Abstract
Host-commensal interactions are critical for the generation of robust inflammatory responses, yet the mechanisms leading to this effect remain poorly understood. Using a murine model of cytokine storm, we identified that host microbiota are required to sustain systemic TLR-driven immune responses. Mice treated with broad-spectrum antibiotics or raised in germ-free conditions responded normally to an initial TLR signal but failed to sustain production of proinflammatory cytokines following administration of repeated TLR signals in vivo. Mechanistically, host microbiota primed JAK signaling in myeloid progenitors to promote TLR-enhanced myelopoiesis, which is required for the accumulation of TLR-responsive monocytes. In the absence of TLR-enhanced monocytopoiesis, antibiotic-treated mice lost their ability to respond to repeated TLR stimuli and were protected from cytokine storm–induced immunopathology. These data reveal priming of TLR-enhanced myelopoiesis as a microbiota-dependent mechanism that regulates systemic inflammatory responses and highlight a role for host commensals in the pathogenesis of cytokine storm syndromes.
Keywords: Inflammation
Keywords: Innate immunity, Monocytes, Mouse models
Normal commensal bacteria are required for inflammatory monocytopoiesis and sustained toll-like receptor responses.
Introduction
Cytokine storm syndromes are devastating clinical conditions that result from dysregulated immune responses to inflammatory and infectious triggers. Failure of the host to clear the underlying inflammatory stimulus perpetuates feed-forward cycles of inflammation, resulting in immune-mediated collateral damage to healthy tissues, life-threatening organ dysfunction, and a cytokine storm mortality rate of 20%–50% (1). Despite the knowledge that immune-mediated damage contributes to cytokine storm pathogenesis, targeted therapeutic interventions to reverse the hyperinflammatory and maladaptive host immune response in cytokine storm syndromes are lacking.
Innate immune cells are central to cytokine storm pathogenesis, as they express a diverse repertoire of pattern recognition receptors including Toll-like receptors (TLRs) that are activated by endogenous and exogenous “danger” signals to drive disease (2). TLR-driven proinflammatory responses are balanced by the induction of negative regulatory mechanisms that dampen innate immune responses. TLR tolerance is a cell-intrinsic regulatory mechanism that prevents TLR-activated cells from producing robust proinflammatory cytokines after repeated TLR stimulation (3). However, murine models of cytokine storm demonstrate that sustained TLR activation is sufficient to amplify proinflammatory cytokine production and induce cytokine storm–mediated immunopathology (4–6). Therefore, mechanisms to bypass TLR tolerance and sustain systemic TLR-driven innate immune responses must exist in vivo.
Using a murine model of cytokine storm, we recently identified a mechanism that bypasses TLR tolerance and sustains TLR-driven immune responses in vivo. Repeated TLR9 activation in WT mice induces clinical manifestations of cytokine storm, including hepatosplenomegaly, hepatitis, cytopenias, and hypercytokinemia (5). Disease induction is dependent on IL-12–induced IFN-γ production, which correlates with the accumulation of TLR9-responsive inflammatory monocytes in this model of cytokine storm (7, 8). Accelerated production of inflammatory monocytes replenishes and enlarges the pool of cells capable of responding to repeated TLR9 activation leading to heightened IL-12–mediated IFN-γ production, which drives cytokine storm–induced immunopathology (7, 8). Identification of inflammatory pathways that regulate the production of TLR-responsive monocytes may reveal attractive therapeutic targets to ameliorate life-threatening immunopathology in cytokine storm syndromes.
Hematopoiesis is a coordinated process dedicated to the continuous production of millions of new blood cells each day. Hematopoietic stem cells (HSCs) are self-renewing cells that form the base of this system, as a single human or mouse HSC can regenerate all hematopoietic lineages and support long-term multilineage hematopoietic engraftment in vivo (9, 10). Cell-intrinsic transcriptional, metabolic, and epigenetic programs limit lineage-committed hematopoietic progenitor cells (HPCs) to produce cells within their prespecified lineage (11–13). The “hard-wired” lineage commitment of HPCs is influenced by cell-extrinsic cues, which allow the hematopoietic system to adjust its cellular output to meet the demands of the host (14–17).
In both mice and humans, extramedullary hematopoietic progenitors increase in frequency during states of systemic inflammation to accelerate the production of myeloid cells through emergency hematopoiesis (18–21). Recent evidence suggests that commensal microbiota regulate steady-state and emergency hematopoiesis, as antibiotic-treated and germ-free mice have altered basal myelopoiesis and fail to induce enhanced myelopoiesis during infectious challenges (22–25). However, the mechanisms leading to microbiota-dependent regulation of myelopoiesis and the relevance of these observations to noninfectious inflammatory diseases are not fully understood.
In this manuscript, we found that commensal microbiota influence susceptibility to cytokine storm by regulating the production of new TLR-responsive myeloid cells and, in turn, control the ability of their host to maintain systemic TLR-driven immune responses. Consistent with this model, antibiotic-treated mice responded normally to an initial TLR9 stimulus but lost the ability to respond to repeated TLR9 stimuli. Loss of responsiveness to repeated TLR9 stimuli correlated with the inability of antibiotic-treated and germ-free mice to induce TLR9-enhanced myelopoiesis. In the absence of TLR9-enhanced myelopoiesis, antibiotic-treated and germ-free mice failed to accumulate TLR9-responsive inflammatory monocytes that produce IL-12 to drive disease. Mechanistically, myeloid progenitors from antibiotic-treated mice had defective responses to JAK-activating cytokines, which are necessary for the induction of TLR9-enhanced myelopoiesis, as mice treated with a JAK1/2 inhibitor failed to develop enhanced myelopoiesis and phenocopy the disease protection seen in antibiotic-treated mice. These data support a model whereby commensal microbiota contribute to the pathogenesis of cytokine storm by priming JAK signaling in myeloid progenitors to enhance inflammation-induced myelopoiesis, which is required to sustain host responses to repeated TLR signals in vivo.
Results
Antibiotic-treated mice are protected from TLR9-enhanced myelopoiesis and cytokine storm–induced immunopathology.
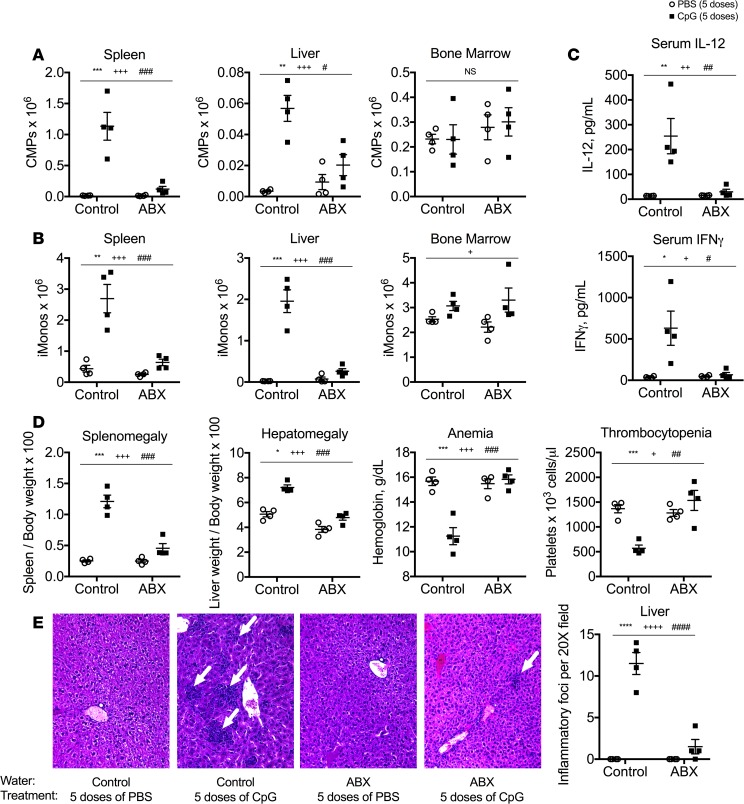
We recently demonstrated that TLR9-enhanced myelopoiesis is implicated in the pathogenesis of cytokine storm in a murine model of disease (7). This TLR9-mediated model of cytokine storm provides a unique opportunity to test whether microbiota-dependent signals prime the accelerated production of TLR9-responsive inflammatory monocytes to drive disease. We predicted that mice pretreated with broad-spectrum antibiotics would be protected from TLR9-enhanced myelopoiesis if host microbiota provide signals necessary for disease induction. Intriguingly, TLR9-mediated accumulation of spleen and liver extramedullary myeloid progenitors (common myeloid progenitors [CMPs]; granulocyte-monocyte progenitors [GMPs]; monocyte-dendritic cell progenitors [MDPs]; committed monocyte progenitors [cMoPs]) occurs after the administration of 5 doses of CpG1826, a TLR9 agonist, over the course of 10 days. The accumulation of extramedullary myeloid progenitors was dramatically reduced in antibiotic-treated mice compared with that in control mice (Figure 1A and Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.124370DS1). As TLR9-induced extramedullary myelopoiesis is required for enhanced production of pathogenic inflammatory monocytes (7), we predicted that repeated TLR9 activation would not lead to the accumulation of inflammatory monocytes in antibiotic-treated mice. Indeed, antibiotic-treated mice failed to accumulate inflammatory monocytes following treatment with repeated TLR9 stimuli (Figure 1B).
Figure 1. Antibiotic-treated mice are protected from TLR9-enhanced myelopoiesis and cytokine storm–induced immunopathology.
C57BL/6 mice were pretreated with antibiotic or control water for 3 weeks. Thereafter, mice were treated with 5 doses of the TLR9 agonist CpG1826 over the course of 10 days. Numbers of bone marrow and extramedullary (spleen and liver) common myeloid progenitors (CMPs) (A) and inflammatory monocytes (iMonos) (B) were enumerated. (C) Serum levels of IL-12 and IFN-γ were measured by ELISA. (D) Clinical manifestations of cytokine storm were evaluated by measuring splenomegaly, hepatomegaly, anemia, and thrombocytopenia. (E) Livers from mice were sectioned and stained with H&E (original magnification, ×20). Each graph displays representative data from 1 of 2–4 independent experiments (n = 4 mice per group). Analysis was performed by 2-way ANOVA (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, interaction; +P < 0.05, ++P < 0.01, +++P < 0.001, ++++P < 0.0001, control vs. antibiotic treated; #P < 0.05, ##P < 0.01, ###P < 0.001, ####P < 0.0001, PBS vs. CpG in vivo treatments). Interaction term is CpG treatment x Antibiotic treatment.
