Abstract
Colorectal cancer (CRC) represents a serious challenge for oncologists due to high incidence and large heterogeneity. Prognostic factors are needed to stratify patients according to risk of disease progression. In this study, we report that high expression of c-Myb protein, determined by immunohistochemistry (IHC), associates with better overall and disease-free survival (OS, DFS) in a cohort of 103 patients. Although MYB has been previously considered to act as oncogene in CRC, our further analysis of datasets deposited in PrognoScan and SurvExpress databases confirmed that high MYB expression largely associates with good prognosis in CRC. As therapies targeting c-Myb have been developed and tested in preclinical studies, we believe that further studies are needed for detailed understanding of c-Myb function in CRC, before the c-Myb-targeted therapy enters clinical trials.
Keywords: c-Myb, colorectal cancer, prognosis, survival
Introduction
Colorectal cancer (CRC) is the third most common type of cancer and its incidence varies significantly across geographic regions. Czech Republic ranks among the countries with highest incidence of CRC worldwide 1. CRC is highly heterogeneous disease with a variable clinical course and response to treatment, even in tumors with similar histopathological characteristics 2. During the last decade, several attempts for comprehensive molecular classifications of CRC have been performed, and a number of molecular signatures with potential clinical usefulness for the stratification of patients with different prognosis and treatment response have been suggested 3-6. However, very few of them were confirmed as valid predictive and prognostic factors for management of CRC 2, 7.
The c-Myb oncoprotein encoded by the MYB gene is an essential transcriptional regulator for the maintenance of stem cells in colon epithelia and it is required for colon crypt homeostasis by regulating cell differentiation, proliferation and crypt integrity 8, 9. Numerous studies linked MYB overexpression and/or mutations to CRC tumorigenesis 10-14. Although MYB was shown to act as an oncogene in many different cancer types including leukemias, breast and colon cancers, recent studies have associated high level of MYB transcript with low incidence of metastasis and good prognosis in breast cancer (BC) patients 15-18. We have recently confirmed a negative association between c-Myb positivity, assessed by IHC, and the presence of distant metastases in CRC patients as well 19.
This study represents a 3-year follow-up survey of the same cohort of CRC patients described in Tichý et al. 19 and bioinformatic analysis of available CRC datasets. Good prognostic value of high MYB transcript was observed through many datasets suggesting that although MYB overexpression may contribute to CRC tumorigenesis, it may have negative impact on tumor cell dissemination and thus represents a good prognostic factor for CRC patients.
Material and Methods
Study group
The study group was described previously in detail 19. It includes 108 patients with resectable CRC who had undergone surgery at St. Anne´s University Hospital Brno in the period April 2012 - February 2015. None of the patients received preoperative chemotherapy or radiotherapy. Follow-up data were summarized at March 1st, 2018 (minimum three years follow-up) and were available for 103 patients. The study was approved by the Ethic Committee of St. Anne´s University Hospital. All patients were informed and they signed written consent allowing the inclusion into this study.
Specimen collections, IHC, RNA isolation, qPCR
All methods were described previously in detail 19.
Survival analysis using PrognoScan and SurvExpress databases
To assess the prognostic significance of MYB in CRCs we used PrognoScan (http://www.prognoscan.org) 20 and SurvExpress databases 21. PrognoScan contains four CRC microarray datasets with different survival endpoints and two Affymetrix MYB probe sets. PrognoScan employs the minimum P-value approach for grouping patients for survival analysis that finds the optimal cutpoint in continuous gene expression measurement.
SurvExpress (http://bioinformatica.mty.itesm.mx:8080/Biomatec/SurvivaX.jsp) is a web-based tool providing survival multivariate analysis and risk assessment based on gene(s) expression. Thirteen independent CRC datasets responding to MYB search were analyzed, eight of them provided significant Cox p-value. Patients of indicated datasets were stratified according to median value of prognostic index. The prognostic index is the linear component of the exponential function used to measure the level of association of a gene in a Cox proportional hazards model. It can be calculated as: PI = βχ in which χ refers to the expression value and β refers to the risk coefficient obtained from the Cox modelling. High risk cases were cases with the prognostic index above the determined median value. The maximum row average was set for duplicated genes. The log-rank test was used to evaluate statistically the equality of survival curves.
