Abstract
Alcohol use prior to and during pregnancy remains a significant societal problem and can lead to developmental fetal abnormalities including compromised myocardia function and increased risk for heart disease later in life. Alcohol-induced cardiac toxicity has traditionally been studied in animal-based models. These models have limitations due to physiological differences from human cardiomyocytes (CMs) and are also not suitable for high-throughput screening. We hypothesized that human-induced pluripotent stem cell-derived CMs (hiPSC-CMs) could serve as a useful tool to study alcohol-induced cardiac defects and/or toxicity. In this study, hiPSC-CMs were treated with ethanol at doses corresponding to the clinically relevant levels of alcohol intoxication. hiPSC-CMs exposed to ethanol showed a dose-dependent increase in cellular damage and decrease in cell viability, corresponding to increased production of reactive oxygen species. Furthermore, ethanol exposure also generated dose-dependent increased irregular Ca2+ transients and contractility in hiPSC-CMs. RNA-seq analysis showed significant alteration in genes belonging to the potassium voltage-gated channel family or solute carrier family, partially explaining the irregular Ca2+ transients and contractility in ethanol-treated hiPSC-CMs. RNA-seq also showed significant upregulation in the expression of genes associated with collagen and extracellular matrix modeling, and downregulation of genes involved in cardiovascular system development and actin filament-based process. These results suggest that hiPSC-CMs can be a novel and physiologically relevant system for the study of alcohol-induced cardiac toxicity.
Keywords: cardiomyocytes, calcium handling, ethanol exposure, induced pluripotent stem cells, oxidative stress, RNA-seq
According to the Center for Disease Control and Prevention, more than 3 million women in the United States are at risk of exposing their fetus to alcohol during early pregnancy (Green et al., 2016). Prenatal alcohol exposure leads to various developmental anomalies including compromised myocardia function (Goh et al., 2011; Kvigne et al., 2004; May et al., 2007; Webster et al., 1984). Heart is the first organ to be established during embryonic development and is highly vulnerable to environmental insults such as alcohol exposure. Impaired heart growth and development in early life can also increase the risk for heart disease later in adulthood (Barker et al., 1989).
Traditionally, studies on alcohol exposure have relied on animal models and primary cardiomyocytes (CMs) from animals. These studies have established a link between the deleterious effect of ethanol and various cardiac malformations (Piano and Phillips, 2014). However, these models have limitations due to physiological differences from human primary CMs which are difficult to obtain and have limited growth capacity. Indeed, key features of alcohol-related human heart disease including congestive heart failure are not consistently recapitulated in animal models although alcohol decreases cardiac contractility in both animals and human (Fernandez-Sola et al., 2007; Hu et al., 2013). In addition, animal models are not suitable for high-throughput screening in drug discovery. Hence there is an unmet need for developing a physiologically relevant model that can reliably capture and reproduce alcohol-induced heart defects. To complement the studies on animal models/cells and bridge the gap between animal models/cells and human primary CMs, we hypothesized that human-induced pluripotent stem cell-derived CMs (hiPSC-CMs) could serve as a useful tool to study alcohol-induced cardiac defects.
hiPSC-CMs can provide an unlimited supply of physiologically relevant cell source with high-throughput capability for disease modeling and drug discovery. Although there are limitations regarding the use of hiPSC-CMs, these cells express the essential cardiac molecular and functional components that are similar to human primary fetal CMs and can be produced in large quantities (Burridge et al., 2012; Hartman et al., 2016; Laflamme and Murry, 2011; Xu, 2012). These functional hiPSC-CMs have been used as a model to study various cardiac diseases, such as long-QT syndromes (Itzhaki et al., 2011; Jha et al., 2016; Moretti et al., 2010), dilated cardiomyopathy (Sun et al., 2012), catecholaminergic polymorphic ventricular tachycardia (Jung et al., 2012; Kujala et al., 2012; Preininger et al., 2016), and cardiac hypertrophy (Ovchinnikova et al., 2018). In addition, hiPSC-CMs represent a novel tool for the evaluation of cardiac toxicity induced by drugs or other factors (Mercola et al., 2013; Savoji et al., 2019). For example, hiPSC-CMs have been leveraged to examine the toxic effect of cigarettes on heart development (Palpant et al., 2015).
In this study, we explored the possibility of utilizing hiPSC-CMs to examine cardiac phenotypes associated with alcohol exposure. Specifically, we examined the effect of ethanol exposure on cellular, molecular, and functional properties of hiPSC-CMs.
MATERIALS AND METHODS
Generation of hiPSC-CMs by directed differentiation and formation of 3D cardiac spheres
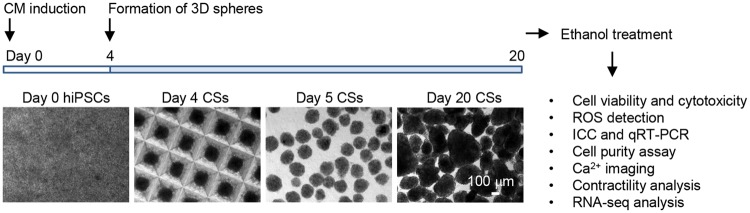
IMR90 hiPSCs (Yu et al., 2007) (by James A. Thomson, University of Wisconsin-Madison, obtained from WiCell Research Institute Inc.) were maintained in a feeder-free condition as described (Xu et al., 2001). Once confluent, cells were induced sequentially with 100 ng/ml activin A on differentiation day 0 and 10 ng/ml BMP4 on differentiation day 1 in RPMI medium containing 2% B27 without insulin (Jha et al., 2016; Preininger et al., 2016). On differentiation day 4, cultures were dissociated using 0.25% trypsin-EDTA, spun for 5 min at 300× g and resuspended for cell counting. Approximately 1750 cells were seeded per microwell in Aggrewell 400 plates (Stem Cell Technologies) cultured in RPMI medium containing 2% B27 and 10 μm Rock inhibitor Y-27632 to facilitate cell survival following dissociation (Nguyen et al., 2014; Watanabe et al., 2007). After 24 h, cardiac spheres were transferred to a suspension culture. Cardiac spheres typically started beating spontaneously by days 10–12 and were cultured until day 20 for subsequent ethanol exposure experiments (Figure 1A).
Figure 1.
Directed differentiation of hiPSCs and 3D culture of cardiac spheres generate enriched CMs and schematic diagram showing the experimental design. Human-induced pluripotent stem cell-derived cardiomyocytes were directed for CM differentiation and cardiac progenitors were forced to aggregate into 3D cardiac spheres (CSs) at differentiation day 4. Cardiac spheres were subsequently cultured in suspension and monitored for their spontaneous beating activities. Cells at day 20 were used for ethanol exposure and subsequent assessments. ROS, reactive oxygen species; ICC, immunocytochemical analysis; qRT-PCR, real-time quantitative reverse transcription polymerase chain reaction.
Rationale for ethanol doses
It is illegal in all jurisdictions within the United States to drive with blood alcohol concentration (BAC) of 0.08% (corresponding to 17 mM ethanol concentration) or more. Blood alcohol concentration higher than 0.2%, 0.3%, and 0.4% progressively corresponds to confusion, stupor and coma leading to alcohol intoxication, respectively. Supplementary Table 1 shows the BAC, corresponding ethanol concentration, and signs and symptoms at that level (adapted from http://www.intox.com/t-physiology.aspx). Several animal studies reported results in response to ethanol exposure with dosages ranging from 25 to 200 mM (Danziger et al., 1991; Delbridge et al., 2000; Mashimo et al., 2003, 2015; Mashimo and Ohno, 2006; Worley et al., 2015). We examined the effects of exposure of hiPSC-CMs to ethanol at the level that was at the legal limit of 17 mM and higher levels that correspond to different levels of alcohol intoxication. Cells were treated with ethanol at 17, 50, 100, and 200 mM in initial experiments. The highest concentration (200 mM) of ethanol had extensive deleterious effects on cell survival and almost all CMs stopped beating at the end of the treatment, hence it was excluded in subsequent experiments.
