Abstract
Medulloblastoma (MB) is the most prevalent malignant brain tumor in children, accounting for 20% of all childhood brain tumors. The molecular profiling of MB into 4 major subgroups (WNT, SHH, Grp3, and Grp4) emphasizes the heterogeneity of MB and opens paths in which treatments may be targeted to molecularly aggressive and distinct tumors. Current therapeutic strategies for Group 3 MB are challenging and can be accompanied by long-term side effects from treatment. The involvement of altered epigenetic machinery in neoplastic transformation in MB has become more evident. Thus, we performed an epigenomic RNAi and chemical screen and identified SETD8/PRE-SET7/KMT5a as a critical player in maintaining proliferation and cell survival of MB cells. We have found that inhibition of SETD8 effects the migration/invasive ability of MB cells. SETD8 alters H4K20me chromatin occupancy at key genes involved in tumor invasiveness and pluripotency. Interestingly, these results link the aggressive and metastatic behavior of MYC-driven MB with SETD8 activity. Based on our results, we suggest that SETD8 has a critical role mediating Group 3 MB tumorigenesis. Establishing a role for SETD8 as a factor in MYC-driven MB has potential to lead to more effective therapies needed to improve outcomes in high-risk patients.
Keywords: Oncology
Keywords: Cancer gene therapy, Cell migration/adhesion, Molecular biology
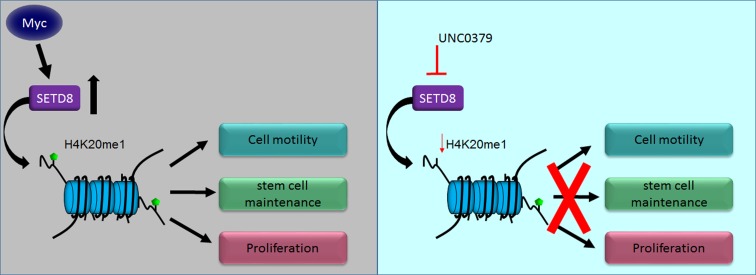
MYC-driven medulloblastoma cells are susceptible to inhibition of SETD8, which regulates genes involved in cell motility, invasion, and pluripotency.
Introduction
Pediatric cancers exhibit a high level of epigenomic dysregulation (1, 2). This is in contrast to a low mutational burden and a genetically quiet cancer genome (3). Pediatric brain tumors, in particular, demonstrate few aberrant genetic events but rather exhibit an altered epigenome (4–7). This aberrant epigenomic regulation often occurs in the face of a singular genetic event such as MYC amplification in medulloblastoma (MB) (7). Because these epigenomic changes are reversible, they represent druggable molecular vulnerabilities that can be therapeutically exploited in these aggressive pediatric brain tumors.
MB, the most common embryonic brain tumor of early childhood, shows remarkable biological and clinical heterogeneity (8).Treatment modalities for MB have remained static, consisting of tumor resection, craniospinal radiation for those >3 years, and combination chemotherapy. In the past decade, intensification of this therapeutic strategy has improved overall survival to 70%, though patients experience neurological complications and long-term sequelae from treatment (9). These detrimental effects make the case for reducing radiation exposure in favor of improved patient-specific targeted therapies. Recent transcriptional profiling studies identify that MB has 4 distinct molecular subgroups characterized by unique genomic and clinical features (10–14). Two subgroups are associated with specific abnormalities in developmental pathways, Wingless (WNT) and Sonic Hedgehog (SHH). The other 2 subgroups, Group 3 and Group 4, have not been characterized, as well (15). More recently, using a combinatorial genomic approach, the number of subtypes of MB was expanded to 12, including 2 WNT, 4 SHH, three Group 3, and three Group 4 subtypes (16). Among these subgroups, Group 3 tumors, and particularly Group 3γ, represent a severe form of the disease characterized by frequent MYC gene amplification, metastasis, relapse, and association with a poor 5-year survival rate of less than 30% (17, 18). Thus, there is a critical need to develop more effective therapies in Group 3 MYC–amplified MB.
The accumulating evidence of epigenetic deregulation during MB tumorigenesis provides an opportunity to identify novel directed therapeutics to improve patient outcome (19). Which epigenetic regulators are functionally critical for MYC-driven MB is, however, unclear. Previously, we performed an epigenome-wide shRNA screen in Group 3 MYC–amplified MB cell lines to identify critical epigenetic regulators necessary for uncontrolled cell growth. We have now performed an epigenome targeted chemical screen to identify therapeutically relevant targets in Group 3 MYC–amplified MB. Our dual screen revealed that the SETD8/PR-SET7/KMT5a gene is crucial for viability of MYC MB cells. SETD8 is a monomethyltransferase with 1 known substrate, histone 4 lysine 20 methylation (H4K20me). H4K20 monomethylation functions to regulate transcription, recruitment of DNA replication machinery, DNA damage repair, and cell cycle progression (20–22). SETD8 is deregulated in some malignancies, including small-cell lung carcinoma, pancreatic cancer, and neuroblastoma, where inhibition of SETD8 can activate p53 proapoptotic programs (23, 24). However, the action of SETD8 in MB is currently unknown, and its connection to MYC-driven tumorigenesis is unclear.
In this study, we characterized SETD8 as a factor that promotes self-renewal, proliferation, and invasion/metastasis of MB cell lines. We report that loss of SETD8 results in decreased occupancy of H4K20me1 at key epithelial to mesenchymal transition (EMT) gene promoter sites, and confers a survival advantage in an orthotopic mouse model. We also establish chemical inhibition of SETD8 as an effective mechanism for reducing self-renewal, growth, and cell viability, indicating that SETD8 is a key therapeutic target in MYC-amplified MB.
Results
Dual genomic and chemical screens identify SETD8 as a critical factor in MB.
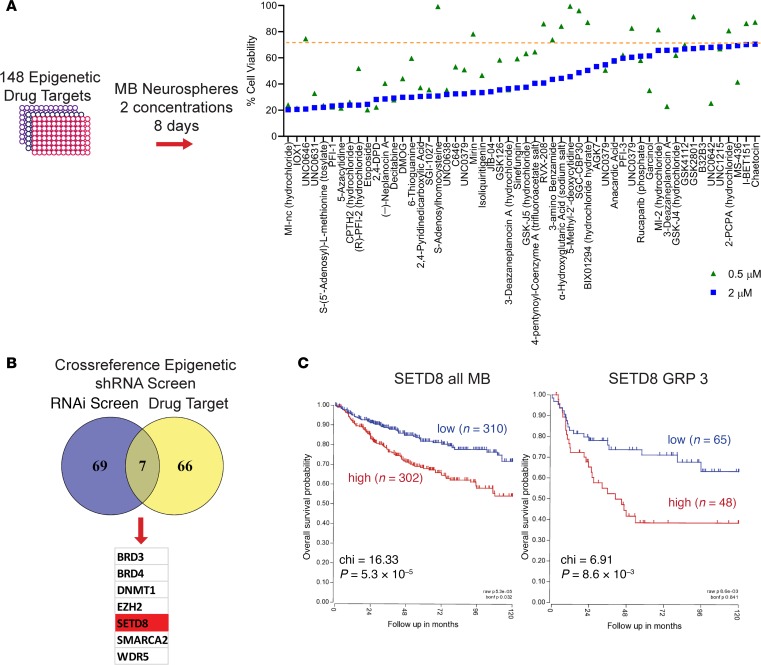
To understand what epigenetic regulators facilitate deregulated growth of MYC-amplified MB, we initially performed a RNAi screen using a pooled shRNA library targeting 410 epigenetic/chromatin regulator genes. Lentiviruses expressing 4399 distinct shRNA (4–12 individual shRNAs targeting each of 408 gene products) were transduced into MYC-driven D458 MB cells. Following puromycin selection, changes in library representation after 18 days of culture were examined in 4 independent replicates using deep sequencing of shRNA amplified from genomic DNA. The initial screen identified 76 epigenetic regulators that, when reduced, resulted in significant cell loss (Gosh et al., University of Colorado Anschutz Medical Campus, personal communication, Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.122933DS1). To complement the shRNA library screen, we evaluated the effectiveness of epigenetic chemical modifiers against MYC-amplified MB neurosphere growth. We examined 148 epigenetic drugs that target the activity of methyltransferases, demethylases, histone acetyl transferase (HATS), Histone deacetylase (HDAC), and acetylated lysine reader proteins. D458 MB neurospheres were treated with 2 μM and 0.5 μM concentrations for 8 days, and cell viability was assessed by 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt; (MTS) assay. Chemical modifiers of HDACs have recently been characterized as potent inhibitors of MYC-amplified MB growth (25). Thus, to focus on potentially novel modifiers, HDAC inhibitors are not represented in our final screen. Chemical inhibitors that reduced cell viability to 70% of the DMSO control well were further examined (Figure 1A). In order to focus on therapeutically relevant targets, we compared the targets of the chemical inhibitors to our original target list from the RNAi screen (Figure 1B). The matched gene list identified 7 epigenetic factors that were strong hits on the RNAi screen and on the chemical screen. These were BRD3/4, DNMT1, SMARCA2, EZH2, SETD8, and WDR5 (Figure 1B). DNMT1 and SMARCA2 are known epigenetic regulators in MB (26–28). Inhibition of BRD3 and -4 through bromodomain inhibitors has been established as an effective treatment in MB, by our laboratory as well as others (29–31). In addition, our laboratory previously identified EZH2 in MB (32) and currently has submitted work on WDR5. SETD8, though known in other cancers, has not been previously linked to MB, to our knowledge. Additionally, SETD8 ranked as a top sequencing hit in the epigenetic shRNA pool (Supplemental Table 1) that was necessary for MB cell growth. SETD8 is a histone methyltransferase that catalyzes monomethylation of histone 4 lysine 20 and is necessary for cell proliferation (33).
Figure 1. Combinatorial screen establishes SETD8 as a factor in medulloblastoma growth.
(A) Epigenetic chemical inhibitor screen. D458 neurospheres were exposed to 148 epigenetic chemical inhibitors at 2 μM and 0.5 μM for 8 days. Graphical representation of cell viability is shown for those chemical inhibitors that met a 70% viable cells threshold. (B) Venn diagram representing the 7 overlapping genes from both shRNA epigenetic library screen and epigenetic chemical inhibitor screen. (C) Kaplan-Meier plots indicating overall survival in relation to SETD8 expression in all MB patient populations (χ2 = 16.33, P = 5.3 × 10–5) or in the Group 3 MB subtype (χ2 = 6.91, P = 8.6 × 10–3). See also Supplemental Table 1 and Supplemental Figure 1.
Initially, we evaluated the expression level of SETD8 in the 4 major subtypes of MB using a large recently published cohort of patients (16). Expression of SETD8 remained moderately consistent in all subtypes (Supplemental Figure 1). However, when we examined overall survival in patient populations, there was a 20% survival disadvantage for patients with high SETD8 mRNA expression (n = 300) compared with those with low levels of SETD8 (n = 310) across all subgroups (Figure 1C). Significantly poorer overall survival was noted in Group 3 patients with high SETD8 (Figure 1C). SETD8 expression was also prognostic in SHH and Group 4 but not WNT (Supplemental Figure 1). A drawback of this analysis is that the patient population is heterogeneous, particularly in terms of treatments for patients. Nevertheless, these results suggest that higher SETD8 levels are associated with poor prognosis in Group 3, sonic hedgehog (Shh), and Group 4 MB patients. Because Group 3 patients have worse outcomes, we chose to focus further on this subtype of MB.
