Although gliomas are thought to arise from a neural-glial lineage of ectodermal origin, many gliomas express a mesenchymal signature. Behnan et al. review the origins of this signature in glioma: it may arise via the tumour stroma, via NF1-mutation in tumour cells and be influenced by the cell of origin, or arise in response to radiotherapy/chemotherapy/anti-angiogenic treatment.
Keywords: glioma, mesenchymal subtype, subtype origin, tumor microenvironment, proneural-mesenchymal transition
Abstract
The complexity of glioblastoma multiforme, the most common and lethal variant of gliomas, is reflected by cellular and molecular heterogeneity at both the inter- and intra-tumoural levels. Molecular subtyping has arisen in the past two decades as a promising strategy to give better predictions of glioblastoma multiforme evolution, common disease pathways, and rational treatment options. The Cancer Genome Atlas network initially identified four molecular subtypes of glioblastoma multiforme: proneural, neural, mesenchymal and classical. However, further studies, also investigated glioma stem cells, have only identified two to three subtypes: proneural, mesenchymal and classical. The proneural–mesenchymal transition upon tumour recurrence has been suggested as a mechanism of tumour resistance to radiation and chemotherapy treatment. Glioblastoma multiforme patients with the mesenchymal subtype tend to survive shorter than other subtypes when analysis is restricted to samples with low transcriptional heterogeneity. Although the mesenchymal signature in malignant glioma may seem at odds with the common idea of the ectodermal origin of neural-glial lineages, the presence of the mesenchymal signature in glioma is supported by several studies suggesting that it can result from: (i) intrinsic expression of tumour cells affected with accumulated genetic mutations and cell of origin; (ii) tumour micro-environments with recruited macrophages or microglia, mesenchymal stem cells or pericytes, and other progenitors; (iii) resistance to tumour treatment, including radiotherapy, antiangiogenic therapy and possibly chemotherapy. Genetic abnormalities, mainly NF1 mutations, together with NF-κB transcriptional programs, are the main driver of acquiring mesenchymal-signature. This signature is far from being simply tissue artefacts, as it has been identified in single cell glioma, circulating tumour cells, and glioma stem cells that are released from the tumour micro-environment. All these together suggest that the mesenchymal signature in glioblastoma multiforme is induced and sustained via cell intrinsic mechanisms and tumour micro-environment factors. Although patients with the mesenchymal subtype tend to have poorer prognosis, they may have favourable response to immunotherapy and intensive radio- and chemotherapy.
Glioma
Malignant primary brain tumours are the leading cause of death in children with cancers, and the third leading cause of death in adults aged 15–34 years (Surawicz et al., 1999; Buckner et al., 2007). Gliomas are tumours of the CNS that make up ∼80% of all malignant brain tumours (Goodenberger and Jenkins, 2012). In the last century, the diagnosis of gliomas was based on microscopic analysis of histology features and the level of differentiation. The WHO grouped gliomas into four main groups (grades I–IV) depending on histological features (cellularity, mitotic figures, necrosis, and vascular proliferation) and astrocytic and oligodendroglial phenotype (Louis et al., 2007). Astrocytoma grade IV, mostly known as glioblastoma multiforme (GBM), represents the most aggressive subtype of malignant brain tumour with a 12–16 month median overall survival (Stupp et al., 2005; Wen and Kesari, 2008). However, this histological classification did not predict the clinical outcomes of gliomas well enough. Major revision was needed to update the century-old diagnostic principle by incorporating the microscopy analysis with the molecular parameters such as IDH status, ATRX loss, H3K27M mutation, TP53 mutation, and 1p/19q co-deletion (Bent, 2010; Louis et al., 2016). The WHO revised classification of brain tumours (2016 CNS WHO) making some major changes. Diffuse astrocytic and oligodendroglial tumours are no longer separated at they were in the 2007 classification. Now, these tumours are grouped together according to their growth pattern and IDH1/IDH2-drived genetic mutations (Louis et al., 2016).
Glioblastoma multiforme molecular subtypes
In 2006, Phillips et al. proposed subtyping of gliomas into three subtypes based on gene expression profiling: proneural, proliferative, and mesenchymal. They found a strong association between tumour grade and subtypes regardless of the oligodendroglial or astrocytic morphology (Phillips et al., 2006). The Cancer Genome Atlas (TCGA) Research Network started a comprehensive genomic study targeting 33 different cancers. GBM and lower grade glioma were among their top targets. Their efforts resulted in deep genomic characterization and molecular subtypes of nearly 600 GBMs and 516 lower grade human gliomas (Wang et al., 2017b).
The first TCGA pilot study identified new GBM genomic alterations such as homozygous deletions of NF1 and PARK2, and amplifications of AKT3, in addition to previously known amplifications and deletions. GBM treated patients had higher mutation rates than untreated, ∼5.8 versus 1.4 somatic silent mutations per sample, respectively. The MGMT promoter methylation, a predictive marker for alkylating agent treatment, induced a hypermutated GBM phenotype (Hegi et al., 2005; Cancer Genome Atlas Research, 2008). Combining sequencing data with gene expression and DNA methylation identified the core biological pathways involved in GBM. RTK/RAS/PI3K was activated in 88% of the samples and p53-signalling was altered in 87% while RB signalling was altered in 78% of GBM samples (Cancer Genome Atlas Research, 2008). TCGA integrated gene expression data from three different platforms into a single unified dataset and identified 840 genes that classified GBM into four subtypes: proneural, mesenchymal, classical, and neural (Verhaak et al., 2010). However, the human GBM tissues that were subcutaneously maintained and propagated in athymic null/null mice (Hodgson et al., 2009), were classified into three subtypes only: proneural, mesenchymal, and classical (Verhaak et al., 2010). The proneural subtype was associated with IDH1 mutation, TP53, PDGFRA amplification and/or mutations (Noushmehr et al., 2010; Verhaak et al., 2010). The classical subtype showed the highest expression of EGFR amplification (95%) compared to other subclasses. Also, 95% of them exhibit CDKN2A (Ink4a/ARF) homozygous deletion. This class lacked IDH1, TP53, PDGFRA and NF1 abnormalities that were common in the proneural and mesenchymal subtypes (Verhaak et al., 2010). Interestingly, chr7 amplification and chr10 deletion, the most common GBM abnormalities, were low in the proneural subtype (20–54%), high in mesenchymal samples (>75%), and highest in the classical subtype of Verhaak et al. (2010) (93%). Phillips et al. (2006) and Verhaak et al. (2010) found that patients with the proneural subtype were younger than patients in other subtypes and tended to survive longer. However, Sturm et al. (2012) showed that the favourable outcome of the proneural GBM subtype was because patients were IDH mutant. When those patients are excluded from analysis, the proneural subtype has a worse prognosis than other subtypes (Sturm et al., 2012). The proneural subtype was also different in terms of having normal EGFR expression, intact PTEN, NOTCH activation (Phillips et al., 2006). Markers of oligodendrocytic development, PDGFRA, NKX2–2, and OLIG2 were highly expressed in this subtype (Verhaak et al., 2010). The mesenchymal subtype was associated with a high frequency of NF1 abnormalities (Phillips et al., 2006; Verhaak et al., 2010), expressed high levels of S100A1, CHI3L1, MET and microglia markers CD68, PTPRC, and TNF, associated with inflammatory, wound healing, and NF-κB signalling pathways, in addition to a higher degree of necrosis (Phillips et al., 2006; Verhaak et al., 2010). Most secondary GBMs and >75% of low grade and grade III gliomas were classified as the proneural subtype (Phillips et al., 2006; Verhaak et al., 2010). Notably, comparing the Phillips classification with 35 genes, to the Verhaak classification with 840 genes, only three mesenchymal genes (CHI3L1/YKL40, SERPINE1 and TIMP1) and five proneural genes (DLL3, KLRC3, SCG3, C20orf42, and NCAM1) were common (Phillips et al., 2006; Verhaak et al., 2010).
Using gene expression and DNA methylation profiles, Brennan et al. (2013) classified 396 GBMs into six methylation groups [clusters M1, M2, M3, M4, glioma CpG island methylator phenotype (G-CIMP), and M6]. The mesenchymal subtype was enriched in the M1 cluster (60%) and classical in the M3 cluster (58%), while the G-CIMP cluster contained mainly the proneural subtype and was associated with somatic mutations (IDH1, TP53, ATRX, MYC). The proneural subtype patients who were G-CIMP+ were younger (41 versus 58 years of age) and survived longer than proneural patients who were G-CIMP−. All IDH mutant tumours were G-CIMP+ (Brennan et al., 2013). This in line with Turcan et al. who established the role of IDH1-mutation in inducing a distinct G-CIMP phenotype commonly seen in lower grade glioma and proneural subtype GBMs through altering the oncometabolite 2-hydroxyglutarate (2-HG) production and reorganizing the methylome and transcriptome landscape (Lu et al., 2012a; Turcan et al., 2012). Nevertheless, the G-CIMP+ patients who lacked IDH mutations were not significantly different, in age and survival, from G-CIMP+ patients who harboured IDH mutations, suggesting that their favourable survival in the proneural subtype is related to G-CIMP rather than IDH status. Although DNA methylation of the MGMT gene promotor (a gene that encodes O-6-methylguanine-DNA methyltransferase) has been associated with longer survival after temozolomide therapy in primary GBM, DNA methylation of this gene was correlated with a treatment response only in the classical subtype, but not proneural or mesenchymal subtypes (Hegi et al., 2005; Brennan et al., 2013). The TCGA proteomic profiling showed that EGFR amplification or mutation and phosphorylation were prominent in the classical subtype. In agreement with Phillips et al., they confirmed that the mesenchymal subtype expressed higher levels of the endothelial markers CD31 and VEGFR2. Furthermore, inflammatory markers such as fibronectin and COX2 were elevated in mesenchymal subtype compared to other subtypes (Brennan et al., 2013).
The last study published by TCGA in 2017 looked at the intrinsic gene signature of brain tumour subtypes, correlating GBM subtypes with the immune micro-environment, and shedding light on treatment-induced phenotypic tumour changes in recurrent tumours (Wang et al., 2017b). This study excluded IDH-mutant GBMs for its favourable survival effect on the proneural subtype (Sturm et al., 2012). Unsupervized clustering, after several steps of gene filtering to exclude environmental factors, resulted in three subtypes: proneural, mesenchymal, and classical, while neural signature was not enriched in any cluster (Wang et al., 2017b). Indeed, the neural subtype was not detected by us and others, and could be due to contamination with normal cells (Sturm et al., 2012; Gill et al., 2014; Behnan et al., 2017a). The final gene signature of the three subtypes consists of 150 genes (50 genes/subtype). Around 42–54% of the classification genes were shared with the old TCGA subtype of 840 genes (Verhaak et al., 2010; Wang et al., 2017b). The subtypes showed a high concordance through different platforms, up to 93% concordance between RNA-seq and Affymetrix-U133A and 85% in a small sample size of 10 GBMs selected randomly. The intra-tumoural transcriptional heterogeneity on the single cell level was captured by the matching tumour bulk signature. The tumour bulks had the same subtype as the majority of their single cells in four of five samples (Wang et al., 2017b). These subtypes were associated with different immune signatures (Bao et al., 2006; Engler et al., 2012; Ye et al., 2012; Gabrusiewicz et al., 2016; Wang et al., 2017b). When subtype stability was compared in 91 wild-type IDH-paired samples of primary and recurrent gliomas, 55% kept the same subtype upon recurrence. The mesenchymal subtype was the most stable subtype (with only 35% changed to another subtype), while the proneural subtype recorded highest subtype shifting (59%). The recurrent tumours showed a decrease in immune signature compared to their primary tumours. Both primary and recurrent tumours with mesenchymal subtype tended to have worse survival compared to proneural and classical subtypes upon restricting sample analysis to those with low transcriptional heterogeneity that activate one subtype (samples with high simplicity score) (Wang et al., 2017b). Behnan et al.rsquo;s classification set was obtained by filtering differentially expressed genes between glioma stem cells (GSCs) of proneural and mesenchymal subtypes that grew adherently under neuro-sphere conditions and crossing them with the TCGA gene set to get the genes shared between GSCs and GBM tissues. Of the 118 differentially expressed genes in GSCs, only 12 genes were common with the GBM-TCGA classification set of 840 genes. Surprisingly, these 12 genes were able to classify all TCGA samples in good concordance with the 840-gene-dependent classification. The 12 genes classified GBM into three subtypes: proneural, identified by P2RX7, STMN4, SOX10 and ERBB3; classical, by ACSBG1 and KCNF1; and mesenchymal, by S100A, DAB2, TGFB1, THBS1, COL1A2 and COL1A1 (Behnan et al., 2017a). The relevance of this 12-gene classification also relies in its intrinsic signature, being identified in cancer stem cells and tumour tissues. However, despite several interesting findings of TCGA consortium in the glioma field, its clinical implications remain debatable (Box 1 presents the main outcomes and weaknesses of large-scale gene expression studies).