Immunocytochemical analysis
Cells were dissociated using 0.25% trypsin-EDTA and plated onto a Matrigel-coated 96-well culture plate at a density of 5× 104 cells/well and cultured for 2 days before fixation. On the day of the immunocytochemical staining, cells were washed with D-PBS and fixed in 2% (vol/vol) paraformaldehyde (Sigma) at room temperature for 15 min and permeabilized in cold methanol for 2 min at room temperature. The cells were then blocked with 5% normal goat serum (NGS) in D-PBS at room temperature for 1–2 h and incubated with the primary antibodies (Supplementary Table 2), including α-actinin (1:800), pan cadherin (1:200), NKX2-5 (1:200), cardiac troponin T (1:400), or cardiac troponin I (1:200) in 1% NGS overnight at 4°C. After the incubation with the primary antibodies, the cells were washed 3 times with D-PBS for 5 min each with gentle agitation to get rid of the unbound primary antibodies. The cells were then incubated with the corresponding conjugated secondary antibodies (1:1000) at room temperature for 1 h in the dark, followed by 3 times wash with D-PBS. The nuclei were counterstained with Vectashield mounting medium containing DAPI (Vector Laboratories, no. H-1200) (Jha et al., 2016; Preininger et al., 2016). Imaging was performed using an inverted microscope (Axio Vert.A1).
xCELLigence-real-time cellular analysis
Impedance-based cell viability and beating activity of hiPSC-CMs were monitored using xCELLigence-real-time cellular analysis (RTCA) CardioECR system (ACEA Biosciences, Inc.). E-Plate Cardio ECR48 (ACEA Biosciences, Inc.) was coated with 100 μl/well of 1:100 fibronectin in DPBS at 37°C for 2 h. After recording baseline reading in RPMI with 2% B27, hiPSC-CMs were dissociated with 0.25% trypsin-EDTA and seeded onto the E-Plate at a density of 6× 104 cells/well and cultured for 48 h in incubator. Cells were treated with ethanol at final concentrations of 50, 100, and 200 mM for 5 days. Mineral oil was added in treated as well as control wells to prevent ethanol evaporation. RTCA recordings were performed before the ethanol treatment at day 0 and thereafter daily from 1 to 5 days after the ethanol treatment. Data were analyzed using xCELLigence software (ACEA Biosciences, Inc.).
Cell viability and cytotoxicity assay
Cardiac spheres were dissociated using 0.25% trypsin-EDTA and plated onto a Matrigel-coated 96-well culture plate at a density of 5 ×104 cells/well and cultured for 2 days to recover spontaneous beating. Treatment groups were maintained in ethanol-containing medium for 5 days and the medium was covered with mineral oil to prevent ethanol evaporation. On day 5, ethanol-containing medium and mineral oil were aspirated and cells were washed 2 times with warm D-PBS and incubated with 0.5 μM calcein-AM (which labels live cells) and 1 μM ethidium homodimer-1 (which labels dead cells) for 30 min at 37°C in the dark. After the incubation, cells were washed 2 times with warm D-PBS and resuspended in warm phenol red free RPMI containing 2% B27 and immediately proceeded with imaging using an inverted microscope (Axio Vert.A1).
Detection of intracellular reactive oxygen species
Cardiac spheres were dissociated using 0.25% trypsin-EDTA and plated onto a Matrigel-coated 96-well culture plate at a density of 5×104 cells/well and cultured for 2 days to recover spontaneous beating. Treatment groups were maintained in ethanol-containing medium which was covered with mineral oil to prevent ethanol evaporation for 5 days. On day 5, ethanol-containing medium and mineral oil were aspirated and cells were washed 2 times with warm D-PBS and incubated with 25 μm carboxy-H2DCFDA (ThermoFisher Scientific) working solution in warm HBSS/Ca2+/Mg2+ for 30 min at 37°C, protected from light. Cells were washed 3 times with warm HBSS/Ca2+/Mg2+ and counter-stained with Hoechst in warm buffer and imaged immediately using Axio Vert.A1 inverted microscope (Zeiss).
Detection of mitochondrial superoxide production
Cardiac spheres were dissociated using 0.25% trypsin-EDTA and plated onto a Matrigel-coated 96-well culture plate at a density of 5×104 cells/well and cultured for 2 days to recover spontaneous beating. Treatment groups were maintained in ethanol-containing medium which was covered with mineral oil to prevent ethanol evaporation for additional 5 days. After the ethanol treatment, ethanol-containing medium and mineral oil were aspirated and cells were washed 2 times with HBSS/Ca2+/Mg2+. The generation of mitochondrial superoxide was assessed using MitoSOX Red (ThermoFisher Scientific). Cells were incubated with 5 µM MitoSOX Red working solution in warm HBSS/Ca2+/Mg2+ for 10 min at 37°C, protected from light. Cells were washed 2 times with warm HBSS/Ca2+/Mg2+ and counter-stained with Hoechst (ThermoFisher Scientific) in warm buffer and imaged immediately using an ArrayScan XTI Live High Content Platform (Life Technologies).
Images of MitoSOX Red and Hoechst were acquired and quantitatively analyzed using ArrayScan XTI Live High Content Platform. Twenty fields/well were selected and 5 replicate wells per condition were imaged using a 10× objective. Acquisition software Cellomics Scan (ThermoFisher Scientific) was used to capture images, and data analysis was performed using Cellomics View Software (ThermoFisher Scientific). Images were analyzed with mask modifier for Hoechst restricted to the nucleus. MitoSOX Red was quantified with a spot mask that extended 7 units from the nucleus. Spot threshold was set to 10 units and detection limit was set at 25 units. Mean MitoSOX Red average fluorescent fluorescence intensity of cells per well and percentage of MitoSOX Red-positive cells in each treatment group were used as readout.
qRT-PCR (real-time quantitative reverse transcription polymerase chain reaction)
RNA was isolated from control and treatment groups (n = 3/group) using Aurum total RNA mini kit (Bio-Rad) as per the manufacturer’s instructions. Briefly, cells were lysed in 350 μl lysis buffer supplemented with 1% β-mercaptoethanol, and stored at −80°C until further processing. Upon collecting RNA, samples were reverse-transcribed using 100 U of Superscript III enzyme and random primers in 20 μl reaction mixture containing Vilo reaction buffer as per the manufacturer’s instructions (SuperScript VILO cDNA Synthesis Kit by Life Technologies). cDNA was further processed in a Bio-Rad thermal cycler upon incubation with the reaction mixture at following temperature cycles: 25°C for 10 min, 42°C for 2 h, and 25°C for 5 min. The reaction mixture was then diluted 15–20 times before further use for real-time PCR (qPCR). Human-specific PCR primers (Supplementary Table 3) for the genes examined were retrieved from Jha et al., 2016 and an open access website (http://pga.mgh.harvard.edu/primerbank/). Thermocycler reaction was set up as follows using the iTaq SyBr green master mix: Initial denaturation step at 95°C for 10 min, 40 cycles of 2 steps with 15 s of denaturation at 95°C followed by 1 min of annealing at 60°C using Applied Biosystems 7500 real-time PCR systems. All samples were normalized to the level of the housekeeping gene GAPDH. Relative expression levels compared with control samples were presented as fold changes calculated by the 2−ΔΔCt method. Data are presented as mean ±SD. Statistical significance was analyzed using paired T-test; p values <.05 were considered significant.
Cell purity assay
Cardiac spheres were dissociated using 0.25% trypsin-EDTA and plated onto a Matrigel-coated 96-well culture plate at a density of 5×104 cells/well and cultured for 2 days to recover spontaneous beating. Treatment groups were maintained in ethanol-containing medium for 5 days and the medium was covered with mineral oil to prevent ethanol evaporation. On day 5, ethanol-containing medium and mineral oil were aspirated and cells were washed with D-PBS and fixed in paraformaldehyde (Sigma) and permeabilized in cold methanol. The cells were then blocked with 5% NGS in D-PBS at room temperature for 1 h and incubated with the primary antibody for NKX2-5 overnight at 4°C. After the incubation with the primary antibody, the cells were washed 3 times with D-PBS and incubated with the corresponding conjugated secondary antibodies followed by 3 times wash with D-PBS. The nuclei were counterstained with Hoechst in warm buffer and imaged using an ArrayScan XTI Live High Content Platform (Life Technologies).
Images of NKX2-5 positive cells and Hoechst were acquired and quantitatively analyzed using ArrayScan XTI Live High Content Platform. Twenty fields/well were selected and 5 replicate wells per condition were imaged using a 10× objective. Acquisition software Cellomics Scan (ThermoFisher Scientific) was used to capture images, and data analysis was performed using Cellomics View Software (ThermoFisher Scientific). Images were analyzed with mask modifier for Hoechst and NKX2-5-positive cells restricted to the nucleus. Percentage of NKX2-5-positive cells and mean average fluorescence intensity of NKX2-5 in each treatment were used as readout.
