Abstract
Arsenic exposure is a worldwide health concern associated with an increased risk of skin, lung, and bladder cancer but arsenic trioxide (AsIII) is also an effective chemotherapeutic agent. The current use of AsIII in chemotherapy is limited to acute promyelocytic leukemia (APL). However, AsIII was suggested as a potential therapy for other cancer types including chronic myeloid leukemia (CML), especially when combined with other drugs. Here, we carried out a genome-wide CRISPR-based approach to identify modulators of AsIII toxicity in K562, a human CML cell line. We found that disruption of KEAP1, the inhibitory partner of the key antioxidant transcription factor Nrf2, or TXNDC17, a thioredoxin-like protein, markedly increased AsIII tolerance. Loss of the water channel AQP3, the zinc transporter ZNT1 and its regulator MTF1 also enhanced tolerance to AsIII whereas loss of the multidrug resistance protein ABCC1 increased sensitivity to AsIII. Remarkably, disruption of any of multiple genes, EEFSEC, SECISBP2, SEPHS2, SEPSECS, and PSTK, encoding proteins involved in selenocysteine metabolism increased resistance to AsIII. Our data suggest a model in which an intracellular interaction between selenium and AsIII may impact intracellular AsIII levels and toxicity. Together this work revealed a suite of cellular components/processes which modulate the toxicity of AsIII in CML cells. Targeting such processes simultaneously with AsIII treatment could potentiate AsIII in CML therapy.
Keywords: arsenic, selenium, CRISPR screen, selenocysteine
Arsenic toxicity is a worldwide public health concern as millions of people are exposed to this metalloid in drinking water (Ravenscroft, 2007) at levels above the 10 μg/l guideline of the World Health Organization (WHO, 1996). In the United States, arsenic is ranked first in the Priority List of Hazardous Substances (ATSDR, 2017). Acute arsenic poisoning can result in severe digestive and neurologic effects and ultimately death while chronic arsenic exposure increases the risk of skin, lung, and bladder cancers (IARC, 2004; Ratnaike, 2003). However, arsenic trioxide (As2O3/ATO; AsIII) has been historically used as a traditional Chinese medicine and was found to be a highly effective treatment of acute promyelocytic leukemia (APL) (Mathews et al., 2006, 2010; Shen et al., 1997; Zhou et al., 2010). In combination with all-trans retinoic acid, AsIII results in remission rates approaching 96% in APL patients (Abaza et al., 2017; Lo-Coco et al., 2016). The major therapeutic effect of AsIII involves degradation of the fusion oncoprotein promyelocytic leukemia (PML)-retinoic acid receptor-α (RARα) (Zhang et al., 2010) and subsequent induction of partial differentiation and apoptosis in APL cells (Emadi and Gore, 2010). AsIII-induced cytotoxic effects that are independent of PML-RARα degradation also exist in APL cells and likely contribute to the observed substantial efficacy of AsIII (Davison et al., 2002; Yedjou et al., 2010).
Despite its exclusive use in APL therapy, AsIII demonstrated a promising therapeutic potential in multiple types of hematologic malignancies (Berenson et al., 2006; Kchour et al., 2009; Wang et al., 2015; Zheng et al., 2008). The cytotoxic effects of AsIII have been studied in various non-APL cancer cells (Akao et al., 1999; Rousselot et al., 1999; Shen et al., 1999; Zhang et al., 1998). The mechanisms of AsIII toxicity remain unclear although a variety of mechanisms that likely vary depending on the cell type have been proposed (Miller et al., 2002). Inside the cell, inorganic AsIII (iAsIII) is typically metabolized to methylated forms including monomethylarsonic acid (MMAV), dimethylarsenic acid (DMAV), monomethylarsonous acid (MMAIII), and dimethylarsinous acid (DMAIII) (Hopenhayn-Rich et al., 1993; Vahter, 2002). MMAIII is likely the primary toxic species (Mass et al., 2001; Styblo et al., 2002) although the other forms may have direct effects. Cytotoxic effects of AsIII are mainly exerted through its reaction with the thiol groups of a variety of intracellular proteins (Watson, 2015) and the diversity of AsIII targets likely underlies the complex nature of its toxicity.
In this study, we employed a genome-wide CRISPR/Cas9 loss-of-function screen to identify genes whose disruption alters sensitivity to AsIII in the erythroleukemic K562 cell line which does not contain the PML-RARα rearrangement. Our study thus focuses on cellular mechanisms of AsIII toxicity independent of PML-RARα degradation. We confirmed the existing pathways that are involved in AsIII toxicity and identified novel pathways that influence sensitivity to AsIII. This work provided new insights into the molecular mechanisms which affect cellular susceptibility to iAsIII.
MATERIALS AND METHODS
Cell culture
Human HEK293T and K562 cells lines were obtained from the biosciences divisional services cell culture facility, UC Berkeley (https://bds.berkeley.edu/facilities/cell-culture; Accessed February 7, 2019). HEK293T cells were cultured in Dulbecco's modified Eagles medium (DMEM; Thermo Fisher) supplemented with 10% fetal bovine serum (FBS; Corning) and 1% penicillin/streptomycin (PS; Thermo Fisher). K562 cells were cultured in RPMI 1640 (Thermo Fisher) supplemented with 10% FBS and 1% PS. Cells were cultured in a humidified incubator with 5% CO2 at 37°C.
Cytotoxicity assays
Cell viability assays were performed using the CellTiter-Glo luminescent cell viability assay kit (Promega) following the manufacturer’s protocol. In brief, cells were seeded at a density of 105 cells/ml (104 cells/well) in opaque 96-well cell culture plates. ATO/As2O3 (STREM Chemicals Inc.) stock of 10 mM was prepared by dissolving 20 mg of As2O3 in 300 μl NaOH (1 M) and completing the volume to 10 ml with PBS. Cells were treated with 0, 0.5, 1, 1.5, and 2 µM ATO for 24, 48, 72, or 96 h as indicated. For experiments involving selenium pretreatment, cells were pretreated with 10 µM sodium selenite (Na2SeO3; MP Biomedicals) for 24 h, washed with PBS, seeded in new plates and treated with 0, 0.5, 1, 1.5, and 2 µM ATO for 48 h. At the end of each incubation period, 100 µl of the CellTiter Glo reagent was added to each well and the cells were lysed at an orbital plate shaker for 2 min. The plate was then incubated for 10 min at room temperature in the dark to stabilize the signal. Luminescent signals from all wells were read on a Synergy H1 microplate reader (BioTek Instruments).
Genome-wide and targeted CRISPR/Cas9 libraries
The human genome-wide CRISPR knockout (GeCKO) version 2 sgRNA library cloned in LentiCRISPR v2 vector (Addgene no. 1000000048, kindly deposited by Dr Feng Zhang) was used for genome-wide screening. The GeCKO v2 library targets 19 050 protein-coding genes and 1864 miRNAs and contains 1000 nontargeting sgRNAs with a total of 123 411 sgRNAs split into 2 half libraries A and B (Sanjana et al., 2014). For primary screening, we used half library A containing 65 383 sgRNAs with an average of 3 sgRNAs targeting each gene. For the focused (validation) library, sgRNA designs targeting each selected gene were picked up from the GeCKO v2 (half-libraries A and B) and the Brunello library (Doench et al., 2016). The validation library targets 307 genes (6–8 sgRNAs/gene) and contains 500 nontargeting sgRNAs, with a total of 2784 sgRNAs. Pooled custom oligonucleotides (79 bp) comprised of the 20 bp sgRNA sequence and the appropriate upstream (5’-cttGTGGAAAGGACGAAACACCg-3’) and downstream (5’-gttttagagctaGAAAtagcaagttaaaataaggct-3’) flanking sequences were synthesized by CustomArray pooled oligo synthesis service (CustomArray Inc., Bothell, Waltham). The obtained full-size oligos were PCR -amplified, gel-purified and cloned into the LentiCRISPR v2 vector (Addgene no. 52961) using Gibson assembly as previously described (Shalem et al., 2014). Both GeCKO v2 and validation libraries were transformed into Endura electrocompetent cells (Lucigen) using previously described protocols for library amplification (Sanjana et al., 2014; Shalem et al., 2014). Transformation efficiency for each library ensured sufficient representation of all constructs (approximately 150-fold library size for GeCKO v2 and approximately 500-fold library size for validation library). Plasmid DNA was isolated from amplified colonies using the Maxiprep plasmid DNA purification kit (Qiagen).