Statistical analysis
To analyze our 103 CRC patients' cohort, Kaplan-Meier methodology was adopted for the visualization of survival data and description of proportion of surviving and its confidence interval in selected time points. The statistical significance of differences in survival among groups of patients was tested using log-rank test; Cox proportional hazard model was used for the description of differences in survival among groups of patients by hazard ratios and their confidence intervals. Statistical analyses were performed using GraphPad Prism 6 (GraphPad Software, La Jolla, USA).
Results
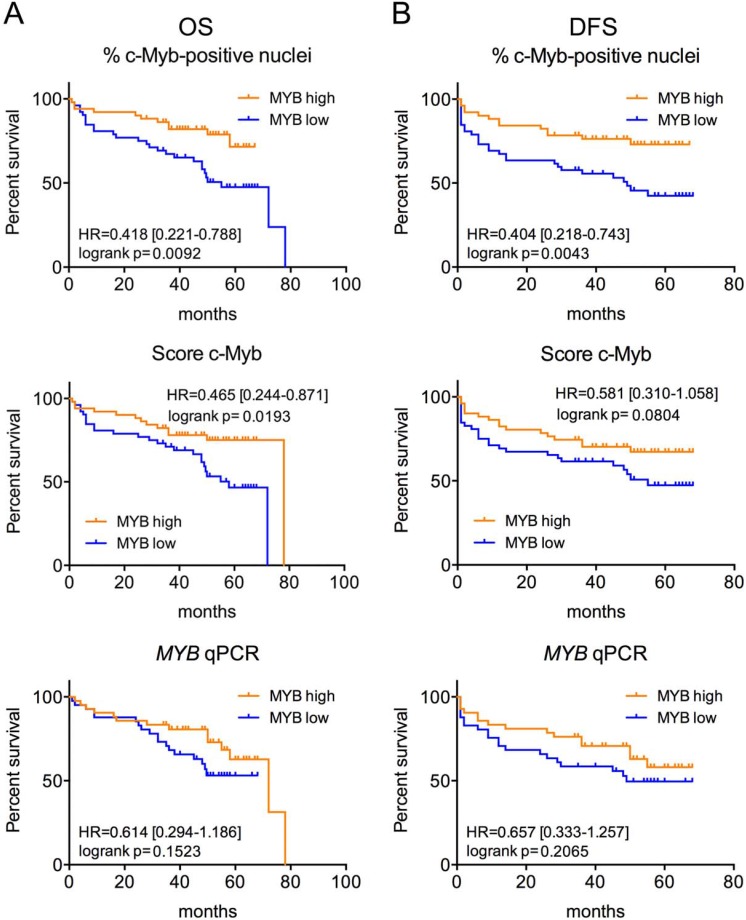
To investigate prognostic significance of c-Myb/MYB, we first performed survival analysis in our cohort of 103 CRC patients. We observed high percent of positive c-Myb nuclei detected by IHC to be significantly associated with better OS and DFS. Another IHC-determined parameter, the c-Myb score, was significantly associated only with OS (Figure 1). The MYB mRNA level was not a significant predictor of OS/DFS in our cohort (Figure 1).
Figure 1.
High c-Myb protein but not MYB transcript is associated with better outcome in our cohort of CRC patients. Kaplan-Meier survival curves for OS (A) and DFS (B) by percent of c-Myb positive nuclei, c-Myb score and MYB qPCR (for all parameters Myb-high means above median value) of our cohort of 103 CRC patients. Log-rank P values, HR and 95% CI are shown.
To validate the effect of MYB on survival of CRC patients observed in our cohort, we used the PrognoScan database which provides a tool to evaluate the association between prognosis and the expression of specific genes 20. In 3 out of 4 CRC cohorts, significant corrected or Cox P values for MYB gene expression were observed. In these 3 cohorts, high MYB expression was associated with favorable prognosis in all cases (Supplementary Table 1).
