Abstract
Background
Promyelocytic leukemia zinc finger (Plzf), a transcriptional regulator involved in a lot of important biological processes during development, has been implied to maintain neural stem cells and inhibit their differentiation into neurons. However, the effects of Plzf on brain structures and functions are still not clarified.
Results
We showed that Plzf expression was detected as early as embryonic day (E) 9.5 in Pax6+ cells in the mouse brain, and was completely disappeared in telencephalon before the initiation of cortical neurogenesis. Loss of Plzf resulted in a smaller cerebral cortex with a decrease in the number of Tbr1+ deep layer neurons due to a decrease of mitotic cell number in the ventricular zone of forebrain at early developmental stage. Microarray, qRT-PCR, and flow cytometry analysis identified dysregulation of Mash1 proneural gene expression. We also observed an impairment of recognition memory in Plzf-deficient mice.
Conclusions
Plzf is expressed at early stages of brain development and involved in the formation of deep layer cortical neurons. Loss of Plzf results in dysregulation of Mash1, microcephaly with reduced numbers of early-born neurons, and impairment of recognition memory.
Electronic supplementary material
The online version of this article (10.1186/s12929-019-0519-8) contains supplementary material, which is available to authorized users.
Keywords: Promyelocytic leukemia zinc finger, Plzf, luxoid mice, Cortical neuron
Background
Promyelocytic leukemia zinc finger (PLZF, also known as ZBTB16, ZNF145) is a kruppel-like C2H2 zinc finger gene containing nine zinc fingers in the C-terminus and a BTB/POZ domain in the N-terminus [1] and has been shown to mediate a lot of important biological processes, especially hematopoiesis, osteogenesis, and spermatogenesis [2–5]. The patient with biallelic loss of PLZF function exhibits abnormality in bone development, genital hypoplasia, and microcephaly with a severe mental retardation, implying the involvement of PLZF in brain development [6, 7]. Actually, Plzf expression was observed in the anterior neuroepithelium at early stage (E7.5), which later extends to the entire neuroectoderm until stage E10 [8]. In zebrafish, Plzf can block neuronal differentiation by inhibiting the expression of proneural gene Ngn1 [9]. A study also showed that Plzf maintains neural progenitors in the spinal cord of chick and mouse by up-regulating FGFR3 expression and through STAT3 activation [10]. Although, these results suggest that PLZF is important for the development of central nervous system, the effects of Plzf on brain structures and functions are still not clarified.
The development of cerebral cortex is an important and complicated process which involves in neural stem cells (NSCs) proliferation, differentiation, cell fate determination, and cell migration [11–13]. During brain development, NSCs can either divide symmetrically to expand the cell pool through the process called self-renewal, or undergo the neurogenesis process to divide asymmetrically and generate neural progenitors that are subsequently differentiated into neurons [14]. The proper balance between NSCs self-renewal and neurogenesis is important to ensure appropriate numbers of neurons being generated. Indeed, defects affecting this balance have been suggested to induce brain malformations [15–17].
To better understand the effect of Plzf in mammalian brain development, we first examined the cell type-restricted expression of Plzf during mouse brain development. We then characterized the developmental brain abnormality and the behavioral phenotypes of the Plzf-deficient mice. In addition, we also performed microarray to examine the differences in gene expression profiles of developmental brain between wild-type and Plzf-deficient mice. Our results revealed that Plzf is expressed at early stages of brain development and involved in the formation of deep layer cortical neurons.
Methods
Animals
The animal protocols used in this study complied with the guidelines of the Taipei Medical University Institutional Animal Care and Use Committee (IACUC). All experiments were performed in accordance with the US Public Health Service Policy on Humane Care and Use of Laboratory Animals. All animals were housed in an air-conditioned vivarium with free access to food and water and a 10/14 h light/dark cycle.
Antibodies
The following antibodies were used, including Satb2, Ctip2, Tbr1, and Mash1 (Abcam, Cambridge, CB, UK), Plzf and PHH3 (Santa Cruz, Santa Cruz, CA, USA), Pax6 (Merck Millipore, Darmstadt, Hessen, Germany), β-actin (Sigma-Aldrich, Louis, MO, USA).
Magnetic resonance imaging
Mouse brains at postnatal day (P) 0 were fixed with 4% paraformaldehyde for 48 h and embedded in 1.5% agarose. All images were acquired using Avance spectrometer (Bruker, Karlsruhe, Baden-Württemberg, Germany) with 9.4 T WB 8.9-cm bore vertical system (Oxford Instrument, Oxford, UK). A 3D fast spin-echo (FSE) T2WI sequence was used to acquire anatomical images for volumetric analysis with the following parameters: a repetition time (TR) of 10,000 ms, an effective echo time (TE) of 91 ms, a RARE factor of 8, a field-of-view (FOV) of 0.9 cm, an acquisition matrix of 256 × 128 × 30 (zero-padded to 512 × 512 × 30), a resolution of 0.0039 × 0.0078 × 0.0073 cm/pixel. Brain volumes were measured using the manual tracing utility in Avizo software (Visualization Sciences Group, Merignac, Aquitaine, France).
Behavioral studies
Open field test
The open field activity was assessed in a chamber (57.4 cm × 59 cm × 95 high cm). Mice were individually placed in the middle of the arena and allowed to explore for 10 min. The moving trace of each mouse was recorded by a video camera and analyzed by the EthoVision XT software (Noldus Information Technology, Leesburg, VA, USA).
Novel object recognition (NOR) test
NOR test was assessed in a chamber (57.4 cm × 59 cm × 95 high cm). The test consists of three sessions: habituation, familiarization (training), and test session. In the habituation session, each mouse was allowed to freely explore the arena in the absence of objects for 10 min. The mice were then removed from the arena and placed in its holding cage. One day after the habituation session, mice were subjected to a single familiarization session of 10 min. During the familiarization session, each mouse was placed in the same arena containing two identical sample objects (A + A). One day after the familiarization session, mice were subjected to a single test session of 10 min: the animal was placed in the arena with two objects, including one sample (A) and one novel object (B). During both the familiarization and test sessions, the time the mice spent exploring each object was recorded.
Immunohistochemistry and immunofluorescence
For preparing embryonic sections, the embryos were fixed with 4% paraformaldehyde (PFA) in phosphate buffered saline (PBS), pH 7.4 at 4 °C for overnight. Fixed embryos were immersed in 30% sucrose in PBS and then frozen in OCT and cryosectioned to 25 μm sections using the Leica CM1950 freezing microtome (Leica Biosystems, Wetzlar, Hesse, Germany). For the preparation of postnatal brain sections, brains were fixed with 4% PFA in PBS, pH 7.4 at 4 °C for 48 h. Fixed brains were then dehydrated with 75% alcohol prior to paraffin embedding. 3 μm thick sections were obtained using Sakura IVS-410 microtome (Sakura Seiki Co, Tokyo, Japan). All the paraffin sections were dewaxed and rehydrated prior to the staining procedures. For both immunohistochemistry and immunofluorescence staining, tissue sections were permeabilized and blocked with blocking buffer (0.2% Triton X-100, 10% goat serum in PBS) at room temperature for 1 h. For immunofluorescence, tissue sections were incubated with primary antibody solution overnight at 4 °C. After extensive washes with PBS, the tissue sections were incubated with the corresponding secondary antibodies at room temperature for 1 h. For immunohistochemistry, the tissue sections were incubated with primary antibodies overnight at 4 °C. After extensive washes with PBS, the tissue sections were incubated with biotinylated secondary antibodies. The colorimetric detection of primary antibodies was developed using the Vectastain ABC Kit (Vector laboratories, Burlingame, California, USA), followed by exposure to peroxidase DAB substrate (Vector laboratories). Cell nuclei were stained with hematoxylin (Leica) or Hoechst 33342 (Sigma-Aldrich) and slides were mounted with anti-fading solution (SouthernBiotech, Birmingham, AL, USA).