Lentiviral production and functional titration
Lentivirus production was performed as previously described (Shalem et al., 2014), with minor modifications. Briefly, HEK293T cells cultured in a T225 flasks were co-transfected with 20 µg of the plasmid library, 15 µg of the packaging plasmid (psPAX2, Addgene no. 12260) and 10 µg of the envelope plasmid (pMD2.G, Addgene no. 12259). Media containing the virus were collected 60-h posttransfection and filtered through a Steriflip-HV 0.45 µm low protein-binding PVDF membrane (Millipore). The lentiviral supernatant was concentrated 50-fold using Lenti-X Concentrator (Clontech) following the manufacturer’s protocol. Viral solutions were aliquoted and stored at −80° C until further use. To perform functional titration of the prepared viral solutions, K562 cells were suspended in transduction medium (RPMI 1640, 10% FBS, 1% PS + 8 μg/ml polybrene) and seeded at a density of 1.25 × 106 cells/ml in 12-well plates (2.5 × 106 cells per well). Different volumes (0, 2.5, 5, 10, 15, and 20 μl) of the virus were mixed with the cell suspension in each well and the plates were centrifuged at 1000 g for 2 h at 33° C. Transduced cells from each well were suspended in fresh media and recovered for 48 h. For each transduction volume, cells were seeded in 96-well plate at a density of 105 cells/ml (104 cells/well; 100 μl) with or without puromycin (2 μg/ml) and maintained for 7 days during which 25 μl of cell suspension from each well were added to 75 μl of fresh media in a new replica plate every 48 h. Following puromycin selection, cell viability in each condition was evaluated by CellTiter Glo and the multiplicity of infection (MOI) corresponding to each transduction volume was calculated by dividing the average luminescence signal from wells with puromycin by the average luminescence signal from wells without puromycin. A transduction volume corresponding to a MOI of 0.25–0.5 was used in the large-scale transduction.
Genome-wide (primary) screening
GeCKO v2 library, packaged in lentiviral particles was transduced into 100 × 106 K562 cells in 12-well plates using the same protocol described for viral titration. Each well, containing 2.5 × 106 cells, was transduced with 10 μl of the GeCKO v2 virus which results in a MOI of 0.45 (determined from titration). Transduced cells from all wells were pooled and the non-infected cells were eliminated by puromycin selection (2 μg/ml) for 7 days during which the infected cells were expanded. The obtained mutant library was split into treatment (1 μM ATO) and control (NaOH vehicle) conditions where at least 25 × 106 cells were maintained resulting in a representation of approximately 400-fold the library size. Selection was applied for 7 days which correspond to approximately 7 K562 cell doublings and the media were replaced every 48 h. Screens were performed in T225 cell culture flasks and each condition was run in duplicate. At the end of the screen, 25 × 106 cells from each replicate were washed with PBS and the pellets were saved for DNA extraction.
Validation (secondary) screening
10 × 106 K562 cells were transduced with the validation library in a 12-well plate using the same described protocol. Each well, containing 2.5 × 106 cells, was transduced with 2.5 μl of the validation library virus which results in a MOI of 0.27 (determined from titration). The same screening conditions described for the primary screen were applied for the validation screen. At least 2.5 × 106 cells were maintained in each condition representing approximately 900-fold the library size. Screens were performed in T25 cell culture flasks and each condition was run in triplicate. At the end of the screen, 2.5 × 106 cells from each replicate were washed with PBS and the pellets were saved for DNA extraction.
DNA extraction, library preparation and next generation sequencing
Genomic DNA was isolated from 25 × 106 cells (primary screen) using the Blood and Cell Culture DNA Midi kit (Qiagen) or 2.5 × 106 cells (secondary screen) using the DNeasy Blood and Tissue kit (Qiagen) following the manufacturer’s protocols. Library preparation for next generation sequencing was performed as previously described (Sanjana et al., 2014), with minor modifications. For each sample, the pool of guide sequences was amplified from genomic DNA by high fidelity PCR using the Herculase II Fusion DNA Polymerase kit (Agilent). For the genome-wide screen, 150 μg genomic DNA were amplified for each sample (10 µg genomic DNA/reaction; 15 reactions/sample). For the secondary screen, 15 μg genomic DNA were amplified for each sample (5 µg genomic DNA/reaction; 3 reactions/sample). In addition to the appropriate amount of genomic DNA template, each PCR reaction contained 20 μl of the 5× reaction buffer, 500 nM of each of the forward and reverse primers, 1 mM deoxyribonucleoside triphosphates (dNTPs), 1 μl polymerase and an appropriate volume of water to reach a final volume of 100 μl. PCR was conducted with the following conditions: 95°C/2 min; 18 cycles of 95°C/20 s, 60°C/20 s, 72°C/30 s; followed by 72°C/3 min. Following amplification, reactions corresponding to the same sample were pooled. To prepare the samples for next generation sequencing, the obtained amplicons were further amplified using primers that include appropriate P5 and P7 illumina adapter sequences. In order to increase the diversity of the libraries, the forward primer used in the second PCR included a 5 N shuffle sequence. To allow multiplexing of samples, multiple reverse primers were used in the second PCR and each primer contained a unique 8 bp index that was used to label each sample. For each PCR2 reaction, 5 μl of the first PCR product were used as a template. Seven PCR2 reactions per sample were performed for the genome-wide screen while a single PCR2 reaction was performed for each sample of the validation screen. PCR2 conditions and amplification protocol were similar to those used for PCR1 but 20 amplification cycles were applied instead of 18. For the genome-wide screen, PCR2 reactions corresponding to the same sample were pooled. Primers used in the first and second PCRs are shown in Supplementary Table 1. The quality of the 358 bp PCR2 amplicon was assessed on a 2% agarose gel and then using a 2100 bioanalyzer (Agilent). If necessary, unincorporated primers and nonspecific products were removed from each sample using pippin prep (Sage Science). Following purification, individual samples labeled with different indices were quantified on a Qubit fluorometer (Thermo Fisher Scientific) and pooled in equimolar amounts. Pooled libraries were deep sequenced using the illumina Hiseq2500 platform (single read 50 bp) with a coverage exceeding 500 folds the size of each library.
Data processing and computational analysis
Raw FASTQ files were demultiplexed using the FASTX-Toolkit (http://hannonlab. cshl.edu/fastx_toolkit/; Accessed February 7, 2019) and processed to contain only the unique 20-bp guide sequences. To align the processed reads with the reference library, guide sequences from the library were assembled into a Burrows-Wheeler index (Li and Durbin, 2009) using the Bowtie build-index function (Langmead et al., 2009). Reads were aligned using the Bowtie aligner and the number of uniquely aligned reads (perfect match) for each guide was calculated. Individual guide counts or the sum of counts of all guides targeting each gene were used as an input into edgeR, where the counts were normalized using the upper-quartile method. Differential abundance of each guide or all guides targeting a gene between treatment and control conditions was determined using the negative binomial generalized linear model (GLM) approach implemented in edgeR (Lun et al., 2016). False discovery rates (FDRs) were estimated to correct for multiple comparisons. Primary candidate selection was based on individual guide sequences displaying differential representation between the 2 conditions with FDR < 0.1. Additional genes were identified by performing a gene-level statistical analysis, based on summing counts from all the guide sequences targeting a given gene. Validation of candidates by secondary screening was based on revealing multiple individual guide sequences per gene that are differentially represented between the 2 conditions with FDR < 0.001. A complementary gene-level confirmatory analysis was performed by fitting a separate linear model to the total sgRNA counts for each gene using the method of limma (Ritchie et al., 2015), alongside the voom transform (Law et al., 2014), implemented in the popular limma R package. Alternatively, gene ranking was performed using Model-based Analysis of Genome-wide CRISPR-Cas9 Knockout (MaGeCK), which combines ranks of individual guides targeting the same gene using a modified version of robust ranking aggregation (Li et al., 2014). Functional enrichments within the list of candidate genes were determined by gene ontology enrichment analysis implemented in the STRING database (http://string-db.org; Accessed February 7, 2019) complemented with literature-based manual curation.
Generation of PSTK knockout and control pools
sgRNAs targeting PSTK or a nontargeting (NTC) sgRNA (sequences shown in Supplementary Table 2) were cloned into the CRISPR lentiviral backbone vector (LentiCRISPR v2) using the Golden Gate method. To produce lentiviral particles, we cotransfected HEK293T cells in T25 culture flasks with 3.4 µg of the target vector, 2.6 µg of the packaging plasmid (psPAX2) and 1.7 µg of the envelope plasmid (pMD2.G) using Lipofectamine 2000 and Plus reagent (Thermo Fisher Scientific) following the manufacturer’s instructions. Viral solutions were collected 60 h posttransfection, filtered through a Steriflip-HV 0.45 µm low protein-binding PVDF membrane (Millipore), and concentrated 50 folds using Lenti-X Concentrator (Clontech) following the manufacturer’s protocol. Cells were transduced with targeting or NTC vectors at a MOI < 0.5 and the transduced cells were enriched by puromycin selection. The obtained cellular pools were used in cytotoxicity assays to evaluate their sensitivity to ATO.
Statistical analysis
Data were presented as the mean ± SD. Statistical significance was determined by 2-tailed, 2-sample t test assuming unequal variance. A p-value < .05 was considered statistically significant.