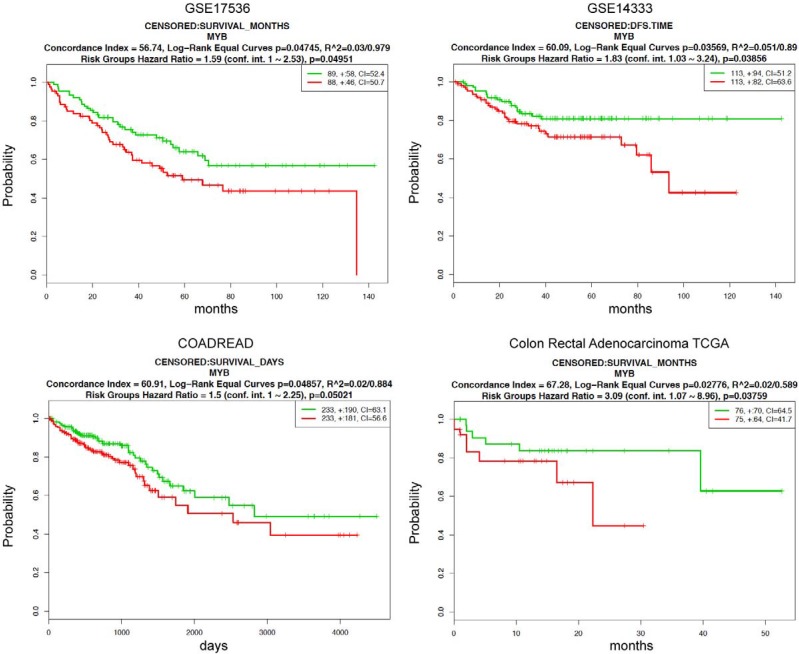
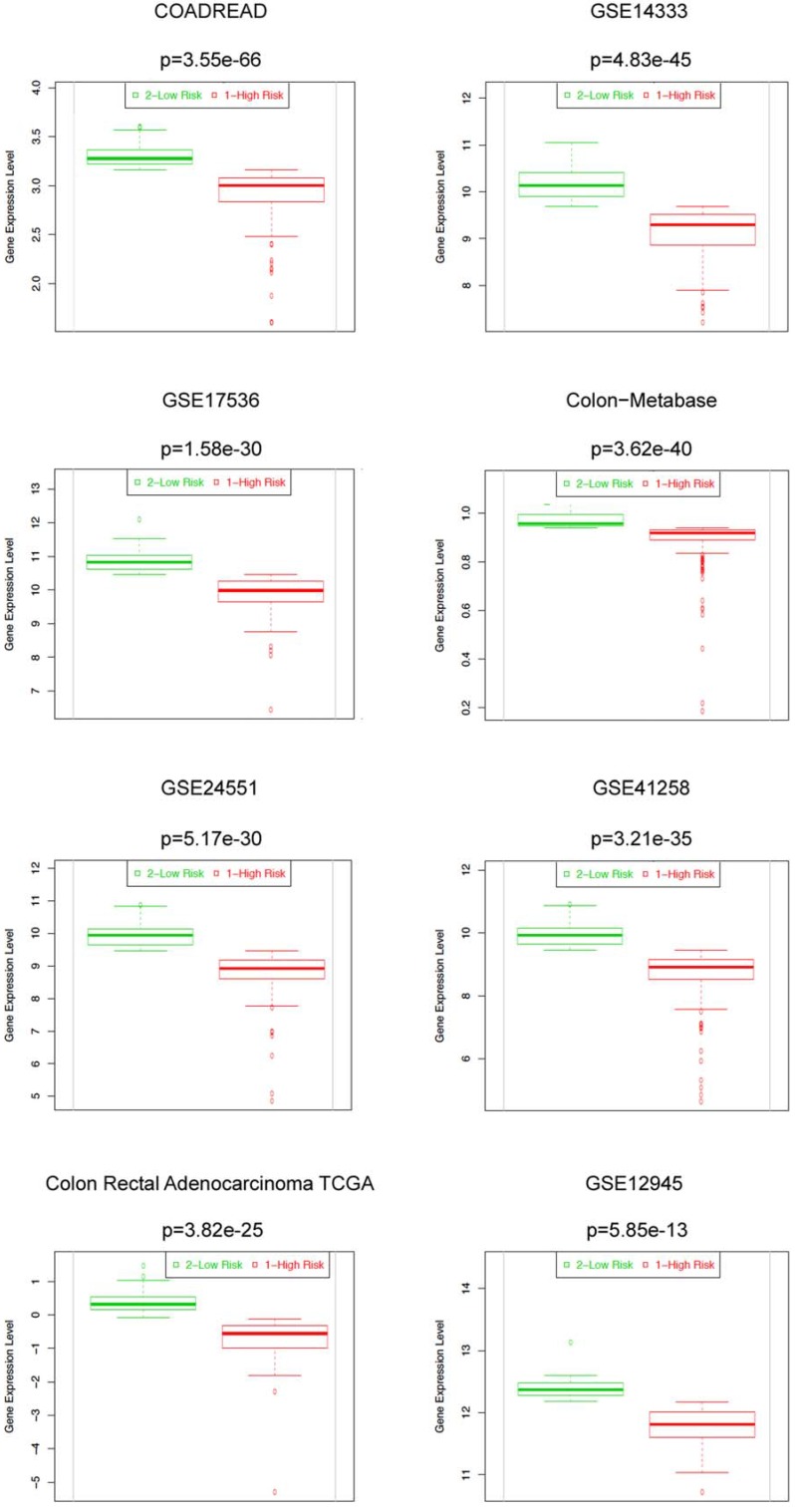
We further evaluated the prognostic significance of MYB in CRCs using a prognostic index to stratify high and low risk groups by SurvExpress tool. Eight out of thirteen datasets of CRCs with significant Cox fitting p-values (Cox p<0.05) were selected. Corresponding beta coefficients from Cox modelling are shown in Supplementary Table 2. Beta can be interpreted as a risk coefficient, positive values are associated with higher risk, negative with lower risk. Beta coefficients for all 8 analyses are negative showing an invariable association of MYB with lower risk. Two datasets (GSE14333 and GSE17536) overlap between PrognoScan and SurvExpress databases providing similar results. MYB can significantly separate low- and high-risk groups (log-rank p for equal curves <0.05) in these 2 datasets and additionally in 2 others cohorts (COADREAD, n=466; and Colon Rectal Adenocarcinoma, n=151) (Figure 2, Supplementary Table 2). Moreover, in all 8 datasets the expression of MYB was significantly lower in high compared to low risk group (Figure 3, Supplementary Table 2). Overall these results underscore MYB as favorable outcome predictor in CRCs.
Figure 2.
