Abstract
Background: Recent studies have highlighted the important roles of long non-coding RNAs (lncRNAs) in pancreatic adenocarcinoma (PCa) prognosis. However, most studies explored a limited number of lncRNAs based on small sample size.
Methods: Systematic and comprehensive analysis of the data from The Cancer Genome Atlas (TCGA) was performed to identify a panel of lncRNA signature for predicting prognosis in PCa.
Results: A total of 160 PCa patients with complete clinical data were included in our study. Twelve lncRNAs were identified to be significantly associated with overall survival (OS) in PCa patients using Cox regression analysis. A risk score formula was constructed to assess the prognostic value of the lncRNA signature in PCa. Patients with high risk score had worse OS than those with low risk score. The multivariate Cox regression analyses revealed that the lncRNA signature was an independent prognostic factor. Additionally, the signature might act as an indicator to predict treatment outcome. Functional enrichment analyses showed that the lncRNAs might involve in several molecular pathways closely related with PCa such as DNA replication, pancreatic cancer and regulation of tor signaling.
Conclusions: Our study demonstrated a lncRNA signature including 12 lncRNAs with the potential to be served as an independent prognostic biomarker of PCa.
Keywords: pancreatic adenocarcinoma, lncRNA, biomarker, prognosis, TCGA
Background
Occupying more than 90% of pancreatic cancer (PC), pancreatic adenocarcinoma (PCa) is one of the most aggressive and lethal malignancies with extremely dismal overall median survival of 6 months and 5-year survival rates of lower than 5% 1. Due to the lack of early symptoms, most PCa patients were diagnosed at an advanced stage, which do not have any opportunity of surgical resection. Even in patients who undergo surgical resection, the disease commonly recurs within 12 months due to the aggressive nature 2. The high resistance to chemotherapy and radiotherapy as well as the limited effectiveness of molecular targeted therapy (MTT) also contributes to exceptionally poor long-term survival 3. Survival analysis of PCa patients with the advantage of providing risk stratification could be utilized to develop effective treatment modalities and improve clinical outcomes.
For the moment, survival for PCa could be indicated according to the traditional pathological characteristics of the tumor such as TNM stage, grade of differentiation, or positive resection margins 4. However, inconsistence often exists between these indicators and survival 5. Carbohydrate antigen 19-9 (CA19-9) is currently confirmed as the best serum marker to establish prognosis for PCa. But its low specificity brings a high number of false positives 6. Additionally, up to 10% of the population cannot synthesize the antigen 7. Therefore, new biomarkers so as to predict PCa progression and prognosis are essential for improving patient survival.
Long non-coding RNAs (lncRNAs) are transcripts with a length of more than 200 nucleotides without protein-coding functions 8. Increasing evidences have shown that lncRNAs play important roles in regulating gene expression at the transcriptional, posttranscriptional and chromosomal levels and are associated with large range of biological processses, such as transcriptional regulation, cell growth and tumorigenesis 9, 10. Some lncRNAs have been found to act as oncogenic or tumor suppressive roles in the development and progression of cancer 11. Over the last few years, lncRNAs have been utilized as biomarkers for diagnosis and prognosis in various cancers 12-18. MALAT-1, HOTAIR, and PVT-1 are some of most widely studied lncRNAs in the prognostic value of PC 19-23. However, most studies focused on a limited number of lncRNAs based on small patient size. In this work, we comprehensively analyzed the data retrieved from The Cancer Genome Atlas (TCGA, http://cancergenome.nih.gov/) to develop a panel of lncRNA signature which might predict the outcome of PCa patients.
Methods and Materials
Expression profiles and sample information
The level 3 RNA-seq files and clinical features of 166 PC patients were retrieved from TCGA data portal. To obtain lncRNAs specific for PCa, six patients with the other types of pancreatic cancer (4 with pancreas-colloid (mucinous non-cystic) carcinoma, 1 with pancreas-undifferentiated carcinoma and 1 with inconsistent histologic type) were excluded. Thus, a total of 160 PCa patients with clinical data and without other malignancies were enrolled in our study. The corresponding clinical information including gender, age, tumor location, tumor size, grade, residual status, AJCC TNM stage, smoking status, drinking status, diabetes status, adjuvant radiotherapy, MTT status and treatment outcome of first course (most patients received gemcitabine based chemotherapy and a few patients with fluorouracil based chemotherapy) was recorded. The lncRNAs derived from TCGA were annotated by Ensemble ID from GENCODE project 24. And the expressed lncRNAs were defined as those with an average reads per kilobase per million mapped reads (RPKM) ≥ 0.3 across all 160 PCa patients. Consequentially, the expression of 2300 lncRNAs was analyzed in our study. As the data were downloaded from TCGA, further approval by an ethics committee was not required. Data procession was performed in accordance with the TCGA human subject protection and data access policies.
Statistical analysis
The expression profiles of lncRNAs were log2 transformed for further analysis. The relationship between the expression level of each lncRNA and PCa patients' OS was evaluated by Cox regression analysis using the BRB array tools package which was developed by Richard Simon and the BRB-Array Tools Development Team 25. Through 10,000 permutations, the lncRNAs with permutation P values < 0.0001 were identified and were classified into risky (with a hazard ratio (HR) for death greater than 1) and protective (based on a HR for death less than 1) types. A risk score formula was constructed for predicting OS based on a linear combination of the expression level multiplied regression coefficient derived from the univariate cox regression model (β) 26, 27: risk score = expgene1*βgene1 + expgene2*βgene2 + …expgenen*βgenen. Using the median risk score as the cutoff point, PCa patients were divided into high score and low score groups.
The prognostic value of clinical variables and the lncRNA risk score on OS of PCa patients were initially assessed by univariate Cox proportional hazards regression analyses. Subsequently, each variable identified via univariate analysis was evaluated by multivariate Cox proportional hazards regression analysis. Survival curves were estimated using the Kaplan- Meier methodology and compared with log-rank test.
