Abstract
Differential artery-vein analysis promises better sensitivity for retinal disease detection and classification. However, clinical optical coherence tomography angiography (OCTA) instruments lack the function of artery-vein differentiation. This study aims to verify the feasibility of using OCT intensity feature analysis to guide artery-vein differentiation in OCTA. Four OCT intensity profile features, including i) ratio of vessel width to central reflex, ii) average of maximum profile brightness, iii) average of median profile intensity, and iv) optical density of vessel boundary intensity compared to background intensity, are used to classify artery-vein source nodes in OCT. A blood vessel tracking algorithm is then employed to automatically generate the OCT artery-vein map. Given the fact that OCT and OCTA are intrinsically reconstructed from the same raw spectrogram, the OCT artery-vein map is able to guide artery-vein differentiation in OCTA directly.
1. Introduction
Early clinical diagnosis and prompt medical intervention are essential for preventing vision loss due to eye diseases. It is known that many eye diseases can target arteries and veins differently. For example, venous loops, venous beading [1–4], and arterial narrowing [5–7] have been reported in diabetic retinopathy (DR) and sickle cell retinopathy (SCR) patients. Therefore, differential artery-vein analysis promises better sensitivity for disease detection and staging classification. Artery-vein ratio of blood vessel caliber, for example, has been reported as a predictor of eye conditions [8–12]. Manual differentiation of retinal artery-vein is time consuming, and it can only be performed by experienced ophthalmologists. Therefore, a number of algorithms have been proposed to explore automated artery-vein differentiation in fundus photography [13–22]. However, a fundus image has limited resolution and sensitivity to reveal microvascular abnormalities associated with eye conditions [23]. Often, microvascular anomalies that occur early on in these ocular diseases cannot be reliably identified in traditional fundus photography [24–26].
By providing depth-resolved capability for visualizing multiple retinal layers with capillary level resolution, quantitative optical coherence tomography (OCT) and OCT angiography (OCTA) contribute to better clinical management of eye diseases. Multiple OCTA features have been developed for quantitative OCTA analysis and objective classification of DR [27], age-related macular degeneration (AMD) [28], glaucoma [29], and SCR [30,31]. Nonetheless, differential artery-vein analysis is not available for existing clinical OCTA instruments. Recently, we demonstrated the feasibility of using color fundus image to guide artery-vein differentiation in OCTA [32]. Two differential artery-vein features, i.e. artery vein ratio of blood vessel caliber (AVR-BVC) and tortuosity (AVR-BVT) improved the performance of OCTA detection and classification of DR. However, clinical deployment of this method is quite difficult due to the requirements of two clinical instruments, i.e., fundus camera and OCTA system, along with a sophisticated image registration.
In this study, we propose to use intensity based feature analysis in OCT to guide artery-vein differentiation in OCTA. In principle, an en face OCT image functions as a near infrared fundus image, thus it retains reflectance intensity based features in the vascular profiles. It is known that the central reflex is larger in arteries; whereas the vessel caliber is larger in veins [33]. The brightness profiles and optical density in arteries are brighter compared to neighboring veins [33]. Therefore, intensity profile based feature analysis promises a feasible method to conduct artery-vein classification in OCT. Because en face OCT and OCTA images can be reconstructed from the same spectrogram data set captured with a single clinical OCT/OCTA device, the artery-vein information can be readily used to guide artery-vein differentiation in OCTA. The proposed artery-vein classification algorithm is validated with a database that consists of a 100 OCT/OCTA image data set and verified by manual classification results prepared by two experienced ophthalmologists. The classification performance is further validated using sensitivity, specificity, and accuracy metrics along with graphical metrics, i.e., receiver operation characteristics (ROC) curve.
2. Methods
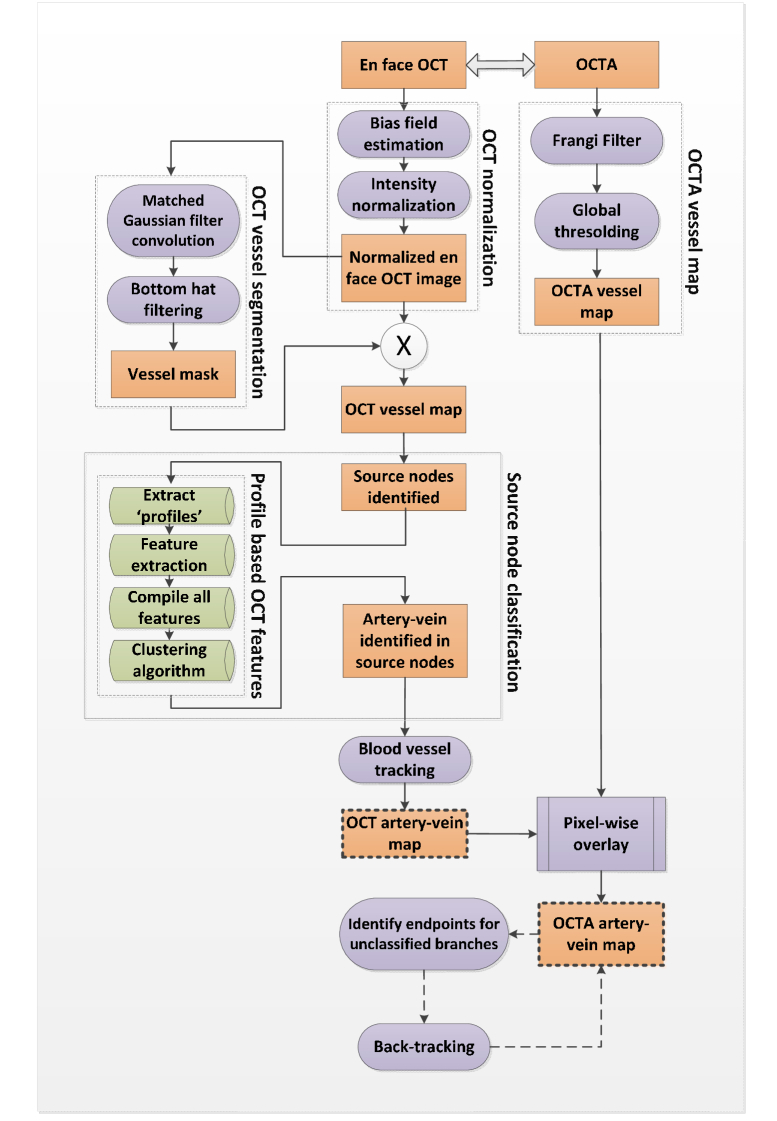
Figure 1 illustrates core steps of OCT guided artery-vein classification in OCTA. Technical details are provided in following sections.