SETD8 knockdown affects MB cell growth.
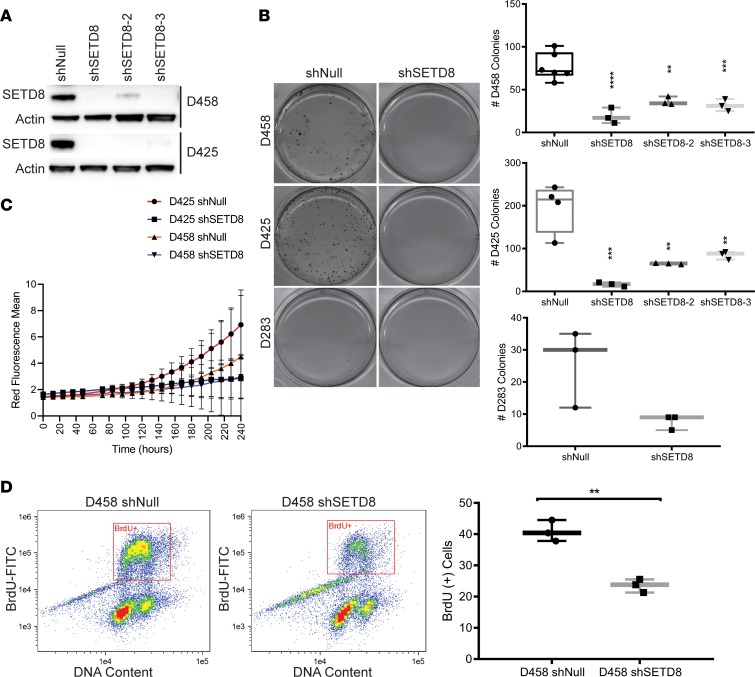
In order to further understand and validate SETD8 as a critical factor in MB tumorigenesis, a stable knockdown of SETD8 was made in the MB cell line D458 using 3 specific shRNA lentiviruses and a shNull negative control. Knockdown of SETD8 by 80% was achieved in D458 cells as compared with control cells (Figure 2A and Supplemental Figure 2). Using the same approach, SETD8 was depleted in additional MYC-amplified cells, D425 and D283; MYC medium expression cell line, DAOY; and a MYC low-expression cell line, ONS76, for a comprehensive evaluation of the relationship between SETD8 and MYC (Figure 2A and Supplemental Figure 2).
Figure 2. SETD8 is a critical mediator of MB cell growth.
(A) Western blot showing SETD8 and actin from stably transduced shNull or shSETD8 D458 and D425 cells. See also Supplemental Figure 2. (B) Methylcellulose assay with D458, D425, and D283 shNull. Representative images are shown. Line and whisker plots represent the mean ± SD from 3 independent replicates. **P < 0.01; ***P < 0.001; ****P < 0.0001; One-way Anova Dunnett’s test and unpaired t test. See also Supplemental Figure 2. (C) Proliferation of D458 and D425 shNull or shSETD8 neurospheres expressing a NucRed fluorescence marker. The mean red fluorescence ± SD vs. time. (D) Flow cytometry analysis of cell proliferation by FITC-conjugated BrdU labeling of actively dividing D458 shNull and D458 shSETD8 cells, costained with 7-AAD. Line and whisker plots represent the mean ± SD from 3 independent replicates. Unpaired t test, **P < 0.01. See also Supplemental Figure 2.
To validate the growth inhibitory phenotype that was observed in our epigenomic screens, we studied the ability of colonies to form in SETD8 knockdown cell lines, D458, D425, and D283, using a methylcellulose assay. The methylcellulose assay measures the ability of a single cell suspension to self-renew and proliferate. shSETD8 cells showed a substantial decrease in the number of colonies when compared with control cell lines (Figure 2B). D458 shNull and D425 cells formed an average of 85 and 200 colonies, respectively, measured as a cluster of 50 or more cells, whereas the D458 and D425 shSETD8 cells were significantly reduced to ≤40 and ≤75 colonies with all 3 shRNAs, respectively (Figure 2B and Supplemental Figure 2). Similar results were observed in the D283 cells, which showed less than 10 colonies formed down from 25 (Figure 2B). Clonogenic assays were conducted for the medium and low MYC expression cell lines DAOY and ONS76, and an analogous decrease in colony formation was observed (Supplemental Figure 2). These data suggest that SETD8 influences self-renewal ability and proliferation in MB cells.
To further examine the role of SETD8 in MB cells, we monitored the growth of D458 and D425 shNull and shSETD8 cell lines, expressing a NucRed live cell fluorescence marker with the Incucyte live cell analysis system. Growth of D458 and D425 shNull cells increased over 10 days as observed by the increase in fluorescence (Figure 2C). In contrast, D458 and D425 shSETD8 cells did not increase in number over the same time period, confirming the growth inhibitory phenotype. Recently, SETD8 loss has been shown to induce senescence in human fibroblasts (34). To determine if the observed growth inhibition in MB cells was a result of increased senescence, we performed a senescence associated β-galactosidase assay. We found an increase in senescence, both in D458 and D425 cells depleted of SETD8 (Supplemental Figure 2). However, the proportion of senescent cells alone could not fully account for the limitation in growth.
Known mechanisms of SETD8 activity suggest that SETD8 controls cell cycle progression at many levels, including mitotic condensation, S phase entry, and progression through DNA damage checkpoint (22). Indeed, expression of SETD8 is tightly regulated throughout the cell cycle but is quickly degraded upon entry into S phase (20). To examine if SETD8 stable knockdown confers similar phenotypes in MB cells, we performed cell cycle analyses with D458, D425, DAOY, and ONS76 cell lines using flow cytometry. Evaluation of cells stained with DRAQ5 DNA stain revealed an overall reduction in the percentage of cells in G1 phase by 10% (Supplemental Figure 2). We also observed an overall increase in the number of cells in S phase by 20% and G2/M by 11% (Supplemental Figure 2). Correspondingly, we evaluated DNA synthesis with bromodexoyuridine staining in D458 stable knockdowns. SETD8 depletion reduced active synthesis by 15% (Figure 2D) in D458 cells, corroborating the reduction of SETD8 impairs MB cell propagation. Taken together, these results indicate that SETD8 is an important factor required for aberrant growth and suggest that SETD8 has a role in self-renewal of MB cancer cells.
RNA sequencing indicates that SETD8 plays roles in cell motility, proliferation, and stemness.
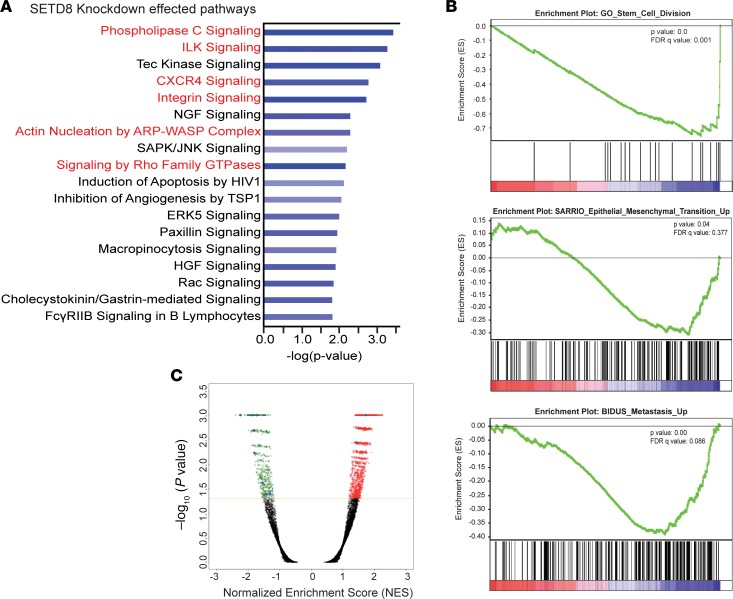
To study the downstream consequences of SETD8 depletion in MB comprehensively, we performed whole transcriptome analyses of SETD8-sufficient and -depleted cells using RNA sequencing (RNA-Seq). We then performed network analysis on the RNA-Seq data to identify pathways that were altered by SETD8 depletion. Ingenuity Pathway Analysis (IPA, https://analysis.ingenuity.com/) revealed that the significantly affected genes are involved in phospholipase C signaling, ILK1/integrin signaling, CXCR4 signaling, Rho GTPase signaling, and actin nucleation (Figure 3A). These pathways maintain cell-to-cell contacts and regulate invasion/metastasis (35, 36). For instance, in clinical samples of MB, elevated levels of IGF1 and insulin-like growth factor binding protein (IGFBP3) were shown to be present in the CSF leptomeningeal surface, and they accelerated migration and adhesion of MYC-driven MB (37). IGFBP2, as well as ITGA5 (integrin α-5), were among the top downregulated genes with SETD8 knockdown (SETD8-KD) (Supplemental Figure 3). Additionally, we found the affected molecular and cellular functions show overlapping factors regulating cellular movement, cell death and survival, and proliferation (Supplemental Figure 3).
Figure 3. RNA-Seq reveals roles for SETD8 in stem cell maintenance, proliferation, and metastatic pathways.
(A) The top pathways from Ingenuity Pathway Analysis effected by SETD8 knockdown. Bars represent significance of gene enrichment for a given pathway on a –log10(P value) scale. Select pathways are highlighted in red. (B) GSEA D458 shNull vs. shSETD8 RNA-Seq. FDR q value and P value are shown for each plot. Negative enrichment of gene sets involved stem cell maintenance, EMT, metastasis. (C) Volcano plot of normalized enrichment score vs. –log10(P value) for genesets effected by knockdown of SETD8 in D458 cells. Selected genesets are highlighted in blue. See also Supplemental Figure 3.
We sought to validate the decrease in ILK1/integrin signaling, CXCR4 signaling, and ITGA5, IGFBP2, and IGFBP3 by assessing RNA transcript levels in D458 and D425 MB cells. A 60% decrease or more in ILK1, ITGA5, IGFBP2, IGFBP3, and CXCR4 was detected in SETD8-reduced D458 and D425 cell lines (Supplemental Figure 3). In order to examine the impact of SETD8 depletion on actin organization, we stained MB cells for phalloidin, an F-actin marker. We identified the presence of podosomes or invadopodia, actin-rich adhesions containing integrins that contact the cytoskeleton and are involved in extracellular matrix degradation (38). These structures are linked to cellular invasiveness and are regulated by Rho GTPase activity, which was found necessary for podosome and invadopodia formation (38). In D458 and D425 shSETD8 cells, we observed a decrease in podosome/invadopodia, indicating a disruption in actin nucleation (Supplemental Figure 3).