The association of the lncRNA signature and clinical variables was evaluated by Chi-square test. The predictive value of the lncRNA risk score for patients' outcome after first course of treatment was assessed by receiver operating characteristic (ROC) curve analysis. All the statistical analyses were conducted with BRB-Array Tools and SPSS 16.0 software (SPSS Inc., Chicago, IL, USA), as appropriate. A two-sided P value < 0.05 was defined as statistical significance unless specifically indicated.
Functional enrichment analysis
Pearson correlation coefficients were computed to assess co-expressed relationships between the risky/protective lncRNAs and protein-coding genes (PCGs). The genes with correlation coefficients higher than 0.4 were identified as the co-expressed PCGs. To evaluate the potential biological processes and pathways the lncRNAs might involve in, functional enrichment analysis based on the co-expressed PCGs was performed for Gene ontology (GO) biological process (BP) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway using the DAVID Bioinformatics Tool (https://david.ncifcrf.gov/, version 6.8) 28. The GO terms and KEGG pathways with P value < 0.01 were considered as significantly enriched function annotations.
Gene set enrichment analysis (GSEA) was carried out to explore the potential altered pathways between the high risk score and low score group with java software GSEA (http://software.broadinstitute. org/gsea/index.jsp) 29. False discovery rate (FDR) value < 0.05 after 1000 random permutations was set as the cutoff criterion.
Results
Characteristics of patients
A total of 160 patients with PCa were included in this study. The detailed clinical features of all patients were shown in Table 1. The mean ± standard deviation (SDV) age for the patients was 64.8 ± 10.9. Among the 160 patients, 73 patients with tumor size less than 35 mm, 99 obtained R0 resection, and 28 and 77 patients received adjuvant radiotherapy and MTT, respectively. Fifty-six patients died during the follow-up (mean ± SDV: 12.8 ± 15.8 months). Moreover, 90 patients' information on treatment outcome was available. According to the RECIST criteria, 38 (42.2%) acquired complete response (CR), 8 (8.9%) partial response (PR), 7 (7.8%) stable disease (SD) and 37 (41.1%) progressive disease (PD).
Table 1.
Characteristics of 160 PCa patients in the study.
| Characteristics | Number |
|---|---|
| All | 160 |
| Gender | |
| male | 88 |
| female | 72 |
| Age (year) | |
| <65 | 74 |
| ≥65 | 86 |
| Location | |
| head | 126 |
| body | 11 |
| tail | 12 |
| others | 11 |
| Size (mm) | |
| <35 | 73 |
| ≥35 | 82 |
| NA | 5 |
| Grade | |
| G1 | 29 |
| G2 | 83 |
| G3 | 47 |
| G4 | 1 |
| Residual tumor | |
| R0 | 99 |
| R1 | 49 |
| R2 | 2 |
| RX | 4 |
| NA | 6 |
| TNM stage | |
| IA | 6 |
| IB | 13 |
| IIA | 23 |
| IIB | 111 |
| III | 3 |
| IV | 3 |
| NA | 1 |
| Smoker | |
| No | 56 |
| Yes | 74 |
| NA | 30 |
| Drinker | |
| No | 27 |
| Yes | 62 |
| NA | 71 |
| Diabetes | |
| No | 97 |
| Yes | 34 |
| NA | 29 |
| Adjuvant radiotherapy | |
| No | 90 |
| Yes | 28 |
| NA | 42 |
| MTT | |
| No | 43 |
| Yes | 77 |
| NA | 40 |
MTT: molecular targeted therapy; NA: not available.
Identification of lncRNAs correlated with OS
According to the criteria, a total of 2300 lncRNAs were initially identified as the expressed lncRNAs and were selected for further analysis on prognostic value in PCa. Twelve lncRNAs (6 protective lncRNAs: CTC-429P9.3, CTD-2186M15.3, RP5-890O3.9, AP000254.8, RP5-1085F17.3, LINC01089, and 6 risky lncRNAs: LINC00941, ABHD11-AS1, CASC8, CYTOR, MIR4435-2HG, UCA1; Supplementary Table S1 online) were additionally proved to be significantly associated with OS in PCa patients.
We also analyzed the relationship between the lncRNAs and clinical characteristics (Table 2). The results showed that the expression level of UCA1 was inversely related with patients' age. RP5-1085F17.3 was up-regulated in no-smoking patients compared with smoking patients. The expression of three protective lncRNAs (CTD-2186M15.3, RP5-890O3.9 and AP000254.8) was higher in patients without diabetes than those with diabetes. Four lncRNAs (a risky lncRNA with positive correlation: LINC00941 and three protective lncRNAs with negative correlation: CTC-429P9.3, AP000254.8 and RP5-1085F17.3) were observed to be significantly associated with tumor grade. Four protective lncRNAs (CTC-429P9.3, CTD-2186M15.3, AP000254.8 and RP5-1085F17.3) showed lower expression level in patients with advanced stages. And four protective lncRNAs (RP5-890O3.9, AP000254.8, RP5-1085F17.3 and LINC01089) were prominently related with better treatment outcome. However, we found no significant correlation else between the lncRNAs and other clinical features.
Table 2.
Correlations of the 12 lncRNAs and the lncRNA signature risk score with clinical features in PCa patients (presented as P value).
