M. (1996). N-nitrosodiethylamine mechanistic data and risk assessment: Bioactivation, DNA-adduct formation, mutagenicity, and tumor initiation. Pharmacol. Ther. 71, 57–81. [DOI] [PubMed] [Google Scholar]
- Weinhold B. (2006). Epigenetics: The science of change. Environ. Health. Perspect. 114, A160–A167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisburger E. K. (1977). Carcinogenicity studies on halogenated hydrocarbons. Environ. Health Perspect. 21, 7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yafune A., Taniai E., Morita R., Hayashi H., Suzuki K., Mitsumori K., Shibutani M. (2013). Aberrant activation of M phase proteins by cell proliferation-evoking carcinogens after 28-day administration in rats. Toxicol. Lett. 219, 203–210. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.