Gene set enrichment analysis (GSEA) using the Molecular Signatures Database (mSigDB, http://software.broadinstitute.org/gsea/index.jsp) corroborated our above findings, and highly significant GSEA genesets (Figure 3C) were examined. A marked decrease in the expression of genes involved in G2/M progression matched the aforementioned cell cycle results (Supplemental Figure 3). As previously reported (24), positive enrichment of hallmark apoptotic and P53 pathways were also observed (Supplemental Figure 3). We examined P53 protein levels in all SETD8 knockdown cells through Western blot but found no changes in P53 protein, suggesting that the apoptotic enrichment may be induced by another mechanism in MB (Supplemental Figure 3). In addition, negative enrichment of gene sets involved in stem cell division, cell-to-cell interactions, cellular movement, and metastasis were detected (Figure 3B). In conjunction with these findings, we found the negative enrichment of genes mediating the EMT, a developmental process known to promote invasion and metastasis of cancer cells. These results, along with IPA, further implicate SETD8 as a factor mediating growth, invasive potential, and stem-like characteristics of MB cells.
Knockdown of SETD8 reduces self-renewal and stem cell marker expression in MB cells.
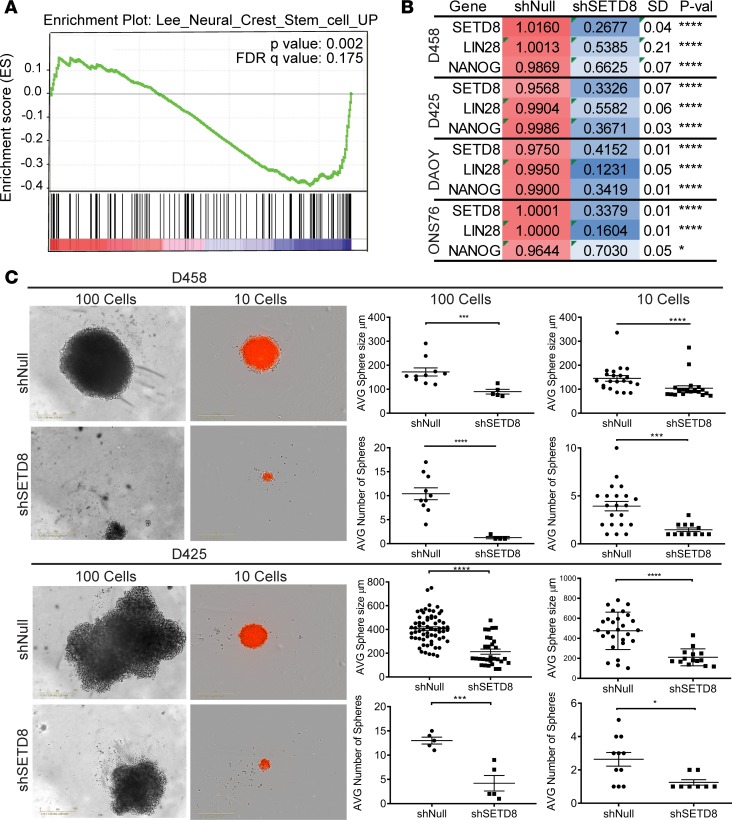
Self-renewal is inherent to cancer stem cells and a major factor in tumor recurrence and resistance to cell death (39, 40). Negative enrichment of genes regulating self-renewing division of stem cells and maintaining neural crest stem cells was observed in the GSEA of our RNA-Seq data (Figure 3B and Figure 4A). To determine if key stem cell factors were affected upon SETD8 depletion, we performed quantitative PCR (qPCR) assays and assessed the levels of expression of factors that maintain neural stem cell lineage. NANOG, a stem cell factor present in the granular precursor cells of the cerebellum; LIN-28, an embryonic stem cell factor; and NESTIN, a neural stem cell factor, were reduced by 30%–60% in most cell lines (Figure 4B and Supplemental Figure 4).
Figure 4. SETD8 knockdown reduces self-renewal.
(A) GSEA negative enrichment plot of genes associated with maintenance of neural crest stem cells. FDR q value and P value are shown. (B) Heatmap indicating the relative mRNA expression of SETD8, Lin-28, and Nanog in shSETD8 cells normalized to shNull in the respective cell line. Three independent replicates are show as mean ± SD. Unpaired t test; *P < 0.05; ****P < 0.0001. See also Supplemental Figure 4. (C) Neurosphere growth assay for shNull and shSETD8 D458 and D425 cells. Representative images from 100 and 10 wells are shown as captured on Incucyte imaging system. ×10 magnification. Dot plots represent the mean ± SD of the number of spheres and size of spheres. Unpaired t test; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. See also Supplemental Figure 4.
We then examined the functional impact of SETD8 on self-renewal of MB cancer cells. SETD8-knockdown MB cells were serially diluted, seeded in serum-free medium, and allowed to grow for 14 days. A significant decrease in the size of spheres was seen, indicating a reduction in proliferation (Figure 4C). In wells where 10 cells were plated, the D458 and D425 shNull cells were ~150–400 μm, respectively, while the shSETD8 counterparts were ≤100 μm (Figure 4C). Single cell suspensions displayed growth inhibition, as well as a reduced ability to form spheres (Supplemental Figure 4). In all conditions, the number of spheres was also greatly reduced, suggesting a defect in self-renewal capacity (Figure 4C). An average of 4 spheres were counted for D458 shNull and 3 for D425 shNull cells in wells where 10 cells were plated. The shSETD8 D458 and D425 cells display a 50% reduction or more in the number of spheres that formed (Figure 4C). Additionally, secondary sphere formation was monitored after 14 days. Notably, D458 and D425 shNull cells formed spheres, while shSETD8 cells remained as single cells (Supplemental Figure 4). To confirm that the neurospheres that grew from shSETD8 MB cells maintained the loss in stemness factors after 14 days, we stained neurospheres for NESTIN. A loss of NESTIN staining was observed, indicating that the knockdown of SETD8 in neurospheres was retained (Supplemental Figure 4). Together, the downregulation of factors maintaining stemness, in addition to decrease in number and size of spheres, supports that SETD8 sustains cancer stem cell self-renewal and growth.
SETD8 regulates expression of EMT genes in MB.
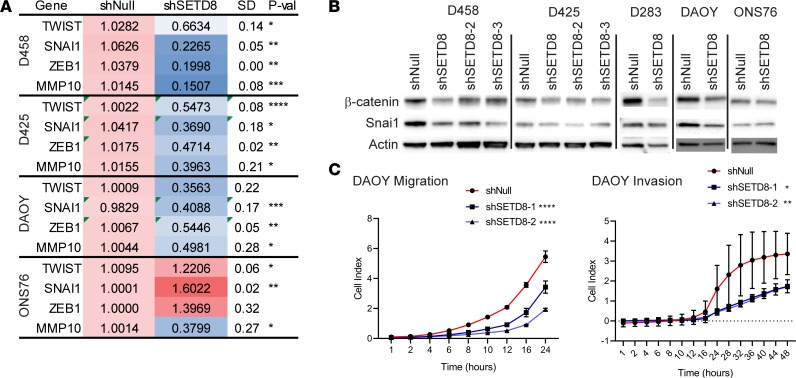
In breast and prostate cancers, SETD8 has been shown to interact with TWIST1 and ZEB1, 2 transcription factors that direct the EMT (41, 42) and affect cell migration. We observed a profound decrease in EMT gene sets in our GSEA, along with negative enrichment of genes that maintain adherens junctions and promote metastasis (Figure 3B). Hence, we sought to further explore the association of SETD8 with EMT genes and migration of MB tumor cells. We first examined the levels of expression of key EMT factors by qPCR. TWIST1, SNAI1, ZEB1, and the matrix metalloproteinase MMP10 were decreased by >50% in most shSETD8 knockdown cells compared with respective control cells (Figure 5A and Supplemental Figure 4). Conversely, no change or an increase in TWIST1, SNAI1, and ZEB1 mRNA expression was detected in the low MYC cell line ONS76.
Figure 5. SETD8 mediates expression of key EMT genes.
(A) Heatmap indicating the relative mRNA expression of TWIST1, SNAI1, ZEB1, and MMP10 in shSETD8 cells normalized to shNull in the respective cell line. Three independent replicates are show as mean ± SD. Unpaired t test; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. See also Supplemental Figure 4. (B) Western blot showing β-catenin, SNAI1, and actin in shSETD8 and shNull MB lines. Three independent replicates were performed. See also Supplemental Figure 4. (C) Migration and invasion assays. Cell index is reflective of the number of cells able to cross the membrane barrier. Line graphs represent the cell index mean vs. time ± SD in 3 independent replicates. Unpaired t test; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. See also Supplemental Figure 4.
Additional proteins that mediate the EMT process include SNAI1, β-catenin, and E- and N-cadherin. SNAI1 transcriptionally represses epithelial promoters through recruitment of PRC2 complexes, including EZH2, to direct chromatin modifications (43). β-Catenin facilitates attachment of E-cadherin (epithelial cell-to-cell junctions) or N-cadherin (mesenchymal cell interactions) to the cytoskeleton (43). Downregulation of E-cadherin and upregulation of N-cadherin is a hallmark of the EMT process. When we examined protein levels of β-catenin and SNAI1 by Western blot, a 50% decrease was detected in SETD8-KD cell lines (Figure 5B and Supplemental Figure 4). A slight decrease was also seen in ONS76 cells. Similarly, we examined the level of N-cadherin and found that it was completely abolished by SETD8-KD (Supplemental Figure 4).
We next examined migratory and invasive phenotypes of SETD8 shNull and -KD MB cancer cells using xCELLigence Cell Migration and Invasion assay. The xCELLigence migration assay is an electronically modified version of the Boyden chamber assay that requires adhesion to the membrane surface; thus, we examined our adherent cell lines DAOY and ONS76 in this assay. The DAOY SETD8-sufficient cells showed increased migration over 24 hours, but SETD8-depleted cells were deficient in their ability to migrate (Figure 5C). DAOY shSETD8 cell invasive capability was also significantly reduced (Figure 5C). On the other hand, the low MYC cell line ONS76 had very little migratory ability and even less when SETD8 was knocked down (Supplemental Figure 4). The ONS76 shNull and shSETD8 cells also had no ability to invade the matrigel basement membrane (Supplemental Figure 4). These results strongly indicate that SETD8 affects the migratory and invasive ability of MB cancer cells with high levels of MYC.
SETD8 mediates cell migration and invasion via H4K20me1 modification.