| ID | Gender (female VS. male) | Age (<65 VS. ≥65) | Grade (G1+G2 VS. G3+G4) | Tumor size (<35 mm VS. ≥35 mm) | Stage (I vs. II Vs. III+IV) | Smoking (smoker VS. non-smoker) | Drinking (drinker VS. non-drinker) | Diabetes (yes VS. no) |
Location (head VS. body VS. tail) |
Treatment outcome (DC VS. PD) |
|---|---|---|---|---|---|---|---|---|---|---|
| LINC00941 | 0.358 | 0.795 | 0.009 | 0.329 | 0.267 | 0.686 | 0.509 | 0.321 | 0.751 | 0.356 |
| ABHD11-AS1 | 0.495 | 0.303 | 0.472 | 0.402 | 0.193 | 0.665 | 0.363 | 0.291 | 0.306 | 0.188 |
| CASC8 | 0.964 | 0.771 | 0.195 | 0.572 | 0.492 | 0.263 | 0.844 | 0.475 | 0.551 | 0.925 |
| CYTOR | 0.373 | 0.24 | 0.644 | 0.864 | 0.884 | 0.377 | 0.407 | 0.416 | 0.945 | 0.673 |
| MIR4435-2HG | 0.97 | 0.454 | 0.693 | 0.246 | 0.735 | 0.595 | 0.437 | 0.908 | 0.975 | 0.331 |
| UCA1 | 0.967 | 0.04 | 0.092 | 0.9 | 0.25 | 0.814 | 0.52 | 0.557 | 0.584 | 0.331 |
| CTC-429P9.3 | 0.781 | 0.748 | 0.019 | 0.06 | 0.003 | 0.269 | 0.175 | 0.358 | 0.645 | 0.058 |
| CTD-2186M15.3 | 0.262 | 0.152 | 0.726 | 0.17 | 0.018 | 0.337 | 0.272 | 0.011 | 0.921 | 0.058 |
| RP5-890O3.9 | 0.3 | 0.235 | 0.864 | 0.634 | 0.316 | 0.686 | 0.459 | 0.047 | 0.277 | 0.027 |
| AP000254.8 | 0.845 | 0.997 | 0.018 | 0.168 | 0.023 | 0.17 | 0.481 | 0.016 | 0.98 | 0.002 |
| RP5-1085F17.3 | 0.693 | 0.782 | 0.008 | 0.26 | <0.001 | 0.013 | 0.701 | 0.208 | 0.35 | 0.014 |
| LINC01089 | 0.146 | 0.735 | 0.652 | 0.14 | 0.101 | 0.843 | 0.218 | 0.737 | 0.201 | 0.007 |
| Risk score | 1 | 0.899 | 0.154 | 0.076 | 0.039 | 0.696 | 0.52 | 0.211 | 0.975 | 0.011 |
DC: disease control; PD: progressive disease.
The lncRNA signature as an independent factor to predict PCa prognosis
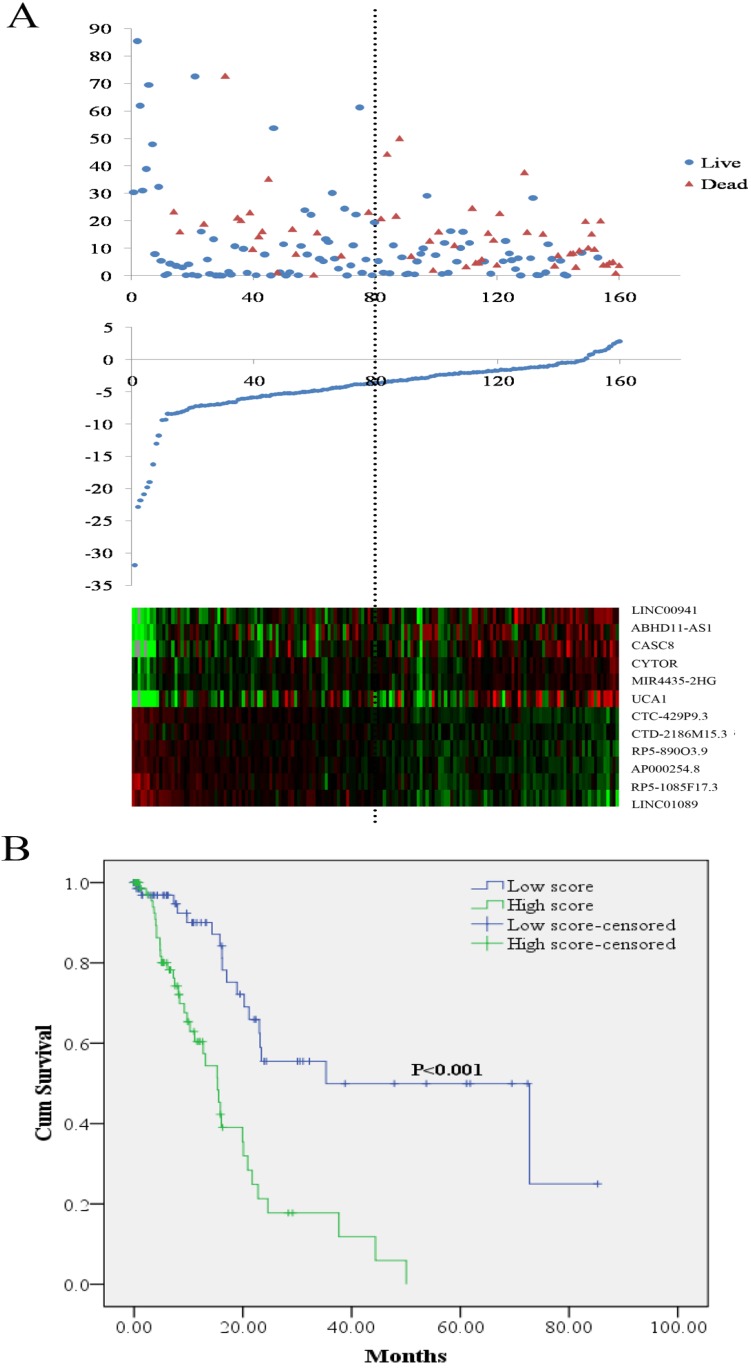
The risk score for predicting OS of each PCa patients was calculated based on the expression levels and the corresponding regression coefficients of the twelve lncRNAs. The equation for the risk score of the 12-lncRNA signature was shown as: risk score = 0.108*LINC00941 + 0.028*ABHD11-AS1+0.198*CASC8+ 0.149 *CYTOR + 0.332*MIR4435-2HG + 0.009* UCA1 - 0.353*CTC-429P9.3 - 0.534*CTD- 2186M15.3 - 0.197*RP5-890 O3.9 - 0.453* AP000254.8 - 0.26*RP5-1085F17.3 - 0.335* LINC01089. According to the median risk score, 160 PCa patients were divided into two groups of low score group (n = 80) and high score group (n = 80). As shown in Figure 1A, higher expression was observed for the protective lncRNAs in low score group, while in high score group higher expression was noted for the risky lncRNAs. Compared with the low score group, the patients in the high score group exhibited obviously worse OS (Figure 1B). Analyses on the relationship between the risk score and clinical features demonstrated that higher risk score was remarkably associated with later stages and worse treatment outcome (Table 2).