High MYB is associated with good prognosis in 4 CRC datasets from the SurvExpress database. Prognostic significance of MYB in CRCs was estimated by SurvExpress online tool. The high (red) and low risk (green) groups were split of the same size depending on the prognostic index. MYB can significantly separate low- and high-risk groups in 4 cohorts (GSE17536, n=177; GSE14333, n=226; COADREAD, n=466; and Colon Rectal Adenocarcinoma, n=151). Outcome event is overall survival for 3 datasets (GSE17536, COADREAD, Colon Rectal Adenocarcinoma) and disease-free survival for GSE14333. Horizontal axis represents time to event. Each panel shows the Kaplan-Meier plot of above indicated dataset, outcome event, the log-rank test of differences between risk groups, the hazard-ratio estimate, and the concordance index, which estimate the probability that subjects with a higher risk will experience the event earlier than subjects with a lower risk. Only the datasets with significant log-rank p values are shown. The insets showed the number of individuals, the number censored (+), and the concordance index (CI) of each risk group.
Figure 3.
High MYB expression levels are found in low risk groups of CRC patients in 8 SurvExpress datasets. Box plots generated by SurvExpress showed the expression levels of MYB in indicated datasets and the P-value resulting from a t-test. Low-risk groups are denoted in green and high-risk groups in red, respectively.
