Abstract
Deposition of amyloid-β protein (Aβ) to form neuritic plaques is the characteristic neuropathology of Alzheimer’s disease (AD). Aβ is generated from amyloid precursor protein (APP) by β- and γ-secretase cleavages. BACE1 is the β-secretase and its inhibition induces severe side effects, whereas its homolog BACE2 normally suppresses Aβ by cleaving APP/Aβ at the θ-site (Phe20) within the Aβ domain. Here, we report that BACE2 also processes APP at the β site, and the juxtamembrane helix (JH) of APP inhibits its β-secretase activity, enabling BACE2 to cleave nascent APP and aggravate AD symptoms. JH-disrupting mutations and clusterin binding to JH triggered BACE2-mediated β-cleavage. Both BACE2 and clusterin were elevated in aged mouse brains, and enhanced β-cleavage during aging. Therefore, BACE2 contributes to AD pathogenesis as a conditional β-secretase and could be a preventive and therapeutic target for AD without the side effects of BACE1 inhibition.
Keywords: Neuroscience
Keywords: Alzheimer’s disease
BACE2 contributes to pathogenesis in murine models of Alzheimer’s disease and could be a preventive and therapeutic target.
Introduction
Alzheimer’s disease (AD) is the most common neurodegenerative disease leading to dementia (1). Deposition of amyloid β protein (Aβ) to form neuritic plaques in the brain is a hallmark of AD neuropathology (2). In the amyloidogenic pathway, amyloid precursor protein (APP) is first cleaved by β-secretases at the β-site within its luminal/extracellular domain to release a C-terminal fragment, C99 (or CTFβ). C99 is then cleaved by γ-secretase at the transmembrane domain to shed the N-terminal fragment Aβ. Mutations enhancing β-cleavage of APP or altering γ-cleavage of C99 result in early-onset AD (3–5). Recently, a missense mutation in the APP gene was shown to suppress Aβ production and reduce the risk of AD (3, 6, 7).
The protease BACE1 is an indispensable β-secretase of APP in vivo (8–10). Genetic ablation of BACE1 in an AD mouse model abolishes Aβ in the brain (11, 12). Inhibition of BACE1 has been considered a valid avenue for AD drug development (13, 14). However, as BACE1 also cleaves other proteins important for brain functions, BACE1 inhibition has been shown to induce severe side effects and is therefore not clinically applicable (15). BACE1 cleaves APP at 2 sites: β-cleavage at Asp1 and β′-cleavage at Glu11 within the Aβ domain produce C99 and C89, respectively. Cleavage of C89 by γ-secretase produces truncated Aβ fragments (Figure 1A) (3, 16). Although BACE1 is the most important β-secretase, most in vivo studies used a mouse model containing the Swedish APP mutant that biasedly enhances β-cleavage by BACE1 (16), and the contribution of other potential and weaker β-secretases, if any, is overshadowed.
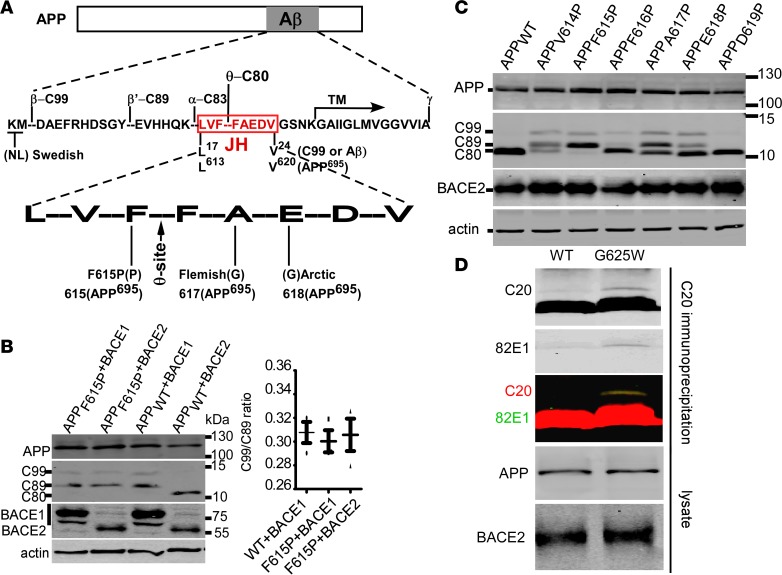
Figure 1. Perturbation of the JH domain by proline causes BACE2-mediated β-cleavages.
(A) Schematic diagram showing the cleavage sites and mutation sites in APP. JH, juxtamembrane helix; TM, transmembrane domain. (B) APP695 wild type (APPWT) and APP F615P mutant (numbering as in APP695 form; APPF615P) were coexpressed in HEK293 cells with Myc-His–tagged BACE1 or BACE2, and cell lysates were blotted for CTFs, APP, and BACEs. Both BACE1 and BACE2 were detected using 9E10 anti-Myc antibody. Ratios of C99 to C89 were plotted. Bars represent mean ± SEM (n = 3). (C) Proline-scanning mutagenesis of the APP JH domain. All residues from V614 to D619 of APP695 were individually replaced by proline, and the mutants were coexpressed in HEK293 with BACE2. Cell lysates were immunoblotted for CTFs, APP, and BACE2. (D) APPWT and the non–membrane-binding APPG625W were coexpressed with BACE2. CTFs were enriched by IP with C20 antibody, and Western blotted with both C20 polyclonal and the C99-specific 82E1 antibody.
BACE2, with 45% of its amino acid residues identical to BACE1, was initially considered as another β-secretase (8, 17, 18). The BACE2 gene is located in the Down syndrome (DS) critical region of chromosome 21 and BACE2 might therefore contribute to neuritic plaque formation and Alzheimer-related dementia in DS (19, 20). Early studies suggested that BACE2 cleaved wild-type APP at the β-site, and 2 APP mutants, i.e., Swedish and Flemish mutants, enhanced this cleavage with unknown reason (18). However, later studies clearly demonstrated that BACE2 is not a β-secretase (21–24). Instead of cleaving APP at the β-site, BACE2 cleaves APP within the Aβ domain, particularly at the θ-site (Phe20) to produce C80 and prevents Aβ generation (21). Overexpression of BACE2 inhibited Aβ production in neurons and transgenic mice (21, 24–27). Moreover, BACE2 expression in the brain is rather weak (28), implying that even if it does cleave APP at the β-site, its contribution to C99/Aβ could be negligible in comparison with BACE1. For these reasons, BACE2 has been less considered an important enzyme for AD pathogenesis or therapy.
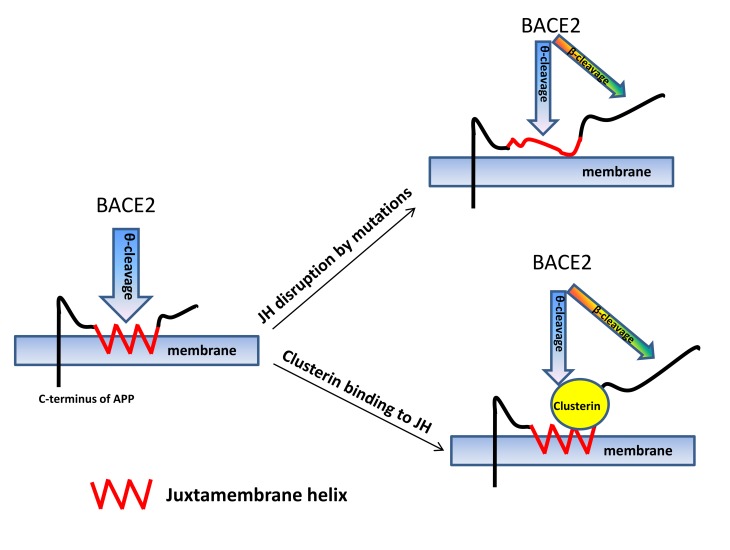
In this report, we found that BACE2 could function as a conditional β-secretase. The juxtamembrane helix (JH) of APP is crucial to inhibit the β-secretase activity of BACE2. Disruption of the JH by artificial and AD-associated mutations triggered the β-secretase activity of BACE2. Suppressing endogenous BACE2 in an AD model mouse carrying the Arctic mutation significantly reduced C99 and Aβ generation, suggesting a high in vivo activity of BACE2 in spite of the low expression. Clusterin, a neuronal chaperone protein (29) that is induced by multiple AD risk factors, upregulated during AD and DS (30–33), and a top 3 sporadic AD risk gene (AlzGene; http://www.alzgene.org/), enabled BACE2-mediated β-cleavage of wild-type APP through binding to the JH. Both BACE2 and clusterin showed enhanced expression in the brains of aged wild-type mice. As β-secretase activity, but not BACE1, increases in aged human, monkey, and nontransgenic mouse brains (34), BACE2 could be the emerging β-secretase in vivo during aging. Moreover, BACE2 displays higher activity in preclinical AD (35). Hence, unlike BACE1 that is constitutively active, BACE2 becomes a β-secretase only conditionally, and may therefore better correlate with AD pathogenesis. BACE2-mediated β-cleavage represents what we believe is a novel mechanism underlying AD pathogenesis, and an early intervention against BACE2 and clusterin may help AD prevention without the anti-BACE1 side effects (15, 36).
Results
BACE2 cleaves the APPF615P mutant at the β-site.