Fig. 1.
Core steps of OCT guided artery-vein classification in OCTA.
2.1 Data acquisition
This study, approved by the Institutional Review Board of the University of Illinois at Chicago (UIC) and in compliance with the tenets of the Declaration of Helsinki, used 100 OCT/OCTA images captured from 50 subjects at the UIC Retina clinic. All OCT and OCTA images were acquired using a SD-OCT angiography system (Angiovue, Optovue, Fremont, CA, USA), with a 70-KHz A-scan rate, and axial and lateral resolutions of ∼5 μm and ~15 μm, respectively. Among the 50 subjects, 20 had DR, 20 had SCR and 10 were control subjects. For every subject, images were acquired from both eyes (OD and OS). All DR and SCR patients underwent a complete anterior and dilated posterior segment examination (JIL). Control data were obtained from healthy volunteers who gave informed consent for OCT and OCTA imaging. Subjects with prior intravitreal injections, vitreous surgery or history of other eye diseases were not included. Eyes with significant ocular pathology, (epiretinal membranes, vein occlusion or macular edema) were also excluded. OCT or OCTA images with severe motion artifacts were excluded.
All analyzed en face OCT and OCTA images were 8 mm × 8 mm scans (Angiovue SD-OCT system, Revue software version 2018.0.0.14). The scan pattern was raster, with 304 A-lines in each B-scan and 304 B-scans in each OCT/OCTA volume. The en face OCT images included all retinal layers and excluded the choroidal layer. OCTA images from the superficial layer, which generally includes 80% of ganglion cell complex (GCC), containing all structures of inner plexiform layer (IPL) up to the border with inner nuclear layer (INL), were used. The segmentation of the superficial layer has been conducted with the commercially available software of Angiovue SD-OCT system (ReVue). The en face OCT and OCTA data were exported from the ReVue, and further image analysis, feature extraction and artery-vein classification tasks were conducted in custom-developed MATLAB (Mathworks, Natick, MA, USA) procedures with graphical user interface (GUI).
2.2 Artery-vein classification in en face OCT image
OCT normalization and vessel segmentation
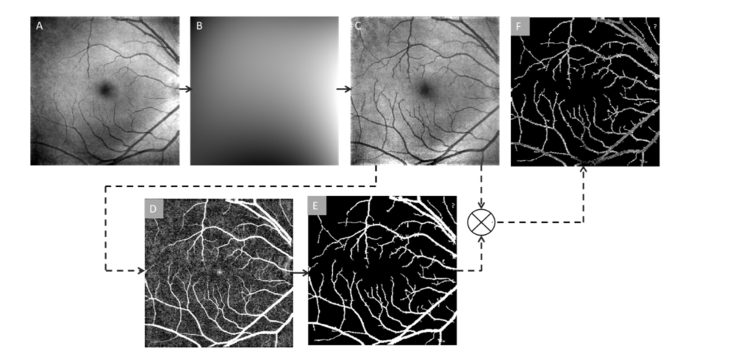
Figure 2 illustrates key procedures of OCT normalization and vessel segmentation. The en face OCT images (Fig. 2(A)) are often affected by intensity inhomogeneity. This artifact can mislead further image analysis. In this study, a bias field correction technique [34,35] is implemented to remove the intensity inhomogeneity before performing OCT feature analysis. This technique estimates the bias field that accounts for the inherent intensity inhomogeneity (Fig. 2(B)) and divides the image by that estimated bias to generate a normalized intensity image (Fig. 2(C)). The en face OCT image can be modeled as,
| (1) |
where I(x, y) is the intensity of the recorded OCT image, b(x, y) is the estimated bias field that accounts for the intensity inhomogeneity of the en face OCT image, and Itrue(x, y) is the intensity normalized image. The bias field b(x, y) can be defined by,
| (2) |
where G is the set of smooth basis functions which ensures the smooth varying property of the bias field. 20 polynomials of the first three degrees are chosen as the basis functions [35,36]. The bias field estimation is performed by calculating the optimal coefficients w = [w1, w2, … … wM]. Then, the intensity normalized image Itrue(x, y) is obtained by dividing I (x, y) by the bias field b(x, y).
Fig. 2.
OCT normalization and vessel segmentation. (A) Original en face OCT image. (B) Bias field estimation generated from the original en face OCT image. (C) Normalized intensity after bias field correction. (D) Edge enhanced using bottom hat filtering. (E) Binary vessel mask. (F) Segmented vessel map by multiplying images C and E.