Sustained responses to repeated TLR9 stimuli are required to drive immunopathology in TLR9-mediated cytokine storm, as systemic administration of a single dose of a TLR9 stimulus fails to induce disease (7). TLR9 responsiveness is maintained by the production of new hematopoietically derived inflammatory monocytes that were not exposed to the initial TLR9 stimulus and maintain full capacity to produce proinflammatory cytokines upon encountering additional TLR9 stimuli. These newly produced monocytes accumulate in the periphery, which leads to increased systemic production of proinflammatory cytokines following each TLR9 stimulus and drives cytokine storm–mediated immunopathology (7). As antibiotic treatment inhibited the ability of mice failed to induce TLR9-enhanced myelopoiesis and accumulate inflammatory monocytes in the periphery, we predicted that antibiotic-treated mice would lose their responsiveness to repeated TLR9 stimuli in vivo. Consistent with our hypothesis, robust induction of serum IL-12 and IFN-γ was observed after the fifth dose of a TLR9 stimulus in control mice, whereas antibiotic treatment inhibited the ability of mice induce IL-12 or IFN-γ after treatment with 5 doses a TLR9 stimulus (Figure 1C). The inability of antibiotic-treated mice to maintain responsiveness to systemic TLR9 stimuli correlated with their protection from cytokine storm–induced immunopathology. Accordingly, antibiotic-treated mice were protected from TLR9-mediated hepatosplenomegaly, hepatitis, and cytopenias compared with control mice (Figure 1, D and E). These data support the hypothesis that microbiota-dependent TLR9-enhanced myelopoiesis is critical for the induction and propagation of TLR9-driven innate immune responses that drive cytokine storm pathogenesis.
Germ-free mice phenocopy the disease protection from TLR9-mediated cytokine storm observed in antibiotic-treated mice.
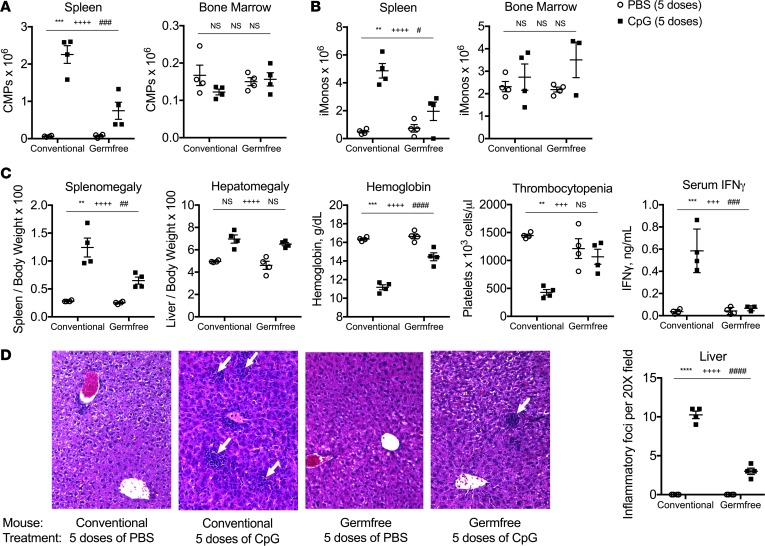
The protection from cytokine storm–induced immunopathology observed in antibiotic-treated mice could occur through antibiotic-induced changes to the microbiota, leading to either the accumulation of microbial-derived inhibitors of TLR9-enhanced myelopoiesis or the loss of microbial-derived activators of TLR9-enhanced myelopoiesis. To differentiate between these possibilities, we treated germ-free mice with repeated TLR9 stimuli (5 doses of CpG1826 over the course of 10 days) to induce cytokine storm. We predicted that germ-free mice would fail to exhibit TLR9-enhanced myelopoiesis and would be protected from TLR9-induced cytokine storm, as previous work demonstrated that germ-free mice have defective basal and inflammation-induced myelopoiesis (22, 25). Consistent with our hypothesis, TLR9-induced accumulation of extramedullary myeloid progenitors (Figure 2A and Supplemental Figure 2) and pathogenic inflammatory monocytes (Figure 2B) was suppressed in germ-free mice, phenocopying our observations in antibiotic-treated mice (Figure 1, A and B). Furthermore, the suppression of TLR9-enhanced myelopoiesis observed in germ-free mice correlated with protection from TLR9-mediated hepatosplenomegaly, hepatitis, cytopenias, and hypercytokinemia (Figure 2, C and D). These data indicate that germ-free mice phenocopy the disease protection observed in antibiotic-treated mice. Collectively, these data support a model whereby host microbiota produce molecules capable of priming TLR9-mediated myelopoiesis, which are depleted during antibiotic treatment and are not made in germ-free mice that are devoid of microbes.
Figure 2. Germ-free mice are protected from TLR9-mediated cytokine storm.
Conventionally housed and germ-free C57BL/6 mice were treated with 5 doses of CpG1826 over the course of 10 days. (A and B) Numbers of medullary (bone marrow) and extramedullary (spleen and liver) common myeloid progenitors (CMPs) and inflammatory monocytes (iMonos) were enumerated. (C) Clinical manifestations of cytokine storm were evaluated by measuring splenomegaly, hepatomegaly, anemia, thrombocytopenia, and serum IFN-γ. (D) Livers from mice were sectioned and stained with H&E (original magnification, ×20). Each graph displays representative data from 1 of 2 independent experiments (n = 4 mice per group). Analysis was performed by 2-way ANOVA (**P < 0.01, ***P < 0.001, ****P < 0.0001, interaction; +++P < 0.001, ++++P < 0.0001, conventionally housed vs. germ free; #P < 0.05, ##P < 0.01, ###P < 0.001, ####P < 0.0001, PBS vs. CpG in vivo treatments). Interaction term is CpG treatment x housing condition.
Initial TLR9 responses are intact in antibiotic-treated mice.
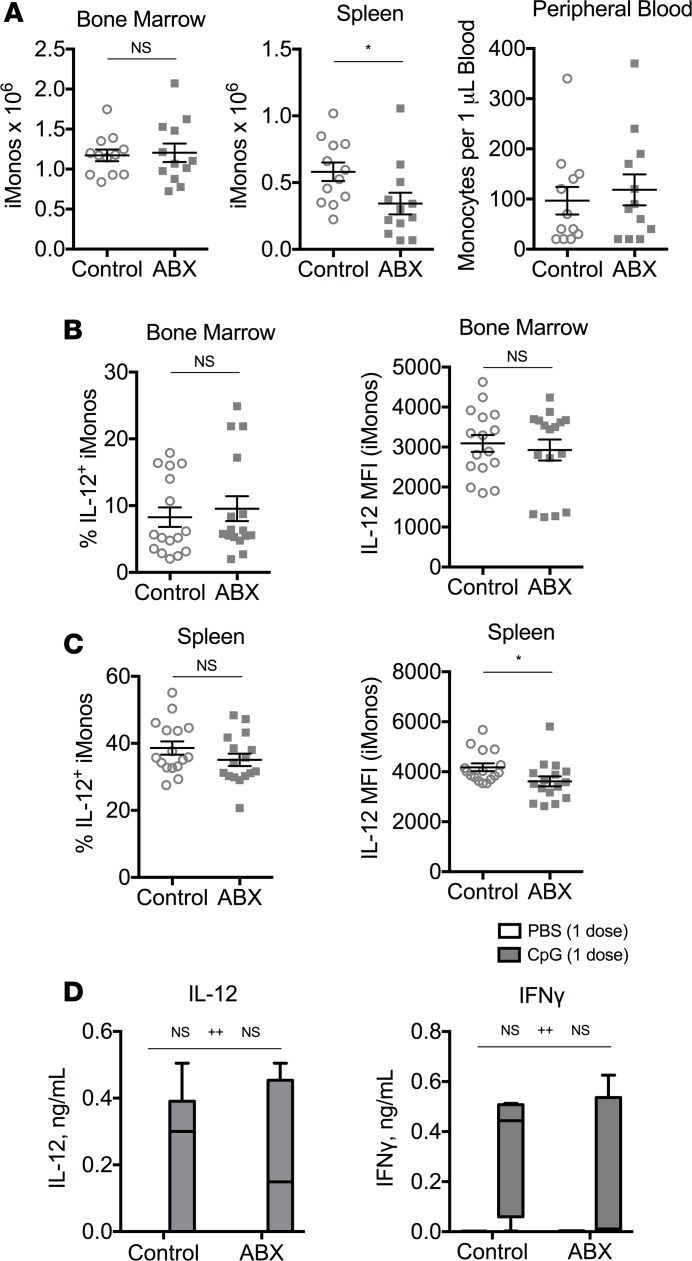
Inflammatory monocytes are found in normal numbers in the bone marrow and peripheral blood of antibiotic-treated mice, with only slightly reduced numbers in the spleen after 3 weeks of antibiotic treatment (Figure 3A). As inflammatory monocytes are the main producers of pathogenic IL-12 in TLR9-mediated cytokine storm (7), we hypothesized that inflammatory monocyte responses to the initial TLR9 stimulus would be intact in antibiotic-treated mice, despite the inability of antibiotic-treated mice to maintain responsiveness to repeated TLR9 stimuli. Consistent with our prediction, inflammatory monocytes from the bone marrow and spleens of antibiotic-treated and control mice were capable of producing IL-12 after TLR9 activation in vitro (Figure 3, B and C). To confirm that the initial response to TLR9 activation was normal in vivo, antibiotic-treated and control mice were stimulated with a single dose of a TLR9 agonist and were found to produce similar levels of serum IL-12 and IFN-γ as control mice (Figure 3D). These data indicate that antibiotic-treated mice respond normally to an initial TLR9 stimulus but fail to respond to repeated TLR9 stimuli in vivo.
