Abstract
Objective:
Interpreting genome-scale genetic association data, particularly for complex diseases and phenotypes, requires extensive use of prior knowledge across a broad range of potential biological and environmental influences, spanning many scientific subdisciplines. We suggest that known or hypothesized disease risk factors, and causal mechanisms, can be represented using an ontology, a computational specification of a set of concepts and the relations between them.
Methods:
We have integrated the expertise of multiple investigators in nicotine pharmacokinetics and pharmacodynamics, nicotine dependence, and clinical smoking cessation outcomes, and represented this knowledge in an ontology-based network model. Our model spans multiple scales, from molecules, genes and cellular pathways, to complex behavioral phenotypes and even environmental factors. To leverage previous and ongoing work in the field of ontology development, we adopt, expand upon and relate elements from existing ontologies whenever possible.
Results:
We discuss several applications of our ontology: to support interdisciplinary research by graphically representing a complex scientific theory, to facilitate meta-analysis across different studies, to highlight potential interactions, and to support statistical analysis and causal modeling. We demonstrate that our ontology can focus hypothesis testing on areas supported by current theory.
Conclusion:
We describe how an ontology-based computational representation can be applied to disease risk factors and mechanisms, enabling the use of prior knowledge in large-scale genetic association studies in general. In specific, we have developed an initial Smoking Behavior Risk Ontology to support studies related to the pharmacogenetics of nicotine addiction and treatment.
Keywords: disease risk, genetic association, nicotine dependence, ontology, smoking cessation treatment
Introduction
Most of the diseases that are highly prevalent in the human population (‘common disease’) are complex in their etiology [1]. Complex disease by definition has no single factor that accounts for the majority of the variation in disease risk among different individuals. Instead, there are contributions from a number of factors, both genetic and environmental, each of which may have a small effect on disease risk. In addition, these factors can interact with each other in complex ways, such that the total disease risk may not simply be the sum of the effects of individual risk factors.
For many complex diseases, considerable research has been carried out to identify the risk factors, and the causal mechanisms by which risk factors affect the probability of developing the disease. This research has often been multidisciplinary, with studies spanning a wide range of time and length scales, from molecular studies to studies of the effects of environment. As a result, the disease risk model may have so many components and interactions, that it becomes difficult to communicate to non-experts, and, importantly, becomes difficult to use in large-scale association studies. Genome-wide genetic association studies are becoming a commonly used tool for discovering genetic variants that affect disease risk [2]. Yet the statistical analyses of most genome-wide genetic association studies performed to date focus on a single phenotype outcome, and make relatively little use of the extensive prior knowledge that has been accumulated about the phenotype. Recent methodological advances such Bayesian model averaging [3] can use prior knowledge to guide the model search toward models that are consistent with known risk factors and mechanisms [4]. However, to be used in large-scale computational analysis, this knowledge must be encoded first in a computer-accessible form.
In this study, we describe the use of formal ontology to represent a risk model for phenotypes related to tobacco smoking behavior. By risk model, we mean how a change in a factor (or exposure) can change the probability of developing a particular phenotype. We call our ontology the Smoking Behavior Risk Ontology (SBRO). This first version of SBRO was designed to support the Pharmacogenetics of Nicotine Addiction and Treatment (PNAT) Consortium, a National Institute on Drug Abuse-funded, multidisciplinary research project that is part of the Pharmacogenetics Research Network [5]. The aim of PNAT is to discover genetic variants that modify either the risk of developing nicotine addiction, or the response to clinical smoking cessation treatments; examples of our discoveries were recently reported [6,7]. We expect that this first version of SBRO will provide the nucleus for a larger formal representation of current theory concerning smoking risk factors, to facilitate communication within this extensive field as well as facilitate analysis of association studies.
Nicotine addiction, and associated tobacco use, is the single greatest cause of preventable, premature death and disability in the United States and much of the world [8,9]. It is also a complex trait, having been shown to be influenced by genetic [10–13] as well as environmental factors [14]. For example, significant associations between nicotine dependence and single nucleotide polymorphism (SNP) genetic markers were recently found in a genome-wide study [11]. There are also a plethora of environmental conditions (such as parental and peer smoking, availability of cigarettes, and tobacco product advertising) that are recognized to play a role in the acquisition of nicotine addiction [15]. Previous genetic investigations of smoking have begun only recently to incorporate environmental measures into their study designs and evidence for gene–environment interaction is beginning to emerge. For example, it has been shown that genetic risk for smoking is lower at higher levels of parental monitoring [16], suggesting that parental monitoring may help curb genetic susceptibility to smoking behavior. McCaffery and others [17] examined the relationship between education level and nicotine dependence in twins. In addition, researchers have identified interactions between genetic variation in the dopamine pathway and physical activity [18].
There has been previous work in areas related to SBRO, most notably in the field of multiscale modeling, and in the rapidly evolving field of biomedical ontology construction. As we are addressing the relationships between genes, environmental factors, and phenotypes (qualities of individuals), our knowledge representation must span a range of time and length scales, from molecules (such as DNA, transcripts, proteins, small biological molecules) to individual organisms and their behavior (such as smoking behaviors), and even societal factors such as smoking-related laws and policies. Furthermore, an event at one scale can influence the probabilities (‘risk’) of events occurring at other scales. Our representation must therefore be multiscale. Multi-scale modeling is a well-established technique in physics that has only recently begun to make inroads in biology [19]. The basic ideas are that (i) microscopic phenomena underlie emergent behavior at more macroscopic levels, (ii) each of these levels can be modeled modularly, and (iii) each level can potentially provide information as an input into models at other levels. The best-established example of multiscale modeling in biology is the human heart [20,21]. Multiscale cardiac models join together modules spanning scales from the molecule (individual proteins and ions) in cardiac muscle cells (myocytes) to heart rate and blood pressure. A multiscale model has recently been proposed for immune system function, spanning molecular events (such as antigen binding and presentation) to observable systemic phenotypes such as pathogen load [22]. Most multiscale modeling approaches have focused on quantitative modeling of observed phenotypes (e.g. heart rate) and lower-level related phenotypes, often referred to as endophenotypes (e.g. intracellular calcium ion concentration). The aim of SBRO, however, is not to simulate the development of disease, but rather to represent prior knowledge and hypotheses about risk factors involved in the development of a phenotype. In fact, analogously to the use of simulation in exploring potential genetic risks in heart failure [23], computational simulations are a potential source of information about potential smoking risk factors, which could then be summarized in SBRO.
Biomedical ontologies have become an essential tool for biology in the era of large ‘omics’ (e.g. genomics, transcriptomics, proteomics, metabolomics) data sets, as a way to represent biological knowledge that is amenable to computation [24,25]. The best-known biomedical ontologies are the Foundational Model of Anatomy [26] for describing anatomical parts of organisms and relationships between these parts, and the gene ontology (GO) [27], which began as a controlled vocabulary for describing the molecular functions, biological roles, and cellular locations of gene products across many different organisms. These ontologies represent concepts and the relationships between them, relative to specific domains of knowledge [24]. More recently, the Open Biomedical Ontologies (OBO) Foundry project has been initiated, with the goal of producing a set of ‘standard’, interoperable ontologies of complementary, nonoverlapping domains of knowledge [28]. New, application-specific ontologies can then be developed that use existing terms from the standard ontologies. The SBRO uses relevant terms from existing ontologies such as the GO and Suggested Ontology for Pharmacogenetics (SO-Pharm) [29] to the extent possible, in a model of risk factors (and relationships between these factors) for smoking behaviors. SO-Pharm, in particular, provides a rich representation for capturing phenotypes, genotypes, drug treatment, and clinical trial information, and terms from SO-Pharm enable the basic data representation for clinical trials of smoking cessation. However, nearly all of the concepts and relationships in SBRO represent the known or hypothesized causal relationships between specific genotypes, drug treatments, environmental exposures, which may contribute to risk for smoking-related phenotypes, or modify the response to smoking cessation treatment. Representation of a disease risk model is, to our knowledge, a novel application of ontologies.