RESULTS
Genome-Wide Screen Reveals Genes Affecting Cellular Sensitivity to AsIII
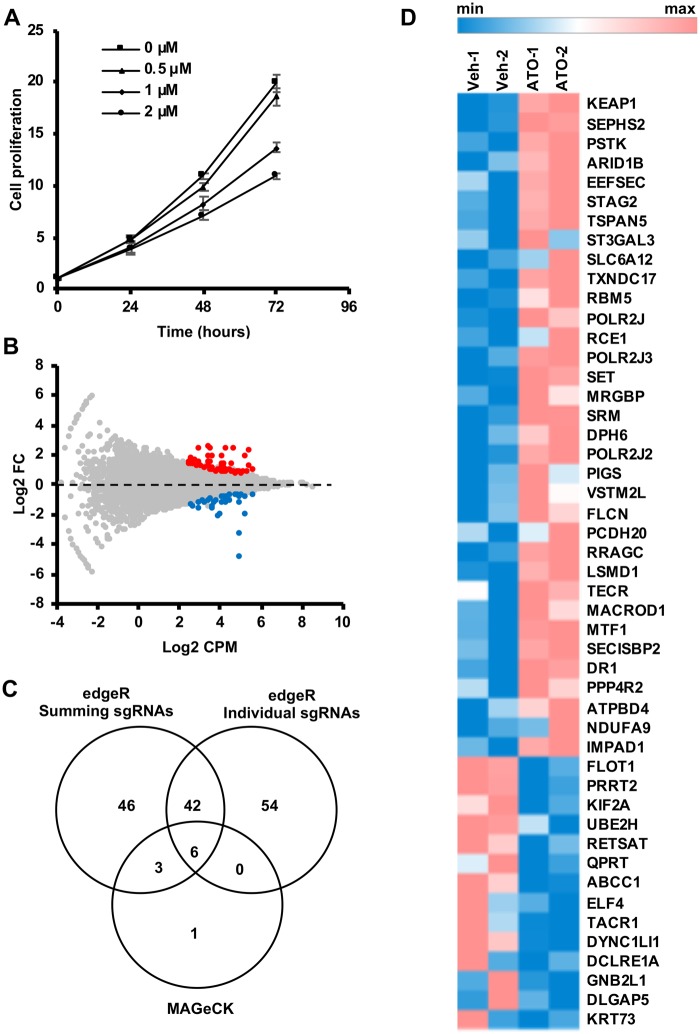
We investigated cellular components of key mechanistic relevance to AsIII toxicity in K562 cells using the CRISPR-Cas9 genome-wide loss-of-function screening approach (Figure 1). We used the GeCKO v2 sgRNA library A containing 65 383 sgRNAs targeting approximately 19 000 human genes to generate a pool of K562 cells with mutations in the corresponding genes. We screened the cellular pool for mutations that alter sensitivity to a sublethal AsIII dose (1 µM of ATO), inhibiting the proliferation of K562 cells by approximately 30% after 72 h, that we determined by cell viability assays (Figure 2A). At the end of the screen, differential growth of mutants between ATO treated and control (vehicle treated) pools was determined by comparing the representation of the corresponding guide sequences between the 2 pools. We selected candidate genes for further evaluation if at least one guide sequence targeting the gene demonstrated differential abundance between ATO and vehicle control at a FDR < 0.1 with average log2 CPM > 2.5 (Figure 2B). Using these criteria, we identified 102 candidate genes implicated in the toxic response to AsIII (Supplementary Table 3). To maximize the number of candidates from the primary screen, we implemented additional analysis methods for candidate selection. An analysis approach which involves summing all guide sequences targeting the same gene identified 97 candidate genes with FDR < 0.1 (Supplementary Table 4). Among these, 49 candidates did not overlap with the list identified from the analysis approach based on individual sgRNAs (Figure 2C). Within the overlapping candidates, there were 34 genes whose disruption potentially confers resistance to AsIII and 14 genes whose loss potentially increases cellular sensitivity to AsIII (Figure 2D). A third, more stringent, analysis method using MAGeCK identified only 10 candidate genes with FDR < 0.1 (Supplementary Table 5), 9 of which were already identified by the other 2 approaches (Figure 2C).
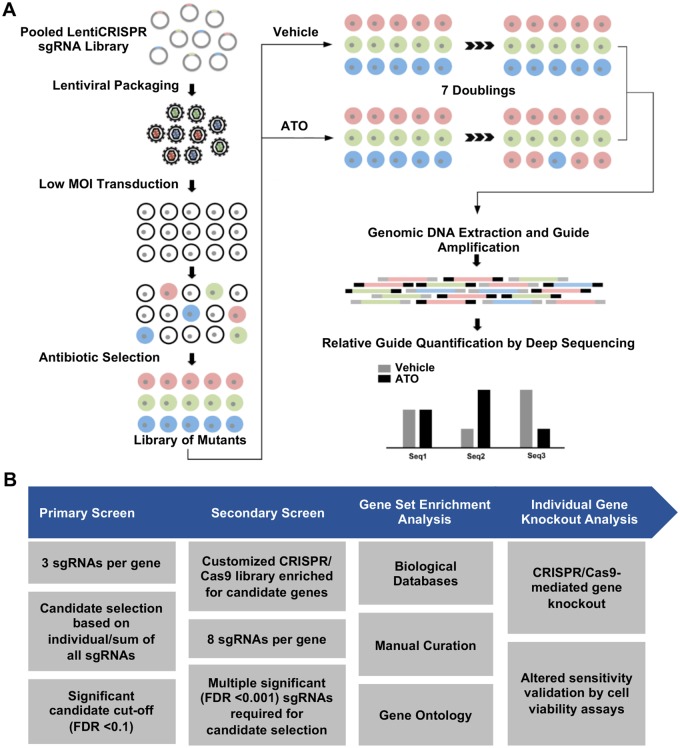
Figure 1.
Overview of the performed CRISPR-based knockout screening approach. A, The Cas9-sgRNA library was packaged into lentiviral particles and transduced into K562 cells. The obtained cellular library was screened to identify mutants with growth advantage or disadvantage in the presence of ATO. Guide sequences, used as barcodes labeling the different mutants, were PCR-amplified and quantified by deep sequencing. B, Study workflow showing screening strategies, candidate gene selection criteria, and validation approaches.
Figure 2.
Identification of multiple candidate genes affecting ATO/AsIII toxicity. A, Cytotoxicity of multiple concentrations of ATO in K562 cells evaluated at 24, 48, and 72 h by CellTiter Glo cell proliferation assay. Data are represented as mean ± SD (n = 3). B, Scatter plot showing relative enrichment (log2 FC > 0) or depletion (Log2 FC < 0) of each gene-specific guide sequence in ATO compared to vehicle control (Veh). Log2 fold changes (FC) are plotted against the average abundance of each guide sequence in the pool represented as Log2 counts per million (CPM). Hits with FDR values < 0.1 are shown in different colors. C, Venn diagram showing unique and common candidates identified by 3 analysis methods. For each method, number of hits with FDR < 0.1 is shown. D, Heatmap of the normalized sum of counts of all guide sequences targeting each gene. Only hits that are in common between the summing and individual sgRNA analysis approaches are shown. The screen was run in duplicate and the values corresponding to each replicate (Veh or ATO) are represented in the heat map.
Simultaneous Validation of Genes Modulating AsIII Cytotoxicity by Secondary Screening
We generated a customized CRISPR-Cas9 validation library, with increased number of sgRNAs targeting each gene (6–8 sgRNAs/gene), that is enriched for the primary hits identified by all the analysis methods implemented for the primary screen. We used the same validation library to confirm hits revealed by other screens which will be reported elsewhere. Thus, the validation library also included multiple sgRNAs targeting genes which were not identified as hits in the primary ATO screen in addition to several nontargeting sgRNAs. We performed a secondary screen in K562 cells with the validation library using conditions identical to those applied for the primary screen but on a smaller scale. We used more stringent criteria for candidate validation which required at least 2 sgRNAs per gene exhibiting significant depletion or enrichment in ATO compared to vehicle control with a FDR < 0.001. Using these criteria, we validated multiple primary candidates. Validated hits included genes whose loss confers either increased or decreased sensitivity to AsIII, with the latter being more prominent (Figure 3A; Supplementary Table 6). The secondary screen validated 20 candidates that were initially revealed by both the summing and the individual guide-based analysis methods, 16 candidates that were exclusively revealed by the summing method and 7 candidates that were uniquely identified by the individual guide-based method. Gene-level confirmatory analysis using the limma package revealed similar enrichment and depletion hits as the main GLM-based method (Supplementary Tables 8 and 9). Additionally, the vast majority of the validated genes were also revealed by MAGeCK analysis (Supplementary Tables 10 and 11). Functional classification of validated genes uncovered multiple biological processes as determinants of cellular sensitivity or tolerance to AsIII including selenocysteine (Sec) metabolism and utilization, oxidative stress response, DNA damage response, cellular transport, intracellular trafficking, histone modification, RNA processing in addition to diverse intracellular signaling processes (Figure 3B; Supplementary Table 7).
Figure 3.