Image analysis
Tissue sections were observed and photographed by Leica STP6000 fluorescent microscope (Leica Biosystems) and scanned by TissueGnostics TissueFAXS & HistoFAXS (TissueGnostics Gmbh, Vienna, Austria). Images were overlaid by MetaMorph software (Universal Imaging Corporation, Philadelphia, PA, USA), and the brightness and contrast of images were adjusted by Photoshop (Adobe, Camarillo CA, USA) as necessary. With regard to the measurement of cerebral cortex area, comparable 3 μm-thick sagittal sections of P7 mice brain were stained by hematoxylin and eosin (H&E). The area of the cortical layers was measured by HistoQuest software (TissueGnostics GmbH). The numbers of Tbr1-, Satb2-, and Ctip2-positive cells was quantified by TissueQuest software (TissueGnostics GmbH). The number of PHH3-positive cells was quantified in 120 μm-wide sampling boxes along the ventricular zone of the telencephalon. The quantification of each experimental group was repeated on two non-adjacent sections for each embryo.
Western blot analysis
Embryos were homogenized by radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 1% NP40, 0.5% Na-deoxycholate, 0.1% sodium dodecylsulfate (SDS), and 50 mM Tris-HCl; pH 8.0) with the addition of a protease inhibitor cocktail used for homogenization. For embryos younger than E11.5, three heads were pooled together as one biological sample. After lysed, the cell debris were removed with centrifugation fall at 12,000 rpm for 5 min at 4 °C, and the supernatants were stored at − 80 °C. Protein concentration was determined using the Bio-Rad dye-binding method with bovine serum albumin (BSA) as the standard. Equal amounts of samples were separated by 8–16% gradient polyacrylamide gel electrophoresis (PAGE). The resolved proteins were then electroblotted onto Immobilon polyvinylidene difluoride (PVDF) membranes (PerkinElmer, Waltham, MA, USA), which was followed by blocking with 5% low-fat milk. Blotted membranes were then incubated with selected primary antibodies overnight at 4 °C. After extensive washes, membranes were incubated with the corresponding secondary antibodies at room temperature for 1 h. After washing, the membranes were processed for visualization using an enhanced chemiluminescence (ECL) system (PerkinElmer, Waltham, MA, USA). Membranes were then exposed to x-ray film to obtain the fluorographic images and quantified by ImageJ (NIH, Bethesda, MD, USA). Values for each lane were normalized to β-actin.
Microarray and gene ontology (GO) analysis
RNA was extracted from forebrain and midbrain regions of mouse E10.5 embryos with TRIzol reagent (Invitrogen). 0.2 μg of total RNA was amplified by a Low Input Quick-Amp Labeling kit (Agilent Technologies, USA) and labeled with Cy3 or Cy5 (CyDye) (Agilent Technologies, California, USA) during the in vitro transcription process. 0.825 μg of Cy-labeled cRNA was fragmented to an average size of about 50–100 nucleotides by incubation with fragmentation buffer at 60 °C for 30 min. Correspondingly fragmented labeled cRNA was then pooled and hybridized to an Agilent SurePrint G3 Mouse Gene Exp v2 Array Kit (Agilent Technologies) at 65 °C for 17 h. After washing and drying by nitrogen gun blowing, microarrays were scanned with an Agilent microarray scanner (Agilent Technologies) at 535 nm for Cy3 and 625 nm for Cy5. Scanned images were analyzed by Feature extraction 10.7.3.1 software (Agilent Technologies), and image analysis and normalization software was used to quantify signal and background intensity for each feature, substantially normalized the data by rank-consistency-filtering LOWESS method. The spots with the processed signal higher than 200 and the signal-to-noise ratio (SNR) greater or equal to 5 were considered positive signals. The raw data were available at http://www.ncbi.nlm.nih.gov/geo/ (accession numbers: GSM3273361~3,273,362, GSE117197). The genes with greater or less than 1.3-fold changes in expression between wild-type and Plzf-deficient mice were selected for further analysis. GO analysis were performed using ToppFun (https://toppgene.cchmc.org/) and Ingenuity Pathways Analysis (IPA) (Ingenuity, Redwood City, CA, USA).
Quantitative RT–PCR
Reverse transcription (RT) was performed with 1.5 μg of total RNA using M-MLV reverse transcriptase (Invitrogen). A real-time qPCR was performed using the iTaq Universal SYBR Green Supermix (Biosystem Applications, Foster City, CA, USA) with the following conditions: step1, 95 °C for 30 s; step2 (repeated for 40 cycles), 95 °C for 5 s, 60 °C for 30 s. Real-time fluorescence monitoring and a melting-curve analysis were performed by StepOnePlus Real-Time PCR System according to the manufacturer’s recommendations (Life Technologies, Carlsbad, CA, USA). Negative controls containing no complementary (c)DNA template were included in each experiment. A melting curve was created at the end of the PCR cycle to confirm that a single product had been amplified. Data were analyzed by StepOne Software version 2.2.2 (Life Technologies) to determine the threshold cycle (Cp) above the background for each reaction. The relative transcript amount of the target gene, calculated using standard curves of serial cDNA dilutions, was normalized to that of Gapdh of the same cDNA. Primers used in PCR assays were as follows: Mash1, forward primer, 5′- TTGAACTCTATGGCGGGTTC-3′, reverse primer, 5′-GGTTGGCTGTCTGGTTTGTT-3′; Gapdh, forward primer, 5′- TGACATCAAGAAGGTGGTGAAG-3′, reverse primer, 5′- AGAGTGGGAGTTGCTGTTGAAG-3′.
Flow cytometry
Forebrain and midbrain tissues were dissected from E10.5 mouse embryos and dissociated by 0.2% Trypsin/EDTA at 37 °C for 5 min, and then filtered through a 70-μm nylon mesh filter (Corning life science, Corning, New York, USA). The isolated cells were fixed by 75% methanol. For cell cycle analysis, the cells were washed once with ice-cold PBS and labeled with propidium iodide (PI) (Sigma) at 25 °C for 1 h. Five thousand events without cell debrides were then analyzed using the Guava EasyCyte system and the InCyte software (Millipore). For analysis of Mash1-expressing cells, the cells were washed once with PBS and incubated with anti-Mash1 antibody (Abcam) with PI solution in blocking buffer (10% normal goat serum, 0.2% RNaseA, and 0.1% Triton-100X in PBS) for 1 h at 25 °C. The cells were washed and incubated with Alexa Fluor 488 anti-mouse IgG (Life Technologies) for 1 h at 25 °C. After washed with ice-cold PBS to remove the unconjugated antibodies, the cells were resuspended in PBS, and 5000 events without cell debrides were analyzed using the Guava EasyCyte system and the InCyte software (Millipore).
Results
Plzf is temporarily expressed in Pax6+ cells at early stages of brain development
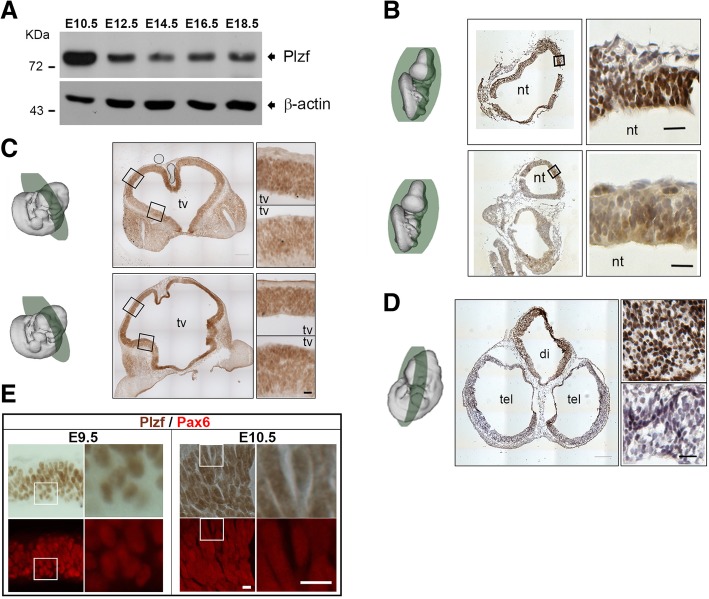
Previous studies have shown that Plzf is expressed in the anterior neuroepithelium of mouse embryo at E7.5 and spreading to the entire neuroectoderm until E10 [8]. We therefore examined the expression pattern of Plzf by Western blotting and immunostaining in the brains of mouse embryos at different stages. Similar to previous study, we found Plzf was highly expressed in the mouse embryonic brains at E10.5, and the expression was decreased after E12.5 (Fig. 1a). Immunostaining results showed that a high expression level of Plzf in the prosencephalon, which later becomes the forebrain, at E9.5 (Fig. 1b) and E10.5 (Fig. 1c). Further examinations showed that Plzf expression was dramatically decreased in telencephalon at E11.5 (Fig. 1d). We also found that Plzf was expressed in Pax6+ cells in prosencephalon at E9.5 and E10.5 (Fig. 1e), implying a role of Plzf on neuroepithelial cells or radial glial cells at early stages of neurogenesis.
