Abstract
Accurate choroidal vessel segmentation with swept-source optical coherence tomography (SS-OCT) images provide unprecedented quantitative analysis towards the understanding of choroid-related diseases. Motivated by the leading segmentation performance in medical images from the use of deep learning methods, in this study, we proposed the adoption of a deep learning method, RefineNet, to segment the choroidal vessels from SS-OCT images. We quantitatively evaluated the RefineNet on 40 SS-OCT images consisting of ~3,900 manually annotated choroidal vessels regions. We achieved a segmentation agreement (SA) of 0.840 ± 0.035 with clinician 1 (C1) and 0.823 ± 0.027 with clinician 2 (C2). These results were higher than inter-observer variability measure in SA between C1 and C2 of 0.821 ± 0.037. Our results demonstrated that the choroidal vessels from SS-OCT can be automatically segmented using a deep learning method and thus provided a new approach towards an objective and reproducible quantitative analysis of vessel regions.
1. Introduction
Choroid is a tissue layer beneath the retina with the most abundant blood flow of all the structures in the eye. It is crucial for the normal function of retinal pigment epithelium and outer retina in terms of the oxygenation and metabolism activity [1]. Structural and functional abnormality of choroid is related to many ocular diseases, such as Age-related Macular Degeneration (AMD) [2], Polypoidal Choroidal Vasculopathy (PCV) [3], Choroidal Neovascularisation (CNV), Multifocal Choroiditis and Panuveitis (MCP). To analyze the choroid, an advanced non-invasive scanner, Swept-Source Optical Coherence Tomography (SS-OCT) for the imaging of eye fundus, has demonstrated great potential in its ability to quantify the choroid layer. Compared to the traditional Spectral Domain OCT (SD-OCT), SS-OCT produces a deeper penetration of the fundus, and therefore it is capable of generating greater detailed acquisitions of the choroid layer and the choroidal vessels within the layer [4].
To quantitatively analyze the choroid layer, it is necessary to segment the individual choroidal vessels which includes measuring the total volume of the vessels, distribution of the vessels within the layer, and the volume ratio of the vessels in relation to its background. This analysis facilitates the understanding of choroid-related diseases, such as AMD, PCV and high myopia. However, current approaches to these analyses rely on tedious, time-consuming, and non-reproducible manual processes. Although there are automated segmentation methods developed for SD-OCT, which with adaptation, may be applicable for choroid vessel quantification from SS-OCT, these methods are however optimized for the visual properties of the SD-OCT which markedly differs from that of SS-OCT’s vessel characterization and thus are not comparable. This study is the first to present quantitatively analysis for choroidal vessels segmentation of SS-OCT images with performance that is consistent to clinician’s manual analysis.
1.1 Related work
We have divided related works into three main categories: SS-OCT segmentation methods, conventional choroidal vessels segmentation methods, and deep learning based OCT segmentation methods. Our separation is mainly attributed to the fact that there are limited number of works on automated segmentation for SS-OCT images. We therefore also included works on the segmentation of SD-OCT, which is the most relevant imaging modality to SS-OCT and have discussed these studies in reflection to how it may potentially relate to SS-OCT.
For SS-OCT segmentation, Li Zhang et al. [5] proposed to use shape model of the Bruch’s membrane with soft-constraint graph-search for segmenting choroidal boundaries. Zhou et al. [6] proposed to use attenuation correction approach to denoise the input SS-OCT images which thereby improved the segmentation of the choroidal boundaries. Although these automated segmentation methods have demonstrated accurate results. Unfortunately, these methods have designed for segmenting layer structures e.g., searching minimum pathways. Consequently, it will be challenging to adopt these methods for segmenting circular choroidal vessel structures.
Conventional choroidal vessels segmentation methods generally require: i) a priori knowledge of the data, ii) the need to tune a large number of parameters, and iii) pre- and/or post-processing techniques to denoise the input image and refine the segmentation results, respectively. Duan et al. [7] and Kajic et al. [8] both applied multi-scale adaptive thresholding methods for choroidal vessels segmentation. Li Zhang et al. [9] proposed the use of 3D tube-like models to fit into the choroidal vessels as a mean of vessel detection. The results were then processed by multi-scale Hessian filter and thresholding to refine the segmented choroidal vessels boundaries. In their experiments, only the reproducibility was demonstrated using repeated scans of the same eye while segmentation accuracy was not reported. In another work, Srinath et al. [10] proposed to initially identify the upper and lower boundaries of the choroid layer and then apply a level set method on the detected choroid layer to iteratively segment the vessels. By isolating the choroid layer, they were able to improve the identification of the vessels and its subsequent segmentation. Unfortunately, in all the above methods, their segmentation performances were only visually demonstrated and did not include quantitatively evaluations with manually annotated ground truth data. Consequently, it is challenging to determine their accuracy and reproducibility.