Simultaneous validation of multiple candidate genes involved in ATO/AsIII toxicity by secondary screening. A, Scatter plot showing relative enrichment (log2 FC > 0) or depletion (Log2 FC < 0) of each gene-specific guide sequence in ATO compared to vehicle control. Log2 fold changes (FC) are plotted against the average abundance of each guide sequence in the pool represented as Log2 counts per million (CPM). Hits with FDR values < 0.001 are shown in different colors. B, Functional enrichment analysis of validated genes showing the top biological processes affecting ATO toxicity. Bars represent numbers of validated genes within each pathway. C, Relative enrichment (Fold change > 1) of each of the guide sequences targeting KEAP1 in ATO-treated compared to control pools. D, Relative depletion (Fold Change < 1) of each of the guide sequences targeting ABCC1 in ATO-treated compared to control pools. Fold changes for each guide sequence are calculated by dividing the normalized abundance of the sequence in ATO-treated pools by that in control pools. The dotted line (y = 1) in each graph represents the baseline abundance of each guide sequence in control pools. Data are represented as mean (n = 3). Only guide sequences displaying differential abundance at FDR < 0.001 are shown.
Oxidative Stress Response and Certain Cellular Transporters Affect Sensitivity to AsIII
Disruption of KEAP1, the negative regulator of the antioxidant response transcription factor Nrf2, resulted in a striking resistance to AsIII (Figure 3C). Alternatively, loss of the multidrug resistance protein ABCC1 drastically increased sensitivity to AsIII (Figure 3D). In contrast, disruption of genes encoding other transporter proteins including aquaporin 3 (AQP3) and the zinc transporter ZnT1(SLC30A1) resulted in AsIII tolerance (Supplementary Table 7).
Cellular Damage Response and DNA Repair Are Important Modulators of AsIII Sensitivity
Disruption of UBE2H involved in ubiquitin-dependent protein catabolism and CNOT2 involved in mRNA degradation increased sensitivity to AsIII (Supplementary Table 7). Multiple components of DNA damage response and double strand DNA repair were also identified in our screen (Supplementary Table 7). Unexpectedly, disruption of these genes increased resistance rather than sensitivity.
Histone Modification, Transcriptional Regulation, and Diverse Signaling Processes Impact AsIII Cytotoxicity
We identified few histone-modifying factors whose disruption increased resistance to AsIII (Supplementary Table 7). These include MRGBP and DR1, components of the NuA4 and ATAC histone acetyltransferase complexes respectively, GFI1B, a member of several histone deacetylase complexes and EED, a member of the PRC2/EED-EZH2 complex involved in histone H3 methylation. Other transcriptional regulators whose loss increased AsIII resistance include 2 mRNA processing/splicing factors, PAPOLA and CCNL2 and 2 positive transcriptional modulators, MEIS2 and RREB1 (Supplementary Table 7). In addition, loss of intracellular signaling components such as FLCN and RRAGC involved in mechanistic target of rapamycin (mTOR) signaling, RCE1 which is required for activation of RAS signaling and CTDNEP1 which suppresses bone morphogenetic proteins (BMP) signaling resulted in AsIII tolerance (Supplementary Table 7).
Modulation of Processes Involved in Protein Translation Affects AsIII Cytotoxicity
Disruption of certain translation components modulated sensitivity to AsIII (Supplementary Table 7). Loss of 2 components of diphthamide biosynthesis (DPH5, DPH6), a process that modifies a histidine residue in the eukaryotic translation elongation factor 2 (eEF-2), resulted in decreased sensitivity to AsIII. Similarly, inactivation of DHPS that uniquely modifies a specific lysine residue in eukaryotic translation initiation factor 5 A (eIF-5A) confers AsIII resistance.
Disruption of Sec Biosynthesis and Utilization Protects against AsIII Cytotoxicity
Inactivation of almost every gene involved in Sec biosynthesis or Sec incorporation into selenoproteins (EEFSEC, SECISBP2, SEPHS2, SEPSECS, and PSTK) conferred resistance to AsIII (Figure 4B). At least 5 distinct sgRNAs targeting each gene, out of the 8 sgRNAs used in the secondary screen, showed significant enrichment in ATO relative to the vehicle control with FDRs < 0.001 (Supplementary Table 6). To further confirm that Sec biosynthesis is a sensitivity determinant for AsIII, we targeted the phosphoseryl tRNA kinase (PSTK) gene in K562 cells using the CRISPR-Cas9 knockout tool. We generated 2 independent PSTK knockout pools by transducing K562 cells with different Cas9-sgRNA vectors. Cell viability assays showed increased resistance of the PSTK mutant pools to AsIII (Figure 4C). Additionally, disruption of TXNDC17 (TRP14), a substrate of the selenoprotein thioredoxin reductase (TxrR), resulted in AsIII tolerance (Figure 4D).
Figure 4.
Disruption of Sec biosynthesis and utilization decreases cellular susceptibility to ATO/AsIII. A, Overview of the Sec biosynthesis pathway and incorporation into selenoproteins. B, Validation of the increased resistance to ATO upon disruption of genes involved in Sec biosynthesis and incorporation into proteins (obtained from secondary screening). Dots represent the different sgRNAs targeting each gene where the relative abundance of each sgRNA in the treated pools is divided by that in the control pools to calculate fold changes. The dotted line (y = 1) represents the baseline abundance of each sgRNA sequence in control pools. Data are represented as mean (n = 3). Only guide sequences displaying differential abundance at FDR < 0.001 are shown. Lines represent median fold enrichment of all guides corresponding to each gene. C, PSTK inactivation results in increased sensitivity to ATO. Cells were transduced with CRISPR/sgRNA vectors targeting PSTK or nontargeting control and treated with multiple doses of ATO for 96 h. Cell viability in each pool is shown as percentage of vehicle-treated control. Data are represented as mean of 3 independent experiments ± SD (n = 3). D, Relative enrichment (Fold Change > 1) of each of the guide sequences targeting TXNDC17 in ATO-treated compared to control pools. Fold changes for each guide sequence are calculated by dividing the normalized abundance of the sequence in ATO-treated pools by that in control pools. The dotted line (y = 1) in represents the baseline abundance of each guide sequence in control pools. Data are represented as mean (n = 3). All the guide sequences shown display higher abundance in ATO at FDR < 0.001. E, Selenium protection against ATO toxicity. K562 cells were pretreated with sodium selenite (10 μM) and ATO cytotoxicity was evaluated in pretreated and control cells at 48 h by CellTiter Glo cell viability assay. Cell viability is represented as percentage of vehicle-treated control. Data are represented as mean of 3 independent experiments ± SD. Statistical significance was determined by Student’s t-test, where *p < .05, **p < .01, ***p < .001.
Selenium Enhances Tolerance of K562 Cells to AsIII
One potential explanation on the role of defective Sec biosynthesis/utilization in enhancing tolerance to AsIII is the accumulation of intracellular selenium. To study the effect of selenium on AsIII toxicity in K562 cells, we pretreated cells with 10 μM sodium selenite (Na2 SeO3) as an exogenous selenium source and assessed cell viability in response to multiple AsIII doses. Pretreatment of cells with sodium selenite significantly improved cell viability in response to AsIII treatment at all the studied doses (Figure 4E).
DISCUSSION
In this study, we used genome-wide CRISPR/Cas9 loss-of-function screening to investigate cellular mechanisms affecting susceptibility to ATO/As2O3; AsIII) in a K562 chronic myeloid leukemia (CML) cell line. To our knowledge, this study is the first comprehensive functional investigation of the genetic components modulating the cellular toxicity of AsIII in mammalian cells. Inorganic AsIII (iAsIII) can be converted intracellularly into methylated metabolites by the arsenic methyltransferase (As3MT) (Khaleghian et al., 2014). K562 cells express low levels of As3MT (Sumi et al., 2011) indicating that these cells can metabolize iAsIII into the methylated forms. Diverse mechanisms of acute and chronic AsIII toxicity have been previously identified in whole organisms and in individual cells (Abdul et al., 2015). In addition, factors influencing susceptibility of different individuals to arsenicals including AsIII were recently reviewed (Minatel et al., 2018). Our work confirmed the importance of previously well studied cellular processes, such as the oxidative stress response, in the cellular response to acute AsIII exposure. We also identified additional functional components and pathways whose role in cellular AsIII toxicity was not clearly appreciated. Both Sec production and its incorporation into selenoproteins were identified as key processes modulating AsIII toxicity in CML cells. Further investigation is needed to assess whether such processes define AsIII toxicity in other cell types. Our work further demonstrates the utility of unbiased functional screens (Gaytan and Vulpe, 2014) to identify components modulating cellular toxicity to exogenous stressors.