Materials and methods
Building the Smoking Behavior Risk Ontology
Building an ontology is a collaborative process between computer scientists and experts in the given domain of knowledge [30,31]. For the SBRO, we have integrated expertise from six different individual scientists, each with over 20 years of experience in smoking-related research, in diverse domains of knowledge that are hypothesized to be causally related to nicotine addiction and treatment phenotypes: neurobiology, nicotine pharmacokinetics, nicotine pharmacodynamics, nicotine dependence, and smoking cessation clinical trials. Of course, even the expertise of these scientists is incomplete with respect to the genetics and neurobiology of nicotine dependence and smoking cessation, and one of the natural directions for the SBRO is to enlist the help of additional experts.
Initially, we tried a number of different approaches to ontology construction, including having each domain expert work directly on the ontology with the Protege [32] software tool and conference calls or in-person meetings to discuss the ontology. We found that, in our case, the best approach to ontology construction was to have only one Protege user, who then iteratively met with each expert one-on-one to revise the ontology in stages. Most experts tended to spend their time revising their own areas of expertise, as well as relationships that connected these areas to other parts of the ontology; whereas only a couple of experts reviewed in detail the entire ontology structure, which at the present time comprises over 100 concepts in web ontology language (OWL) and well over 100 genes and proteins in systems biology markup language (SBML).
Use of existing ontologies
Most of the concepts in the SBRO are taken from existing ontologies, or are specified using the concept definitions given by existing ontologies (Table 1). For example, SBRO:bupropion_treatment and SBRO:varenicline_treatment are children of the concept SOPHARM:drug_treatment (SOPHARM_23000). Biological processes involved in nicotine response are children of the GO biological process concept response_to_nicotine (GO:0035094). For all concepts other than those pertaining to molecular pathways, we have encoded SBRO using the OWL specification, using Protege-OWL[32].The choice of OWL was motivated primarily to facilitate the use of existing ontology terms, as both the GO and SO-Pharm (as well as the 19 other ontologies that are referenced in SO-Pharm) are available in OWL format.
Table 1.
Some of the concepts from existing ontologies used by SBRO
| Ontology:term | Use in SBRO |
|---|---|
| GO:cellular_process | Parent class for all molecular pathways |
| GO:nicotine_catabolic_process | Parent class for nicotine metabolism pathway in human hepatic cells |
| GO:behavior | Parent class for smoking-related behaviors |
| GO:response_to_chemical_stimulus | Parent class for all drug responses |
| GO:response_to_nicotine | Parent class for acute and chronic physiological response to nicotine |
| PATO:quality | Parent class for behavioral qualities |
| SO-Pharm:smoking_behavior | Class for smoking behavior |
| SO-Pharm: genotype item | Individual genotypes from a clinical trial |
| SO-Pharm: phenotype item | Individual phenotypes from a clinical trial |
| SO-Pharm: clinical trial event | Parent class for time points in a clinical trial |
| SBML: reaction | Parent class for all chemical reactions in cellular pathways |
| SBML: species | Parent class for all molecules in cellular pathways (including macromolecules and small molecules) |
| SBML: modifier | Catalyzes relation |
GO, gene ontology; PATO, Phenotype and Trait Ontology; SBML, systems biology markup language; SBRO, Smoking Behavior Risk Ontology; SO-Pharm, Suggested Ontology for Pharmacogenetics.
For molecular interactions in pathways, we used the SBML specification [33]. In SBML, the primary concepts are chemical reactions and molecules, such as proteins and small molecules (e.g. nicotine), linked together by relations. We have encoded several nicotine dependence-related pathways into SBML using the PANTHER pathway system, following a previously described process [34]. This system is available for public use at http://curation.pantherdb.org All 20 pathways can be searched, browsed, and downloaded at http://www.pantherdb.org. The molecular pathway representation (in SBML) is linked to the rest of SBRO (in OWL) by cross-referencing each OWL pathway concept with the corresponding PANTHER pathway identifier for SBML representation of the pathway.
Novel concepts and relationships in Smoking Behavior Risk Ontology
Many concepts in SBRO, such as nicotine_dependence, nicotine_withdrawal and smoking_abstinence, are specific to the knowledge domain of smoking research, and are not represented in any existing ontologies. These concepts are thus novel to SBRO. In addition, SBRO contains a relationship type that is not found in any currently existing ontologies, but which is necessary to represent effects on risk. Fortunately, however, this relationship can be viewed as a generalization of the newly adopted regulates relationship in the GO. The ‘regulates’ relationship is defined as follows: ‘When a biological process E regulates a function or a process F, it modulates the occurrence of F. If F is a biological quality, then E modulates the value of F’ (http://www.geneontology.org/GO.doc.shtml#term-term-relationships). This definition captures the main attributes of the relationship needed to represent the effect that one concept (a ‘risk factor’) may have on another concept in SBRO. Using this definition, the statement nicotine_withdrawal regulates smoking_relapse would mean that the severity of nicotine withdrawal modulates the probability of smoking relapse, which has been demonstrated for smokers who are trying to quit [35]. Furthermore, the fact that regulates is transitive also applies to risk models. However, because the word regulates has very different connotations outside the realm of molecular biology, SBRO uses the term influences instead, though even this term has common usages that may cause some confusion. In biology, the statement process_A positively_regulates process_B means that if A (or its activity) increases, B (or its activity) also increases; and process_A negatively_ regulates process_B means that if A (or its activity) increases, B (or its activity) decreases. Similarly, for risk models, we adopt the relations positively_influences and negatively_influences. For example, stating that nicotine_withdrawal positively_influences smoking_relapse means that an increase in withdrawal severity would tend to increase the probability of smoking relapse, all other things being equal.
Ontology representation of phenotypes
To represent phenotypes, SBRO adopts the concept phenotype_item (SOPHARM_15000) from the SO-Pharm Ontology. This concept is defined as ‘measured on a patient during a particular clinical event according to a measuring method’ and having a particular measured value. Each phenotype_item must be specified using the bipartite ‘entity: quality’ syntax (http://www.bioontology.org/wiki/index.php/PATO:Main_page) recommended by the Phenotype and Trait Ontology (PATO). This syntax has two distinct advantages. First, it enables the SBRO ontology to remain as simple as possible; new ontology concepts do not have to be invented for every different phenotype that is measured. Second, and most important for our purposes, each phenotype_item measured in a study refers to a concept that is represented in the risk model. Table 2 gives the ontology representation for a number of phenotypes used in the first PNAT study, from a clinical trial of the smoking cessation drug bupropion [36]. As an example, consider two of the key behavioral phenotypes used to assess the level of nicotine dependence: number of cigarettes smoked per day, and time to first cigarette after waking up in the morning. The number of cigarettes smoked per day is encoded as: daily_cigarette_smoking:count, which combines the behavioral concept daily_cigarette_smoking with the quality concept count (from the PATO ontology, available at http://bioportal.bioontology.org/ontologies/39480). Again, for important smoking behavior-related phenotypes, SBRO contains additional quality concepts. For example, time to first cigarette is encoded as: regular_tobacco_use:time_delay_before_first_daily_behavior. As daily_cigarette_smoking is a subclass of regular_tobacco_use, the ontology specifies the relationship between number of cigarettes per day and time to first cigarette.
Table 2.