Calcium imaging
Human-induced pluripotent stem cell-derived CMs were dissociated with 0.05% trypsin-EDTA, seeded onto Matrigel-coated 25× 25 mm glass coverslips and cultured for 2–3 days until they recovered beating. The cells were then treated with 0, 17, and 50 mM of ethanol for 5 days in sealed chambers to prevent ethanol evaporation (Polikandriotis et al., 2005). For live cell imaging of intracellular Ca2+, the cells were incubated with 10 μM of Fluo-4 AM (ThermoFisher Scientific) for 30 min at 37°C in culture medium, washed for 30 min, and then transferred to an inverted laser confocal microscope (Olympus FV1000) equipped with FluoView software (Olympus), where they were perfused with normal tyrode solution (140 mM NaCl, 4 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 10 mM HEPES, 5 mM glucose, pH 7.4 with NaOH) (Lian et al., 2012). Fluo-4 was excited by the 488 nm laser and emitted fluorescence was captured at >505 nm. Recordings of Fluo-4 fluorescence were acquired in line-scan mode. Regions exhibiting heterogeneous fluorescence of Fluo-4 were avoided, so as to exclude artifacts (eg, endoplasmic reticulum, mitochondria, vesicles, etc.). Data were analyzed with ClampFit 10.0 software (Molecular Devices) (Burridge et al., 2014).
Video-based analyses of contractility
Contractility of hiPSC-CMs was recorded using a phase contrast inverted microscope (Axio Vert.A1) equipped with Zeiss AxioCam digital camera system, 20× magnification, 30 s for each sample, 100 ms interval between frames. Videos were processed and exported using Zeiss AxioVision LE imaging software. Video-based analysis of contractility parameters was performed with Matlab R2016b software as described (Huebsch et al., 2015).
RNA-seq analysis
RNA was isolated from control and ethanol-treated hiPSC-CMs (n = 3/group) using Aurum total RNA mini kit (Bio-Rad) as per the manufacturer’s instructions. RNA concentration was measured using a NanoDrop ND-1000 spectrophotometer (ThermoFisher Scientific). At the high-throughput DNA Sequencing Core, Parker H. Petit Institute for Bioengineering and Bioscience of Georgia Tech, total RNA quality was tested using a 2100 Bioanalyzer, and RNA 6000 Nano Chip (Agilent Technologies) and all samples had RNA integrity number ≥9.4. The Illumina TruSeq technology was then used to prepare RNA-Seq libraries, and next-generation sequencing was done through an Illumina HiSeq 2500, 75 bp paired end sequencing. RNA sequence reads were aligned to the human reference genome (GRCh38) (Kent et al., 2002) using HISAT2 (Kim et al., 2015), followed by using UCSC reference annotation and HTSeq (Anders et al., 2015) to estimate gene abundance. In total, there were 26 485 genes detected, though only 17 671 genes with greater than 5 reads in at least 3 samples were retained for downstream analyses. All downstream analyses were carried out in R v3.4.0 (R Development Core Team, 2014). The DESeq2 R package was used for normalization and detection of differentially expressed genes (Anders and Huber, 2010; Love et al., 2014). The up- and downregulated genes in ethanol-treated samples were identified following Benjamini-Hochberg corrected p-value at a significance threshold of .05 (Benjamini and Hochberg, 1995). Functional analysis of the up-/downregulated differentially expressed genes was completed using the ToppFun software package (Chen et al., 2009). Visualization was performed for upregulated gene ontology (GO) terms with FDR values of less than 0.05 and downregulated GO terms with FDR values of less than 0.01.
RESULTS
Directed Differentiation of hiPSCs and 3D Culture Generate Highly Enriched CMs
To generate hiPSC-CMs, we treated hiPSCs sequentially with growth factors activin A and BMP4 at differentiation days 0 and 1, respectively, and then generated cardiac spheres through microscale tissue engineering at day 4 (Figure 1). After suspension culture of the spheres for additional 16 days, cells were characterized for the expression of CM markers by immunocytochemical analysis. As shown in Supplementary Figure 1, the majority of the cells were positive for cardiac transcription factor NKX2-5 and structural proteins including cardiac troponin I, cardiac troponin T and cadherin, suggesting that the 3D cultures contained enriched CMs. These cells were then used for ethanol treatment and subsequent cellular, molecular, and functional assessments.
Ethanol Exposure of hiPSC-CMs Reduces Cell Viability
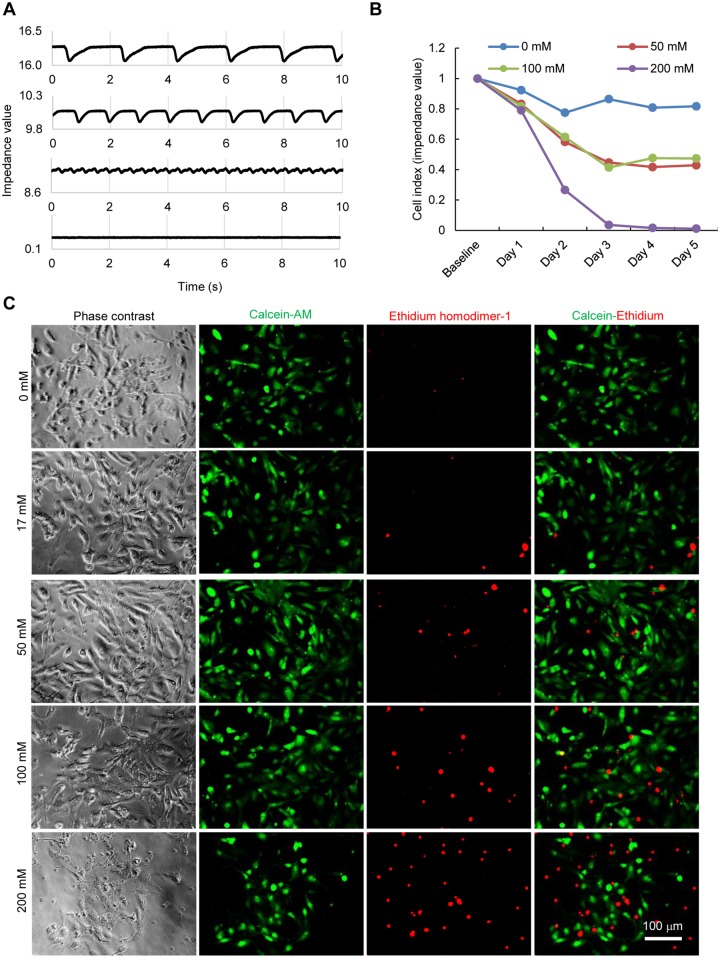
To examine the effect of ethanol exposure on the survival of hiPSC-CMs, we treated hiPSC-CMs with ethanol at doses corresponding to different levels of alcohol intoxication observed in clinic. We first monitored cellular impedance continuously for over a period of 5 days after the ethanol treatment using xCELLigence-RTCA system. The impedance of electron flow caused by adherent cells was measured noninvasively using microelectrode biosensors and then converted into a parameter called cell index which is positively correlated to the cell number, cell size and morphology, and the quality of cell-substrate attachment. After 3 days, the cell index in untreated group was 86.4% compared with initially seeded cells, whereas the cell index in the 50, 100, and 200 mM ethanol-treated groups decreased to 61.4%, 58.2%, and 3.6%, respectively (Figs. 2A and B). By day 5, the cell index was almost 0 in the group treated with 200 mM ethanol. These results indicate a progressive cell loss in hiPSC-CMs upon ethanol treatment.
Figure 2.
Ethanol exposure of hiPSC-CMs reduces cell viability and increases cytotoxicity. A, Representative trace showing cellular impedance based on xCELLigence-real-time cellular analysis. B, Graphical representation of progressive loss of cell viability upon ethanol exposure for 5 days. C, Images of LIVE/DEAD staining showing viable cells with green fluorescence from calcein AM staining and dead cells with red fluorescence from ethidium homodimer staining.
Next, we examined cell viability by the LIVE/DEAD staining (which segregates live or calcein-AM positive from dead or ethidium homodimer-1 positive cells) in cultures after 5 days of the treatment with ethanol at 0, 17, 50, 100, and 200 mM. As shown in Figure 2C, ethanol had dose-dependent deleterious effect on cellular viability. The cultures treated with ethanol contained more dead cells than did untreated cultures. In the culture treated with 200 mM ethanol, we observed cell detachment, and the remaining cells on the dish stopped beating and showed reduced cell viability (hence 200 mM treatment was excluded in subsequent experiments). These results were consistent with the cell index results acquired independently, showing that ethanol induced a dose-dependent cellular damage and cell loss in hiPSC-CMs.