Figure 3. Initial TLR9 responses are preserved in antibiotic-treated mice.
(A) Numbers of monocytes from the bone marrow, spleen, and peripheral blood were enumerated from mice treated with antibiotic or control water for 3 weeks. (B and C) Whole bone marrow and spleen cells were isolated from antibiotic-treated and control mice. CpG-responsive bone marrow (B) and spleen (C) inflammatory monocytes (iMonos) were calculated by the percentage of Ly6G–CD11b+ monocytes staining positive for intracellular IL-12 after stimulation with brefeldin A and CpG1826 for 12–18 hours (left). Median fluorescence intensity (MFI) of IL-12 was measured on IL-12+ monocytes (right). (D) Antibiotic-treated (ABX) and control mice were treated with a single dose of PBS or CpG1826, and sera were harvested 20 hours later to measure IL-12 and IFN-γ. Data are graphed as box-and-whiskers plots showing minimum/maximum, and the line in the box shows the median value. Data from A and B are compiled from 3 experiments (N = 12–16 per group) and analyzed by the Mann-Whitney test. Data from C are representative of 2 independent experiments (N = 6 per group) and analyzed by 2-way ANOVA (*P < 0.05,interaction; ++P < 0.01, control vs. antibiotic treated). Interaction term is CpG treatment x Antibiotic treatment.
Basal myelopoiesis is altered in antibiotic-treated mice.
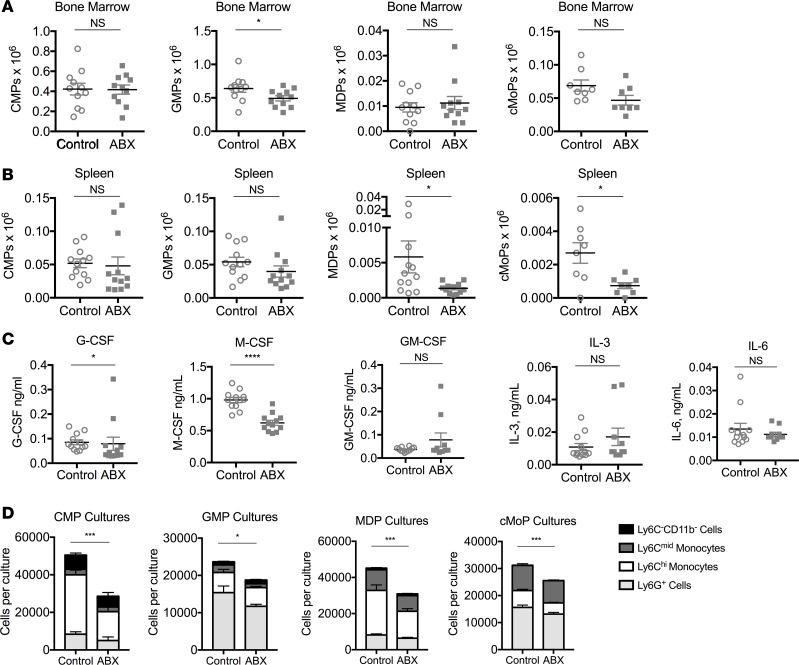
We predicted that antibiotic-treated mice were unable to mount continued responses to TLR9 activation because they fail to accumulate new inflammatory monocytes in the periphery during systemic TLR9-driven inflammatory responses. To characterize basal myelopoiesis, numbers of myeloid progenitors were enumerated in antibiotic-treated mice and were found to be largely intact in the bone marrow (Figure 4A), while baseline numbers of spleen MDPs and cMoPs were reduced (Figure 4B). Serum concentrations of G-CSF and M-CSF were also reduced in antibiotic-treated mice, although levels of IL-3, IL-6, and GM-CSF were unchanged (Figure 4C). Basal changes in myelopoiesis and myeloid-specifying cytokines correlated with reduced function of myeloid progenitors in liquid in vitro myelopoiesis assays when isolated from antibiotic-treated mice (Figure 4D). These data suggest that myeloid progenitors from antibiotic-treated mice have cell-intrinsic functional defects that correlate with defective TLR9-enhanced myelopoiesis in vivo.
Figure 4. Basal myelopoiesis and myeloid progenitor cell function is altered in antibiotic-treated mice.
Numbers of bone marrow (A) and spleen (B) myeloid progenitors were enumerated from mice that were treated with antibiotic (ABX) or control water for 3 weeks. (C) Baseline levels of serum myeloid-specifying cytokines were measured by ELISA from antibiotic-treated and control mice. (D) Myeloid progenitors were sorted from the bone marrow of antibiotic-treated and control mice and cultured in vitro for 7 days in media containing M-CSF, GM-CSF, IL-3, and SCF. Mature myeloid cells were enumerated by flow cytometry. Data from A–C are compiled from 3 independent experiments (n = 10–12 mice per group) and analyzed by the Mann-Whitney U test. Data from D are representative of 4 independent experiments (n = 4 mice per group) and analyzed by 2-way ANOVA (*P < 0.05, ***P < 0.001, ****P < 0.0001, antibiotic-treated vs. control mice).
Disease protection and TLR9-enhanced myelopoiesis are associated with gut bacterial load and alteration in species composition.
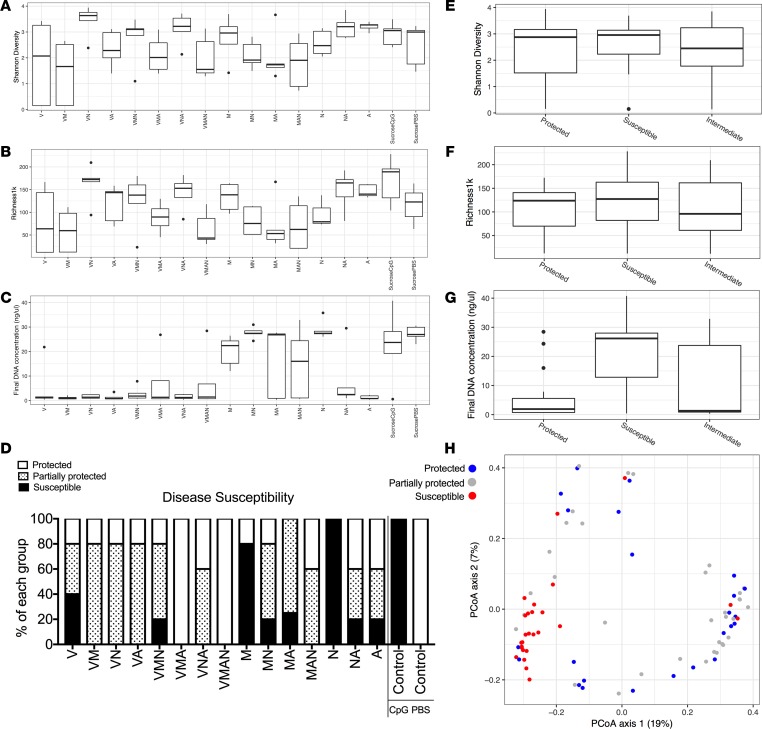
In order to better characterize how changes in gut bacterial content relate to disease susceptibility and TLR9-enhanced myelopoiesis, we treated a cohort of mice with every possible combination of the 4 antibiotics used in the preceding experiments (Table 2). The ability of any individual combination of antibiotics to suppress disease activity was correlated with the amount of extramedullary monocytopoiesis present (Supplemental Figures 3 and 4). We measured the α-diversity of the bacteria contained in the stool samples of these mice using Shannon diversity and the richness per 1000 reads metrics (Figure 5, A and B) as well as the total bacterial load in the stool using total stool DNA content (Figure 5C) for each of the antibiotic combinations. We then summarized the susceptibility to CpG-induced disease using a composite score of multiple parameters for each antibiotic combination (Figure 5D). Using these groupings of disease susceptibility, low levels of stool bacterial content correlated with disease protection, while measures of α-diversity did not differ between protected and susceptible mice (Figure 5, E–G). While total bacterial load may correlate with disease protection, this does not preclude the possibility that an alteration in species content is also important, even if α-diversity is maintained. In order to address this, we measured β-diversity in the stool samples using principle component analysis to look for clusters based on disease susceptibility (Figure 5H). The species composition in the stools of mice treated with antibiotic combinations that fully or partially protected from disease clustered separately from the species composition in the stools of mice that remained susceptible to disease. These data suggest that both total bacterial load and β-diversity associate with the protective effect of antibiotics.
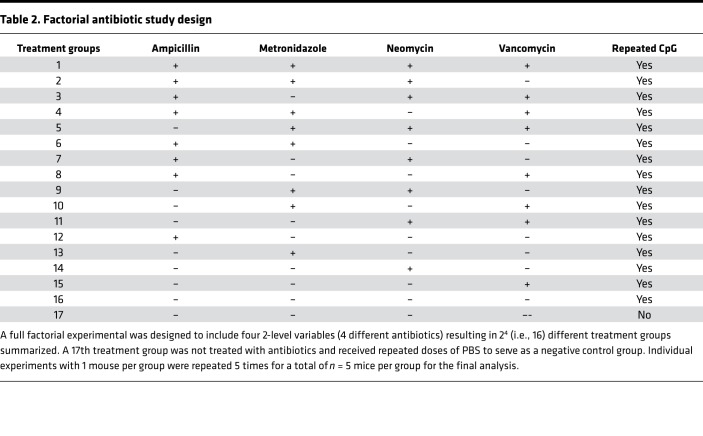
Table 2. Factorial antibiotic study design.
Figure 5. Various antibiotic combinations result in protection from TLR9 cytokine storm, which correlates with low-stool DNA concentrations and distinct stool bacterial compositions.