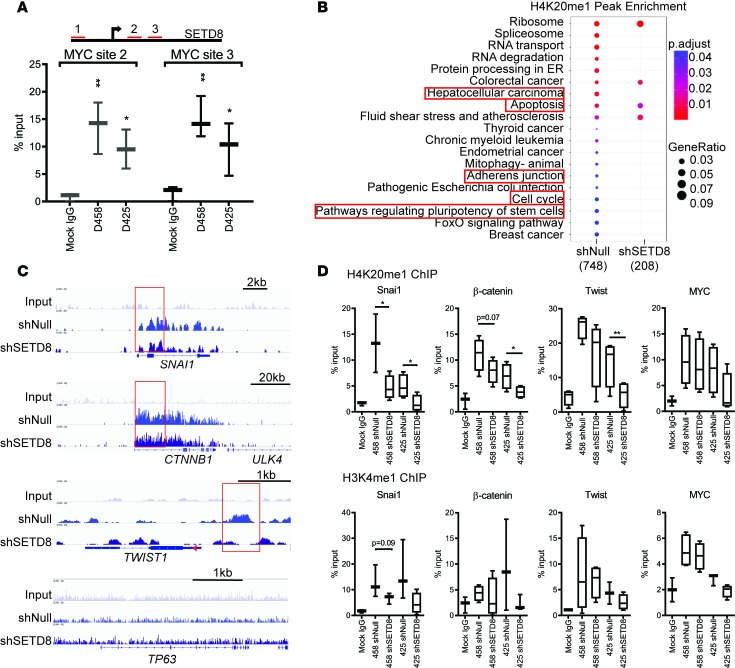
MYC amplification in Group 3 MB increases the propensity for metastasis and recurrence (44). Our SETD8 knockdowns show reduced metastatic potential due to depletion of factors that induce movement and invasion. Previous studies have shown that MYC can associate with the SETD8 promoter and that SETD8 can mediate MYC function in epidermal cells (45). Though, there was no significant change in MYC expression with SETD8-KD in our studies (Supplemental Figure 5), we examined MYC for its potential to bind the SETD8 promoter in MB cells. The SETD8 promoter region contains 3 prospective noncanonical MYC binding sites (Figure 6A). We performed ChIP assays on the SETD8 promoter with MYC. Compared with the mock, we observed a considerable enrichment of MYC at 2 of the 3 sites within the SETD8 promoter in 2 separate cell lines (Figure 6A).
Figure 6. Directed modification of H4K20 by SETD8 on key EMT gene promoters.
(A) ChIP of MYC at SETD8 promoter regions highlighted in red. Dot plots shown indicate the percent input detected by qPCR. Three independent replicates represented as mean ± SD. Unpaired t test; *P < 0.05; **P < 0.01. (B) Gene ontology H4K20me1 peak enrichment plot. Color indicates adjusted P < 0.05, and circle diameter indicates gene ratio. Selected functions are boxed. (C) ChIP-seq on H4K20me1. Genome browser view of H4K20me1 profile on SNAI1, CTNNB1, TWIST, and P63 promoter regions in D458 shNull and shSETD8 cells. Areas of interest are boxed. (D) ChIP of H4K20me1 and H3K4me1 on SNAI1, β-catenin, TWIST1, and MYC promoter regions in D458 and D425 shNull and shSETD8 cells. Box and whisker plots show minimum and maximum values from 3 independent replicates. Unpaired t test; *P < 0.05; **P < 0.01. See also Supplemental Figure 5 and Supplemental Table 2.
SETD8 methylation of H4K20 has been shown to be associated with transcriptionally active genes (46). To determine if H4K20me1 was altered at distinct promoter regions in MB cells with sufficient SETD8 compared with reduced SETD8, we performed ChIP-seq on D458 cells. H4K20me1 enrichment was mapped to promoter regions within 1 kb upstream and downstream of the transcriptional start site (TSS) (Supplemental Figure 5). Using gene ontology analysis, statistically significant alterations in peak enrichment of H4K20me1 were mapped to promoters. In total, 748 genes were significantly enriched in D458 shNull cells, whereas 208 were detected in shSETD8 cells (Figure 6B). Genes affiliated with posttranscriptional processing, including spliceosome, RNA transport, and degradation, showed the strongest enrichment (Figure 6B). Consistent with our gene expression data, genes involving adherens junctions, cell cycle, and signaling regulation of stem cell pluripotency exhibited significant reduction in H4K20me1 (Figure 6B). Furthermore, genes mediating hepatocellular carcinoma, including MYC, TGFβR1, β-CATENIN, PIK3CB, IGF1R, PLCG2, SMARC1B, and ACTB displayed decreased enrichment of H4K20me1 (Supplemental Figure 5) corresponding with the top affected pathways of SETD8 knockdown. Notably, TGFβR1, β-CATENIN, and IGF1R facilitate EMT progression in hepatocellular carcinoma (47).
To further determine if SETD8 methylation activity distinctly affects expression of EMT genes, we examined the H4K20me1 profile of SNAI1, β-catenin, and TWIST1, in addition to genes not related to EMT. The broad H4K20me1 profile spanning the length of SNAI1 and β-CATENIN was diminished following SETD8 knockdown (Figure 6C). Likewise, enrichment on TWIST1 localized to 1 kb upstream of the TSS, and upon SETD8 depletion, a considerable decrease in H4K20me1 was identified. In contrast, TP63 — a gene not linked with EMT progression — was examined for H4K20me1 enrichment. No significant enrichment was detected on TP63, and little to no enrichment was identified on ULK4 (Figure 6C), a gene adjacent to β-CATENIN.
We further validated the ChIP-seq results by ChIP-PCR assays in D458 and D425 shNull and shSETD8 cell lysates. Enrichment of H4K20me1 was reduced on SNAI1- in both SETD8-KD cells by 65% and 67%, respectively, when compared with shNull cells (Figure 6D). The β-CATENIN promoter region also showed a substantial decrease, 40% in D425 and 30% in D458 cells, when compared with respective shNull cells. Deposits of H4K20me1 was also decreased on TWIST1 in D425 cells but was less prominent in the D458 cell line. When enrichment of H4K20me1 on MYC was examined, we observed no significant change (Figure 6D), consistent with our previous findings. As a control methylation modification, we examined H3K4me1, which has been associated with gene activation on enhancer elements. Interestingly, a significant decrease in H3K4me1 enrichment was seen with SNAI1 (Figure 6D), while no noticeable difference was observed on the β-CATENIN, TWIST, and MYC sites. Taken together, these results suggest that SETD8 targets distinct EMT promoter regions and that depletion of SETD8 correlates with a decrease in active-promoter enrichment for H4K20me1 on SNAI1, β-CATENIN, and TWIST1.
Reduction in SETD8 enhances survival in an orthotopic mouse model.
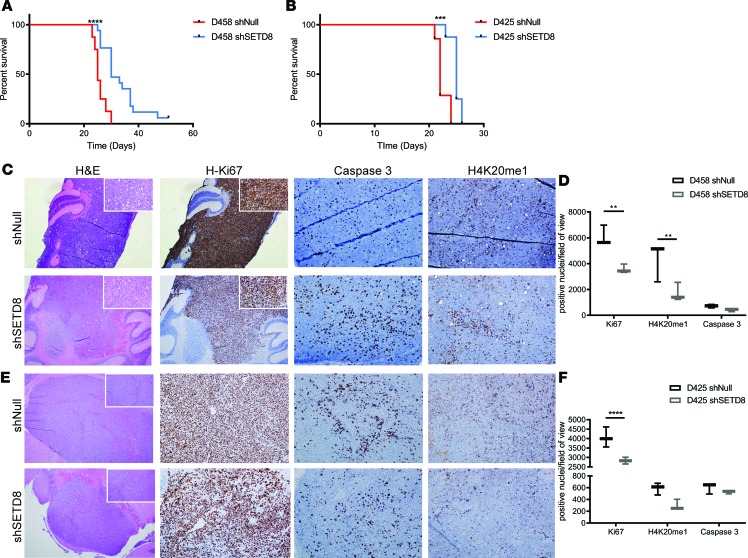
We sought to examine whether depletion of SETD8 would alter MB tumor growth in vivo. We injected SETD8 shNull and -KD D458 cells into murine cerebella and monitored tumors in vivo. Strikingly, mice bearing SETD8-depleted D458 cells demonstrated an enhanced survival, from 30–51 days, compared with mice bearing the same cells with WT SETD8 (Figure 7A). After euthanizing the animals, we collected cerebellar tissue and examined tumor histology and cell proliferation by staining with H&E and an antibody against Ki67, respectively. Compared with the shNull D458 tumors, the shSETD8 D458 tumors had decreased staining for the cell proliferative marker Ki67, closely matching our in vitro data (Figure 7, C and D). Examination of caspase 3 staining showed a slight increase in caspase 3+ cells, indicating an increase in apoptotic cell death. Additionally, H4K20me1-stained tumor cells were decreased in D458-KD cells compared with shNull, indicating that the depletion in SETD8 was maintained throughout tumor growth in vivo (Figure 7, C and D). Murine cerebella injected with D425 shNull and SETD8-KD cells similarly displayed an increase in survival, as well as a decrease in Ki67 and H4K20me1 methylation (Figure 7, B, E, and F).
Figure 7. In vivo mouse model shows reduced proliferation and enhancement of survival.
(A) Kaplan-Meier survival plot from intracranial orthotopic mouse model. D458 shNull (n = 17) and shSETD8 (n = 20) and (B) D425 shNull (n = 7) and shSETD8 (n = 8). Log-rank Mantel-Cox test; ***P < 0.001; ****P < 0.0001. (C and E) Immunohistochemical staining of cerebellar tumors from D458 shNull and shSETD8 (C) and D425 shNull and shSETD8 injected mice (E). H&E, Ki67, caspase 3, and H4K20me1 stains at 4× and 20× magnifications are shown. (D and F) Quantification of C and E. Box and whisker plots show minimum and maximum values from quantification of 6 representative images by ImageJ analysis. Unpaired t test; *P < 0.05; **P < 0.01.
Chemical inhibition of SETD8 phenocopies genetic manipulation.
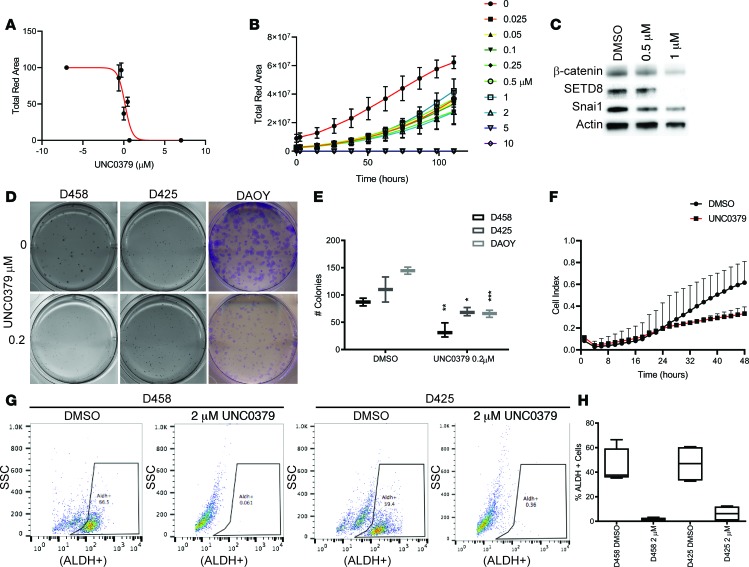
Epigenetic therapies are being investigated as an alternative in resilient tumors that have evaded other therapeutic approaches. The chemical inhibitor UNC0379 is a synthetic small molecule inhibitor that is selective for SETD8 over 15 other methyltransferases and (48, 49) has been shown as an inhibitor of SETD8 in neuroblastoma (24); however, its efficacy in MB has not been determined. To evaluate the effect of UNC0379 in MB, we first determined an in vitro IC50. NucRed DAOY cells were exposed to UNC0379 at increasing concentrations and monitored for cell growth over 5 days on the Incuctye live imaging system. Analysis of the red fluorescence as a reflection of cell viability with increasing concentration of UNC0379 determined an IC50 of ~2 μM (Figure 8A). To examine if UNC0379 has a similar effect on proliferation as the SETD8 knockdown, we monitored growth of NucRed-expressing D458 neurospheres over 10 days with increasing concentrations of UNC0379. Reduced neurosphere growth was observed with increasing concentration of UNC0379. Concentrations lower than 2 μM were most effective at limiting neurosphere growth (Figure 8B). Methylcellulose (Sigma Aldrich) and colony formation was explored over the same concentrations in D458, D425, and DAOY MB cell lines. More than a 50% decrease in colonies formed was observed with 0.2 μM in D458 cells, and most colonies were gone by 0.5 μM for all cell lines (Figures 8, D and E, and Supplemental Figure 6). These results suggest that UNC0379 restricts proliferation and colony formation, consistent with genetic depletion of SETD8.