Ethanol Exposure Generates Cellular Oxidative Stress and Increases Production of ROS
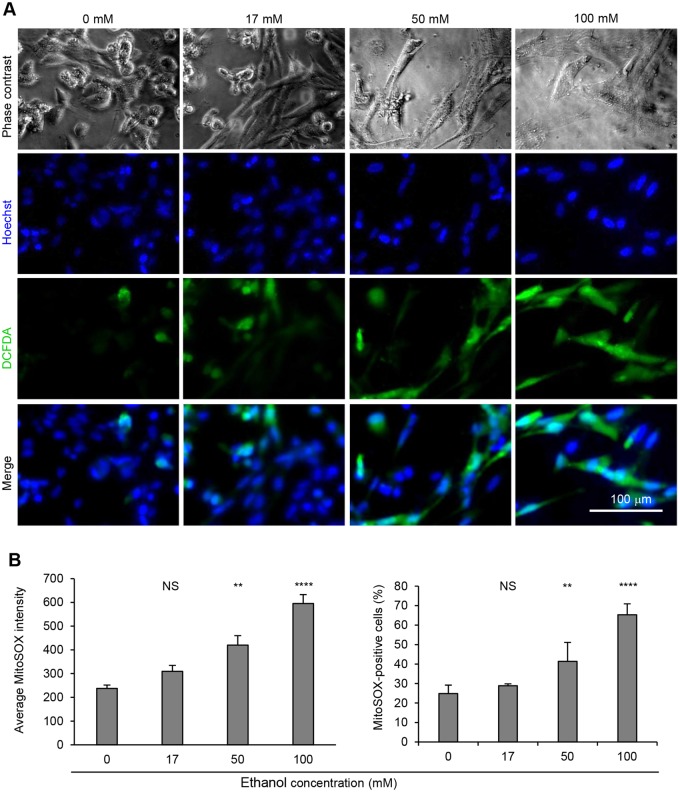
To examine if the reduced cell viability in ethanol-treated cells was associated with increased cellular oxidative stress, we treated hiPSC-CMs with various doses of ethanol (0, 17, 50, and 100 mM ethanol) for 5 days and initially measured reactive oxygen species (ROS) by H2DCFDA probe. As shown in Figure 3A, increased ROS signals were detected in the cells treated with ethanol, suggesting exposure of ethanol induces cellular oxidative stress in hiPSC-CMs. To further confirm these results, we quantitatively measured mitochondrial ROS production through ArrayScan XTI Live High Content Platform. An algorithm software was used to detect all nuclei stained with Hoechst, and masks were generated in the area of cells stained with MitoSOX Red around the nuclei to determine the mean average fluorescence intensity of MitoSOX Red per well and the percentage of MitoSOX Red-positive cells in cultures. Treatment of hiPSC-CMs with 50 and 100 mM ethanol significantly increased mitochondrial ROS production as indicated by both mean average fluorescence intensity of MitoSOX Red and the percentage of MitoSOX Red-positive cells (Figure 3B and Supplementary Figure 2).
Figure 3.
Ethanol exposure of hiPSC-CMs increases ROS production. A, Detection of intracellular ROS production. Human-induced pluripotent stem cell-derived cardiomyocytes were stained with DCFDA to detect ROS production upon ethanol exposure for 5 days. Reactive oxygen species-positive cells exhibited bright green cytosolic fluorescence and nuclei were stained with Hoechst. B, Detection of mitochondrial ROS production through ArrayScan. Human-induced pluripotent stem cell-derived cardiomyocytes were stained with MitoSOX Red to detect mitochondrial ROS production upon ethanol exposure for 5 days. Data presented as average fluorescence intensity per well and percentage of MitoSOX Red-positive cells, shown as mean±SD (n = 5). NS, no significant difference compared with control (0 mM). **p-value<.01; ****p-value < .0001. Representative images are presented in Figure S2.
Effect of Ethanol Exposure on the Expression of CM-Associated Markers
To assess the effect of ethanol treatment on the expression of cardiac markers, we analyzed the expression of α-actinin and NKX2-5 in treated (17, 50, and 100 mM ethanol) and untreated hiPSC-CMs by immunocytochemical analysis. As shown in Supplementary Figure 3A, the expression of α-actinin was detected in the cytoplasm of untreated and ethanol-treated hiPSC-CMs and NKX2-5 was detected in the nuclei of α-actinin+ cells in all groups. The signal intensities of the expression of α-actinin and NKX2-5 were comparable among untreated and ethanol-treated groups, and all groups contained cells with muscle striations as detected by the expression of α-actinin (Supplementary Figure 3A). We did not observe discernable loss of sarcomeric organization in hiPSC-CMs treated with ethanol at 100 mM. In addition, we observed no significant difference among these groups in the expression of cardiac markers including α-actinin, cardiac troponin I, and cardiac troponin T as examined by qRT-PCR (Supplementary Figure 3B).
In addition, we quantitatively measured NKX2-5 expression through ArrayScan XTI Live High Content Platform. An algorithm software was used to detect all nuclei stained with Hoechst, and masks restricted to the nuclei were generated in cells stained for NKX2-5, to determine the mean average fluorescence intensity per well and the percentage of NKX2-5-positive cells in cultures. Human-induced pluripotent stem cell-derived CMs in all ethanol-treated conditions showed no significant difference in NKX2-5 expression compared with untreated cells (Supplementary Figure 4).
Ethanol Exposure Alters Intracellular Ca2+ Transients and Contractility in hiPSC-CMs
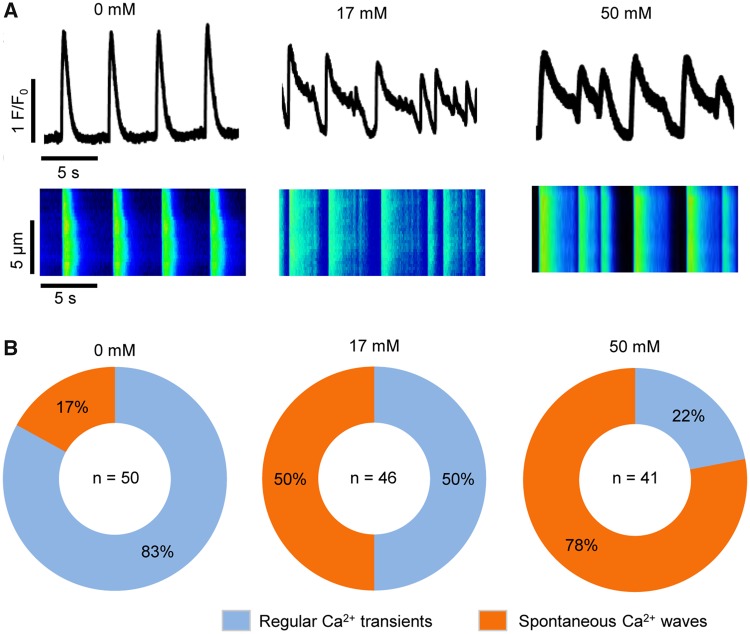
To test the effect of exposure of hiPSC-CMs to ethanol on Ca2+ handling, we assessed intracellular Ca2+ transients in untreated hiPSC-CMs (n = 50) and hiPSC-CMs treated with ethanol at 17 mM (n = 46) and 50 mM final concentrations (n = 41) using confocal line-scan imaging of cytosolic Ca2+. Two categories of whole cell Ca2+ release events were observed: Regular Ca2+ transients and spontaneous Ca2+ waves (SCW or irregular Ca2+ transients). Ca2+ transients were categorized as “regular” if the Ca2+ transients recorded over 1 min had mostly consistent amplitudes and beat periods, typical cardiac transient morphology (ie, rapid upstroke and decay kinetics), and no detectable instances of spontaneous Ca2+ release in-between transients (eg, representative transient trace of untreated cells in Figure 4A). Ca2+ transients were categorized as “SCWs” (Preininger et al., 2016) if they exhibited wavelets—oscillations of diastolic cytosolic Ca2+ in specific regions of interests along a cell, or waves—whole-cell oscillations of diastolic cytosolic Ca2+ (eg, representative Ca2+ transient traces of cells treated with 17 and 50 mM ethanol in Figure 4A). The number of cells exhibiting regular or irregular Ca2+ transients was counted, and percentages of the cells in each category were calculated for each condition (Figure 4B). For untreated hiPSC-CMs, the majority (83%) of the cells exhibited regular Ca2+ transients, with only 17% of the cells exhibiting SCWs. When the cells were treated with 17 mM ethanol, 50% of the cells had regular transients, and 50% of the cells had SCWs. When the cells were treated with 50 mM ethanol, only 22% of the cells had regular Ca2+ transients, and 78% of the cells had SCWs. These results suggest that exposure of hiPSC-CMs to ethanol increases the incidence of irregular Ca2+ transients in a dose-dependent manner. In addition, treatment of hiPSC-CMs with ROS scavenger N-acetyl cysteine (NAC) reduced both ethanol-induced ROS production (Supplementary Figs. 5A and B) and the incidence of abnormal Ca2+ transients in ethanol-treated hiPSC-CMs (Supplementary Figs. 5C and D).
Figure 4.
Ethanol exposure of hiPSC-CMs alters intracellular Ca2+ handling. A, Representative confocal images showing cytosolic calcium traces and line-scan of intercellular Ca2+ transients in cells from control (0 mM ethanol) and the treatment groups (17 and 50 mM ethanol). B, Pie chart showing the percentage of cells exhibiting regular Ca2+ transients or Ca2+ transients with spontaneous Ca2+ waves (SCWs) under each condition. Sample sizes (n) are denoted in the center of the graphs for the control and treatment groups.