An artificial APP mutant (APPF615P), in which phenylalanine 615 (F615 of APP695 or F19 of Aβ) is replaced by a proline, inhibits the nonamyloidogenic α-cleavage of APP to generate C83 (Figure 1A) (37). As F615 flanks the θ-site (Figure 1A), we tested BACE2 cleavage of this mutant. Surprisingly, the F615P mutation enabled BACE2 to process APP at BACE1 cleavage sites to generate C99 and C89, and abolished the θ-cleavage–derived C80 produced by BACE2 (Figure 1B and Figure 2, A and B). The C-terminal fragment (CTF) patterns generated by BACE2 and BACE1 cleavage of APPF615P were indistinguishable (Figure 1B). To examine if this is due to the destruction of the θ-site by F615P, which forced BACE2 to cleave at the β- and β′-sites, recombinant BACE2 substrates with or without F615P substitution were generated for in vitro BACE2 cleavage (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.123431DS1). These substrates contained only θ-site but no β- or β′-sites. Circular dichroism revealed a reduction of α-helix feature bands at 210 nm and 220 nm in the F615P-containing substrate (Supplemental Figure 1A). When incubated with recombinant BACE2, the wild-type and mutant substrates decreased at similar rates (Supplemental Figure 1A). Moreover, a double mutation abolishing BACE1-catalyzed CTF shedding (3) partially restored C80 from APPF615P by BACE2 (Supplemental Figure 1B). These results suggested that rather than destroying the θ-site, F615P shifted BACE2 cleavage preference to β- and β′-sites.
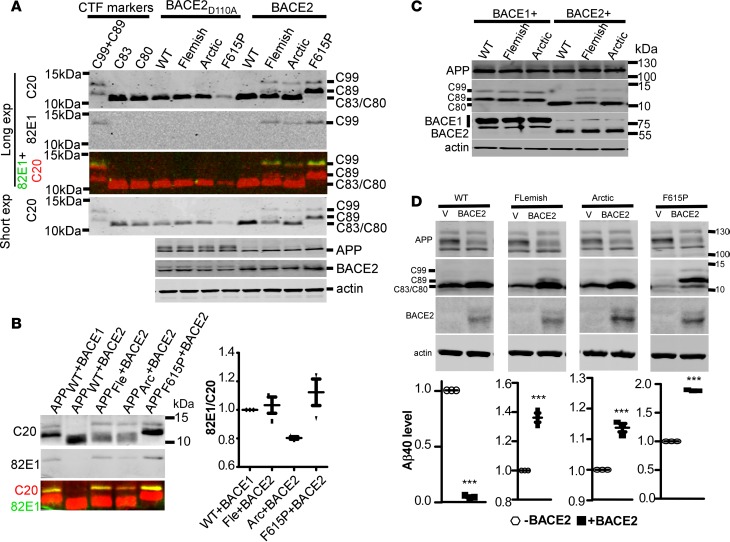
Figure 2. AD-associated APP mutations perturbed the JH domain and induced β-cleavage by BACE2.
(A) APP variants were coexpressed in HEK293 cells with BACE2 or inactive BACE2 mutant (D110A). Cell lysates were blotted for CTFs, APP, and BACE2. CTF markers expressed in HEK293 cells were loaded to indicate the mobility of CTFs. CTFs were blotted with rabbit polyclonal C20 antibody against the last 20 residues of APP, and the same membrane was also blotted with C99/Aβ(1–x)–specific mouse monoclonal antibody 82E1. The color panel is the merged images of C20 and 82E1 on the same gel. For clear visualization of C99 and C89 from APP mutants, Western blot membranes were exposed for a longer time. For the comparison of C83/C80, membranes were exposed for a shorter time. (B) APP variants were coexpressed with BACE1 or BACE2 in HEK293 cells, and CTFs were simultaneously blotted with the rabbit polyclonal C20 antibody and the mouse monoclonal 82E1 antibody. Images of C20 (red) and 82E1 (green) bands were merged in the bottom panel. Ratios of 82E1 signals to C20 signals were plotted, and comparison was by paired t tests. (C) APP wild type, Flemish, and Arctic mutants were coexpressed with either BACE1-Myc or BACE2-Myc in HEK293 cells. Cell lysates were blotted for APP, CTFs, and BACEs. (D) APPWT, Flemish, Arctic, and APPF615P mutants were coexpressed with BACE2-Myc-His in PC12 cells. Cell lysates were immunoblotted for indicated proteins and the conditioned media were measured for Aβ1–40. All bars represent mean ± SEM (n = 3). ***P < 0.001 (paired t tests). WT, APP wild type; APPFle and Fle, APP Flemish mutant; APPArc and Arc, APP Arctic mutant.
Perturbation of the JH domain of APP by proline causes BACE2-mediated β-cleavages.
F615P is within the JH of C99 (L17–V24)/APP (L613–V620 in APP695) (38) (Figure 1A). As the substituting residue proline is α-helix breaking, structural perturbation may underlie the altered BACE2 cleavage. Proline-scanning mutagenesis of APP from V614 to D619 within the JH demonstrated that all these substitutions led to C99 production by BACE2 (Figure 1C). As this helix interacts with cholesterol and organelle membranes (38), the loss of these 2 bindings upon the perturbation of JH might be a more direct cause of BACE2-mediated β-cleavage. Non–cholesterol-binding APPG625A and non–membrane-associating APPG625W (39) with the mutation site G625 outside the JH were therefore tested for BACE2 cleavage. Only APPG625W was very weakly processed into C99 (Figure 1D), suggesting that membrane binding of JH plays an important role in inhibiting β-cleavage by BACE2. However, as C99 derived from APPG625W by BACE2 was so weak that an enrichment by immunoprecipitation was required for clear detection, and C80 remained the predominant product of BACE2, the affected membrane binding by the F615P mutation, if any, is unlikely the major cause of the shifted BACE2 cleavage site preference.
BACE2 cleaves APP Flemish and Arctic mutants at the β-site.
To confirm that the JH inhibits β-cleavage by BACE2, we coexpressed APP Flemish (A617G, APPFlemish) and Arctic (E618G, APPArctic) mutants with BACE2 in HEK293 cells. These 2 APP mutants both have the mutation sites in the JH, with glycine as the substituting residue (Figure 1A). Like proline, glycine is α-helix breaking. Both APPFlemish and APPArctic were cleaved by BACE2 into C99 and C89 (Figure 2, A–D). By contrast, endogenous α-secretases did not produce C99 from either wild-type or mutant APPs (Figure 2A). Meanwhile, the θ-cleavage product C80 from APPFlemish and APPArctic by BACE2 were reduced compared with C80 from APPWT (Figure 2A). Western blot using C99/Aβ(1–x)–specific antibody 82E1 and C20 antibody against the last 20 amino acids of APP/CTFs demonstrated that C99 bands from APP F165P, Flemish, and Arctic mutants by BACE2, as detected by C20, contained similar percentages of bona fide C99 (recognized by 82E1) as the C99 from APPWT by BACE1 (Figure 2B), indicating that BACE2 produced little or no non-C99 CTFs with the same gel mobility as C99 (e.g., C100, C98, etc.). BACE1 cleavage of Flemish and Arctic mutants was not apparently affected (Figure 2C). Three other AD-associated mutations at the Arctic site, i.e., Dutch E618Q, Osaka E618del, and Italian E618K, did not induce detectable C99 through BACE2 (data not shown). Usually, Q (glutamine) and K (lysine) are not α-helix disrupting, and the structural effect of E618 deletion is unclear. Decreased Aβ production from APPWT and increased Aβ generation from APPF615P, Flemish, and Arctic mutants upon BACE2 overexpression (Figure 2D) further corroborated that BACE2 is nonamyloidogenic for wild-type APP, but becomes amyloidogenic for these APP mutants.
F615P, Flemish, and Arctic mutations of APP abolish BACE2 cleavage of the nascent APP.
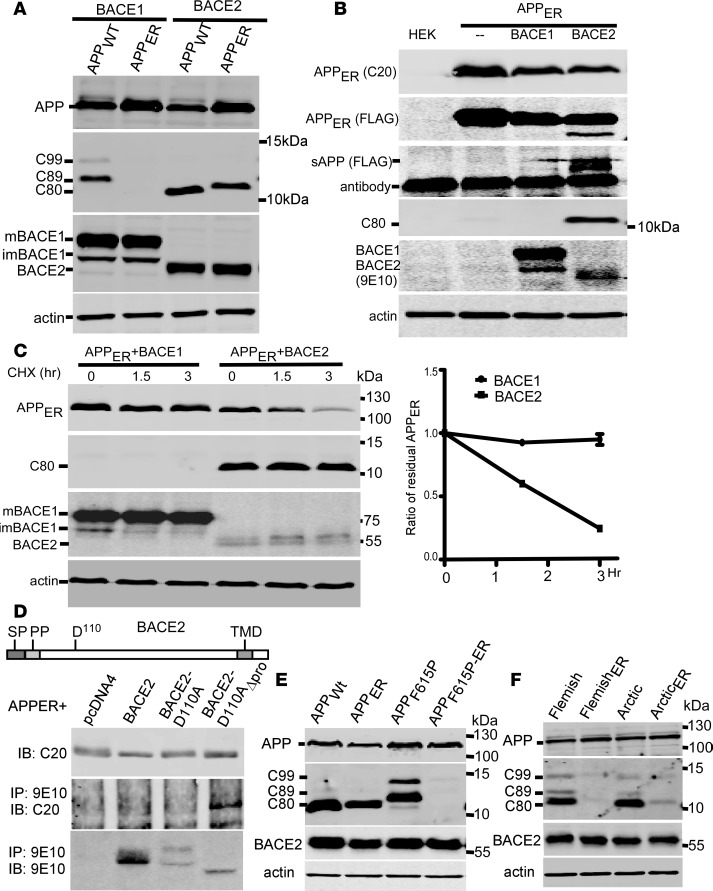
When nascent APP was arrested in the endoplasmic reticulum (APPER) to prevent maturation, BACE2 still produced C80 (Figure 3A) and a secreted APP N-terminal fragment (Figure 3B) from APPER, and decreased APPER in a time-dependent manner upon the inhibition of protein synthesis (Figure 3C). APPER coimmunoprecipitated with inactive D110A-mutant BACE2 without the inhibitory propeptide (Figure 3D), but not with wild-type BACE2 or inactive mutant BACE2, suggesting that APPER quickly separates from BACE2 after cleavage, and does not interact with BACE2 containing the propeptide that must be removed by autocleavage before substrate recognition. In contrast, overexpressed BACE1 and endogenous α-secretase did not cleave APPER (Figure 3, A–C). Therefore, BACE2 can cleave APP earlier than other secretases do, and may as such reduce the supply of APP as BACE1 substrate. However, retaining nascent APPF615P (Figure 3E), APPFlemish, and APPArctic (Figure 3F) in the ER abolished the cleavage by BACE2. Thus, these mutants can both trigger BACE2-dependent C99/Aβ generation and abolish the protective θ-cleavage of the nascent APP by BACE2.