Figure 1.
LncRNA risk score analysis and outcome in PCa patients. (A) Patient survival status and duration (Top; y axis: survival time; x axis: patient id sorted by risk score); risk score (y axis) of the 12 lncRNAs in PCa patients (Middle; x axis: patient id sorted by risk score); heatmap of the 12 lncRNAs expression in PCa patients (Bottom). (B) Kaplan-Meier curve for the low score and high score group. PCa: pancreatic adenocarcinoma
The univariate Cox regression analyses were performed to evaluate various factors correlated with patients' prognosis. The results showed that tumor size (P = 0.037), tumor grade (P = 0.001), residual tumor (P = 0.006), TNM stage (P < 0.001), adjuvant radiotherapy (P = 0.006), MTT (P = 0.002), treatment outcome of the first course (P = 0.001) and risk score (P < 0.001) were significantly associated with PCa patients' OS. Subsequently, multivariate Cox regression analyses suggested that tumor grade (P = 0.002), TNM stage (P = 0.033), MTT (P < 0.001) and risk score (P = 0.002) were independent prognostic indictors (Table 3).
Table 3.
Cox proportional regression analysis for assessing the correlation of clinical factors and the lncRNA signature risk score with OS in PCa patients.
| Variables | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|
| HR (95%CI) | P value | HR (95%CI) | P value | ||
| Gender (female VS. male) | 0.79 (0.462, 1.353) | 0.391 | |||
| Age (≥65 VS. <65) | 1.493 (0.876, 2.546) | 0.141 | |||
| Location (tail VS. body VS. head) | 0.582 (0.335, 1.014) | 0.056 | |||
| Size (≥35 VS. <35; mm) | 1.799 (1.035, 3.124) | 0.037 | 0.846 (0.426, 1.679) | 0.633 | |
| Grade (G4 vs. G3 VS. G2 VS. G1) | 1.808 (1.269, 2.575) | 0.001 | 1.89 (1.255, 2.846) | 0.002 | |
| Residual tumor (yes Vs. no) | 2.217 (1.262, 3.896) | 0.006 | 1.308 (0.648, 2.64) | 0.453 | |
| TNM stage (IV VS. III VS. IIB VS. IIA VS. IB VS. IA) | 1.764 (1.267, 2.455) | <0.001 | 1.606 (1.037,2.487) | 0.033 | |
| Smoking (smoker VS. non-smoker) | 1.201 (0.699, 2.066) | 0.507 | |||
| Alcohol (drinker VS. none) | 0.632 (0.295, 1.353) | 0.238 | |||
| Diabetes (yes VS. no) | 0.992 (0.509, 1.933) | 0.981 | |||
| Adjuvant radiotherapy (yes VS. no) | 0.33 (0.149, 0.733) | 0.006 | 0.853 (0.304, 2.389) | 0.762 | |
| Molecular targeted therapy (yes VS. no) | 0.428 (0.249, 0.735) | 0.002 | 0.203 (0.091, 0.454) | <0.001 | |
| Treatment outcome (PD VS. DC) | 2.731 (1.489, 5.009) | 0.001 | 1.308 (0.67, 2.552) | 0.432 | |
| Risk score (high VS. low) | 3.732 (2.075, 6.714) | <0.001 | 3.561 (1.58, 8.024) | 0.002 | |
HR: hazard ratio; CI: confidence interval; DC: disease control; PD: progressive disease.
The lncRNA signature risk score as an indicator to predict treatment outcome
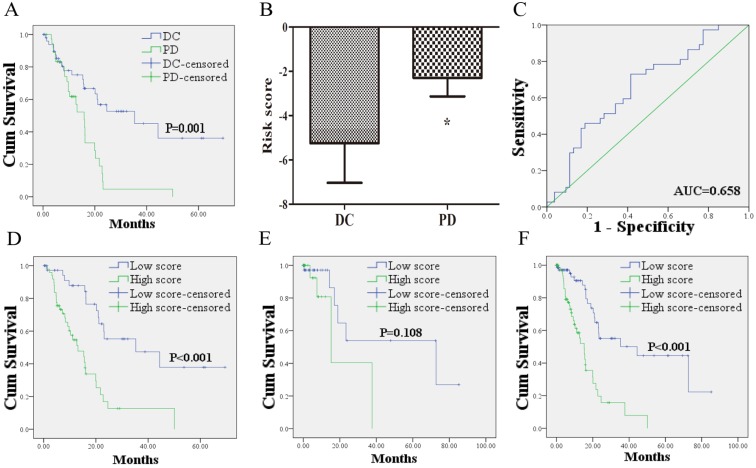
Kaplan-Meier curves showed that the 53 PCa patients who acquired disease control (DC: CR + PR + SD) after treatment of first course had better OS than the 37 patients with PD (Figure 2A). To confirm whether the risk score has the power to predict treatment outcome of first course, the difference of risk score in the two groups (DC VS. PD) was compared. And the ability of risk score in distinguishing the two groups was evaluated with ROC curves. As shown in Figure 2B, risk score of PCa patients in DC group was markedly lower than those in PD group. The area under the curve (AUC) was 0.658 (95% confidence interval (CI): 0.545 - 0.772). The optimal cutoff value to predict treatment outcome was set at -3.38. Using this threshold, the risk score achieved a sensitivity of 72.9% and a specificity of 58.5%.
Figure 2.
The relationship between the lncRNA signature risk score and treatment outcome. (A): 53 PCa patients who acquired DC after treatment of first course suffered better OS than the 37 patients with PD. (B) risk score of PCa patients in DC group was lower than those in PD group. (C) ROC curve analysis of the risk score to discriminate patients with DC from those with PD. (D) 49 PCa patients with risk score higher than the cutoff value exhibited worse OS than the 41 cases lower than the score. (E) Kaplan-Meier curve for patients without information of treatment outcome. (F) Kaplan-Meier curve for the 160 patients form the entire dataset with the same cutoff value. DC: disease control; PD: progressive disease; OS: overall survival; ROC: receiver-operating characteristic.