From the intensity normalized image (Fig. 2(C)), only the vessel information is required for following analysis. A matched filtering method is combined with bottom hat filtering to enhance and extract the blood vessel map from the en face OCT image. For the match filtering, 2D Gaussian kernels of 12 different orientations and 10 different sizes are implemented to match blood vessels. These kernels cover all the blood vessel diameters and directions. They are defined as [36],
| (3) |
where x, y are pixel coordinates; θ = rotation angle of each kernel, θ ranges from 0 to π; and σ = kernel width. The en face OCT image is convolved with 120 kernels after subtracting the mean from each kernel. In the process of cross-correlation, the features matching the kernels (representing blood vessels of various widths) produce larger coefficient values. Larger coefficient values representing the vessel structures are selected to produce the segmented blood vessel map (Fig. 2(D)) from a maximum intensity projection of all the convolved images. A bottom hat filter with dimension of 20 × 20 pixels is then used to reduce further background variance due to uneven illumination. A global thresholding method is used to extract the binary blood vessel map (Fig. 2(E)). The binary map is multiplied with the intensity normalized en face OCT image to generate the segmented vessel map of en face OCT image (Fig. 2(F)). This en face OCT vessel map is used for further source node identification and artery-vein tracking.
Source node identification and classification
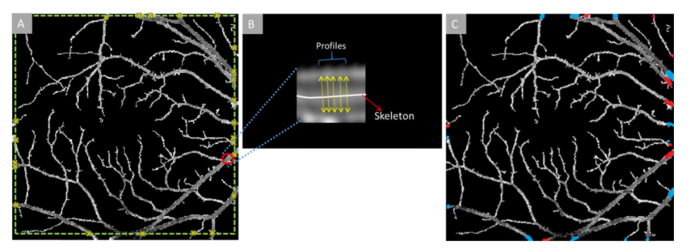
The first step is to identify source nodes at the boundary, and extract features to classify them to artery or vein. Around each of the boundaries of the vessel map, a gradient line is drawn (green dotted lines in Fig. 3(A)). These gradient lines can identify blood pixels from the background based on intensity information. The identified source node points are marked with yellow crosses in Fig. 3(A).
Fig. 3.
(A) En face OCT vessel map with source nodes identified with yellow crosses. (B) An enlarged sample source node segment (marked with red circle in (A)). Sample profiles (marked with yellow arrows) are extracted from each segment like this for further feature extraction and artery-vein classification. (C) En face OCT vessel map with identified source nodes as artery (red) or vein (blue).
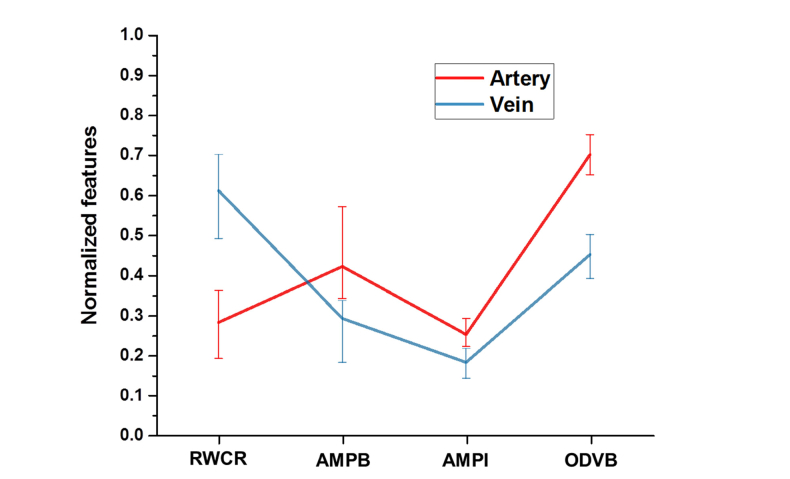
After the vessel source nodes along the boundary of the vessel map are selected, multi-feature analysis is employed on the vessel source nodes to classify them into artery and veins (red/blue nodes at the boundary of Fig. 3(C)). The features are extracted from the source node segments and are based on the concept of ‘profile’ [37]. From each source node segment, multiple profiles perpendicular to the vessel skeleton along the vessel are extracted. The number of profiles depends on the size of the source node segments. The profiles are set to have a distance of two pixels among each other along the vessel skeleton. For example, if a vessel skeleton within the source node segment is 40 pixels long, the first profile will be extracted at the middle point, pixel 20. Then the profiles will be extracted at 2 pixel intervals in both directions. The maximum number of profiles is set at 20 and minimum is 5. In case of a segment smaller than 5 pixels long, profiles are taken from all of the pixels. From each source node segments (S), N numbers (range, 5 to 20) of profiles are extracted. So, each segment will have a profile set ‘PSi’ equal to S × N, where i is the number of source nodes in one image. Total profile set PS for one image is then [PS1, PS2, PS3, … … …PSi]. Four features are measured from each of the profiles: i) ratio of vessel width to central reflex, ii) average of maximum profile brightness, iii) average of median profile intensity, and iv) optical density of vessel boundary intensity compared to background intensity. The average normalized feature distribution in artery and vein is shown in Fig. 4.
Fig. 4.
Normalized feature distribution in artery and vein vessels. RWCR: ratio of width to central reflex; AMPB: average of maximum profile brightness; AMPI: average of median profile intensity; ODVB: optical density of vessel boundary.