Box 1.
The main outcomes and impact of TCGA large-scale gene expression studies
| Main outcomes of TCGA consortium in glioma | Impact and weaknesses |
|---|---|
| In general | |
|
|
| Key findings | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
The mesenchymal signature in glioblastoma multiforme
About 30–49% of GBM tissues have been classified as the mesenchymal subtype. Patients with this subtype, both with primary and recurrent tumours, tend to have the worst survival rates compared to other subtypes. Despite this signature having been identified in GBM tissues at the beginning, it is far from being a tissue artefact, as single cell GBM, GSCs, and circulating brain tumour cells expressed the mesenchymal signature as well.
The mesenchymal signature in glioblastoma multiforme tissue
The mesenchymal signature in GBM tissue was suggested by Phillips et al. (2006), where 49% of the study cohort samples were classified as mesenchymal subtype and were associated with poor survival compared to the proneural subtype. Later, TCGA classified GBM tissues into four molecular subtypes: proneural, neural, mesenchymal and classical, where mesenchymal patients constituted 29–30% of GBM samples in both the primary and validation set (200 and 246 GBMs, respectively) (Verhaak et al., 2010). The mesenchymal subtype is one of the most consistent subtypes described in the literature, in both GBM tissues and culture-enriched GSCs (Verhaak et al., 2010; Bhat et al., 2013; Behnan et al., 2017a; Wang et al., 2017b). Upon integrating micro-environmental factors and using single cell analysis, the new TCGA classification confirmed the subtype segregation into three subtypes rather than four, with mesenchymal subtype constituting ∼34% of the samples (Wang et al., 2017b). Furthermore, these three subtypes were relevant to subtype even lower grade gliomas (Guan et al., 2014; Ceccarelli et al., 2016). The mesenchymal subtype samples express mesenchymal markers and are negative/downregulate proneural markers (Supplementary Table 1) (Phillips et al., 2006; Verhaak et al., 2010; Riddick and Fine, 2011; Bhat et al., 2013; Jin et al., 2017). However, some mesenchymal tumours express the proneural marker, OLIG2, and others do not express the mesenchymal marker, CHI3L1 (Bhat et al., 2013). Mesenchymal tumours are predominantly IDH wild-type and G-CIMP−, contrary to the proneural subtype (Noushmehr et al., 2010; Verhaak et al., 2010). GBMs that express high mesenchymal signature, even when the analysis was limited to wild-type IDH, were associated with poor radiation response and worse survival (Bhat et al., 2013). Mesenchymal tumours expressed higher levels of angiogenic markers, in addition to higher levels of necrosis (Phillips et al., 2006; Verhaak et al., 2010). Genetic abnormalities and tumour micro-environment have been shown to drive mesenchymal subtype tumours (Verhaak et al., 2010; Wang et al., 2017b). Furthermore, the epigenetic chromatin modifiers EZH2 and BMI1 were shown to have differential expression between the mesenchymal and proneural subtypes, keeping in mind that 40% of GBMs harbour mutations in BMI1 and EZH2 (Brennan et al., 2013; Jin et al., 2017). The proneural subtype tumours expressed a higher level of EZH2 compared to mesenchymal and classical subtypes. This expression was positively correlated with OLIG2 and mature vascular signature expression (proneural marker). Mesenchymal tumours expressed a higher level of BMI1 and its expression was positively correlated with CD44 expression and micro-environment in mesenchymal and classical subtypes (Jin et al., 2017). Correlating gene expression with the survival data showed that GBM patients with low EZH2 and BMI1 expression have the best survival, while those with upregulated expression of both genes have the worst survival, and those who express either EZH2 or BMI1 have intermediate survival (Jin et al., 2017). This indicates that patients with more heterogeneous tumours have unfavourable survival. Also, recently it was shown that ZBTB18 promotor methylation, which is strongly correlated with wild-type IDH1, acts as a negative regulator of the mesenchymal subtype, and ZBTB18 loss is associated with poor prognosis and an aggressive tumour phenotype (Fedele et al., 2017).
The mesenchymal signature in glioma stem cells
GSCs or glioma initiating cells, were first reported in 2003 (Singh et al., 2003). GSCs are known as a small population of stem cells that exist in fresh tumours and are characterized by self-renewal and expression of stemness markers and multi-lineage differentiation properties that give rise to other types of cells in the tumour, in addition to in vivo tumour formation ability (Lathia et al., 2015). While the origin and clear definition of GSCs are disputable, it is well established that these cells are responsible for maintaining tumour growth and invasion, and cause tumour recurrence because of their chemo- and radio-resistant properties (Bao et al., 2006; Liu et al., 2006; Chen et al., 2012). Vascular niche in addition to other micro-environmental factors, such as hypoxia, nutrients, and acid, affect the distribution of these cells (Bao et al., 2006; Calabrese et al., 2007; Li et al., 2009; Hjelmeland et al., 2011; Flavahan et al., 2013). Chemo- and radiotherapy are thought to enrich for GSCs (Bao et al., 2006; Liu et al., 2006). Because of the heterogeneity among patient tumours and within the same tumour, there is no unique marker that can distinguish GSCs from non-GSCs. We have recently suggested a panel of markers that distinguish GSCs from non-tumorigenic stromal cells and cover inter-tumoural heterogeneity: CD56+SOX2+SOX9+CD133+CD15+CD248−CD105−αSMA− (Behnan et al., 2017a). Molecular subtyping has shown that GSCs have two to three subtypes, proneural, mesenchymal and classical, while the neural subtype is absent in GSCs. In fact, only normal oligodendrocytes were classified as neural (Huse et al., 2011; Zheng et al., 2012; Bhat et al., 2013; Patel et al., 2014; Behnan et al., 2017a). Also, it was found that GSCs tend to express the transcriptional subtype of the dominant cell population in the parental fresh tumour (Joo et al., 2013; Mao et al., 2013). Furthermore, GSCs expressed proneural to mesenchymal transition (PMT) properties in response to chemo- and radiotherapy (Bhat et al., 2013; Halliday et al., 2014). GSCs subtype-specific markers have been suggested by several investigators (Lottaz et al., 2010; Bhat et al., 2013; Behnan et al., 2017a). The mesenchymal subtype GSCs were mainly negative for CD133 and highly positive for CD44, YKL40, and BMI1, while the proneural subtype GSCs were positive for CD133, OLIG2, SOX2 and EZH2 (Bhat et al., 2013; Jin et al., 2017). Lottaz et al. (2010) reported the subtyping of 17 GSCs into two groups: group I, which expresses the proneural signature, is CD133+, while group II expresses the mesenchymal signature and is CD133− (Lottaz et al., 2010). GSCs of the mesenchymal subtype expressed ALDH1A3, had aggressive in vitro and in vivo tumour phenotypes, and were more resistant to radiation treatment than the proneural subtype. Furthermore, the radiation treatment of proneural subtype induced PMT, while ALDH1A3 inhibition blocked this process (Mao et al., 2013). Cusulin et al. (2015) classified 20 GSCs according to their biological features including sphere formation, marker expression, asymmetric cell division, and label retention. They were divided into two groups: (i) a stem-like subtype that expressed the proneural signature and was associated with longer in vivo survival after transplantation; and (ii) a progenitor-like subtype that expressed mesenchymal-signature and was associated with shorter in vivo survival (Cusulin et al., 2015). Bhat et al. (2013) subtyped 14 GSCs into two clusters. Cluster I enriched for the mesenchymal signature with ontology enrichment of genes that play a role in wound response, vasculature formation, and cell motility. Cluster 2 enriched for the proneural signature with ontology enrichment of genes that play a role in neural and glial function and homeostatic activity (Bhat et al., 2013). Our subtyping also showed that the mesenchymal subtype upregulated genes related to epithelial to mesenchymal transition (EMT) such as TWIST1, SNAI1, SNAI2, TGFB1, STAT3, and CD248. Also, the in vitro and in vivo growth pattern and morphology seemed different in proneural and mesenchymal subtypes, where the mesenchymal subtype contained several GSCs that grow adherently in sphere culture (a serum and adherent free condition) (Behnan et al., 2017a). Furthermore, GSCs with proneural and mesenchymal subtypes were found to have different distribution within the tumour. Proneural subtype GSCs were SOX2+OLIG2+ and were perivascular localized, whereas mesenchymal subtype GSCs were CD44+YKL40+ and were exclusively localized to the necrotic hypoxic regions (Jin et al., 2017). Mimicking the necrotic region milieu through in vitro stress conditions such as hypoxia and nutrient deprivation revealed that mesenchymal subtype GSCs survived the stress, upregulated mesenchymal markers and maintained BMI1 expression, while proneural subtype GSCs and normal neural precursors lost BMI1 and EZH2 expression and eventually died. Also, GSCs from proneural and mesenchymal subtypes responded differentially to BMI1 and EZH2 inhibitors (Jin et al., 2017). Further, the epigenetic signature of mesenchymal and proneural subtype GSCs were different from their parental tumour tissue (Bhat et al., 2013). Contrary to their parental tumour, proneural subtype GSCs became hypermethylated in vitro (G-CIMP+) even in the absence of IDH mutation. However, mesenchymal subtype GSCs maintained the G-CIMP− profile and some mesenchymal tumours switched to the proneural subtype upon in vivo transplantation (Bhat et al., 2013). This does not sound surprising as the selective pressure of cell culture conditions and growth factor supplements favour the enrichment of GSCs with the proneural subtype (Bhat et al., 2013). Also, this phenomenon can be explained through the newly acquired understanding of intra-tumoural subtype heterogeneity generated by single cell analysis and multisampling techniques collected from different anatomical locations in the same patient (Patel et al., 2014; Puchalski et al., 2018). We expect that the subtype with the most proliferative GSCs that take best advantage of culture conditions, will dominate the cell culture, and other cell subtypes will gradually disappear.
The mesenchymal signature in single cell glioblastoma multiforme
Although the TCGA subtype approach has been useful to characterize the molecular diversity of tumour bulk among GBM patients (inter-patient heterogeneity), it has limited insight into the intra-tumoural heterogeneity within individual GBM patients (Cancer Genome Atlas Research, 2008; Verhaak et al., 2010). Patel et al. performed the first study that investigated the DNA and RNA molecular diversity of neoplastic cells within individual GBM patients at the single cell resolution level, keeping in mind that both single cells and bulk were depleted of CD45+ cells (Patel et al., 2014). Using RNA-seq (96–192 single cells per patient from five GBM wild-type IDH patients) and comparing single cells to bulk tumours and their derived cultures, showed a broad correlation between neoplastic cells from single GBM biopsies (0.2–0.7 correlation coefficient) indicating intra-tumoural heterogeneity and that cells were more correlated to each other than were cells from different patients. Also, intra-tumoural heterogeneity was indicated by mosaic expression of RTK and their signalling molecules within the same tumour and across different patients. Interestingly, individual cells of single tumours have heterogeneous subtype expression. Although three of five tumour bulks were the mesenchymal subtype, all tumours contained a small number of cells from the proneural subtype and very few cells from the neural or classical subtypes. As the mesenchymal subtype was the dominant subtype in these tumour cells, this led to tumour bulks to be scored as mesenchymal subtype only. Importantly, the increased intra-tumoural subtype heterogeneity was associated with worse survival only in proneural subtype patients (Patel et al., 2014). However, the most well-known genetic alterations of GBM, gain of chr7 and loss of chr10, were expressed in all single cells from different patients (Patel et al., 2014; Darmanis et al., 2017). Although neuro-sphere culture conditions are thought to enrich the culture for cells that mirror the parental tumour cells (Lee et al., 2006), cell diversity was highly reduced in cultured cells compared to parental tumour cells. Furthermore, only three subtypes were expressed in vitro, and none scored for the neural subtype (Patel et al., 2014). Darmanis et al. (2017) profiled invasive glioma cells isolated from peri-tumoural areas at the single cell level and compared them to those of from the tumour core, in addition to profiling a variety of other normal cells within individual GBM patients. The single cell transcriptome analysis of 3589 cells from four paired samples of tumour cores and peri-tumoural samples revealed a large degree of diversity between cells within the same tumour and between different tumours. Their classification resulted in 12 clusters, only 3 of 12 were neoplastic cells, 3 of 12 contained vascular cells, 3 of 12 contained immune cells, one cluster were oligodendrocyte precursor cells, one cluster were oligodendrocytes, and one cluster were astrocytes. Notably, intra-tumoural subtype heterogeneity was seen in the three neoplastic clusters: neoplastic clusters 1 and 2 consisted of proneural and classical cells, while neoplastic cluster 3 consisted of mesenchymal and classical cells (Darmanis et al., 2017). All this information together suggests that despite the intra-tumoural subtype heterogeneity, the mesenchymal subtype exists on a single cell transcriptomic level in neoplastic cells. This indicates that this signature is induced by an intrinsic driver of the mesenchymal signature within these cells rather than by environmental factors.