Figure 3. F615P, Flemish, and Arctic mutations abolished BACE2 cleavage of nascent APP.
(A) APPWT and APP with the KKXX ER retention signal (APPER) were coexpressed with BACE1 or BACE2 in HEK293 cells. CTFs and APP were blotted with C20 and BACE1 and BACE2 were probed with anti-Myc antibody. mBACE1 and imBACE1, mature and immature BACE1, respectively. (B) APPER with a FLAG tag inserted into APP after the signal peptide was coexpressed with BACE1 or BACE2 in HEK293 cells. Cell lysates were blotted for APP, CTF, and BACEs. Secreted APP (sAPP) in the conditioned media was enriched by immunoprecipitation using FLAG-agarose, and Western blotted using anti-FLAG antibody. (C) APPER was coexpressed with BACE1 or BACE2 in HEK293 cells, and treated with the translation inhibitor cycloheximide (CHX, 100 μM) for the indicated times. Full-length APPER, CTFs, and BACE2 were blotted. Immature BACE1 decreased due to ceased protein synthesis and continuous BACE1 maturation. Residual full-length APPER relative to time 0 was plotted. Curves represent mean ± SEM. (D) Schematic diagram indicates domains and the first active site in BACE2. SP, signal peptide; PP, propeptide; D110, the first active site; TMD, transmembrane domain. APPERwas coexpressed with Myc-tagged BACE2, inactive BACE2 mutant (BACE2D110A), or BACE2D110A without the propeptide (BACE2D110A-Δpro). BACE2 variants were immunoprecipitated (IP) with anti-Myc 9E10 antibody, and the precipitates were immunoblotted (IB) using the indicated antibodies. APPF615P (E) and APP Flemish and Arctic mutants (F) with the ER retention signal (APPF615P-ER, FlemishER, and ArcticER, respectively) were coexpressed with BACE2 in HEK293 cells. CTFs, APP, and BACE2 in lysates were blotted.
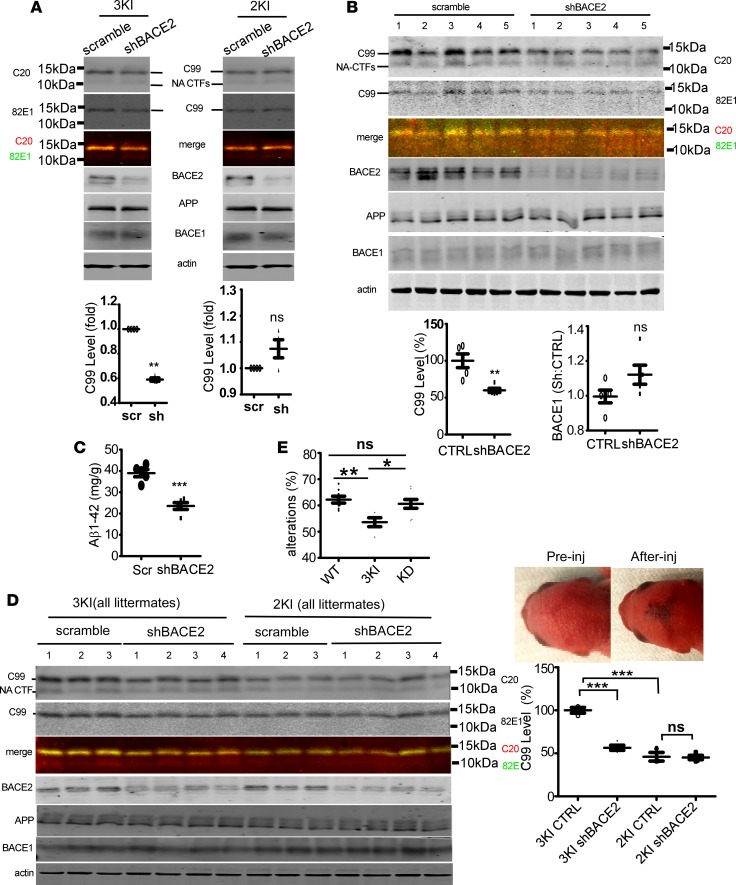
BACE2 knockdown reduces C99 and Aβ accumulation in the brains of APP Arctic mutation–knockin mice.
An APP Swedish-Arctic-Iberian triple mutation–knockin mouse (3KI) displayed stronger C99 and faster Aβ and plaque accumulation than the Swedish-Iberian mutation KI mouse (2KI) (40). Since in both human and mouse BACE2 and APP are located in the same chromosome (21 in human and 16 in mouse), and the 2 genes are very close to each other on the chromosome, obtaining the APP3KI/3KI;BACE2–/– double-mutant mice by mouse line crossing is not realistic. We intraventrically injected adeno-associated virus (AAV) expressing BACE2 shRNA into the brains of 3KI mice. Although the Swedish mutation greatly increases C99 and Aβ generation by enhancing BACE1-mediated β-cleavage at the Asp1 site (3, 16), knockdown of BACE2 reduced the C99 level in primary cortical neurons of 3KI to 59.63% ± 2.99% (P < 0.01; Figure 4A and Supplemental Figure 2A). In the brains of 11-week-old 3KI mice, BACE2 knockdown significantly lowered C99 (100% ± 20.95% vs. 59.88% ± 6.35%; P < 0.01) (Figure 4B) and Aβ levels (38.96 ± 3.81 μg/g vs. 23.54 ± 3.58 μg/g brain tissue; P < 0.001) (Figure 4C) without apparently affecting BACE1 (Figure 4B). No apparent C99 change was found for primary neurons of 2KI or brains of 2-week-old 2KI expressing BACE2 shRNA (Figure 4, A and D). Furthermore, BACE2 suppression slowed the growth of neuritic plaques in 3KI mouse brains (Supplemental Figure 2B). To determine if Aβ and plaques derived from BACE2-mediated β-cleavage contribute to memory deficits in 3KI mice, BACE2-suppressed 3KI mice were subjected to behavioral test at the age of 6 months. At this age, the 3KI mice display cognitive impairment only in the Y-maze test, whereas the 2KI mice whose APP is not a β-cleavage substrate of BACE2 develop similar impairment at 18 months (40). Knocking down of BACE2 significantly improved the performance of 3KI mice (Figure 4E). Hence, despite its weak expression, BACE2 exhibited remarkable β-secretase activity, and contributed to AD pathogenesis in Arctic mutation–containing brains.
Figure 4. BACE2 suppression reduces C99 and Aβ accumulation in the brains of APP Arctic mutation–knockin mice.
(A) Primary neurons from E18 APPSwedish-Arctic-Iberian/Swedish-Arctic-Iberian–knockin mice (3KI) or APPSwedish-Iberian/Swedish-Iberian–knockin mice (2KI) were transduced with adeno-associated virus-9 (AAV9) carrying scrambled shRNA (scr) or shRNA against mouse BACE2 (sh). Cells were analyzed by Western blot for CTFs (C-terminal fragments of APP), APP, BACE2, and BACE1.NA-CTF, nonamyloidogenic C-terminal fragments of APP. (B) Viruses containing scrambled shRNA or BACE2 shRNA were intraventricularly injected into the brains of neonatal 3KI mice (all littermates, n = 5 for each). Mice were sacrificed at 11 postnatal weeks. Half brains were Western blotted for the indicated proteins. C99 on the same membrane was also blotted with C99-specific antibody 82E1, and merged with C20 blots. Nonamyloidogenic CTFs (NA-CTFs) include C89, C83, and C80. CTRL, control scrambled shRNA. (C) The other brain halves were analyzed by Aβ1–42 ELISA. ELISA readout for Aβ1–40 was no higher than background noise (n = 5). (D) AAV9 carrying scrambled shRNA or shBACE2 were intraventricularly injected into the brains of neonatal 3KI (all littermates) or 2KI (all littermates) mice. Successful injection was indicated by fast dye (blue). Mice were sacrificed 2 weeks after virus injection, and brain homogenates in RIPA buffer were Western blotted for the indicated proteins. C99 levels were expressed as percentages of C99 in scramble shRNA–injected 3KI mice. n = 4 for 3KI Ctrl, 3KI shBACE2, and 2KI shBACE2, and n = 3 for 2KI control. (E) Y-maze test for 6-month-old 3KI mice with or without BACE2 suppression. n = 8 for wild-type mice (WT), 4 for 3KI, and 8 for 3KI with suppressed BACE2 expression (KD). *P < 0.05; **P < 0.01; ***P < 0.001 by unpaired t test. ns, non-significant. All bars represent mean ± SEM.
BACE2 isoform 1 and isoform 2 function differently in APP processing.
A BACE2 splicing variant without exon-7 (isoform-2; Supplemental Figure 3A) has been reported to be upregulated in AD (35). This BACE2 isoform extremely weakly cleaved APPWT into a C99-like CTF. More strikingly, it did not cleave nascent APPWT, and its β-cleavage of APPFlemish was much weaker than that by BACE2 (i.e., isoform-1; Supplemental Figure 3B). Thus, deletion of BACE2 exon-7 and disruption of the JH of APP showed similar but nonsynergistic/additive effects on APP processing by BACE2, implying a common structural or regulatory basis.