Figure 8. UNC0379 treatment mimics SETD8 knockdown in medulloblastoma cell lines.
(A) Cell viability (total red fluorescence) of NucRed-expressing DAOY cells vs. UNC0379 (μM). Three independent replicates are shown as mean ± SD. IC50 of 2 μM was calculated with GraphPad prism. (B) NucRed-expressing D458 neurospheres treated with increasing concentration of UNC0379. Three independent replicates are represented in the line graph of cell viability (total red fluorescence) mean ± SD vs. time. (C) Western blot for β-catenin, SETD8, Snai1, and Actin from D458 cells treated with 0.5 μM or 1 μM UNC0379 for 48 hours. See also Supplemental Figure 6 for quantification. (D) Methylcellulose assay with D458, D425, and clonogenic assay with DAOY cells treated with 0.2 μM UNC0379. Representative images are shown. See also Supplemental Figure 6. (E) Box and whisker plots from D represent the mean ± SD from 3 independent replicates. Two-way Anova; *P < 0.05, **P < 0.01, ***P < 0.001. (F) Invasion assay of DAOY cells treated with 2 μM UNC0379 over 48 hours. Line graphs represent the cell index mean vs. time ± SD in 3 independent replicates. (G) Representative plots of Aldefluor+ cells vs. side scatter are shown for D458 or D425 cells with or without 2 μM UNC0379 treatment. (H) Box and whisker plot represents 3 independent replicates of percent ALDH+ cells with and without treatment. See also Supplemental Figure 6.
To determine if treatment with UNC0379 reduced EMT factors, we examined β-catenin and SNAI1 by Western blot at 0.5 μM and 1 μM in the D458 cell line. Diminished SNAI1 and β-catenin was observed at 0.5 μM and was stronger at 1 μM (Figure 8C and Supplemental Figure 6). Additionally, SETD8 levels were less pronounced at 0.5 μM and gone by 1 μM. We then examined the invasive ability of DAOY cells exposed to 2 μM UNC0379 over 48 hours. We found a reduced ability of DAOY cells to transverse the matrigel matrix, restricting the invasive ability of the DAOY cell line when compared with the DMSO control (Figure 8F). To examine if UNC0379 similarly affects stemness, we performed an Aldefluor stain on D458 and D425 neurospheres exposed to 2 μM UNC0379 for 48 hours. High levels of the ALDH enzyme is present in stem and progenitor cells, enabling their detection. We costained D458 and D425 neurospheres with DRAQ5 to isolate live cells for analysis (Supplemental Figure 6). D458 and D425 displayed 45%–40% ALDH+ cells prior to treatment, whereas — after treatment — a dramatic shift of cells was observed with less than 5% ALDH+ cells remaining (Figure 8, G and H). The chemical inhibition of SETD8 with UNC0379 recapitulates our SETD8-KD studies, affirming that SETD8 mediates MB cell growth and stem cell maintenance and that it can affect SNAI1 and β-catenin protein levels. Additionally, these results further support that inhibition of SETD8 limits the invasive ability of MB cells.
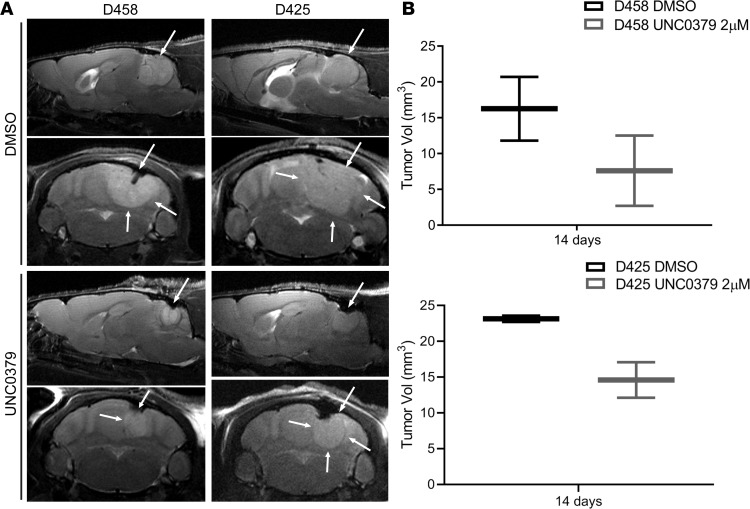
We proceeded to determine the effect of UNC0379 on intracranial tumor growth in vivo. In the absence of validated pharmocokinetic and animal toxicology studies on UNC0379, we treated D458 and D425 cells with 2 μM UNC0379 for 24 hours in vitro. Equal numbers of viable cells (control and UNC0379 treated) were then injected into murine cerebella, and mice were monitored for tumor growth, as before. At 14 days after injection, tumor volumes were assessed by high-resolution T2-weighted MRI. Chemical inhibition of SETD8 slowed growth of tumors at 14 days compared with cells treated with DMSO, as seen in sagittal and axial T2 MRI images in Figure 9A. Evaluation of tumor volume by T2-MRI displayed smaller overall sizes in UNC0379-treated D458 and D425 cells (Figure 9B).
Figure 9. Pharmacological inhibition of SETD8 inhibits in vivo growth.
(A) Representative sagittal and axial T2-weighted turboRARE (rapid acquisition with relaxation enhancement) MR images on cerebellar D458 and D425 tumor lesions with and without UNC0379 treatment. White arrows point to tumor lesions. (B) Differences in tumor volumes (in mm3) calculated from T2-weighted MRI scans. All images were acquired at a Bruker 9.4 Tesla MRI scanner. Three mice from each treatment group were analyzed. See also Supplemental Figure 7.
These data show that chemical inhibition of SETD8 restricts tumor development in vivo. The overall survival of mice with treated cells was prolonged, though a significant difference from control mice was not determined (Supplemental Figure 7). It is probable that, without continual treatment, the D458 and D425 cells restored expression of SETD8, enabling rapid proliferation of tumor cells. While these results are in line with our previous results, these data demonstrate the limitations to a 24-hour in vitro treatment and further indicate the need to develop improved SETD8 chemical inhibitors that can be more thoroughly validated in vivo.
Discussion
Advances in whole-genome sequencing reveal increasing prevalence of altered epigenetic machinery that can drive tumorigenesis (1, 19). Pediatric cancers, in particular, demonstrate high prevalence of epigenetic dysregulation (1, 6). Given the role of epigenetic changes in normal development they may be especially important in pediatric cancer patients, representing normal development gone wrong (50). Epigenetic changes are reversible, unlike genetic changes, and are thus attractive targets for drug design (51). Previous studies of MB have identified epigenetic regulators as key mechanisms of tumorigenesis (52). We have previously demonstrated that EZH2, which methylates H3K27, is necessary for oncogenic transformation of neural stem cells and is a critical regulator of MB cell growth (32). Our epigenome-wide RNAi screen, combined with an epigenetic drug–focused chemical screen, identified molecular vulnerabilities in MYC-driven MB that are highly druggable in this very high-risk tumor. Our studies identified 7 potential targets. Among these targets, EZH2 and BRD4 have previously been described by us and others (29–31, 53, 54). SETD8 is a potentially novel target identified by our combinatorial approach for which a new chemical inhibitor has recently been developed.
Our studies demonstrate that SETD8 inhibition results in attenuation of cell self-renewal and is accompanied by a significant decrease in invasive properties. Mechanistically, SETD8 inhibition results in altered H4K20me chromatin occupancy at EMT gene promoters, thus altering expression of these critical proinvasive and promoting prodifferentiation genomic programs. Genetic ablation of SETD8 suppresses growth of MYC-driven MB in vivo and prolongs survival of these xenograft-bearing mice. Furthermore, high expression of SETD8 is associated with a poor prognosis, even in the high-risk MYC-amplified MB patients, identifying a patient population that would benefit from SETD8 inhibition.
SETD8 is a methyltransferase that monomethylates H4K20me. During development, SETD8 is expressed in early embryogenesis, and deletion of SETD8 is embryonic lethal (55, 56). In epidermal stem cells, loss of SETD8 leads to decreased proliferation and defects in differentiation (45). Despite an association with normal stem cell maintenance in the epidermis, SETD8 has not been established as a factor that sustains cancer stem cell growth. Here, we show evidence that loss of SETD8 results in downregulation of stem cell–associated genomic programs, including the lineage markers NANOG, LIN-28, and NESTIN. These genomic changes were accompanied by a functional decrease in cell self-renewal upon SETD8 depletion.
Analysis of our RNA-Seq results reveal several cellular processes in which SETD8 activity has a definitive effect. IPA and GSEA indicate that the top functions are involved in 3 major processes: cell death, proliferation, and cell movement. While methylcellulose assays confirm defects in MB cancer cell proliferation, recent studies (24) reveal that SETD8 blocks apoptotic signals through inactivation of P53 in neuroblastoma. Our GSEA found correlative enhancement of P53 pathway–involved genes; however, the examination of P53 protein levels in MB cell lines revealed no enhancement. Additionally, though we found enhanced apoptotic gene enrichment through GSEA, the in vivo xenograft model showed only a slight increase in apoptosis-associated caspase 3 IHC staining. These results suggest that, in our MB cell models, apoptosis is not the primary mechanism by which SETD8 inhibition restricts MB cell growth.
Interestingly, the top pathways effected by SETD8 knockdown are ILK1/integrin signaling as well as phospholipase C signaling. ILK1/integrin signaling and phospholipase C signaling are major regulatory nodes that influence proliferation, invasion, and metastasis mechanisms (57). In this context, genomic programs associated with EMT were also significantly attenuated in SETD8-depleted MB cells. Specifically, there was a >50% decrease in expression of TWIST, ZEB1, SNAI1, and β-catenin, all key components of the proinvasive EMT switch (43, 58). Consistent with this, SETD8 inhibition restricted migratory and invasive abilities of MB cells. In breast and prostate cancers, SETD8 was found to directly interact with either TWIST or ZEB1, both transcriptional regulators of EMT (41, 42). SETD8 and TWIST were found to transcriptionally repress E-cadherin, while activating N-cadherin (41). Similarly, SETD8 and ZEB1 were shown to repress E-cadherin, while activating vimentin (42). We found that the H4K20me1 mark was significantly depleted at promoter sites for SNAI1, β-catenin, and TWIST1 in SETD8-KD cells. These results demonstrate that SETD8 can target H4K20me1 to EMT promoter sites, thereby modulating their transcriptional activity. In addition to mediating EMT gene expression, our GSEA results suggest that SETD8 appears to have broader regulatory control over cell motility and migration.