Discussion
Our data and data extracted from public databases (PrognoScan, SurvExpress) suggest that high expression of MYB, a widely accepted oncogene in numerous types of cancer including CRC, associates with good prognosis of CRC patients. Data obtained in our cohort suggest that the c-Myb protein level has stronger predictive potential than the MYB transcript. This could be explained by rather weaker correlation of both parameters (correlation coefficient 0.326) observed in the same samples by us previously 19. Association between high MYB RNA level and good prognosis has been previously described by us and others in BC as well 15-18 suggesting that albeit c-Myb is considered as an important factor contributing to early mammary and colon tumorigenesis 14, 22 it has a negative impact on CRC and BC progression. Several explanation of positive role of c-Myb in later phases of tumor progression can be postulated. First, c-Myb may negatively regulate tumor cell dissemination and formation of distant metastases. An association between low c-Myb level and the presence of distant metastases in CRC patients has been described by us previously in the same cohort of patients 19. Moreover, we have recently found that MYB expression inhibits spontaneous lung metastasis in BC mouse models and predicts lung metastasis relapse in BC patients 18. Second, as c-Myb is considered to act as a driver of cancer cell proliferative growth, Hugo et al. suggested that non-quiescent, actively cycling (c-Myb-positive) tumor cells could be more amenable to treatment with cytotoxic drugs targeting highly proliferating cells 23. As we and others previously reported that high c-Myb expression in CRC cell lines enhanced their chemorezistance 24-26, the „metastasis hypothesis“ seems to be more relevant. It should be noted, however, that high c-Myb protein level was associated with poor prognosis in gallbladder adenocarcinoma 27, 28 and esophageal squamous cell carcinoma 29 and two previous studies by Ramsay group reported that high c-Myb associates with poor prognosis in CRC patients 30, 31. While Millen et al. found an association between c-Myb level and relapse-free survival in a small cohort of preselected CRC patients (only T2 patients, cases with deficient mismatch repair excluded) 30, Biroccio et al. reported a relation between c-Myb, DFS and OS in a 91 CRC patients (70 colon, 21 rectum) without any preselection 31. Although there are some differences in design of our study and that of Biroccio group (inclusion of patients with tumor localized in rectum by Biroccio et al., different proportions of patients in group T1/T2 vs T3/T4, the length of follow-up, immunohistochemistry protocol etc.) it is difficult to identify the reason of such opposite results. Nevertheless, based on our data and data retrieved from public resources it seems clear that the view of c-Myb as a negative prognostic factor in colon cancer should be reconsidered. Future effort is required to uncover yet unidentified mechanisms of c-Myb action in CRC tumor progression. It has been recently suggested that c-Myb-modulated interaction between cancer cells and immune system within tumor- and metastasis-specific microenvironment may be crucial in this context 16, 18, 30. As therapies targeting c-Myb have been already tested in preclinical studies 32, 33 and undergo further development, the understanding of c-Myb function is of great importance before they enter clinical trials.
Supplementary Material
Supplementary tables.
Acknowledgments
This work was supported by project LQ1605 from the National Program of Sustainability II (Ministry of Education, Youth and Sports of the Czech Republic), 17-08985Y from Czech Science Foundation and NV18-07-00073 from Ministry of Health of the Czech Republic.
Abbreviations
- BC
breast cancer
- CI
confidence interval
- CRC
colorectal cancer
- DFS
disease-free survival
- HR
hazard ratio
- IHC
immunohistochemistry
- qPCR
quantitative real-time polymerase chain reaction
- OS
overall survival.