The mesenchymal signature in circulating glioblastoma multiforme cells
Despite the aggressive nature and highly invasive cells of GBM, this tumour rarely develops extracranial metastasis, with an incidence of <0.4% (Smith et al., 1969). This disease property, supported by the seed-soil hypothesis, made it unreasonable to question the existence of circulating brain tumour cells (CTCs) in GBM until recently (Sullivan et al., 2014; Gao et al., 2016). CTCs were identified in blood samples of GBM patients and a patient-derived xenograft GBM mouse model. CTCs were isolated using a microfluidic platform (CTC-iChip). These assays deplete blood cells and excessive magnetic beads, then sort CTC cells positive for SOX2, TUBB3, EGFR, A2B5, and c-MET. Thirty-nine per cent of GBM patients scored positively for CTCs (13 of 33 GBM patients). Patients with progressive GBM had a median of 11.8 cells/ml while patients with stable GBM had 2.1 cells/ml. Although CTCs expressed a similar pattern of EFGR amplification as tumour cells from the tumour core biopsy, the CTCs were less proliferative. To compare the molecular subtype of CTCs to tumour core cells, a 25-gene panel was used: ASCL1, SOX2, OLIG2, and DLL3 for proneural subtype; GFAP, AKT2, and EGFR for classical; SYT1 and SLC12A5 for neural; SERPINE1, TGFB1, and RELB for mesenchymal subtype, in addition to other embryonic stem cells and self-renewing markers (Sullivan et al., 2014). Comparing the single cell analysis of CTCs with their parental tumours showed that all patient-derived CTCs and xenograft-derived CTCs expressed an elevated mesenchymal transcriptional profile (of SERPINE1, TGFB1, TGFBR2, and VIM), and downregulated neural and oligodendrocyte markers (ASCL1, GFAP, NCAM1, and SOX9). However, CTCs that were positive for PROM1/CD133 (a proneural marker) also retained expression of this stem cell marker transcriptionally, similar to parental tumours. Cells with similar CTC mesenchymal signature were detected mainly around the necrotic foci and palisading cells in GBM tissues. A GBM patient with recurrent brain lesions and rare condition of extracranial metastases to the pulmonary nodules and hilar lymphadenopathy, had high CTCs after 12 months of initial diagnosis (48 cells/ml) that were similar to his primary brain GBM regarding EGFR amplification. Although his recurrent brain tumours had cells from different subtypes (neural: 19.0%, neural/mesenchymal: 65.3%, and mesenchymal: 15.7%), his metastatic lesions were mainly the mesenchymal subtype (∼62% and ∼54% of the cells of the metastatic left hilar lymph node and the pulmonary metastases, respectively, were mesenchymal). Furthermore, PDGFRB mutation increased from 3.5% in primary GBM to 50% in all metastatic lesions. Also, the metastatic lesions acquired mutations in EGFR, RB1, and SETD2 that were not detected in the primary tumour (Sullivan et al., 2014).
The shift towards mesenchymal subtype upon glioma recurrence
High grade gliomas, especially GBMs, tend almost always to recur, often at the site of the initial tumour, but sometimes recur away from this site or even in the other hemisphere (Wen and Kesari, 2008; Kim et al., 2015). The subtype of a primary and recurrent tumour might be different. In total, two-thirds of primary GBMs switch transcriptional subtypes at tumour recurrence, while secondary GBM seems more stable (2/7 switched subtype) (Wang et al., 2016). In another study, 45% of wild-type IDH tumours switched subtype upon recurrence (Wang et al., 2017b). Phillips and colleagues reported that ∼30% of proneural tumours shifted to mesenchymal-signature upon recurrence. This shift appears to reflect tumour progression and was marked by losing OLIG2 expression and the upregulation of YKL40 expression. Furthermore, these tumours upregulated CD44, STAT3, and vimentin (VIM) expression upon recurrence (Phillips et al., 2006). It is thought that the unidirectional shift proneural→mesenchymal might happen through accumulating genetic and epigenetic abnormalities with the time. This hypothesis is supported by the fact that mesenchymal subtype patients are older than proneural subtype patients. It is also supported by another hypothesis that suggests the proneural subtype as an origin for all subtypes (Phillips et al., 2006; Ozawa et al., 2014). In vitro proneural→mesenchymal reprogramming of GSCs and neural stem cells was achieved by overexpression of master transcription factors such as TAZ or STAT3 and CEBPB (Carro et al., 2010; Bhat et al., 2011). Later, it was found that these master transcription factors of mesenchymal transition were under the control of NF-κB (Bhat et al., 2013). Furthermore, TAZ overexpression in the proneural mouse model of RCAS-PDGFB transformed the tumours into more aggressive mesenchymal-like tumours (Bhat et al., 2011). Bhat et al. suggested that the mechanism of PMT upon radiation treatment was through NF-κB activation. Macrophages and microglia were suggested as the potential driver of this transition in vivo. Also, when TNF-α treatment induced mesenchymal differentiation in proneural subtype GSCs and was followed by radiation treatment, it resulted in the development of radioresistant GSCs with stronger mesenchymal signature. However, some proneural subtype GSCs were resistant to TNF-α treatment and retained the proneural signature, indicating that not all proneural cultures have a similar response (Bhat et al., 2013). The PMT could be induced in proneural mouse model (PDGFA/shP53) and GBM cell lines with PDGFRA amplification through subsequent knockdown of NF1. The NF1 deletion resulted in significantly shorter survival in the triple infected mice with mesenchymal-like subtype compared to the double infected proneural-like subtype. This PMT was converted by mTOR inhibitor rapamycin in vitro (Ozawa et al., 2014).
Although later studies showed that the mesenchymal subtype was the most stable subtype (55–65% kept the same subtype), the shift towards the mesenchymal subtype was not significant upon tumour recurrence with some tumours shifted toward proneural and others toward classical (Verhaak et al., 2010; Wang et al., 2016, 2017b). Interestingly, the mesenchymal subtype at tumour recurrence tended to have a worse overall survival (Wang et al., 2016). However, the frequency of mesenchymal and proneural subtypes was highest among recurrent tumours, confirming previous findings about classical being more sensitive to treatment (Wang et al., 2017b). Also, the loss of EGFR expression in ∼80% of tumours upon recurrence supports the loss of the classical signature upon recurrence (van den Bent et al., 2015). Another study showed that the loss of EGFRvIII was associated with the transition from classical to other subtypes (Wang et al., 2016). Additionally, the increased transcriptional heterogeneity was associated with a higher subtype switch upon tumour recurrence, suggesting an important role of tumour heterogeneity in this phenomenon (Wang et al., 2017b). Using multi-cancer computational analysis, 64 genes were identified as a signature of mesenchymal transition. These genes were markers of aggressive and invasive stage tumour expressed in several solid tumours, including neuroblastoma and glioma (Kim et al., 2010; Cheng et al., 2012). The low expression of the 64 mesenchymal metagene signature was associated with preferable survival and longer time to recurrence in GBM with an extremely low expression level in GBM patients with exceptionally long time to recurrence. Seven of the 64 genes (POSTN, COLA1A1, COLA3A1, COLA1A2, LOX, COLA6A2, and LUM) were among the top differentially expressed genes in GBM versus low grade gliomas. The metagene was strongly associated with CD44 expression (Cheng et al., 2012).
The tumour evolution theory in response to treatment, adapted a Darwinian model to explain the clonal shift in recurrent tumour after treatment. This theory suggests that the tumorigenic clones that are vulnerable to treatment will disappear, while positive selection will favour resistant clones to expand and dominate the recurrent tumour (Nordling, 1953; Nowell, 1976). Despite having >45% of the mutation pool shared between primary and recurrent tumours, the dominant clones at recurrence are not lineal ancestors of the dominant clone at first disease diagnosis. Rather both types of clones seem to have been branched from a common ancestor a decade or more before diagnosis. The whole-exome and transcriptome analyses of untreated and recurrent GBM from 114 patients, in addition to their matched normal tissue, found some genetic markers and tumour evolutionary trajectories in recurrent tumours. The primary tumours had 60 mutations on average, whereas the hypermutated recurrent tumours (six primary GBM and 11 secondary GBM) were all temozolomide-treated and harboured >500 mutant genes per tumour, whereas none of temozolomide-untreated tumours were hypermutants (0/14). The mutational average of non-hypermutant tumours was <50 mutations/tumour. Also, hypermutated tumours were associated with MGMT promoter methylation (Wang et al., 2016). The tumour evolution mutational dynamic seems to have a similar alteration rate for a given time, for both primary and recurrent tumours after treatment (∼0.03 substitutions/megabase/year), except for those with a hypermutated profile. The replacement of dominant clones before treatment with new clones at relapse is a frequent event in GBM. The main GBM-driver mutations including TP53, PTEN, EGFR, PIK3CA, ATRX, IDH1, PIK3R1, and PDGFRA were shared between both primary and recurrent tumours. Interestingly, EGFR, EGFRvIII, TP53, PDGFRA, PTEN, ATRX, NF1, and RB1 seem to express differentially mutated versions of the same gene, indicating their important roles in both primary and recurrent tumours. Also, significant associations were observed between co-deletion of RB1 and PTEN, co-mutation of NF1 and TP53, and MGMT promoter methylation and hypermutation (Wang et al., 2016).
Origin and role of the mesenchymal signature in brain tumours
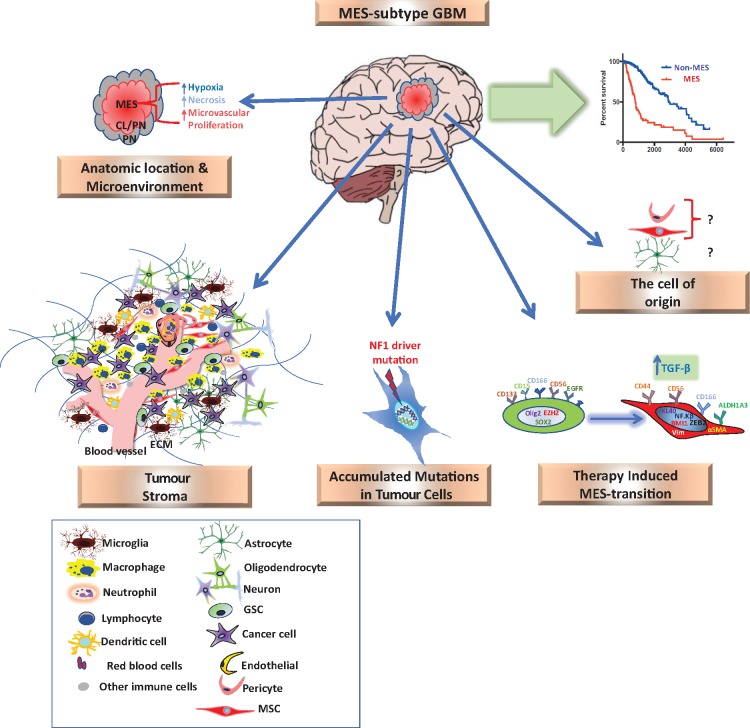
From neurosurgical and neuropathological point of view, gliomas are thought to arise from glial cells, and preliminary understanding of embryonic development suggests that the brain and spinal cord are developed from the ectoderm. These concepts made researchers hesitant to address the mesenchymal components in malignant brain tumours until recently. So, what is the origin of mesenchymal signature in brain tumours and what is its role? (Fig. 1 illustrates the factors that induce the mesenchymal signature in GBM).
Figure 1.
The origin of the mesenchymal signature in malignant glioma. GBM patients with the mesenchymal (MES) subtype tend to have worse survival than non-mesenchymal subtype patients. The mesenchymal signature in glioma can be induced by several factors: (i) stromal cells of recruited macrophages/microglia, MSCs/pericytes, and other progenitors; (ii) intrinsic expression of tumour cells, NF1 as main driver mutation; (iii) the cell of origin; (iv) anatomical location and tumour micorenviroments; and (v) therapy-induced mesenchymal-signature. Radiotherapy, antiangiogenic therapy and chemotherapy might induce the mesenchymal signature. ECM = extracellular matrix.
Mesenchymal stem cells/pericytes and other cell progenitors in glioma
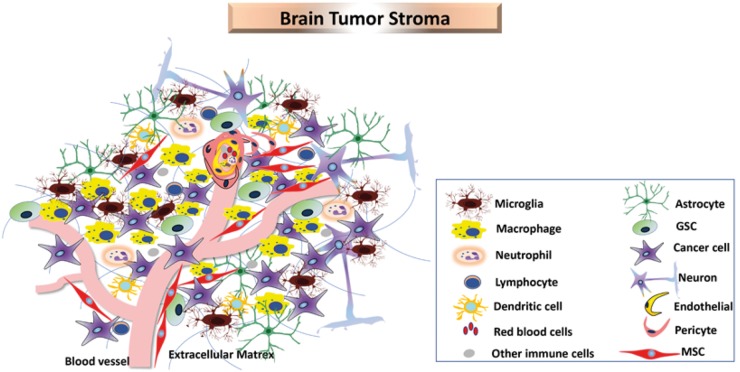
The current understanding of brain tumour biology suggests that the tumour stroma of non-neoplastic origin make up ∼50% of GBM tumour mass (Darmanis et al., 2017). Several stromal factors including vasculature cells such as endothelial cells, perictyes/mesenchymal stem cell (MSCs), immune cells, and glial cells are recruited into and play a role in tumour formation and progression (Fig. 2) (Charles et al., 2011).