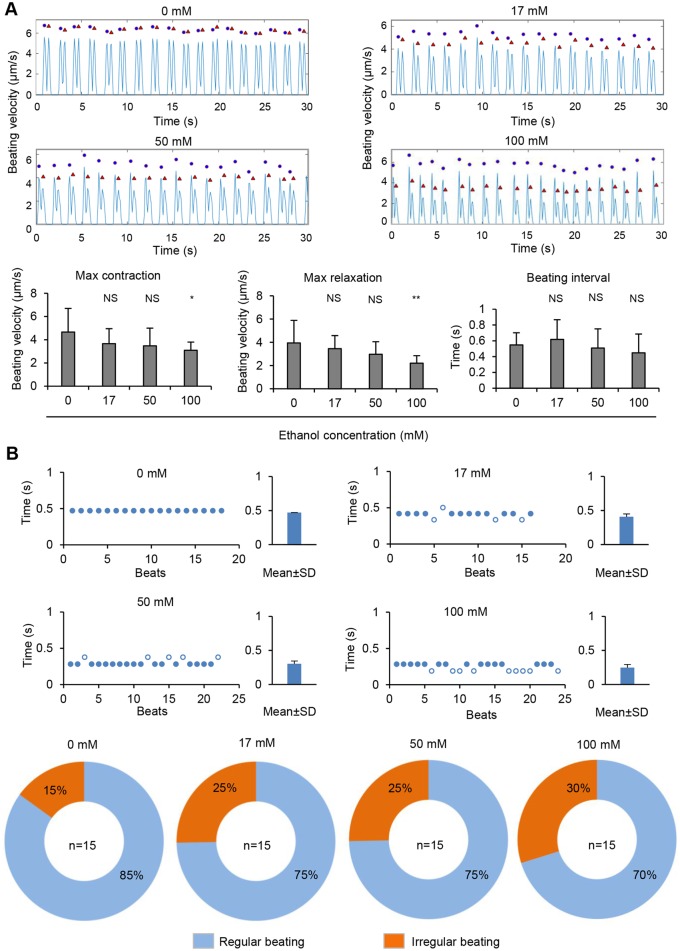
We next compared cardiac contractility between untreated and ethanol-treated hiPSC-CMs using a video-based method (Huebsch et al., 2015). Treatment of hiPSC-CMs with ethanol at 100 mM significantly decreased maximum contraction and maximum relaxation, although cells treated with ethanol at 17 and 50 mM did not alter these parameters (Figure 5A). Treatment of hiPSC-CMs with ethanol did not significantly alter the average of beating interval (Figure 5A). We observed, however, an increase in the incidence of irregular beating, from 15% in untreated hiPSC-CMs to 25% in cells treated with 17 and 50 mM ethanol and 30% in cells treated with 100 mM ethanol (Figure 5B). These results suggest that exposure of hiPSC-CMs to ethanol affect cardiac contractility.
Figure 5.
Ethanol exposure of hiPSC-CMs alters cells contractility. A, Video-based analysis of contractility. Representative traces show beating velocity recording of cardiomyocytes under each condition. Dots denote contraction, and triangles denote relaxation. Contraction parameters are presented as mean±SD of n = 15 biological samples. NS, no significant difference compared with control (0 mM). *p-value < .05; **p-value < .01. B, Scatter charts show representative traces of beating for hiPSC-CMs under each condition. The irregular beating (hollow dots) are those with beating period larger than mean+SD of beat to beat time or smaller than mean – SD of beat to beat time. Bar graphs of beating periods are presented as mean±SD of beat to beat time. Pie chart showing the percentage of cells with regular beating or irregular beating under each condition. Sample sizes (n) are denoted in the center of the graphs for the control and treatment groups.
RNA-seq Analysis Showed Genes Significantly Altered in hiPSC-CMs Upon Treatment With Ethanol
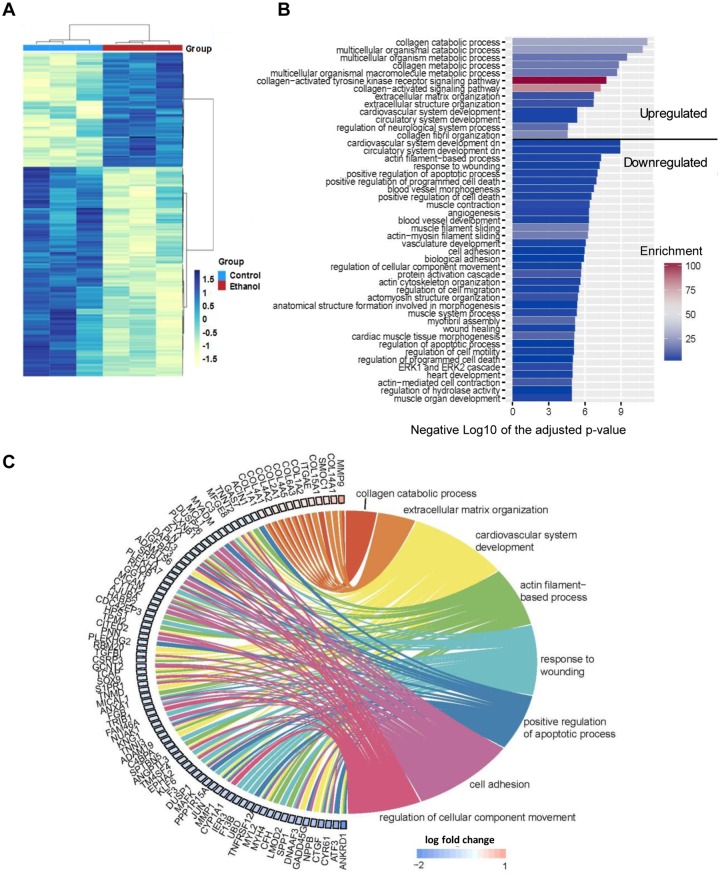
To further evaluate the molecular alteration of hiPSC-CMs induced by the ethanol treatment, we performed RNA-seq analysis to compare global gene expression profiles of hiPSC-CMs treated with ethanol with those without ethanol treatment. As detected by RNA-seq, 83 genes were significantly upregulated and 152 downregulated in ethanol-treated hiPSC-CMs (Figure 6A and Supplementary Figure 6). Among the top 60 genes significantly altered by ethanol exposure, 13.3% (8 genes) are members either of the potassium voltage-gated channel family or solute carrier family (Supplementary Table 4). The changes in expression of genes involved in ion channels may be in part contributing to the abnormal intracellular Ca2+ transients we observed in ethanol-treated hiPSC-CMs.
Figure 6.
Differentially expressed genes and gene ontology (GO) terms identified by RNA-seq analysis of hiPSC-CMs upon treatment with ethanol. A, Heatmap of differentially expressed genes between control and ethanol-treated hiPSC-CMs (n = 3). B, GO results bar plot. The GO terms are summarized using REVIGO. Length of bar indicates the gene enrichment score and color of bar is –log10 (p-value). C, Chord diagram showing the relationship between interested GO clusters and corresponding differentially expressed genes. In each chord diagram, enriched GO terms were presented on the right, and genes contributing to these enrichments were drawn on the left. Colored squares on the left are indicated LFC value and ordered from highest to lowest LFC.
Gene ontology showed that ethanol upregulated the expression of genes associated with collagen metabolism and extracellular matrix (ECM) modeling in hiPSC-CMs (Figs. 6B and C). Among the top 5 upregulated genes (Supplementary Table 4), 3 were related to ECM or the reorganization of ECM (MMP9, EMID1, and COL14A1). Gene ontology terms also showed that most of the downregulated genes are involved in cardiovascular system development (NPPB, DNAAF3), actin filament-based process (LMOD2, MYH4), and muscle contraction (MYL2) (Figs. 6B and C). These findings are consistent with previous studies showing a correlation between alcohol exposure and defects in heart and circulatory system development (Caputo et al., 2016; Sarmah and Marrs, 2013).
DISCUSSION
Our results demonstrate that exposure of hiPSC-CMs to ethanol reduces cell viability, increases cell loss, and leads to overproduction of ROS. In functional and molecular aspects, ethanol exposure results in deranged Ca2+ handling in hiPSC-CMs and altered expression of genes involved in cardiovascular system development and ECM remodeling. These results indicate that hiPSC-CMs can serve as a novel, human cell-based model for the characterization of cardiac defects induced by ethanol exposure and these cells have the potential to be used as a platform for high-throughput screening and discovery of novel drugs to treat alcohol-induced cardiac defects. Furthermore, we provide a unique resource of human whole transcriptomic dataset for the study of alcohol-induced cardiotoxicity (the RNA-seq data reported in this manuscript are available on the GEO database with accession number GSE125917).