References
- 1.Ferlay J, Soerjomataram I, Ervik M, GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11. Lyon, France: International Agency for Research on Cancer; 2013. http://globocan.iarc.fr. [Google Scholar]
- 2.Blanco-Calvo M, Concha Á, Figueroa A. et al. Colorectal Cancer Classification and Cell Heterogeneity: A Systems Oncology Approach. Int J Mol Sci. 2015;16:13610–32. doi: 10.3390/ijms160613610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan SK, Griffith OL, Tai IT. et al. Meta-analysis of colorectal cancer gene expression profiling studies identifies consistently reported candidate biomarkers. Cancer Epidemiol Biomark Prev. 2008;17:543–52. doi: 10.1158/1055-9965.EPI-07-2615. [DOI] [PubMed] [Google Scholar]
- 4.Marisa L, de Reyniès A, Duval A. et al. Gene expression classification of colon cancer into molecular subtypes: Characterization, validation, and prognostic value. PLoS Med. 2013;10:e1001453. doi: 10.1371/journal.pmed.1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roepman P, Schlicker A, Tabernero J. et al. Colorectal cancer intrinsic subtypes predict chemotherapy benefit, deficient mismatch repair and epithelial-to-mesenchymal transition. Int J Cancer. 2013;134:552–62. doi: 10.1002/ijc.28387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Müller MF, Ibrahim AE, Arends MJ. Molecular pathological classification of colorectal cancer. Virchows Arch. 2016;469:125–34. doi: 10.1007/s00428-016-1956-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sagaert X. Prognostic biomarkers in colorectal cancer: where do we stand? Virchows Arch. 2014;464:379–91. doi: 10.1007/s00428-013-1532-z. [DOI] [PubMed] [Google Scholar]
- 8.Malaterre J, Carpinelli M, Ernst M. et al. c-Myb is required for progenitor cell homeostasis in colonic crypts. Proc Natl Acad Sci U S A. 2007;104:3829–34. doi: 10.1073/pnas.0610055104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zorbas M, Sicurella C, Bertoncello I. et al. c-Myb is critical for murine colon development. Oncogene. 1999;18:5821–30. doi: 10.1038/sj.onc.1202971. [DOI] [PubMed] [Google Scholar]
- 10.Meltzer SJ, Ahnen DJ, Battifora H. et al. Protooncogene abnormalities in colon cancers and adenomatous polyps. Gastroenterology. 1987;92:1174–80. doi: 10.1016/s0016-5085(87)91074-2. [DOI] [PubMed] [Google Scholar]
- 11.Torelli G, Venturelli D, Coló A. et al. Expression of c-myb protooncogene and other cell cycle-related genes in normal and neoplastic human colonic mucosa. Cancer Res. 1987;47:5266–9. [PubMed] [Google Scholar]
- 12.Ramsay RG, Thompson MA, Hayman JA. et al. Myb expression is higher in malignant human colonic carcinoma and premalignant adenomatous polyps than in normal mucosa. Cell Growth Differ. 1992;3:723–30. [PubMed] [Google Scholar]
- 13.Hugo H, Cures A, Suraweera N. et al. Mutations in the MYB intron I regulatory sequence increase transcription in colon cancers. Genes Chromosomes Cancer. 2006;45:1143–54. doi: 10.1002/gcc.20378. [DOI] [PubMed] [Google Scholar]
- 14.Malaterre J, Pereira L, Putoczki T. et al. Intestinal-specific activatable Myb initiates colon tumorigenesis in mice. Oncogene. 2016;35:2475–84. doi: 10.1038/onc.2015.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thorner AR, Parker JS, Hoadley KA. et al. Potential tumor suppressor role for the c-Myb oncogene in luminal breast cancer. PLoS One. 2010;5:e13073. doi: 10.1371/journal.pone.0013073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nicolau M, Levine AJ, Carlsson G. Topology based data analysis identifies a subgroup of breast cancers with a unique mutational profile and excellent survival. Proc Natl Acad Sci USA. 2011;108:7265–70. doi: 10.1073/pnas.1102826108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu LY, Chang LY, Kuo WH. et al. A supervised network analysis on gene expression profiles of breast tumors predicts a 41-gene prognostic signature of the transcription factor MYB across molecular subtypes. Comput Math Methods Med. 2014;2014:813067. doi: 10.1155/2014/813067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knopfová L, Biglieri E, Volodko N. et al. Transcription factor c-Myb inhibits breast cancer lung metastasis by suppression of tumor cell seeding. Oncogene. 2018;37:1020–30. doi: 10.1038/onc.2017.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tichý M, Knopfová L, Jarkovský J. et al. Overexpression of c-Myb is associated with suppression of distant metastases in colorectal carcinoma. Tumour Biol. 2016;37:10723–9. doi: 10.1007/s13277-016-4956-7. [DOI] [PubMed] [Google Scholar]