After the feature extraction, a clustering algorithm is used to classify source nodes as artery or vein. K-means algorithm [34] is chosen as the clustering algorithm due to its computational simplicity and efficiency. The centroids of each class (artery and vein in our case) are set initially to the maximum and the minimum of the K-means input set, as the cluster centers of two classes should be as far as possible. The cluster centers of both the classes are calculated by the algorithm, and feature vectors are classified thereafter using the standardized Euclidian distance metric. Theoretically, the empirical probability of a certain source node to be classified as an artery (PSA) or a vein (PSV) is defined by [37],
| (4) |
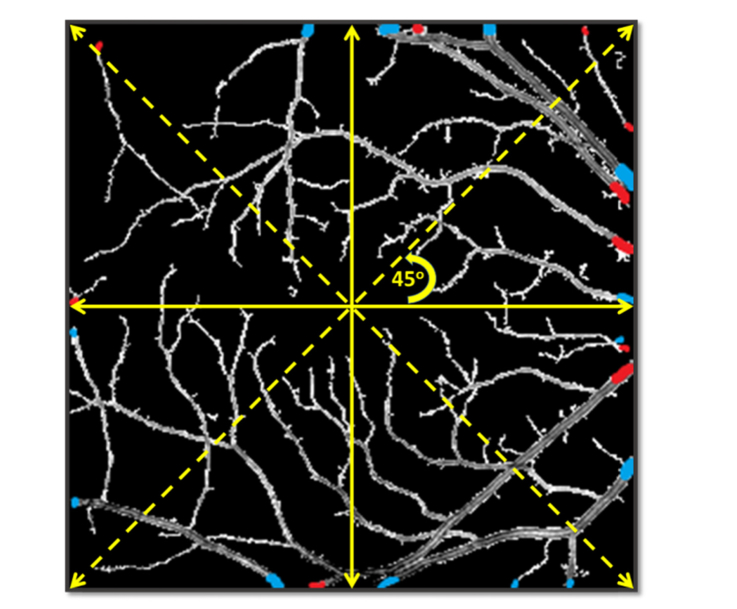
The clustering algorithm consists of three approaches [37]: 1) the K-means algorithm is applied to all the source node segments; 2) the image is divided into four quadrants centered at the fovea, and the K-means of each quadrant is calculated separately.
Separate estimation of these four quadrants can minimize the effect of the uneven intra-image intensity distribution among the quadrants; 3) Four quadrants are rotated (Fig. 5), and the clustering algorithm is applied to each rotated quadrant. This allows overlapped areas and a vessel can be classified eight times (360 degrees, rotated each time by 45 degrees). These classification results are combined for taking the final classification decision so that the influences of different outliers are reduced. In this approach, the average classification probability P for an artery or vein source node is the mean of classification probability of all the quadrants where that vessel source node was found.
Fig. 5.
Illustration of quadrant rotation relative to the fovea center.
Blood vessel tracking in en face OCT
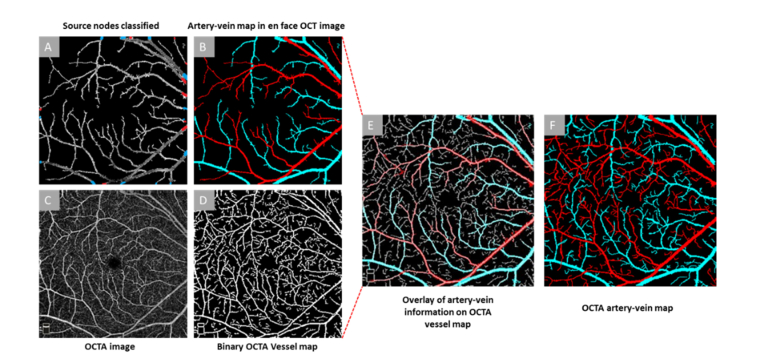
Once the source nodes are identified as artery and vein, the whole vessel map (skeleton) is tracked from the source nodes using a blood vessel tracking algorithm [38], which uses curvature angle information, to classify the rest of the vasculature in to artery or vein. The skeleton map was generated from vessel map using MATLAB morphological functions (‘bwmorph’). Further morphological functions were used to remove spurs and unwanted pixels. The final artery-vein map derived from the en face OCT image is shown as OCT artery-vein map in Fig. 6(B).
Fig. 6.
Artery-vein classification in OCTA. (A) En face OCT vessel map with artery-vein classified source nodes (B) En face OCT artery-vein map. (C) Original OCTA image, (D) OCTA binary vessel map, (E) En face OCT artery-vein map overlaid onto the OCTA binary vessel map, (F) Final OCTA artery-vein map.
2.3 OCT guided artery-vein classification in OCTA
The en face OCT artery-vein map (Fig. 6(B)) is overlaid with the OCTA vessel map to guide the artery-vein classification in OCTA. For generating the OCTA vessel map, a Hessian based multi-scale Frangi filter is first used on the original OCTA image (Fig. 6(C)) to enhance the vascular flow information. Frangi filtering methodology uses the Eigen vectors of Hessian matrices and computes the likeliness of an OCTA region being a vascular structure. Adaptive thresholding and morphological functions are further used for cleaning the vessel map and removing small capillary mesh structures that are not feasible for vessel tracking algorithms. The final vessel map (Fig. 6(D)) is then overlaid with the en face OCT artery-vein map (Fig. 6(E)). The en face OCT and OCTA images are generated from 3D projection of OCT B-scans and B-Scan speckle variance images, respectively. This means the structural coordinates of both images are same. Therefore, any process of image registration is not required for the overlaying process. So, artery-vein classification information of en face OCT (XOCT,YOCT) are transferred to corresponding OCTA(XOCTA,YOCTA) coordinates. The OCTA vessel map has some additional vascular structures compared to the en face OCT map. For each of the additional branches, the endpoints and corresponding branch points are identified in the skeleton using morphological functions in MATLAB. The endpoints are then back-tracked and linked to the already identified artery-vein branches. The tracking employs textural, morphological and optical density information and includes protocols for two, three or four-way intersections, gaps in the tracking path and overlapping of artery or vein. Further details of this tracking algorithm are explained in our recent publication [38]. The final OCTA artery-vein map is shown in Fig. 6(F). The performances of the artery-vein classification in en face OCT and OCTA images have been validated with ground truths prepared by two independent graders (JIL and DT). The ground truths only referred to vessel areas with identical artery-vein assignments by these two independent graders.