The 49 PCa patients with risk score higher than the cutoff value suffered worse OS than the 41 cases with lower score (Figure 2D). To confirm the prognostic power of the 12 lncRNA signature for survival, the other 70 patients without information of treatment outcome were divided into a high score group (n = 26) and a low score group (n = 44) with the same cutoff value. As shown in Figure 2E, the patients in high score group suffered shorter survival than patients in low score group with borderline significance (P = 0.108). Similar results were found when the 12 lncRNA signature was further applied to the entire dataset. Based on the cutoff value, the low score group (n = 85) exhibited longer survival, whereas the high score group exhibited shorter survival (n = 75; Figure 2F).
Assessment of the biological processes and pathways related with the 12-lncRNA signature
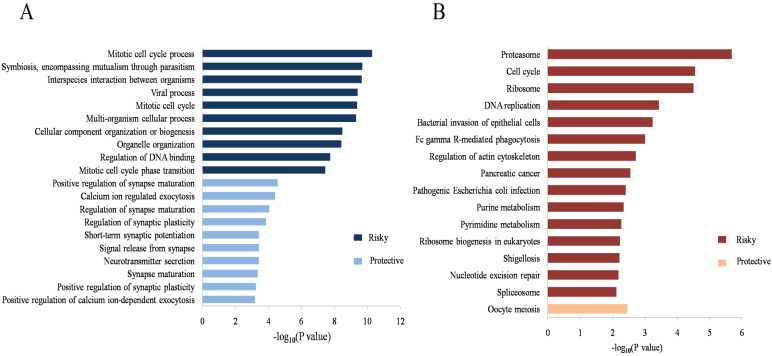
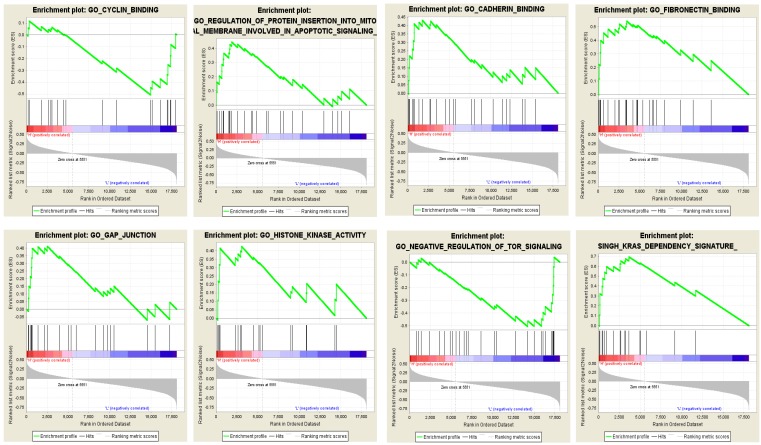
To evaluate the potential biological processes and pathways the identified lncRNAs might involve in, GO and KEGG functional enrichment analyses based on co-expressed PCGs were conducted for the risky and protective lncRNAs, respectively. The results showed that co-expressed PCGs of the risky lncRNAs were enriched in 629 GO BP terms including mitotic cell cycle process, DNA metabolic process, positive regulation of ubiquitin protein ligase activity and so on. The protective lncRNAs might also play important roles in 66 GO BP terms (Supplementary Table S2 online; Figure 3A: the top 10 GO BP terms for the risky and protective lncRNAs, respectively). Fifteen and one KEGG pathways for the risky and protective lncRNAs were enriched, respectively. Among them, the risky lncRNAs related pathways such as proteasome, cell cycle, DNA replication, regulation of actin cytoskeleton and PC were more closely related with PCa (Figure 3B). In addition, GSEA was performed according to the risk score of the 12-lncRNA signature. And a total of 304 biological processes or pathways were identified (Supplementary Table S2 online). Among these pathways, cyclin binding and regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling might be associated with cell proliferation; cadherin binding, fibronectin binding and gap junction might play important roles in migration of cancer cells; histone kinase activity, regulation of tor signaling and kras dependency signature should be noted for their roles in carcinogenesis and cancer maintenance (Figure 4; Supplementary Table S3 online).
Figure 3.
GO and KEGG functional enrichment analyses of the 12-lncRNA signature. (A) Top 10 GO BP terms for the risky and protective lncRNAs. (B) Significant KEGG pathways for the risky and protective lncRNAs. GO: gene ontology; BP: biological process; KEGG: Kyoto Encyclopedia of Genes and Genomes.
Figure 4.
GSEA showed eight typical pathways and processes which might be closely associated with PCa. Red bar: positively related with risky lncRNAs; Blue bar: positively related with protective lncRNAs; GSEA: gene set enrichment analysis; PCa: pancreatic adenocarcinoma.
Discussion
PCa is the main types of PC and remains one of the most lethal of malignancies with extremely poor prognosis all around the world. Over the past decades, efforts in clinical treatment did not satisfactorily improved survival time 1. It is of great urgency to develop an optimized individualized therapeutic approach for PCa. Specific prognostic indicators may identify subsets of patients and are more powerful when used to help make therapeutic decisions 30.