Our screen revealed an expected role for oxidative stress response in acute cellular toxicity of AsIII in K562 cells. AsIII induces cellular oxidative stress likely through multiple mechanisms (Flora, 2011). We found that loss of KEAP1, which negatively regulates the primary antioxidant transcription factor Nrf2, conferred substantial resistance to AsIII (Figure 3C). Nrf2 is responsible for the transcriptional activation of a complex antioxidant response pathway (Jaramillo and Zhang, 2013). Our finding is consistent with the reported activation of the Nrf2-KEAP1 pathway by arsenical compounds (Lau et al., 2013), and the increased resistance of cells to arsenic through treatment with Nrf2 activators (Du et al., 2008). The Nrf2-KEAP1 pathway was also shown to modulate AsIII toxicity in NB4 APL cells (Nishimoto et al., 2016) indicating that the protective effect of KEAP1 loss against AsIII is not limited to CML cells. In addition to KEAP1, we also identified TRP14 (TXNDC17), a reported reductase of L-cystine to L-cysteine in conjunction with TxrR (Pader et al., 2014), as an important determinant of AsIII sensitivity. L-cysteine levels are critical for GSH synthesis as well as for maintaining cytosolic GSH levels (Stipanuk et al., 2006; Yu and Long, 2016). However, contrary to a role in protecting against oxidative stress via maintaining GSH levels, loss of TXNDC17, decreased rather than increased sensitivity to AsIII (Figure 4D) suggesting a potentially different role in AsIII toxicity.
Sec biosynthesis and incorporation into selenoproteins was revealed by the genome-wide CRISPR screen as a key process influencing AsIII toxicity in K562 CML cells. Production of Sec is initiated by charging the Sec tRNA with serine (Ser) which is converted to Sec in a multi-step process catalyzed by several enzymes (Schmidt and Simonović, 2012) (Figure 4A). Binding of the Sec tRNA (tRNAsec) into the translational complex and subsequent insertion of Sec into the polypeptide chain requires a UGA stop codon and a downstream Sec insertion sequence (SECIS) in the corresponding mRNA and is facilitated by the SECIS-binding protein and the Sec-specific elongation factor (Hoffmann and Berry, 2006). We found that the loss of almost any enzyme involved in Sec biosynthesis or factors involved in its incorporation into the polypeptide chain results in substantial AsIII tolerance (Figs. 4B and 4C). It is well-known that selenoproteins, including glutathione peroxidase and TxrR play an important role in protecting against oxidative damage (Arbogast and Ferreiro, 2010; Benhar, 2018). However, our finding that the disruption of selenoprotein synthesis increases rather than decreases tolerance to AsIII suggests an alternative role in AsIII toxicity that needs further investigation. The effect of Sec metabolism on AsIII toxicity could be specific to CML cells although a previous report indicated that AsIII can inhibit selenoprotein synthesis in A549 lung cancer cells (Talbot et al., 2008). Further investigation is required to determine whether selenoprotein synthesis is a general determinant of AsIII cytotoxicity.
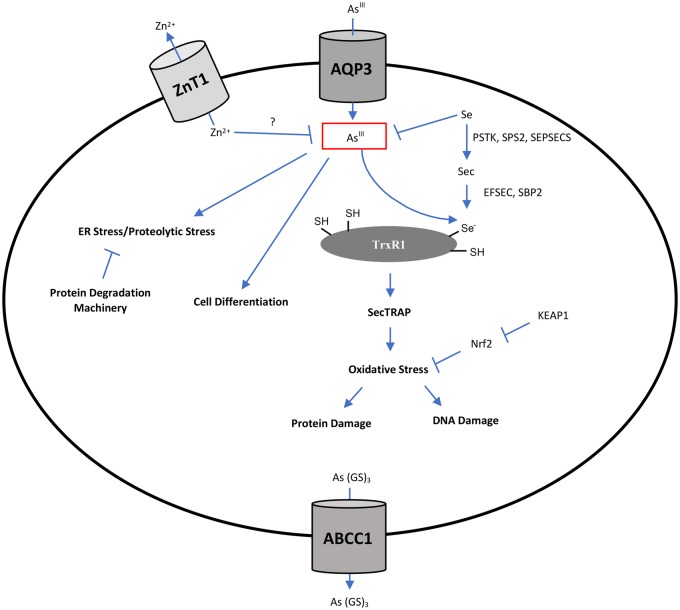
The selenoprotein, TxrR was previously identified as a direct molecular target of AsIII (Lu et al., 2007). The Sec residue of TrxR is essential for Trx reduction as well as AsIII binding (Anestål et al., 2008; Lu et al., 2007). Several studies indicated that binding of electrophiles, including AsIII, to TrxR simultaneously inhibits Trx reduction and enhances NADPH oxidase activity and thus converts the anti-oxidant protein into a pro-oxidant termed selenium compromised TxrR-derived apoptotic protein (SeCTRAP) (Anestål and Arnér, 2003; Anestål et al., 2008) although others did not find evidence of increased NADPH oxidase activity after AsIII binding to TrxR in vitro (Lu et al., 2007). These findings, in addition to our observation that defective Sec production and insertion into selenoproteins decreases cellular sensitivity to AsIII, suggest that targeting of TrxR and generation of SeCTRAPs could be one of the mechanisms underlying AsIII cytotoxicity. Therefore, resistance of Sec biosynthesis mutants to AsIII could be due to absence of SecTRAP formation. Additionally, it has been suggested that the apoptotic activity of the compromised TrxR requires an endogenous cellular substrate (Anestål et al., 2008). Intriguingly, the thioredoxin (Trx)-like TrxR substrate TRP14 (TXNDC17), was revealed by our screen as a determinant of sensitivity to AsIII (Figure 4D). This finding suggests that TRP14 could potentially serve as an endogenous substrate for SeCTRAPs. The role of TRP14 in the apoptotic function of SecTRAPs requires further investigation. A proposed model for AsIII-derived SecTRAPs is shown in Figure 5A.
Figure 5.
Proposed models illustrating potential roles of selenocysteine metabolism in AsIII toxicity. (A) Selenium Compromised Thioredoxin Reductase-derived Apoptotic Proteins (SeCTRAPs) model. Binding of inorganic AsIII (iAsIII) or its metabolite to the selenol group of Sec in the C-terminal active site of thioredoxin reductase (TrxR1) modifies the enzyme disrupting its ability to reduce thioredoxin (Trx). The modified enzyme gains an NADPH oxidase activity that produces superoxide. TRP14, the thioredoxin-related protein encoded by the TXNDC17 gene is a potential endogenous substrate required for the NADPH oxidase activity of SeCTRAPs. (B) Seleno-bis (S-glutathionyl) arsinium ion ([GS]2AsSe)- export model. AsIII forms a complex with selenium and glutathione that can be exported outside the cell through multidrug resistance proteins. (Left) Utilization of selenium (Se) for selenoprotein biosynthesis may result in depletion of intracellular selenium levels and subsequent AsIII accumulation leading to increased cytotoxicity. (Right) Disruption of the biosynthesis of selenocysteine or its incorporation into selenoproteins can spare intracellular selenium levels resulting in efficient AsIII detoxification through formation of the ([GS]2AsSe)- complex and its export outside the cell.
An alternative but not mutually exclusive hypothesis to explain the role of Sec biosynthesis and utilization in AsIII toxicity is the reduction of intracellular selenium bioavailability. The clear interrelationship between selenite (SeIV) and AsIII is well documented in animals and people (Carew and Leslie, 2010; Levander, 1977; Srivastava et al., 2010; Sun et al., 2014). In mammalian cells including erythrocytes, the Seleno-bis (S-glutathionyl) arsinium ion ([GS]2AsSe)− can form (Manley et al., 2006) and was shown to be exported by the multidrug resistance-associated protein ABCC2 (MRP2) (Carew and Leslie, 2010). The production of Sec may reduce the bioavailable intracellular selenium pool. Thus, defective Sec biosynthesis could spare bioavailable selenium for use in the formation of the ([GS]2AsSe)− complex and hence detoxification of AsIII (Figure 5B). This suggestion is further supported by our finding that preloading the cells with selenium protects against subsequent AsIII toxicity (Figure 4E). Additionally, we identified the multidrug transporter ABCC1 (MRP1) as a major determinant of AsIII tolerance in K562 CML cells as its loss markedly increased AsIII sensitivity (Figure 3D). However, previous studies on membrane vesicles prepared from human erythrocytes indicate that ([GS]2AsSe)− complex can be exported through ABCC2 (MRP2) and not ABCC1 (Carew and Leslie, 2010). ABCC1 has been demonstrated to transport As(GS)3 complexes (Leslie, 2012; Leslie et al., 2004) so it is possible that the observed increased sensitivity upon ABCC1 loss is due to a defect in As(GS)3 export rather than ([GS]2AsSe)−. Given our findings, it may be worthwhile to further investigate the role of ABCC1 in transporting ([GS]2AsSe)− in K562 cells. The role of ABCC1 in AsIII detoxification might be cell-type specific as its overexpression in HL60 APL cells did not result in resistance to AsIII (Sertel et al., 2012). Overall, our results indicate a clear relationship between intracellular Sec metabolism and cellular AsIII toxicity.