Fig. 1.
Plzf temporarily expressed in Pax6+ cells at early stages of brain development. a, Western blot analysis of Plzf expression in whole brain lysates of mouse embryo at different stages. b-d, 20 μm frozen sections were prepared from the E9.5 (b), E10.5 (c), and E11.5 (d) embryos and immunohistochemistry for Plzf was performed with DAB (brown) followed by Hematoxylin-eosin (HE) counterstaining. The diagrams of mouse embryo were obtained from e-Mouse Atlas Project (http://www.emouseatlas.org/emap/ema/home.php) and the green circles indicate the approximate plane of the cross section. The higher magnification images of the black boxes were shown in the right panels. Scale bar, 20 μm. e, The expressions of Pax6 (red) and Plzf (brown) were examined by immunostaining in the same brain sections. The higher magnification images of the white boxes were shown in the right panels. E, embryonic day; nt, neural tube; tv, third ventricle; tel., telencephalon; di, diencephalon. Scale bar, 10 μm
Plzf-deficient mice display a decrease in cerebral cortex size and the number of deep-layer cortical neurons
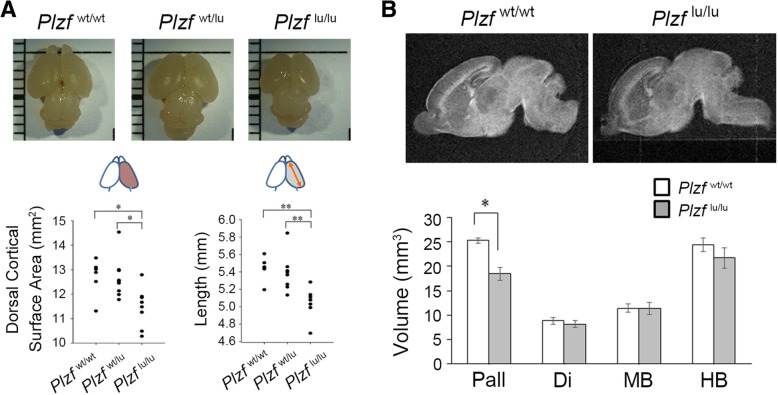
To explore the effect of Plzf in mammalian brain development, we examined the possible structural and functional alterations in the brain of Plzf-deficient (luxoid; lu) mice, Plzf-mutant mice which arose spontaneously in 1950’s and was characterized with a single nucleotide change in the first coding exon of Plzf gene that resulted a severely truncated protein [3]. We first found that the dorsal cortical surface area and hemisphere length were significantly reduced in Plzf-deficient mice (Plzflu/lu) at postnatal day (P) 0 when compared to wild type (Plzfwt/wt) and heterozygous (Plzfwt/lu) littermates (Fig. 2a). At P0, the average of dorsal cortical area were 12.717 ± 0.338 mm2 in Plzfwt/wt, 12.659 ± 0.288 mm2 in Plzfwt/lu, and 11.467 ± 0.305 mm2 in Plzflu/lu, respectively, and the average lengths of hemisphere were 5.441 ± 0.061 mm in Plzfwt/wt, 5.390 ± 0.071 mm in Plzfwt/lu, and 5.076 ± 0.071 mm in Plzflu/lu, respectively (Fig. 2a). Using 3D MRI imaging, we accurately measured the volumes of brain regions at P0 and found that when compared to wild-type littermates, Plzflu/lu mice showed a significantly decrease in the volume of pallium (the cortex and hippocampus) but not in other brain areas (Fig. 2b).
Fig. 2.
Plzf-deficient mice exhibit smaller cerebral cortex. a, Dorsal view of whole-mount brains at P0. For the scale size, each of the thin line indicates 1 mm, and the thick line indicates 5 mm. Quantifications of dorsal cortical surface area and cortical length shown in the lower panels. Statistical analysis performed using one-way ANOVA with appropriate post hoc tests: *p < 0.05 and **p < 0.01; Plzfwt/wt, n = 6; Plzfwt/lu, n = 9; Plzflu/lu, n = 8. b, Representative sagittal views of 9.4 T MRI sections of Plzfwt/wt and Plzflu/lu mice brain. Quantification of the volume of brain regions shown in the lower panel. Statistics performed with Student’s t test: *p < 0.05; n = 4 per genotype. Data are represented as mean ± SEM. P, postnatal day; Plzfwt/wt, wild-type; Plzfwt/lu, heterozygous; Plzflu/lu, homozygous luxoid mice; Pall, Pallium; Di, diencephalon; MB, midbrain; HB, hindbrain
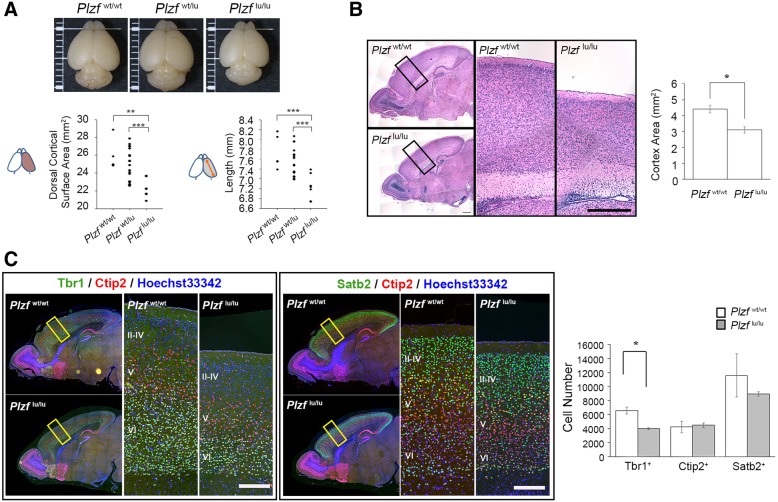
Similar to observations at P0, the dorsal cortical surface area and hemisphere length were also significantly reduced in Plzflu/lu mice at P7 (Fig. 3a). Our data showed that the average of dorsal cortical area at P7 is 25.202 ± 0.748 mm2 in Plzfwt/wt, 24.852 ± 0.434 mm2 in Plzfwt/lu, and 21.747 ± 0.564 mm2 in Plzflu/lu, respectively, and the average of hemisphere length is 7.619 ± 0.146 mm in Plzfwt/wt, 7.521 ± 0.068 mm in Plzfwt/lu, and 6.984 ± 0.103 mm in Plzflu/lu, respectively (Fig. 3a). We also evaluated the cerebral cortical area from haematoxylin and eosin (H&E) stained brain sections, and found that the cortical area was significantly decreased in Plzflu/lu mice (Fig. 3b). Our data showed that the Plzflu/lu mice have a thinner cortex. Using immunostaining, we further evaluated the alteration of cortical structure in Plzflu/lu mice labelled with specific cortical layer markers, such as Tbr1 (a marker of cortical deep-layer neurons), Ctip2 (a marker of layer 5 neurons), and Satb2 (a marker of layer 2/3 neurons). Our result showed a significantly decreased number of Tbr1+ neurons in layer VI of Plzflu/lu mouse brain cortex (Fig. 3c). On the other hand, the numbers of Ctip2+ neurons and Satb2+ neurons were not different between Plzfwt/wt and Plzflu/lu mice. Our results thus indicate that loss of Plzf results in a decrease in the numbers of early-born neurons, suggesting the requirement of Plzf in early stage of neurogenesis.
Fig. 3.