Our results of ethanol-induced dose-dependent cytotoxicity, as demonstrated by ethidium homodimer staining and quantitative assessment of cell index by xCELLigence-RTCA, are consistent with previous findings in animal studies. A strong association between ethanol exposure and cellular apoptosis leading to cellular loss has been observed in various cell types including CMs (Capasso et al., 1992; Chen et al., 2000; Li et al., 2006; Tan et al., 2012). Furthermore, ethanol exposure-induced cell loss ultimately progresses to heart failure in an animal model (Capasso et al., 1992).
Ethanol exposure of hiPSC-CMs also results in overproduction of ROS in a dose-dependent manner. This observation in hiPSC-CMs mirrors other animal studies that report extensive ROS generation and cell death upon ethanol exposure (Shi et al., 2016; Wang et al., 2015). Overproduction of ROS can lead to cellular oxidative stress which in turn results in abnormal embryogenesis (Guan et al., 2004; Jing et al., 2012; Kannan et al., 2004; Sarmah and Marrs, 2013; Umoh et al., 2014). The ROS generation has also been reported in cultured cells, including cardiac cells, upon ethanol exposure (Guan et al., 2004; Jing et al., 2012; Kannan et al., 2004). For example, ethanol-induced ROS generation in H9c2 CMs results in apoptosis, highlighting the vital role of cellular antioxidants in attenuating ethanol-induced cytotoxicity (Shi et al., 2016).
The sources of ROS generation we detected also show similarities to the observation in animal and human primary CMs upon ethanol exposure. In CMs, ROS can be generated through mitochondrial and nonmitochondrial sources (Santos et al., 2011). Ethanol exposure of hiPSC-CMs results in overproduction of ROS from both mitochondrial and nonmitochondrial sources, as we detected intracellular ROS (nonmitochondrial hydrogen peroxide) with the DCFDA probe and mitochondrial ROS (mitochondrial superoxide anion) with the MitoSOX Red probe. This observation is consistent with studies showing that ethanol exposure in mice increased the production of both intracellular ROS and mitochondrial ROS in CMs (Guo and Ren, 2010; Wang et al., 2015). In addition, extensive intracellular ROS production was also observed in human fetal CMs treated with ethanol or an ethanol metabolite (Li et al., 2006).
Intracellular Ca2+ signaling plays an important role in the regulation of contraction of cardiac cells (Berridge et al., 2003), and alcohol exposure increases the risk of arrhythmias (Kodama et al., 2011; Mandyam et al., 2012) even for individuals with modest levels of alcohol intake (Voskoboinik et al., 2016). Our results show that ethanol exposure to hiPSC-CMs increases the incidence of irregular intracellular Ca2+ transients in the form of SCWs in a dose-dependent manner. Previous studies report intracellular Ca2+ dysregulation including transient irregular Ca2+ transients (SCWs), altered rates of decay time and rise time, or amplitude of Ca2+ transients, in mouse or rat CMs exposed to ethanol or its metabolite acetaldehyde (Ge et al., 2011; Guo and Ren, 2010; Oba et al., 2008; Zhang et al., 2010). A recent study also reports that ethanol induces severe Ca2+ leak from sarcoplasmic reticulum (SR) in human atrial CMs and murine ventricular CMs (Mustroph et al., 2018). Similarly, we observed persistent irregular Ca2+ transients indicated by oscillations of diastolic cytosolic Ca2+ (Ca2+ waves) in hiPSC-CMs. These results suggest that hiPSC-CMs can be used as a new human CM model for investigating the effect of ethanol exposure on Ca2+ transients and examining whether these irregular Ca2+ transients are responsible for ethanol-induced arrhythmogenic events.
In many cardiac disorders, deranged ion channels and Ca2+ signaling are linked to the overproduction of ROS (Giordano, 2005). Reactive oxygen species is known to modulate the activity of several key Ca2+ handling proteins, including RyR2 (cardiac ryanodine receptor) and SERCA (SR calcium transport ATPase) which are responsible for SR Ca2+ release and uptake, respectively (Santos et al., 2011). Such modifications contribute to abnormal Ca2+ handling and increased risk of arrhythmia (Santos et al., 2011). Consistent with the role of ROS in regulating Ca2+ handling, our results indicate that ROS scavenger NAC reduced the ethanol-induced ROS production and abnormal Ca2+ transients in hiPSC-CMs.
Our RNA-seq results reveal gene expression signatures of cardiac toxicity in hiPSC-CMs that are consistent with alcohol-induced pathophysiology observed in animal models and the clinic. For example, 3 of the top 5 upregulated genes (MMP9, EMID1, and COL14A1) in ethanol-treated hiPSC-CMs are related to ECM or the reorganization of ECM. Similarly, MMP9 concentrations are significantly higher in human sera of chronic alcohol abusers (Sillanaukee et al., 2002), and MMP9 mRNA and protein levels are increased in the myocardium of rats following acute ethanol exposure (Li et al., 2012). Collagen content in myocardium is approximately 10-fold higher in patients with alcoholic dilated cardiomyopathy than in controls without heart disease (Soufen et al., 2008), and collagen mRNA and/or protein expression in the heart are significantly increased in rats following long-term exposure to alcohol (El Hajj et al., 2014; Steiner et al., 2015).
In summary, our results suggest that exposure of hiPSC-CMs to ethanol triggers myocyte loss, oxidative stress, and altered Ca2+ handling. These findings support the utility of hiPSC-CMs as a novel, physiologically relevant in vitro system to explore the molecular underpinnings of prenatal alcohol exposure. In addition, this cell model can be leveraged for high-throughput screening of potential drugs to treat alcohol-induced cardiac defects. For example, the irregular Ca2+ transients observed in ethanol-treated hiPSC-CMs may be served as a functional readout for screening antiarrhythmic drugs using high-throughput Ca2+ imaging. Such a study will not only advance the application of hiPSCs in drug discovery for treatment of alcohol exposure-induced heart disease but also facilitate establishment of hiPSC models for studies of alcohol-induced damages in other organs because hiPSCs have the ability to differentiate into various cell types. Moreover, hiPSC lines of variable genetic backgrounds (eg, different activities of enzymes for detoxification) may also facilitate investigation on the effects of genetic variation on the susceptibility to alcohol-induced injury as well as the development of effective treatments.
Supplementary Material
ACKNOWLEDGMENTS
We thank the staff at the high-throughput DNA Sequencing Core of the Parker H. Petit Institute for Bioengineering & Bioscience, Georgia Institute of Technology for their help with the preparation of the RNA-seq library.
FUNDING
Center for Pediatric Technology Center at Emory/Georgia Tech, NIH/NIEHS (P30ES019776), Woodruff Health Sciences Center, NIH/NIAAA (R21AA025723), and NIH/NHLBI (R01HL136345).