We identified 2 transmembrane transporters as determinants of AsIII sensitivity. Loss of AQP3 decreased sensitivity of K562 cells to AsIII consistent with a role of AQP3 in AsIII uptake. AQPs were previously reported to import trivalent arsenicals into mammalian cells (Rosen, 2002) and polymorphisms of AQP3 were reported to increase the risk of bladder cancer in individuals with high AsIII exposure (Lesseur et al., 2012). Similarly, in human lung adenocarcinoma cells, AsIII resistance is associated with a decrease in AQP3 expression and the knockdown of AQP3 in nonresistant cells confers AsIII resistance (Lee et al., 2006). Another AQP involved in AsIII uptake, AQP9, is not intrinsically expressed in K562 cells (Bhattacharjee et al., 2004) and hence was not identified as a modulator of AsIII toxicity in our screen. In contrast, the expression of AQP3 was previously detected in K562 cells (http://www.proteinatlas.org/; Accessed February 7, 2019). These findings indicate that multiple AQPs are involved in cellular AsIII uptake and that the expression of a specific AQP in a cell type defines its role in modulating AsIII sensitivity. We also identified the zinc exporter ZnT1 (SLC30A1) as a sensitivity factor for AsIII in K562 cells. Inactivation of ZnT1 or its transcriptional activator metal regulatory transcription factor 1 (MTF1) resulted in considerable AsIII resistance. Several groups have suggested a protective role of zinc in AsIII toxicity (Kreppel et al., 1994; Milton et al., 2004) which would be consistent with AsIII tolerance due to increased intracellular zinc levels as a result of ZnT1 dysfunction. Interestingly, SNP polymorphisms in a different zinc transporter, SLC39A2, have been associated with increased bladder cancer risk after arsenic exposure (Karagas et al., 2012).
Various cellular stress response components were identified in our study as key modulators of AsIII toxicity in K562 cells. For example, we found that loss of the ubiquitin conjugating enzyme UBE2H results in higher AsIII sensitivity suggesting a role for proteolytic stress in AsIII cytotoxicity. AsIII was previously shown to impair protein folding leading to the formation of protein aggregates in yeast (Jacobson et al., 2012). In addition, ER stress was shown to synergize with AsIII to induce apoptosis in APL cells (Masciarelli et al., 2018). Collectively, these findings suggest that degradation of misfolded proteins partially mitigates AsIII cytotoxicity. Although AsIII was previously shown to induce DNA damage in APL cells (Kumar et al., 2014), our results do not support a role for DNA repair in mitigating AsIII toxicity. We found that the disruption of selected DNA damage response and DNA repair components in K562 cells confers increased, rather than decreased, AsIII tolerance. It is possible that impairment of DNA damage response results in bypassing the mitotic check point, hence leading to a cellular growth advantage in the presence of AsIII (Bartkova et al., 2005).
Our study revealed multiple cellular processes that can modulate AsIII cytotoxicity (Figure 6) and hence influence its anti-cancer therapeutic potential. AsIII is a standard effective therapy for APL, and its major effects in APL cells involve the degradation of the PML-RARα fusion oncoprotein (Lallemand-Breitenbach et al., 2012). However, AsIII can also induce apoptosis in APL cells through other mechanisms (Davison et al., 2002; Yedjou et al., 2010). AsIII resistance can arise in APL leading to poor prognosis (Zhu et al., 2014). PML-RARα mutations account for some but not all of the observed resistance (Chendamarai et al., 2015) and our screens, although performed in CML cells, suggest potential AsIII resistance mechanisms that could be explored. Importantly, AsIII was suggested as a potential therapy for other cancer types including CML when combined with other agents such as interferons (El Eit et al., 2014) or tyrosine kinase inhibitors (Wang et al., 2015). Revealing the molecular determinants of AsIII sensitivity and tolerance in a CML cell line provided insight into cellular processes which can modulate the efficacy of AsIII in CML. Targeting such processes simultaneously with AsIII treatment could potentiate AsIII in CML therapy, with the caveat of potentially increased cytotoxicity in nontumor cells.
Figure 6.
Summary of the potential cellular processes modulating AsIII toxicity in K562 cells revealed by CRISPR-based loss-of-function screening. AsIII enters the cell through Aquaporin 3 (AQP3). AsIII or its metabolites can have multiple cellular effects. AsIII-induced oxidative stress could result from mitochondrial damage and/or from a direct effect on thioredoxin reductase (TrxR) and generation of SecTRAPs. AsIII can have anti-proliferative effects by driving cell differentiation that is mediated by multiple cellular regulatory processes. Nrf2-based antioxidant response, intracellular selenium, intracellular zinc and export through ABCC1 can protect against AsIII cytotoxicity.
Our work demonstrated the capability of CRISPR-based functional genomics in deciphering molecular mechanisms modulating susceptibility to AsIII and our findings could have implications for a broader use of AsIII in chemotherapy beyond the treatment of APL. However, we recognize that the mechanisms influencing sensitivity to AsIII identified in K562 cells may not extend to other cell types. Hence, further studies are needed to validate the relevance of our findings in other cells and tissues. Further, additional modulators of AsIII sensitivity or tolerance might not have been identified by our screen due to lack of expression/activity of such determinants in K562 cells, existing compensatory mechanisms in these cells resulting in functional redundancy, or cell line specific genetic changes which abrogate the need for these modulators. Therefore, a more comprehensive approach requires complementary genetic screens in multiple cell lines to determine common as well as context-specific modulators of AsIII toxicity. Another limitation of our genome-wide approach could arise from poor on-target activity of the library sgRNAs which can result in false negatives. Since the genome-wide library used in our study involves only 3 sgRNAs targeting each gene, it can be expected that several determinants of AsIII sensitivity or tolerance in the studied cell type were not revealed. Using genome-wide libraries with improved sgRNA designs and more sgRNAs targeting each gene would likely reveal additional novel modulators of susceptibility to AsIII. Overall, our work is an important first step that will enable additional studies that could ultimately define all the molecular components involved in exacerbating or mitigating AsIII cytotoxicity.
DECLARATION OF CONFLICTING INTERESTS
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
FUNDING
This work was supported by grant from the Superfund Hazardous Substance Research and Training Program (P42ES004705 to M.T.S.) and funding to C.V. from the University of Florida Research Foundation at the University of Florida, Gainesville. A.S was a trainee in the Superfund Research Program (University of California, Berkeley). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Environmental Health Sciences.
Supplementary Material
REFERENCES
- Abaza Y., Kantarjian H., Garcia-Manero G., Estey E., Borthakur G., Jabbour E., Faderl S., O’Brien S., Wierda W., Pierce S., et al. (2017). Long-term outcome of acute promyelocytic leukemia treated with all-trans-retinoic acid, arsenic trioxide, and gemtuzumab. Blood 129, 1275–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdul K. S., Jayasinghe S. S., Chandana E. P., Jayasumana C., De Silva P. M. (2015). Arsenic and human health effects: A review. Environ. Toxicol. Pharmacol. 40, 828–846. [DOI] [PubMed] [Google Scholar]
- Agency for Toxic Substances and Disease Registry. (2017). Priority List of Hazardous Substances. https://www.atsdr.cdc.gov/spl/#2017spl. Accessed February 7, 2019.