Recently, deep learning methods have achieved great success in medical image segmentation tasks [11,12]. Such success is primarily attributed to the ability of the deep learning methods to leverage large data sets to hierarchically learn the image features that best correspond to the appearances, as well as the semantics, of the images. Motivated by this success, many investigators have also attempted to adapt deep learning based methods for OCT images. Fang et al. [13] adopted an 8-layer convolutional neural networks (CNN) to classify whether each pixel is located in the retinal layer. However, this patch based method is inefficient, where accurate segmentation requires a prediction for every pixel in the image. In addition, as the patches are independently trained and used in the prediction, this resulted in a loss of spatial context, meaning that the segmentation results lacked consistency with inconsistent labelling of neighborhood pixels. To overcome these limitations, fully convolutional networks (FCN) based methods have been proposed. FCN was derived from CNN (VGGNet [14]) to provide efficient dense inference, where the classification modules (fully connected layers) were replaced with deconvolutional layers (transposed convolutional layers) to upsample the learned features and output the segmentation results. For instance, Sui et al. [15] embedded segmentation results from three FCNs for choroid layer segmentation. Venhuizen et al. [16] introduced a modified U-shaped FCN (U-net) for retina thickness segmentation. Xu et al. [17] used a dual stage FCN to progressively segment the pigment epithelium detachment (PED) structures from SD-OCT images. Although these FCN based methods have demonstrated accurate segmentation results for layer structures, unfortunately, they have not been validated for small choroidal vessels segmentation.
1.2 Our contributions
In this study, we propose the adoption of an FCN variant named RefineNet proposed by Lin et al. [18] which achieved the state-of-the-art segmentation results on natural images. RefineNet differs from commonly used VGG-FCN (FCN using the VGGNet as the backbone [14]) in two aspects: (1) RefineNet propagates and integrates the multi-scale intermediary results in an end-to-end manner. In contrast, VGG-CNN propagates in a single-scale. The multi-scale capability allows for improved segmentation of regions of various sizes that are evident in our OCT images; (2) RefineNet uses deeper ResNet backbone architecture (101 layers) [19] instead of the conventional VGGNet (usually less than 20 layers). In our experiments, the deeper layers and residual connection only used in ResNet backbone allowed for the extraction of more meaningful image features that resulted in improved segmentation. In this study, we introduce the following contributions:
-
•
Development of a deep learning based choroidal vessels segmentation method for SS-OCT for use in choroidal vessels analysis.
-
•
Our choroidal vessels segmentation method is an end-to-end model for training and testing, which means that it does not require any pre- and post-processing steps e.g., image denoising and filtering, or manual feature / parameter selections. Hence, our approach is practically suitable for choroid vessels which usually consist of large number of regions with complex patterns that are difficult to manually analyze.
-
•
We evaluated our segmentation results with manual annotations derived from two clinicians, and further compared our results with implementations of conventional vessels segmentation methods. Furthermore, we present the first inter-observer variabilities analysis of the manual choroidal vessels segmentation from SS-OCT images.
2. Methods
2.1 Image acquisition and manual annotation
The images used in our study were acquired using SS-OCT (model DRI OCT-1 Atlantis; Topcon) with 12-line radial scan patterns having a resolution of 1024 × 12. Each image is an average of 32 overlapped consecutive scans focused on the fovea and has a resolution of 1024 × 992 representing an actual area of 12 mm × 2.6 mm. This study was approved by the Institutional Review Board of Shanghai General Hospital, Shanghai Jiao Tong University and was conducted under the tenets of the Declaration of Helsinki. Informed consent was obtained from each participant.
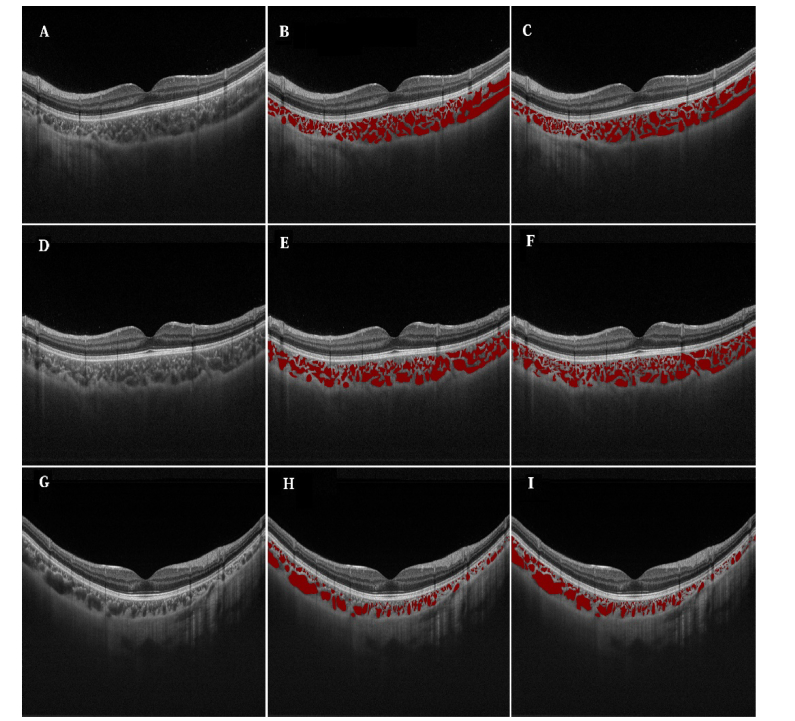
10 subjects were randomly selected, 5 emmetropes and 5 high myopes (refractive error≤-5.0), respectively. For each subject, 4 slices were selected, which were 45°, 90°, 135°and 180° from the horizontal view, representing the four main directions of eye fundus scanning. In total, 40 images were selected for the manual annotation for use as the ground truth. Those images were annotated by two clinicians independently. For each image, there were 97.9 ± 28.2 annotated regions. In total, approximately 3900 regions were annotated. Figure 1 shows 3 example images from 3 studies and their annotations (annotated by two clinicians). In total, each clinician spent about 10 hours to annotate these 40 images.