Ontology representation of key phenotypes
| Clinical trial phenotype |
Ontology mapping |
||||
|---|---|---|---|---|---|
| Variable | Description | Smoking behavior (SBRO) | Quality (PATO, SBRO) | Clinical trial event
baseline (SBRO) |
Measurement type (SBRO) |
| CIGDAY | Number of cigarettes smoked on a typical day | daily_cigarette_ smoking | count | baseline | self_report_ from_ diagnostic_ tool |
| FTND02 | Cigarettes per day | daily_cigarette_ smoking | numerical | baseline | FTND |
| FTND03 | Time to first cigarette of day | regular_tobacco_use | time_delay_ before_first_ occurrence_ each_day | baseline | FTND |
| FTND04 | Hard not to smoke in forbidden places? | regular_tobacco_use | occurrence_in_ prohibited_ places | baseline | FTND |
| FTND05 | Which cigarette would you hate most to give up? | regular_tobacco_use | increased_ priority_of_ first_ occurrence_ each_day | baseline | FTND |
| FTND06 | Do you smoke more during first hours of day? | regular_tobacco_use | increased_ priority_early_ in_ day | baseline | FTND |
| FTND07 | Do you smoke if you are ill in bed? | regular_tobacco_use | occurrence_ when_ill | baseline | FTND |
| FAGER | FTND summary score | nicotine_dependence | numerical | baseline | FTND |
| FAGERD6 | FTND summary score (dichotomized) | nicotine_dependence | presence | baseline | FTND |
| ADDIC | RFS addiction score | nicotine_dependence | numerical | baseline | RFS |
| CES-D_0 | CES-D total score (baseline) | depression | numerical | baseline | CES-D |
| PPS_EOT | Verification of 7-day point-prevalence success at EOT | point-prevalence_ tobacco_abstinence | presence | end_of_ treatment | biochemical_ measurement |
| PPS_6MO | Verification of 7-day point-prevalence success at 6 months | point-prevalence_ tobacco_abstinence | presence | six_months_ post_quit_date | biochemical_ measurement |
| PASVEOT | 7-day abstinence success (no relapse) by EOT | continuous_tobacco_ abstinence_without_ relapse | presence | end_of_ treatment | biochemical_ measurement |
| PASV6M | 7-day abstinence success (no relapse) by 6 months | continuous_tobacco_ abstinence_without_ relapse | presence | six_months_ post_quit_date | biochemical_ measurement |
| CAFDZEOT | Days to first lapse | continuous_complete_ tobacco_abstinence | duration | end_of_ treatment | self_report_ from_ diagnostic_tool |
| PAFDZ_EOT | Days to first relapse | continuous_tobacco_ abstinence_without_ relapse | duration | end_of_ treatment | self_report_ from_ diagnostic_tool |
| CES-D_9 | CES-D at EOT | depression | numerical | end_of_ treatment | CES-D |
| CES-D_10 | CES-D at 6 months | depression | numerical | six_months_ post_quit_date | CES-D |
| CAS_6M | Continuous abstinence success (no smoke)
by 6 months |
continuous_complete_ tobacco_abstinence | presence | six_months_ post_quit_date | biochemical_ measurement |
| CASEOT | Continuous abstinence success (no smoke) by EOT | continuous_complete_ tobacco_abstinence | presence | end_of_ treatment | biochemical_ measurement |
The first column gives the clinical variable names from Ref. [36] (a randomized, placebo-controlled clinical trial cohort recently genotyped by PNAT to study the pharmacogenetics of smoking cessation treatments).
CES-D, center for epidemiological studies depression scale; EOT, end of treatment; FTND, Fagerstrom test of nicotine dependence; PATO, Phenotype and Trait Ontology; PNAT, Pharmacogenetics of Nicotine Addiction and Treatment; RFS, reasons for smoking test; SBRO, Smoking Behavior Risk Ontology.
The SO-Pharm concept phenotype_item has two associated properties, the time at which the measurement was taken (clinical_trial_event, SOPHARM_61000) and the method used for measurement (measurement_method, SOPHARM_62100). The SBRO includes subclasses that refer to specific events in clinical trials of smoking cessation treatments, such as end_of_treatment, and six_months_post_quit_date; and specific measurement methods, including self_report_from_diagnostic_tool (many smoking-related phenotypes are captured by standardized questionnaires) and biochemical_measurement (e.g. to verify smoking status).
Availability
The SBRO ontology is available as an OWL file from the National Center for Biomedical Ontology BioPortal (http://bioportal.bioontology.org/ontologies/39939) and SBML files from the PANTHER pathway resource (ftp://ftp.pantherdb.org/pathway/SBRO).
Results
A risk model for smoking behavior and related phenotypes
It is critical to incorporate prior knowledge into both study design and data analysis in genetic studies. However, the biological and environmental knowledge relevant to most disease phenotypes can span many research fields, which together define a complex, multidisciplinary scientific theory. We suggest that computational representations of a complex theory, such as formal ontology, provide a useful platform for application to large-scale genetic studies.
Our aim is to generate hypotheses about how variation in human genes might ultimately impact individual organism-level phenotypes such as smoking cessation and subsequent relapse behavior. A great body of research relevant to this aim already exists in the fields of nicotine dependence, nicotine pharmacokinetics and pharmacodynamics, smoking behavior, neurobiology, and cell and molecular biology. Within and between each of these fields, research results have been used by experts to define theories, and these theories are used to guide further research in expanding or revising the prevailing theories. However, because of the sheer breadth of these research fields, no one individual scientist will typically develop models of the entire field, but rather will focus on a specific part of the field. One solution is to construct a formal, computational representation of this prevailing theory in terms of a network model that integrates the expertise from multiple individual researchers, so that a more complete model (or set of models) can be used in genetic association studies.
The ultimate goal of the PNAT is to improve the efficacy of smoking cessation treatment by using genetic information to identify novel targets for pharmacotherapy and potentially to assign patients to specific therapies. Therefore, we focus here on smoking relapse behavior, the return to smoking behavior after an attempt to quit of sufficient duration, and subsequent sustained nonsmoking (smoking abstinence). Of course, relapse behavior is an emergent property of a complex system and is not a simple causal manifestation of one or a handful of genetic variants. Nevertheless, a great deal of research has already been done in the areas of nicotine dependence, neurobiology, and smoking cessation, and there is a prevailing model (or set of models) for the biological and environmental underpinnings of these complex phenotypes. Individual researchers in the field may differ in their opinions about the relative importance of different parts of the model, but qualitatively there is substantial agreement in the field. Cigarette smoking is agreed to be a behavior that is influenced by many factors, both genetic and environmental. In addition, a number of different concepts (often called ‘constructs’ in behavioral fields) have been shown to be useful in predicting relapse behavior [35]. Many of these concepts are measurable, or related to other concepts that are measurable. One of the most useful of these concepts is nicotine dependence. Nicotine dependence is a psycho-physiological state induced in many, but not all, individuals in response to long-term, chronic nicotine exposure. Like other drug dependence, nicotine dependence is a physical change in the brain and corresponding cellular pathways that manifests as a craving for nicotine and its effects, and associated drug-seeking behavior. Other key related concepts are nicotine’s positive reinforcing (acute exposure) effects, such as cognitive enhancement and mood (affect) enhancement; and negative reinforcing effects, such as avoidance of withdrawal. Each of these concepts can be treated as a basis for a measurable phenotype, and there is evidence that each is to some degree the result of a unique combination of neural pathways in the brain, and corresponding cellular pathways. For example, dopaminergic neurons (and intracellular and intercellular dopamine pathways) have been shown to be involved in both acute nicotine response and chronic nicotine dependence. The corticotropin releasing hormone pathway, in contrast, has been shown to be involved in withdrawal-induced behavior, which is an element of chronic nicotine response but not acute response. All of these concepts are part of the SBRO.
Another critical element in SBRO is to describe what happens when nicotine is administered to an individual through cigarette smoking, that is, nicotine pharmacokinetics and pharmacodynamics. Both nicotine pharmacokinetics (the time course of the amount of nicotine present at its site of action in the brain) and nicotine pharmacodynamics (the effect of nicotine on the brain and behavior) are known to play a role in nicotine addiction and smoking cessation. Nicotine pharmacokinetics is dominated by the metabolism of nicotine in hepatic cells to other, essentially non-bioactive, chemicals (http://www.pharmgkb.org/search/pathway/nicotine/nicotine.jsp). One hypothesis is that individuals with faster metabolism will need to smoke more to maintain the desired level of nicotine and the desired pharmacodynamic effects such as improved cognition, enhancement of mood, and avoidance of withdrawal. The slow metabolizers, in contrast, need to smoke less to achieve and maintain the same effects as well as to avoid toxic effects of too much nicotine (e.g. nausea) [37,38]. In this way, nicotine metabolism rates are thought to affect behavioral phenotypes such as cigarettes per day and time to first cigarette, which are used to assess the degree of nicotine dependence [39], and predict quitting success with alternate medications [40,41]. Figure 1 shows the major nicotine-metabolizing reactions, and the gene products acting as catalysts, in typical human liver cells.
Fig. 1. Human hepatic nicotine metabolism reactions.