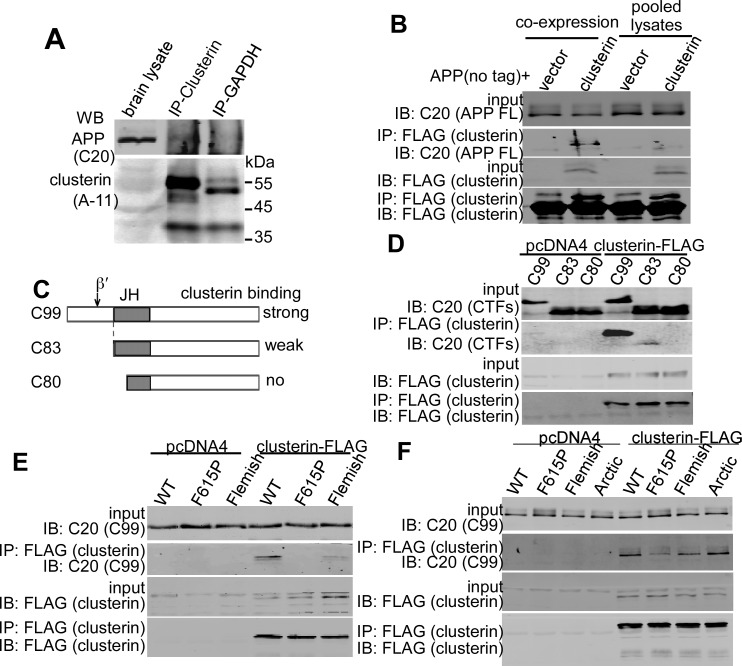
Clusterin binds APP with intact and wild-type JH intracellularly.
To explore BACE2’s involvement in sporadic AD and DS, we hypothesized that an Aβ-binding protein could also bind APP to the JH (Figure 1A) to create a structural change or spatial hindrance, hence initiating β-cleavage of APPWT by BACE2. Clusterin/ApoJ is a secreted Aβ-binding chaperone and is upregulated in AD and DS (30–32). It coimmunoprecipitates with transgenic APP in the brain (41). We found that endogenous-level APP coimmunoprecipitated with endogenous clusterin in mouse brain lysates (Figure 5A). Clusterin coimmunoprecipitated with APP in cells coexpressing exogenous clusterin and APP in vitro, but when APP and clusterin were separately expressed and the lysates were pooled, there was no binding of the 2 proteins, suggesting that the intracellular environment is required for the binding (Figure 5B). Clusterin coimmunoprecipitated with APP, C99, and C83, but not with C80 (Figure 5, C and D). APP, C99, and C83 all contain the intact JH, whereas C80 contains only half of the JH (Figure 5C). Hence, the N-terminal part of the JH is crucial for APP/C99 binding with clusterin. The binding of clusterin with C83 is much weaker than that with C99, presumably because C83 shares the first residue with the JH. Without flanking residues, the helix may become unstable. Furthermore, the JH-disrupting F615P and Flemish mutations abolished or significantly reduced the binding of clusterin and C99, respectively, indicating that residues F615 and A617 (Flemish mutation site) are indispensable for C99-clusterin interaction (Figure 5E). In full-length APP, however, F615P and Flemish mutations only partially inhibited APP-clusterin interaction (Figure 5F), implying that full-length APP may have multiple clusterin-binding domains and the JH is one of them. As the Arctic mutation in APP did not affect APP-clusterin coimmunoprecipitation (Figure 5F), the integrity of the entire JH is not necessary for the binding.
Figure 5. Clusterin binds APP with intact and wild-type JH intracellularly.
(A) Coimmunoprecipitation (CoIP) of endogenous APP and clusterin from brain homogenate. Anti-GAPDH was used as control IP antibody. (B) Clusterin-FLAG and APPWT without tag were either coexpressed or separately expressed in HEK293 cells. Cells with both clusterin and APP were directly lysed (coexpression), and cells with only clusterin or APP overexpression were lysed and the lysates were combined (pooled lysates). After IP with FLAG-agarose, the precipitated proteins were blotted with C20 antibody for APP and FLAG antibody for clusterin. (C) Schematic diagram showing the position of the JH in different CTFs. (D) C99, C83, and C80 were coexpressed with clusterin-FLAG in HEK293, and the lysates were subjected to anti-FLAG CoIP. C20 antibody was used to detect CTFs in the precipitates. (E) Clusterin-FLAG was coexpressed with wild-type C99 (C99WT), F615P containing C99 (C99F615P), and Flemish mutation containing C99 (C99Fle) in HEK293 cells, and cell lysates were immunoprecipitated using anti-FLAG antibody. Precipitates were Western blotted using C20 for C99 variants and anti-FLAG for clusterin. (F) CoIP of clusterin-FLAG with APP variants in HEK293 cells. IP was by anti-FLAG antibody, and APP detection was by C20 antibody.
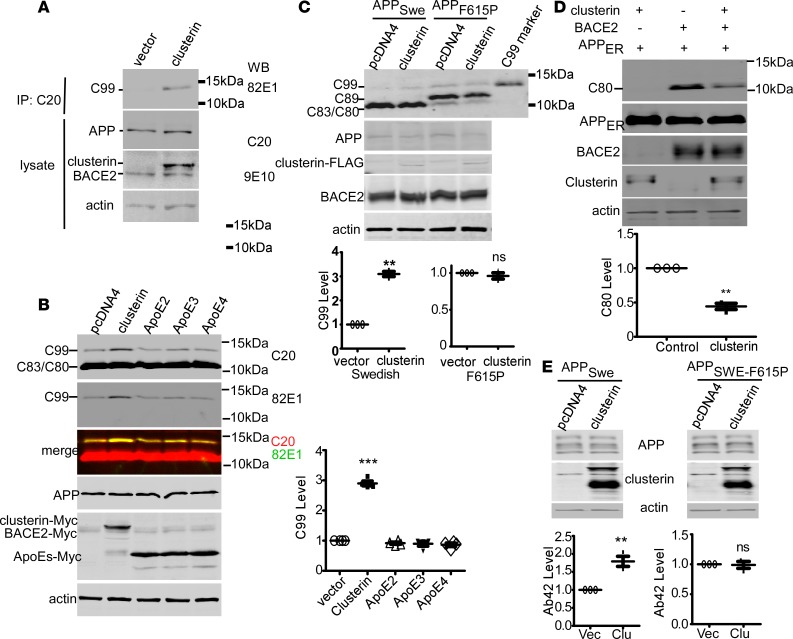
The binding of clusterin to the JH facilitates BACE2-mediated β-cleavages.
To assess if clusterin binding to the JH may trigger BACE2-mediated β-cleavage of APP, we coexpressed APPWT and BACE2 with or without clusterin in HEK293 cells. It was previously shown that the overexpressed clusterin in COS7 cells forms aggregates in the perinuclear region, a pattern closely resembling that in aged rat brains (42). Overexpression of clusterin triggered C99 generation by BACE2 from APPWT (Figure 6A). In 4EB2 cells stably expressing both human BACE2 and APPSwe (wild-type JH), BACE2 weakly produced C99, which could be because the Swedish mutation makes the β-site more susceptible to potential β-secretases. Overexpression of clusterin but not another Aβ-binding protein, ApoE, increased C99 production in 4EB2 (Figure 6, B and C). In HEK293 cells stably expressing the APPSwe mutation (20E2, without endogenous BACE2 expression; see ref. 43), clusterin did not increase C99 generated by endogenous BACE1. Instead, there was a slight reduction in C99 with clusterin overexpression (Supplemental Figure 4), suggesting that clusterin specifically induced the β-secretase activity of BACE2 but not endogenous BACE1 or α-secretases. Meanwhile, clusterin overexpression also suppressed BACE2 cleavage of nascent APP (Figure 6C). Both of the induced β-cleavages by BACE2 and the reduced θ-cleavage of nascent APP upon clusterin overexpression are reminiscent of the effects of JH-disrupting mutations. Clusterin did not increase BACE2-produced C99 from APPF615P whose JH region does not bind clusterin (Figures 5E and 6D). Consistently, clusterin overexpression increased Aβ from APPSwe but not from APPSwe-F615P (Figure 6E). Therefore, the binding of clusterin to the JH facilitates BACE2-catalyzed β-cleavage of APPWT and APPSwe.
Figure 6. The binding of clusterin to the JH facilitates BACE2-mediated β-cleavages.
(A) APPWT (no tag) and BACE2 (Myc-His tagged) were expressed in HEK293 in the presence or absence (vector) of coexpressed clusterin (Myc-His tagged). CTFs were enriched by immunoprecipitation (IP) with C20 antibody, and blotted with 82E1 antibody specific for C99. (B) Clusterin or ApoEs whose binding motif in Aβ resides in the JH region as well were overexpressed in 4EB2 cells, a HEK293 cell stably expressing human APP Swedish mutant (APPSwe) and BACE2. Total CTFs were blotted with C20 antibody and C99 was also blotted with 82E1. C99s were quantified (using C20 polyclonal) and expressed as the ratio to C99 in vector-expressing cells (n = 4 replicates). ***P < 0.001 (1-way ANOVA, Tukey’s post hoc test). (C) APPSwe or APPF615P were coexpressed with either pcDNA4 or clusterin-FLAG in 4B25 cells, a HEK293 cell line stably expressing Myc-tagged human BACE2. C99 levels were expressed as the ratio to C99 in pcDNA4-APP–expressing cells (n = 3 replicates). **P < 0.01 (paired t tests). (D) Nascent APP (APPER) and BACE2 were coexpressed with pcDNA4 (vector, clusterin “–”) or clusterin (clusterin “+”) in HEK293 cells. C80 bands were quantified and plotted as the ratio to C80 in vector-expressing (clusterin “–”) cells (n = 3 replicates). **P < 0.01 (paired t tests). (E) Clusterin was coexpressed in PC12 cells with APPSwe or the APP Swedish-F615P double mutant (APPSwe-F615P). Forty-eight hours after transfection, cell lysates were blotted for the indicated proteins and conditioned media were analyzed by Aβ1–42 ELISA. The amounts of Aβ1–42 were expressed as the ratio to Aβ1–42 from cells with pcDNA4 transfection (Vec) (n = 3 replicates). **P < 0.01 (paired t tests). All bars represent mean ± SEM.