2.4 Performance metrics
The dataset used to test and validate the proposed en face OCT image guided artery-vein classification in OCTA consisted of 100 en face OCT and OCTA images from 50 subjects (20 DR, 20 SCR, 10 controls, both eyes imaged). For evaluating the classification performance, sensitivity, specificity and accuracy metrics were calculated. A graphical metric, receiver operation characteristic (ROC) curve and corresponding area under the ROC curve (AUC) was also measured. ROC curve plots the ‘sensitivity’ (true positive rate) as a function of ‘1-specificity’ (false positive rate) at different cutoff points. The closer the ROC curve is to the upper left corner, the more accurate the prediction is. AUC measures how well the classifier is able to identify the two classes (artery and vein). A 100% AUC represents a perfect prediction whereas lower than 50% represents bad prediction. Separate evaluation metrics for artery and vein are measured with respect to the labeled ground truths.
3. Results
The performances of artery-vein classification in OCT and OCTA are summarized in Table 1 and Table 2, respectively. These artery-vein classification results with the automated feature analysis are validated with the ground truth vessel maps manually labeled by the two experienced observers (JIL and DT). The ground truths only refer to vessel map areas with identical artery and vein assignments by these two independent graders. The two observers had 97.01% and 94.56% agreements respectively on the marked artery-vein vessel maps for en face OCT and OCTA images. Manual labelling is performed on the binary vessel maps in both cases. The observers manually traced the binary vessel maps with red (artery) and blue (vein) markings and identified each branchpoint with yellow markings. All of the manually labelled source nodes, branchpoints and artery-vein branches are matched pixel by pixel with the classified artery-vein map to generate the performance metrics. Specific areas in the vessel map for which the two observers do not agree on are identified as unclassified.
Table 1. Performance of Artery-Vein Classification: En Face OCT Image.
| Performance Measure | Source nodes | Whole vessel map | ||||
|---|---|---|---|---|---|---|
| Arteries | Veins | All vessels | Arteries | Veins | All vessels | |
| Sensitivity (%) | 97.16 | 96.64 | 96.86 | 97.07 | 96.52 | 96.79 |
| Specificity (%) | 95.73 | 96.15 | 95.94 | 95.29 | 96.14 | 95.72 |
| Classification Accuracy (%) | 96.89 | 96.59 | 96.74 | 96.81 | 96.33 | 96.57 |
| AUC (%) | 98.29 | 98.03 | 98.21 | 98.16 | 97.93 | 98.05 |
| Classification Error rate (%) | 3.11 | 3.41 | 3.26 | 3.19 | 3.67 | 3.43 |
Table 2. Performance of Artery-Vein Classification: OCTA Image.
| Performance Measure | Arteries | Veins | All vessels |
|---|---|---|---|
| Sensitivity (%) | 97.02 | 96.3 | 96.66 |
| Specificity (%) | 94.98 | 95.01 | 95.00 |
| Classification Accuracy (%) | 96.77 | 96.25 | 96.51 |
| AUC (%) | 95.47 | 96.18 | 95.83 |
| Classification Error rate (%) | 3.23 | 3.75 | 3.49 |
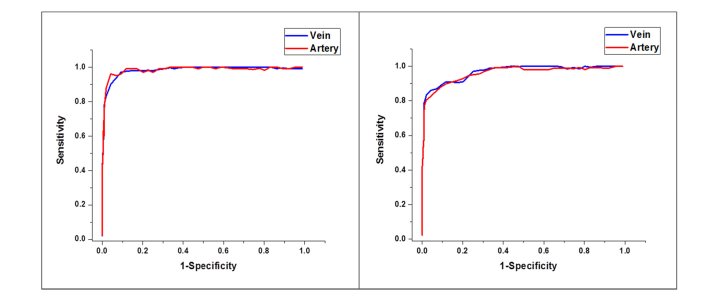
Figure 7 illustrates the ROC curves for artery-vein classification in en face OCT and OCTA images respectively.
Fig. 7.
Mean ROC curves for artery-vein classification. (A) En face OCT, (B) OCTA.
Our algorithm demonstrates 96.89% and 96.59% accuracies in identifying artery and vein source nodes, respectively, in the en face OCT images (ICC 0.98 and 0.97 for 2-repeat measurement, 95% CI 0.93-1). There are 97.16% sensitivity and 95.73% specificity for artery identification; 96.64% sensitivity and 96.15% specificity for vein identification. For the whole en face OCT vessel map, we observe 96.81% and 96.33% accuracies in identifying artery and vein source nodes respectively, in the en face OCT images (ICC 0.96 and 0.97 for 2-repeat measurement, 95% confidence interval (CI) 0.91-.97). There are 97.07% sensitivity and 95.29% specificity for artery identification; 96.52% sensitivity and 96.14% specificity for vein identification. In the case of classifying OCTA images, average accuracy is 96.51% for all vessels. 97.08% sensitivity and 94.98% specificity for artery identification; 96.30% sensitivity and 95.01% specificity for vein identification are observed. The accuracies are 96.77% and 96.25%, respectively, for identifying a blood vessel as artery and vein in the OCTA images (ICC 0.94 and 0.91 for 2-repeat measurement, 95% CI 0.87-0.96). The performance metrics were averaged for all OCTA images. The artery-vein classification accuracy for OCTA images from DR, SCR and control cohorts were 96.82%, 94.37% and 98.34% respectively.