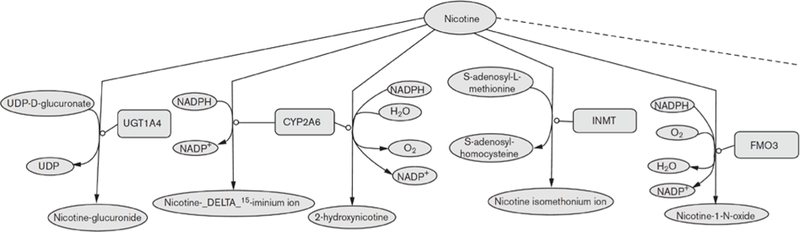
Small molecules are represented as green ovals; proteins are represented as light red boxes. The diagram was drawn with Cell Designer [65]. The full pathway can be found at http://www.pantherdb.org/pathway/pathwayDiagram.jsp?catAccession=P05914, and at http://biocyc.org/HUMAN/NEW-IMAGE?type=PATHWAY&object=PWY66–201. NADPH, nicotinamide adenine dinucleotide phosphate (reduced); UDP, uridine diphosphate.
Nicotine pharmacodynamics are believed to arise primarily from the direct binding of nicotine to neuronal/ neuroendocrine nicotinic acetylcholine receptors (nAchRs), leading to cell depolarization and increased vesicle release. The two most highly studied nicotine effects are on the release of adrenaline by chromaffin cells in the medulla [42–44] (http://www.pharmgkb.org/search/pathway/nicotine/nicotine-pd-chromaffin.jsp), and on the release of dopamine by dopaminergic neurons in the nucleus accumbens, the so-called ‘reward pathway’ [45] (see: http://www.pharmgkb.org/search/pathway/nicotine/nicotine-pd-dopaminergic.jsp, and http://www.pantherdb.org/pathway/pathwayDiagram.jsp?catAccession=P05912). Nicotine effects on the peripheral nervous system, such as increases in heart rate, blood pressure, and blood glucose, result from adrenaline release into the bloodstream. In dopaminergic neurons of the central nervous system, nAChRs are found in neurons in presynaptic, postsynaptic, and nonsynaptic sites [46]. With regard to nicotine addiction, research has been primarily focused on the effects of presynaptic nAChRs on dopamine release in the nucleus accumbens. nAChRs are multiple-subunit receptors encoded by a total of 18 genes in humans. Nine of these genes are known to be expressed in the brain, in several different combinations, and we consider these nine genes as candidates for involvement in nicotine addiction and treatment. For example, through a combination of genome-wide and individual candidate gene association studies, acetylcholine nicotinic receptor candidate genes have been associated with a wide variety of smoking behaviors, including nicotine dependence, smoking quantity, age of initiation, and subjective responses to smoking [11,12,47–56]. Genetic variation in the dopamine-mediated reward pathway has been implicated in cigarette smoking and cessation [57] and this pathway is thought to play a key role in development of addiction to numerous drugs including alcohol, opiates, and nicotine [58,59].
To build an initial model of the network of relationships between genes, environment and smoking behavior, and related phenotypes, we have constructed a formal ontology [60]. Ontologies are entering widespread use in biology to create structured, computationally accessible representations of complex biological systems and related domains of knowledge [27,61]. Ontology, in its computer science sense, is a specification of a formal model of a knowledge domain in terms of concepts (things that exist, or processes that occur) and the relationships between them; this category-based model of reality was adopted, along with the name, from the branch of philosophy called ontology, which has its basis in Aristotelian metaphysics [60]. In computer science, an ontology is one way of formally representing a large and complex scientific theory [62]. We describe our process for building the SBRO ontology, as well as the software tools to facilitate this process, in Materials and methods.
The model must encode the known or hypothesized relationships between genes and genetic variation on the one hand, and smoking-related phenotypes on the other. Our model is multiscale, from the molecular (genes, genetic variation, proteins) to biochemical pathways (chemical reactions and processes requiring multiple molecular steps), cell–cell interactions, and effects on the brain and behavior. Figure 2 shows an example of how genes and genetic variation in one gene (CYP2A6, which encodes a protein that metabolizes nicotine) are connected by a path of concepts and relationships at increasingly larger biological scales, and ultimately to smoking-related behaviors. These causal relationships (such as catalyzes and influences) and hierarchical class–subclass relationships (such as between nicotine_catabolic_process and specific chemical reactions) specify the mechanism by which genetic variation in nicotine-metabolizing genes is believed to affect phenotypes that concern regular_tobacco_use.
Fig. 2. Example ontology representation of candidate genotype–phenotype relationships.
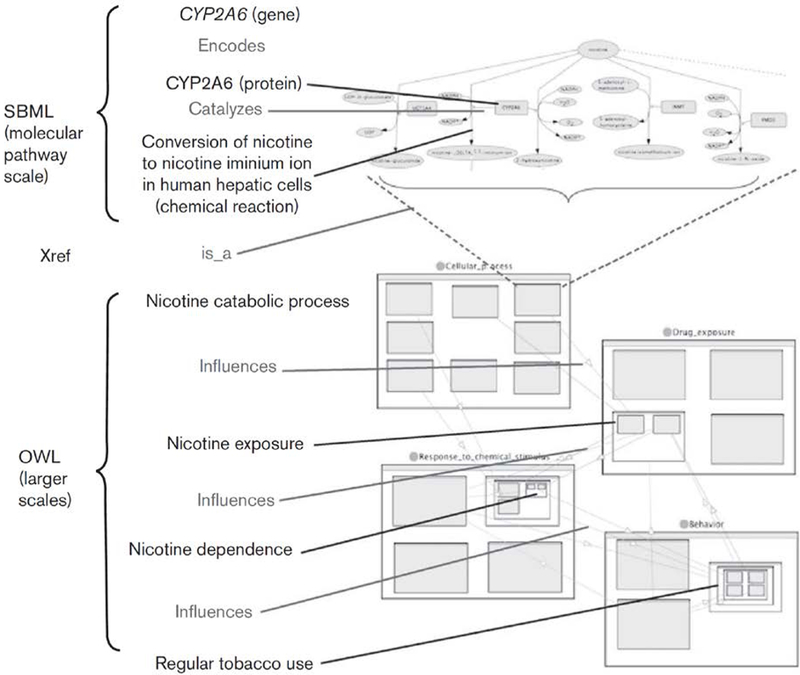
A chain of concepts (black) and relationships (blue) connects CYP2A6 genotype to the phenotypes involving the concept regular_tobacco_use, such as time to first cigarette and number of cigarettes per day (see Ontology representation of phenotypes). Molecular reactions and interactions are represented using systems biology markup language (SBML); higher-level concepts and relationships are represented using web ontology language (OWL). The hierarchical, nested relationships in OWL are depicted with nested rectangles; for example, nicotine_dependence is a nested subclass of response_to_chemical_stimulus.
In this case, the genetic variant is a difference in the molecular sequence of the DNA of the CYP2A6 gene (either a regulatory or coding element). If the sequence difference results in a difference in the amount or activity of CYP2A6 in catalyzing the conversion of nicotine, this difference may result in a difference in the rate of metabolism of nicotine. Thus, physical interactions can be considered a causal mechanism for differences to propagate to subsequent steps in the pathway. In the larger scales of the system, the ontology asserts that changes in a nicotine_catabolic_process (occurring in the liver) can be propagated to changes in nicotine_exposure, which is defined as the amount of nicotine at the site of pharmacological effect (the brain); then to an effect on nicotine_dependence and then regular_tobacco_use. For clarity, Fig. 2 traces only one of the paths leading from CYP2A6 genotype to possible phenotypic effects, but in the entire risk model, paths of potential causal influence fork and intersect to form a network structure. A depiction of part of the larger network structure is shown in Fig. 3.
Fig. 3. Major concepts and relationships in the Smoking Behavior Risk Ontology.
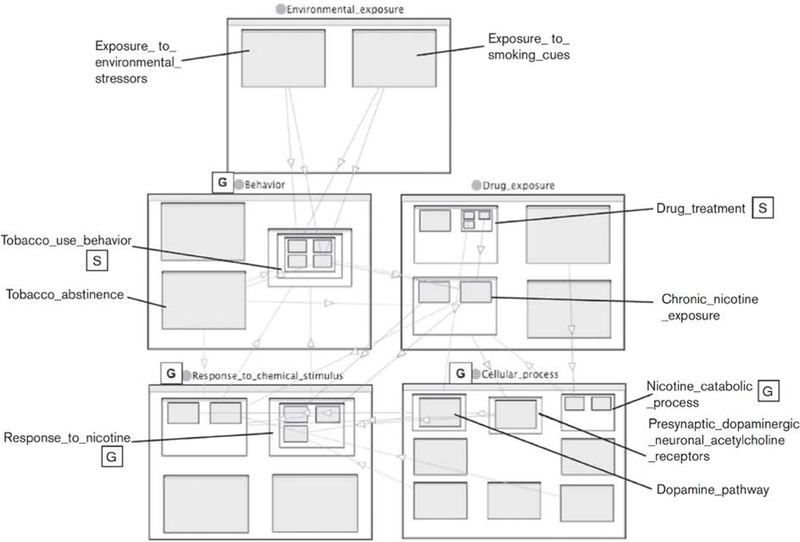
The main highest-level classes are environmental_exposure, behavior, chemical_exposure, cellular_process and response_to_chemical_stimulus. Subclasses are shown as nested boxes. Relations are represented as directed lines. The nested representation was constructed using the Jambalaya viewer [63] in the Protege ontology editor [32]. Classes taken from other ontologies are labeled with boxed letters: G, Gene Ontology; S, SO-Pharm.