Fig. 1.
Examples of raw SS-OCT images and their annotations by the two clinicians. The left column (A, D, G) shows the raw images; Middle column (B, E, H) is the annotations by clinician 1; and the right column (C, F, I) is the annotations by clinician 2.
2.2 Fully convolutional networks (FCN)
The traditional FCN architecture [11] was converted from convolutional neural networks (CNNs) (VGGNet [14]) for efficient dense inference. It contains downsampling and upsampling path. The downsampling path has stacked convolutional layers to extract high-level abstract information and has been widely used in convolutional neural networks (CNN) for image classification related tasks [20]. The upsampling part has stacked deconvolutional layers, which are transposed convolutional layers that upsample the feature maps derived from downsampling part to output the segmentation results.
Then an FCN network can be defined as:
| (1) |
where is the output prediction, is the input image, denotes the feature map produced by the stacked convolutional layers with a list of downsampling factors , denotes the deconvolutional layers that upscale the feature map by a list of factors to ensure both the output and input have the same size (height and width). and are the learned parameters. To explore the fine-scaled local appearance of the input image, the skip architecture was employed to combine the output feature maps of both lower convolution layers and the higher convolutional layer for more accurate inference [11,12].
For training of FCN for choroidal vessels segmentation, the whole architecture can be defined as minimizing the overall loss between the predicted results and the ground truth annotation:
| (2) |
Here, calculates the loss (per-pixel cross entropy loss) of the ground truth annotation and the predicted results. The network parameters and can then be iteratively updated using stochastic gradient descent (SGD) algorithm. For inference, FCN takes an image of arbitrary size and outputs a probability map of the same size that indicates the choroidal vessels area.
2.3 RefineNet for choroidal vessels segmentation
RefineNet is another variation of FCN and has two major differences when compared with the traditional FCN (VGG-FCN) architecture. On the downsampling path, traditional FCN architecture is based on the VGGNet architecture [14] and therefore, its downsampling path usually has limited capacity to add additional layers. Experimental data has shown that beyond certain depths, adding extra layers results in higher training errors and therefore, it is challenging to optimize very deep networks with many layers [21]. In addition, traditional FCN usually relies on skip connection with deconvolution operation on the upsampling path for producing the final segmentation results. Unfortunately, the deconvolution operations are not able to recover the smaller objects e.g., choroidal vessels which are lost after the downsampling operations on the downsampling path [18]. Therefore, they are unable to output accurate segmentation for individual small choroidal vessel regions.
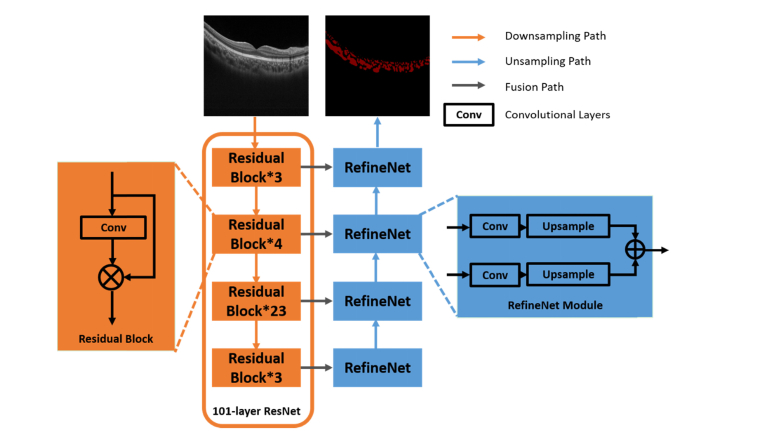
To overcome the limitation on the down-sampling path, a 101-layer residual network (ResNet) was used [19] for visual features learning and representations. The ResNet architecture consists of a number of residual blocks with shortcuts that bypasses few convolutional layers at a time (as exemplified in Fig. 2). Therefore, the ResNet architecture enables having multiple down-sampling paths and thus allow deriving optimal results by averaging the output of different paths. For improving the segmentation results on the upsampling path, multiple RefineNet modules were used to fuse high-level features by incorporating low-level features derived from downsampling path in a step-wise manner (Fig. 2). Compared with the skip connection with the deconvolution operations, the upsampling and fusion process allows to retain the image resolution while accurately segment the choroidal vessels.
Fig. 2.
Overview of the RefineNet for choroidal vessels segmentation.
2.4 Implementation details
There is a scarcity of medical images with annotations for use as training data due to the cost and complexity of the acquisition procedures [22,23]. In contrast to the limited data in the medical domain, there are greater availability of general image data [24]. Existing works [22] have shown evidence that the problem of insufficient training data can be alleviated by fine-tuning (continue training the model trained on general images), where the lower layers of the fine-tuned network are more general filters (trained on general images) while those in the higher layers are more specific to the target problem (trained with specific medical images) [22,25]. Therefore, we used the pre-trained 101-layer-ResNet (trained on ImageNet) for initialization. The implementation was based on the MatConvNet library [26].