4. Discussions
In summary, we have demonstrated the feasibility of using OCT intensity profile feature analysis to guide artery-vein differentiation in OCTA. In comparison with ground truths prepared by experienced ophthalmologists, this automated method has been able to differentiate individual arteries and veins in clinical OCTA with 96.51% accuracy.
It is known that subtle distortions of retinal arteries and veins can occur at the onset of various retinovascular diseases. Accurate identification of these subtle microvascular changes may provide early disease detection and may serve as biomarkers for treatment assessment. OCTA has been commercially available to provide better visualization of microvascular distortions, compared to traditional fundus photography. However, current clinical OCTA instruments do not have the function of differential artery-vein analysis. Recently, we demonstrated color fundus image guided artery-vein classification in OCTA to improve the sensitivity of DR detection and classification [32]. Because this method requires two separate devices, clinical deployment of the color fundus image guided artery-vein classification is challenging. In this study, we demonstrated the OCT feature analysis guide artery-vein differentiation in OCTA, with all required information obtained from the same OCT/OCTA instrument.
It is established that the central reflex, brightness profile and optical density in arteries are larger compared to that in veins; whereas the vessel caliber is larger in veins. These intensity reflectance properties have been used to classify arteries and veins in traditional fundus photography [33]. An en face OCT image is in principle a fundus image, with improved axial resolution compared to traditional fundus photography. Therefore, the near infrared light intensity profile features were chosen to be quantified to classify arteries and veins in en face OCT.
For reliable artery-vein classification in OCT, we have identified vessel source nodes at the boundaries, i.e., perifoveal retina where the vessels are relatively large, first in each en face OCT image. In order to ensure reliable artery-vein source node classification, a bias field correction technique has been implemented before quantitative feature extraction and analysis. This procedure is helpful to minimize the effect of intensity inhomogeneity in en face OCT to enhance the coherent reflectance intensity properties of vessel profiles, improving the overall classification performance of the K-means clustering algorithm. As a robust classifier, K-means algorithm also provided reliable performance with high computational efficiency, without the requirement of extensive parameter tuning. Additionally, the K-means clustering algorithm used to classify vessel profile features was performed using three approaches. First strategy was to apply the K-means algorithm for all source nodes in the image. This resulted in relatively lower classification accuracy (79.38% and 82.61% for artery and vein respectively), as different regions of the en face OCT images often suffered from uneven illumination and background contrast. The second approach was to divide the image into four quadrants (Fig. 4). The K-means clustering of each quadrant was conducted separately. This significantly improved the classification performance (92.23% and 91.05% for artery and vein respectively). To further improve the clustering performance, we rotated the four quadrants by 45 degrees and applied K-means algorithm on overlapped quadrants within the en face OCT image. The overlapped quadrant strategy compensated for the effect of uneven intra-image intensity distribution among the quadrants and further improved the artery-vein classification performance. From Table 1, we can observe that the final accuracies of artery and vein source node classification by the K-means algorithm are 96.89% and 96.59%, respectively. This robust classification performance on source nodes facilitated artery-vein identification of the whole vessel map to produce a robust OCT/OCTA artery-vein map.
Because the en face OCT and OCTA are naturally reconstructed from the same spectrogram data sets, the OCT artery-vein vessel map can be readily used to guide artery-vein differentiation in corresponding OCTA. Once the OCTA artery-vein map is generated, further tracking is used to classify remaining smaller capillary arteriole and venule branches. Each of identified artery-vein maps are validated by ground truths prepared by two experienced ophthalmologists. Only the blood vessels on which the two graders have an agreement on artery-vein assignment are selected to verify the performance of the automated artery-vein classification. For testing the robustness of the algorithm, OCT/OCTA images from control, DR and SCR patients were verified. With only a single device required for OCT and OCTA image acquisition, this approach can be easily integrated with existing commercial OCTA devices to foster clinical deployment of differential artery-vein analysis.
5. Conclusion
In conclusion, robust artery-vein classification in OCTA is demonstrated by incorporating OCT feature analysis and blood vessel tracking. This provides a feasible solution to facilitate differential artery-vein analysis in OCTA for clinical management of eye diseases.
Acknowledgment
The authors acknowledge Mr. Mark Janowicz and Ms. Andrea Degillio for their valuable support in OCT/OCTA data acquisition.
Funding
National Eye Institute (R01 EY023522, R01 EY024628, P30 EY001792); Research to Prevent Blindness; Richard and Loan Hill Endowment; Marion H. Schenk Chair Endowment; CCTS-PECTS Fellowship; Provost’s Graduate Research Award.
Disclosures
X. Yao, pending patent application; M, Alam, pending patent application. No other conflicting relationship exists for any author.