Targeted small molecule inhibitors are currently being developed for SETD8, and UNC0379 has been verified as a viable option for continued development as a clinical therapy (24, 48). Our results with UNC0379 affirm that the chemical inhibition of SETD8 reduces proliferation, colony formation, and Snai1 and β-catenin protein levels. Furthermore, treatment with UNC0379 also limited the ability of MB cells to invade a matrigel matrix, suggesting a reduction in invasive potential.
In vivo, using a mouse model of MYC-amplified tumors, the overall survival of mice increased substantially with SETD8 depletion. This was accompanied by reduced number of proliferating cells and reduced H4K20me1, indicative of sustained loss in SETD8. Furthermore, chemical inhibition of SETD8 in vivo displayed diminished growth of MB tumors at 14 days. Although genetic and chemical inhibition of SETD8 prolonged overall survival, prior xenograft models with limited SETD8 expression have shown a larger impact on survival when mice were injected s.c. To closely recapitulate the brain tumor–growing environment, we measure tumor growth within the cerebellum where the tumor originated. Furthermore, it is worthy to note that brain tumors are mechanistically divergent from tumors that originate in the rest of the body. Hence, the difference in tumor microenvironment and varying tumor type may have contributed to the difference in survival times. Overall, our data are consistent with previous studies that implicate SETD8 in enhanced tumorigenesis of breast, prostate, and neuroblastoma (24, 42, 59, 60). In the context of MB, our results validate the finding that SETD8 is an important regulator of MB growth and reinforce the notion that reducing SETD8 could improve patient outcomes.
In high-risk patients, the amplification of MYC increases the propensity for metastatic disease, with the overall survival in the combined Group 3 and Group 4 tumors decreasing from 73%–27% with the presence of metastases (61). Additionally, 39% of recurrent tumors in Group 3 usually present with metastatic dissemination at relapse (62). These metastatic lesions are genetically divergent from the primary tumor and unresponsive to traditional therapies given at the primary site (44). Thus, the need to develop a specific therapy that targets molecules such as SETD8 that confer the ability of tumor cells to self-renew, recur, and metastasize is crucial. Our finding further underscores the involvement of epigenetic modifiers in neoplastic transformation and emphasizes SETD8 as a putative target for treatment in MB.
Methods
Cell lines and cell culture.
The DAOY and D283 MB cell lines are from American Type Cell Culture. DAOY, D283, and ONS-76 cells were cultured in DMEM (Thermo Fisher Scientific) supplemented with 10% FBS (MilliporeSigma), 1 mM sodium pyruvate (Thermo Fisher Scientific), 1× penicillin/streptomycin solution (Cellgro), and 1× nonessential amino acids (MilliporeSigma). D425 and D458 cell lines were provided by Darell D. Bigner (Duke University Medical Center, Durham, North Carolina, USA). Cells were cultured in DMEM, but without nonessential amino acids and with 1× L-glutamine (Cellgro). All cell lines were cultured at 37°C with 95% air and 5% CO2.
Stable cell lines were established using gene specific mission shRNAs (catalog 359304, 130139, 148268) or nontargeting control (MilliporeSigma). Cells were plated at 5 × 105 in a 6-well dish in growth medium containing 8 μg/ml polybrene. Viral particles were added with 8 μg/ml polybrene and incubated for 7 hours. Transduced cells were disaggregated, grown, and selected with 1 μg/ml puromycin for 14 days. Transformants were confirmed by qPCR and Western blot.
Methylcellulose assays.
Five hundred cells/3 ml were plated in a 1:1 mixture of 2.6% methylcellulose and complete growth medium. Cells were allowed to grow for 10 days. Colonies were stained with nitrotetrazolium blue chloride (MilliporeSigma) at 1.5 mg/ml in PBS for 24 hours at 37°C and were then counted.
Neurosphere assay.
D458 or D425 shNull and shSETD8 cells were serially diluted at 100-, 10-, and single-cell suspensions in neurosphere growth media (neurobasal medium, B-27 + vitamin A, L-glutamine, penicillin/streptomycin [Gibco], EGF, and FGF [Shenandoah biotechnology]). Cells were grown for 14 days, with media replacement every 3 days at 37°C. At 14 days, spheres were collected, disassociated, and replated at 100-, 10-, and single-cell suspensions and were allowed to grow for an additional 14 days. Spheres were imaged on an Olympus U-CMAD3 microscope and measured with Qcapture software. Additional neurosphere proliferation and real-time monitoring was done using the Incucyte S3 live cell imaging system (Essen Bioscience).
Epigenetic chemical inhibitor screen.
Epigenetics chemical screening library catalog 11076 (Cayman Chemical) was used at 2 μM or 0.5 μM to treat D458 cells plated at 1000 cells/well in a 96-well dish in neurosphere growth media. Cells were grown for 8 days, with media and drug replenishment every 3 days. Cell viability was measured via CellTiter 96 AQueous One Solution Cell Proliferation Assay (Promega, Thermo Fisher Scientific) as per manufacturer’s instructions. Chemical inhibitors that reduced cell viability to <70% of control were then selected.
BrdU Flow assay.
DNA synthesis was analyzed with the FITC conjugated-BrdU Flow kit (catalog 559619, BD Pharmingen). D458 cells were plated at 1 × 106 and labeled with 10 μM bromodeoxyuridine for 1 hour. Cells were collected, fixed, and analyzed as per manufacturer’s instructions on the Amnis Flowsight Imaging Flow Cytometer (MilliporeSigma) and analyzed by Amnis IDEAs software.
qPCR.
cDNA was synthesized from 2 μg of total RNA with High-Capacity cDNA Reverse transcription kit (Thermo Fisher Scientific). qPCR was performed using gene specific TaqMan probes and TaqMan Universal PCR Mastermix as per manufacturer’s instructions. qPCR for ChIP analysis was performed using Power Sybr-Green PCR mastermix (Thermo Fisher Scientific). Primers were designed to specific loci flanking the promoter region occupied by H3K4me1 and H4K20me1 using University of California, Santa Cruz Genome Browser (https://genome.ucsc.edu/). Similarly, primers were designed for sequence flanking the 3 potential MYC-binding sites in the SETD8 promoter for the MYC ChIP. For primer sequences, refer to Supplemental Table 2. qPCR was performed on the StepOnePlus Real-Time PCR system (Thermo Fisher Scientific).
RNA-Seq.
Total RNA was isolated from 2 separate D458 shSETD8 and shNull clones using RNeasy Mini kit (Qiagen). RNA-Seq was performed at Genomics and Microarray Core Facility, Anschutz Medical Campus, using the Illumina HiSEQ 4000 HT for 1 × 50 sequencing. High-quality base calls at 95% ≥ Q30 were obtained with 53–69 M single reads. Q30 (base call accuracy) is the maximum quality score achieved if all bases are called accurately. Statistical analysis of count data was performed with DESeq2 R package (63). Further analysis by GSEA was performed using the MSigDB (64, 65) and IPA (Qiagen). Graphical representation of the molecular and cellular functions was made with Venny 2.1 (66). All original microarray data were deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE121247).
Western blot.
Cells were lysed in RIPA buffer (Pierce, Thermo Fisher Scientific) containing an EDTA-free protease inhibitor (Roche Diagnostics), and protein concentrations were determined with the BCA Protein Assay Kit (Pierce, Thermo Fisher Scientific). Protein (30 μg of total) was separated on a 4-20% gradient SDS-PAGE (Bio-Rad). Primary antibodies α–β-catenin D10A8 (catalog 8480), α–snai1 C15D3 (catalog 3879), or α–SETD8 C18B7 (catalog 2996; Cell Signaling Technology) were exposed overnight at 4°C. Secondary antibody, α–mouse-HRP (catalog 7076) and α–rabbit-HRP (catalog 7074) or α–actin-HRP (catalog 12262; Cell Signaling Technology), was exposed for 1 hour at room temperature. Blots were developed with Luminata Forte Western HRP (MilliporeSigma) and imaged using Syngene GBox Chemi-SL1.4 gel doc. Western blots were quantified using ImageJ (67). See supplemental materials for full uncut blots.
xCELLigence migration and invasion assay.
ACEA Biosciences CIM-plate 16 was assembled as per manufacturer’s instructions with 100 μl of 3 × 105/ml cell suspension in serum free media added to the upper chambers. For invasion assays, matrigel (Corning, catalog 354234) was thawed on ice, diluted in cold serum free media to 800 μg/ml, and added to the upper chambers to polymerize at 37°C for 4 hours. Following polymerization, the CIM-plate 16 was assembled as per manufacturer’s instructions. Measurements were taken every 15 minutes for 24 hours during migration assays and 48 hours for invasion assays.
ChIP and ChIP-seq.
Cells were collected at confluency or approximately 1 million cells/immunoprecipitation. Cells were washed with 1× PBS, cross-linked with 1% paraformaldehyde (ChemCruz) in PBS at room temperature, and quenched with 2.5 M glycine (Sigma Aldrich). Cells were then washed with ice cold PBS and lysed in 1 ml RIPA buffer (150 mM NaCl [Alfa Aesar], 1% v/v Nonidet P-40, 0.5% w/v deoxycholate [Amresco], 0.1% w/v SDS [Amresco], 50 mM Tris [Corning Cellgro] pH 8.0, 5 mM EDTA [Amresco]) plus inhibitors (leupeptin 1 μg/ml [Sigma Aldrich], aprotinin 1 μg/ml [Sigma Aldrich], pepstatin 1 μg/ml [Sigma Aldrich], benzamidine 1 mM [Sigma Aldrich], and PMSF 1 mM [Sigma Aldrich]). Lysates were sonicated using Diagenode Bioruptor with 25 cycles of 30-second pulses and 90-second intervals to shear DNA to ~500-bp fragments. Lysates were then cleared by centrifugation at 18,000 g for 15 minutes at 4°C. Protein A/G Sepharose beads (40 μl; MilliporeSigma) were added, and the lysate was precleared for 1 hour at 4°C. Protein A/G beads (20 μl) blocked with 1 mg/ml BSA was mixed with the precleared lysate, and immunoprecipitation was carried out by adding 5–10 μg of antibody (H4K20me1 [catalog 39727], H3K4me1 [catalog 39297]; Active Motif) rotating overnight at 4°C. Beads were washed 2× with RIPA, 4× with IP wash buffer (100 mM TrisHCL pH8.5, 500 mM LiCl, 1% v/v Nonidet-P-40, 1% w/v deoxycholic acid), and 2× with TE buffer. Immunocomplexes were eluted at 65°C with elution buffer (70 mM TrisHCl pH 8, 1 mM EDTA, 1.5% w/v SDS). The eluate was brought to a final concentration of 200 mM NaCl and reverse crosslinked at 65°C. The eluate was then treated with proteinase K (Invitrogen), and DNA was isolated by phenol/chloroform extraction (Invitrogen) and ethanol precipitation. ChIP-DNA was quantified using the Qubit dsDNA High Sensitivity Assay kit (Thermo Fisher Scientific).