We used a 5-fold cross-validation approach to evaluate the proposed method. For each fold, 8 studies (32 images, ~2880 annotated regions) were used as the training data and 2 studies (8 images, ~720 annotated regions) were used as the testing data. Equal number of randomly sampled manual annotations from C1 and C2 were used as part of training data. We then rotated the training and testing data for 5 times to cover all 10 studies. Data augmentation techniques, including random crops and flips, were used to further improve the robustness [27,28]. Due to the limited GPU memory, we have to reduce the size of the input image instead of using the full resolution. However, downsampling the input image will result in losing all the small choroidal vessels. Therefore, input image was cropped into 500 × 500 smaller patches and the cropped small patches were then resized to fit into the pre-trained model input size. It took an average of ~10 hours to train one fold, where each fold was trained for 100 epochs at a learning rate of 0.0005 and batch size of 1. The framework was implemented in MATLAB R2017a running on a desktop PC with a 11GB NVidia GTX 1080Ti GPU with Intel Core i7 2.60 GHz. The average running time of RefineNet per image is 0.72 seconds.
2.5 Experimental setup
We performed the following experiments: (a) evaluation of the segmentation accuracy of the RefineNet method compared to manual ground truth (from observers); (b) comparison of inter-observer variability; and (c) comparison of our RefineNet with other choroidal vessels segmentation methods. These comparison methods are: (1) VGG-FCN – traditional fully convolutional network based on VGGNet architecture [11]; (2) LS – level set based segmentation method, which was used in [10]; and (3) AT – adaptive thresholding based segmentation method, which was used in [7].
The VGG-FCN was trained with the same 5-fold cross-validation training process as used in RefineNet with pre-trained ImageNet [24] model. It took an average of ~6 hours to train one fold, where each fold was trained for 200 epochs at a learning rate of 0.0001 (decrease by 10 times for every 100 epochs) and batch size of 20. We used the same patch-based method (the image was cropped into 500 × 500 small patches. The cropped small patches were then resized to fit into the pre-trained model input size) at both training and inferencing stages. Both LS and AT methods required sophisticated choroid layer detection approach as the pre-processing step. In order to minimize the pre-processing errors and also for fair comparison purpose, one clinician manually delineated the choroid layer for these two comparison methods.
For evaluation, we used segmentation agreement (SA), mean absolute differences (MAD), intra-class correlation coefficient (ICC) and Bland-Altman plots to measure the segmentation performance and the variability between Our method (Ours) and clinician 1 (C1), Ours and clinician 2 (C2), and C1 and C2. SA was defined as:
| (3) |
where represents the pixels both agreed to annotate (e.g., both C1 and C2 annotated), represents the pixels both agreed not to annotate (background) and are the pixels which are not agreed.
MAD was defined as:
| (4) |
where is the number of total pixels annotated by observer 1 (e.g., clinician 1), is the number of total pixels annotated by observer 2 (e.g., Our method), and R is the zoom ratio of the image to the actual tissue. In this study, eye tissue of 12.00 × 2.60 mm was scanned into an image of 1024 × 992 pixels, which equates to a zoom ratio of: 12.00 × 2.60/(1024 × 992). Mean absolute differences (MAD) and intraclass correlation coefficient (ICC) were used to access the interobserver reproducibility of the measurements. Bland-Altman plots were used to graphically derive interobserver variability measurements [29].
3. Results
Table 1 shows that our method achieved an average SA of 0.840 ± 0.035 with C1 and 0.823 ± 0.027 with C2. The inter-observer variabilities with C1 and C2 of SA resulted in an average of 0.821 ± 0.037. In comparison, VGG-FCN had an average SA of 0.780 ± 0.057 with C1 and 0.785 ± 0.044 with C2. AT and LS achieved a lower SA, where AT had an average SA of 0.720 ± 0.051 with C1 and 0.679 ± 0.045 with C2, respectively, while LS has an average SA of 0.670 ± 0.113 with C1 and 0.625 ± 0.124 with C2, respectively.
Table 1. Segmentation results and inter-observer variabilities among different methods.
| SA | MAD | Bland-Altman | ICC | |
|---|---|---|---|---|
| C1 vs C2 | 0.821 ± 0.037 | 0.121 ± 0.126 | 0.082 ± 0.155 | 0.941 |
| C1 vs RefineNet | 0.840 ± 0.035 | 0.126 ± 0.102 | -0.006 ± 0.163 | 0.941 |
| C2 vs RefineNet | 0.823 ± 0.027 | 0.125 ± 0.108 | 0.075 ± 0.148 | 0.945 |
| C1 vs VGG-FCN | 0.780 ± 0.057 | 0.335 ± 0.310 | −0.335 ± 0.310 | 0.679 |
| C2 vs VGG-FCN | 0.785 ± 0.044 | 0.254 ± 0.247 | −0.253 ± 0.248 | 0.752 |
| C1 vs AT | 0.720 ± 0.051 | 0.608 ± 0.336 | −0.607 ± 0.336 | 0.854 |
| C2 vs AT | 0.679 ± 0.045 | 0.689 ± 0.375 | −0.689 ± 0.375 | 0.805 |
| C1 vs LS | 0.670 ± 0.113 | 0.804 ± 0.626 | −0.801 ± 0.630 | 0.709 |
| C2 vs LS | 0.625 ± 0.124 | 0.883 ± 0.696 | −0.883 ± 0.696 | 0.631 |
SA: segmentation agreement; MAD: mean absolute differences; ICC: intraclass correlation coefficient; C1: clinician 1; C2: clinician 2; AT: adaptive thresholding; LS: level set. The best results are highlighted in bold.