Reduced numbers of neurons in deep-layer cortex of Plzflu/lu mice. a, Dorsal view of whole-mount brains at P7. For the scale size, each of the thin lines indicates 1 mm, and the thick line indicates 5 mm. Quantifications of dorsal cortical surface area and cortical length shown in the lower panels. Statistical analysis performed using one-way ANOVA with appropriate post hoc tests: **p < 0.01 and ***p < 0.001. Plzfwt/wt, n = 7; Plzfwt/lu, n = 17; Plzflu/lu, n = 9. b, HE staining of sagittal brain sections, the higher magnification images of the black boxes were shown in the middle and right sides of the panel. Quantification of the area of cerebral cortex from rostral to caudal shown in the right panel. Statistics performed with Student’s t test: *p < 0.05; n = 3 per genotype. c, The left panel showed immunofluorescence images of Plzfwt/wt and Plzflu/lu mice brain sections. Tbr1 as layer VI neuron marker (green), Ctip2 as layer V neuron marker (red) and Hoechst33342 for nucleus staining (blue). For the middle panels, Satb2 as layer II-III marker (green). The higher magnification images of the yellow boxes were shown in the middle and right sides of panels. Quantification of the Tbr1+, Satb2+, and Ctip2+ neuron numbers in cerebral cortex shown in the right panel. Statistics performed with Student’s t test: *p < 0.05; n = 3 per genotype. Values represent the mean ± SEM. P, postnatal day; Plzfwt/wt, wild-type; Plzfwt/lu, heterozygous; Plzflu/lu, homozygous; Scale bar, 200 μm
Cell proliferation and gene expression changes in Plzf-deficient mice
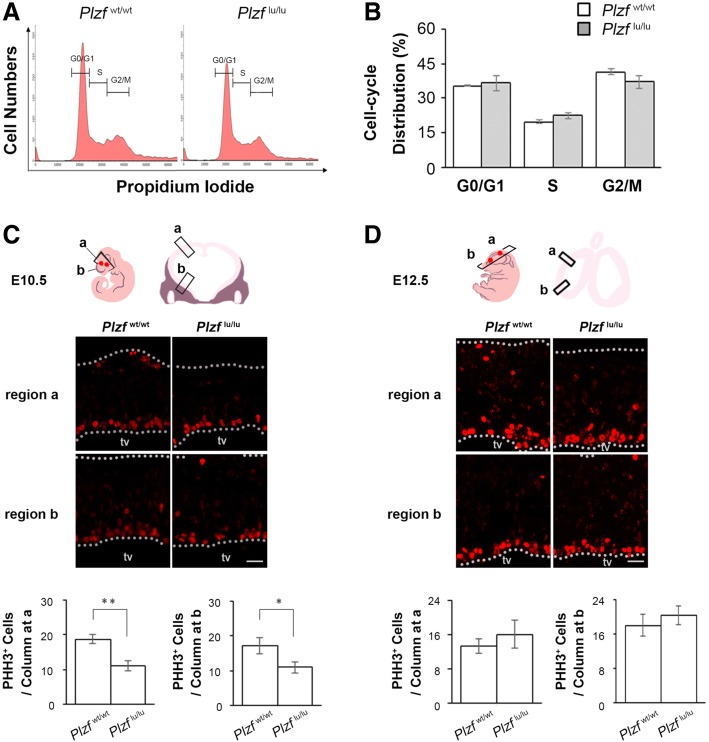
To evaluate the possible mechanism of cortical thinning in Plzf-deficient mice, we first analyzed the cell proliferation in the brain of wild type and Plzflu/lu mice at E10.5 by flow cytometry. Although not significant, our result showed that there was a slight decrease in G2/M phase cells in Plzflu/lu mice (Fig. 4a and b). To accurately measure the cell proliferation in the ventricular zone of the prosencephalon, we further analyzed the amount of mitotic cells in the ventricular zone of prosencephalon in wild-type and Plzflu/lu mice by using phospho-histone H3 (PHH3) immunostaining. As shown in Fig. 4c and d, the number of mitotic cell in the ventricular zone of prosencephalon is significantly decreased in Plzflu/lu mice at E10.5 (Fig. 4c) but not at E12.5 (Fig. 4d), indicating cell proliferation change at early stage of neurogenesis may cause the abnormal cortical development in Plzflu/lu mice.
Fig. 4.
Cell proliferation change at early stage of neurogenesis in Plzflu/lu mice brain. a, Representative flow cytometric data showing the cell cycle distribution. X-axis is the intensity of propidium iodide, and Y-axis is the cell numbers. b, Quantification of proportions of cells in each phase of the cell cycle. Plzfwt/wt, n = 4; Plzflu/lu, n = 3. c and d, E10.5 (c) and E12.5 (d) brain sections were immunstained by anti-PHH3 antibodies. The PHH3+ cells were calculated in the rostral (b) and caudal (a) regions, respectively. The white dashed lines represent the apical and basal borders of cerebral cortex. Quantification of the PHH3+ cell numbers in the region shown in the lower panels. n = 4 embryos per genotype at E10.5; n = 3 embryos per genotype at E12.5. Statistics were performed with Student’s t test. Values represent the mean ± SEM. E, embryonic day; Plzfwt/wt, wild-type; Plzflu/lu, homozygous luxoid mice; tv, third ventricle; Scale bar 20 μm
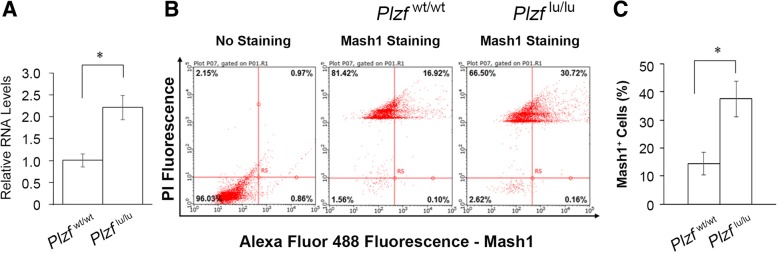
Next, we compared the differences in gene expression profiles of forebrain between wild type and Plzflu/lu mice at E10.5 by microarray. Compared with the wild type mice, 42 genes were upregulated and 22 genes were downregulated more than 1.3-fold in the Plzflu/lu mice (Additional file 1: Table S1). Functional annotation of these genes was performed by ToppFun, a web database provides users to explore the functions of genes. The five highest-ranking processes in the category of ‘biological process’ were neurogenesis, generation of neurons, neuron differentiation, CNS development, and cell morphogenesis involved in differentiation (Fig. 5a and Additional file 2: Table S2). This result thus further supports the role of Plzf in brain development and neurogenesis. We also used the ingenuity pathway analysis (IPA) to further analyze these genes and found that the function of ASCL1 (MASH1), ARX, and SHH were associated with the formation of neural precursor cells (Fig. 5b). Among these genes, the expression of Mash1 was related to the numbers of early-born neurons [18, 19]. Therefore, we used quantitative RT-PCR to analyze the Mash1 RNA expression. Similar to the microarray data, the quantitative RT-PCR analysis demonstrated a significant increase in Mash1 mRNA in Plzflu/lu mice when compared with wild type littermates at E10.5 (Fig. 6a). We further analyzed the cell populations by flow cytometry from the forebrain of E10.5 embryos. Our results showed a significant increase of Mash1+ cells in the embryos of Plzflu/lu mice (Fig. 6b,c), suggesting that loss of Plzf causes the early expression of Mash1.
Fig. 5.