Figure 2.
Cellular heterogeneity in brain tumours. GBM consists of heterogeneous cell populations including cancer cells, GSCs, macrophages, microglia, neutrophils, lymphocytes, dendritic cells, red blood cells, astrocytes, neurons, endothelial cells, pericytes and MSCs.
It is thought that the expression of mesenchymal lineage markers of smooth muscle, endothelial cells, and cartilage by mesenchymal subtype brain tumours is related to the reported multipotentiality of NSCs from adult brain (Bani-Yaghoub et al., 2004; Phillips et al., 2006). NSCs are suggested as the common cell of origin for brain tumours and tumour subtypes could be driven by the plasticity and multi-differentiation pathways of these cells (Galli et al., 2004; Verhaak et al., 2010). Another source for the mesenchymal signature in glioma could be the recruited stromal cells including endogenous MSCs and other stem cell-like populations that accumulate in the tumour during disease progression (Behnan et al., 2014). It was also speculated that the growth of mesenchymal subtype tumours might be supported by the increased level of VEGF and other endothelial derived factors in brain tumours (Jin et al., 2002; Phillips et al., 2006). It is thought that tumour-associated stromal cells might be one of the main drivers in acquiring the mesenchymal signature (Behnan et al., 2017a; Wang et al., 2017b). This has been seen to be the case in colorectal cancer, but no definitive proof of these phenomena yet exists in glioma (Isella et al., 2015). However, the stromal signature has been correlated to reduced tumour purity and is associated with the mesenchymal signature in different solid tumours including GBM (Martinez et al., 2015). We have previously reported that endogenous MSCs are recruited into glioma and contribute to tumour progression in the GL261 glioma model, which was defined as mesenchymal-like subtype. Furthermore, MSC-like cells were identified in both fresh human GBM tissues and their derived cell cultures (Behnan et al., 2014). Svensson et al. (2015) using the same GL261 model, confirmed that cells expressing pericyte markers infiltrated into the tumour and mainly localized around the blood vessels (Svensson et al., 2015). However, perivascular localization of MSCs was described in several tissues indicating an MSC/pericyte dualism (Crisan et al., 2008). So far, there is no single unique marker that characterizes MSCs or pericytes, which makes it difficult to distinguish between them. Furthermore, perivascular MSCs which were isolated from different tissue types and expanded in culture showed similarity to each other and to bone marrow MSCs more than to their original tissue. This suggests a common developmental origin for the perivascular MSCs (Crisan et al., 2008).
The question presented here is that if the tumour stroma and recruited cells are behind the acquisition of the mesenchymal signature in brain tumours, how do the tumour cells per se, even at a single cell level, express the mesenchymal signature? And how do propagated GSCs under neurosphere culture conditions express the mesenchymal signature, keeping in mind that such culture conditions do not favour the expansion of stromal cell components. Additionally, macrophages/microglia markers (ITGAM and AIF1) that were highly expressed in GBM tissues were absent in GSCs (Behnan et al., 2017a; Wang et al., 2017b). One proposed explanation is that the tumour cells acquire the mesenchymal signature in response to radiotherapy (Bhat et al., 2013). Bhat et al. showed that cultured GSCs with the proneural subtype acquired the mesenchymal signature through activation of the NF-κB pathway. However, the general practice treatment of GBM is surgery followed by radio- and chemotherapy. Thus, most tissue samples in the subtype’s dataset of primary GBM are left over from biopsies dissected before being exposed to radio- or chemotherapy stress. This means that the mesenchymal signature was already present before the either radio- or chemotherapy stress.
The mesenchymal subtype can be driven by genetic mutation
The main known driver mutation of mesenchymal subtype is NF1 deletion or mutation (Bhat et al., 2013; Herting et al., 2017; Wang et al., 2017b). NF1 is best known to also be expressed in WHO grade I pilocytic astrocytoma. These patients have a higher risk of progression to higher grade infiltrating gliomas (Rodriguez et al., 2008). NF1 encodes for neurofibromin protein, which plays a role as a GTPase activating protein that negatively regulates Ras and Ras-associated downstream signalling pathways (Ratner and Miller, 2015). However, NF1 deletion or mutation was detected in 24–37% of mesenchymal samples in GBM (Bhat et al., 2013; Herting et al., 2017; Wang et al., 2017b). Also, GSCs that had normal NF1 expression, released from the influence of macrophage/microglia micro-environment, maintained the mesenchymal expression profile indicating that these cells have an intrinsic and NF1-independent mesenchymal signature (Bhat et al., 2013). This means that approximately two-thirds of mesenchymal subtype samples cannot be explained by NF1 abnormalities. Our subtyping with 12 genes showed that mesenchymal subtype samples were separated into two clusters. Cluster I was named tissue-mesenchymal and was characterized by overexpression of COL1A1, COL1A2 and THBS1, which are known as matrix and connective tissue factors. Cluster II was named cell-mesenchymal. GBM tissues in this second cluster showed similarity to cultured MSCs derived from bone marrow, adipose tissue, and brain, and expressed high levels of TGFBI, S100A4, and DAB2 (Behnan et al., 2017a). Recently, single cell analysis of immune cells in GBM showed that TGFBI and other S100 family members including S100A8/9 (macrophages markers) were highly expressed in immune cells in the tumour core (Darmanis et al., 2017). This finding supports our hypothesis that there are at least two subclasses within the mesenchymal subtype, one subclass induced by recruited cells and the other by tumour cells. Furthermore, in our experience, the mesenchymal subtype cells from GBM patients, enriched under neuro-sphere culture conditions, also have two subclasses depending on their in vitro growth morphology (Behnan et al., 2016, 2017a). One type grows as floating spheres and the other grows adherently in serum-free non-adherent conditions (Behnan et al., 2017a).
The glioma subtype driving mutations can be explained in glioma transgenic mouse models. While RCAS-PDGFB-amplification can drive a proneural-like transcriptome pattern, RCAS-NF1 silencing in a paediatric or adult glioma mouse model drive a mesenchymal-like subtype (Hambardzumyan et al., 2009; Liu et al., 2011; Halliday et al., 2014; Ozawa et al., 2014; Herting et al., 2017). The PDGFB model exhibits some features of oligodendroglioma such as OLIG2 and DCX expression, while NF1-silenced models exhibit more astrocytoma-like features represented by high expression of GFAP and CD44. Similar to human mesenchymal subtype tumours, the NF1 tumour model contains significantly higher macrophage numbers compared to the proneural PDGFB model. Interestingly, the NF1 model had significantly longer survival than the proneural PDGFB model. However, by reanalysing the survival of proneural versus mesenchymal subtypes in the TCGA dataset, and after limiting the mesenchymal subtype to only NF1-deleted or mutant samples, no adverse survival was seen for mesenchymal subtype. Also, the NF1 model had less proliferating cells, smaller vessel size, and less permeability than the proneural model. All these together suggest that genetic driver mutations may impact tumour vasculature and structure (Herting et al., 2017).
The cell of origin dictates the tumour subtype
The cell of origin might play an important role in determining GBM phenotype. The proneural subtype PDGFB overexpressing model has 100% tumour formation with GBM characteristics when RCAS-PDGFB is injected into the left or right hemisphere or subventricular zone. However, the TP53/NF1 model requires co-delivery of RCAS shNF1, shTp53, and Cre-Pten specifically to the subventricular zone of adult mice to get GBM-like tumour formation (Hambardzumyan et al., 2009; Herting et al., 2017). This suggests that each tumour subtype might have a different cell of origin (Alcantara Llaguno et al., 2009; Liu et al., 2011). The oligodendrocyte precursor cells (OPCs) were identified as the cell of origin for the Tp53/Nf1 tumour model that have dramatic growth expansion prior to malignancy (Liu et al., 2011). Alcantara Llaguno et al. (2015) showed that two populations of adult CNS progenitors, with different cell of origin, gave rise to molecularly and biological distinct gliomas despite being hit with the same tumour suppressor mutation in Nf1, Trp53, and Pten. The two transformed progenitor populations aligned either with normal OPCs or astrocytes, where this last one enriched for mesenchymal signature (Alcantara Llaguno et al., 2015). These data suggest an astrocytic lineage and OPCs as origin of the mesenchymal and proneural subtypes, respectively (Alcantara Llaguno et al., 2015; Herting et al., 2017).
Contrary to this, Mazor et al. (2015) showed that the differences between primary and recurrent tumours that progressed from low grade to GBM, were more likely attributed to tumour progression rather than cell of origin. The malignant progression was associated with strong promoter hypomethylation (1953 CpG sites) leading to alteration in gene transcription that enriched for cell cycle genes. The authors suggested an interdependency between genetics and epigenetics during the tumour evolution process and that phyloepigenetic tree recapitulated the early divergence between primary and recurrent tumours similar to somatic phylogenetic tree (Mazor et al., 2015).
The mesenchymal subtype can be shaped by immune cells
The main characteristic of the mesenchymal subtype is its association with immune-related genes and pathway analysis showed enrichment of immunological processes and inflammation. This subtype scored the lowest purity score compared to proneural and classical subtypes indicating the infiltration of non-neoplastic cells into this subtype (Wang et al., 2017b). Macrophages/microglial cells that constituted the largest stromal cell populations in GBMs and their infiltration was associated with the mesenchymal subtype, were suggested to induce PMT (Engler et al., 2012; Li and Graeber, 2012; Bhat et al., 2013; Gabrusiewicz et al., 2016; Wang et al., 2017b). The mesenchymal subtype enriched for macrophage/microglia of M1 and M2 signatures, in addition to neutrophil cell signature, while the classical subtype included activated dendritic cells (Bao et al., 2006; Engler et al., 2012; Ye et al., 2012; Gabrusiewicz et al., 2016; Wang et al., 2017b). Also, activated natural killer cells were less expressed in mesenchymal subtype samples than in classical and proneural samples (Wang et al., 2017b). Furthermore, tumour-infiltrating lymphocytes (TILs) were enriched in mesenchymal tumours versus non-mesenchymal tumours (Engler et al., 2012; Li and Graeber, 2012; Bhat et al., 2013; Gabrusiewicz et al., 2016; Wang et al., 2017b).
Although the mesenchymal subtype has a significantly higher level of necrosis compared to other subtypes (Verhaak et al., 2010; Cooper et al., 2012), the mesenchymal tumours with abundant TILs did not have significantly higher necrosis than those with no TILs. This indicates that TILs might represent an adaptive immune response against the tumour rather than a response against necrosis. TILs were also associated with specific histopathological features including sarcomatous regions, giant cell, epithelioid, and gemistocytic components (Rutledge et al., 2013). Not surprising to us, tumours that were enriched with sarcomatic regions, pathologically known as gliosarcoma, were also classified as the mesenchymal subtype recently (Brain tumor conference, Warsaw, 2018).
Forty-two per cent of mesenchymal tumours in TCGA set have TILs, indicating its immunogenic nature. Interestingly, classical subtype tumours were depleted of TILs. TIL infiltration was associated with NF1 and RB1 mutations, a common alteration in the mesenchymal subtype (Rutledge et al., 2013). It was shown recently that NF1 abnormalities were associated with less purity and higher stromal signature indicated by the infiltration of macrophages/microglia, which were supported by higher expression of M1 and M2 macrophages in the mesenchymal subtype. Further, NF1 knockdown increased the in vitro recruitment of human macrophages isolated from GBM patients (Wang et al., 2017b). Additionally, TILs were strongly associated with NF1 deletion or mutation, and absent in both EGFR-amplified tumours and those with homozygous PTEN deletion (Rutledge et al., 2013). Recurrent glioma tumours showed a decreased monocyte gene signature compared to primary tumours; the recurrent tumours with mesenchymal transition exhibited an increased immune cell infiltration. M2 macrophages were higher in recurrent tumours that underwent mesenchymal transition. Non-polarized M0-macrophages were also increased in recurrent tumours with the mesenchymal profile, compared to primary tumours from the same subtype (Wang et al., 2017b).