References
- 1.Fonseca R. A., Dantas M. A., “Retinal venous beading associated with recurrent branch vein occlusion,” Can. J. Ophthalmol. 37(3), 182–183 (2002). 10.1016/S0008-4182(02)80062-X [DOI] [PubMed] [Google Scholar]
- 2.Gregson P. H., Shen Z., Scott R. C., Kozousek V., “Automated grading of venous beading,” Comput. Biomed. Res. 28(4), 291–304 (1995). 10.1006/cbmr.1995.1020 [DOI] [PubMed] [Google Scholar]
- 3.Kozousek V., Shen Z., Gregson P., Scott R. C., “Automated detection and quantification of venous beading using Fourier analysis,” Can. J. Ophthalmol. 27(6), 288–294 (1992). [PubMed] [Google Scholar]
- 4.Piguet B., Gross-Jendroska M., Holz F. G., Bird A. C., “Inherited venous beading,” Eye (Lond.) 8(1), 84–88 (1994). 10.1038/eye.1994.16 [DOI] [PubMed] [Google Scholar]
- 5.Pedersen L., Jeppesen P., Knudsen S. T., Poulsen P. L., Bek T., “Improvement of mild retinopathy in type 2 diabetic patients correlates with narrowing of retinal arterioles. A prospective observational study,” Graefes Arch. Clin. Exp. Ophthalmol. 252(10), 1561–1567 (2014). 10.1007/s00417-014-2614-6 [DOI] [PubMed] [Google Scholar]
- 6.Klein R., Klein B. E., Moss S. E., Wang Q., “Hypertension and retinopathy, arteriolar narrowing, and arteriovenous nicking in a population,” Arch. Ophthalmol. 112(1), 92–98 (1994). 10.1001/archopht.1994.01090130102026 [DOI] [PubMed] [Google Scholar]
- 7.Cheung N., Bluemke D. A., Klein R., Sharrett A. R., Islam F. M., Cotch M. F., Klein B. E., Criqui M. H., Wong T. Y., “Retinal arteriolar narrowing and left ventricular remodeling: the multi-ethnic study of atherosclerosis,” J. Am. Coll. Cardiol. 50(1), 48–55 (2007). 10.1016/j.jacc.2007.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niemeijer M., Xu X., Dumitrescu A. V., Gupta P., van Ginneken B., Folk J. C., Abramoff M. D., “Automated measurement of the arteriolar-to-venular width ratio in digital color fundus photographs,” IEEE Trans. Med. Imaging 30(11), 1941–1950 (2011). 10.1109/TMI.2011.2159619 [DOI] [PubMed] [Google Scholar]
- 9.Hubbard L. D., Brothers R. J., King W. N., Clegg L. X., Klein R., Cooper L. S., Sharrett A. R., Davis M. D., Cai J., “Methods for evaluation of retinal microvascular abnormalities associated with hypertension/sclerosis in the Atherosclerosis Risk in Communities Study,” Ophthalmology 106(12), 2269–2280 (1999). 10.1016/S0161-6420(99)90525-0 [DOI] [PubMed] [Google Scholar]
- 10.Ikram M. K., Witteman J. C., Vingerling J. R., Breteler M. M., Hofman A., de Jong P. T., “Retinal vessel diameters and risk of hypertension: the Rotterdam Study,” Hypertension 47(2), 189–194 (2006). 10.1161/01.HYP.0000199104.61945.33 [DOI] [PubMed] [Google Scholar]
- 11.Liew G., Wong T. Y., Mitchell P., Cheung N., Wang J. J., “Retinopathy predicts coronary heart disease mortality,” Heart 95(5), 391–394 (2008). 10.1136/hrt.2008.146670 [DOI] [PubMed] [Google Scholar]
- 12.Wong T. Y., Knudtson M. D., Klein R., Klein B. E., Meuer S. M., Hubbard L. D., “Computer-assisted measurement of retinal vessel diameters in the Beaver Dam Eye Study: methodology, correlation between eyes, and effect of refractive errors,” Ophthalmology 111(6), 1183–1190 (2004). 10.1016/j.ophtha.2003.09.039 [DOI] [PubMed] [Google Scholar]
- 13.Aguilar W., Martinez-Perez M. E., Frauel Y., Escolano F., Lozano M. A., Espinosa-Romero A., “Graph-based methods for retinal mosaicing and vascular characterization,” Lect. Notes Comput. Sci. 4538, 25–36 (2007). 10.1007/978-3-540-72903-7_3 [DOI] [Google Scholar]
- 14.Chrástek R., Wolf M., Donath K., Niemann H., Michelson G., “Automated Calculation of Retinal Arteriovenous Ratio for Detection and Monitoring of Cerebrovascular Disease Based on Assessment of Morphological Changes of Retinal Vascular System,” in MVA, 2002), 240–243. [Google Scholar]
- 15.E. Grisan and A. Ruggeri, “A divide et impera strategy for automatic classification of retinal vessels into arteries and veins,” in Engineering in medicine and biology society, 2003. Proceedings of the 25th annual international conference of the IEEE, (IEEE, 2003), 890–893. 10.1109/IEMBS.2003.1279908 [DOI] [Google Scholar]
- 16.Jelinek H., Depardieu C., Lucas C., Cornforth D., Huang W., Cree M., “Towards vessel characterization in the vicinity of the optic disc in digital retinal images,” in Image Vis Comput Conf, 2005), 2–7. [Google Scholar]
- 17.H. Li, W. Hsu, M.-L. Lee, and H. Wang, “A piecewise Gaussian model for profiling and differentiating retinal vessels,” in Image Processing, 2003. ICIP 2003. Proceedings. 2003 International Conference on, (IEEE, 2003), I-1069. 10.1109/ICIP.2003.1247151 [DOI] [Google Scholar]
- 18.M. Niemeijer, B. van Ginneken, and M. D. Abràmoff, “Automatic classification of retinal vessels into arteries and veins,” Medical imaging, 72601F–72601F (2009). 10.1117/12.813826 [DOI]
- 19.Rothaus K., Jiang X., Rhiem P., “Separation of the retinal vascular graph in arteries and veins based upon structural knowledge,” Image Vis. Comput. 27(7), 864–875 (2009). 10.1016/j.imavis.2008.02.013 [DOI] [Google Scholar]
- 20.Simó A., de Ves E., “Segmentation of macular fluorescein angiographies. A statistical approach,” Pattern Recognit. 34(4), 795–809 (2001). 10.1016/S0031-3203(00)00032-7 [DOI] [Google Scholar]