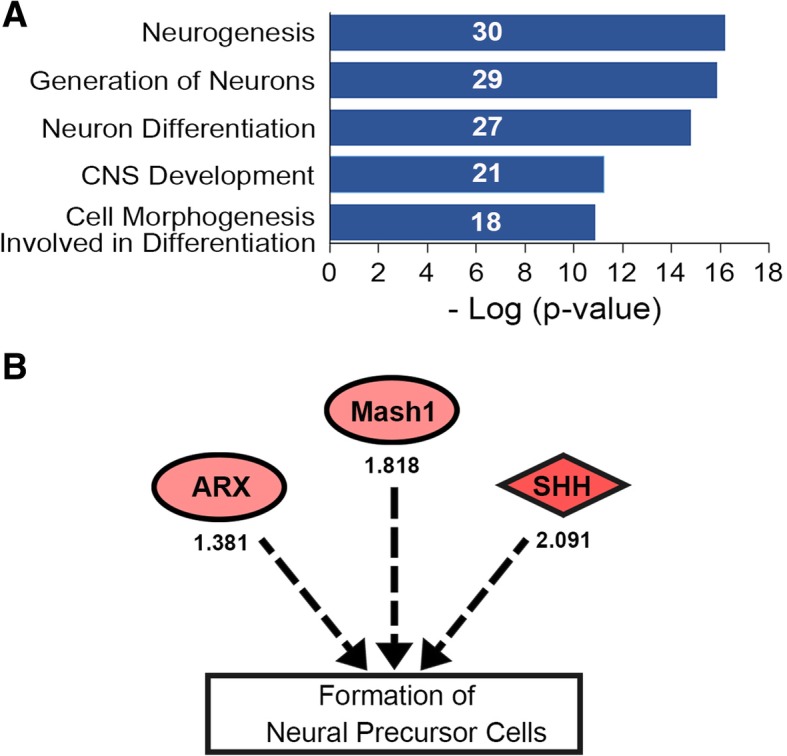
Differentially expressed genes in the embryonic brain of Plzflu/lu mice with functions related to neurogenesis. a, E10.5 forebrain and midbrain were collected for gene expression profiling by microarray. The 64 candidate genes with > or < 1.3-fold changes in expression by Plzflu/lu mice were further analyzed by ToppFun (https://toppgene.cchmc.org/). Using the default parameters to annotate target genes for GO Biological Process, the top five physiological system development and function categories were identified. Significance referred to the –log (p-value). The numbers in blue bars represent the numbers of genes involved in the category. b, Potential genes involved in formation of neural precursor cells. Arx: Aristaless related homeobox; Ascl1 (Mash1): Achaete-scute family bHLH transcription factor 1; Shh: Sonic hedgehog. The numbers under the circles represent the fold changed between Plzflu/lu to Plzfwt/wt
Fig. 6.
The increase of Mash1 expression and Mash1+ cell population in Plzflu/lu mice at E10.5. a, qRT-PCR analysis of Mash1 mRNA levels, normalized to GAPDH, from Plzfwt/wt and Plzflu/lu mice. Plzfwt/wt, n = 3; Plzflu/lu, n = 4. b, Flow cytometry dot plot shows the levels of Mash1+ cells in E10.5 Plzfwt/wt and Plzflu/lu mice brain. Numbers at upper-right quadrant indicate the proportions of Mash1+ cells. c, Quantification of the proportions of Mash1+ cells. n = 4 per genotype. Values represent the mean ± SEM. Statistics performed with Student’s t test: *p < 0.05
Plzf-deficient mice show a deficit in recognition memory
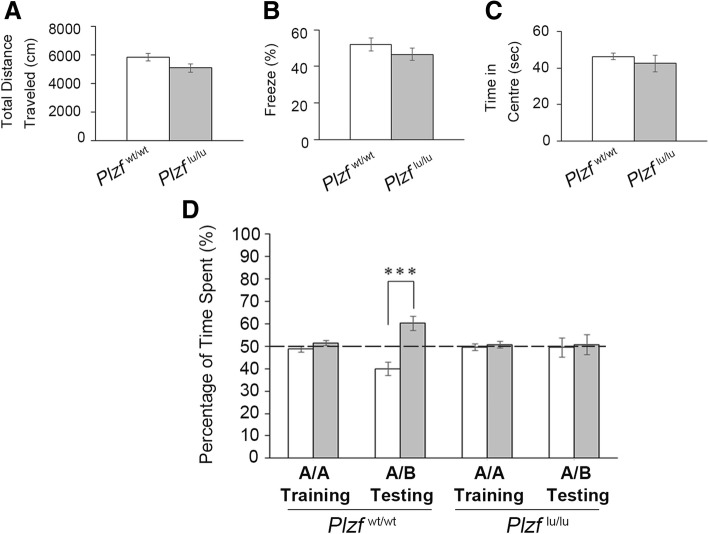
To further analyze the consequence of Plzf loss postnatally, we examined the functional defects in Plzf-deficient mice. We first performed open-field tests to assess anxiety behavior and measure basal activity. Our results showed that, although Plzflu/lu mice exhibited skeletal abnormalities [20], they had similar locomotor activity levels as the wild-type littermates (Fig. 7a). We also found that the duration of freezing and staying in the central zone of open field were not significantly different when compared wild-type with Plzf-deficient mice, suggesting that the Plzf-deficient mice did not exhibit more anxiety-associated behavior than wild-type littermates (Fig. 7b,c). We then analyzed the recognition memory function in wild type and Plzf-deficient mice using the novel object recognition test. Plzfwt/wt and Plzflu/lu both explored two identical objects “A” equally during the training session (Fig. 7d). After 24 h, we placed mice with one familiar object “A” and one novel object “B” and found that wild-type mice spent significantly more time exploring the novel object “B”, suggesting that they were able to remember and recognize the familiar object (Fig. 7d). On the other hand, Plzflu/lu mice displayed no preference for the displaced object as opposed to the wild-type littermates (Fig. 7d). Our results thus suggest that loss of Plzf results in structural and functional abnormalities of the brain in mice.
Fig. 7.
Impairment of novel object recognition memory in Plzflu/lu mice. A-C, Open field test results of Plzflu/lu and Plzfwt/wt mice. a, Quantification of the total distance traveled; Plzfwt/wt, n = 16; Plzflu/lu, n = 7. b, Quantification of the percentage of freezing time (the duration with which the mouse was completely stationary); Plzfwt/wt, n = 12; Plzflu/lu, n = 5. c, Quantification of the time spent in the center zone; Plzfwt/wt, n = 16; Plzflu/lu, n = 7. d, Novel object recognition test results of Plzflu/lu and Plzfwt/wt mice. The % of total time the mice spent exploring each object during the training and testing sessions was recorded. The dashed line indicates 50% exploration time if none of the objects is preferred. Plzfwt/wt, n = 16; Plzflu/lu, n = 7. Values represent the mean ± SEM. Statistical analysis was performed using Student’s t-test: *** p < 0.001
Discussion
The function of Plzf in brain development is less known as compared to the studies of Plzf in other biological processes such as hematopoiesis, osteogenesis, and spermatogenesis. This study demonstrated that Plzf expression was strictly regulated during brain development. Loss of Plzf resulted in microcephaly with thinner cortex and reduced numbers of Tbr1+ neurons in layer VI but not in other layers. PHH3 immunostaining revealed a significant decrease of mitotic cell number in Plzflu/lu mice at early stage of neurogenesis. Microarray, qRT-PCR, and flow cytometry analysis identified Mash1 expression was up-regulated in the embryonic brain of Plzf-deficient mice at E10.5. Impairment of recognition memory was also observed in Plzf-deficient mice. Together, these findings clarify the effects of Plzf on the formation and function of the brain.
Plzf has been proposed to mediate a lot of essential biological processes and the Plzf-deficient mice, including both Plzf knockout and mutant mice, have been characterized in these contexts [2–4, 20, 21]. Although these studies have demonstrated defects of testis and limb homeosis and patterning in Plzf-deficient mice, whether the brain is also affected has not been investigated in these mouse models. Our results showing decreased cerebral cortex size and deficits in recognition memory in the Plzf-mutant mice (Figs. 2, 3 and 7d) are consistent with previous reports in a patient, who was identified with biallelic loss of function of the PLZF gene [6, 7]. In these reports, the patient was diagnosed with severe skeletal defects and genital hypoplasia. In addition, his head circumference (33 cm) was 2.4 S.D. below the mean size at birth, and the patient had severe mental retardation.
A previous study has identified an extremely dynamic expression pattern of Plzf mRNA during CNS development by in situ hybridization [8]. In that study, Plzf mRNA expression can be first detected at E7.5 in the anterior neuroepithelium and extended to the entire neuroectoderm until stage E10. The study also revealed that although Plzf was still expressed abundantly in CNS at E10.5, the expression was no longer detectable in many regions of the forebrain at E12.5. In contrast, Plzf expression in hindbrain and spinal cord remains until at least E16.5. Consistent with the result of in situ hybridization, our Western blotting results showed that Plzf protein was highly expressed in the mouse embryonic brain at E10.5 (Fig. 1a). The expression was then decreased and maintained at lower levels until E18.5. Using immunostaining, we also confirmed the dynamic expression pattern of Plzf protein in forebrain. Similar to the mRNA expression, we showed that the protein expression of Plzf was highly abundant in the forebrain at E10.5 but dramatically decreased at E11.5 (Fig. 1b-d).