DECLARATION OF CONFILICTING INTERESTS
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
REFERENCES
- Anders S., Huber W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11, R106.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P. T., Huber W. (2015). HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker D. J., Winter P. D., Osmond C., Margetts B., Simmonds S. J. (1989). Weight in infancy and death from ischaemic heart disease. Lancet 2, 577–580. [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300. [Google Scholar]
- Berridge M. J., Bootman M. D., Roderick H. L. (2003). Calcium signalling: Dynamics, homeostasis and remodelling. Nat. Rev. Mol. Cell Biol. 4, 517–529. [DOI] [PubMed] [Google Scholar]
- Burridge P. W., Keller G., Gold J. D., Wu J. C. (2012). Production of de novo cardiomyocytes: Human pluripotent stem cell differentiation and direct reprogramming. Cell Stem Cell 10, 16–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge P. W., Matsa E., Shukla P., Lin Z. C., Churko J. M., Ebert A. D., Lan F., Diecke S., Huber B., Mordwinkin N. M., et al. (2014). Chemically defined generation of human cardiomyocytes. Nat. Methods 11, 855–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capasso J. M., Li P., Guideri G., Malhotra A., Cortese R., Anversa P. (1992). Myocardial mechanical, biochemical, and structural alterations induced by chronic ethanol ingestion in rats. Circ. Res. 71, 346–356. [DOI] [PubMed] [Google Scholar]
- Caputo C., Wood E., Jabbour L. (2016). Impact of fetal alcohol exposure on body systems: A systematic review. Birth Defects Res. C Embryo Today 108, 174–180. [DOI] [PubMed] [Google Scholar]
- Chen D. B., Wang L., Wang P. H. (2000). Insulin-like growth factor I retards apoptotic signaling induced by ethanol in cardiomyocytes. Life Sci. 67, 1683–1693. [DOI] [PubMed] [Google Scholar]
- Chen J., Bardes E. E., Aronow B. J., Jegga A. G. (2009). ToppGene suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 37, W305–W311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danziger R. S., Sakai M., Capogrossi M. C., Spurgeon H. A., Hansford R. G., Lakatta E. G. (1991). Ethanol acutely and reversibly suppresses excitation-contraction coupling in cardiac myocytes. Circ. Res. 68, 1660–1668. [DOI] [PubMed] [Google Scholar]
- Delbridge L. M., Connell P. J., Harris P. J., Morgan T. O. (2000). Ethanol effects on cardiomyocyte contractility. Clin. Sci. (Lond.) 98, 401–407. [PubMed] [Google Scholar]
- El Hajj E. C., El Hajj M. C., Voloshenyuk T. G., Mouton A. J., Khoutorova E., Molina P. E., Gilpin N. W., Gardner J. D. (2014). Alcohol modulation of cardiac matrix metalloproteinases (MMPs) and tissue inhibitors of MMPs favors collagen accumulation. Alcohol Clin. Exp. Res. 38, 448–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Sola J., Preedy V. R., Lang C. H., Gonzalez-Reimers E., Arno M., Lin J. C., Wiseman H., Zhou S., Emery P. W., Nakahara T., et al. (2007). Molecular and cellular events in alcohol-induced muscle disease. Alcohol Clin. Exp. Res. 31, 1953–1962. [DOI] [PubMed] [Google Scholar]
- Ge W., Guo R., Ren J. (2011). AMP-dependent kinase and autophagic flux are involved in aldehyde dehydrogenase-2-induced protection against cardiac toxicity of ethanol. Free Radic. Biol. Med. 51, 1736–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano F. J. (2005). Oxygen, oxidative stress, hypoxia, and heart failure. J. Clin. Invest. 115, 500–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh J. M., Bensley J. G., Kenna K., Sozo F., Bocking A. D., Brien J., Walker D., Harding R., Black M. J. (2011). Alcohol exposure during late gestation adversely affects myocardial development with implications for postnatal cardiac function. Am. J. Physiol. Heart Circ. Physiol. 300, H645–H651. [DOI] [PubMed] [Google Scholar]
- Green P. P., McKnight-Eily L. R., Tan C. H., Mejia R., Denny C. H. (2016). Vital signs: Alcohol-exposed pregnancies–United States, 2011-2013. MMWR Morb. Mortal Wkly. Rep. 65, 91–97. [DOI] [PubMed] [Google Scholar]
- Guan Z., Lui C. Y., Morkin E., Bahl J. J. (2004). Oxidative stress and apoptosis in cardiomyocyte induced by high-dose alcohol. J. Cardiovasc. Pharmacol. 44, 696–702. [DOI] [PubMed] [Google Scholar]
- Guo R., Ren J. (2010). Alcohol dehydrogenase accentuates ethanol-induced myocardial dysfunction and mitochondrial damage in mice: Role of mitochondrial death pathway. PLoS One 5, e8757.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman M. E., Dai D. F., Laflamme M. A. (2016). Human pluripotent stem cells: Prospects and challenges as a source of cardiomyocytes for in vitro modeling and cell-based cardiac repair. Adv. Drug Deliv. Rev. 96, 3–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C., Ge F., Hyodo E., Arai K., Iwata S., Lobdell H. t., Walewski J. L., Zhou S., Clugston R. D., Jiang H., et al. (2013). Chronic ethanol consumption increases cardiomyocyte fatty acid uptake and decreases ventricular contractile function in C57BL/6J mice. J. Mol. Cell Cardiol. 59, 30–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huebsch N., Loskill P., Mandegar M. A., Marks N. C., Sheehan A. S., Ma Z., Mathur A., Nguyen T. N., Yoo J. C., Judge L. M., et al. (2015). Automated video-based analysis of contractility and calcium flux in human-induced pluripotent stem cell-derived cardiomyocytes cultured over different spatial scales. Tissue Eng. Part C Methods 21, 467–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzhaki I., Maizels L., Huber I., Zwi-Dantsis L., Caspi O., Winterstern A., Feldman O., Gepstein A., Arbel G., Hammerman H., et al. (2011). Modelling the long QT syndrome with induced pluripotent stem cells. Nature 471, 225–229. [DOI] [PubMed] [Google Scholar]
- Jha R., Wu Q., Singh M., Preininger M. K., Han P., Ding G., Cho H. C., Jo H., Maher K. O., Wagner M. B., et al. (2016). Simulated microgravity and 3D culture enhance induction, viability, proliferation and differentiation of cardiac progenitors from human pluripotent stem cells. Sci Rep. 6, 30956.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing L., Jin C. M., Li S. S., Zhang F. M., Yuan L., Li W. M., Sang Y., Li S., Zhou L. J. (2012). Chronic alcohol intake-induced oxidative stress and apoptosis: Role of CYP2E1 and calpain-1 in alcoholic cardiomyopathy. Mol. Cell Biochem. 359, 283–292. [DOI] [PubMed] [Google Scholar]
- Jung C. B., Moretti A., Mederos y Schnitzler M., Iop L., Storch U., Bellin M., Dorn T., Ruppenthal S., Pfeiffer S., Goedel A., et al. (2012). Dantrolene rescues arrhythmogenic RYR2 defect in a patient-specific stem cell model of catecholaminergic polymorphic ventricular tachycardia. EMBO Mol. Med. 4, 180–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan M., Wang L., Kang Y. J. (2004). Myocardial oxidative stress and toxicity induced by acute ethanol exposure in mice. Exp. Biol. Med. (Maywood) 229, 553–559. [DOI] [PubMed] [Google Scholar]
- Kent W. J., Sugnet C. W., Furey T. S., Roskin K. M., Pringle T. H., Zahler A. M., Haussler D. (2002). The human genome browser at UCSC. Genome Res. 12, 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Langmead B., Salzberg S. L. (2015). HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama S., Saito K., Tanaka S., Horikawa C., Saito A., Heianza Y., Anasako Y., Nishigaki Y., Yachi Y., Iida K. T., et al. (2011). Alcohol consumption and risk of atrial fibrillation: A meta-analysis. J. Am. Coll. Cardiol. 57, 427–436. [DOI] [PubMed] [Google Scholar]
- Kujala K., Paavola J., Lahti A., Larsson K., Pekkanen-Mattila M., Viitasalo M., Lahtinen A. M., Toivonen L., Kontula K., Swan H., et al. (2012). Cell model of catecholaminergic polymorphic ventricular tachycardia reveals early and delayed afterdepolarizations. PLoS One 7, e44660.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvigne V. L., Leonardson G. R., Neff-Smith M., Brock E., Borzelleca J., Welty T. K. (2004). Characteristics of children who have full or incomplete fetal alcohol syndrome. J. Pediatr. 145, 635–640. [DOI] [PubMed] [Google Scholar]
- Laflamme M. A., Murry C. E. (2011). Heart regeneration. Nature 473, 326–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Korkmaz S., Loganathan S., Weymann A., Radovits T., Barnucz E., Hirschberg K., Hegedus P., Zhou Y., Tao L., et al. (2012). Acute ethanol exposure increases the susceptibility of the donor hearts to ischemia/reperfusion injury after transplantation in rats. PLoS One 7, e49237.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. Y., Li Q., Shen J. J., Dong F., Sigmon V. K., Liu Y., Ren J. (2006). Attenuation of acetaldehyde-induced cell injury by overexpression of aldehyde dehydrogenase-2 (ALDH2) transgene in human cardiac myocytes: Role of MAP kinase signaling. J. Mol. Cell Cardiol. 40, 283–294. [DOI] [PubMed] [Google Scholar]