While tumour cells with NF1 deletion exert their effect by recruiting macrophages/microglial cells to the tumour, the macrophage/microglia cells also affect the tumour and create a micro-environment that shapes mesenchymal subtype tumour cells (Wang et al., 2017b). This predicts that a two-way interaction might exist between immune cells and tumour cells, represented by macrophages/microglia cells within the tumour expressing a unique transcription profile that demonstrates a direct effect of the tumour milieu on these cells. On the other hand, the macrophages/microglia cells express anti-inflammatory, pro-angiogenic, and extracellular matrix remodelling factors that are known to enhance tumour growth, survival, and invasion (Szulzewsky et al., 2016; Darmanis et al., 2017; Wang et al., 2017b). Furthermore, the stroma heterogeneity investigated on single cell level has yield a precise quantification of cellular compartments within the tumour core and invasive edges of GBM patients. This study showed that only 44% of the core cells were tumour cells, ∼50% were infiltrative immune cells (CD45+ cells), ∼1.5% clustered with oligodendrocytes (GalC+ cells), 2% clustered with oligodendrocyte precursors (O4+ cells), 2% clustered with endothelial cells (sorted as BSL1+ cells), and <0.05% with neurons (sorted as CD90+ cells). Macrophages and microglia were the main cell populations in the immune cell cluster, ∼95%, whereas dendritic cells made up 4.5% (Darmanis et al., 2017).
The mesenchymal subtype can be shaped by anatomical location and micro-environment
One important criticism for the TCGA subtyping is that each sample was dissected from only one single random spot of the tumour bulk, assuming that GBMs are homogenous tumours and intra-tumoural heterogeneity is not big issue. This was shown not to be the case when a fluorescence-guided multiple sampling (FGMS) approach was used to dissect four to six tumour fragments from spatially distinct regions within an individual tumours from each GBM patient. In 6 of 10 patients analysed, samples from the same patient were classified into two to three different subtypes revealing intra-tumoural heterogeneity (Sottoriva et al., 2013). Recently, Puchalski et al. correlated the inter- and intra-tumour heterogeneity with GBM anatomical regions. Their data showed that samples from the same anatomical region, regardless of whether they were from the same patient or different patients, were more similar to each other than to samples from different anatomical regions from the same patient. While the mesenchymal signature was expressed by perinecrotic/hypoxic regions and microvascular proliferative areas, the vascular areas and invasive edges were proneural. Patients who expressed both vascular and hypoxic signatures had worse survival than those did not (Jin et al., 2017; Puchalski et al., 2018).
Surprisingly, the three or four TCGA subtypes can exist within the same tumour. The leading edges were subtyped as neural, the infiltrative tumours were subtyped as neural/proneural, the central tumour regions were either the classical or neural/proneural subtype, and the pseudopalisading cells around necrosis and microvascular proliferation were almost exclusively mesenchymal subtype (Puchalski et al., 2018). Also, single cell analysis revealed that the tumour bulk consists of individual tumour cells that belong to at least three subtypes and that cell diversity impacted patient survival. This study concluded that a subtype label of glioma sample is the same subtype signature of the dominant cell population within the tumour bulk (Patel et al., 2014; Darmanis et al., 2017). Furthermore, the vascular and hypoxic signatures associated with tumour grade and histology were also associated with different GBM subtypes. Including the gene expression profiles of human microvascular endothelial cells (HMVEC), endothelial cells (HUVEC), and GBM cells that underwent hypoxia and normoxia showed that proneural subtype expressed markers of mature vessels, while mesenchymal subtype expressed markers of hypoxia and microvascularity. Patients that expressed high hypoxic and vasculature signature simultaneously, exhibited the worst survival (Jin et al., 2017).
Macrophages and microglia that constitute the largest stromal population in GBM, had a variable distribution between the tumour core and the invasive edge. While macrophages were dominant within the tumour core (813 single cell macrophages versus 365 microglial), microglia were the main population in the invasive edge (85 macrophages versus 574 microglia). This distribution was associated with a specific cytokine profile with myeloid cells in the tumour core express anti-inflammatory cytokines and pro-angiogenic markers suggesting an essential role in tumour growth, survival, extracellular matrix remodelling and promoting angiogenesis. On the other hand, immune cells with pro-inflammatory cytokine profile were dominant in the tumour periphery (Darmanis et al., 2017). However, this study has major limitations as the analysis of stromal cells depended on single cells sorted after labelling with known but unspecific markers and most importantly, the leftover stromal cells were discarded. Addressing cells like neurons, endothelial cells, and astrocytes with single marker does not mean that these cells are what they are thought to be, because many other cell types can express these markers. This can be seen when a single marker picked three different cell types clustered under what was supposed to be an endothelial cluster. Also, CD90, which was used as marker for neurons, is highly expressed by brain derived-MSCs/pericytes (Behnan et al., 2017a, b).
All these together highlight the complexity of GBM heterogeneity and subtypes and suggest that NF1 abnormality and tumour micro-environment with its recruited cells, vasculature and anatomic location induce the mesenchymal criteria.
Therapy-induced mesenchymal transition
The EMT, a critical biological process in embryonic development and wound healing, has been shown to play essential roles in tumour cell migration, metastasis and therapy resistance (Lee et al., 2014). In brain tumours, this process could have another name such as glial-mesenchymal transition (GMT) or PMT, because of the believed glial origin of these tumours. Antiangiogenic treatments, DNA damaging agents and ionizing radiation might affect cancer cell behaviour through EMT/GMT/PMT and, eventually, patient prognosis (Fig. 1). The mesenchymal subtype GBM patients, of both primary and recurrent tumours, tended to have worse survival than other subtypes (Phillips et al., 2006; Verhaak et al., 2010; Wang et al., 2017b). As the mesenchymal subtype was associated with higher expression of angiogenesis and endothelial markers, the mesenchymal subtype patients were predicted to benefit from the antiangiogenic treatments. However, antiangiogenic drugs, DNA damaging agents and ionizing radiation impact the biology of the tumour and its subsequent behaviour (Phillips et al., 2006; Piao et al., 2012, 2013).
In combination with radiotherapy and chemotherapy, the antiangiogenic drugs, including the U.S. Food and Drug Administration approved anti-VEGF (bevacizumab) for recurrent GBM and many other solid tumours, have been shown to decrease vascular permeability resulting in preliminary radiographic response associated with prolongation of progression free survival, but not overall survival (Batchelor et al., 2007; Friedman et al., 2009; Port et al., 2010; Lai et al., 2011; Chinot et al., 2014; Taal et al., 2014). The preclinical studies showed that a combined bevacizumab and sunitinib (anti-VEGFR) treatment improved animal survival, in addition to delayed hypoxia and decreased the recruited myeloid cells compared to bevacizumab single treatment (Piao et al., 2012, 2013). However, the clinical outcomes of both anti-VEGF and/or anti-VEGFR or the combined treatments with chemo- and radiotherapy, did not achieve significant overall survival despite the first one showed better tumour oxygenation at early stage associated with improvement of progression free survival, while the anti-VEGFR induced rapid hypoxia and tumour progression (Friedman et al., 2009; Galanis et al., 2013). Upon disease progression after anti-VEGF failure or combined anti-VEGF and anti-VEGFR, treated tumours became even more aggressive and treatment-resistant (Quant et al., 2009; de Groot et al., 2010; Scott et al., 2010; Piao et al., 2012, 2013). The preclinical data showed a significant increase in recruited macrophages/microglia and aggressive mesenchymal features at the time of tumour progression in treated groups (Piao et al., 2012, 2013). EMT or PMT has been suggested as one of the main mechanisms behind the development of resistant phenotype tumours after antiangiogenic treatments (Lu et al., 2012b; Piao et al., 2012, 2013). Comparing the recurrent tumours in GBM patients who developed bevacizumab resistance to their original untreated tumours, the recurrent tumours showed increased MET activation and elevation in mesenchymal markers including VIM, CD44, YKL-40, N-cadherin and downregulation of T-cadherin (Lu et al., 2012b). Piao et al. (2013) showed an increase in cell migration or invasion, elevation in pro-inflammatory cytokine secretion, and enrichment of genes associated with mesenchymal signature and EMT transcription factors in tumour cells that developed resistance to bevacizumab. The histology analysis of these specimens revealed regions containing sarcoma and spindle-shaped cells (Piao et al., 2013). Bevacizumab treatment (in a GBM xenograft model derived from patient tumour spheroids) resulted in significant reduction in vascularity and blood flow revealed by dynamic contrast enhanced MRI, and reduced blood vessel size by histology analysis. This resulted in a glycolytic hypoxic micro-environment that enhanced tumour cell invasion (Keunen et al., 2011). Lu et al. (2012b) showed that knockout of VEGF enhanced MET activation (phospho-MET), which induced highly aggressive and invasive mesenchymal-like GBM. Importantly, combining the ablation of VEGF with knocking down MET expression reduced vascularity and invasion, converted cell phenotype into epithelial-like cells, and extended animal survival 3-fold compared to the control group (Lu et al., 2012b).
The current standard of care for GBM treatment includes maximal tumour resection followed by radiotherapy (whole brain radiation or stereotactic radiosurgery) plus adjuvant temozolomide chemotherapy (Stupp et al., 2005). Preclinical data has shown that GBM samples contain radioresistant GSCs (specifically CD133+ cells) that activate DNA damage checkpoint signals and increase DNA repair capacity upon radiation treatment. This cell population expanded after radiation treatment, both in vitro and in vivo. Using checkpoint inhibitors (Chk1 and Chk2) sensitized CD133+ cells to radiation treatment, consequently, impaired tumour growth (Bao et al., 2006). Bhat et al. (2013) and others showed that GSCs with mesenchymal subtype are more radio-resistant than proneural subtype (Segerman et al., 2016; Pencheva et al., 2017). Upon radiation treatment, proneural subtype cells pretreated with TNF-α displayed an upregulation of CD44 expression and activation of NF-κB pathways to undergo PMT (Bhat et al., 2013). PMT was detected as early as 6 h post radiation treatment. NF-κB, STAT3, Snail, P53, E2F, and ALDH1A3 were defined as the main mediators and regulators of PMT and their inhibition blocked this phenomenon. Radiation treatment upregulated CEBPB and mesenchymal markers (CD44, α-SMA/ACTA2, VIM, FN1, COL1A1 and COL1A2, MMP2, MMP9, and YKKL-40), while downregulating proneural markers (SOX10 and PDGFR-α) (Mao et al., 2013; Halliday et al., 2014; Lau et al., 2015). Murine and human GBM cells that underwent PMT post-radiation treatment increased cell motility, invasion, reduced cellular stiffness, and significantly increased temozolomide-resistance compared to non-irradiated cells (Lau et al., 2015). Clinical data showed that GBM patients with high CD44 expression and NF-κB activation were associated with poor response rate to radiation treatment and have lower survival than those with low mesenchymal metagene expression (Bhat et al., 2013). It is thought that the proneural tumours contain a small population of mesenchymal subtype cells, marked as SOX2−CD44+ cells, which are radioresistant and they dominate the culture after radiation inducing the mesenchymal signature (Mao et al., 2013).
Despite alkylating agents such as temozolomide has been considered as first-line treatment for GBM patients since 2005, it offers limited survival benefit for those patients (Stupp et al., 2005; Wick et al., 2012). In fact, it has been recommended to avoid temozolomide in elderly GBM patients, and retain it for patients harbouring a methylated MGMT promoter gene that has a good response to this treatment (Wick et al., 2012). However, the outcomes of a recent clinical trial confirmed some survival benefit in elderly patients when hypofractionated radiotherapy was combined with temozolomide (Perry et al., 2017). MGMT has been associated with developing resistance to the standard of care treatment with temozolomide. Direct inhibition of MGMT expression or increasing the promotor methylation of this gene decreased glioma chemoresistance (Hegi et al., 2005; Siebzehnrubl et al., 2013). Chen et al. (2012) suggested that temozolomide targets the active proliferating progenitors, not the quiescent chemoresistant GSCs that recapitulate the tumour and no survival benefit was achieved. Even after combining temozolomide with ablation of quiescent GSCs, a less invasive circumscribed tumour recurs with different histology from the primary tumour expressing high level of S100B and PDGFR-α, and very low GFAP, resembling oligodendroglioma (Chen et al., 2012). Temozolomide-resistant glioma cell lines were established by treating cultured cells with temozolomide for 6 months. The temozolomide-resistant cells expressed some mesenchymal properties such as: reduced cell polarity, increased pseudopodia formation, cell motility and invasion, upregulation of EMT markers (VIM, N-cadherin, and Slug), and CDC20. Inhibition of CDC20 abolished the EMT features and enhanced temozolomide sensitivity (Wang et al., 2017a). ZEB1—a tumour formation regulator, and EMT- and stemness-inducer—is expressed in 45% of GBM patients with 50% overlapping with MGMT expression. Its expression is correlated with glioma grade, patient survival, and tumour invasion. Surprisingly, ZEB1, and not MGMT, correlated with shorter temozolomide response and reduced GBM patient survival. ZEB1 knock-down not only decreased GSC marker expression (SOX2, OLIG2 and CD133), it also reduced tumour invasion and chemo-resistance. On the other hand, over expression of ZEB1 induced c-MYB, miR‐200c and MGMT elevation, which made GBM cells more chemo-resistant. However, even in the absence of ZEB1, overexpression of c-MYB increased MGMT expression and chemo-resistance (Siebzehnrubl et al., 2013). Also, upregulation of TWIST1 and loss of CDH1 was associated with poor temozolomide response and short survival in GBM patients, indicating the role of EMT in tumour progression (Velpula et al., 2011; Siebzehnrubl et al., 2013). Further, recent outcomes from drug screening on clonal levels showed that the in vitro expanded clones with mesenchymal characteristics are more radio- and chemo-resistant than those with proneural characteristics and changed the methylation pattern of the mesenchymal subtype master regulators upon treatment. Interestingly, the clones that showed resistance to one drug, were mostly resistant to all other drugs in the panel, and this multidrug resistance was correlated with radio-resistance (Segerman et al., 2016). Finally, it is speculated that tumour surgery might activate EMT as a response to wound healing signals triggered by the injured tissue. However, only few reports from breast cancer, prostate, and oral squamous cell carcinoma addressed this issue (Kast et al., 2017). To our knowledge, such studies have not been performed in glioma. However, more research is needed to know whether PMT/EMT/GMT is caused by accumulated genetic alterations upon disease progression or by stromal cells and accelerated by treatment regimes.