We found that Plzf was expressed in the neuroepithelium before the time when neurogenesis starts (Fig. 1d), implicating that the effect of Plzf on neurogenesis may be more important in early stage than in late stage. Consistent with our speculation, we found that a significant decrease of mitotic cell number in Plzflu/lu mice at E10.5 (Fig. 4c) but not at E12.5 (Fig. 4d). In the spinal cord of chick and mouse, Plzf is able to promote neural progenitors proliferation and decrease neuronal differentiation [10]. In zebrafish, Plzf can block neuronal differentiation by inhibiting the expression of proneural gene Ngn1 [9]. Our result further reveals the effect of Plzf in the proliferation of neural stem cells in the brain. Loss of Plzf may result in a reduction of the neural stem/progenitor cell pool and lead to reduce the size of cerebral cortex. In mammalian brain, cerebral cortex is a highly organized, six-layered structure [22]. Among these layers, layer VI is the deepest one and the neurons in layer VI are born as early as E11.5 in mouse [23]. Two models are used to explain the mechanisms that establish cellular diversity in the cortex [24]. One is progressive restriction model, in which a single kind of radial glia cells sequentially generates all different subtypes of projection neurons and astrocytes in a defined temporal order. Base on this model, a reduction of cell proliferation in early neural progenitors should affect the number of progenitor cells in later embryogenesis, because it might reduce total number of progenitor pools. The other is lineage restriction model, in which distinct subtypes of radial glia cells co-exist and are pre-specified to generate different subtypes of projection neurons and astrocytes. Base on this model, early and later-born neurons are derived from distinct progenitor pools. We found that loss of Plzf resulted in a smaller cerebral cortex with a specific decrease in the number of neurons in layer VI but not in other layers (Fig. 3), suggesting Plzf is required for the early progenitor pools. Our results are thus in agreement with the lineage restriction model.
Our result showed an impairment of recognition memory in Plzf-deficient mice (Fig. 7d). Previous studies reveal that prefrontal cortex supports the hippocampus for both long-tern and short-term memory [25, 26]. And, recognition memory, in general, depends on interactions within a circuit involving the insular cortex, perirhineal cortex, ventromedial prefrontal cortex, and hippocampus [26–29]. More importantly, layer VI of visual cortex area is specifically implicated in object recognition memory formation [30]. Therefore, the impairment of recognition memory in Plzf-deficient mice may relate to the decrease in cerebral cortex size and the number of deep-layer cortical neurons.
Brain development is a complicated process involving neural stem cell proliferation, differentiation, and survival. Human autosomal recessive primary microcephaly (MCPH) is a congenital brain disorder caused by mutations in at least 12 different genes [31]. Patients with MCPH exhibit marked reduction in brain size and intellectual disability. To date, most of MCPH genes identified are centrosomal proteins involved in cell cycle regulation. For example, CDK5RAP2 is required to maintain centriole engagement and cohesion [32], WDR62 is related to mitotis spindle assembly and stability [33], CENPJ and PLK4 are involved in centrosome biogenesis [34, 35], and ASPM is related to spindle regulation [36]. In animal models, deficiency of MCPH genes causes brain size reduction as observed in patients [33, 37–40]. However, our data showed no change of expressions of MCPH genes in the embryonic brain of Plzf-deficient mice at E10.5 (Additional file 1: Table S1), implying that the effect of Plzf in cerebral cortex formation is not related to MCPH genes dysregulation.
Impaired or premature neurogenesis also affects nervous system development and causes brain size change. For example, RP58 is highly expressed in differentiating neurons to repress the expressions of proneural genes and essential for neuronal differentiation from progenitors. Loss of RP58 results in impaired neurogenesis and smaller brain size [41]. Our previous studies also reveal that Rnf112/Znf179 expresses in differentiating neurons to modulate cell cycle exit, and loss of Rnf112/Znf179 also causes smaller brain size [42, 43]. In addition to impaired neurogenesis, premature neurogenesis caused by up-regulation of proneural genes in NSCs leads to depletion of progenitor pool and reduction of brain size [44, 45]. Our result showed up-regulation of proneural gene Mash1 expression and increase of Mash1+ cells in the embryonic brain of Plzf-deficient mice at E10.5 (Fig. 6). Mash1, also known as Ascl1, is a proneural gene regulating neurogenesis in the ventral telencephalon and is also critical for laminar fate determination of cortical neurons [18, 19, 46, 47]. As shown in the study of Hatakeyama et al., Hes genes including Hes1, Hes3, and Hes5 are expressed in NSCs to repress the expressions of proneural genes such as Mash1 and contribute to the maintenance of NSCs [44]. In the absence of Hes genes, proneural genes (including Mash1) are up-regulated, leading to premature neurogenesis and concomitant a wide range of defects in brain formation. Therefore, the Mash1 dysregulation may be related to the brain malformation in Plzf-deficient mice.
In addition to Mash1, Shh and Arx are also suggested to mediate the generation of neural precursor cells and are up-regulated in the embryonic brain of Plzf-deficient mice at E10.5 (Fig. 5b, Additional file 1: Table S1). Shh gene encodes a secreted protein, sonic hedgehog, and is required for expansion of neuron precursors in cerebellum and also in neocortex [48, 49]. Arx is a homeobox-containing gene and regulates cell expansion of cortical intermediate progenitor [50]. Whether the Shh and Arx genes dysregulation also contributes to the defect of cerebral cortex formation in Plzf-deficient mice will be further examined.
Conclusions
In conclusion, Plzf is expressed at early stages of brain development and required for the early progenitor pools. Loss of Plzf results in dysregulation of Mash1, microcephaly with reduced numbers of early-born neurons, and impairment of recognition memory.
Additional files
Table S1. Gene expression changes identified from a microarray of E10.5 Plzfwt/wt and Plzflu/lu mice forebrain and midbrain. (DOCX 20 kb)
Table S2. List of genes associated with the GO term. (DOCX 19 kb)
Acknowledgements
This work was supported by the Ministry of Science and Technology (MOST), Taiwan (MOST 105-2320-B-038-061, 106-2320-B-038-022, and 107-2320-B-038-050-MY3). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We thank the Taiwan Animal Consortium (MOST 106-2319-B-001-004)-Taiwan Mouse Clinic which is funded by the Ministry of Science and Technology (MOST) of Taiwan for technical support in MRI experiment. We also thank National Laboratory Animal Center (NLAC), NARLabs, Taiwan, for technical support in contract breeding and testing services.
Funding
This work was supported by the Ministry of Science and Technology (MOST), Taiwan. (MOST 105–2320-B-038-061, 106–2320-B-038-022, and 107–2320-B-038-050-MY3).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- E
embryonic day
- IPA
ingenuity pathway analysis
- lu
luxoid
- NOR
Novel Object Recognition
- NSCs
neural stem cells
- PHH3
phospho-histone H3
- Plzf
promyelocytic leukemia zinc finger
Authors’ contributions
Conceived and designed the experiments: HL, WC, TK* and YL*. Performed the experiments: HL, CH, PP and YL. Analyzed the data: HL, YC, TK* and YL*. Contributed reagents/materials/analysis tools: YC. Wrote the paper: HL, TK* and YL*. All authors read and approved the final manuscript.
Ethics approval
Experimental protocols used in this study complied with the guidelines of the Taipei Medical University Institutional Animal Care and Use Committee (IACUC). All experiments were performed in accordance with the US Public Health Service Policy on Humane Care and Use of Laboratory Animals.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Hsin-Chuan Lin, Email: m120100023@tmu.edu.tw.
Yung-Hao Ching, Email: yching@mail.tcu.edu.tw.
Chi-Chen Huang, Email: hcc0609@tmu.edu.tw.
Ping-Chieh Pao, Email: jennypao@mit.edu.
Yi-Hua Lee, Email: tuberevalee@gmail.com.
Wen-Chang Chang, Email: wcchang@tmu.edu.tw.
Tzu-Jen Kao, Phone: +886 2 27361661, Email: geokao@tmu.edu.tw.
Yi-Chao Lee, Phone: +886 2 27361661, Email: yclee@tmu.edu.tw.