- Lian X., Hsiao C., Wilson G., Zhu K., Hazeltine L. B., Azarin S. M., Raval K. K., Zhang J., Kamp T. J., Palecek S. P. (2012). Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc. Natl. Acad. Sci. U.S.A. 109, E1848–E1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandyam M. C., Vedantham V., Scheinman M. M., Tseng Z. H., Badhwar N., Lee B. K., Lee R. J., Gerstenfeld E. P., Olgin J. E., Marcus G. M. (2012). Alcohol and vagal tone as triggers for paroxysmal atrial fibrillation. Am. J. Cardiol. 110, 364–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashimo K., Arthur P. G., Ohno Y. (2015). Ethanol dose- and time-dependently increases alpha and beta subunits of mitochondrial ATP synthase of cultured neonatal rat cardiomyocytes. J. Nippon Med. Sch. 82, 237–245. [DOI] [PubMed] [Google Scholar]
- Mashimo K., Ohno Y. (2006). Ethanol hyperpolarizes mitochondrial membrane potential and increases mitochondrial fraction in cultured mouse myocardial cells. Arch. Toxicol. 80, 421–428. [DOI] [PubMed] [Google Scholar]
- Mashimo K., Sato S., Ohno Y. (2003). Chronic effects of ethanol on cultured myocardial cells: Ultrastructural and morphometric studies. Virchows. Arch. 442, 356–363. [DOI] [PubMed] [Google Scholar]
- May P. A., Gossage J. P., Marais A. S., Adnams C. M., Hoyme H. E., Jones K. L., Robinson L. K., Khaole N. C., Snell C., Kalberg W. O., et al. (2007). The epidemiology of fetal alcohol syndrome and partial FAS in a South African community. Drug Alcohol Depend. 88, 259–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercola M., Colas A., Willems E. (2013). Induced pluripotent stem cells in cardiovascular drug discovery. Circ. Res. 112, 534–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti A., Bellin M., Welling A., Jung C. B., Lam J. T., Bott-Flugel L., Dorn T., Goedel A., Hohnke C., Hofmann F., et al. (2010). Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N. Engl. J. Med. 363, 1397–1409. [DOI] [PubMed] [Google Scholar]
- Mustroph J., Wagemann O., Lebek S., Tarnowski D., Ackermann J., Drzymalski M., Pabel S., Schmid C., Wagner S., Sossalla S., et al. (2018). SR Ca(2+)-leak and disordered excitation-contraction coupling as the basis for arrhythmogenic and negative inotropic effects of acute ethanol exposure. J. Mol. Cell Cardiol. 116, 81–90. [DOI] [PubMed] [Google Scholar]
- Nguyen D. C., Hookway T. A., Wu Q., Jha R., Preininger M. K., Chen X., Easley C. A., Spearman P., Deshpande S. R., Maher K., et al. (2014). Microscale generation of cardiospheres promotes robust enrichment of cardiomyocytes derived from human pluripotent stem cells. Stem Cell Rep. 3, 260–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oba T., Maeno Y., Nagao M., Sakuma N., Murayama T. (2008). Cellular redox state protects acetaldehyde-induced alteration in cardiomyocyte function by modifying Ca2+ release from sarcoplasmic reticulum. Am. J. Physiol. Heart Circ. Physiol. 294, H121–H133. [DOI] [PubMed] [Google Scholar]
- Ovchinnikova E., Hoes M., Ustyantsev K., Bomer N., de Jong T. V., van der Mei H., Berezikov E., van der Meer P. (2018). Modeling human cardiac hypertrophy in stem cell-derived cardiomyocytes. Stem Cell Rep., 10, 794–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palpant N. J., Hofsteen P., Pabon L., Reinecke H., Murry C. E. (2015). Cardiac development in zebrafish and human embryonic stem cells is inhibited by exposure to tobacco cigarettes and e-cigarettes. PLoS One 10, e0126259.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piano M. R., Phillips S. A. (2014). Alcoholic cardiomyopathy: Pathophysiologic insights. Cardiovasc. Toxicol. 14, 291–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polikandriotis J. A., Rupnow H. L., Hart C. M. (2005). Chronic ethanol exposure stimulates endothelial cell nitric oxide production through PI-3 kinase-and hsp90-dependent mechanisms. Alcohol Clin. Exp. Res. 29, 1932–1938. [DOI] [PubMed] [Google Scholar]
- Preininger M. K., Jha R., Maxwell J. T., Wu Q., Singh M., Wang B., Dalal A., McEachin Z. T., Rossoll W., Hales C. M., et al. (2016). A human pluripotent stem cell model of catecholaminergic polymorphic ventricular tachycardia recapitulates patient-specific drug responses. Dis. Model Mech. 9, 927–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. (2014). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Santos C. X., Anilkumar N., Zhang M., Brewer A. C., Shah A. M. (2011). Redox signaling in cardiac myocytes. Free Radic. Biol. Med. 50, 777–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarmah S., Marrs J. A. (2013). Complex cardiac defects after ethanol exposure during discrete cardiogenic events in zebrafish: Prevention with folic acid. Dev. Dyn. 242, 1184–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savoji H., Mohammadi M. H., Rafatian N., Toroghi M. K., Wang E. Y., Zhao Y., Korolj A., Ahadian S., Radisic M. (2019). Cardiovascular disease models: A game changing paradigm in drug discovery and screening. Biomaterials, 198, 3–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X., Li Y., Hu J., Yu B. (2016). Tert-butylhydroquinone attenuates the ethanol-induced apoptosis of and activates the Nrf2 antioxidant defense pathway in H9c2 cardiomyocytes. Int. J. Mol. Med. 38, 123–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillanaukee P., Kalela A., Seppa K., Hoyhtya M., Nikkari S. T. (2002). Matrix metalloproteinase-9 is elevated in serum of alcohol abusers. Eur. J. Clin. Invest. 32, 225–229. [DOI] [PubMed] [Google Scholar]
- Soufen H. N., Salemi V. M., Aneas I. M., Ramires F. J., Benicio A. M., Benvenuti L. A., Krieger J. E., Mady C. (2008). Collagen content, but not the ratios of collagen type III/I mRNAs, differs among hypertensive, alcoholic, and idiopathic dilated cardiomyopathy. Braz. J. Med. Biol. Res. 41, 1098–1104. [DOI] [PubMed] [Google Scholar]
- Steiner J. L., Pruznak A. M., Navaratnarajah M., Lang C. H. (2015). Alcohol differentially alters extracellular matrix and adhesion molecule expression in skeletal muscle and heart. Alcohol Clin. Exp. Res. 39, 1330–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N., Yazawa M., Liu J., Han L., Sanchez-Freire V., Abilez O. J., Navarrete E. G., Hu S., Wang L., Lee A., et al. (2012). Patient-specific induced pluripotent stem cells as a model for familial dilated cardiomyopathy. Sci. Transl. Med. 4, 130ra47.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y., Li X., Prabhu S. D., Brittian K. R., Chen Q., Yin X., McClain C. J., Zhou Z., Cai L. (2012). Angiotensin II plays a critical role in alcohol-induced cardiac nitrative damage, cell death, remodeling, and cardiomyopathy in a protein kinase C/nicotinamide adenine dinucleotide phosphate oxidase-dependent manner. J. Am. Coll. Cardiol. 59, 1477–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umoh N. A., Walker R. K., Al-Rubaiee M., Jeffress M. A., Haddad G. E. (2014). Acute alcohol modulates cardiac function as PI3K/Akt regulates oxidative stress. Alcohol Clin. Exp. Res. 38, 1847–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voskoboinik A., Prabhu S., Ling L. H., Kalman J. M., Kistler P. M. (2016). Alcohol and atrial fibrillation: A sobering review. J. Am. Coll. Cardiol. 68, 2567–2576. [DOI] [PubMed] [Google Scholar]
- Wang Y., Zhao J., Yang W., Bi Y., Chi J., Tian J., Li W. (2015). High-dose alcohol induces reactive oxygen species-mediated apoptosis via PKC-beta/p66Shc in mouse primary cardiomyocytes. Biochem. Biophys. Res. Commun. 456, 656–661. [DOI] [PubMed] [Google Scholar]
- Watanabe K., Ueno M., Kamiya D., Nishiyama A., Matsumura M., Wataya T., Takahashi J. B., Nishikawa S., Nishikawa S.-I., Muguruma K., et al. (2007). A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat. Biotechnol. 25, 681–686. [DOI] [PubMed] [Google Scholar]
- Webster W. S., Germain M. A., Lipson A., Walsh D. (1984). Alcohol and congenital heart defects: An experimental study in mice. Cardiovasc. Res. 18, 335–338. [DOI] [PubMed] [Google Scholar]
- Worley S. L., Vaughn B. J., Terry A. I., Gardiner C. S., DeKrey G. K. (2015). Time- and dose-dependent effects of ethanol on mouse embryonic stem cells. Reprod. Toxicol. 57, 157–164. [DOI] [PubMed] [Google Scholar]
- Xu C. (2012). Differentiation and enrichment of cardiomyocytes from human pluripotent stem cells. J. Mol. Cell Cardiol. 52, 1203–1212. [DOI] [PubMed] [Google Scholar]
- Xu C., Inokuma M. S., Denham J., Golds K., Kundu P., Gold J. D., Carpenter M. K. (2001). Feeder-free growth of undifferentiated human embryonic stem cells. Nat. Biotechnol. 19, 971–974. [DOI] [PubMed] [Google Scholar]
- Yu J., Vodyanik M. A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J. L., Tian S., Nie J., Jonsdottir G. A., Ruotti V., Stewart R., et al. (2007). Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920. [DOI] [PubMed] [Google Scholar]
- Zhang B., Turdi S., Li Q., Lopez F. L., Eason A. R., Anversa P., Ren J. (2010). Cardiac overexpression of insulin-like growth factor 1 attenuates chronic alcohol intake-induced myocardial contractile dysfunction but not hypertrophy: Roles of Akt, mTOR, GSK3beta, and PTEN. Free Radic. Biol. Med. 49, 1238–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.