Recent studies highlighted the important roles of lncRNAs in PC prognosis. Overexpression of MALAT1 was reported as an unfavorable prognostic biomarker for PC patients in several studies 19-21. Kim et al. showed that HOTAIR was a negative prognostic factor for PC patients 22. Another study reported that increased expression of PVT1 was associated with poor prognosis in PC patients 23. TCGA, a database with large-scale genomic analyses, has the power to evaluate the molecular features associated with cancer outcomes 31, 32. In the previous study, we have identified a 13-miRNA signature which could predict prognosis in PC using the database 33. To better understand molecular markers of PCa, we comprehensively analyzed the database and identified lncRNAs which could predict OS in the main types of PC patients. This is the first study to identify a panel of lncRNAs signature based on TCGA data for the survival analysis of PCa patients. To reduce heterogeneity, patients with other types of PC were not enrolled in the study. Using careful exclusion criteria, a total of 160 PCa patients with 2300 lncRNAs were selected for further evaluation. Through 10,000 permutations with P values < 0.0001, 12 lncRNAs (6 protective and 6 risky lncRNAs) significantly related to OS were identified. The relationship between the 12 lncRNAs and clinical features was also analyzed. LINC00941, CTC-429P9.3, CTD-2186M15.3, AP000254.8 and RP5-1085F17.3 might be more closely related with PCa itself. Besides AP000254.8 and RP5-1085F17.3, RP5- 890O3.9 and LINC01089 might also play important roles in treatment of the disease. A panel of lncRNAs is more sensitive and specific than a single lncRNA. Thus, we constructed a risk score with the 12 lncRNAs. The risk score was proved to be an independent prognostic indicator for OS in PCa patients. Moreover, the risk score could also serve as a predictor for treatment outcome of first course. The prognostic power of the risk score for treatment outcome was further validated in patients without information of treatment outcome. By applying the same cutoff value, relatively less cases (26/70) were divided into the high score group. Meanwhile, few patients (10/70) reached the end- point might (due to relatively short follow-up: 9.2 ± 16.8 months; the 90 cases with information of treatment outcome: 15.5 ± 14.4) affect the statistical significance. However, the lncRNA signature could also act as a prognostic indicator with borderline significance which could also validate our findings to some extent. While extend to the whole cohort, the signature could exactly predict OS of 160 PCa patients with the cutoff value induced from the treatment outcome. These findings further confirmed our results.
Among the 12 lncRNAs, UCA1 has been demonstrated to be correlated with PC. It has been reported to function as an oncogene in multiple types of cancers 34-36. Moreover, numerous studies have demonstrated that overexpression of UCA1 is an unfavorable prognostic indicator in cancer patients, including the cases with PC 37-39. Chen et al. recently found that the expression of UCA1 in 128 PC tissue specimens was significantly higher than their matched adjacent non-tumor tissues. High UCA1 expression was an independent predictor of poor survival in PC. Besides, up-regulation of UCA1 was significantly correlated with malignant factors such as tumor size, depth of invasion, CA19-9 level and tumor stage 40. Fu et al. analyzed the lncRNA expression profiles in GEO and identified 34 dysregulated lncRNAs in pancreatic ductal adenocarcinoma (PDAC). In addition, UCA1 was found to be involved in PDAC progression and could serve as an independent prognostic biomarker for OS of PDAC 41. In our study, the prognostic value of UCA1 was in concordance with recent reports. However, UCA1 expression was only found to be associated with age, its relationship with other clinicopathological features did not show any significance. No study has evaluated the role of the other 11 lncRNAs in PC so far. But some lncRNAs have been reported to be involved in other types of cancers. Recently, Zhang et al. identified a 4-lncRNA signature (SPRY4-IT1, LINC00941, GPR158-AS1 and KCNK15-AS1) displaying prognostic values for lung adenocarcinoma (LUAD) based on TCGA dataset. Among the 4 lncRNAs, LINC00941 with higher expression was associated with longer survival 42. The findings were not consistent with our results that LINC00941 acted as an indicator of worse prognosis in PCa. Chen et al. showed that ABHD11-AS1 played oncogenic roles in the development and progression of bladder cancer 43. Another study reported that ABHD11-AS1 was up-regulated in gastric cancer (GC) tissues and also significantly related with clinical features 44. Grembergen et al. suggested that CYTOR could indicate poor relapse-free survival of breast cancer patients, regulate genes involved in the EGFR/ mammalian target of rapamycin pathway and was required for cell proliferation, cell migration, and cytoskeleton organization 45. MIR4435-2HG was reported to be related to cell-cycle in lung cancer cells and up-regulated in GC tissue and plasma samples 46. A recent study showed that LINC01089 might represent a new class of EGF-controlled and ERK-mediated inhibitors of breast cancer metastasis, functioning as a tumor-suppressor lncRNA 47. These results were in concordance with our study. As for the other 6 lncRNAs, no study has explored their roles to date and further researches are warranted. In the present study, we conducted GO and KEGG analyses based on co-expressed PCGs and found some molecular pathways might be closely related with PCa such as cell cycle, DNA replication and pancreatic cancer. The results of GSEA showed that the 12-lncRNA signature might play important roles in proliferation and migration of cancer cells, thus leading to maintenance of cancer and carcinogenesis. The significantly positive relationship of tumor stage and the risk score might support the findings. However, whether these lncRNAs play exactly important roles in the development and progression of PCa remains to be further investigated.
Some limitations of the study should be addressed. First, to reduce heterogeneity, patients with other types of PC were excluded. Thus, a total of 160 PCa patients were enrolled in our study. The mean time of follow-up for the whole cohort, patients with information of treatment outcome and those without was 12.8, 15.5 and 9.2 months, respectively. The research included more patients with longer follow-up time is warranted to confirm our findings in the future. Second, among the 12 lncRNAs, some lncRNAs were closely related with specific features (e.g. LINC00941, CTC-429P9.3, AP000254.8 and RP5-1085F17.3 to tumor grade; RP5-890O3.9, AP000254.8, RP5-1085F17.3 and LINC01089 to treatment outcome). In the study, we focused on the relationship of 12-lncRNA signature and OS, thus not further assessed the combination of some lncRNAs in indicating specific features of PCa. Third, the exact roles and mechanisms of the identified lncRNAs in the development, progression and treatment of PCa were not assessed with in vitro/vivo experiment and needed to be further studied.
Conclusions
In summary, we assessed the genome-wide lncRNA expression profiles with 160 cases of PCa patients from TCGA, and constructed a 12-lncRNA signature, which could be served as a predictor for treatment outcome and an independent prognostic indicator for PCa. Our study could also complement clinical and pathological parameters in the effort to further understand the mechanisms of PCa.
Supplementary Material
Supplementary tables.
Acknowledgments
This work was supported by the National Natural Science Foundation of China [Grant number: 81702364; 81672400; 81672788]; the Natural Science Foundation of Jiangsu Province [Grant number: BK20171085]; Science and Technology Project of Jiangsu Province in China [Grant number: KS1643]; and Jiangsu Province Clinical science and technology projects [Clinical Research Center, BL2012008].