A factorial study design was conducted to treat cohorts of mice with every combination of the 4-antibiotic cocktail found to protect mice from murine TLR9 cytokine storm (V, vancomycin; M, metronidazole; N, neomycin; A, ampicillin). C57BL/6 mice were pretreated with each antibiotic combination for 3 weeks prior to treatment of mice with 5 doses of the TLR9 agonist CpG1826 over the course of 10 days to induce TLR9 cytokine storm. Mice were sacrificed 24 hours after the fifth injection, and markers of disease were measured (see Supplemental Figures 3 and 4 for details). Stool was collected for 16S rRNA marker gene analysis prior to the first dose of CpG1826. The α-diversity of stool samples was measured using the Shannon index (A and E) and richness per 1000 reads (B and F). The DNA concentration of the stool samples was measured using the Quant-iT PicoGreen Assay Kit from Thermo Fisher Scientific (C and G). (D) Individual mice were categorized based on disease outcomes (splenomegaly, hepatomegaly, anemia, leukopenia, and thrombocytopenia) into groups of protected mice (4 of 5 disease parameters within 20% of PBS-treated mice or better), susceptible mice (4 of 5 disease parameters within 20%–30% of CpG-treated mice or worse), or partially protected mice (those mice with intermediate phenotypes). (E–H) Data display the indicated stool sample analysis by disease susceptibility grouping. (H) The β-diversity was assessed using unweighted UniFrac distances to compare the species composition of each sample principal coordinates analysis. The percentage in each axis represents the proportion of the total variance captured along the axis.
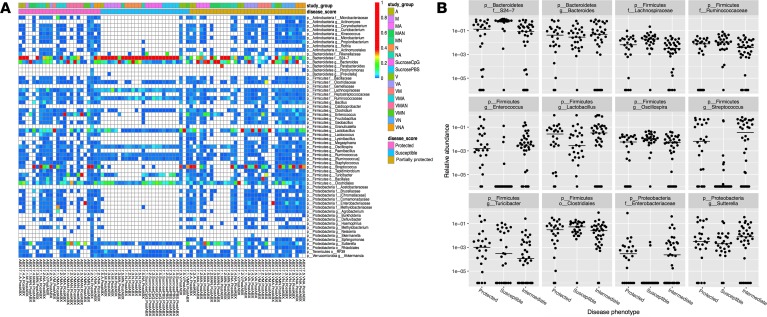
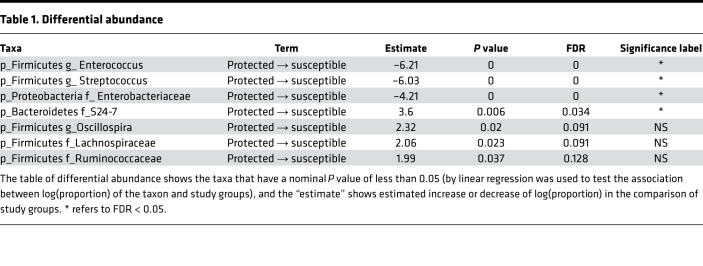
In order to better identify which species are driving this association, we used 16S sequencing from these stool samples to identify bacterial species associated with protection (Figure 6 and Table 1). Enterococcus, Streptococcus, and Enterobacteriaceae were the 3 top statistically significantly (P <0.01 and FDR <0.05, Table 1) enriched bacteria in mice that had any element of protection from disease, whereas taxonomic members of the Bacteroidetes S24-7 family were depleted in protected mice compared with susceptible mice. Given the decrease in total bacterial load and the loss of LPS-containing members of the Bacteroidetes S24-7 family in protected mice, we considered whether a decrease in the concentration of LPS in the gut might contribute to disease protection. To test this hypothesis, we predicted that mice incapable of sensing LPS would be protected from TLR9-induced cytokine storm. However, mice triply deficient in the LPS-sensing pathways TLR4/caspase-1/caspase-11 were not protected from murine MAS (Supplemental Figure 5), suggesting that LPS is not the bacterial-derived factor driving TLR9 cytokine storm susceptibility.
Figure 6. Unique bacterial taxa are differentially abundant between protected and susceptible mice.
The stool samples described in Figure 5 were analyzed to determine if unique microbiota taxa were differentially abundant between the identified groups of protected, partially protected, and susceptible mice. (A) A taxonomic heatmap was created to display the abundance of individual microbial taxa identified in the 16S rRNA marker gene sequencing analysis. Each column of the heatmap represents 1 sample, and each row represents 1 taxon, typically a genus. Taxa were included in the chart if the abundance in any sample exceeded 1%. The chart is colored white if taxa were not observed in the sample, dark blue if taxa were observed at very low abundance, and abundance values exceeding 40% are colored red, indicating an extremely dominant species. This allows the reader to quickly survey presence/absence of individual microbial taxa in addition to identifying dominant species within individual samples. (B and C) Taxa were tested for differential abundance if their mean abundance was greater than 1%. Each of the 12 taxa tested in this analysis are displayed in individual plots separating samples based on whether they were obtained from a mouse classified as protected, partially protected, or susceptible to TLR9 cytokine storm (B). Linear regression was employed to test the association between log(proportion) of the taxon and study groups.
Table 1. Differential abundance.
JAK signaling is altered in myeloid progenitors from antibiotic-treated mice.
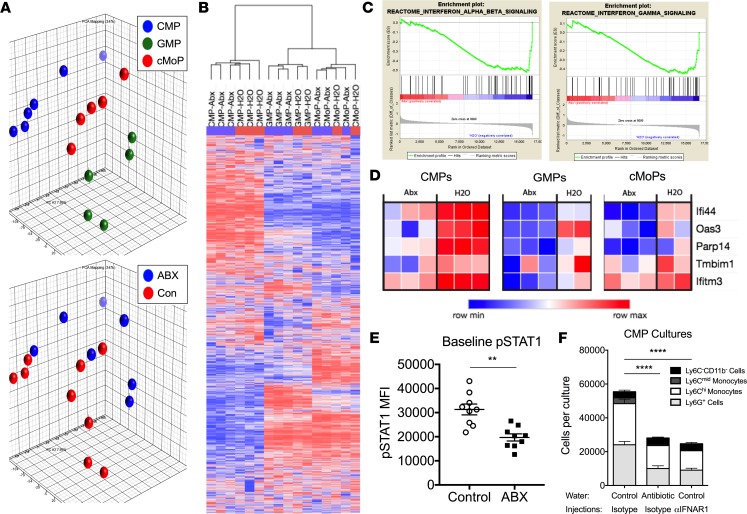
Since we did not have a definitive bacterial product to link to monocyte production and disease susceptibility, we next focused on cell-intrinsic pathways that could be altered in myeloid precursors by antibiotics. Microarray analysis of the transcriptomes of CMPs, GMPs, and cMoPs was performed to identify candidate mechanisms leading to the cell-intrinsic functional defect observed in myeloid progenitors from antibiotic-treated mice. Principal component analysis (PCA) and unsupervised hierarchical clustering analysis revealed clustering of samples by cell type and by mouse treatment, confirming the presence of transcriptomic differences between myeloid progenitors from antibiotic-treated and control mice (Figure 7, A and B). Gene set enrichment analysis (GSEA) indicated that CMPs from antibiotic-treated mice were negatively enriched for type I and type II IFN-regulated genes (Figure 7C and Supplemental Table 1). Similar results were observed for GSEA of GMPs and cMoPs (data not shown). Further analysis of the microarray data set identified 91 differentially expressed genes (DEGs) among CMPs, GMPs, and cMoPs from antibiotic-treated mice and their counterparts from control mice (Supplemental Table 2). DEGs were subjected to Ingenuity Pathway Analysis (IPA), which predicted decreased IRF7 function as an upstream regulator responsible for the altered transcriptional profile observed in myeloid progenitors from antibiotic-treated mice (P = 3.69 × 10–4, Z-score = –2.191). Five IRF7-regulated genes identified within the list of DEGs were suppressed in myeloid progenitors from antibiotic-treated mice compared with myeloid progenitors from control mice, supporting the IPA predictions (Figure 7D).
Figure 7. JAK signaling is altered in myeloid progenitors from antibiotic-treated mice.
(A–D) C57BL/6 mice were treated with antibiotic (ABX) or control water for 3 weeks prior to sacrifice and extraction of whole bone marrow cells. Bone marrow CMPs, GMPs, and cMoPs were sorted to isolate RNA for microarray analysis. (A) Principle component analysis was used to visualize the variance between samples in a 3-dimensional PCA plot. (B) Unsupervised hierarchical clustering analysis was used to compare the transcriptomes of each sample of the microarray. (C) Gene set enrichment analysis plots demonstrate negative enrichment of type I (left) and type II (right) IFNs genes in CMPs from antibiotic-treated mice. (D) Ingenuity Pathway Analysis (IPA) of differentially expressed genes identified suppression of IRF7 as an upstream regulator of the transcriptional changes observed between myeloid progenitors from antibiotic-treated and control mice. Heatmaps of the 5 IRF7-regulated genes identified by IPA analysis from the microarray data set are displayed. (E) Whole bone marrow cells were isolated from mice treated with antibiotic or control water for 3 weeks. Baseline levels of pSTAT1 in myeloid progenitors were measured by flow cytometry. Data were compiled from 2 separate experiments, for a total of 9 mice per group, and analyzed by a Mann-Whitney U test. (F) CMPs were sorted from mice treated with antibiotic or control water in combination with every-other-day dosing of an isotype antibody or anti-mouse IFNAR1 antibody for 3 weeks (n = 4 mice/group). Sorted CMPs were cultured for 7 days in media containing M-CSF, GM-CSF, IL-3, and SCF before enumeration of mature myeloid cell progeny by flow cytometry. Data were analyzed by 2-way ANOVA (***P < 0.01, ****P < 0.0001, antibiotic-treated vs. control mice and antibiotic-treated vs. anti-IFNAR–treated mice).