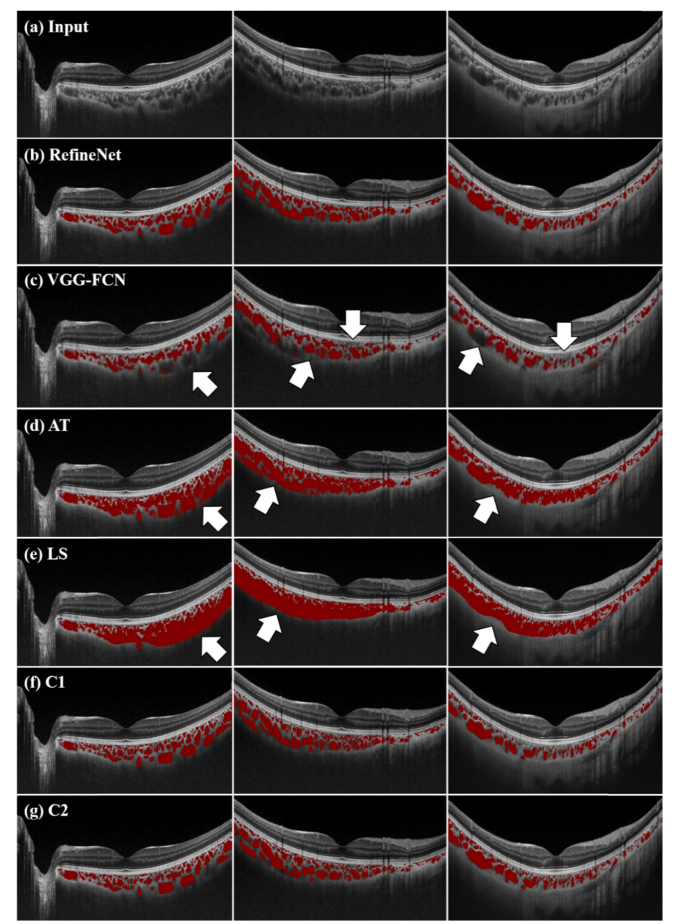
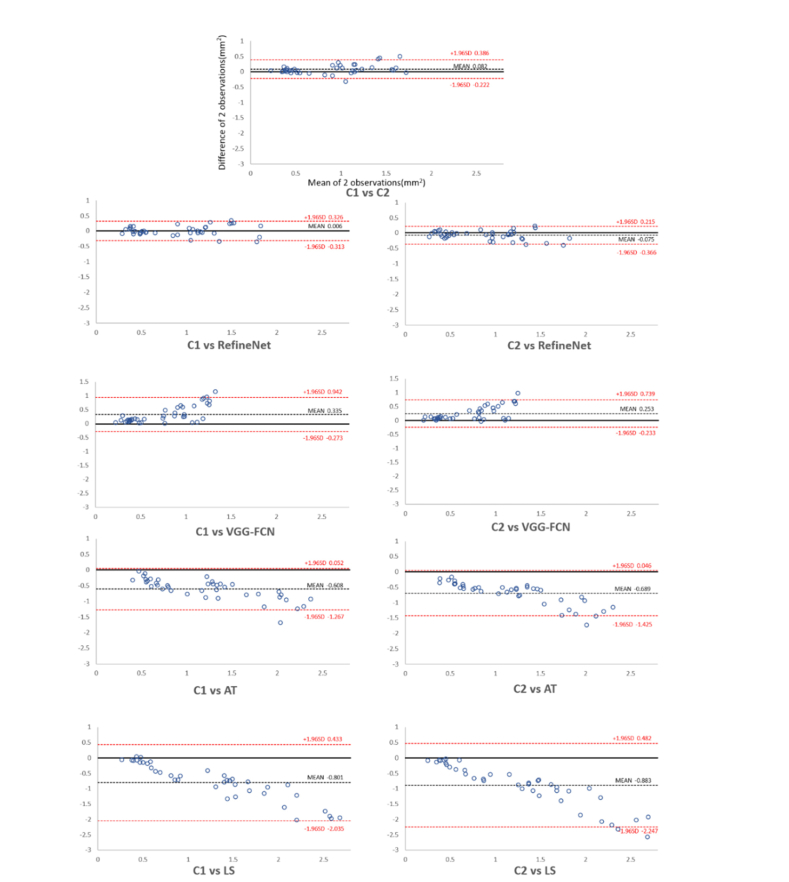
Figure 3 shows the results from various segmentation methods (rows) derived from three randomly selected patient studies (columns) for visual qualitative assessment. Figure 4 shows the Bland-Altman plots comparing inter-observer variability among different segmentation approaches.