References
- 1.Li JY, English MA, Ball HJ, Yeyati PL, Waxman S, Licht JD. Sequence-specific DNA binding and transcriptional regulation by the promyelocytic leukemia zinc finger protein. J Biol Chem. 1997;272(36):22447–22455. doi: 10.1074/jbc.272.36.22447. [DOI] [PubMed] [Google Scholar]
- 2.Barna M, Hawe N, Niswander L, Pandolfi PP. Plzf regulates limb and axial skeletal patterning. Nat Genet. 2000;25(2):166–172. doi: 10.1038/76014. [DOI] [PubMed] [Google Scholar]
- 3.Buaas FW, Kirsh AL, Sharma M, McLean DJ, Morris JL, Griswold MD, de Rooij DG, Braun RE. Plzf is required in adult male germ cells for stem cell self-renewal. Nat Genet. 2004;36(6):647–652. doi: 10.1038/ng1366. [DOI] [PubMed] [Google Scholar]
- 4.Costoya JA, Hobbs RM, Barna M, Cattoretti G, Manova K, Sukhwani M, Orwig KE, Wolgemuth DJ, Pandolfi PP. Essential role of Plzf in maintenance of spermatogonial stem cells. Nat Genet. 2004;36(6):653–659. doi: 10.1038/ng1367. [DOI] [PubMed] [Google Scholar]
- 5.Vincent-Fabert C, Platet N, Vandevelde A, Poplineau M, Koubi M, Finetti P, Tiberi G, Imbert AM, Bertucci F, Duprez E. PLZF mutation alters mouse hematopoietic stem cell function and cell cycle progression. Blood. 2016;127(15):1881–1885. doi: 10.1182/blood-2015-09-666974. [DOI] [PubMed] [Google Scholar]
- 6.Fischer S, Kohlhase J, Bohm D, Schweiger B, Hoffmann D, Heitmann M, Horsthemke B, Wieczorek D. Biallelic loss of function of the promyelocytic leukaemia zinc finger (PLZF) gene causes severe skeletal defects and genital hypoplasia. J Med Genet. 2008;45(11):731–737. doi: 10.1136/jmg.2008.059451. [DOI] [PubMed] [Google Scholar]
- 7.Wieczorek D, Koster B, Gillessen-Kaesbach G. Absence of thumbs, a/hypoplasia of radius, hypoplasia of ulnae, retarded bone age, short stature, microcephaly, hypoplastic genitalia, and mental retardation. Am J Med Genet. 2002;108(3):209–213. doi: 10.1002/ajmg.10271. [DOI] [PubMed] [Google Scholar]
- 8.Avantaggiato V, Pandolfi PP, Ruthardt M, Hawe N, Acampora D, Pelicci PG, Simeone A. Developmental analysis of murine Promyelocyte leukemia zinc finger (PLZF) gene expression: implications for the neuromeric model of the forebrain organization. J Neurosci. 1995;15(7 Pt 1):4927–4942. doi: 10.1523/JNEUROSCI.15-07-04927.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sobieszczuk DF, Poliakov A, Xu Q, Wilkinson DG. A feedback loop mediated by degradation of an inhibitor is required to initiate neuronal differentiation. Genes Dev. 2010;24(2):206–218. doi: 10.1101/gad.554510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaber ZB, Butler SJ, Novitch BG. PLZF regulates fibroblast growth factor responsiveness and maintenance of neural progenitors. PLoS Biol. 2013;11(10):e1001676. doi: 10.1371/journal.pbio.1001676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8(6):427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- 12.Ohtaka-Maruyama C, Okado H. Molecular pathways underlying projection neuron production and migration during cerebral cortical development. Front Neurosci. 2015;9(447). [DOI] [PMC free article] [PubMed]
- 13.Paridaen JT, Huttner WB. Neurogenesis during development of the vertebrate central nervous system. EMBO Rep. 2014;15(4):351–364. doi: 10.1002/embr.201438447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gotz M, Huttner WB. The cell biology of neurogenesis. Nat Rev Mol Cell Biol. 2005;6(10):777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- 15.Faedo A, Tomassy GS, Ruan Y, Teichmann H, Krauss S, Pleasure SJ, Tsai SY, Tsai MJ, Studer M, Rubenstein JL. COUP-TFI coordinates cortical patterning, neurogenesis, and laminar fate and modulates MAPK/ERK, AKT, and beta-catenin signaling. Cereb Cortex. 2008;18(9):2117–2131. doi: 10.1093/cercor/bhm238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lian G, Lu J, Hu J, Zhang J, Cross SH, Ferland RJ, Sheen VL. Filamin a regulates neural progenitor proliferation and cortical size through Wee1-dependent Cdk1 phosphorylation. J Neurosci. 2012;32(22):7672–7684. doi: 10.1523/JNEUROSCI.0894-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pucilowska J, Puzerey PA, Karlo JC, Galan RF, Landreth GE. Disrupted ERK signaling during cortical development leads to abnormal progenitor proliferation, neuronal and network excitability and behavior, modeling human neuro-cardio-facial-cutaneous and related syndromes. J Neurosci. 2012;32(25):8663–8677. doi: 10.1523/JNEUROSCI.1107-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Casarosa S, Fode C, Guillemot F. Mash1 regulates neurogenesis in the ventral telencephalon. Development. 1999;126(3):525–534. doi: 10.1242/dev.126.3.525. [DOI] [PubMed] [Google Scholar]
- 19.Yun K, Fischman S, Johnson J, Hrabe de Angelis M, Weinmaster G, Rubenstein JL. Modulation of the notch signaling by Mash1 and Dlx1/2 regulates sequential specification and differentiation of progenitor cell types in the subcortical telencephalon. Development. 2002;129(21):5029–5040. doi: 10.1242/dev.129.21.5029. [DOI] [PubMed] [Google Scholar]
- 20.Green MC. Luxoid-a new hereditary leg and foot abnormality. J Hered. 1955;46(3):91–99. [Google Scholar]
- 21.Savage AK, Constantinides MG, Han J, Picard D, Martin E, Li B, Lantz O, Bendelac A. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 2008;29(3):391–403. doi: 10.1016/j.immuni.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramón y Cajal S . Histology of the nervous system of man and vertebrates. New York: Oxford University Press; 1995. [Google Scholar]
- 23.Polleux F, Dehay C, Kennedy H. The timetable of laminar neurogenesis contributes to the specification of cortical areas in mouse isocortex. J Comp Neurol. 1997;385(1):95–116. [PubMed] [Google Scholar]
- 24.Franco SJ, Muller U. Shaping our minds: stem and progenitor cell diversity in the mammalian neocortex. Neuron. 2013;77(1):19–34. doi: 10.1016/j.neuron.2012.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ennaceur A, Neave N, Aggleton JP. Spontaneous object recognition and object location memory in rats: the effects of lesions in the cingulate cortices, the medial prefrontal cortex, the cingulum bundle and the fornix. Exp Brain Res. 1997;113(3):509–519. doi: 10.1007/pl00005603. [DOI] [PubMed] [Google Scholar]
- 26.Euston DR, Gruber AJ, McNaughton BL. The role of medial prefrontal cortex in memory and decision making. Neuron. 2012;76(6):1057–1070. doi: 10.1016/j.neuron.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clarke JR, Cammarota M, Gruart A, Izquierdo I, Delgado-Garcia JM. Plastic modifications induced by object recognition memory processing. Proc Natl Acad Sci U S A. 2010;107(6):2652–2657. doi: 10.1073/pnas.0915059107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hammond RS, Tull LE, Stackman RW. On the delay-dependent involvement of the hippocampus in object recognition memory. Neurobiol Learn Mem. 2004;82(1):26–34. doi: 10.1016/j.nlm.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 29.Vogel-Ciernia A. and Wood M.A. Examining object location and object recognition memory in mice. Curr Protoc Neurosci.2014;69:8 31 31–17. [DOI] [PMC free article] [PubMed]