Basal levels of phosphorylated STAT1 (pSTAT1), a known signal downstream of type I and type II IFN receptor activation, were also reduced in myeloid progenitors from antibiotic-treated mice (Figure 7E). Furthermore, chronic in vivo blockade of type I IFN signaling using an anti–IFNAR-1 antibody resulted in CMPs having reduced output of mature myeloid cells in liquid in vitro myelopoiesis assays, which is similar to the defective function observed for CMPs isolated from antibiotic-treated mice (Figure 7F). Collectively, our data suggest host microbiota regulate tonic type I IFN signals that are sensed by myeloid progenitors, prime myeloid progenitor responses to inflammatory stimuli, and promote emergency hematopoiesis.
Inhibition of individual myeloid-specifying cytokines does not impair TLR9-enhanced myelopoiesis or cytokine storm pathology.
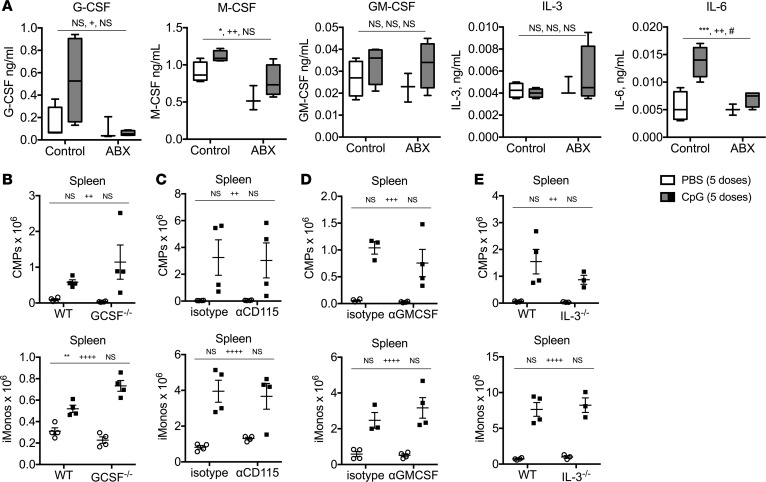
Previous work demonstrated that IFN-γ–/– and IFNAR-1–/– mice accumulate similar numbers of inflammatory monocytes downstream of repeated TLR9 activation in vivo, suggesting that these two cytokines are not required for induction of TLR9-enhanced myelopoiesis (7). Furthermore, IFNAR-1–/– mice are not protected from TLR9 cytokine storm. Neutralization of IL-6 was also insufficient to protect against TLR9-induced immunopathology (Supplemental Figure 6A). Therefore, we sought to identify alternative myeloid-specifying cytokines that may be required for induction of TLR9-enhanced myelopoiesis and cytokine storm. Systemic activation of TLR9 induces the production of multiple myeloid-specifying cytokines, levels of which were altered in antibiotic-treated mice (Figure 8A). However, to our surprise, genetic deficiency or cytokine neutralization of G-CSF, M-CSF, GM-CSF, or IL-3 individually was insufficient to mitigate the accumulation of extramedullary myeloid progenitors and inflammatory monocytes in the spleens of TLR9-activated mice (Figure 8, B–E). Furthermore, these mice were fully susceptible to TLR9-induced cytokine storm immunopathology, as they developed similar manifestations of disease compared with their control counterparts (Supplemental Figure 6, B–E). These data indicate that TLR9-enhanced myelopoiesis and cytokine storm immunopathology are not affected by the genetic loss or neutralization of type I IFNs, type II IFNs, or individual myeloid-specifying cytokines.
Figure 8. Inhibition of individual myeloid-specifying cytokines does not impair TLR9-enhanced myelopoiesis.
(A) WT C57BL/6 mice were treated with antibiotic (ABX) or control water for 3 weeks prior to receiving 5 doses of PBS or CpG1826. Serum cytokines were measured by ELISA 24 hours after the last injection. Data are graphed as box-and-whiskers plots showing minimum/maximum, and the line in the box shows the median value. (B) WT C57BL/6 and G-CSF–/– mice were treated with 5 doses of PBS or CpG1826, and numbers of spleen CMPs and spleen iMonos were enumerated. (C) WT C57BL/6 mice were treated with anti-CD115 (the M-CSF receptor) or isotype control antibodies in combination with 5 doses of PBS or CpG1826 injections over the course of 10 days, and numbers of spleen CMPs and spleen iMonos were enumerated. (D) WT C57BL/6 mice were treated with anti–GM-CSF or isotype control antibodies in combination with 5 doses of PBS or CpG1826 injections over the course of 10 days, and numbers of spleen CMPs and spleen iMonos were enumerated. (E) WT C57BL/10J and IL-3–/– mice were treated with 5 doses of PBS or CpG1826, and numbers of spleen CMPs and spleen iMonos were enumerated. Analysis was performed by 2-way ANOVA (*P < 0.05, **P < 0.01, ***P < 0.001, interaction; ++P < 0.01, ++++P < 0.0001, control vs. antibiotic treated; #P < 0.05, ##P < 0.01, ###P < 0.001, PBS vs. CpG in vivo treatments). In A the interaction terms term is CpG treatment x Antibiotic treatment. In B–E, the interaction terms are CpG treatment x antibody treatment or CpG treatment x genotype.
JAK1/2 signaling is required for induction of TLR9-enhanced myelopoiesis and cytokine storm immunopathology.
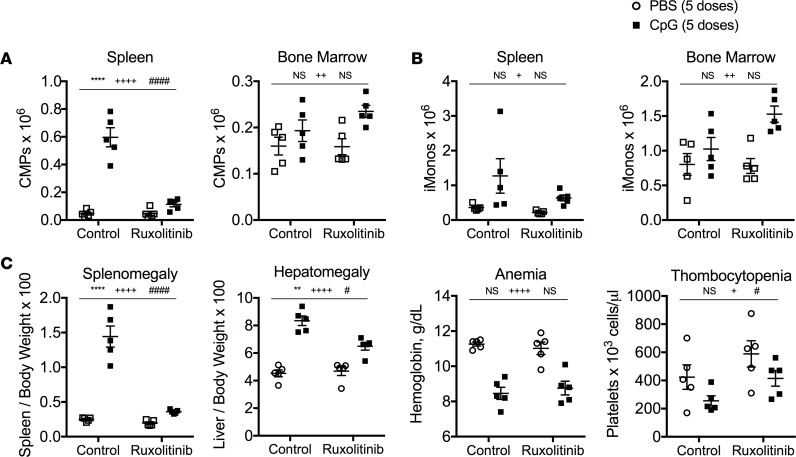
We hypothesized that induction of multiple myeloid-specifying cytokines downstream of systemic TLR9 activation could complement the loss of any single myeloid-specifying cytokine and induce TLR9-enhanced myelopoiesis leading to cytokine storm immunopathology. To test this hypothesis, we took advantage of the JAK1/2 inhibitor ruxolitinib that nonselectively inhibits signaling downstream of JAK-activating cytokines (26) and has previously been shown to mitigate inflammation in murine models of cytokine storm (27). Consistent with our hypothesis, inhibition of JAK1/2 was sufficient to suppress TLR9-enhanced myelopoiesis, inflammatory monocyte accumulation, and cytokine storm–induced immunopathology, which phenocopies the disease protection observed in antibiotic-treated mice (Figure 9 and Supplemental Figure 7). These data are consistent with a model whereby host microbiota prime JAK signaling in myeloid progenitors to promote TLR9-enhanced myelopoiesis and cytokine storm–induced immunopathology.
Figure 9. Pharmacologic inhibition of JAK1/2 signaling phenocopies the protection from TLR9-inudced immunopathology observed in antibiotic-treated mice.
C57BL/6 mice were treated with 5 doses of PBS or CpG1826 over the course of 10 days. Mice received control chow or chow containing the JAK1/2 inhibitor ruxolitinib starting with the third injection. Numbers of bone marrow and spleen common myeloid progenitors (CMPs) (A) and inflammatory monocytes (iMonos) (B) were enumerated. (C) Clinical manifestations of cytokine storm were evaluated by measuring splenomegaly, hepatomegaly, anemia, and thrombocytopenia. Each graph displays representative data from one of 2 independent experiments (N = 5 mice per group). Analysis was performed by 2-way ANOVA (**P < 0.01, ****P < 0.0001, interaction; +P < 0.05, ++++P < 0.0001, control vs. ruxolitinib chow; #P < 0.05, ##P < 0.01, ####P < 0.0001, PBS vs. CpG in vivo treatments). The interaction term is CpG treatment x Ruxolitinib treatment.
Discussion
Host-commensal interactions are emerging as key regulators of systemic immune responses, which have important consequences for health and disease. We provide evidence to support a model whereby commensal microbiota regulate systemic TLR-driven immune responses by priming JAK signaling in host myeloid progenitors. Microbiota-dependent priming of JAK signaling is necessary for the induction of TLR-enhanced myelopoiesis, which sustains the accumulation of new inflammatory monocytes and amplifies systemic TLR-driven cytokine production, leading to immunopathology in a murine model of cytokine storm. These data provide mechanistic insight into how host-commensal interactions regulate systemic innate immune responses and provide rationale for targeting microbiota-dependent innate immune responses for therapeutic benefit.
Innate immune cells are central mediators of systemic inflammation that drive immunopathology in cytokine storm syndromes (2). Noninfectious triggers of cytokine storm include autoimmune and autoinflammatory conditions that predispose to the development of macrophage activation syndrome (MAS), a cytokine storm syndrome that develops in patients with underlying chronic inflammatory diseases (1). However, only a minority of patients with autoimmune and autoinflammatory diseases develop MAS, indicating that an unidentified variable predisposes susceptible patients to cytokine storm (28). Our data using a murine model of TLR9-driven cytokine storm suggest that composition of the host microbiota may be one factor that contributes to disease susceptibility, as antibiotic-treated and germ-free mice are protected from TLR-driven cytokine storm. Future efforts will be needed to determine if MAS susceptible patients harbor unique microbiota or microbial-derived molecules that predispose them to the development of MAS. We note that antibiotic treatment must precede the TLR9 stimulus for it to offer protection from cytokine storm (data not shown), which correlates with the impression that antibiotics are not useful for the treatment of MAS once it has developed. It is intriguing to note that patients with MAS-predisposing conditions have been described as having a gene transcription signature predicting active hematopoiesis (29), consistent with our notion of enhanced myelopoiesis in our animal model system. Furthermore, a patient with NLRC4-MAS has been found to have elevated serum levels of myelopoiesis enhancing cytokines, consistent with this notion (30).