ChIP-seq libraries were sequenced on the Illumina Novaseq 6000 platform. 62–69 M reads with high-quality base calls at 90%≥ Q30 were obtained. Bowtie2 was used to align the 150-bp paired-end sequencing reads to a reference human genome (hg19) downloaded from the UCSC Genome Browser. Unmapped and nonuniquely mapped reads were removed, and PCR duplicate reads were removed using SAMtools version 1.5. Peaks were called using MACS2 (v2.1.1.20160309) (68) with default parameters. Peak locations were further annotated according to the known genes in hg19, and 3000 bp of upstream and downstream of transcription start sites were considered as promoter regions using the R/Bioconductor package ChIPseeker (69). All original microarray data were deposited in the NCBI’s GEO (GSE121556).
IHC.
Tissue was fixed in 10% formalin and submitted to the University of Colorado Denver Tissue Histology Shared Resource for sectioning and staining. Tissue was stained with α-H4K20me1 (catalog 39727; Active Motif), α–caspase 3 (catalog 9661; Cell Signaling Technology), or α-Ki67 (catalog RM-9106; Thermo Fisher Scientific). Images were captured on an Olympus Bx43 light microscope and quantified with ImageJ.
Chemical Inhibition.
UNC0379 was provided in house. For all assays, drug concentrations are as follows: DMSO, 0.025, 0.05, 0.1, 0.25, 0.5, 1, 2, 5, and 10 μM. IC50 determination NucRed-expressing DAOY cells were plated at 1000 cells/well in a 96-well dish. Neurosphere proliferation assay NucRed–expressing D458 cells were plated at 10 cells/well in neurosphere medium in a 96-well dish. Growth was monitored as previously described. Methylcellulose and clonogenic assay were set up as previously described with UNC0379 mixed in media. Total number of colonies were counted, and the percent of control was determined. For Western blots, D458 cells were plated at 5 × 105 in a 6-well dish and treated for 48 hours at 0.5 and 1 μM before harvesting. Lysates were prepared as previously described. For the invasion assay, DAOY cells were pretreated for 48 hours with 2 μM UNC0379; they were then added to the CIM-plate 16 with matrigel and monitored over 48 hours on the xCELLligence as previously described.
Aldefluor assay.
D458 and D425 cells were seeded at 2 × 105 in a 6-well dish with DMSO or 2 μM UNC0379 and allowed to grow for 48 hours. The ALDEFLUOR Kit (catalog 01700, Stem Cell Technologies) was used for detection of ALDH+ cells as per manufacturer’s instructions. Data was collected on the Guava easyCyte HT Flow Cytometer and analyzed using FlowJo software.
MRI.
For in vivo MRI acquisitions, mice were anesthetized shortly before and during the MR session using 1.5% of isoflurane/ oxygen mixture. Anesthetized mice were placed on a temperature-controlled mouse bed below a mouse head array coil and inserted into Bruker 9.4 Tesla BioSpec MR scanner (Bruker Medical). First, T2-weighted turboRARE images were acquired using the following parameters: repetition time (TR) = 3268 ms; echo time (TE) = 60 ms; RARE factor = 12, 8 averages; FOV = 20 mm; matrix size = 350 × 350; slice thickness = 700 μm, 24 sagittal and axial slices; in-plane spatial resolution = 51 μm. Then, diffusion weighted EPI sequence with 6 b values was used using 4 axial slices covering the entire tumor lesions and unaffected brain tissue. Tumor regions were manually segmented on T2-weighted images by placing hand-drawing regions of interest (ROI), and the volume was calculated as mm3. The apparent diffusion coefficients (ADC, s/mm2) were calculated from diffusion-weighted imaging maps as a criterion for tumor cellularity. All acquisitions and image analysis were performed using Bruker ParaVision NEO software.
Animal experiments.
D458 or D425 shNull and shSETD8 cells were collected and resuspended as a single cell suspension of 20,000 cells/3 μl in serum free media. Intracranial injection of cells into outbred athymic Nude-Foxn1nu mice (The Jackson Laboratory, strain 07850) was done at 1.5 mm lateral and 2 mm posterior of lambda at 400 nanoliters/minute. Mice were monitored for tumor growth daily and euthanized when 15% weight loss was reached. A Kaplan-Meier plot was used for survival calculations. For in vivo chemical inhibition, treatment with UNC0379 at 2 μM for 24 hours was done in vitro. Treated D458 or D425 cells were then counted, and viability was assessed. A total of 20,000 cells/3 μl in serum free media was then injected as before into outbred athymic Nude-Foxn1nu mice (The Jackson Laboratory, strain 07850).
Statistics.
All experiments were performed with at least 3 independent replications. All data was collected in Excel, and Graphpad Prism 7 statistical software was used for analysis. The mean ± SD is graphed. P < 0.05 was considered significant, where *P < 0.05, **P < 0.01,***P < 0.001, and ****P < 0.0001. Unpaired, 2-tailed t tests were used for 2-group comparisons, and one- and two-way ANOVA and Dunnett’s test were used for multiple group comparisons.
Study approval.
All animal procedures were performed in accordance with the National Research Council’s Guide for the Care and Use of Laboratory Animals and were approved by the University of Colorado Anschutz Medical Campus IACUC.
Author contributions
RV and BV designed the study and wrote the manuscript. BV conducted the experiments, performed data analysis, and prepared figures. ED performed ChIP-seq data processing and alignments (Figure 6 and Supplemental Figure 5), SV performed epigenomic screen (Supplemental Figure 1), AP performed mouse injections, DW performed volcano plots (Supplemental Figure 3C), IS conducted GSEA Supplemental Figure 3B), and NS performed MRI analysis (Supplemental Figure 9). JJ and AM provided UNC0379. NKF developed idea with RV and edited manuscript.
Supplementary Material
Acknowledgments
We would like to thank Darell D. Bigner (Duke University) for providing the D458 cell line used in this study. Thank you Eric Prince, Daniel Moreira, Amber Johnson, Kate Wallerius, and Aaron Abai for their assistance over the course of study. The authors appreciate the contribution made by the University of Colorado Denver Tissue Histology Shared Resource, supported in part by the Cancer Center Support grant (P30CA046934). We thank the University of Colorado Cancer Center Functional Genomics core facility for lentiviral constructs and the Genomics and Microarray Shared Resource for their assistance with RNA sequencing and ChIP sequencing. We also thank Wen Hua for support with bioinformatics analysis. The authors thank Jenna Steiner from the Colorado Animal Imaging Shared Resource (AISR) for acquiring all mouse MRI scans. The AISR is supported by the University of Colorado Cancer Center, the P30CA046934 Cancer Center, and S10 OD023485 High-End Shared Instrumentation grants (NJS). This work was supported by the Morgan Adams Foundation Pediatric Research Program and NIH grant R01NS088283.
Version 1. 01/10/2019
Electronic publication
Footnotes
Conflict of interest: The authors have declared no conflict of interest exists.
License: Copyright 2019, American Society for Clinical Investigation.
Reference information: JCI Insight. 2019;4(1):e122933. https://doi.org/10.1172/jci.insight.122933.
Contributor Information
Etienne Danis, Email: Etienne.Danis@ucdenver.edu.
Angela Pierce, Email: ANGELA.PIERCE@UCDENVER.EDU.
Ismail Sola, Email: Ismail.sola@ucdenver.edu.
Dong Wang, Email: Dong.2.wang@ucdenver.edu.
Jian Jin, Email: jian.jin@mssm.edu.
Anqi Ma, Email: anqi.ma@mssm.edu.
Natalie Serkova, Email: natalie.serkova@ucdenver.edu.
Sujatha Venkataraman, Email: Sujatha.venkataraman@ucdenver.edu.
Rajeev Vibhakar, Email: Rajeev.Vibhakar@ucdenver.edu.