- 30.Lopez-Aranda MF, Lopez-Tellez JF, Navarro-Lobato I, Masmudi-Martin M, Gutierrez A, Khan ZU. Role of layer 6 of V2 visual cortex in object-recognition memory. Science. 2009;325(5936):87–89. doi: 10.1126/science.1170869. [DOI] [PubMed] [Google Scholar]
- 31.Barbelanne M, Tsang WY. Molecular and cellular basis of autosomal recessive primary microcephaly. Biomed Res Int. 2014;547986, 2014. [DOI] [PMC free article] [PubMed]
- 32.Barrera JA, Kao LR, Hammer RE, Seemann J, Fuchs JL, Megraw TL. CDK5RAP2 regulates centriole engagement and cohesion in mice. Dev Cell. 2010;18(6):913–926. doi: 10.1016/j.devcel.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen JF, Zhang Y, Wilde J, Hansen KC, Lai F, Niswander L. Microcephaly disease gene Wdr62 regulates mitotic progression of embryonic neural stem cells and brain size. Nat Commun. 2014;5(3885). [DOI] [PMC free article] [PubMed]
- 34.Martin CA, Ahmad I, Klingseisen A, Hussain MS, Bicknell LS, Leitch A, Nurnberg G, Toliat MR, Murray JE, Hunt D, Khan F, Ali Z, Tinschert S, Ding J, Keith C, Harley ME, Heyn P, Muller R, Hoffmann I, Cormier-Daire V, Dollfus H, Dupuis L, Bashamboo A, McElreavey K, Kariminejad A, Mendoza-Londono R, Moore AT, Saggar A, Schlechter C, Weleber R, Thiele H, Altmuller J, Hohne W, Hurles ME, Noegel AA, Baig SM, Nurnberg P, Jackson AP. Mutations in PLK4, encoding a master regulator of centriole biogenesis, cause microcephaly, growth failure and retinopathy. Nat Genet. 2014;46(12):1283–1292. doi: 10.1038/ng.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McIntyre RE, Lakshminarasimhan Chavali P, Ismail O, Carragher DM, Sanchez-Andrade G, Forment JV, Fu B, Del Castillo Velasco-Herrera M, Edwards A, van der Weyden L, Yang F, Sanger Mouse Genetics P, Ramirez-Solis R, Estabel J, Gallagher FA, Logan DW, Arends MJ, Tsang SH, Mahajan VB, Scudamore CL, White JK, Jackson SP, Gergely F, Adams DJ. Disruption of mouse Cenpj, a regulator of centriole biogenesis, phenocopies Seckel syndrome. PLoS Genet. 2012;8(11):e1003022. doi: 10.1371/journal.pgen.1003022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fish JL, Kosodo Y, Enard W, Paabo S, Huttner WB. Aspm specifically maintains symmetric proliferative divisions of neuroepithelial cells. Proc Natl Acad Sci U S A. 2006;103(27):10438–10443. doi: 10.1073/pnas.0604066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gruber R, Zhou Z, Sukchev M, Joerss T, Frappart PO, Wang ZQ. MCPH1 regulates the neuroprogenitor division mode by coupling the centrosomal cycle with mitotic entry through the Chk1-Cdc25 pathway. Nat Cell Biol. 2011;13(11):1325–1334. doi: 10.1038/ncb2342. [DOI] [PubMed] [Google Scholar]
- 38.Lizarraga SB, Margossian SP, Harris MH, Campagna DR, Han AP, Blevins S, Mudbhary R, Barker JE, Walsh CA, Fleming MD. Cdk5rap2 regulates centrosome function and chromosome segregation in neuronal progenitors. Development. 2010;137(11):1907–1917. doi: 10.1242/dev.040410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pulvers JN, Bryk J, Fish JL, Wilsch-Brauninger M, Arai Y, Schreier D, Naumann R, Helppi J, Habermann B, Vogt J, Nitsch R, Toth A, Enard W, Paabo S, Huttner WB. Mutations in mouse Aspm (abnormal spindle-like microcephaly associated) cause not only microcephaly but also major defects in the germline. Proc Natl Acad Sci U S A. 2010;107(38):16595–16600. doi: 10.1073/pnas.1010494107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang YJ, Baltus AE, Mathew RS, Murphy EA, Evrony GD, Gonzalez DM, Wang EP, Marshall-Walker CA, Barry BJ, Murn J, Tatarakis A, Mahajan MA, Samuels HH, Shi Y, Golden JA, Mahajnah M, Shenhav R, Walsh CA. Microcephaly gene links trithorax and REST/NRSF to control neural stem cell proliferation and differentiation. Cell. 2012;151(5):1097–1112. doi: 10.1016/j.cell.2012.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiang C, Baubet V, Pal S, Holderbaum L, Tatard V, Jiang P, Davuluri RV, Dahmane N. RP58/ZNF238 directly modulates proneurogenic gene levels and is required for neuronal differentiation and brain expansion. Cell Death Differ. 2012;19(4):692–702. doi: 10.1038/cdd.2011.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pao PC, Huang NK, Liu YW, Yeh SH, Lin ST, Hsieh CP, Huang AM, Huang HS, Tseng JT, Chang WC, Lee YC. A novel RING finger protein, Znf179, modulates cell cycle exit and neuronal differentiation of P19 embryonal carcinoma cells. Cell Death Differ. 2011;18(11):1791–1804. doi: 10.1038/cdd.2011.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsou JH, Yang YC, Pao PC, Lin HC, Huang NK, Lin ST, Hsu KS, Yeh CM, Lee KH, Kuo CJ, Yang DM, Lin JH, Chang WC, Lee YC. Important roles of Ring finger protein 112 in embryonic vascular development and brain functions. Mol Neurobiol. 2017;54(3):2286–2300. doi: 10.1007/s12035-016-9812-7. [DOI] [PubMed] [Google Scholar]
- 44.Hatakeyama J, Bessho Y, Katoh K, Ookawara S, Fujioka M, Guillemot F, Kageyama R. Hes genes regulate size, shape and histogenesis of the nervous system by control of the timing of neural stem cell differentiation. Development. 2004;131(22):5539–5550. doi: 10.1242/dev.01436. [DOI] [PubMed] [Google Scholar]
- 45.Ishibashi M, Ang SL, Shiota K, Nakanishi S, Kageyama R, Guillemot F. Targeted disruption of mammalian hairy and enhancer of split homolog-1 (HES-1) leads to up-regulation of neural helix-loop-helix factors, premature neurogenesis, and severe neural tube defects. Genes Dev. 1995;9(24):3136–3148. doi: 10.1101/gad.9.24.3136. [DOI] [PubMed] [Google Scholar]
- 46.Dennis DJ, Wilkinson G, Li S, Dixit R, Adnani L, Balakrishnan A, Han S, Kovach C, Gruenig N, Kurrasch DM, Dyck RH, Schuurmans C. Neurog2 and Ascl1 together regulate a postmitotic derepression circuit to govern laminar fate specification in the murine neocortex. Proc Natl Acad Sci U S A. 2017;114(25):E4934–E4943. doi: 10.1073/pnas.1701495114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dixit R, Zimmer C, Waclaw RR, Mattar P, Shaker T, Kovach C, Logan C, Campbell K, Guillemot F, Schuurmans C. Ascl1 participates in Cajal-Retzius cell development in the neocortex. Cereb Cortex. 2011;21(11):2599–2611. doi: 10.1093/cercor/bhr046. [DOI] [PubMed] [Google Scholar]
- 48.Dahmane N, Sanchez P, Gitton Y, Palma V, Sun T, Beyna M, Weiner H, Ruiz i, Altaba A. The sonic hedgehog-Gli pathway regulates dorsal brain growth and tumorigenesis. Development. 2001;128(24):5201–5212. doi: 10.1242/dev.128.24.5201. [DOI] [PubMed] [Google Scholar]
- 49.Lewis PM, Gritli-Linde A, Smeyne R, Kottmann A, McMahon AP. Sonic hedgehog signaling is required for expansion of granule neuron precursors and patterning of the mouse cerebellum. Dev Biol. 2004;270(2):393–410. doi: 10.1016/j.ydbio.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 50.Colasante G, Simonet JC, Calogero R, Crispi S, Sessa A, Cho G, Golden JA, Broccoli V. ARX regulates cortical intermediate progenitor cell expansion and upper layer neuron formation through repression of Cdkn1c. Cereb Cortex. 2015;25(2):322–335. doi: 10.1093/cercor/bht222. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Gene expression changes identified from a microarray of E10.5 Plzfwt/wt and Plzflu/lu mice forebrain and midbrain. (DOCX 20 kb)
Table S2. List of genes associated with the GO term. (DOCX 19 kb)
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.