- Akao Y., Nakagawa Y., Akiyama K. (1999). Arsenic trioxide induces apoptosis in neuroblastoma cell lines through the activation of caspase 3 in vitro. FEBS Lett. 455, 59–62. [DOI] [PubMed] [Google Scholar]
- Anestål K., Arnér E. S. J. (2003). Rapid induction of cell death by selenium-compromised thioredoxin reductase 1 but not by the fully active enzyme containing selenocysteine. J. Biol. Chem. 278, 15966–15972. [DOI] [PubMed] [Google Scholar]
- Anestål K., Prast-Nielsen S., Cenas N., Arnér E. S. J. (2008). Cell death by sectraps: Thioredoxin reductase as a prooxidant killer of cells. PLoS One 3, e1846.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbogast S., Ferreiro A. (2010). Selenoproteins and protection against oxidative stress: Selenoprotein n as a novel player at the crossroads of redox signaling and calcium homeostasis. Antioxid. Redox. Signal. 12, 893–904. [DOI] [PubMed] [Google Scholar]
- Bartkova J., Horejsí Z., Koed K., Krämer A., Tort F., Zieger K., Guldberg P., Sehested M., Nesland J. M., Lukas C., et al. (2005). DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434, 864.. [DOI] [PubMed] [Google Scholar]
- Benhar M. (2018). Roles of mammalian glutathione peroxidase and thioredoxin reductase enzymes in the cellular response to nitrosative stress. Free Radic. Biol. Med. 127, 160–164. [DOI] [PubMed] [Google Scholar]
- Berenson J. R., Boccia R., Siegel D., Bozdech M., Bessudo A., Stadtmauer E., Talisman Pomeroy J., Steis R., Flam M., Lutzky J., et al. (2006). Efficacy and safety of melphalan, arsenic trioxide and ascorbic acid combination therapy in patients with relapsed or refractory multiple myeloma: A prospective, multicentre, phase ii, single-arm study. Br. J. Haematol. 135, 174–183. [DOI] [PubMed] [Google Scholar]
- Bhattacharjee H., Carbrey J., Rosen B. P., Mukhopadhyay R. (2004). Drug uptake and pharmacological modulation of drug sensitivity in leukemia by aqp9. Biochem. Biophys. Res. Commun. 322, 836–841. [DOI] [PubMed] [Google Scholar]
- Carew M. W., Leslie E. M. (2010). Selenium-dependent and -independent transport of arsenic by the human multidrug resistance protein 2 (mrp2/abcc2): Implications for the mutual detoxification of arsenic and selenium. Carcinogenesis 31, 1450–1455. [DOI] [PubMed] [Google Scholar]
- Chendamarai E., Ganesan S., Alex A. A., Kamath V., Nair S. C., Nellickal A. J., Janet N. B., Srivastava V., Lakshmi K. M., Viswabandya A., et al. (2015). Comparison of newly diagnosed and relapsed patients with acute promyelocytic leukemia treated with arsenic trioxide: Insight into mechanisms of resistance. PLoS One 10, e0121912.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison K., Mann K. K., Miller W. H. (2002). Arsenic trioxide: Mechanisms of action. Semin. Hematol. 39, 3–7. [DOI] [PubMed] [Google Scholar]
- Doench J. G., Fusi N., Sullender M., Hegde M., Vaimberg E. W., Donovan K. F., Smith I., Tothova Z., Wilen C., Orchard R., et al. (2016). Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34, 184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y., Villeneuve N. F., Wang X.-J., Sun Z., Chen W., Li J., Lou H., Wong P. K., Zhang D. D. (2008). Oridonin confers protection against arsenic-induced toxicity through activation of the Nrf2-mediated defensive response. Environ. Health Perspect. 116, 1154–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Eit R. M., Iskandarani A. N., Saliba J. L., Jabbour M. N., Mahfouz R. A., Bitar N. M., Ayoubi H. R., Zaatari G. S., Mahon F. X., De The H. B., et al. (2014). Effective targeting of chronic myeloid leukemia initiating activity with the combination of arsenic trioxide and interferon alpha. Int. J. Cancer 134, 988–996. [DOI] [PubMed] [Google Scholar]
- Emadi A., Gore S. D. (2010). Arsenic trioxide - an old drug rediscovered. Blood Rev. 24, 191–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flora S. J. S. (2011). Arsenic-induced oxidative stress and its reversibility. Free Radic. Biol. Med. 51, 257–281. [DOI] [PubMed] [Google Scholar]
- Gaytan B. D., Vulpe C. D. (2014). Functional toxicology: Tools to advance the future of toxicity testing. Front. Genet. 5, 110.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann P. R., Berry M. J. (2006). The Importance of Subcellular Localization of SBP2 and EFsec for Selenoprotein Synthesis. Selenium, pp. 73–82. Springer, Boston, MA. [Google Scholar]
- Hopenhayn-Rich C., Smith A. H., Goeden H. M. (1993). Human studies do not support the methylation threshold hypothesis for the toxicity of inorganic arsenic. Environ. Res. 60, 161–177. [DOI] [PubMed] [Google Scholar]
- International Agency for Research on Cancer. (2004). Some Drinking-Water Disinfectants and Contaminants, Including Arsenic. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. Lyon, France: International Agency for Research on cancer. 84, 1–477. [PMC free article] [PubMed] [Google Scholar]
- Jacobson T., Navarrete C., Sharma S. K., Sideri T. C., Ibstedt S., Priya S., Grant C. M., Christen P., Goloubinoff P., Tamás M. J. (2012). Arsenite interferes with protein folding and triggers formation of protein aggregates in yeast. J. Cell Sci. 125, 5073–5083. [DOI] [PubMed] [Google Scholar]
- Jaramillo M. C., Zhang D. D. (2013). The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev. 27, 2179–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karagas M. R., Andrew A. S., Nelson H. H., Li Z., Punshon T., Schned A., Marsit C. J., Morris J. S., Moore J. H., Tyler A. L., et al. (2012). SLC39A2 and FSIP1 polymorphisms as potential modifiers of arsenic-related bladder cancer. Hum. Genet. 131, 453–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kchour G., Tarhini M., Kooshyar M. M., El Hajj H., Wattel E., Mahmoudi M., Hatoum H., Rahimi H., Maleki M., Rafatpanah H., et al. (2009). Phase 2 study of the efficacy and safety of the combination of arsenic trioxide, interferon alpha, and zidovudine in newly diagnosed chronic adult t-cell leukemia/lymphoma (ATL). Blood 113, 6528–6532. [DOI] [PubMed] [Google Scholar]
- Khaleghian A., Ghaffari S. H., Ahmadian S., Alimoghaddam K., Ghavamzadeh A. (2014). Metabolism of arsenic trioxide in acute promyelocytic leukemia cells. J Cell Biochem. 115, 1729–1739. [DOI] [PubMed] [Google Scholar]
- Kreppel H., Liu J., Liu Y., Reichl F. X., Klaassen C. D. (1994). Zinc-induced arsenite tolerance in mice. Toxicol. Sci. 23, 32–37. [DOI] [PubMed] [Google Scholar]
- Kumar S., Yedjou C. G., Tchounwou P. B. (2014). Arsenic trioxide induces oxidative stress, DNA damage, and mitochondrial pathway of apoptosis in human leukemia (hl-60) cells. J. Exp. Clin. Cancer Res. 33, 42.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V., Zhu J., Chen Z., de Thé H. (2012). Curing APL through PML/RARA degradation by As2O3. Trends Mol. Med. 18, 36–42. [DOI] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S. L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau A., Whitman S. A., Jaramillo M. C., Zhang D. D. (2013). Arsenic-mediated activation of the nrf2-keap1 antioxidant pathway. J. Biochem. Mol. Toxicol. 27, 99–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law C. W., Chen Y., Shi W., Smyth G. K. (2014). Voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, R29.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T.-C., Ho I. C., Lu W.-J., Huang J-d. (2006). Enhanced expression of multidrug resistance-associated protein 2 and reduced expression of aquaglyceroporin 3 in an arsenic-resistant human cell line. J. Biol. Chem. 281, 18401–18407. [DOI] [PubMed] [Google Scholar]
- Leslie E. M. (2012). Arsenic–glutathione conjugate transport by the human multidrug resistance proteins (MRPs/ABCCs). J. Inorg. Biochem. 108, 141–149. [DOI] [PubMed] [Google Scholar]
- Leslie E. M., Haimeur A., Waalkes M. P. (2004). Arsenic transport by the human multidrug resistance protein 1 (MRP1/ABCC1). Evidence that a tri-glutathione conjugate is required. J. Biol. Chem. 279, 32700–32708. [DOI] [PubMed] [Google Scholar]
- Lesseur C., Gilbert-Diamond D., Andrew A. S., Ekstrom R. M., Li Z., Kelsey K. T., Marsit C. J., Karagas M. R. (2012). A case-control study of polymorphisms in xenobiotic and arsenic metabolism genes and arsenic-related bladder cancer in New Hampshire. Toxicol. Lett. 210, 100–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levander O. A. (1977). Metabolic interrelationships between arsenic and selenium. Environ. Health Perspect. 19, 159–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. (2009). Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Xu H., Xiao T., Cong L., Love M. I., Zhang F., Irizarry R. A., Liu J. S., Brown M., Liu X. S. (2014). Mageck enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 5, 554.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo-Coco F., Di Donato L., Gimema, Schlenk R. F., German-Austrian A. M., Leukemia S. G., Study A. L. (2016). Targeted therapy alone for acute promyelocytic leukemia. N. Engl. J. Med. 374, 1197–1198. [DOI] [PubMed] [Google Scholar]
- Lu J., Chew E.-H., Holmgren A. (2007). Targeting thioredoxin reductase is a basis for cancer therapy by arsenic trioxide. Proc. Natl. Acad. Sci. U.S.A. 104, 12288–12293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun A. T., Chen Y., Smyth G. K. (2016). It’s DE-licious: A recipe for differential expression analyses of RNA-seq experiments using quasi-likelihood methods in edger. Methods Mol. Biol. 1418, 391–416. [DOI] [PubMed] [Google Scholar]
- Manley S. A., George G. N., Pickering I. J., Glass R. S., Prenner E. J., Yamdagni R., Wu Q., Gailer J. (2006). The seleno bis(s-glutathionyl) arsinium ion is assembled in erythrocyte lysate. Chem. Res. Toxicol. 19, 601–607. [DOI] [PubMed] [Google Scholar]
- Masciarelli S., Capuano E., Ottone T., Divona M., De Panfilis S., Banella C., Noguera N. I., Picardi A., Fontemaggi G., Blandino G., et al. (2018). Retinoic acid and arsenic trioxide sensitize acute promyelocytic leukemia cells to ER stress. Leukemia 32, 285–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mass M. J., Tennant A., Roop B. C., Cullen W. R., Styblo M., Thomas D. J., Kligerman A. D. (2001). Methylated trivalent arsenic species are genotoxic. Chem. Res. Toxicol. 14, 355–361. [DOI] [PubMed] [Google Scholar]
- Mathews V., George B., Chendamarai E., Lakshmi K. M., Desire S., Balasubramanian P., Viswabandya A., Thirugnanam R., Abraham A., Shaji R. V., et al. (2010). Single-agent arsenic trioxide in the treatment of newly diagnosed acute promyelocytic leukemia: Long-term follow-up data. J. Clin. Oncol. 28, 3866–3871. [DOI] [PubMed] [Google Scholar]
- Mathews V., George B., Lakshmi K. M., Viswabandya A., Bajel A., Balasubramanian P., Shaji R. V., Srivastava V. M., Srivastava A., Chandy M. (2006). Single-agent arsenic trioxide in the treatment of newly diagnosed acute promyelocytic leukemia: Durable remissions with minimal toxicity. Blood 107, 2627–2632. [DOI] [PubMed] [Google Scholar]
- Miller W. H., Schipper H. M., Lee J. S., Singer J., Waxman S. (2002). Mechanisms of action of arsenic trioxide. Cancer Res. 62, 3893–3903. [PubMed] [Google Scholar]
- Milton A. G., Zalewski P. D., Ratnaike R. N. (2004). Zinc protects against arsenic-induced apoptosis in a neuronal cell line, measured by DEVD-caspase activity. Biometals 17, 707–713. [DOI] [PubMed] [Google Scholar]
- Minatel B. C., Sage A. P., Anderson C., Hubaux R., Marshall E. A., Lam W. L., Martinez V. D. (2018). Environmental arsenic exposure: From genetic susceptibility to pathogenesis. Environ. Int. 112, 183–197. [DOI] [PubMed] [Google Scholar]
- Nishimoto S., Suzuki T., Koike S., Yuan B., Takagi N., Ogasawara Y. (2016). Nrf2 activation ameliorates cytotoxic effects of arsenic trioxide in acute promyelocytic leukemia cells through increased glutathione levels and arsenic efflux from cells. Toxicol. Appl. Pharmacol. 305, 161–168. [DOI] [PubMed] [Google Scholar]
- Pader I., Sengupta R., Cebula M., Xu J., Lundberg J. O., Holmgren A., Johansson K., Arnér E. S. J. (2014). Thioredoxin-related protein of 14 kda is an efficient l-cystine reductase and S-denitrosylase. Proc. Natl. Acad. Sci. U.S.A. 111, 6964–6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnaike R. (2003). Acute and chronic arsenic toxicity. Postgrad. Med. J. 79, 391–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravenscroft P. (2007). Predicting the global distribution of natural arsenic pollution in groundwater. Royal Geographical Society Annual International Conference. https://www.geog.cam.ac.uk/research/projects/arsenic/symposium/S1.2_P_Ravenscroft.pdf. Accessed February 7, 2019.