Fig. 3.
Randomly selected segmentation results (rows) from three patient studies (in columns). (a) input images, (b, c, d, e) segmentation results derived from RefineNet, VGG-FCN, AT and LS; and (f, g) manual annotations from clinician 1 (C1) and clinician 2 (C2).
Fig. 4.
Bland–Altman plots comparing inter-observer variability. C1: clinician 1; C2: clinician 2; AT: adaptive thresholding; LS: level set.
4. Discussion
We presented the results from a deep learning based segmentation analysis, as well as the inter-observer segmentation variability analysis for choroidal vessels from SS-OCT. Our experiments in Table 1 indicate that our RefineNet method resulted in higher segmentation performance for choroidal vessels when compared to the other comparison methods. In general, AT and LS performed poorly with results ~17% lower compared to RefineNet in SA measure, on challenging choroidal vessels that have inhomogeneous variations. This is as expected as they are not able to understand image-wide semantics, e.g., the relationships between the choroidal vessels and the surrounding structures (as exemplified in Fig. 3). It is important to note that AT and LS are not fully automatic and that they are reliant on pre-and post-processing techniques and various parameter selections (i.e. handcrafted features) that restricts them from generalizability. In contrast, the deep learning approaches of VGG-FCN and RefineNet achieved higher segmentation accuracy in an end-to-end manner without pre- or post-processing techniques or parameter selections.
Table 1, and Figs. 3 and 4 validates that the RefineNet achieved higher segmentation accuracy compared with the VGG-FCN method in both the qualitative and quantitative measurements. We attributed the improvements of RefineNet compared to the VGG-FCN to the fusion of multi-scale intermediary segmentation results, which thereby can better capture the small choroidal vessel representations. In contrast, the VGG-FCN’s inherent dependence of a single-scale processing means that it cannot provide accurate segmentation results of the choroidal vessels which widely vary in sizes. In addition, VGG-FCN only supports limited number of convolutional layers and therefore is unable to derive the same amount of meaningful feature representations, i.e., high-level semantics, compared to RefineNet. We suggest that it is these deeper layers that produced better segmentation on challenging choroidal vessels e.g., vessels that are closer to the choroid layer boundaries. This effect is exemplified in Fig. 3 which shows several examples where VGG-FCN fails to segment several choroidal vessels (indicated by the arrows). In contrast, RefineNet was able to accurately segment the choroidal vessels and had higher correlation with the segmentation results derived from two clinicians (as exemplified in Fig. 4).
The results also demonstrate that RefineNet achieved competitive segmentation accuracy with low inter-observer variability compared with two clinicians (C1 and C2). The results in Table 1 shows that RefineNet resulted in a lower inter-observer variability with higher segmentation agreement, when compared with the inter-observer variability among C1 and C2. More specifically, RefineNet was ~2% higher in terms of segmentation agreement (SA) with the two clinicians, which suggests that RefineNet as a fully automated method is able to segment the choroidal vessels that are consistent to what clinicians would produce in a manual manner. In addition, the Bland-Altman plot in Fig. 4 shows that both RefineNet with C1 and RefineNet with C2 have better reproducibility compared to the reproducibility of C1 with C2. We attribute this better reproducibility to the training process which allows to retain important characteristics of the choroidal vessels structures and resulted in consistent segmentation results.
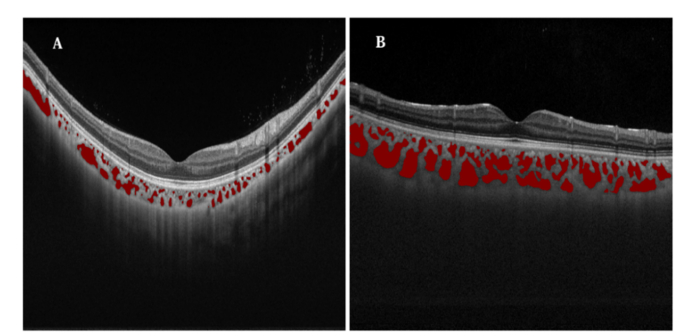
Automatic segmentation of choroidal vessels on SS-OCT, which removes the need for manual, time consuming and error-prone annotation process, provides opportunities to visualize the full thickness (e.g. volumetric rendering) of the choroidal blood flow in a non-invasive manner. Figure 5 exemplifies the segmented choroidal vessels of high myopia and emmetropia. With the segmentation derived from RefineNet, we can clearly visualize that the abnormal high myopia studies (A) have markedly less segmented regions compared to the normal emmetropia studies in (B). Our analysis thus makes it practical and facilitates future researches about choroidal vessels, such as in clarifying discrepancy about the status of choroid and choroidal vessels in ocular diseases, creating population based choroidal disease models.
Fig. 5.
Difference of high myopia and normal study. A: High Myopia; B: Normal (Emmetropia).