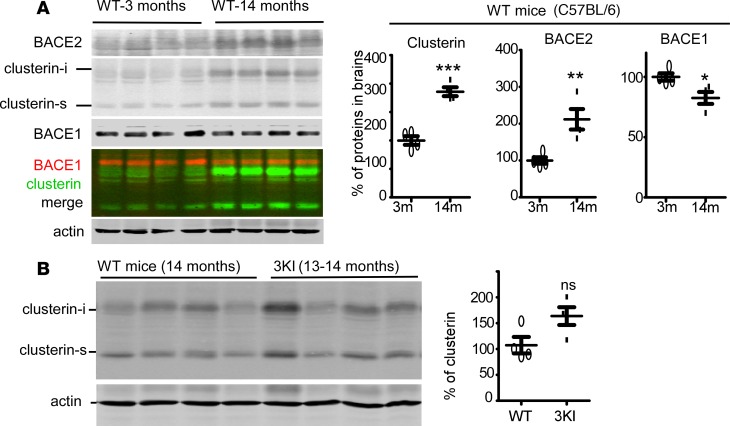
Altered expression of clusterin, BACE1, and BACE2 in aged brains.
Aging is the biggest risk factor of AD. Clusterin has been shown to be upregulated in some peripheral tissues or cells during aging, and proposed to be a marker for senescence (44, 45). In a DS model mouse, clusterin is also increased in older brains (46). Although this study did not observe any change in clusterin in aged wild-type mouse brains, only mice up to 200 days (<7 months) were tested.
To assess the possibility that aging may facilitate Aβ production through clusterin and BACE2, we compared clusterin, BACE1, and BACE2 protein levels in the brains of 3- and 14-month-old wild-type mice. Clusterin increased from 100% ± 15.13% in 3-month-old brains to 185% ± 15.53% in 14-month-old brains; BACE2 also displayed a dramatic increase in older mouse brains (100% ± 17.93% at 3 months vs. 212% ± 55.44% at 14 months). By contrast, brain BACE1 in older mice was slightly decreased (100% ± 6.20% vs. 83% ± 9.89%) (Figure 7A). Additionally, clusterin in the brains of 13- to 14-month-old 3KI mice did not show a significant change as compared with that in age-matched wild-type mice (Figure 7B). As the 3KI mice only display mild deficit in some cognition tests (40), the real AD pathology inducing clusterin upregulation in the human AD brain may not be triggered in the 3KI mouse model. Alternatively, the difference in clusterin levels in 3KI and wild-type mice is not sufficiently apparent, and given the variations of clusterin in 3KI, a larger sample pool would be required to reveal the difference.
Figure 7. Altered expression of clusterin, BACE1, and BACE2 in aged brains.
(A) Brains from C57BL/6 mice at the age of 3 months and 14 months were homogenized in RIPA buffer and the brain lysates were Western blotted for clusterin, BACE1, and BACE2. Protein bands were quantified and plotted. For the quantification of clusterin, the intracellular form (clusterin-i) and the secreted form (clusterin-s) were combined. (B) Brain lysates of C57BL/6 mice at the age of 14 months and 13- to 14-month-old 3KI (APPSwedish-Arctic-Iberian/Swedish-Arctic-Iberian) were blotted for clusterin. Both forms of clusterin were quantified and combined for plotting. *P < 0.05; **P < 0.01; ***P < 0.001 (paired t tests). ns, non-significant. All bars represent mean ± SEM.
Discussion
β-Cleavage of APP to generate C99 is the first and rate-limiting step for Aβ generation (47, 48). BACE1 is the β-secretase catalyzing β-cleavage of APP, and therefore inhibiting BACE1 has been long believed to be a therapeutic strategy for AD (49, 50). However, other indispensable functions of BACE1 in the brain have limited the clinical application of BACE1 inhibitors. As BACE1 is the β-secretase generating the most, if not all, C99, there seem to be no other targets available for β-cleavage suppression, unless a method can be developed to inhibit only BACE1 cleavage of APP without affecting BACE1 cleavage of other proteins. In addition to BACE1 as a constitutive β-secretase, there could be one or more other β-secretases that are activated during the initiation or progression of AD. Due to the high basal C99 production by BACE1, however, the contribution by the conditional β-secretase(s) may not be readily discernable, especially in animal models where APP mutants preferentially favoring BACE1 cleavage are used. Nevertheless, the abnormally induced β-secretases may better correlate with AD than BACE1.
We found that the artificial F615P mutant of APP could be cleaved by BACE2 at the β-site, and then further identified Flemish and Arctic mutants of APP from AD patients as β-cleavage substrates of BACE2. The substituting amino acids of these 3 mutants and the location of the mutations in the JH of APP led us to the hypothesis that structural perturbation of the JH enabled β-cleavage by BACE2. This hypothesis was further validated by proline scanning of the JH. Hence, BACE2 is a conditional β-secretase whose β-secretase activity is normally suppressed by the JH during the interaction with APP.
The F615P, Flemish, and Arctic mutations in APP also abolished the θ-cleavage of nascent APP by BACE2 in the ER. It is of interest that both BACE1 and BACE2 are suggested to be dependent on low pH for activity, but the ER is a neutral-pH environment. Hence, although BACEs themselves may require low pH for highest activity in vitro when the substrates are peptides with cleavage sites always exposed, the interaction with full-length APP could alter BACEs’ mode of action.
It is also interesting to note that overexpressed BACE1 and endogenous α-secretase only cleave mature APP after ER export. Since there is always sufficient APP, both mature and immature, in the brain (see full-length APP in Figure 4), it is unlikely that α-secretase, BACE2, and BACE1 have substantial competition for APP processing under normal conditions. The excessive APP expression (relative to the combined activities of all these secretases) may ensure maximal cleavage by all these enzymes. If this is the case, any enzyme gaining the ability to catalyze β-cleavage could contribute to Aβ, regardless of other products from APP. On the other hand, high-level overexpression of any of these secretases to deplete mature APP may create competition. For instance, 7-fold overexpression of BACE1 in the brain greatly diminishes mature APP, and further BACE1 overexpression (10- to 20-fold) decreases Aβ production (51). One possible reason is that without enough mature APP as substrate, BACE1 may even compete with itself to decide which β- and β′-sites to cut.
We have shown that disruption of the JH triggered β-cleavage by BACE2. Since this observation highlighted a structural factor regulating BACE2 cleavage of APP, events in neuronal cells resulting in protein misfolding or structural perturbation may induce BACE2’s activity as a β-secretase. These events may include ER stress, defects in intracellular trafficking and posttranslational modifications, oxidative stress, impaired protein degradation, etc. Additionally, although the dissociation of the JH and organelle/plasma membranes (as embodied by the G625W substitution) only slightly induced C99 production by BACE2, it might serve as a starting point to initiate a cascade at an early stage, or given sufficient time, contribute to C99/Aβ accumulation in the brain. Defective lipid metabolism may affect JH and membrane interaction as well.
We also found that BACE2 isoform 2 without exon-7 functions differently from isoform 1 during APP processing. Isoform 2 cleaving APPWT or APPER was on some level similar to isoform 1 cleaving APPF615P, Flemish, and Arctic mutants, but isoform 2 and Flemish mutation did not synergistically enhance C99 production. Therefore, the sequence encoded by exon-7 of BACE2 may play an important role in regulating the interaction between BACE2 and APP. Sequence changes in either of the BACE2 and APP proteins may cause cleavage alterations towards the amyloidogenic direction.
As a θ-secretase with very low abundance in the brain, BACE2 is no longer deemed a key player in the amyloidogenic pathway. Nonetheless, our results indicated that BACE2 can be induced into a β-secretase when the inhibition by the JH is lifted. Endogenous BACE2 vigorously produced C99 from Arctic mutation–containing APP in the mouse brain, and the C99 production by BACE2 was comparable with that by BACE1. Therefore, BACE2 may be a weakly expressed protein in the brain, but its β-secretase activity, at least for the APP mutant in the model mouse, was not as low as expected. However, this does not necessarily mean that under physiological conditions BACE2 is also an active θ-secretase in vivo. Spatial, temporal, and intracellular trafficking factors need to be taken into consideration when cleavage activity is assessed in cells or in vivo.
To identify nongenetic factors that could induce β-cleavage by BACE2, we aimed at potential JH-binding proteins, and found clusterin/ApoJ. Clusterin is a known APP- and Aβ-binding protein, and we further showed it bound full-length APP in the JH motif. We also tested different ApoE isoforms that are known to bind Aβ in the JH region as well. However, these ApoE isoforms appeared to be irrelevant to BACE2 cleavage of APP in our experimental settings. Presumably, ApoE does not bind full-length APP and only regulates Aβ metabolism, or, its binding to the JH in APP does not cause structural or spatial effects that can affect BACE2 cleavage.
The gene CLU ranks third among sporadic AD risk genes (AlzGene) in GWAS studies, and clusterin protein is greatly increased in AD and DS brains. However, its role in AD is elusive. Clusterin facilitates Aβ clearance (52), but an AD model mouse with CLU knocked out does not show Aβ change at the age of 12 months (53). Presumably, Aβ clearance is mediated by extracellular secreted clusterin. With initial Aβ upregulation, clusterin accumulates intraneuronally (54) and promotes Aβ production through mechanisms such as BACE2-mediated β-cleavage. Consistent with this hypothesis, clusterin mutations identified in AD patients retain clusterin in the intracellular form (55). Hence, the role of clusterin in sporadic AD may be time/stage dependent.