Practical uses of the ontology
Shared representation of a complex scientific theory
As described above, the aim of the SBRO is a formal representation of the current hypotheses of how genes and genetic variation causally relate to observed phenotypes at the individual level. Building an ontology is not trivial, but we believe that the benefits outweigh these costs, especially in the long run. The main point is that complex diseases and traits are exactly that: complex. Complex traits will be influenced by more than one risk factor, and these factors may interact with each other. As more and more knowledge becomes available about the biological factors as well as environmental factors, it becomes increasingly difficult for one individual scientist to have deep expertise in all the areas with relevance to a given complex trait. As a result, genetic analysis of complex traits can benefit from computational models of the system, and these models must integrate knowledge from multiple experts.
For both domain experts and non-experts alike, one practical use of an ontology is to provide a condensed representation of a broad field of knowledge; in the case of the SBRO, known and hypothesized relationships connecting genetic and environmental factors to smoking behavior. Ontology visualization tools such as Jambalaya [63] or OBO-Edit [64], and molecular pathway visualization tools such as CellDesigner [65], are a tremendous aid in this use. In this way, an ontology can play an important role in systems biology research, which because of its breadth must be a collaborative exercise. It is a way to share expertise across a collaborative, interdisciplinary group.
Controlled vocabulary for comparing data across studies
An ontology has other useful applications beyond integration and visualization of a complex network of causal relationships. On the more mundane side, an ontology provides a controlled vocabulary for annotating the genotype and phenotype data collected and analyzed in genetic association studies. In the semantic web paradigm, these annotations provide metadata tags that describe the data and provide a basis for improved data searches and integration [66]. Specifically for genetic association studies, the use of controlled annotations can greatly facilitate meta-analysis (analysis spanning multiple studies) [67], by allowing a computer to recognize when different studies address the same, or similar, phenotypes.
Phenotypes are represented using the phenotype_item from SO-Pharm, as described in Materials and methods. Each phenotype_item is a combination of two terms, an ‘entity’ and a ‘quality,’ which together describe what is being measured. The entity is the object or process of interest, and the quality is the attribute of the entity that is being measured. For example, daily_cigarette_smoking is an entity (the behavior of regular smoking), and presence is a quality that can be measured (in this case, by a ‘yes’ or ‘no’). The entity is taken from SBRO, so that it can be related to the risk model. The quality should be taken from the PATO ontology (http://bioontology.org/wiki/index.php/PATO:Main_Page), or additional quality terms in SBRO. In addition, phenotype_item is associated with a measurement_method (how it was measured) and a clinical_trial_event (when it was measured).
In Table 3, we give examples of four smoking-related variables available from three different studies from dbGAP (http://www.ncbi.nlm.nih.gov/sites/entrez?db=gap). There are only two smoking phenotypes that span all three studies: current smoker versus current nonsmoker. However, there is an additional phenotype across both the National Eye Institute and Framingham studies; current nonsmokers can be subdivided into former smokers (the behavior smoking_abstinence is defined as following a period of regular smoking) and ‘never smokers’ (smoking_naive). This information is encoded quite differently in variables from the three studies, and mapping to the ontology allows us to recognize not only identical phenotypes, but also relationships between different phenotypes (e.g. all phenotypes in Table 3 are subclasses of regular_ tobacco_use).
Table 3.
Some smoking-related phenotypes from three genetic studies deposited in dbGAP, mapped to a combination of ontology terms
| dbGAP phenotype |
Ontology mapping |
||||
|---|---|---|---|---|---|
| Study (dbGAP accession) |
Variable (dbGAP accession) |
Value | Entity (SBRO concept) | Quality (PATO concept) |
Clinical trial event (SO-Pharm, SBRO) |
| GAIN: major depression (phv00020.v1.p1) | Smoker (phv00020565.v1.p1) | 0 | daily_cigarette_smoking | presence | baseline |
| GAIN: major depression (phv00020.v1.p1) | Smoker (phv00020565.v1.p1) | 1 | daily_cigarette_smoking | presence | baseline |
| NEI age-related eye disease study (phv000001.v1.p1) | smk00 (phv00000072.v1.p1) | 1 | smoking_naive | presence | baseline |
| NEI age-related eye disease study (phv000001.v1.p1) | smk00 (phv00000072.v1.p1) | 2 | smoking_abstinence | presence | baseline |
| NEI age-related eye disease study (phv000001.v1.p1) | smk00 (phv00000072.v1.p1) | 3 | daily_cigarette_smoking | presence | baseline |
| Framingham SHARe Main Exams (phv000008.v2.p1) | Smoking now (phv00000072.v1.p1) | 0 | daily_cigarette_smoking | presence | baseline |
| Framingham SHARe Main Exams (phv000008.v2.p1) | Smoking now (phv00000072.v1.p1) | 1 | daily_cigarette_smoking | presence | baseline |
| Framingham SHARe Main Exams (phv000008.v2.p1) | Stopped smoking last year or longer (phv00007749.v1.p1) |
0 | daily_cigarette_smoking | presence | baseline |
| Framingham SHARe Main Exams (phv000008.v2.p1) | Stopped smoking last year or longer (phv00007749.v1.p1) |
1 | continuous_tobacco_ abstinence_without_ relapse | presence | baseline |
| Framingham SHARe Main Exams (phv000008.v2.p1) | Stopped smoking last year or longer (phv00007749.v1.p1) |
8 | smoking_naive | presence | baseline |
Phenotypes with exactly the same SBRO concept, PATO quality concept, and clinical_trial_event concept are the same, and could potentially be combined in a meta-analysis across the respective studies.
NEI, National Eye Institute; PATO, Phenotype and Trait Ontology; SBRO, Smoking Behavior Risk Ontology; SO-Pharm, Suggested Ontology for Pharmacogenetics.
Mapping phenotypes to an ontology does present challenges. Similar to other ‘ontology annotation’ projects, such as the GO annotation [68,69], it is not trivial to choose concepts from the ontology consistently between different individual annotators. Even with consistent usage of concepts, there is inevitably loss of information when using a restricted set of discrete concepts. In the limited number of ontology annotations we have attempted for smoking-related phenotypes, a serious challenge was how to relate clinical time points over different studies, except for the baseline (pretrial) measurements.
Hypothesis generation in large-scale genetic analysis
On the more cutting-edge side, an ontology enables a computer to automatically generate and prioritize hypotheses to be tested in association studies [70]. This is important for overcoming the multiple-testing issues that affect analysis of multiple genetic markers, for example, genome-wide association studies or large candidate gene studies. A number of similarly motivated approaches have been proposed, in which some combination of association P value and ‘biological plausibility’ is used to prioritize candidate markers for validation studies. For example, Saccone et al. [12] used a weighted P value approach to prioritize genes that are good biological candidates for involvement in nicotine dependence, relative to genes with no a priori evidence of such involvement. The problem of prioritizing which hypotheses to test becomes even more pronounced when interactions between different factors are considered [for N genes there are N(N–1)/2 potential gene–gene interactions]. One application of the SBRO is to guide such prioritization.