Availability of data and materials
The datasets supporting the conclusions of this article were retrieved from TCGA, (http://cancergenome.nih.gov/).
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA: a cancer journal for clinicians. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Lennon AM, Wolfgang CL, Canto MI, Klein AP, Herman JM, Goggins M. et al. The early detection of pancreatic cancer: what will it take to diagnose and treat curable pancreatic neoplasia? Cancer Res. 2014;74:3381–9. doi: 10.1158/0008-5472.CAN-14-0734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heestand GM, Murphy JD, Lowy AM. Approach to patients with pancreatic cancer without detectable metastases. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2015;33:1770–8. doi: 10.1200/JCO.2014.59.7930. [DOI] [PubMed] [Google Scholar]
- 4.Hidalgo M. Pancreatic cancer. The New England journal of medicine. 2010;362:1605–17. doi: 10.1056/NEJMra0901557. [DOI] [PubMed] [Google Scholar]
- 5.Ferrone CR, Pieretti-Vanmarcke R, Bloom JP, Zheng H, Szymonifka J, Wargo JA. et al. Pancreatic ductal adenocarcinoma: long-term survival does not equal cure. Surgery. 2012;152:S43–9. doi: 10.1016/j.surg.2012.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goonetilleke KS, Siriwardena AK. Systematic review of carbohydrate antigen (CA 19-9) as a biochemical marker in the diagnosis of pancreatic cancer. European journal of surgical oncology: the journal of the European Society of Surgical Oncology and the British Association of Surgical Oncology. 2007;33:266–70. doi: 10.1016/j.ejso.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 7.Kawai S, Suzuki K, Nishio K, Ishida Y, Okada R, Goto Y. et al. Smoking and serum CA19-9 levels according to Lewis and secretor genotypes. International journal of cancer Journal international du cancer. 2008;123:2880–4. doi: 10.1002/ijc.23907. [DOI] [PubMed] [Google Scholar]
- 8.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–41. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 9.Kornienko AE, Guenzl PM, Barlow DP, Pauler FM. Gene regulation by the act of long non-coding RNA transcription. BMC biology. 2013;11:59. doi: 10.1186/1741-7007-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nature reviews Genetics. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 11.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Molecular cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Serghiou S, Kyriakopoulou A, Ioannidis JP. Long noncoding RNAs as novel predictors of survival in human cancer: a systematic review and meta-analysis. Molecular cancer. 2016;15:50. doi: 10.1186/s12943-016-0535-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen C, Lu Z, Yang J, Hao W, Qin Y, Wang H. et al. MiR-17-5p promotes cancer cell proliferation and tumorigenesis in nasopharyngeal carcinoma by targeting p21. Cancer medicine. 2016;5:3489–99. doi: 10.1002/cam4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Z, Guo X, Li G, Shi Y, Li L. Long noncoding RNAs as potential biomarkers in gastric cancer: Opportunities and challenges. Cancer letters. 2016;371:62–70. doi: 10.1016/j.canlet.2015.11.011. [DOI] [PubMed] [Google Scholar]
- 15.Ricciuti B, Mencaroni C, Paglialunga L, Paciullo F, Crino L, Chiari R. et al. Long noncoding RNAs: new insights into non-small cell lung cancer biology, diagnosis and therapy. Medical oncology. 2016;33:18. doi: 10.1007/s12032-016-0731-2. [DOI] [PubMed] [Google Scholar]
- 16.Zhou M, Zhao H, Xu W, Bao S, Cheng L, Sun J. Discovery and validation of immune-associated long non-coding RNA biomarkers associated with clinically molecular subtype and prognosis in diffuse large B cell lymphoma. Mol Cancer. 2017;16:16. doi: 10.1186/s12943-017-0580-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tu Z, He D, Deng X, Xiong M, Huang X, Li X. et al. An eight-long non-coding RNA signature as a candidate prognostic biomarker for lung cancer. Oncol Rep. 2016;36:215–22. doi: 10.3892/or.2016.4817. [DOI] [PubMed] [Google Scholar]
- 18.Zhou M, Guo M, He D, Wang X, Cui Y, Yang H. et al. A potential signature of eight long non-coding RNAs predicts survival in patients with non-small cell lung cancer. J Transl Med. 2015;13:231. doi: 10.1186/s12967-015-0556-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pang EJ, Yang R, Fu XB, Liu YF. Overexpression of long non-coding RNA MALAT1 is correlated with clinical progression and unfavorable prognosis in pancreatic cancer. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2015;36:2403–7. doi: 10.1007/s13277-014-2850-8. [DOI] [PubMed] [Google Scholar]
- 20.Liu JH, Chen G, Dang YW, Li CJ, Luo DZ. Expression and prognostic significance of lncRNA MALAT1 in pancreatic cancer tissues. Asian Pacific journal of cancer prevention: APJCP. 2014;15:2971–7. doi: 10.7314/apjcp.2014.15.7.2971. [DOI] [PubMed] [Google Scholar]
- 21.Li L, Chen H, Gao Y, Wang YW, Zhang GQ, Pan SH. et al. Long Noncoding RNA MALAT1 Promotes Aggressive Pancreatic Cancer Proliferation and Metastasis via the Stimulation of Autophagy. Molecular cancer therapeutics. 2016;15:2232–43. doi: 10.1158/1535-7163.MCT-16-0008. [DOI] [PubMed] [Google Scholar]
- 22.Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R. et al. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 2013;32:1616–25. doi: 10.1038/onc.2012.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang C, Yu W, Wang Q, Cui H, Wang Y, Zhang L. et al. Increased expression of the lncRNA PVT1 is associated with poor prognosis in pancreatic cancer patients. Minerva medica. 2015;106:143–9. [PubMed] [Google Scholar]
- 24.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F. et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome research. 2012;22:1760–74. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simon R, Lam A, Li MC, Ngan M, Menenzes S, Zhao Y. Analysis of gene expression data using BRB-ArrayTools. Cancer informatics. 2007;3:11–7. [PMC free article] [PubMed] [Google Scholar]