The concept of the proneural origin of all glioblastoma multiforme subtypes
Ozawa et al. (2014) suggested that all non-G-CIMP GBMs, including both proneural and mesenchymal subtypes, arise from common proneural-like precursors and that disease lethality is increased by accumulated mutations such as TP53, PTEN, or CDKN2A in PDGFA-driven gliomas. Their computational model identified cells harbouring both chr7-gain, driven by PDGFA amplification, and chr10-loss, driven by PTEN loss, as the most likely cells of origin for gliomas. The mesenchymal subtype might evolve from the proneural subtype and be driven by accumulated mutations (NF1 loss in this case) in tumour cells that overexpress PDGFA. These tumours have been shown to upregulate mesenchymal-related markers including CD44, pSTAT3, STAT5, STAT6, C/EBPb.IRF1 and RUNX1 (Ozawa et al., 2014). However, the hypothesis that one subtype, such as the proneural subtype, is the origin of all other subtypes, and it is just matter of disease progression that shifts tumours from one subtype to another, can be also criticized. The mesenchymal subtype does exist in lower grade gliomas. Furthermore, upon tumour recurrence, ∼50% of the tumours keep the same subtype and some tumours shift in the opposite direction, from mesenchymal→proneural upon disease progression (Murat et al., 2008; Verhaak et al., 2010; Wang et al., 2016, 2017b). Also, simultaneous deletion of NF1 and Tp53 without PDGFA amplification background induced GBM with the mesenchymal profile in an RCAS/tv-a model (Ozawa et al., 2014). Altogether, these results suggest that the mesenchymal subtype might have a different origin than the proneural subtype. Despite having clear proof of the effect of macrophages/microglia, inflammatory cytokines, and radiation on subtype plasticity, it is still difficult to confirm the hypothesis that suggests all GBM subtypes evolve from a proneural origin through the exposure to different micro-environmental factors (Bhat et al., 2013). Also, current understanding of GBM indicates that all primary GBMs (not secondary), arise de novo without previous history of low grade tumour. Therefore, it is challenging to draw tumour evolution maps based on this hypothesis and to predict when the subtype shift happened in response to micro-environmental factors.
In spite of Ozawa et al.rsquo;s suggestion that the proneural-like precursors are the origin of all subtypes, the clonal evolution of GBM does not support this conclusion (Gerstung et al., 2011; Ozawa et al., 2014; Kim et al., 2015; Wang et al., 2016). However, the clonal evolution concept is indeed controversial and does not have clear agreement on the early GBM-driving mutational events. Sottoriva and colleagues (2013) have suggested that mutations in EGFR and CDKN2A/B/p14ARF are early event drivers, while PDGFRA and PTEN mutations are later events (Sottoriva et al., 2013). A study by Wang et al. (2016) described mutations in IDH1, PIK3CA, and ATRX as early events while mutations in TP53, NF1, and PTEN occur later during tumour evolution (Wang et al., 2016). Ozawa et al. (2014) suggested that PDGFA amplification and PTEN deletion are the common and leading events in all subtypes of non-GCIMP GBMs. NF1 loss is a late event based on the subsequent targeting of NF1 in the proneural (PDGFA/shP53) mouse model that results in mesenchymal-like tumour formation (Ozawa et al., 2014). However, these studies have several limitations. First, IDH1 mutation, which is a common mutation in the proneural subtype, is detected in fewer than 10% of GBM patients. Thus, it would be difficult to make the assertion that this mutation is the driver for all tumour subtypes. Second, the NF1 mutation, which is associated with the mesenchymal subtype, and IDH1, which is the common driver mutation of the proneural subtype, are expressed in different groups explicitly. Third, a mesenchymal-like subtype has a different cell of origin in other mouse model and can arise de novo by Nf1/Tp53 loss. Finally, NF1 is a driver mutation in primary and recurrent tumours, even among a subgroup of low grade tumours (Rodriguez et al., 2008; Ozawa et al., 2014; Cancer Genome Atlas Research et al., 2015; Eckel-Passow et al., 2015; Wang et al., 2016). All these together suggest that it is unlikely that the proneural subtype is the origin of all other subtypes.
The polyclonal hypothesis of glioblastoma multiforme origin
The concept of a single clone derived from one type of cell that gives rise to cancer can be accepted in some haematological cancers, but it is highly unlikely to stand for the origin of highly heterogeneous tumours such as GBM (McGranahan and Swanton, 2017). Although the cancer stem cell model and the stochastic clonal evolution model might explain the intra-tumoural heterogeneity, it can’t ascertain the clonal origin of the tumour (Parsons, 2018). Furthermore, recent in vitro outcomes of GBM clonal analysis showed a high clonal heterogeneity among GBM patients, even within the same tumour. These clones vary in their chemo- and radio-sensitivity. Contrary to previous studies, the therapy resistance was not associated with increasing GSC marker expression (Auffinger et al., 2014; Segerman et al., 2016). This clonal heterogeneity might be behind therapeutic failure where minor untargeted clones can drive treatment resistance upon GBM recurrence. However, we should keep in mind that such an in vitro system is influenced by clonal selection dynamics during cell culture and xenografting (Patel et al., 2014; deCarvalho et al., 2018). The multiple cell of origin hypothesis was suggested a long time ago in hereditary neurofibromas, a disease related to NF1 mutation/deletion (Fialkow et al., 1971). In medulloblastoma, genetic analysis of dissected primary and recurrent tumours, from both transgenic mice and patient samples, revealed a high degree of genetic divergence with <5% overlap. The conventional treatment of medulloblastoma including surgery and radiation treatment, imposed an evolutionary pressure that selected highly divergent clones at recurrence, while the sensitive clones that were dominant in the primary tumour tended to disappear (Morrissy et al., 2016). The GBM heterogeneity at the mutational, clonal, and transcriptional subtype level, in addition to the cell of origin, suggests a polyclonal evolution of GBM origin rather than monoclonal one. The polyclonal hypothesis of GBM origin can be supported by several arguments: (i) the glioma animal models of the mesenchymal and proneural subtypes suggest that different cell populations are the cell of origin of these two different subtypes (Liu et al., 2011; Alcantara Llaguno et al., 2015; Herting et al., 2017); (ii) the clonal divergence between primary and recurrent glioma, especially in multifocal tumours, long-term recurrence and distant recurrence (Wang et al., 2016; Lee et al., 2017); (iii) <45% of the mutations are shared between primary and recurrent glioma (Johnson et al., 2014; Wang et al., 2016); (iv) switching the transcriptional subtype in recurrent GBM (Wang et al., 2016); and (v) the intra-tumoural heterogeneity among geographically-distinct tumour parts obtained from a single patient, in addition to the outcomes of single cell analysis, showed that an individual GBM patient has cells that belong to the three different subtypes (Sottoriva et al., 2013; Patel et al., 2014). Altogether, these results suggest a polyclonal origin of GBM rather than a monoclonal one. However, more research is needed to confirm this hypothesis in glioma field.
Our hypothesis is that specific genetic mutations that hit specific cell types give rise either to proneural, classical or mesenchymal subtypes, and the polyclonal evolution of GBM, which is influenced by accumulated genetic and epigenetic changes, the cell of origin, recruited stromal cells with their secreted factors, and the dynamic interactions between these variable clones, in addition to their interaction with micro-environment, drive molecular subtype heterogeneity that leads to intra- and inter-tumoural heterogeneity.
The clinical implication of subtypes
The subtype driving mutations exhibit differential therapy responses in clinical and preclinical tests. The proneural-PDGFB overexpressing model was significantly sensitive to both temozolomide and radiation treatment treatments with preferable survival achieved with radiation treatment over temozolomide. On the other hand, the NF1-silencing model was in general less sensitive than the proneural-PDGFB model and significant survival was achieved with radiation treatment treatment alone but not temozolomide treatment. And recurrent tumours displayed a trend toward more aggressive tumours (Herting et al., 2017). It has been suggested that GBM patients with pure proneural subtype that have minimum transcriptional signal of other subtypes have significantly better survival rates than patients with mesenchymal subtype (Patel et al., 2014). However, we want to keep in mind that patients with mesenchymal subtype tumours are older than proneural patients, and age by itself is negative predictor in GBM survival (Wick et al., 2012). It is speculated that the tumour micro-environment with its immune cell and vasculature components plays an essential role in the differential treatment response between different subtypes (Herting et al., 2017). Another reason for treatment sensitivity could be the intrinsic nature of tumour cells in each subtype. The oligodendroglioma characters observed in the proneural subtype model could explain the preferable response to temozolomide treatment (Schindelin et al., 2012; Schmid et al., 2016). The sensitivity of proneural subtype tumours to chemo and radiation treatment could be the reason for the induced PMT upon tumour recurrence (Segerman et al., 2016; Pencheva et al., 2017). It is expected that tumour cells with proneural subtype die in response to treatment, while cells with the mesenchymal subtype proliferate and dominate the recurrent tumour. It is worthy to note that patients from the classical and mesenchymal subtypes, but not those from the proneural subtype, get a survival benefit from aggressive treatment protocols with chemo- and radiotherapy or more than three repeated cycles of chemotherapy (Verhaak et al., 2010). Preliminary data from some immunotherapy targets showed promising outcomes in mesenchymal subtype tumours. Animal data of adoptive transfer of engineered T-cells expressing IL13-zetakine. T-cells displayed potent anti-tumour activity in the mesenchymal subtype (Brown et al., 2012). Recently it was shown that CD4+ chimeric antigen receptor (CAR) T cells targeting IL13Rα2 antigen induced a long term anti-oncogenic effect and outperformed CD8+ CAR T cells or a combination of CD4+ and CD8+ CAR T cells (Wang et al., 2018).
Further, the proneural and mesenchymal subtypes showed a differentiational response to epigenetic therapeutic targets, such as EZH2 and BMI1 inhibitors, with proneural being more sensitive to EZH2 inhibitor and mesenchymal more sensitive to BMI1. However, a heterogeneous tumour showed a better response to a combination therapy of EZH2 and BMI inhibition (Jin et al., 2017). These agents are suggested to reverse the radio and chemo-resistance properties of cancer cells (Facchino et al., 2010; Fan et al., 2014). However, although tumour subtype might give a better prediction for treatment response, intra-tumoural heterogeneity remains one of the main obstacles that contributes to GBM treatment resistance and recurrence (Patel et al., 2014).
Supplementary Material
Acknowledgements
We thank Marianne Hanna for participating in the graphic drawing, Joe Herbert and Jan Helge Solbakk for proof reading, and The University of Oslo Medical Library for logistic support.
Glossary
Abbreviations
- CTC
circulating brain tumour cell
- EMT
epithelial to mesenchymal transition
- G-CIMP
glioma CpG island methylator phenotype
- GBM
glioblastoma multiforme
- GSC
glioma stem cell
- MSC
mesenchymal stem cell
- PMT
proneural to mesenchymal transition
- TCGA
The Cancer Genome Atlas
- TIL
tumour-infiltrating lymphocyte
Funding
This work was supported by Knut and Alice Wallenbergs Foundation.
Competing interests
The authors report no competing interests.