There are many ways one might use knowledge of the system network to prioritize hypotheses. We briefly discuss two relatively intuitive possibilities. The first is in prioritization of main effects of genes on an outcome. This is quite straightforward. Each gene can be defined as on path or off path, where on path genes can be connected by causal relationships and other concepts to the outcome of interest. These types of prioritization can also make use of additional information that can be encoded into the ontology about the relative importance of different paths. Genes could then be weighted according to the weights of the relationships connecting the intervention and outcome. For example, the likelihood of relapse in a smoker can be influenced both by a desire to avoid withdrawal – an effect of chronic nicotine exposure – as well as by a desire for acute nicotine effects such as mild cognitive enhancement. However, most experts would hypothesize that the likelihood of relapse tends to be influenced more by nicotine’s chronic effects on the smoker than by the generally mild acute effects of nicotine.
The prioritization of interaction effects is more complicated. However, some simple rules for prioritizing potential nonadditive effects have recently been suggested from analysis of synthetic lethal gene knockouts in yeast, together with known protein–protein interaction networks [71]. One rule is that direct physical interaction of two gene products in a multi-subunit protein complex increases the probability of a genetic interaction (epistasis). Perhaps less obviously, this study also found that more genetic interactions were explained by ‘between pathway’ mechanisms than ‘within pathway’ mechanisms. This result makes sense if there is partial redundancy in the system; for example, where one pathway can partially compensate for another in single knockouts, but knockouts in both pathways cause system failure. This leads to the general prioritization guideline that two genetic variants in parallel or alternate paths through the network may be more likely to result in nonadditive effects than two variants along a single, serial path.
Validating the ontology
To assess the utility of the causal hypotheses (‘statements’) made by the SBRO, we performed an evaluation of these statements. We first generated two sets of statements: one comprising all of the actual statements made by the SBRO, and one comprising all of the statements made by a random ontology containing the same terms and relational connectivity as the SBRO. Each statement has the structure concept_A relation concept_B. We created a random ontology using the method of Kelley et al. [72], randomly relabeling both concepts (nodes) and relationships (edges) separately. We then randomly selected 50 statements from each ontology, mixed them together randomly, and gave the resulting list of 100 statements to two experts in both smoking behavior and genetics (only one of whom was involved in the development of SBRO), who were blinded to whether the statements came from the actual SBRO, or the randomized ontology. Each expert rated each statement as either true or false.
The two experts were in agreement for 90% of the statements. Of the 10 disagreements, nine were from the randomized ontology, and only one from the actual SBRO. The disagreements for the randomized ontology statements were split between the two experts, with neither of them showing a bias toward making more true or false judgments than the other. The larger number of disagreements for the randomized statements may reflect the fact that some of these statements could be construed as being possibly or indirectly true. For instance, one of the experts marked age influences pharmacological_treatment_compliance as true. This was not encoded in the SBRO, but this is certainly a plausible statement. The only SBRO-encoded statement that was marked false by either expert was chronic_nicotine_exposure negatively_influences response_to_lack_of_nicotine. This is a confusing statement to a human, but follows from the two SBRO statements: (i) nicotine_exposure negatively_ influences response_to_lack_of_nicotine and (ii) chronic_ nicotine_exposure subclass_of nicotine_exposure.
Using the expert ratings to provide a gold standard for true and false statements, we can calculate both the true positive rate (TPR) and false discovery rate (FDR). The TPR is how often the actual SBRO statements were judged to be true, whereas the FDR can be approximated by how often the random statements were judged to be true. The difference between these measures quantifies how much information the actual relationships provide compared with randomized relationships between the same concepts. A high TPR is expected of an expert-curated ontology; however, the FDR can also be high if the ontology is small, or if it is highly connected. The new discovery rate, or difference between TPR and FDR, is therefore much more meaningful than either alone.
If we consider only those statements for which there was complete agreement between experts, the FDR (estimated from random statements) was 44%, and the TPR (for the actual SBRO) was 100%. If we consider any statement to be false that was earlier judged false by at least one expert, the FDR was 36% and the TPR was 98%. Together, these yield a new discovery rate estimate of approximately 54–62%. For statistical model searching, use of the SBRO would enrich the true statements tested in a main effect model by approximately two to three-fold, relative to a random selection process.
Discussion
Our work represents a first attempt to build a computational infrastructure for studying a disease in a systems biology paradigm, specifically the pharmacogenetics of nicotine addiction and treatment. We have made steps toward a computational model of potentially causal relationships between genetic variation and both nicotine addiction-related phenotypes and clinical smoking cessation outcomes. This model includes biological pathways and hypotheses about how these pathways, in addition to environmental factors, may influence these phenotypes. We have encoded a disease risk model, capturing the underlying biological system and interactions with the environment, in an ontology; this is, to our knowledge, a novel application of an ontology. One of the main motivations for creating the SBRO ontology was to apply expert human knowledge to large-scale, computational genotype–phenotype association analysis.
Our work suggests that ontologies can play a role in qualitative modeling of complex systems, with a number of practical applications to genetic association studies: integrating expertise in a multidisciplinary collaborative study; communication and visualization of a succinct model of the complex network of potential relationships between genetic variation and phenotypic variation; facilitating meta-analysis over different studies; and computational hypothesis generation and prioritization for statistical tests of associations.
In addition, ontologies can be used in a more quantitative manner when combined with data obtained from observational studies. An ontology of the type we have described here, explicitly states the beliefs by domain experts regarding causal relationships between different factors in a complex system. Translation of these causal beliefs to observational studies can be as simple as annotating which factors are measured and unmeasured within the dataset. Moreover, because an ontology is a structure of the relationships that defines a directed acyclic graph, it can feed naturally into more specific causal diagrams and associated causal theory [73,74]. Causal graphs derived from the ontology can be used to identify confounders (both measured and unmeasured) and to construct algebraic formulations of assumptions and results. This, in turn, can be informative as to the most appropriate analysis technique to be used, such as generalized linear models [75], marginal structural models [76], or structural nested models [77]. Finally, ontologies integrate well with statistical methods that leverage prior external information in the analysis [70]. Bayesian hierarchical modeling [78] and Bayesian model averaging [3] offer many advantages over more conventional approaches by providing improved estimation and reflection of model uncertainty while analyzing highly correlated variables or many more variables than observations. One of the keys to these Bayesian techniques is the availability of high-quality and reliable external information [4]. Well-constructed ontologies created by the experts in the field can be a valuable source in this regard.
In the systems biology view, disease results from one or more perturbations to the ‘normal’ system [79]. These perturbations are propagated through the network comprising the parts of the system and the interactions between parts; in other words, the change in one part results in changes in one or more functions at the system level. Some genetic variations, and some environmental changes, will perturb the system so as to influence the development or progression of disease. A formal model of the network can be of help in developing hypotheses regarding the types of perturbations that may be relevant to a particular disease, particularly when the system is very complex, and how perturbations in different parts of the system may interact.
Our model of the roles of genetic variants in nicotine addiction and treatment is incomplete and will likely prove to be wrong, at least in part, as additional research improves our understanding; nevertheless, it may serve as a useful starting point for iterative refinement. We have constructed an ontology description of the genotype–phenotype network (pathways as well as higher-level terms and relations) of many risk factors for relapse during a smoking cessation clinical trial. It is hoped that our ontology can serve as both a starting point for further development by a wider community of researchers in the pharmacogenetics of nicotine addiction and treatment, as well as a model for representing causal risk factors in other diseases.
Acknowledgements
The authors thank Russ Altman, and the anonymous reviewers, for helpful comments on the manuscript. This work was supported by the National Institute on Drug Abuse, grant U01 DA020830.