In addition, our experiments have demonstrated the usefulness and potential broader adaptation of SS-OCT for quantitative analysis of choroid vessels. Currently, researchers have limited imaging modalities to obtain information about the choroidal vessels. Traditional indocyanine green angiography (ICGA) is the gold standard in clinical practice for detecting abnormality in the choroidal vessels. ICGA provide 2D images of the choroid vasculature, which can show the exudation or filling defects. However, ICGA does not provide 3D choroidal structure or the volume of the whole choroidal vessel networks, and the ICGA images overlap retinal vessels and choroidal vessels together, thereby making it hard to independently observe and analyze the choroidal vessels quantitatively. OCT Angiography (OCTA) can clearly show the blood flow from superficial and deep retinal capillary network, as well as retinal pigment epithelium to superficial choroidal vascular network; however, it cannot show the blood flow in deep choroidal vessels.
In this work, we focused on the automated choroidal vessels segmentation and therefore, we didn’t explore different visualization options that are possible with the segmentation results, such as the face projection and 3D volume rendering. Our experiments demonstrate that 2D approach is sufficient for the purpose of segmentation analysis (both quantitative and qualitative). Nevertheless, as future work, we will explore the usefulness of our segmentation with 3D visualizations.
5. Conclusions
We proposed a RefineNet deep learning method to automatically segment the choroidal vessels on SS-OCT images. Our experiments demonstrated that RefineNet achieved higher segmentation accuracy with low inter-observer variabilities compared to the measurements between the two clinicians. Further, quantitative evaluation to baseline VGG-FCN resulted in higher segmentation performance. The RefineNet improvements were attributed to the iteratively propagation of multi-scale intermediary segmentation results, and the usage of deeper architecture. As future work, we will investigate the adaptation of RefineNet for quantitative analysis and as well as potential clinical applications.
Funding
National Natural Science Foundation of China (NSFC) (81570851, 81800878); Project of the National Key Research Program on Precision Medicine (2016YFC0904800); University of Sydney – Shanghai Jiao Tong University Joint Research Alliance (USYD-SJTU JRA).
Disclosures
Authors declare that there is no conflict of interest related to this article.
References
- 1.Nickla D. L., Wallman J., “The multifunctional choroid,” Prog. Retin. Eye Res. 29(2), 144–168 (2010). 10.1016/j.preteyeres.2009.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mullins R. F., Johnson M. N., Faidley E. A., Skeie J. M., Huang J., “Choriocapillaris vascular dropout related to density of drusen in human eyes with early age-related macular degeneration,” Invest. Ophthalmol. Vis. Sci. 52(3), 1606–1612 (2011). 10.1167/iovs.10-6476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jirarattanasopa P., Ooto S., Nakata I., Tsujikawa A., Yamashiro K., Oishi A., Yoshimura N., “Choroidal thickness, vascular hyperpermeability, and complement factor H in age-related macular degeneration and polypoidal choroidal vasculopathy,” Invest. Ophthalmol. Vis. Sci. 53(7), 3663–3672 (2012). 10.1167/iovs.12-9619 [DOI] [PubMed] [Google Scholar]
- 4.Lavinsky F., Lavinsky D., “Novel perspectives on swept-source optical coherence tomography,” Int. J. Retina Vitreous 2(1), 25 (2016). 10.1186/s40942-016-0050-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou H., Chu Z., Zhang Q., Dai Y., Gregori G., Rosenfeld P. J., Wang R. K., “Attenuation correction assisted automatic segmentation for assessing choroidal thickness and vasculature with swept-source OCT,” Biomed. Opt. Express 9(12), 6067–6080 (2018). 10.1364/BOE.9.006067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou H., Chu Z., Zhang Q., Dai Y., Gregori G., Rosenfeld P. J., Wang R. K., “Attenuation correction assisted automatic segmentation for assessing choroidal thickness and vasculature with swept-source OCT,” Biomed. Opt. Express 9(12), 6067–6080 (2018). 10.1364/BOE.9.006067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duan L., Hong Y. J., Yasuno Y., “Automated segmentation and characterization of choroidal vessels in high-penetration optical coherence tomography,” Opt. Express 21(13), 15787–15808 (2013). 10.1364/OE.21.015787 [DOI] [PubMed] [Google Scholar]