References
- 1.Huether R, et al. The landscape of somatic mutations in epigenetic regulators across 1,000 paediatric cancer genomes. Nat Commun. 2014;5:3630. doi: 10.1038/ncomms4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feinberg AP, Koldobskiy MA, Göndör A. Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat Rev Genet. 2016;17(5):284–299. doi: 10.1038/nrg.2016.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gröbner SN, et al. The landscape of genomic alterations across childhood cancers. Nature. 2018;555(7696):321–327. doi: 10.1038/nature25480. [DOI] [PubMed] [Google Scholar]
- 4.Mack SC, et al. Epigenomic alterations define lethal CIMP-positive ependymomas of infancy. Nature. 2014;506(7489):445–450. doi: 10.1038/nature13108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee RS, et al. A remarkably simple genome underlies highly malignant pediatric rhabdoid cancers. J Clin Invest. 2012;122(8):2983–2988. doi: 10.1172/JCI64400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maury E, Hashizume R. Epigenetic modification in chromatin machinery and its deregulation in pediatric brain tumors: Insight into epigenetic therapies. Epigenetics. 2017;12(5):353–369. doi: 10.1080/15592294.2016.1278095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dubuc AM, et al. Aberrant patterns of H3K4 and H3K27 histone lysine methylation occur across subgroups in medulloblastoma. Acta Neuropathol. 2013;125(3):373–384. doi: 10.1007/s00401-012-1070-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leary SE, Olson JM. The molecular classification of medulloblastoma: driving the next generation clinical trials. Curr Opin Pediatr. 2012;24(1):33–39. doi: 10.1097/MOP.0b013e32834ec106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramaswamy V, et al. Medulloblastoma subgroup-specific outcomes in irradiated children: who are the true high-risk patients? Neuro-oncology. 2016;18(2):291–297. doi: 10.1093/neuonc/nou357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thompson MC, et al. Genomics identifies medulloblastoma subgroups that are enriched for specific genetic alterations. J Clin Oncol. 2006;24(12):1924–1931. doi: 10.1200/JCO.2005.04.4974. [DOI] [PubMed] [Google Scholar]
- 11.Kool M, et al. Integrated genomics identifies five medulloblastoma subtypes with distinct genetic profiles, pathway signatures and clinicopathological features. PLoS ONE. 2008;3(8):e3088. doi: 10.1371/journal.pone.0003088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho YJ, et al. Integrative genomic analysis of medulloblastoma identifies a molecular subgroup that drives poor clinical outcome. J Clin Oncol. 2011;29(11):1424–1430. doi: 10.1200/JCO.2010.28.5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Northcott PA, et al. Medulloblastoma comprises four distinct molecular variants. J Clin Oncol. 2011;29(11):1408–1414. doi: 10.1200/JCO.2009.27.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Northcott PA, et al. Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature. 2012;488(7409):49–56. doi: 10.1038/nature11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kool M, et al. Molecular subgroups of medulloblastoma: an international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, Group 3, and Group 4 medulloblastomas. Acta Neuropathol. 2012;123(4):473–484. doi: 10.1007/s00401-012-0958-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cavalli FMG, et al. Intertumoral Heterogeneity within Medulloblastoma Subgroups. Cancer Cell. 2017;31(6):737–754.e6. doi: 10.1016/j.ccell.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ellison DW, et al. Definition of disease-risk stratification groups in childhood medulloblastoma using combined clinical, pathologic, and molecular variables. J Clin Oncol. 2011;29(11):1400–1407. doi: 10.1200/JCO.2010.30.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ryan SL, et al. MYC family amplification and clinical risk-factors interact to predict an extremely poor prognosis in childhood medulloblastoma. Acta Neuropathol. 2012;123(4):501–513. doi: 10.1007/s00401-011-0923-y. [DOI] [PubMed] [Google Scholar]
- 19.Northcott PA, Pfister SM, Jones DT. Next-generation (epi)genetic drivers of childhood brain tumours and the outlook for targeted therapies. Lancet Oncol. 2015;16(6):e293–e302. doi: 10.1016/S1470-2045(14)71206-9. [DOI] [PubMed] [Google Scholar]
- 20.Beck DB, Oda H, Shen SS, Reinberg D. PR-Set7 and H4K20me1: at the crossroads of genome integrity, cell cycle, chromosome condensation, and transcription. Genes Dev. 2012;26(4):325–337. doi: 10.1101/gad.177444.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tardat M, et al. The histone H4 Lys 20 methyltransferase PR-Set7 regulates replication origins in mammalian cells. Nat Cell Biol. 2010;12(11):1086–1093. doi: 10.1038/ncb2113. [DOI] [PubMed] [Google Scholar]
- 22.Houston SI, et al. Catalytic function of the PR-Set7 histone H4 lysine 20 monomethyltransferase is essential for mitotic entry and genomic stability. J Biol Chem. 2008;283(28):19478–19488. doi: 10.1074/jbc.M710579200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takawa M, et al. Histone lysine methyltransferase SETD8 promotes carcinogenesis by deregulating PCNA expression. Cancer Res. 2012;72(13):3217–3227. doi: 10.1158/0008-5472.CAN-11-3701. [DOI] [PubMed] [Google Scholar]
- 24.Veschi V, et al. Epigenetic siRNA and Chemical Screens Identify SETD8 Inhibition as a Therapeutic Strategy for p53 Activation in High-Risk Neuroblastoma. Cancer Cell. 2017;31(1):50–63. doi: 10.1016/j.ccell.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pei Y, et al. HDAC and PI3K Antagonists Cooperate to Inhibit Growth of MYC-Driven Medulloblastoma. Cancer Cell. 2016;29(3):311–323. doi: 10.1016/j.ccell.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoffman GR, et al. Functional epigenetics approach identifies BRM/SMARCA2 as a critical synthetic lethal target in BRG1-deficient cancers. Proc Natl Acad Sci USA. 2014;111(8):3128–3133. doi: 10.1073/pnas.1316793111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi X, Wang Q, Gu J, Xuan Z, Wu JI. SMARCA4/Brg1 coordinates genetic and epigenetic networks underlying Shh-type medulloblastoma development. Oncogene. 2016;35(44):5746–5758. doi: 10.1038/onc.2016.108. [DOI] [PubMed] [Google Scholar]
- 28.Ecke I, et al. Antitumor effects of a combined 5-aza-2’deoxycytidine and valproic acid treatment on rhabdomyosarcoma and medulloblastoma in Ptch mutant mice. Cancer Res. 2009;69(3):887–895. doi: 10.1158/0008-5472.CAN-08-0946. [DOI] [PubMed] [Google Scholar]
- 29.Venkataraman S, et al. Inhibition of BRD4 attenuates tumor cell self-renewal and suppresses stem cell signaling in MYC driven medulloblastoma. Oncotarget. 2014;5(9):2355–2371. doi: 10.18632/oncotarget.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bandopadhayay P, et al. BET bromodomain inhibition of MYC-amplified medulloblastoma. Clin Cancer Res. 2014;20(4):912–925. doi: 10.1158/1078-0432.CCR-13-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henssen A, et al. BET bromodomain protein inhibition is a therapeutic option for medulloblastoma. Oncotarget. 2013;4(11):2080–2095. doi: 10.18632/oncotarget.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alimova I, et al. Targeting the enhancer of zeste homologue 2 in medulloblastoma. Int J Cancer. 2012;131(8):1800–1809. doi: 10.1002/ijc.27455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu S, et al. Dynamic regulation of the PR-Set7 histone methyltransferase is required for normal cell cycle progression. Genes Dev. 2010;24(22):2531–2542. doi: 10.1101/gad.1984210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanaka H, et al. The SETD8/PR-Set7 Methyltransferase Functions as a Barrier to Prevent Senescence-Associated Metabolic Remodeling. Cell Rep. 2017;18(9):2148–2161. doi: 10.1016/j.celrep.2017.02.021. [DOI] [PubMed] [Google Scholar]
- 35.Tharmalingam S, Wu C, Hampson DR. The calcium-sensing receptor and integrins modulate cerebellar granule cell precursor differentiation and migration. Dev Neurobiol. 2016;76(4):375–389. doi: 10.1002/dneu.22321. [DOI] [PubMed] [Google Scholar]
- 36.Kassis J, Moellinger J, Lo H, Greenberg NM, Kim HG, Wells A. A role for phospholipase C-gamma-mediated signaling in tumor cell invasion. Clin Cancer Res. 1999;5(8):2251–2260. [PubMed] [Google Scholar]
- 37.Svalina MN, et al. IGF1R as a Key Target in High Risk, Metastatic Medulloblastoma. Sci Rep. 2016;6:27012. doi: 10.1038/srep27012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Linder S. The matrix corroded: podosomes and invadopodia in extracellular matrix degradation. Trends Cell Biol. 2007;17(3):107–117. doi: 10.1016/j.tcb.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 39.Wang X, et al. The role of cancer stem cells in cancer metastasis: new perspective and progress. Cancer Epidemiol. 2013;37(1):60–63. doi: 10.1016/j.canep.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 40.Wang J, Wechsler-Reya RJ. The role of stem cells and progenitors in the genesis of medulloblastoma. Exp Neurol. 2014;260:69–73. doi: 10.1016/j.expneurol.2012.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang F, et al. SET8 promotes epithelial-mesenchymal transition and confers TWIST dual transcriptional activities. EMBO J. 2012;31(1):110–123. doi: 10.1038/emboj.2011.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hou L, Li Q, Yu Y, Li M, Zhang D. SET8 induces epithelial‑mesenchymal transition and enhances prostate cancer cell metastasis by cooperating with ZEB1. Mol Med Rep. 2016;13(2):1681–1688. doi: 10.3892/mmr.2015.4733. [DOI] [PubMed] [Google Scholar]
- 43.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15(3):178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shih DJ, et al. Cytogenetic prognostication within medulloblastoma subgroups. J Clin Oncol. 2014;32(9):886–896. doi: 10.1200/JCO.2013.50.9539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Driskell I, Oda H, Blanco S, Nascimento E, Humphreys P, Frye M. The histone methyltransferase Setd8 acts in concert with c-Myc and is required to maintain skin. EMBO J. 2012;31(3):616–629. doi: 10.1038/emboj.2011.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Talasz H, Lindner HH, Sarg B, Helliger W. Histone H4-lysine 20 monomethylation is increased in promoter and coding regions of active genes and correlates with hyperacetylation. J Biol Chem. 2005;280(46):38814–38822. doi: 10.1074/jbc.M505563200. [DOI] [PubMed] [Google Scholar]
- 47.Giannelli G, Koudelkova P, Dituri F, Mikulits W. Role of epithelial to mesenchymal transition in hepatocellular carcinoma. J Hepatol. 2016;65(4):798–808. doi: 10.1016/j.jhep.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 48.Ma A, et al. Discovery of a selective, substrate-competitive inhibitor of the lysine methyltransferase SETD8. J Med Chem. 2014;57(15):6822–6833. doi: 10.1021/jm500871s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ma A, Yu W, Xiong Y, Butler KV, Brown PJ, Jin J. Structure-activity relationship studies of SETD8 inhibitors. Medchemcomm. 2014;5(12):1892–1898. doi: 10.1039/C4MD00317A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ghantous A, Hernandez-Vargas H, Byrnes G, Dwyer T, Herceg Z. Characterising the epigenome as a key component of the fetal exposome in evaluating in utero exposures and childhood cancer risk. Mutagenesis. 2015;30(6):733–742. doi: 10.1093/mutage/gev010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miozzo M, Vaira V, Sirchia SM. Epigenetic alterations in cancer and personalized cancer treatment. Future Oncol. 2015;11(2):333–348. doi: 10.2217/fon.14.237. [DOI] [PubMed] [Google Scholar]
- 52.Northcott PA, et al. Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma. Nat Genet. 2009;41(4):465–472. doi: 10.1038/ng.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alimova I, et al. Inhibition of EZH2 suppresses self-renewal and induces radiation sensitivity in atypical rhabdoid teratoid tumor cells. Neuro-oncology. 2013;15(2):149–160. doi: 10.1093/neuonc/nos285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Knutson SK, et al. Durable tumor regression in genetically altered malignant rhabdoid tumors by inhibition of methyltransferase EZH2. Proc Natl Acad Sci USA. 2013;110(19):7922–7927. doi: 10.1073/pnas.1303800110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Assou S, et al. Transcriptome analysis during human trophectoderm specification suggests new roles of metabolic and epigenetic genes. PLoS ONE. 2012;7(6):e39306. doi: 10.1371/journal.pone.0039306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oda H, et al. Monomethylation of histone H4-lysine 20 is involved in chromosome structure and stability and is essential for mouse development. Mol Cell Biol. 2009;29(8):2278–2295. doi: 10.1128/MCB.01768-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Park JB, et al. Phospholipase signalling networks in cancer. Nat Rev Cancer. 2012;12(11):782–792. doi: 10.1038/nrc3379. [DOI] [PubMed] [Google Scholar]
- 58.Puisieux A, Brabletz T, Caramel J. Oncogenic roles of EMT-inducing transcription factors. Nat Cell Biol. 2014;16(6):488–494. doi: 10.1038/ncb2976. [DOI] [PubMed] [Google Scholar]
- 59.Dhami GK, et al. Dynamic methylation of Numb by Set8 regulates its binding to p53 and apoptosis. Mol Cell. 2013;50(4):565–576. doi: 10.1016/j.molcel.2013.04.028. [DOI] [PubMed] [Google Scholar]
- 60.Milite C, et al. The emerging role of lysine methyltransferase SETD8 in human diseases. Clin Epigenetics. 2016;8:102. doi: 10.1186/s13148-016-0268-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schwalbe EC, et al. Novel molecular subgroups for clinical classification and outcome prediction in childhood medulloblastoma: a cohort study. Lancet Oncol. 2017;18(7):958–971. doi: 10.1016/S1470-2045(17)30243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ramaswamy V, et al. Recurrence patterns across medulloblastoma subgroups: an integrated clinical and molecular analysis. Lancet Oncol. 2013;14(12):1200–1207. doi: 10.1016/S1470-2045(13)70449-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 65.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Oliveros JC. Venny 2.1. http://bioinfogp.cnb.csic.es/tools/venny/index.html Accessed December 5, 2018.
- 67.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu G, Wang LG, He QY. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015;31(14):2382–2383. doi: 10.1093/bioinformatics/btv145. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.