- Ritchie M. E., Phipson B., Wu D., Hu Y., Law C. W., Shi W., Smyth G. K. (2015). Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen B. P. (2002). Biochemistry of arsenic detoxification. FEBS Lett. 529, 86–92. [DOI] [PubMed] [Google Scholar]
- Rousselot P., Labaume S., Marolleau J. P., Larghero J., Noguera M. H., Brouet J. C., Fermand J. P. (1999). Arsenic trioxide and melarsoprol induce apoptosis in plasma cell lines and in plasma cells from myeloma patients. Cancer Res. 59, 1041–1048. [PubMed] [Google Scholar]
- Sanjana N. E., Shalem O., Zhang F. (2014). Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R. L., Simonović M. (2012). Synthesis and decoding of selenocysteine and human health. Croat. Med. J. 53, 535–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sertel S., Tome M., Briehl M. M., Bauer J., Hock K., Plinkert P. K., Efferth T. (2012). Factors determining sensitivity and resistance of tumor cells to arsenic trioxide. PLoS One 7, e35584.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O., Sanjana N. E., Hartenian E., Shi X., Scott D. A., Mikkelsen T. S., Heckl D., Ebert B. L., Root D. E., Doench J. G., et al. (2014). Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z. X., Chen G. Q., Ni J. H., Li X. S., Xiong S. M., Qiu Q. Y., Zhu J., Tang W., Sun G. L., Yang K. Q., et al. (1997). Use of arsenic trioxide (As2O3) in the treatment of acute promyelocytic leukemia (apl): II. Clinical efficacy and pharmacokinetics in relapsed patients. Blood 89, 3354–3360. [PubMed] [Google Scholar]
- Shen Z. Y., Tan L. J., Cai W. J., Shen J., Chen C., Tang X. M., Zheng M. H. (1999). Arsenic trioxide induces apoptosis of oesophageal carcinoma in vitro. Int. J. Mol. Med. 4, 33–37. [DOI] [PubMed] [Google Scholar]
- Srivastava D., Subramanian R. B., Madamwar D., Flora S. J. S. (2010). Protective effects of selenium, calcium, and magnesium against arsenic-induced oxidative stress in male rats. Arh. Hig. Rada Toksikol. 61, 153–159. [DOI] [PubMed] [Google Scholar]
- Stipanuk M. H., Dominy J. J. E., Lee J.-I., Coloso R. M. (2006). Mammalian cysteine metabolism: New insights into regulation of cysteine metabolism. J. Nutr. 136, 1652S–1659S. [DOI] [PubMed] [Google Scholar]
- Styblo M., Drobna Z., Jaspers I., Lin S., Thomas D. J. (2002). The role of biomethylation in toxicity and carcinogenicity of arsenic: A research update. Environ. Health Perspect 110(Suppl 5), 767–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumi D., Fukushima K., Miyataka H., Himeno S. (2011). Alternative splicing variants of human arsenic (+3 oxidation state) methyltransferase. Biochem. Biophys. Res. Commun. 415, 48–53. [DOI] [PubMed] [Google Scholar]
- Sun H. J., Rathinasabapathi B., Wu B., Luo J., Pu L. P., Ma L. Q. (2014). Arsenic and selenium toxicity and their interactive effects in humans. Environ. Int. 69, 148–158. [DOI] [PubMed] [Google Scholar]
- Talbot S., Nelson R., Self W. (2008). Arsenic trioxide and auranofin inhibit selenoprotein synthesis: Implications for chemotherapy for acute promyelocytic leukaemia. Br. J. Pharmacol. 154, 940–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahter M. (2002). Mechanisms of arsenic biotransformation. Toxicology 181–182, 211–217. [DOI] [PubMed] [Google Scholar]
- Wang W., Lv F. F., Du Y., Li N., Chen Y., Chen L. (2015). The effect of nilotinib plus arsenic trioxide on the proliferation and differentiation of primary leukemic cells from patients with chronic myoloid leukemia in blast crisis. Cancer Cell Int. 15, 10.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson WH. (2015). Chapter two - molecular mechanisms in arsenic toxicity. In: Fishbein JC, Heilman JM, editors. Advances in molecular toxicology. Waltham, MA: Elsevier. p. 35–75. [Google Scholar]
- World Health Organization & International Program on Chemical Safety. (1996). Guidelines for drinking-water quality. Vol. 2, Health criteria and other supporting information, 2nd ed. Geneva: World Health Organization. https://apps.who.int/iris/handle/10665/38551. Accessed February 7, 2019.
- Yedjou C., Tchounwou P., Jenkins J., McMurray R. (2010). Basic mechanisms of arsenic trioxide (ato)-induced apoptosis in human leukemia (hl-60) cells. J. Hematol. Oncol. 3, 28.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Long Y. C. (2016). Crosstalk between cystine and glutathione is critical for the regulation of amino acid signaling pathways and ferroptosis. Sci. Rep. 6, 30033.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Ohnishi K., Shigeno K., Fujisawa S., Naito K., Nakamura S., Takeshita K., Takeshita A., Ohno R. (1998). The induction of apoptosis and cell cycle arrest by arsenic trioxide in lymphoid neoplasms. Leukemia 12, 1383–1391. [DOI] [PubMed] [Google Scholar]
- Zhang X. W., Yan X. J., Zhou Z. R., Yang F. F., Wu Z. Y., Sun H. B., Liang W. X., Song A. X., Lallemand-Breitenbach V., Jeanne M., et al. (2010). Arsenic trioxide controls the fate of the pml-raralpha oncoprotein by directly binding pml. Science 328, 240–243. [DOI] [PubMed] [Google Scholar]
- Zheng W. L., Zhang G. S., Xu Y. X., Shen J. K., Dai C. W., Pei M. F. (2008). Arsenic trioxide, thalidomide and retinoid acid combination therapy in higher risk myelodysplastic syndrome patients. Leuk. Res. 32, 251–254. [DOI] [PubMed] [Google Scholar]
- Zhou J., Zhang Y., Li J., Li X., Hou J., Zhao Y., Liu X., Han X., Hu L., Wang S., et al. , (2010). Single-agent arsenic trioxide in the treatment of children with newly diagnosed acute promyelocytic leukemia. Blood 115, 1697–1702. [DOI] [PubMed] [Google Scholar]
- Zhu H.-H., Qin Y.-Z., Huang X.-J. (2014). Resistance to arsenic therapy in acute promyelocytic leukemia. N. Engl. J. Med. 370, 1864–1866. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.