References
- Alcantara Llaguno S, Chen J, Kwon CH, Jackson EL, Li Y, Burns DK, et al. Malignant astrocytomas originate from neural stem/progenitor cells in a somatic tumor suppressor mouse model. Cancer Cell 2009; 15: 45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcantara Llaguno SR, Wang Z, Sun D, Chen J, Xu J, Kim E, et al. Adult lineage-restricted CNS progenitors specify distinct glioblastoma subtypes. Cancer Cell 2015; 28: 429–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auffinger B, Tobias AL, Han Y, Lee G, Guo D, Dey M, et al. Conversion of differentiated cancer cells into cancer stem-like cells in a glioblastoma model after primary chemotherapy. Cell Death Differ 2014; 21: 1119–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bani-Yaghoub M, Kendall SE, Moore DP, Bellum S, Cowling RA, Nikopoulos GN, et al. Insulin acts as a myogenic differentiation signal for neural stem cells with multilineage differentiation potential. Development 2004; 131: 4287–98. [DOI] [PubMed] [Google Scholar]
- Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 2006; 444: 756–60. [DOI] [PubMed] [Google Scholar]
- Batchelor TT, Sorensen AG, di Tomaso E, Zhang WT, Duda DG, Cohen KS, et al. AZD2171, a pan-VEGF receptor tyrosine kinase inhibitor, normalizes tumor vasculature and alleviates edema in glioblastoma patients. Cancer Cell 2007; 11: 83–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnan J, Isakson P, Joel M, Cilio C, Langmoen IA, Vik-Mo EO, et al. Recruited brain tumor-derived mesenchymal stem cells contribute to brain tumor progression. Stem Cells 2014; 32: 1110–23. [DOI] [PubMed] [Google Scholar]
- Behnan J, Stangeland B, Hosainey SA, Joel M, Olsen TK, Micci F, et al. Differential propagation of stroma and cancer stem cells dictates tumorigenesis and multipotency. Oncogene 2017a; 36: 570–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnan J, Stangeland B, Langella T, Finocchiaro G, Murrell W, Brinchmann JE. Ultrasonic surgical aspirate is a reliable source for culturing glioblastoma stem cells. Sci Rep 2016; 6: 32788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnan J, Stangeland B, Langella T, Finocchiaro G, Tringali G, Meling TR, et al. Identification and characterization of a new source of adult human neural progenitors. Cell Death Dis 2017b; 8: e2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent MJVD. Interobserver variation of the histopathological diagnosis in clinical trials on glioma: a clinician's perspective. Acta Neuropathologica 2010; 120: 297–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat KP, Salazar KL, Balasubramaniyan V, Wani K, Heathcock L, Hollingsworth F, et al. The transcriptional coactivator TAZ regulates mesenchymal differentiation in malignant glioma. Genes Dev 2011; 25: 2594–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat KPL, Balasubramaniyan V, Vaillant B, Ezhilarasan R, Hummelink K, Hollingsworth F, et al. Mesenchymal differentiation mediated by NF-kappaB promotes radiation resistance in glioblastoma. Cancer Cell 2013; 24: 331–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, et al. The somatic genomic landscape of glioblastoma. Cell 2013; 155: 462–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CE, Starr R, Aguilar B, Shami AF, Martinez C, D’Apuzzo M, et al. Stem-like tumor-initiating cells isolated from IL13Ralpha2 expressing gliomas are targeted and killed by IL13-zetakine-redirected T Cells. Clin Cancer Res 2012; 18: 2199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner JC, Brown PD, O'Neill BP, Meye FB, Wetmore CJ, Uhm JH. Central nervous system tumors. Mayo Clin Proc 2007; 82: 1271–86. [DOI] [PubMed] [Google Scholar]
- Calabrese C, Poppleton H, Kocak M, Hogg TL, Fuller C, Hamner B, et al. A perivascular niche for brain tumor stem cells. Cancer Cell 2007; 11: 69–82. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008; 455: 1061–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research N, Brat DJ, Verhaak RG, Aldape KD, Yung WK, Salama SR, et al. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med 2015; 372: 2481–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carro MS, Lim WK, Alvarez MJ, Bollo RJ, Zhao X, Snyder EY, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature 2010; 463: 318–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccarelli M, Barthel FP, Malta TM, Sabedot TS, Salama SR, Murray BA, et al. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell 2016; 164: 550–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles NA, Holland EC, Gilbertson R, Glass R, Kettenmann H. The brain tumor microenvironment. Glia 2011; 59: 1169–80. [DOI] [PubMed] [Google Scholar]
- Chen J, Li Y, Yu TS, McKay RM, Burns DK, Kernie SG, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature 2012; 488: 522–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng WY, Kandel JJ, Yamashiro DJ, Canoll P, Anastassiou D. A multi-cancer mesenchymal transition gene expression signature is associated with prolonged time to recurrence in glioblastoma. PLoS One 2012; 7: e34705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinot OL, Wick W, Mason W, Henriksson R, Saran F, Nishikawa R, et al. Bevacizumab plus radiotherapy-temozolomide for newly diagnosed glioblastoma. N Engl J Med 2014; 370: 709–22. [DOI] [PubMed] [Google Scholar]
- Cooper LA, Gutman DA, Chisolm C, Appin C, Kong J, Rong Y, et al. The tumor microenvironment strongly impacts master transcriptional regulators and gene expression class of glioblastoma. Am J Pathol 2012; 180: 2108–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisan M, Yap S, Casteilla L, Chen CW, Corselli M, Park TS, et al. A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell 2008; 3: 301–13. [DOI] [PubMed] [Google Scholar]
- Cusulin C, Chesnelong C, Bose P, Bilenky M, Kopciuk K, Chan JA, et al. Precursor states of brain tumor initiating cell lines are predictive of survival in xenografts and associated with glioblastoma subtypes. Stem Cell Rep 2015; 5: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darmanis S, Sloan SA, Croote D, Mignardi M, Chernikova S, Samghababi P, et al. Single-Cell RNA-Seq analysis of infiltrating neoplastic cells at the migrating front of human glioblastoma. Cell Rep 2017; 21: 1399–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot JF, Fuller G, Kumar AJ, Piao Y, Eterovic K, Ji Y, et al. Tumor invasion after treatment of glioblastoma with bevacizumab: radiographic and pathologic correlation in humans and mice. Neuro Oncol 2010; 12: 233–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deCarvalho AC, Kim H, Poisson LM, Winn ME, Mueller C, Cherba D, et al. Discordant inheritance of chromosomal and extrachromosomal DNA elements contributes to dynamic disease evolution in glioblastoma. Nat Genet 2018; 50: 708–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckel-Passow JE, Lachance DH, Molinaro AM, Walsh KM, Decker PA, Sicotte H, et al. Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med 2015; 372: 2499–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler JR, Robinson AE, Smirnov I, Hodgson JG, Berger MS, Gupta N, et al. Increased microglia/macrophage gene expression in a subset of adult and pediatric astrocytomas. PLoS One 2012; 7: e43339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facchino S, Abdouh M, Chatoo W, Bernier G. BMI1 confers radioresistance to normal and cancerous neural stem cells through recruitment of the DNA damage response machinery. J Neurosci 2010; 30: 10096–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan TY, Wang H, Xiang P, Liu YW, Li HZ, Lei BX, et al. Inhibition of EZH2 reverses chemotherapeutic drug TMZ chemosensitivity in glioblastoma. Int J Clin Exp Pathol 2014; 7: 6662–70. [PMC free article] [PubMed] [Google Scholar]
- Fedele V, Dai F, Masilamani AP, Heiland DH, Kling E, Gatjens-Sanchez AM, et al. Epigenetic regulation of ZBTB18 promotes glioblastoma progression. Mol Cancer Res 2017; 15: 998–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fialkow PJ, Sagebiel RW, Gartler SM, Rimoin DL. Multiple cell origin of hereditary neurofibromas. N Engl J Med 1971; 284: 298–300. [DOI] [PubMed] [Google Scholar]
- Flavahan WA, Wu Q, Hitomi M, Rahim N, Kim Y, Sloan AE, et al. Brain tumor initiating cells adapt to restricted nutrition through preferential glucose uptake. Nat Neurosci 2013; 16: 1373–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman HS, Prados MD, Wen PY, Mikkelsen T, Schiff D, Abrey LE, et al. Bevacizumab alone and in combination with irinotecan in recurrent glioblastoma. J Clin Oncol 2009; 27: 4733–40. [DOI] [PubMed] [Google Scholar]
- Gabrusiewicz K, Rodriguez B, Wei J, Hashimoto Y, Healy LM, Maiti SN, et al. Glioblastoma-infiltrated innate immune cells resemble M0 macrophage phenotype. JCI Insight 2016; 1. doi: 10.1172/jci.insight.85841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanis E, Anderson SK, Lafky JM, Uhm JH, Giannini C, Kumar SK, et al. Phase II study of bevacizumab in combination with sorafenib in recurrent glioblastoma (N0776): a north central cancer treatment group trial. Clin Cancer Res 2013; 19: 4816–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S, et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res 2004; 64: 7011–21. [DOI] [PubMed] [Google Scholar]
- Gao F, Cui Y, Jiang H, Sui D, Wang Y, Jiang Z, et al. Circulating tumor cell is a common property of brain glioma and promotes the monitoring system. Oncotarget 2016; 7: 71330–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstung M, Eriksson N, Lin J, Vogelstein B, Beerenwinkel N. The temporal order of genetic and pathway alterations in tumorigenesis. PLoS One 2011; 6: e27136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill BJ, Pisapia DJ, Malone HR, Goldstein H, Lei L, Sonabend A, et al. MRI-localized biopsies reveal subtype-specific differences in molecular and cellular composition at the margins of glioblastoma. Proc Natl Acad Sci USA 2014; 111: 12550–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenberger ML, Jenkins RB. Genetics of adult glioma. Cancer Genet 2012; 205: 613–21. [DOI] [PubMed] [Google Scholar]
- Guan X, Vengoechea J, Zheng S, Sloan AE, Chen Y, Brat DJ, et al. Molecular subtypes of glioblastoma are relevant to lower grade glioma. PLoS One 2014; 9: e91216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday J, Helmy K, Pattwell SS, Pitter KL, LaPlant Q, Ozawa T, et al. In vivo radiation response of proneural glioma characterized by protective p53 transcriptional program and proneural-mesenchymal shift. Proc Natl Acad Sci USA 2014; 111: 5248–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hambardzumyan D, Amankulor NM, Helmy KY, Becher OJ, Holland EC. Modeling adult gliomas using RCAS/t-va technology. Transl Oncol 2009; 2: 89–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med 2005; 352: 997–1003. [DOI] [PubMed] [Google Scholar]
- Herting CJ, Chen Z, Pitter KL, Szulzewsky F, Kaffes I, Kaluzova M, et al. Genetic driver mutations define the expression signature and microenvironmental composition of high-grade gliomas. Glia 2017; 65: 1914–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjelmeland AB, Wu Q, Heddleston JM, Choudhary GS, MacSwords J, Lathia JD, et al. Acidic stress promotes a glioma stem cell phenotype. Cell Death Differ 2011; 18: 829–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson JG, Yeh RF, Ray A, Wang NJ, Smirnov I, Yu M, et al. Comparative analyses of gene copy number and mRNA expression in glioblastoma multiforme tumors and xenografts. Neuro Oncol 2009; 11: 477–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse JT, Phillips HS, Brennan CW. Molecular subclassification of diffuse gliomas: seeing order in the chaos. Glia 2011; 59: 1190–9. [DOI] [PubMed] [Google Scholar]
- Isella C, Terrasi A, Bellomo SE, Petti C, Galatola G, Muratore A, et al. Stromal contribution to the colorectal cancer transcriptome. Nat Genet 2015; 47: 312–9. [DOI] [PubMed] [Google Scholar]
- Jin K, Zhu Y, Sun Y, Mao XO, Xie L, Greenberg DA. Vascular endothelial growth factor (VEGF) stimulates neurogenesis in vitro and in vivo. Proc Natl Acad Sci USA 2002; 99: 11946–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X, Kim LJY, Wu Q, Wallace LC, Prager BC, Sanvoranart T, et al. Targeting glioma stem cells through combined BMI1 and EZH2 inhibition. Nat Med 2017; 23: 1352–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BE, Mazor T, Hong C, Barnes M, Aihara K, McLean CY, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science 2014; 343: 189–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo KM, Kim J, Jin J, Kim M, Seol HJ, Muradov J, et al. Patient-specific orthotopic glioblastoma xenograft models recapitulate the histopathology and biology of human glioblastomas in situ. Cell Rep 2013; 3: 260–73. [DOI] [PubMed] [Google Scholar]