- 26.Dave SS, Wright G, Tan B, Rosenwald A, Gascoyne RD, Chan WC. et al. Prediction of survival in follicular lymphoma based on molecular features of tumor-infiltrating immune cells. The New England journal of medicine. 2004;351:2159–69. doi: 10.1056/NEJMoa041869. [DOI] [PubMed] [Google Scholar]
- 27.Zhao Q, Sun J. Cox survival analysis of microarray gene expression data using correlation principal component regression. Statistical applications in genetics and molecular biology. 2007;6:Article16. doi: 10.2202/1544-6115.1153. [DOI] [PubMed] [Google Scholar]
- 28.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 29.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang J, Wang B, Chen X, Bi J. The prognostic value of RASSF1A promoter hypermethylation in non-small cell lung carcinoma: a systematic review and meta-analysis. Carcinogenesis. 2011;32:411–6. doi: 10.1093/carcin/bgq266. [DOI] [PubMed] [Google Scholar]
- 31.Gore J, Craven KE, Wilson JL, Cote GA, Cheng M, Nguyen HV. et al. TCGA data and patient-derived orthotopic xenografts highlight pancreatic cancer-associated angiogenesis. Oncotarget. 2015;6:7504–21. doi: 10.18632/oncotarget.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen X, Guo W, Xu XJ, Su F, Wang Y, Zhang Y. et al. Melanoma long non-coding RNA signature predicts prognostic survival and directs clinical risk-specific treatments. Journal of dermatological science. 2017;85:226–34. doi: 10.1016/j.jdermsci.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 33.Zhou X, Huang Z, Xu L, Zhu M, Zhang L, Zhang H. et al. A panel of 13-miRNA signature as a potential biomarker for predicting survival in pancreatic cancer. Oncotarget. 2016;7:69616–24. doi: 10.18632/oncotarget.11903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Na XY, Liu ZY, Ren PP, Yu R, Shang XS. Long non-coding RNA UCA1 contributes to the progression of prostate cancer and regulates proliferation through KLF4-KRT6/13 signaling pathway. International journal of clinical and experimental medicine. 2015;8:12609–16. [PMC free article] [PubMed] [Google Scholar]
- 35.Han Y, Yang YN, Yuan HH, Zhang TT, Sui H, Wei XL. et al. UCA1, a long non-coding RNA up-regulated in colorectal cancer influences cell proliferation, apoptosis and cell cycle distribution. Pathology. 2014;46:396–401. doi: 10.1097/PAT.0000000000000125. [DOI] [PubMed] [Google Scholar]
- 36.Ni B, Yu X, Guo X, Fan X, Yang Z, Wu P. et al. Increased urothelial cancer associated 1 is associated with tumor proliferation and metastasis and predicts poor prognosis in colorectal cancer. International journal of oncology. 2015;47:1329–38. doi: 10.3892/ijo.2015.3109. [DOI] [PubMed] [Google Scholar]
- 37.Gao J, Cao R, Mu H. Long non-coding RNA UCA1 may be a novel diagnostic and predictive biomarker in plasma for early gastric cancer. International journal of clinical and experimental pathology. 2015;8:12936–42. [PMC free article] [PubMed] [Google Scholar]
- 38.Li JY, Ma X, Zhang CB. Overexpression of long non-coding RNA UCA1 predicts a poor prognosis in patients with esophageal squamous cell carcinoma. International journal of clinical and experimental pathology. 2014;7:7938–44. [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang L, Cao X, Zhang L, Zhang X, Sheng H, Tao K. UCA1 overexpression predicts clinical outcome of patients with ovarian cancer receiving adjuvant chemotherapy. Cancer chemotherapy and pharmacology. 2016;77:629–34. doi: 10.1007/s00280-016-2963-4. [DOI] [PubMed] [Google Scholar]
- 40.Chen P, Wan D, Zheng D, Zheng Q, Wu F, Zhi Q. Long non-coding RNA UCA1 promotes the tumorigenesis in pancreatic cancer. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2016;83:1220–6. doi: 10.1016/j.biopha.2016.08.041. [DOI] [PubMed] [Google Scholar]
- 41.Fu XL, Liu DJ, Yan TT, Yang JY, Yang MW, Li J. et al. Analysis of long non-coding RNA expression profiles in pancreatic ductal adenocarcinoma. Scientific reports. 2016;6:33535. doi: 10.1038/srep33535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang X, Chi Q, Zhao Z. Up-regulation of long non-coding RNA SPRY4-IT1 promotes tumor cell migration and invasion in lung adenocarcinoma. Oncotarget. 2017;8:51058–65. doi: 10.18632/oncotarget.16918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen M, Li J, Zhuang C, Cai Z. Increased lncRNA ABHD11-AS1 represses the malignant phenotypes of bladder cancer. Oncotarget. 2017;8:28176–86. doi: 10.18632/oncotarget.14945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin X, Yang M, Xia T, Guo J. Increased expression of long noncoding RNA ABHD11-AS1 in gastric cancer and its clinical significance. Medical oncology. 2014;31:42. doi: 10.1007/s12032-014-0042-4. [DOI] [PubMed] [Google Scholar]
- 45.Van Grembergen O, Bizet M, de Bony EJ, Calonne E, Putmans P, Brohee S. et al. Portraying breast cancers with long noncoding RNAs. Science advances. 2016;2:e1600220. doi: 10.1126/sciadv.1600220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ke D, Li H, Zhang Y, An Y, Fu H, Fang X. et al. The combination of circulating long noncoding RNAs AK001058, INHBA-AS1, MIR4435-2HG, and CEBPA-AS1 fragments in plasma serve as diagnostic markers for gastric cancer. Oncotarget. 2017;8:21516–25. doi: 10.18632/oncotarget.15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sas-Chen A, Aure MR, Leibovich L, Carvalho S, Enuka Y, Korner C. et al. LIMT is a novel metastasis inhibiting lncRNA suppressed by EGF and downregulated in aggressive breast cancer. EMBO molecular medicine. 2016;8:1052–64. doi: 10.15252/emmm.201606198. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary tables.
Data Availability Statement
The datasets supporting the conclusions of this article were retrieved from TCGA, (http://cancergenome.nih.gov/).