Aging is the biggest risk factor for AD. Clusterin has been shown to be induced during senescence and therefore a marker for senescent cells (44, 45, 56). We found that both clusterin and BACE2 were increased in the brains of aged mice. Such a change in expression highlighted the possibility that clusterin and BACE2 may collaborate to enhance Aβ production in aging brains. It was previously shown that β-secretase activity instead of BACE1 is remarkably elevated in aged brains of humans, monkeys, and nontransgenic mice (34). The extra β-secretase activity could result from BACE1 being superactivated for unknown reasons, or because of the upregulation of other β-secretases. It is worth noting that an artificial substrate was used for the in vitro cleavage assay in this study, and the substrate does not contain the JH to suppress BACE2’s β-secretase activity. Hence, the increased β-secretase activity in aged brains might simply be due to higher BACE2 expression. Moreover, as there was no significant difference in clusterin levels between the brains of aged 3KI mice (artificial AD) and age-matched wild-type mice, clusterin upregulation in sporadic AD brains is unlikely a necessary consequence of AD (at least in the 3KI AD model mouse). The correlation between sporadic AD and clusterin upregulation may therefore imply a 1-directional causal relationship, even though a positive feedback loop could not be excluded. If such a causal relationship is true, some AD risk factors such as injury could impose a risk of AD by enhancing clusterin expression (57), and AD could be a long-term side effect of acute clusterin-mediated protection. However, owing to the great variations of clusterin protein level and the Arctic mutation in Aβ in 3KI mice, whether or not clusterin increase could also be a result of sporadic AD still needs to be demonstrated.
In summary, we discovered that BACE2 transforms from a θ-secretase to a β-secretase of APP upon disruption of the JH by mutations or upon clusterin binding to the JH. An AD risk factor could trigger AD pathogenesis through altering BACE2’s function to an induced β-cleavage.BACE2-mediated β-cleavage may account for a subtype of AD or disease progression at certain stages of AD and DS. Inhibition of BACE2 and clusterin or correcting the JH structure could be an alternative preventive and therapeutic strategy for BACE2-mediated AD cases. Considering that the function of BACE2 in the central nervous system is not well documented, and the BACE2-knockout mice do not show apparent defects, inhibiting BACE2 might not cause the side effects of BACE1 inhibition. Meanwhile, BACE2 suppression also reduces the risk of diabetes that is a strong risk factor for AD (43), and therefore BACE2 inhibition could benefit AD patients in multiple aspects. We recently reported that BACE2 may cleave a potassium channel and as such counter neuronal apoptosis (58), and inhibition of BACE2 might compromise this protection. In this case, repressing the factors that trigger the β-secretase activity of BACE2 could be a better strategy, and retaining the θ-cleavage by BACE2 is desired for Aβ suppression.
Methods
Plasmid constructs.
cDNAs coding for APP695, BACE1, BACE2, and clusterin were PCR amplified from human brain cDNAs reverse transcribed from total mRNA. Mutations were introduced by PCR-based site-directed mutagenesis. PCR products were subcloned into the pcDNA4/myc-HisA expression vector (Invitrogen). An ER retention signal was introduced into pcDNA4-APPWT by substituting QMQN in the C-terminal tail of APP to KKQN. To generate plasmid pFLAG-APPER expressing N-terminally FLAG-tagged APP protein, the coding sequence for the FLAG tag was inserted between the signal peptide and the luminal/extracellular domain of APP695, and the resulting DNA was subcloned into pcDNA4/myc-HisA. The FLAG tag was fused to the C-terminus of clusterin to generate clusterin-FLAG cDNA with an in-frame stop codon immediately following the FLAG tag. The cDNA was inserted into pcDNA4/myc-HisA for expression.
Cell culture and transfection.
HEK293 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS), 1 mM sodium pyruvate, 2 mM L-glutamine, 50 U/ml penicillin G sodium, and 50 μg/ml streptomycin sulfate (Invitrogen). PC12 cells were cultured in DMEM supplemented with 15% FBS. Stable cell lines were maintained in media containing zeocin (50 μg/ml) and/or G418 (100 μg/ml). All cells were maintained at 37°C in an incubator containing 5% CO2. Plasmid transfection was performed using Lipofectamine 2000 Reagent (Invitrogen) according to the manufacturer’s instructions. The 4EB2 cells stably expressing human APP Swedish mutant and BACE2 in HEK293 cells were as previously described (21).
Primary mouse cortical neuron culture.
Cortical neuron cultures were prepared from embryonic day 18 mouse embryos using previously described methods (59). Briefly, the mouse cortices were dissected out and the meninges were removed completely under a dissection microscope. The cortices were pooled in a 15-ml Falcon tube and digested with papain at 37°C with gentle rotation for 20 minutes. Then, the digestion solution was removed and cells were dissociated by pipetting with inactivation solution (MEM containing 0.6% D-[+]-glucose, 1 mM pyruvate, 10% horse serum, 2.5% bovine serum albumin [BSA], and 2.5% trypsin inhibitor). Cells were plated on poly-D-lysine–coated 35-mm dishes with 4 ml Neurobasal Media (Invitrogen) containing B27, GlutaMAX (Invitrogen), and insulin. For shRNA-mediated knockdown experiments, neurons were exposed to 2 × 1010 genomic copies (GC) of AAV9 per dish overnight at 3 days in vitro (DIV 3) and the medium was changed on DIV 4. Neurons were lysed in 1× RIPA buffer on DIV 7 for Western blot.
Immunoblot analysis.
Cell lysates or brain homogenates in RIPA buffer (1% Triton X-100, 1% sodium deoxycholate, 0.4% SDS, 0.15 M NaCl, 0.05 M Tris-HCl, pH 7.2, 5 mM NaF, 1 mM Na3VO4) supplemented with Complete Mini Protease Inhibitor Cocktail tablet (Roche Diagnostics) were resolved by SDS-PAGE in 8% Tris-glycine or 16% Tris-tricine gels (for CTFs), and then transferred to nitrocellulose membranes. Membranes were blocked for 1 hour in phosphate-buffered saline (PBS) containing 5% nonfat dried milk, and sequentially blotted with primary antibodies and IRDye fluorescence-labeled secondary antibodies. To specifically blot C99, biotin-labeled C99/Aβ(1–x)–specific antibody 82E1 (IBL) was used as primary antibody, and detection was by IRDye800CW-labeled streptavidin (LI-COR Biosciences). Membranes were scanned with an Odyssey system (LI-COR Biosciences). The following rabbit polyclonal antibodies were used: C20 (with the last 20 amino acids of APP as epitope, was generated in house; see ref. 49) for APP and CTFs; anti-BACE1 (D10E5, Cell Signaling Technology), and anti-FLAG (SAB1306078, Sigma-Aldrich). The following mouse monoclonal antibodies were used: anti-Myc (9E10, Abcam), anti-FLAG (M2, Sigma-Aldrich), anti-clusterin (B-5, A-11, Santa Cruz Biotechnology), anti-BACE2 (H3, Santa Cruz Biotechnology), and anti–Aβ N-terminus (82E1, IBL).
Immunoprecipitation and coimmunoprecipitation.
Cells were lysed in lysis buffer (for coimmunoprecipitation, 15 mM Tris pH 7.4, 150 mM NaCl, 1 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 5 mM NaF, 1 mM Na3VO4, complete protease inhibitor) or RIPA buffer (for immunoprecipitation to enrich target protein). For precipitations with C20 (for APP or CTFs), A-11 (for clusterin), or 9E10 (for Myc-tagged proteins), antibodies were first incubated with protein A/G beads (Santa Cruz Biotechnology) for at least 3 hours at 4°C with shaking, and added to cell lysates after washing with lysis buffer to remove unbound antibodies. For the precipitation of FLAG-tagged proteins, FLAG-agarose (Sigma-Aldrich) was added to the cell lysates after washing. Lysates were incubated with antibody-conjugated beads for 3–5 hours at 4°C with gentle shaking, and unbound proteins were removed by washing with buffer. Precipitated proteins were eluted from the beads by boiling in 1× SDS sample buffer.
AAV injection into neonatal mouse brains.
Newborn pups within 10 hours of birth were transferred to new cages for anesthesia with isoflurane. AAV9 viruses containing mouse BACE2 shRNA or scramble shRNA at a titer of approximately 2 × 1013 GC/ml were intracranially injected into bilateral ventricles. A previously validated shRNA sequence (60) targeting mouse BACE2 (5′-ccggccgcagaaggtacagattcttctcgagaagaatctgtaccttctgcggttttt) was packaged into an AAV9 expression vector by Virovek. The control scramble shRNA containing AAV9 at approximately 2 × 1013 GC/ml was provided by Virovek.
Intracranial injection of AAV into adult mice.
APP-KI3 mice were obtained from Takaomi C. Saido of the RIKEN Brain Science Institute (Wako, Japan). Twelve-week-old APP-KI3 mice were anesthetized with isoflurane (2%) and secured on a stereotaxic frame (Kopf Instruments). Meloxicam (2 mg/kg) was then subcutaneously injected. A small section of the skull was removed using a dental drill (Fine Science Tools, 0.5-mm tip diameter) at the coordinates of 0.5 mm rostral and 1.2 mm lateral to the bregma. Two microliters of AAV-shBACE2-GFP or AAV-scramble shRNA-GFP (~2.5 × 1013 GC) was injected unilaterally (2.1 mm ventral from the dura) at a rate of 0.2 μl/min with a microinjection system (UltraMicroPump, UMP3-1, WPI). Four weeks after AAV intracranial injection, mice were euthanized for subsequent experiments.
ELISA.
The concentrations of Aβ1–42 and Aβ1–40 were measured using a β-amyloid Colorimetric ELISA kit (Invitrogen) according to the manufacturer’s instructions. Briefly, half brains were weighed and homogenized in 10-fold excess (v/w) 5 M guanidine buffer (pH 8.0). The homogenates were gently shaken at room temperature for 4 hours, and clarified by centrifugation. Supernatants after clarification were diluted to reduce the guanidine concentration to 0.1 M before application to ELISA.
Neuritic plaque staining and quantification.