Microbiota-dependent priming of TLR-enhanced myelopoiesis may also be important in patients with sepsis, a cytokine storm syndrome induced by infection. In sepsis, systemic activation of TLRs results in an initial state of hyperinflammation followed by induction of TLR tolerance throughout the body. This leaves the host in a state of TLR hyporesponsiveness, which can be overcome by a compensatory acceleration in the production of new myeloid cells through TLR-enhanced myelopoiesis (7). As patients with sepsis are empirically administered broad-spectrum antibiotics, we hypothesize that this may suppress microbiota-dependent TLR-enhanced myelopoiesis and lead to a chronic state of TLR hyporesponsiveness that predisposes septic patients to opportunistic infections and increased mortality. Intriguingly, a subset of up to fifty percent of patients with sepsis develop a state of “immunoparalysis,” defined by the inability of their peripheral blood monocytes to respond to exogenous TLR activation ex vivo (31, 32). Both pediatric and adult patients with sepsis who develop immunoparalysis are at increased risk of developing secondary infections, multisystem organ failure, and death, consistent with our hypothesis (31, 32). Our data suggest that antibiotic-induced depletion of host microbiota that prime TLR-enhanced myelopoiesis may contribute to the development of immunoparalysis and poor clinical outcomes in patients with sepsis by suppressing the production of new myeloid cells that are critical for maintenance of TLR-driven protective immune responses.
Other inflammatory diseases may also be influenced by host-commensal interactions that regulate the production of innate immune cells. For example, intestinal dysbiosis is associated with states of chronic autoimmunity (33–36), and animal models demonstrate that host microbiota are critical for disease pathogenesis, as antibiotic-treated and germ-free animals are protected from murine models of chronic autoimmunity (37–40). Despite these apparent associations, there is a paucity of mechanistic data connecting how changes in the host microbiota influence the pathogenesis of acute and chronic inflammatory diseases in mice and humans. As myeloid cells play important roles in the initiation and propagation of chronic inflammation, it is intriguing to speculate that differences in the composition of the host microbiota may influence microbiota-dependent priming of TLR-enhanced myelopoiesis. If this notion is correct, those individuals who harbor microbes that support more effective TLR-enhanced myelopoiesis would be inherently more susceptible to the initiation and propagation of chronic inflammatory diseases and vice versa. Future work will be needed to confirm whether the dysbiosis of autoimmunity supports the accelerated production of myeloid effector cells that fuel maladaptive inflammatory responses to promote chronic inflammation in autoimmune diseases.
Our data support a model whereby commensal microbiota produce molecules that prime TLR-enhanced myelopoiesis. However, the individual microbe(s) and the microbial-derived molecule(s) that promote the accelerated production of monocytes upon systemic TLR activation remain unknown. Recent evidence points to both microbial-associated molecular patterns (MAMPs) and microbial-derived metabolites as putative candidate molecules that regulate hematopoiesis. In favor of MAMP-mediated priming of inflammation-induced myelopoiesis, gavage of bacterial-derived peptidoglycans rescues basal myelopoiesis defects observed in germ-free mice (22). Furthermore, sera from conventionally housed mice rescue hematopoietic defects in germ-free WT mice but not germ-free MyD88–/–TRIF–/– double-deficient mice that cannot signal downstream of TLRs (41). These data suggest that MAMPs are constituently present in the circulation of conventionally housed mice and provide tonic signals to the hematopoietic compartment. In contrast, reports have identified microbial-derived metabolites that also regulate hematopoiesis, including short-chain fatty acids (42), bile acids (43), and amino acids (44). Future research will need to confirm whether one or more of these microbial-derived molecules is capable of priming microbiota-dependent inflammation-induced myelopoiesis to yield insight into the molecular mechanisms governing how host-commensal interactions tune systemic TLR-driven innate immune responses by regulating the production of new hematopoietically derived myeloid cells.
It is intriguing to note that IFN-γ is not required for the myelopoiesis seen with repeated TLR9 exposure and yet IFN-γ is critical for the disease manifestations of MAS (7). This suggests that IFN-γ is downstream of the myelopoiesis that subsequently results in its generation and pathogenicity but that it is not part of the enhanced myelopoiesis process itself. Thus, targeting IFN-γ interrupts MAS manifestations but does not affect myelopoiesis, while disrupting myelopoiesis interrupts IFN-γ and therefore the downstream disease processes. It is therefore not the myelopoiesis directly that is problematic, but rather the IFN-γ that results from the newly generated TLR9-responsive monocytes derived by myelopoiesis and exposed to repeated TLR9 stimulation that is pathogenic. Interestingly, a broader JAK/STAT blockade with ruxolitinib does suppress myelopoiesis where IFN-γ blockade does not, suggesting that a number of different receptors converge on the same IRF7 signaling cascade to support the inflammatory myelopoiesis program in this model of murine cytokine storm.
Signals released by host microbiota may interface with a myriad of host receptors or signaling pathways that regulate myelopoiesis. Recent evidence suggests that signaling through the JAK/STAT pathway is critical for microbiota-dependent priming of myelopoiesis, as STAT1-decificient mice have basal defects in myelopoiesis similar to those of antibiotic-treated mice, which are not further suppressed by antibiotic treatment (23). Furthermore, in agreement with our results, others have shown that peripheral monocytes from germ-free or antibiotic-treated mice display a decreased IFN-regulated gene response (45, 46). We extend these findings to demonstrate that myeloid progenitors from antibiotic-treated mice also have an altered transcriptional program suggestive of impaired expression of IFN-regulated genes, reduced IRF7 function, and diminished basal phosphorylation of STAT1. These data support a model whereby commensal microbiota-derived signals prime JAK signaling in myeloid progenitors to promote TLR-enhanced myelopoiesis. Since total bacterial DNA load is correlated with this phenomenon, it is intriguing to speculate that there is a common bacterial motif responsible for priming TLR-enhanced myelopoiesis. Perhaps this is the bacterial DNA itself, whose pattern recognition receptors are well known to initiate type I IFN/IRF7 programs. Furthermore, our data suggest that impaired JAK/STAT signaling in myeloid progenitors from antibiotic-treated mice may contribute to the defective TLR9-enhanced myelopoiesis seen in these mice, as JAK1/2 inhibition in conventionally housed mice suppresses TLR9-enhanced myelopoiesis and results in disease protection similar to that observed in antibiotic-treated mice.
Our data raise important questions about how host-commensal interactions alter systemic innate immune responses by regulating the production of new hematopoietically derived innate immune cells that are required to sustain systemic TLR-driven immune responses. We provide proof of principle that manipulation of the host microbiota to suppress TLR-enhanced myelopoiesis may be an effective strategy to mitigate maladaptive innate immune responses in noninfectious inflammatory diseases. Future research will be needed to clarify the molecular mechanisms responsible for mediating microbiota-dependent priming of TLR-enhanced myelopoiesis and to determine the extent to which cross talk between commensal microbiota and the host’s hematopoietic system affects clinically relevant inflammatory and infectious diseases in humans.
Methods
Experimental design.
The goal of this study was to determine how commensal microbiota regulate systemic TLR-driven immune responses in a murine model of cytokine storm. Sample size was determined based on pilot experiments. Each experimental group contained 3–5 biological replicates per in vivo experiment. Randomization and blinding were not performed. Mice used in individual experiments were age and sex matched. Efforts were made to ensure equal use of male and female mice in all experiments. Five- to eight-week-old mice were used in experiments. Sources for key reagents are listed in Supplemental Table 3.
TLR9-mediated cytokine storm.
TLR9-mediated cytokine storm was induced by injecting 50 μg CpG1826 into the peritoneal cavity of mice every other day for a total of 5 doses. For Figure 3, a single dose of 50 μg CpG1826 was administered intraperitoneally to mice. For Figure 9, ruxolitinib and control chow was administered to mice ad libitum starting with the third injection. Splenomegaly and hepatomegaly was calculated by dividing organ weight by body weight of the mouse and multiplying by 100. Cheek bleeds were performed to obtain whole blood for complete blood count analysis on a Sysmex XT-2000iV Automated Hematology Analyzer. Serum cytokines were measured by ELISA from sera obtained by cardiac puncture after euthanasia. Whole livers were fixed in 10% formaldehyde, embedded in paraffin, and stained with H&E. All histology images were taken using a Leica DM4000B microscope and SPOT Software 5.1. Foci of inflammatory cell infiltrates were counted per high-power field using a ×20 objective.
Antibiotic treatment.
Mice were treated with antibiotic or control water for 3 weeks prior to the start of experiments, and antibiotic treatment was continued for the duration of the experiments involving induction of TLR9-mediated cytokine storm. Antibiotic-treated mice received a combination of 4 antibiotics (ampicillin, metronidazole, neomycin, and vancomycin, 1 g/l/antibiotic, in 2% sucrose) ad libitum. Antibiotic and control water was changed twice weekly.
Cytokine and cytokine receptor neutralization.
For Figure 7F, mice were treated with intraperitoneal injections of 500 μg of control antibody (clone MOPC-21) or anti-mouse IFNAR-1 every other day for 3 weeks. For Figure 8C and Supplemental Figure 6C, mice were treated with intraperitoneal injections of 1 mg of the control antibody (clone LFT-2) or anti-mouse CD115 (M-CSF receptor) for 3 doses every other day the week prior to the induction of TLR9-mediated cytokine storm. An additional 500 μg of control antibody or anti-mouse CD115 was administered concomitantly with each of the 5 doses of PBS or CpG1826. For Figure 8D and Supplemental Figure 6C, mice were treated with intraperitoneal injections of 500 μg of control antibody (clone LFT-2) or anti-mouse GM-CSF concomitantly with each of the 5 doses of PBS or CpG1826. For Supplemental Figure 6A, mice were treated with intraperitoneal injections of 200 μg of control antibody (clone SFR8-B6) or anti-mouse IL-6 concomitantly with each of the 5 doses of PBS or CpG1826. The SFR8-B6 control antibody was produced using a B lymphocyte hybridoma from ATCC and purified over a protein G column.