- 8.Kajić V., Esmaeelpour M., Glittenberg C., Kraus M. F., Honegger J., Othara R., Binder S., Fujimoto J. G., Drexler W., “Automated three-dimensional choroidal vessel segmentation of 3D 1060 nm OCT retinal data,” Biomed. Opt. Express 4(1), 134–150 (2013). 10.1364/BOE.4.000134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang L., Lee K., Niemeijer M., Mullins R. F., Sonka M., Abràmoff M. D., “Automated segmentation of the choroid from clinical SD-OCT,” Invest. Ophthalmol. Vis. Sci. 53(12), 7510–7519 (2012). 10.1167/iovs.12-10311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.N. Srinath, A. Patil, V. K. Kumar, S. Jana, J. Chhablani, and A. Richhariya, “Automated detection of choroid boundary and vessels in optical coherence tomography images,” in Engineering in Medicine and Biology Society (EMBC), 2014 36th Annual International Conference of the IEEE, (IEEE, 2014), 166–169. 10.1109/EMBC.2014.6943555 [DOI] [PubMed] [Google Scholar]
- 11.Shelhamer E., Long J., Darrell T., “Fully Convolutional Networks for Semantic Segmentation,” IEEE Trans. Pattern Anal. Mach. Intell. 39(4), 640–651 (2017). 10.1109/TPAMI.2016.2572683 [DOI] [PubMed] [Google Scholar]
- 12.Dolz J., Desrosiers C., Ben Ayed I., “3D fully convolutional networks for subcortical segmentation in MRI: A large-scale study,” Neuroimage 170, 456–470 (2018). 10.1016/j.neuroimage.2017.04.039 [DOI] [PubMed] [Google Scholar]
- 13.Fang L., Cunefare D., Wang C., Guymer R. H., Li S., Farsiu S., “Automatic segmentation of nine retinal layer boundaries in OCT images of non-exudative AMD patients using deep learning and graph search,” Biomed. Opt. Express 8(5), 2732–2744 (2017). 10.1364/BOE.8.002732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simonyan K., Zisserman A., “Very Deep Convolutional Networks for Large-Scale Image Recognition,” Comput. Sci. (2014). [Google Scholar]
- 15.Sui X., Zheng Y., Wei B., Bi H., Wu J., Pan X., Yin Y., Zhang S., “Choroid segmentation from OpticalCoherence Tomography with graph-edge weights learned from deep convolutional neural networks,” Neurocomputing 237, 332–341 (2017). 10.1016/j.neucom.2017.01.023 [DOI] [Google Scholar]
- 16.Venhuizen F. G., van Ginneken B., Liefers B., van Grinsven M. J. J. P., Fauser S., Hoyng C., Theelen T., Sánchez C. I., “Robust total retina thickness segmentation in optical coherence tomography images using convolutional neural networks,” Biomed. Opt. Express 8(7), 3292–3316 (2017). 10.1364/BOE.8.003292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu Y., Yan K., Kim J., Wang X., Li C., Su L., Yu S., Xu X., Feng D. D., “Dual-stage deep learning framework for pigment epithelium detachment segmentation in polypoidal choroidal vasculopathy,” Biomed. Opt. Express 8(9), 4061–4076 (2017). 10.1364/BOE.8.004061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.G. M. Lin, Anton; Shen, Chunhua; Reid, Ian, “RefineNet: Multi-Path Refinement Networks for High-Resolution Semantic Segmentation,” eprint arXiv:1611.06612 (2016).
- 19.He K., Zhang X., Ren S., Sun J., “Deep Residual Learning for Image Recognition,” 770–778 (2015).
- 20.Chatfield K., Simonyan K., Vedaldi A., Zisserman A., “Return of the Devil in the Details: Delving Deep into Convolutional Nets,” Comput. Sci. (2014). [Google Scholar]
- 21.Veit A., Wilber M., Belongie S., “Residual Networks Behave Like Ensembles of Relatively Shallow Networks,” Adv. Neural Inf. Process. Syst. (2016). [Google Scholar]
- 22.Shin H. C., Roth H. R., Gao M., Lu L., Xu Z., Nogues I., Yao J., Mollura D., Summers R. M., “Deep Convolutional Neural Networks for Computer-Aided Detection: CNN Architectures, Dataset Characteristics and Transfer Learning,” IEEE Trans. Med. Imaging 35(5), 1285–1298 (2016). 10.1109/TMI.2016.2528162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bi L., Kim J., Ahn E., Kumar A., Fulham M., Feng D., “Dermoscopic Image Segmentation via Multistage Fully Convolutional Networks,” IEEE Trans. Biomed. Eng. 64(9), 2065–2074 (2017). 10.1109/TBME.2017.2712771 [DOI] [PubMed] [Google Scholar]
- 24.J. Deng, W. Dong, R. Socher, L. J. Li, K. Li, and F. F. Li, “ImageNet: A large-scale hierarchical image database,” in Computer Vision and Pattern Recognition, 2009. CVPR 2009. IEEE Conference on, 2009), 248–255. 10.1109/CVPR.2009.5206848 [DOI] [Google Scholar]
- 25.Chen H., Qi X., Yu L., Dou Q., Qin J., Heng P. A., “DCAN: Deep contour-aware networks for object instance segmentation from histology images,” Med. Image Anal. 36, 135–146 (2017). 10.1016/j.media.2016.11.004 [DOI] [PubMed] [Google Scholar]
- 26.Vedaldi A., Lenc K., “MatConvNet: Convolutional Neural Networks for MATLAB,” in ACM International Conference on Multimedia, 2015), 689–692. [Google Scholar]
- 27.Krizhevsky A., Sutskever I., Hinton G. E., “ImageNet classification with deep convolutional neural networks,” in International Conference on Neural Information Processing Systems, 2012), 1097–1105. [Google Scholar]
- 28.Kumar A., Kim J., Lyndon D., Fulham M., Feng D., “An Ensemble of Fine-Tuned Convolutional Neural Networks for Medical Image Classification,” IEEE J. Biomed. Health Inform. 21(1), 31–40 (2017). 10.1109/JBHI.2016.2635663 [DOI] [PubMed] [Google Scholar]
- 29.Bland J. M., Altman D. G., “Statistical methods for assessing agreement between two methods of clinical measurement,” Lancet 1(8476), 307–310 (1986). 10.1016/S0140-6736(86)90837-8 [DOI] [PubMed] [Google Scholar]