Mouse brains were fixed in 4% paraformaldehyde and dehydrated with 30% sucrose solution, and sectioned to 30-μm thickness. Every 12th slice with the same reference position was stained. After antigen retrieval with formic acid, the slices were stained using biotinylated monoclonal 6E10 antibody, and then Alex 594–conjugated streptavidin. Brain slices were mounted and images were acquired with a Zeiss Axio Zoom Microscope at the same settings. Plaques (cutoff = 100 μm2) were quantified by ImageJ (NIH), and data were analyzed with GraphPad Prism 5.
Y-maze behavioral test.
Mice were placed in the center of the maze facing toward one of the arms and then allowed to explore freely for 8 minutes (Y-maze from Stoelting). We recorded and analyzed the activity and spontaneous behavioral alternations of the mice using Any-Maze (Stoelting).
In vitro BACE2 cleavage.
GFP-APP607–625 (as in APP695)-FLAG-His fusion proteins with or without the F615P substitution were expressed in bacteria, and purified using Ni2+ beads. Purified proteins were dialyzed in PBS containing 10% glycerol. Proteins at 0.2 mg/ml were analyzed by circular dichroism using a Jasco J-815 spectrophotometer. For in vitro BACE2 cleavage assay, 1 μg of recombinant protein substrate was mixed with 125 ng BACE2 (R&D Systems) in 90 μl of 50 mM NaOAc pH 4 containing 1% BSA. After incubation at room temperature for the indicated times, an aliquot was transferred to 4× SDS sample buffer supplemented with 1.5 M Tris (pH 8.8) to neutralize pH, and boiled before Western blotting.
Statistics.
The numbers of replicates for each experiment are indicated in the figure legends. Statistical analyses were performed using GraphPad Prism 5.0. Results are expressed as mean ± SEM. A 2-tailed Student’s t test was used to analyze the difference between 2 groups, and multiple-comparisons were analyzed by ANOVA followed by post hoc Newman–Keuls test, unless otherwise specified. P less than 0.05 was considered statistically significant.
Study approval.
Animal experiments were approved by and conducted in accordance with the University of British Columbia Animal Care and Use Committee and the Canadian Institutes of Health Research (CIHR) guidelines.
Author contributions
WS and ZW conceived and designed the experiments. ZW, QX, FC, XL, YW, and WS performed the experiments. ZW, QX, FC, XL, and WS analyzed and contributed reagents, materials, and analytical tools. ZW and WS wrote the manuscript. All authors reviewed the manuscript.
Supplementary Material
Acknowledgments
We thank Takaomi C. Saido of the RIKEN Brain Science Institute for providing us APP mutation knockin mice. This work was supported by CIHR Operating Grant MOP-142487 to WS, and by National Science Foundation of China (no. 81870832) to ZW. WS is the holder of the Tier 1 Canada Research Chair in Alzheimer’s Disease. ZW was supported by the Alzheimer Society of Canada Postdoctoral Fellowship. XL was the recipient of the Chinese Scholarship Council award.
Version 1. 01/10/2019
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
License: Copyright 2019, American Society for Clinical Investigation.
Reference information: JCI Insight. 2019;4(1):e123431. https://doi.org/10.1172/jci.insight.123431.
Contributor Information
Zhe Wang, Email: wangz@xwhosp.org.
Qin Xu, Email: qxu@signalchem.com.
Fang Cai, Email: caifang@mail.ubc.ca.
Xi Liu, Email: dr.liuxi120@gmail.com.
Yili Wu, Email: wu_yili@aliyun.com.
Weihong Song, Email: weihong@mail.ubc.ca.
References
- 1.Kang Y, et al. Nutritional deficiency in early life facilitates aging-associated cognitive decline. Curr Alzheimer Res. 2017;14(8):841–849. doi: 10.2174/1567205014666170425112331. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y, Song W. Islet amyloid polypeptide: Another key molecule in Alzheimer’s pathogenesis? Prog Neurobiol. 2017;153:100–120. doi: 10.1016/j.pneurobio.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 3.Zhang S, et al. BACE1 cleavage site selection critical for amyloidogenesis and Alzheimer’s pathogenesis. J Neurosci. 2017;37(29):6915–6925. doi: 10.1523/JNEUROSCI.0340-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. doi: 10.1038/s41380-018-0101-x. Zhang S, et al. A presenilin-1 mutation causes Alzheimer disease without affecting Notch signaling [published online ahead of print]. Mol Psychiatry. doi: 10.1038/s41380-018-0101-x. [DOI] [PubMed] [Google Scholar]
- 5.Zeng J, et al. Marginal vitamin A deficiency facilitates Alzheimer’s pathogenesis. Acta Neuropathol. 2017;133(6):967–982. doi: 10.1007/s00401-017-1669-y. [DOI] [PubMed] [Google Scholar]
- 6.Jonsson T, et al. A mutation in APP protects against Alzheimer’s disease and age-related cognitive decline. Nature. 2012;488(7409):96–99. doi: 10.1038/nature11283. [DOI] [PubMed] [Google Scholar]
- 7.Kokawa A, et al. The A673T mutation in the amyloid precursor protein reduces the production of β-amyloid protein from its β-carboxyl terminal fragment in cells. Acta Neuropathol Commun. 2015;3:66. doi: 10.1186/s40478-015-0247-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan R, et al. Membrane-anchored aspartyl protease with Alzheimer’s disease beta-secretase activity. Nature. 1999;402(6761):533–537. doi: 10.1038/990107. [DOI] [PubMed] [Google Scholar]
- 9.Sinha S, et al. Purification and cloning of amyloid precursor protein beta-secretase from human brain. Nature. 1999;402(6761):537–540. doi: 10.1038/990114. [DOI] [PubMed] [Google Scholar]
- 10.Vassar R, et al. Beta-secretase cleavage of Alzheimer’s amyloid precursor protein by the transmembrane aspartic protease BACE. Science. 1999;286(5440):735–741. doi: 10.1126/science.286.5440.735. [DOI] [PubMed] [Google Scholar]
- 11.Luo Y, et al. Mice deficient in BACE1, the Alzheimer’s beta-secretase, have normal phenotype and abolished beta-amyloid generation. Nat Neurosci. 2001;4(3):231–232. doi: 10.1038/85059. [DOI] [PubMed] [Google Scholar]
- 12.Cai H, et al. BACE1 is the major beta-secretase for generation of Abeta peptides by neurons. Nat Neurosci. 2001;4(3):233–234. doi: 10.1038/85064. [DOI] [PubMed] [Google Scholar]
- 13.Chen CH, et al. Increased NF-κB signalling up-regulates BACE1 expression and its therapeutic potential in Alzheimer’s disease. Int J Neuropsychopharmacol. 2012;15(1):77–90. doi: 10.1017/S1461145711000149. [DOI] [PubMed] [Google Scholar]
- 14.Ly PT, et al. Inhibition of GSK3β-mediated BACE1 expression reduces Alzheimer-associated phenotypes. J Clin Invest. 2013;123(1):224–235. doi: 10.1172/JCI64516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan R, Vassar R. Targeting the β secretase BACE1 for Alzheimer’s disease therapy. Lancet Neurol. 2014;13(3):319–329. doi: 10.1016/S1474-4422(13)70276-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng Y, et al. Amyloid-β protein (Aβ) Glu11 is the major β-secretase site of β-site amyloid-β precursor protein-cleaving enzyme 1(BACE1), and shifting the cleavage site to Aβ Asp1 contributes to Alzheimer pathogenesis. Eur J Neurosci. 2013;37(12):1962–1969. doi: 10.1111/ejn.12235. [DOI] [PubMed] [Google Scholar]
- 17.Lin X, Koelsch G, Wu S, Downs D, Dashti A, Tang J. Human aspartic protease memapsin 2 cleaves the beta-secretase site of beta-amyloid precursor protein. Proc Natl Acad Sci USA. 2000;97(4):1456–1460. doi: 10.1073/pnas.97.4.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farzan M, Schnitzler CE, Vasilieva N, Leung D, Choe H. BACE2, a beta-secretase homolog, cleaves at the beta site and within the amyloid-beta region of the amyloid-beta precursor protein. Proc Natl Acad Sci USA. 2000;97(17):9712–9717. doi: 10.1073/pnas.160115697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saunders AJ, et al. BACE maps to chromosome 11 and a BACE homolog BACE2 reside in the obligate Down syndrome region of chromosome 21. Science. 1999;286(5443):1255a. doi: 10.1126/science.286.5443.1255a. [DOI] [Google Scholar]
- 20.Solans A, Estivill X, de La Luna S. A new aspartyl protease on 21q22.3, BACE2, is highly similar to Alzheimer’s amyloid precursor protein beta-secretase. Cytogenet Cell Genet. 2000;89(3–4):177–184. doi: 10.1159/000015608. [DOI] [PubMed] [Google Scholar]
- 21.Sun X, He G, Song W. BACE2, as a novel APP theta-secretase, is not responsible for the pathogenesis of Alzheimer’s disease in Down syndrome. FASEB J. 2006;20(9):1369–1376. doi: 10.1096/fj.05-5632com. [DOI] [PubMed] [Google Scholar]
- 22.Fluhrer R, et al. A non-amyloidogenic function of BACE-2 in the secretory pathway. J Neurochem. 2002;81(5):1011–1020. doi: 10.1046/j.1471-4159.2002.00908.x. [DOI] [PubMed] [Google Scholar]
- 23.Basi G, et al. Antagonistic effects of beta-site amyloid precursor protein-cleaving enzymes 1 and 2 on beta-amyloid peptide production in cells. J Biol Chem. 2003;278(34):31512–31520. doi: 10.1074/jbc.M300169200. [DOI] [PubMed] [Google Scholar]
- 24.Yan R, Munzner JB, Shuck ME, Bienkowski MJ. BACE2 functions as an alternative alpha-secretase in cells. J Biol Chem. 2001;276(36):34019–34027. doi: 10.1074/jbc.M105583200. [DOI] [PubMed] [Google Scholar]