Processing of organs, whole blood, and sera.
Bone marrow cells were flushed from the leg bones of mice with cold PBS and single-cell suspensions were generated by mechanical disruption through a 70-μm strainer. Whole spleens were digested with 30 mg/ml DNase I (Roche) and 1 mg/ml collagenase (Roche) at 37°C for 30 minutes. Single-cell suspensions of splenocytes were generated by mechanical disruption through a 70-μm strainer. Single-cell suspensions of liver cells were generated by mechanical disruption of the whole liver through a 70-μm strainer. After centrifugation, the liver cell pellet was resuspended in 30% Percoll and layered over 70% Percoll. Liver cells were spun at room temperature at 1000 g for 20 minutes with no brake and harvested at the Percoll interface. Red blood cell lysis was performed on all cells using ACK lysis buffer (Lonza). Total cells per organ were counted on a Countess Automated Cell Counter from Thermo Fisher Scientific.
Cellular immunophenotyping.
Flow cytometry and sorting was performed on a Miltenyi MacsQuant and BD FACS Aria II, respectively. FACS data were analyzed using FlowJo software. All cellular immunophenotyping staining was performed at 4°C under conditions protected from ambient light. Forward and side scatter was used to gate out debris and doublets. Live cells were identified by excluding cells staining positive for LIVE/DEAD Fixable Aqua Dead Cell Stain from Thermo Fisher Scientific. Fc block (anti-CD16/32, clone 2.4G2) was used prior to antibody staining, except for myeloid progenitor panels. Inflammatory monocytes were identified as Ly6G–Ly6Chi cells that expressed CD11b, CD115, or Ccr2. Myeloid progenitors were identified as CMPs (Lin–c-Kit+CD105– CD16/32midCD115–), GMPs (Lin–c-Kit+CD105–CD16/32hiCD115–), MDPs (Lin–cKit+CD105–CD115+Ly6C–), and cMoPs (Lin–c-Kit+CD105–CD115+Ly6C+). The lineage panel included antibodies against B220, CD4, CD5, CD8a, CD11b, CD11c, CD49b, CD90.2, Ly6G, NK1.1, Sca-1, and Ter119. Sca-1 was excluded from the lineage cocktail for experiments involving in vivo CpG1826 treatments that induce the upregulation of Sca-1 on myeloid progenitors, as Sca-1 is induced by type I and type II IFNs. Representative flow plots depicting the gating strategy used to identify these various populations can be found in our previous publication (7).
Activation of TLR9 in vitro.
Whole bone marrow cells and splenocytes were isolated from antibiotic-treated and control mice and cultured in DMEM (Gibco) supplemented with 10% heat-inactivated FBS (Atlanta Biologicals) and Penicillin-Streptomycin–L-Glutamine (Cellgro) at 37°C in 6% CO2. Cells were stimulated with CpG1826 (10 μg/ml) and brefeldin A (2 μg/ml) for 12–18 hours. TLR9 responsiveness was measured in inflammatory monocytes by flow cytometry after staining for intracellular IL-12 using the BD Cytofix/Cytoperm reagent per the manufacturer’s instructions.
Analysis of STAT1 phosphorylation.
Whole bone marrow cells were isolated and rested in MEM-α media (Gibco) supplemented with 20% heat-inactivated FBS and Penicillin-Streptomycin–L-Glutamine at 37°C in 6% CO2 for 15 minutes. After surface staining for myeloid progenitors, 40 μl ice-cold 4% paraformaldehyde was added to each sample for 20 minutes prior to washing samples with FACS buffer (1× PBS, 2% FBS, 0.02% sodium azide). Samples were treated with 50 μl ice-cold 90% methanol and stored at –20°C for 30 minutes to 12 hours. Cells were washed with FACS buffer and incubated with 40 μl anti-mouse pSTAT1 diluted 1:50 in FACS buffer at 4°C for at least 6 hours. Samples were washed with FACS buffer and analyzed for pSTAT1 levels in total myeloid progenitors (Lin–cKit+CD105– cells) by flow cytometry.
Myelopoiesis assays.
Bone marrow myeloid progenitors were sorted from antibiotic-treated and control mice or from mice treated with anti-IFNAR1 antibodies for 3 weeks. Myeloid progenitors (500–2000 per well) were cultured in MEM-α media supplemented with 20% heat-inactivated FBS and penicillin–streptomycin–L-glutamine at 37°C in 6% CO2. GM-CSF, M-CSF, IL-3, and stem cell factor were obtained from Peprotech and were used to stimulate myeloid progenitor cell division and differentiation. All cytokines were used at 5 ng/ml, except GM-CSF, which was used at 3.3 ng/ml. After 7 days in culture, myeloid progenitor cell progeny were collected and analyzed for acquisition of mature myeloid cell surface markers and total cell counts by flow cytometry, as previously described (7).
Factorial antibiotic study design.
C57BL/6 mice were housed in the specific pathogen–free animal facility at Children’s Hospital of Philadelphia. A full factorial experimental was designed to include four 2-level variables (4 different antibiotics) resulting in 24 (i.e., 16) different treatment groups summarized in Table 2.
Five-week-old mice were housed separately in 17 different cages and pretreated with the above combinations of antibiotics for 3 weeks. A stool sample from each mouse was collected for 16S rRNA marker gene sequencing prior to treating mice with 5 doses of CpG1826 over the course of 10 days to induce murine MAS. Antibiotic treatments were continued for the duration of the experiment. Disease parameters were measured 24 hours after the fifth injection. Individual mice were categorized into 1 of 3 groups based on the severity of their disease outcomes (splenomegaly, hepatomegaly, WBC count, hemoglobin, and platelet count). Mice were categorized as “protected” from disease if 4 of 5 of their disease outcomes were within 20% of the mean values for PBS-treated mice or better. Mice were categorized as “susceptible” to disease if 4 of 5 of their disease outcomes were within 20%–30% of the mean values for CpG-treated mice or worse. All other mice were categorized as “partially protected.”
16S rRNA marker gene sequencing and analysis.
Stool samples collected from the 17 different groups of mice in the factorial antibiotic study design experiment were subjected to 16S rRNA marker gene sequencing (Table 2). DNA was extracted from the stool samples using the Qiagen DNeasy PowerSoil kit. Primers to amplify the bacterial 16S rRNA gene region were barcoded to label each sample (47). Primers annealing to the V1V2 region of the 16S bacterial gene were used for amplification (48). Amplified 16S rDNA was purified and sequenced using Illumina MiSeq. Sequence data were processed using QIIME (49). Read pairs were quality filtered and joined to form a complete V1V2 amplicon sequence. Operational taxonomic units (OTUs) were selected by clustering reads at 97% sequence similarity (50). Taxonomic assignments were generated by comparison to the Greengenes reference database (51), using the consensus method implemented in QIIME. A phylogenetic tree was inferred from the OTU data using FastTree (52). α-Diversity were measured using Shannon diversity and richness per 1,000 reads, as implemented in the R (version 3.2.3) vegan package (ver 2.5-2). Stool DNA concentration was measured by the Quant-iT PicoGreen Assay Kit from Thermo Fisher Scientific. Global differences in bacterial community composition were visualized using unweighted UniFrac distances, as previously described (53, 54). Community-level differences between sample groups were visualized using a taxonomic heatmap for each sample, which displayed all taxa exceeding 1% of any individual sample. The heatmap was colored white if the taxa were not observed in a sample, dark blue if taxa were observed at very low abundance, and red if abundance exceeded 40% to indicate an extremely dominant taxon. Taxa were tested for differential abundance if their mean abundance across all samples was greater than 1%. Twelve taxa were identified that fit this criterion. Linear regression was used to test the association between the log(proportion) of each of these 12 taxa and the study groups (mice classified into groups of protected, partially protected, and susceptible mice).
Myeloid progenitor microarray.
Bone marrow CMPs, GMPs, and cMoPs were sorted from antibiotic-treated and control mice directly into Qiagen RLT Lysis Buffer and frozen at –80°C prior to RNA extraction. Too few MDPs were obtained for RNA extraction. The Qiagen RNeasy Micro kit was used to isolate RNA per the manufacturer’s instructions. Based on poor RNA quality, one GMP sample and one cMoP sample were discarded with all other groups containing 3 replicate samples. NuGen Ovation Pico WTA System V2 (PN: 3302) was used to generate single stranded SPIA cDNA from 2.6 ng of total RNA. 5 μg SPIA cDNA was fragmented and labeled using the NuGen Encore Biotin Module (PN: 4200). 2 μg fragmented and labeled SPIA cDNA was hybridized to an Affymetrix GeneChip Mouse Gene 2.0ST Array for 17 hours at 50°C and with gentle stirring. Arrays were washed on an Affymetrix GeneChip Fluidics Station 450 using fluidics protocol FS450_0002 and scanned on Affymetrix GeneChip Scanner 3000.
Transcriptional analyses.
Normalization of the individual microarray chips was performed with robust multichip averaging using Transcriptome Analysis Console software. The microarray data set was filtered using Partek Genomics Suite software to include probe sets with annotations to gene names in the Refseq and ENSEMBL databases. PCA was performed for dimensionality reduction. The gene list was filtered on transcripts that had an absolute value fold difference of >1.5 between myeloid progenitors from antibiotic-treated mice and control mice using GenePattern modules from the Broad Institute. The filtered gene list was subjected to hierarchical clustering analysis using the GenePattern software from the Broad Institute. DEGs were identified by 2-way ANOVA and corrected for multiple testing using the Benjamini-Hochberg procedure to calculate a FDR of <0.2 using Partek Genomics Suite software. Ninety-one transcripts met an FDR cutoff of <0.2 and were defined as DEGs between myeloid progenitors from antibiotic-treated and control mice (Supplemental Table 2). DEGs were analyzed by IPA, which identified suppression of IRF7 function in myeloid progenitors from antibiotic-treated mice (P < 0.0005). Z-scores were calculated as fold-change differences between CMPs from antibiotic-treated and control mice. Fold-change values for GMPs and cMoPs between antibiotic-treated and control mice were similar (data not shown). The microarray expression data of the 5 IRF7-regulated transcripts identified using IPA software are displayed as a heatmap in Figure 7D. GSEA was performed using gene sets identified by searching the Molecular Signatures database for pathways involved in hematopoiesis or innate immunity. The 12 identified gene sets were analyzed using gene set permutation, and, prior to analysis, the following threshold for significant enrichment was chosen: a nominal P value of less than or equal to 0.05 and FDR of less than 0.05. A full list of the gene sets analyzed is provided as Supplemental Table 2.