References
- 1.Williamson R The molecular genetics of complex inherited diseases. Br J Cancer Suppl 1988; 9:14–16. [PMC free article] [PubMed] [Google Scholar]
- 2.Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature 2007; 449:851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Viallefont V, Raftery AE, Richardson S. Variable selection and Bayesian model averaging in case-control studies. Stat Med 2001; 20:3215–3230. [DOI] [PubMed] [Google Scholar]
- 4.Thomas DC, Witte JS, Greenland S. Dissecting effects of complex mixtures: who’s afraid of informative priors? Epidemiology 2007; 18:186–190. [DOI] [PubMed] [Google Scholar]
- 5.Giacomini KM, Brett CM, Altman RB, Benowitz NL, Dolan ME, Flockhart DA, et al. The pharmacogenetics research network: from SNP discovery to clinical drug response. Clin Pharmacol Ther 2007; 81:328–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergen AW, Conti DV, Berg DVD, Lee W, Liu J, Li D et al. Dopamine genes and nicotine dependence in treatment seeking and community smokers. Neuropsychopharmacology 2009. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conti DV, Lee W, Li D, Liu J, Van Den Berg D, Thomas PD, et al. Nicotinic acetylcholine receptor beta2 subunit gene implicated in a systems-based candidate gene study of smoking cessation. Hum Mol Genet 2008; 17:2834–2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mathers CD, Loncar D. Projections of global mortality and burden of disease from 2002 to 2030. PLoS Med 2006; 3:e442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pollock JD, Koustova E, Hoffman A, Shurtleff D, Volkow ND. Treatments for nicotine addiction should be a top priority. Lancet 2009; 24:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Swan GE, Hops H, Wilhelmsen KC, Lessov-Schlaggar CN, Cheng LS, Hudmon KS, et al. A genome-wide screen for nicotine dependence susceptibility loci. Am J Med Genet B Neuropsychiatr Genet 2006; 141B:354–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bierut LJ, Madden PA, Breslau N, Johnson EO, Hatsukami D, Pomerleau OF, et al. Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum Mol Genet 2007; 16:24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saccone SF, Hinrichs AL, Saccone NL, Chase GA, Konvicka K, Madden PA, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet 2007; 16:36–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lessov-Schlaggar CN, Pergadia ML, Khroyan TV, Swan GE. Genetics of nicotine dependence and pharmacotherapy. Biochem Pharmacol 2008; 75:178–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Swan GE, Hudmon KS, Jack LM, Hemberger K, Carmelli D, Khroyan TV, et al. Environmental and genetic determinants of tobacco use: methodology for a multidisciplinary, longitudinal family-based investigation. Cancer Epidemiol Biomarkers Prev 2003; 12:994–1005. [PMC free article] [PubMed] [Google Scholar]
- 15.USDHHS, preventing tobacco use among young people: a report of the surgeon general. 1994, Atlanta, GA: US: Department of Health and Human Services, Public Health Service, Centers for Disease Control and Prevention, National Center for Chronic Disease Prevention and Health Promotion, Office on Smoking and Health. [Google Scholar]
- 16.Dick DM, Viken R, Purcell S, Kaprio J, Pulkkinen L, Rose RJ. Parental monitoring moderates the importance of genetic and environmental influences on adolescent smoking. J Abnorm Psychol 2007; 116:213–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCaffery JM, Papandonatos GD, Lyons MJ, Koenen KC, Tsuang MT, Niaura R. Educational attainment, smoking initiation and lifetime nicotine dependence among male Vietnam-era twins. Psychol Med 2007; 22:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Audrain-McGovern J, Rodriguez D, Wileyto EP, Schmitz KH, Shields PG. Effect of team sport participation on genetic predisposition to adolescent smoking progression. Arch Gen Psychiatry 2006; 63:433–441. [DOI] [PubMed] [Google Scholar]
- 19.Coveney PV, Fowler PW. Modelling biological complexity: a physical scientist’s perspective. J R Soc Interface 2005; 2:267–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cortassa S, Aon MA, O0 Rourke B, Jacques R, Tseng HJ, Marban E, Winslow RL. A computational model integrating electrophysiology, contraction, and mitochondrial bioenergetics in the ventricular myocyte. Biophys J 2006; 91:1564–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gennari JH, Neal ML, Carlson BE, Cook DL. Integration of multi-scale biosimulation models via light-weight semantics. Pac Symp Biocomput 2008; 13:414–425. [PMC free article] [PubMed] [Google Scholar]
- 22.Kirschner DE, Chang ST, Riggs TW, Perry N, Linderman JJ. Toward a multiscale model of antigen presentation in immunity. Immunol Rev 2007; 216:93–118. [DOI] [PubMed] [Google Scholar]
- 23.Gao Z, Barth AS, DiSilvestre D, Akar FG, Tian Y, Tanskanen A, et al. Key pathways associated with heart failure development revealed by gene networks correlated with cardiac remodeling. Physiol Genomics 2008; 35:222–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rubin DL, Shah NH, Noy NF. Biomedical ontologies: a functional perspective. Brief Bioinform 2008; 9:75–90. [DOI] [PubMed] [Google Scholar]
- 25.Simon J, Dos Santos M, Fielding J, Smith B. Formal ontology for natural language processing and the integration of biomedical databases. Int J Med Inform 2006; 75:224–231. [DOI] [PubMed] [Google Scholar]
- 26.Rosse C, Mejino JL Jr. A reference ontology for biomedical informatics: the Foundational Model of Anatomy. J Biomed Inform 2003; 36:478–500. [DOI] [PubMed] [Google Scholar]
- 27.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000; 25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, et al. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat Biotechnol 2007; 25:1251–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coulet A, Smail-Tabbone M, Napoli A, Devignes M. Suggested ontology for pharmacogenomics (SO-Pharm): modular construction and preliminary testing. Proc Workshop Knowl Syst Bioinform 2006; 648–657. [Google Scholar]
- 30.Noy NF, McGuinness D. Ontology development 101: a guide to creating your first ontology. Stanford Medical Informatics Technical Report SMI-2001-0880, 2001. [Google Scholar]
- 31.Diehl AD, Lee JA, Scheuermann RH, Blake JA. Ontology development for biological systems: immunology. Bioinformatics 2007; 23:913–915. [DOI] [PubMed] [Google Scholar]
- 32.Noy NF, Crubezy M, Fergerson RW, Knublauch H, Tu SW, Vendetti J, Musen MA. Protege-2000: an open-source ontology-development and knowledge-acquisition environment. AMIA Annu Symp Proc 2003; 953:953. [PMC free article] [PubMed] [Google Scholar]
- 33.Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, Kitano H, et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics 2003; 19:524–531. [DOI] [PubMed] [Google Scholar]
- 34.Mi H, Guo N, Kejariwal A, Thomas PD. PANTHER version 6: protein sequence and function evolution data with expanded representation of biological pathways. Nucleic Acids Res 2007; 35:D247–D252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Piper ME, McCarthy DE, Baker TB. Assessing tobacco dependence: a guide to measure evaluation and selection. Nicotine Tob Res 2006; 8:339–351. [DOI] [PubMed] [Google Scholar]
- 36.Lerman C, Jepson C, Wileyto EP, Epstein LH, Rukstalis M, Patterson F, et al. Role of functional genetic variation in the dopamine D2 receptor (DRD2) in response to bupropion and nicotine replacement therapy for tobacco dependence: results of two randomized clinical trials. Neuropsychopharmacology 2006; 31:231–242. [DOI] [PubMed] [Google Scholar]
- 37.Malaiyandi V, Sellers EM, Tyndale RF. Implications of CYP2A6 genetic variation for smoking behaviors and nicotine dependence. Clin Pharmacol Ther 2005; 77:145–158. [DOI] [PubMed] [Google Scholar]
- 38.Benowitz NL, Perez-Stable EJ, Herrera B, Jacob P III. Slower metabolism and reduced intake of nicotine from cigarette smoking in Chinese-Americans. J Natl Cancer Inst 2002; 94:108–115. [DOI] [PubMed] [Google Scholar]
- 39.Heatherton TF, Kozlowski LT, Frecker RC, Fagerstrom KO. The fagerstrom test for nicotine dependence: a revision of the fagerstrom tolerance questionnaire. Br J Addict 1991; 86:1119–1127. [DOI] [PubMed] [Google Scholar]