- 25.Azkona G, Levannon D, Groner Y, Dierssen M. In vivo effects of APP are not exacerbated by BACE2 co-overexpression: behavioural characterization of a double transgenic mouse model. Amino Acids. 2010;39(5):1571–1580. doi: 10.1007/s00726-010-0662-8. [DOI] [PubMed] [Google Scholar]
- 26.Azkona G, et al. Characterization of a mouse model overexpressing beta-site APP-cleaving enzyme 2 reveals a new role for BACE2. Genes Brain Behav. 2010;9(2):160–172. doi: 10.1111/j.1601-183X.2009.00538.x. [DOI] [PubMed] [Google Scholar]
- 27.Abdul-Hay SO, Sahara T, McBride M, Kang D, Leissring MA. Identification of BACE2 as an avid β-amyloid-degrading protease. Mol Neurodegener. 2012;7:46. doi: 10.1186/1750-1326-7-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bennett BD, et al. Expression analysis of BACE2 in brain and peripheral tissues. J Biol Chem. 2000;275(27):20647–20651. doi: 10.1074/jbc.M002688200. [DOI] [PubMed] [Google Scholar]
- 29.McGeer PL, Kawamata T, Walker DG. Distribution of clusterin in Alzheimer brain tissue. Brain Res. 1992;579(2):337–341. doi: 10.1016/0006-8993(92)90071-G. [DOI] [PubMed] [Google Scholar]
- 30.Beeg M, et al. Clusterin binds to Aβ1-42 oligomers with high affinity and interferes with peptide aggregation by inhibiting primary and secondary nucleation. J Biol Chem. 2016;291(13):6958–6966. doi: 10.1074/jbc.M115.689539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kida E, Choi-Miura NH, Wisniewski KE. Deposition of apolipoproteins E and J in senile plaques is topographically determined in both Alzheimer’s disease and Down’s syndrome brain. Brain Res. 1995;685(1–2):211–216. doi: 10.1016/0006-8993(95)00482-6. [DOI] [PubMed] [Google Scholar]
- 32.May PC, Lampert-Etchells M, Johnson SA, Poirier J, Masters JN, Finch CE. Dynamics of gene expression for a hippocampal glycoprotein elevated in Alzheimer’s disease and in response to experimental lesions in rat. Neuron. 1990;5(6):831–839. doi: 10.1016/0896-6273(90)90342-D. [DOI] [PubMed] [Google Scholar]
- 33.Lidström AM, Bogdanovic N, Hesse C, Volkman I, Davidsson P, Blennow K. Clusterin (apolipoprotein J) protein levels are increased in hippocampus and in frontal cortex in Alzheimer’s disease. Exp Neurol. 1998;154(2):511–521. doi: 10.1006/exnr.1998.6892. [DOI] [PubMed] [Google Scholar]
- 34.Fukumoto H, Rosene DL, Moss MB, Raju S, Hyman BT, Irizarry MC. Beta-secretase activity increases with aging in human, monkey, and mouse brain. Am J Pathol. 2004;164(2):719–725. doi: 10.1016/S0002-9440(10)63159-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holler CJ, et al. BACE2 expression increases in human neurodegenerative disease. Am J Pathol. 2012;180(1):337–350. doi: 10.1016/j.ajpath.2011.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barão S, Moechars D, Lichtenthaler SF, De Strooper B. BACE1 physiological functions may limit its use as therapeutic target for Alzheimer’s disease. Trends Neurosci. 2016;39(3):158–169. doi: 10.1016/j.tins.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 37.Sisodia SS. Beta-amyloid precursor protein cleavage by a membrane-bound protease. Proc Natl Acad Sci USA. 1992;89(13):6075–6079. doi: 10.1073/pnas.89.13.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barrett PJ, et al. The amyloid precursor protein has a flexible transmembrane domain and binds cholesterol. Science. 2012;336(6085):1168–1171. doi: 10.1126/science.1219988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lemmin T, Dimitrov M, Fraering PC, Dal Peraro M. Perturbations of the straight transmembrane α-helical structure of the amyloid precursor protein affect its processing by γ-secretase. J Biol Chem. 2014;289(10):6763–6774. doi: 10.1074/jbc.M113.470781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saito T, et al. Single App knock-in mouse models of Alzheimer’s disease. Nat Neurosci. 2014;17(5):661–663. doi: 10.1038/nn.3697. [DOI] [PubMed] [Google Scholar]
- 41.Norstrom EM, Zhang C, Tanzi R, Sisodia SS. Identification of NEEP21 as a β-amyloid precursor protein-interacting protein in vivo that modulates amyloidogenic processing in vitro. J Neurosci. 2010;30(46):15677–15685. doi: 10.1523/JNEUROSCI.4464-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Debure L, Vayssiere JL, Rincheval V, Loison F, Le Drean Y, Michel D. Intracellular clusterin causes juxtanuclear aggregate formation and mitochondrial alteration. J Cell Sci. 2003;116(Pt 15):3109–3121. doi: 10.1242/jcs.00619. [DOI] [PubMed] [Google Scholar]
- 43.Esterházy D, et al. Bace2 is a β cell-enriched protease that regulates pancreatic β cell function and mass. Cell Metab. 2011;14(3):365–377. doi: 10.1016/j.cmet.2011.06.018. [DOI] [PubMed] [Google Scholar]
- 44.Gonos ES, et al. Cloning and identification of genes that associate with mammalian replicative senescence. Exp Cell Res. 1998;240(1):66–74. doi: 10.1006/excr.1998.3948. [DOI] [PubMed] [Google Scholar]
- 45.Petropoulou C, Trougakos IP, Kolettas E, Toussaint O, Gonos ES. Clusterin/apolipoprotein J is a novel biomarker of cellular senescence that does not affect the proliferative capacity of human diploid fibroblasts. FEBS Lett. 2001;509(2):287–297. doi: 10.1016/S0014-5793(01)03150-7. [DOI] [PubMed] [Google Scholar]
- 46.Shapiro LA, Marks A, Whitaker-Azmitia PM. Increased clusterin expression in old but not young adult S100B transgenic mice: evidence of neuropathological aging in a model of Down Syndrome. Brain Res. 2004;1010(1–2):17–21. doi: 10.1016/j.brainres.2003.12.057. [DOI] [PubMed] [Google Scholar]
- 47.Bodendorf U, et al. Expression of human beta-secretase in the mouse brain increases the steady-state level of beta-amyloid. J Neurochem. 2002;80(5):799–806. doi: 10.1046/j.0022-3042.2002.00770.x. [DOI] [PubMed] [Google Scholar]
- 48.Li Y, Zhou W, Tong Y, He G, Song W. Control of APP processing and Abeta generation level by BACE1 enzymatic activity and transcription. FASEB J. 2006;20(2):285–292. doi: 10.1096/fj.05-4986com. [DOI] [PubMed] [Google Scholar]
- 49.Ly PT, et al. Inhibition of GSK3β-mediated BACE1 expression reduces Alzheimer-associated phenotypes. J Clin Invest. 2013;123(1):224–235. doi: 10.1172/JCI64516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun X, et al. Hypoxia facilitates Alzheimer’s disease pathogenesis by up-regulating BACE1 gene expression. Proc Natl Acad Sci USA. 2006;103(49):18727–18732. doi: 10.1073/pnas.0606298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee EB, et al. BACE overexpression alters the subcellular processing of APP and inhibits Abeta deposition in vivo. J Cell Biol. 2005;168(2):291–302. doi: 10.1083/jcb.200407070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zlokovic BV, et al. Glycoprotein 330/megalin: probable role in receptor-mediated transport of apolipoprotein J alone and in a complex with Alzheimer disease amyloid beta at the blood-brain and blood-cerebrospinal fluid barriers. Proc Natl Acad Sci USA. 1996;93(9):4229–4234. doi: 10.1073/pnas.93.9.4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DeMattos RB, et al. Clusterin promotes amyloid plaque formation and is critical for neuritic toxicity in a mouse model of Alzheimer’s disease. Proc Natl Acad Sci USA. 2002;99(16):10843–10848. doi: 10.1073/pnas.162228299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Killick R, et al. Clusterin regulates β-amyloid toxicity via Dickkopf-1-driven induction of the wnt-PCP-JNK pathway. Mol Psychiatry. 2014;19(1):88–98. doi: 10.1038/mp.2012.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bettens K, et al. Reduced secreted clusterin as a mechanism for Alzheimer-associated CLU mutations. Mol Neurodegener. 2015;10:30. doi: 10.1186/s13024-015-0024-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trougakos IP, Gonos ES. Clusterin/apolipoprotein J in human aging and cancer. Int J Biochem Cell Biol. 2002;34(11):1430–1448. doi: 10.1016/S1357-2725(02)00041-9. [DOI] [PubMed] [Google Scholar]
- 57.Wehrli P, et al. Inhibition of post-ischemic brain injury by clusterin overexpression. Nat Med. 2001;7(9):977–979. doi: 10.1038/nm0901-977. [DOI] [PubMed] [Google Scholar]
- 58.Liu F, et al. Cleavage of potassium channel Kv2.1 by BACE2 reduces neuronal apoptosis. Mol Psychiatry. 2018;23(7):1542–1554. doi: 10.1038/s41380-018-0060-2. [DOI] [PubMed] [Google Scholar]
- 59.Graf ER, Zhang X, Jin SX, Linhoff MW, Craig AM. Neurexins induce differentiation of GABA and glutamate postsynaptic specializations via neuroligins. Cell. 2004;119(7):1013–1026. doi: 10.1016/j.cell.2004.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stützer I, Selevsek N, Esterházy D, Schmidt A, Aebersold R, Stoffel M. Systematic proteomic analysis identifies β-site amyloid precursor protein cleaving enzyme 2 and 1 (BACE2 and BACE1) substrates in pancreatic β-cells. J Biol Chem. 2013;288(15):10536–10547. doi: 10.1074/jbc.M112.444703. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.