- Kast RE, Skuli N, Karpel-Massler G, Frosina G, Ryken T, Halatsch ME. Blocking epithelial-to-mesenchymal transition in glioblastoma with a sextet of repurposed drugs: the EIS regimen. Oncotarget 2017; 8: 60727–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keunen O, Johansson M, Oudin A, Sanzey M, Rahim SA, Fack F, et al. Anti-VEGF treatment reduces blood supply and increases tumor cell invasion in glioblastoma. Proc Natl Acad Sci USA 2011; 108: 3749–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Watkinson J, Varadan V, Anastassiou D. Multi-cancer computational analysis reveals invasion-associated variant of desmoplastic reaction involving INHBA, THBS2 and COL11A1. BMC Med Genomics 2010; 3: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Zheng S, Amini SS, Virk SM, Mikkelsen T, Brat DJ, et al. Whole-genome and multisector exome sequencing of primary and post-treatment glioblastoma reveals patterns of tumor evolution. Genome Res 2015; 25: 316–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai A, Tran A, Nghiemphu PL, Pope WB, Solis OE, Selch M, et al. Phase II study of bevacizumab plus temozolomide during and after radiation therapy for patients with newly diagnosed glioblastoma multiforme. J Clin Oncol 2011; 29: 142–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CL, Rich JN. Cancer stem cells in glioblastoma. Genes Dev 2015; 29: 1203–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau J, Ilkhanizadeh S, Wang S, Miroshnikova YA, Salvatierra NA, Wong RA, et al. STAT3 blockade inhibits radiation-induced malignant progression in glioma. Cancer Res 2015; 75: 4302–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell 2006; 9: 391–403. [DOI] [PubMed] [Google Scholar]
- Lee JK, Joo KM, Lee J, Yoon Y, Nam DH. Targeting the epithelial to mesenchymal transition in glioblastoma: the emerging role of MET signaling. Onco Targets Ther 2014; 7: 1933–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JK, Wang J, Sa JK, Ladewig E, Lee HO, Lee IH, et al. Spatiotemporal genomic architecture informs precision oncology in glioblastoma. Nat Genet 2017; 49: 594–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Graeber MB. The molecular profile of microglia under the influence of glioma. Neuro Oncol 2012; 14: 958–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Bao S, Wu Q, Wang H, Eyler C, Sathornsumetee S, et al. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer Cell 2009; 15: 501–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Sage JC, Miller MR, Verhaak RG, Hippenmeyer S, Vogel H, et al. Mosaic analysis with double markers reveals tumor cell of origin in glioma. Cell 2011; 146: 209–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Yuan X, Zeng Z, Tunici P, Ng H, Abdulkadir IR, et al. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol Cancer 2006; 5: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lottaz C, Beier D, Meyer K, Kumar P, Hermann A, Schwarz J, et al. Transcriptional profiles of CD133+ and CD133- glioblastoma-derived cancer stem cell lines suggest different cells of origin. Cancer Res 2010; 70: 2030–40. [DOI] [PubMed] [Google Scholar]
- Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol 2007; 114: 97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis DN, Perry A, Reifenburger G, von Deimling A, Figarella-Branger D, Cavanee WK, et al. The 2016 world health organization classification of tumors of the central nervous system: a summary. Acta Neuropathol 2016; 131: 803–20. [DOI] [PubMed] [Google Scholar]
- Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 2012a; 483: 474–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu KV, Chang JP, Parachoniak CA, Pandika MM, Aghi MK, Meyronet D, et al. VEGF inhibits tumor cell invasion and mesenchymal transition through a MET/VEGFR2 complex. Cancer Cell 2012b; 22: 21–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao P, Joshi K, Li J, Kim SH, Li P, Santana-Santos L, et al. Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc Natl Acad Sci USA 2013; 110: 8644–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez E, Yoshihara K, Kim H, Mills GM, Trevino V, Verhaak RG. Comparison of gene expression patterns across 12 tumor types identifies a cancer supercluster characterized by TP53 mutations and cell cycle defects. Oncogene 2015; 34: 2732–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazor T, Pankov A, Johnson BE, Hong C, Hamilton EG, Bell RJA, et al. DNA methylation and somatic mutations converge on the cell cycle and define similar evolutionary histories in brain tumors. Cancer Cell 2015; 28: 307–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGranahan N, Swanton C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell 2017; 168: 613–28. [DOI] [PubMed] [Google Scholar]
- Morrissy AS, Garzia L, Shih DJ, Zuyderduyn S, Huang X, Skowron P, et al. Divergent clonal selection dominates medulloblastoma at recurrence. Nature 2016; 529: 351–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murat A, Migliavacca E, Gorlia T, Lambiv WL, Shay T, Hamou MF, et al. Stem cell-related “self-renewal” signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J Clin Oncol 2008; 26: 3015–24. [DOI] [PubMed] [Google Scholar]
- Nordling CO. A new theory on cancer-inducing mechanism. Br J Cancer 1953; 7: 68–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 2010; 17: 510–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowell PC. The clonal evolution of tumor cell populations. Science 1976; 194: 23–8. [DOI] [PubMed] [Google Scholar]
- Ozawa T, Riester M, Cheng YK, Huse JT, Squatrito M, Helmy K, et al. Most human non-GCIMP glioblastoma subtypes evolve from a common proneural-like precursor glioma. Cancer Cell 2014; 26: 288–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons BL. Multiclonal tumor origin: evidence and implications. Mutat Res 2018; 777: 1–18. [DOI] [PubMed] [Google Scholar]
- Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 2014; 344: 1396–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pencheva N, de Gooijer MC, Vis DJ, Wessels LFA, Wurdinger T, van Tellingen O, et al. Identification of a druggable pathway controlling glioblastoma invasiveness. Cell Rep 2017; 20: 48–60. [DOI] [PubMed] [Google Scholar]
- Perry JR, Laperriere N, O’Callaghan CJ, Brandes AA, Menten J, Phillips C, et al. Short-course radiation plus temozolomide in elderly patients with glioblastoma. N Engl J Med 2017; 376: 1027–37. [DOI] [PubMed] [Google Scholar]
- Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 2006; 9: 157–73. [DOI] [PubMed] [Google Scholar]
- Piao Y, Liang J, Holmes L, Henry V, Sulman E, de Groot JF. Acquired resistance to anti-VEGF therapy in glioblastoma is associated with a mesenchymal transition. Clin Cancer Res 2013; 19: 4392–403. [DOI] [PubMed] [Google Scholar]
- Piao Y, Liang J, Holmes L, Zurita AJ, Henry V, Heymach JV, et al. Glioblastoma resistance to anti-VEGF therapy is associated with myeloid cell infiltration, stem cell accumulation, and a mesenchymal phenotype. Neuro Oncol 2012; 14: 1379–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port RE, Bernstein LJ, Barboriak DP, Xu L, Roberts TP, van Bruggen N. Noncompartmental kinetic analysis of DCE-MRI data from malignant tumors: application to glioblastoma treated with bevacizumab. Magn Reson Med 2010; 64: 408–17. [DOI] [PubMed] [Google Scholar]
- Puchalski RB, Shah N, Miller J, Dalley R, Nomura SR, Yoon JG, et al. An anatomic transcriptional atlas of human glioblastoma. Science 2018; 360: 660–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quant EC, Norden AD, Drappatz J, Muzikansky A, Doherty L, Lafrankie D, et al. Role of a second chemotherapy in recurrent malignant glioma patients who progress on bevacizumab. Neuro Oncol 2009; 11: 550–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratner N, Miller SJ. A RASopathy gene commonly mutated in cancer: the neurofibromatosis type 1 tumour suppressor. Nat Rev Cancer 2015; 15: 290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddick G, Fine HA. Integration and analysis of genome-scale data from gliomas. Nat Rev Neurol 2011; 7: 439–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez FJ, Perry A, Gutmann DH, O’Neill BP, Leonard J, Bryant S, et al. Gliomas in neurofibromatosis type 1: a clinicopathologic study of 100 patients. J Neuropathol Exp Neurol 2008; 67: 240–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutledge WC, Kong J, Gao J, Gutman DA, Cooper LA, Appin C, et al. Tumor-infiltrating lymphocytes in glioblastoma are associated with specific genomic alterations and related to transcriptional class. Clin Cancer Res 2013; 19: 4951–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 2012; 9: 676–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid RS, Simon JM, Vitucci M, McNeill RS, Bash RE, Werneke AM, et al. Core pathway mutations induce de-differentiation of murine astrocytes into glioblastoma stem cells that are sensitive to radiation but resistant to temozolomide. Neuro Oncol 2016; 18: 962–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott BJ, Quant EC, McNamara MB, Ryg PA, Batchelor TT, Wen PY. Bevacizumab salvage therapy following progression in high-grade glioma patients treated with VEGF receptor tyrosine kinase inhibitors. Neuro Oncol 2010; 12: 603–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segerman A, Niklasson M, Haglund C, Bergstrom T, Jarvius M, Xie Y, et al. Clonal variation in drug and radiation response among glioma-initiating cells is linked to proneural-mesenchymal transition. Cell Rep 2016; 17: 2994–3009. [DOI] [PubMed] [Google Scholar]
- Siebzehnrubl FA, Silver DJ, Tugertimur B, Deleyrolle LP, Siebzehnrubl D, Sarkisian MR, et al. The ZEB1 pathway links glioblastoma initiation, invasion and chemoresistance. EMBO Mol Med 2013; 5: 1196–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res 2003; 63: 5821–8. [PubMed] [Google Scholar]
- Smith DR, Hardman JM, Earle KM. Contiguous glioblastoma multiforme and fibrosarcoma with extracranial metastasis. Cancer 1969; 24: 270–6. [DOI] [PubMed] [Google Scholar]
- Sottoriva A, Spiteri I, Piccirillo SG, Touloumis A, Collins VP, Marioni JC, et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci USA 2013; 110: 4009–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 2005; 352: 987–96. [DOI] [PubMed] [Google Scholar]
- Sturm D, Witt H, Hovestadt V, Khuong-Quang DA, Jones DT, Konermann C, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell 2012; 22: 425–37. [DOI] [PubMed] [Google Scholar]
- Sullivan JP, Nahed BV, Madden MW, Oliveira SM, Springer S, Bhere D, et al. Brain tumor cells in circulation are enriched for mesenchymal gene expression. Cancer Discov 2014; 4: 1299–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surawicz TS, McCarthy BJ, Kupelian V, Jukich PJ, Bruner JM, Davis FG. Descriptive epidemiology of primary brain and CNS tumors: Results from the Central Brain Tumor Registry of the United States, 1990-1994. Neuro-Oncology 1999; 1: 14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson A, Ozen I, Genove G, Paul G, Bengzon J. Endogenous brain pericytes are widely activated and contribute to mouse glioma microvasculature. PLoS One 2015; 10: e0123553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulzewsky F, Arora S, de Witte L, Ulas T, Markovic D, Schultze JL, et al. Human glioblastoma-associated microglia/monocytes express a distinct RNA profile compared to human control and murine samples. Glia 2016; 64: 1416–36. [DOI] [PubMed] [Google Scholar]
- Taal W, Oosterkamp HM, Walenkamp AM, Dubbink HJ, Beerepoot LV, Hanse MC, et al. Single-agent bevacizumab or lomustine versus a combination of bevacizumab plus lomustine in patients with recurrent glioblastoma (BELOB trial): a randomised controlled phase 2 trial. Lancet Oncol 2014; 15: 943–53. [DOI] [PubMed] [Google Scholar]
- Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 2012; 483: 479–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Bent MJ, Gao Y, Kerkhof M, Kros JM, Gorlia T, van Zwieten K, et al. Changes in the EGFR amplification and EGFRvIII expression between paired primary and recurrent glioblastomas. Neuro Oncol 2015; 17: 935–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velpula KK, Dasari VR, Tsung AJ, Dinh DH, Rao JS. Cord blood stem cells revert glioma stem cell EMT by down regulating transcriptional activation of Sox2 and Twist1. Oncotarget 2011; 2: 1028–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010; 17: 98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Aguilar B, Starr R, Alizadeh D, Brito A, Sarkissian A, et al. Glioblastoma-targeted CD4+ CAR T cells mediate superior antitumor activity. JCI Insight 2018; 3. doi: 10.1172/jci.insight.99048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Cazzato E, Ladewig E, Frattini V, Rosenbloom DI, Zairis S, et al. Clonal evolution of glioblastoma under therapy. Nat Genet 2016; 48: 768–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhou F, Li Y, Li Q, Wu Z, Yu L, et al. Cdc20 overexpression is involved in temozolomide-resistant glioma cells with epithelial-mesenchymal transition. Cell Cycle 2017a; 16: 2355–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Hu B, Hu X, Kim H, Squatrito M, Scarpace L, et al. Tumor evolution of glioma-intrinsic gene expression subtypes associates with immunological changes in the microenvironment. Cancer Cell 2017b; 32: 42–56. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med 2008; 359: 492–507. [DOI] [PubMed] [Google Scholar]
- Wick W, Platten M, Meisner C, Felsberg J, Tabatabai G, Simon M, et al. Temozolomide chemotherapy alone versus radiotherapy alone for malignant astrocytoma in the elderly: the NOA-08 randomised, phase 3 trial. Lancet Oncol 2012; 13: 707–15. [DOI] [PubMed] [Google Scholar]
- Ye XZ, Xu SL, Xin YH, Yu SC, Ping YF, Chen L, et al. Tumor-associated microglia/macrophages enhance the invasion of glioma stem-like cells via TGF-beta1 signaling pathway. J Immunol 2012; 189: 444–53. [DOI] [PubMed] [Google Scholar]
- Zheng S, Chheda MG, Verhaak RG. Studying a complex tumor: potential and pitfalls. Cancer J 2012; 18: 107–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.