- 21.S. Vázquez, N. Barreira, M. Penedo, M. Penas, and A. Pose-Reino, “Automatic classification of retinal vessels into arteries and veins,” in 7th international conference biomedical engineering (BioMED 2010), 2010), 230–236. 10.2316/J.2010.216.680-0094 [DOI] [Google Scholar]
- 22.Vázquez S., Cancela B., Barreira N., Penedo M. G., Rodríguez-Blanco M., Seijo M. P., de Tuero G. C., Barceló M. A., Saez M., “Improving retinal artery and vein classification by means of a minimal path approach,” Mach. Vis. Appl. 24(5), 919–930 (2013). 10.1007/s00138-012-0442-4 [DOI] [Google Scholar]
- 23.Zahid S., Dolz-Marco R., Freund K. B., Balaratnasingam C., Dansingani K., Gilani F., Mehta N., Young E., Klifto M. R., Chae B., Yannuzzi L. A., Young J. A., “Fractal Dimensional Analysis of Optical Coherence Tomography Angiography in Eyes With Diabetic Retinopathy,” Invest. Ophthalmol. Vis. Sci. 57(11), 4940–4947 (2016). 10.1167/iovs.16-19656 [DOI] [PubMed] [Google Scholar]
- 24.Gramatikov B. I., “Modern technologies for retinal scanning and imaging: an introduction for the biomedical engineer,” Biomed. Eng. Online 13(1), 52 (2014). 10.1186/1475-925X-13-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mendis K. R., Balaratnasingam C., Yu P., Barry C. J., McAllister I. L., Cringle S. J., Yu D.-Y., “Correlation of histologic and clinical images to determine the diagnostic value of fluorescein angiography for studying retinal capillary detail,” Invest. Ophthalmol. Vis. Sci. 51(11), 5864–5869 (2010). 10.1167/iovs.10-5333 [DOI] [PubMed] [Google Scholar]
- 26.Cheng S.-C., Huang Y.-M., “A novel approach to diagnose diabetes based on the fractal characteristics of retinal images,” IEEE Trans. Inf. Technol. Biomed. 7(3), 163–170 (2003). 10.1109/TITB.2003.813792 [DOI] [PubMed] [Google Scholar]
- 27.Kim A. Y., Chu Z., Shahidzadeh A., Wang R. K., Puliafito C. A., Kashani A. H., “Quantifying Microvascular Density and Morphology in Diabetic Retinopathy Using Spectral-Domain Optical Coherence Tomography Angiography,” Invest. Ophthalmol. Vis. Sci. 57(9), OCT362 (2016). 10.1167/iovs.15-18904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palejwala N. V., Jia Y., Gao S. S., Liu L., Flaxel C. J., Hwang T. S., Lauer A. K., Wilson D. J., Huang D., Bailey S. T., “Detection of non-exudative choroidal neovascularization in age-related macular degeneration with optical coherence tomography angiography,” Retina 35(11), 2204–2211 (2015). 10.1097/IAE.0000000000000867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holló G., “Vessel density calculated from OCT angiography in 3 peripapillary sectors in normal, ocular hypertensive, and glaucoma eyes,” Eur. J. Ophthalmol. 26(3), e42–e45 (2016). 10.5301/ejo.5000717 [DOI] [PubMed] [Google Scholar]
- 30.Alam M., Thapa D., Lim J. I., Cao D., Yao X., “Quantitative characteristics of sickle cell retinopathy in optical coherence tomography angiography,” Biomed. Opt. Express 8(3), 1741–1753 (2017). 10.1364/BOE.8.001741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alam M., Thapa D., Lim J. I., Cao D., Yao X., “Computer-aided classification of sickle cell retinopathy using quantitative features in optical coherence tomography angiography,” Biomed. Opt. Express 8(9), 4206–4216 (2017). 10.1364/BOE.8.004206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alam M., Toslak D., Lim J. I., Yao X., “Color fundus image guided artery-vein differentiation in optical coherence tomography angiography (Accepted, in production),” Invest. Ophthalmol. Vis. Sci. 59(12), 4953 (2018). 10.1167/iovs.18-24831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kondermann C., Kondermann D., Yan M., “Blood vessel classification into arteries and veins in retinal images,” inMedical Imaging 2007: Image Processing, (International Society for Optics and Photonics, 2007), 651247. [Google Scholar]
- 34.Vovk U., Pernus F., Likar B., “A review of methods for correction of intensity inhomogeneity in MRI,” IEEE Trans. Med. Imaging 26(3), 405–421 (2007). 10.1109/TMI.2006.891486 [DOI] [PubMed] [Google Scholar]
- 35.Li C., Gore J. C., Davatzikos C., “Multiplicative intrinsic component optimization (MICO) for MRI bias field estimation and tissue segmentation,” Magn. Reson. Imaging 32(7), 913–923 (2014). 10.1016/j.mri.2014.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li C., Xu C., Anderson A. W., Gore J. C., “MRI tissue classification and bias field estimation based on coherent local intensity clustering: A unified energy minimization framework,” in International conference on information processing in medical imaging, (Springer, 2009), 288–299. 10.1007/978-3-642-02498-6_24 [DOI] [PubMed] [Google Scholar]
- 37.Vázquez S., Barreira N., Penedo M. G., Saez M., Pose-Reino A., “Using retinex image enhancement to improve the artery/vein classification in retinal images,” in International Conference Image Analysis and Recognition, (Springer, 2010), 50–59. 10.1007/978-3-642-13775-4_6 [DOI] [Google Scholar]
- 38.Alam M., Son T., Toslak D., Lim J. I., Yao X., “Combining ODR and Blood Vessel Tracking for Artery-Vein Classification and Analysis in Color Fundus Images,” Transl. Vis. Sci. Technol. 7(2), 23 (2018). 10.1167/tvst.7.2.23 [DOI] [PMC free article] [PubMed] [Google Scholar]