- 20.Mizuno H, Kitada K, Nakai K. et al. PrognoScan: a new database for meta-analysis of the prognostic value of genes. BMC Med Genomics. 2009;2:18. doi: 10.1186/1755-8794-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aguirre-Gamboa R, Gomez-Rueda H, Martínez-Ledesma E. et al. SurvExpress: an online biomarker validation tool and database for cancer gene expression data using survival analysis. PLoS One. 2013;8:e74250. doi: 10.1371/journal.pone.0074250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miao RY, Drabsch Y, Cross RS. et al. MYB is essential for mammary tumorigenesis. Cancer Res. 2011;71:7029–37. doi: 10.1158/0008-5472.CAN-11-1015. [DOI] [PubMed] [Google Scholar]
- 23.Hugo HJ, Saunders C, Ramsay RG. et al. New Insights on COX-2 in Chronic Inflammation Driving Breast Cancer Growth and Metastasis. J Mammary Gland Biol Neoplasia. 2015;20:109–19. doi: 10.1007/s10911-015-9333-4. [DOI] [PubMed] [Google Scholar]
- 24.Abaza MS, Al-Attiyah RJ, Al-Saffar AM. et al. Antisense oligodeoxynucleotide directed against c-myb has anticancer activity and potentiates the antiproliferative effect of conventional anticancer drugs acting by different mechanisms in human colorectal cancer cells. Tumour Biol. 2003;24:241–57. doi: 10.1159/000076139. [DOI] [PubMed] [Google Scholar]
- 25.Funato T, Satou J, Kozawa K. et al. Use of c-myb antisense oligonucleotides to increase the sensitivity of human colon cancer cells to cisplatin. Oncol Rep. 2001;8:807–10. doi: 10.3892/or.8.4.807. [DOI] [PubMed] [Google Scholar]
- 26.Pekarčíková L, Knopfová L, Beneš P. et al. c-Myb regulates NOX1/p38 to control survival of colorectal carcinoma cells. Cell Signal. 2016;28:924–36. doi: 10.1016/j.cellsig.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 27.Cai W, Li Q, Yang Z. et al. Expression of p53 upregulated modulator of apoptosis (PUMA) and C-myb in gallbladder adenocarcinoma and their pathological significance. Clin Transl Oncol. 2013;15:818–24. doi: 10.1007/s12094-013-1010-8. [DOI] [PubMed] [Google Scholar]
- 28.Shu GS, Lv F, Yang ZL. et al. Immunohistochemical study of PUMA, c-Myb and p53 expression in the benign and malignant lesions of gallbladder and their clinicopathological significances. Int J Clin Oncol. 2013;18:641–50. doi: 10.1007/s10147-012-0431-7. [DOI] [PubMed] [Google Scholar]
- 29.Lu H, Wang Y, Huang Y. et al. Expression and prognostic role of c-Myb as a novel cell cycle protein in esophageal squamous cell carcinoma. Clin Transl Oncol. 2013;15:796–801. doi: 10.1007/s12094-013-1009-1. [DOI] [PubMed] [Google Scholar]
- 30.Millen R, Malaterre J, Cross RS. et al. Immunomodulation by MYB is associated with tumor relapse in patients with early stage colorectal cancer. Oncoimmunology. 2016;5:e1149667. doi: 10.1080/2162402X.2016.1149667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Biroccio A, Benassi B, D'Agnano I. et al. c-Myb and Bcl-x overexpression predicts poor prognosis in colorectal cancer: clinical and experimental findings. Am J Pathol. 2001;158:1289–99. doi: 10.1016/S0002-9440(10)64080-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu X, Xu Y, Han L, Yi Y. Reassessing the Potential of Myb-targeted Anti-cancer Therapy. J Cancer. 2018;9:1259–1266. doi: 10.7150/jca.23992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cross RS, Malaterre J, Davenport AJ. et al. Therapeutic DNA vaccination against colorectal cancer by targeting the MYB oncoprotein. Clin Transl Immunology. 2015;4:e30. doi: 10.1038/cti.2014.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary tables.