- 40.Lerman C, Tyndale R, Patterson F, Wileyto EP, Shields PG, Pinto A, Benowitz N. Nicotine metabolite ratio predicts efficacy of transdermal nicotine for smoking cessation. Clin Pharmacol Ther 2006; 79:600–608. [DOI] [PubMed] [Google Scholar]
- 41.Patterson F, Schnoll R, Wileyto E, Pinto A, Epstein L, Shields P, et al. Toward personalized therapy for smoking cessation: a randomized placebo-controlled trial of bupropion. Clin Pharmacol Ther 2008; 2:2. [DOI] [PubMed] [Google Scholar]
- 42.Yokotani K, Okada S, Nakamura K. Characterization of functional nicotinic acetylcholine receptors involved in catecholamine release from the isolated rat adrenal gland. Eur J Pharmacol 2002; 446:83–87. [DOI] [PubMed] [Google Scholar]
- 43.Tachikawa E, Mizuma K, Kudo K, Kashimoto T, Yamato S, Ohta S. Characterization of the functional subunit combination of nicotinic acetylcholine receptors in bovine adrenal chromaffin cells. Neurosci Lett 2001; 312:161–164. [DOI] [PubMed] [Google Scholar]
- 44.Sala F, Nistri A, Criado M. Nicotinic acetylcholine receptors of adrenal chromaffin cells. Acta Physiol (Oxf) 2008; 192:203–212. [DOI] [PubMed] [Google Scholar]
- 45.Grady SR, Salminen O, Laverty DC, Whiteaker P, McIntosh JM, Collins AC, Marks MJ. The subtypes of nicotinic acetylcholine receptors on dopaminergic terminals of mouse striatum. Biochem Pharmacol 2007; 74:1235–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dani JA, Bertrand D. Nicotinic acetylcholine receptors and nicotinic cholinergic mechanisms of the central nervous system. Annu Rev Pharmacol Toxicol 2007; 47:699–729. [DOI] [PubMed] [Google Scholar]
- 47.Berrettini W, Yuan X, Tozzi F, Song K, Francks C, Chilcoat H, et al. Alpha-5/alpha-3 nicotinic receptor subunit alleles increase risk for heavy smoking. Mol Psychiatry 2008; 13:368–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Caporaso N, Gu F, Chatterjee N, Sheng-Chih J, Yu K, Yeager M, et al. Genome-wide and candidate gene association study of cigarette smoking behaviors. PLoS ONE 2009; 4:e4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ehringer MA, Clegg HV, Collins AC, Corley RP, Crowley T, Hewitt JK, et al. Association of the neuronal nicotinic receptor beta2 subunit gene (CHRNB2) with subjective responses to alcohol and nicotine. Am J Med Genet B Neuropsychiatr Genet 2007; 144B:596–604. [DOI] [PubMed] [Google Scholar]
- 50.Hoft NR, Corley RP, McQueen MB, Schlaepfer IR, Huizinga D, Ehringer MA. Genetic association of the CHRNA6 and CHRNB3 genes with tobacco dependence in a nationally representative sample. Neuropsychopharmacology 2009; 34:698–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hutchison KE, Allen DL, Filbey FM, Jepson C, Lerman C, Benowitz NL, et al. CHRNA4 and tobacco dependence: from gene regulation to treatment outcome. Arch Gen Psychiatry 2007; 64:1078–1086. [DOI] [PubMed] [Google Scholar]
- 52.Li MD, Beuten J, Ma JZ, Payne TJ, Lou XY, Garcia V, et al. Ethnic- and gender-specific association of the nicotinic acetylcholine receptor alpha4 subunit gene (CHRNA4) with nicotine dependence. Hum Mol Genet 2005; 14:1211–1219. [DOI] [PubMed] [Google Scholar]
- 53.Schlaepfer IR, Hoft NR, Collins AC, Corley RP, Hewitt JK, Hopfer CJ, et al. The CHRNA5/A3/B4 gene cluster variability as an important determinant of early alcohol and tobacco initiation in young adults. Biol Psychiatry 2008; 63:1039–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature 2008; 452:638–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weiss RB, Baker TB, Cannon DS, von Niederhausern A, Dunn DM, Matsunami N, et al. A candidate gene approach identifies the CHRNA5-A3-B4 region as a risk factor for age-dependent nicotine addiction. PLoS Genet 2008; 4:e1000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zeiger JS, Haberstick BC, Schlaepfer I, Collins AC, Corley RP, Crowley TJ, et al. The neuronal nicotinic receptor subunit genes (CHRNA6 and CHRNB3) are associated with subjective responses to tobacco. Hum Mol Genet 2008; 17:724–734. [DOI] [PubMed] [Google Scholar]
- 57.Lerman C, Caporaso NE, Audrain J, Main D, Bowman ED, Lockshin B, Boyd NR. Evidence suggesting the role of specific genetic factors in cigarette smoking. Health Psychol 1999; 18:14–20. [DOI] [PubMed] [Google Scholar]
- 58.Wise RA. Dopamine and reward: the anhedonia hypothesis 30 years on. Neurotox Res 2008; 14:169–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Le Foll B, Gallo A, Le Strat Y, Lu L, Gorwood P. Genetics of dopamine receptors and drug addiction: a comprehensive review. Behav Pharmacol 2009; 20:1–17. [DOI] [PubMed] [Google Scholar]
- 60.Ontology Smith B.. In: Floridi L, editor. Blackwell guide to the philosophy of computing and information. Oxford: Blackwell; 2003. pp. 155–166. [Google Scholar]
- 61.Rubin DL, Lewis SE, Mungall CJ, Misra S, Westerfield M, Ashburner M, et al. National Center for Biomedical Ontology: advancing biomedicine through structured organization of scientific knowledge. Omics 2006; 10:185–198. [DOI] [PubMed] [Google Scholar]
- 62.Karp PD. Pathway databases: a case study in computational symbolic theories. Science 2001; 293:2040–2044. [DOI] [PubMed] [Google Scholar]
- 63.Storey MA, Musen MA, Noy NF, Best C, Fergerson RW, Silva J. Jambalaya: an interactive environment for exploring ontologies. Int Conf Intell User Interfaces 2002; 1. [Google Scholar]
- 64.Day-Richter J, Harris MA, Haendel M, Lewis S. OBO-Edit–an ontology editor for biologists. Bioinformatics 2007; 23:2198–2200. [DOI] [PubMed] [Google Scholar]
- 65.Kitano H, Funahashi A, Matsuoka Y, Oda K. Using process diagrams for the graphical representation of biological networks. Nat Biotechnol 2005; 23:961–966. [DOI] [PubMed] [Google Scholar]
- 66.Splendiani A RDFScape: semantic web meets systems biology. BMC Bioinformatics 2008; 9 (Suppl 4):S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lohmueller KE, Pearce CL, Pike M, Lander ES, Hirschhorn JN. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat Genet 2003; 33:177–182. [DOI] [PubMed] [Google Scholar]
- 68.Barrell D, Dimmer E, Huntley RP, Binns D, O0Donovan C, Apweiler R. The GOA database in 2009 – an integrated gene ontology annotation resource. Nucleic Acids Res 2009; 37:D396–D403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hong EL, Balakrishnan R, Dong Q, Christie KR, Park J, Binkley G, et al. Gene ontology annotations at SGD: new data sources and annotation methods. Nucleic Acids Res 2008; 36:D577–D581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Conti DV, Lewinger JP, Swan GE, Tyndale RF, Benowitz NL, Thomas PD. Using ontologies in hierarchical modeling of genes and exposures in biologic pathways, in NCI monograph 22: phenotypes, endophenotypes, and genetic studies of nicotine dependence. In: Swan GE, editor. 2009. (in press). [Google Scholar]
- 71.Kelley R, Ideker T. Systematic interpretation of genetic interactions using protein networks. Nat Biotechnol 2005; 23:561–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kelley BP, Sharan R, Karp RM, Sittler T, Root DE, Stockwell BR, Ideker T. Conserved pathways within bacteria and yeast as revealed by global protein network alignment. Proc Natl Acad Sci U S A 2003; 100:11394–11399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Greenland S, Pearl J, Robins JM. Causal diagrams for epidemiologic research. Epidemiology 1999; 10:37–48. [PubMed] [Google Scholar]
- 74.Pearl J Causality: models, reasoning and inference. London: Cambridge University Press; 2000. [Google Scholar]
- 75.McCullagh P, Nelder JA. Generalized linear models. Boca Raton: Chapman; 1989. [Google Scholar]
- 76.Robins JM, Hernan MA, Brumback B. Marginal structural models and causal inference in epidemiology. Epidemiology 2000; 11:550–560. [DOI] [PubMed] [Google Scholar]
- 77.Robins JM, Blevins D, Ritter G, Wulfsohn M. G-estimation of the effect of prophylaxis therapy for Pneumocystis carinii pneumonia on the survival of AIDS patients. Epidemiology 1992; 3:319–336. [DOI] [PubMed] [Google Scholar]
- 78.Greenland S Principles of multilevel modelling. Int J Epidemiol 2000; 29:158–167. [DOI] [PubMed] [Google Scholar]
- 79.Hood L, Heath JR, Phelps ME, Lin B. Systems biology and new technologies enable predictive and preventative medicine. Science 2004; 306:640–643. [DOI] [PubMed] [Google Scholar]