Data and materials availability.
The microarray data set was deposited in the Gene Expression Omnibus (accession GSE112453). A materials transfer agreement from Incyte Corporation was required to obtain ruxolitinib-containing and control chow.
Statistics.
Statistical analyses were performed using Prism v7.0a (GraphPad). The Mann-Whitney U test was used for all single comparisons. A 2-way ANOVA was used to analyze the interaction between 2 independent variables on a dependent variable. P < 0.05 was considered significant. For graphs, individual symbols represent 1 mouse and horizontal lines represent mean values.
Study approval.
All animals were cared for in accordance with institutional animal facility guidelines of Children’s Hospital of Philadelphia, Perelman School of Medicine, and St. Jude Children’s Research Hospital. Procedures were performed after ethics board review and approval from Children’s Hospital of Philadelphia, Perelman School of Medicine, and St. Jude Children’s Research Hospital.
Author contributions
Author contributions are based on the CRediT taxonomy (https://casrai.org/credit/). LKW and EMB conceived the study. LKW, JJL, KB, EMB, and KEN provided study methodology. LKW, DM, CB, NC, JJL, KB, and SA provided investigation. LKW wrote the original draft. LKW, KEN, and EMB reviewed and edited the manuscript. LKW, EMB, and KEN acquired funding. LKW, JJL, and KB provided visualization. LKW and EMB supervised the study.
Supplementary Material
Acknowledgments
The authors thank Hamid Bassiri, Martha Jordan, Taku Kambayashi, Gary Koretzky, Paula Oliver, Michael Silverman, and their laboratories for helpful discussions; the flow cytometry core at the University of Pennsylvania for cell sorting; the Penn Gnotobiotic Mouse Facility for experiments involving germ-free mice; the Children’s Hospital of Philadelphia NAPCore facility for performing the microarray; the Children’s Hospital of Philadelphia Pathology Core Laboratories for processing histology slides; and the Children’s Hospital of Philadelphia Clinical and Translational Research Center for performing complete blood counts. Funding for this research was provided by the National Heart, Lung, and Blood Institute grant R01-HL112836 and a Howard Hughes Medical Institute early career investigator award to EMB, a Rheumatology Research Foundation Scientist Development Award and a Penn Children’s Hospital of Philadelphia Microbiome Pilot & Feasibility Grant to LKW, and the National Institute of Allergy and Infectious Diseases (R21-AI113490) and the American Lebanese Syrian Associated Charities to KEN. KEN is funded by a grant from Incyte Corporation and Alpine Immune Sciences.
Version 1. 01/10/2019
Electronic publication
Footnotes
Conflict of interest: KEN is funded by a grant from Incyte Corporation and Alpine Immune Sciences.
License: Copyright 2019, American Society for Clinical Investigation.
Reference information: JCI Insight. 2019;4(1):e124370. https://doi.org/10.1172/jci.insight.124370.
Contributor Information
Danielle Minichino, Email: edani@pennmedicine.upenn.edu.
Chhanda Biswas, Email: biswasc1@email.chop.edu.
Niansheng Chu, Email: chun1@email.chop.edu.
Jung-Jin Lee, Email: LEEJ39@EMAIL.CHOP.EDU.
Kyle Bittinger, Email: bittingerk@email.chop.edu.
Sabrin Albeituni, Email: Sabrin.Albeituni@STJUDE.ORG.
Kim E. Nichols, Email: Kim.Nichols@stjude.org.
References
- 1.Weaver LK, Behrens EM. Weathering the storm: Improving therapeutic interventions for cytokine storm syndromes by targeting disease pathogenesis. Curr Treatm Opt Rheumatol. 2017;3(1):33–48. doi: 10.1007/s40674-017-0059-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chousterman BG, Swirski FK, Weber GF. Cytokine storm and sepsis disease pathogenesis. Semin Immunopathol. 2017;39(5):517–528. doi: 10.1007/s00281-017-0639-8. [DOI] [PubMed] [Google Scholar]
- 3.Arbibe L, Sansonetti PJ. Epigenetic regulation of host response to LPS: causing tolerance while avoiding Toll errancy. Cell Host Microbe. 2007;1(4):244–246. doi: 10.1016/j.chom.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 4.Avau A, et al. Systemic juvenile idiopathic arthritis-like syndrome in mice following stimulation of the immune system with Freund’s complete adjuvant: regulation by interferon-gamma. Arthritis Rheumatol. 2014;66(5):1340–1351. doi: 10.1002/art.38359. [DOI] [PubMed] [Google Scholar]
- 5.Behrens EM, et al. Repeated TLR9 stimulation results in macrophage activation syndrome-like disease in mice. J Clin Invest. 2011;121(6):2264–2277. doi: 10.1172/JCI43157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strippoli R, et al. Amplification of the response to Toll-like receptor ligands by prolonged exposure to interleukin-6 in mice: implication for the pathogenesis of macrophage activation syndrome. Arthritis Rheum. 2012;64(5):1680–1688. doi: 10.1002/art.33496. [DOI] [PubMed] [Google Scholar]
- 7.Weaver LK, Chu N, Behrens EM. TLR9-mediated inflammation drives a Ccr2-independent peripheral monocytosis through enhanced extramedullary monocytopoiesis. Proc Natl Acad Sci USA. 2016;113(39):10944–10949. doi: 10.1073/pnas.1524487113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Canna SW, Wrobel J, Chu N, Kreiger PA, Paessler M, Behrens EM. Interferon-γ mediates anemia but is dispensable for fulminant toll-like receptor 9-induced macrophage activation syndrome and hemophagocytosis in mice. Arthritis Rheum. 2013;65(7):1764–1775. doi: 10.1002/art.37958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Notta F, Doulatov S, Laurenti E, Poeppl A, Jurisica I, Dick JE. Isolation of single human hematopoietic stem cells capable of long-term multilineage engraftment. Science. 2011;333(6039):218–221. doi: 10.1126/science.1201219. [DOI] [PubMed] [Google Scholar]
- 10.Osawa M, Hanada K, Hamada H, Nakauchi H. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science. 1996;273(5272):242–245. doi: 10.1126/science.273.5272.242. [DOI] [PubMed] [Google Scholar]
- 11.Moignard V, et al. Characterization of transcriptional networks in blood stem and progenitor cells using high-throughput single-cell gene expression analysis. Nat Cell Biol. 2013;15(4):363–372. doi: 10.1038/ncb2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lara-Astiaso D, et al. Immunogenetics. Chromatin state dynamics during blood formation. Science. 2014;345(6199):943–949. doi: 10.1126/science.1256271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oburoglu L, Romano M, Taylor N, Kinet S. Metabolic regulation of hematopoietic stem cell commitment and erythroid differentiation. Curr Opin Hematol. 2016;23(3):198–205. doi: 10.1097/MOH.0000000000000234. [DOI] [PubMed] [Google Scholar]
- 14.Essers MA, et al. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature. 2009;458(7240):904–908. doi: 10.1038/nature07815. [DOI] [PubMed] [Google Scholar]
- 15.Burberry A, et al. Infection mobilizes hematopoietic stem cells through cooperative NOD-like receptor and Toll-like receptor signaling. Cell Host Microbe. 2014;15(6):779–791. doi: 10.1016/j.chom.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takizawa H, et al. Pathogen-induced TLR4-TRIF innate immune signaling in hematopoietic stem cells promotes proliferation but reduces competitive fitness. Cell Stem Cell. 2017;21(2):225–240.e5. doi: 10.1016/j.stem.2017.06.013. [DOI] [PubMed] [Google Scholar]
- 17.Zhao JL, et al. Conversion of danger signals into cytokine signals by hematopoietic stem and progenitor cells for regulation of stress-induced hematopoiesis. Cell Stem Cell. 2014;14(4):445–459. doi: 10.1016/j.stem.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Massberg S, et al. Immunosurveillance by hematopoietic progenitor cells trafficking through blood, lymph, and peripheral tissues. Cell. 2007;131(5):994–1008. doi: 10.1016/j.cell.2007.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cortez-Retamozo V, et al. Origins of tumor-associated macrophages and neutrophils. Proc Natl Acad Sci USA. 2012;109(7):2491–2496. doi: 10.1073/pnas.1113744109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dutta P, et al. Myocardial infarction accelerates atherosclerosis. Nature. 2012;487(7407):325–329. doi: 10.1038/nature11260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manz MG, Boettcher S. Emergency granulopoiesis. Nat Rev Immunol. 2014;14(5):302–314. doi: 10.1038/nri3660. [DOI] [PubMed] [Google Scholar]
- 22.Iwamura C, Bouladoux N, Belkaid Y, Sher A, Jankovic D. Sensing of the microbiota by NOD1 in mesenchymal stromal cells regulates murine hematopoiesis. Blood. 2017;129(2):171–176. doi: 10.1182/blood-2016-06-723742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Josefsdottir KS, Baldridge MT, Kadmon CS, King KY. Antibiotics impair murine hematopoiesis by depleting the intestinal microbiota. Blood. 2017;129(6):729–739. doi: 10.1182/blood-2016-03-708594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deshmukh HS, et al. The microbiota regulates neutrophil homeostasis and host resistance to Escherichia coli K1 sepsis in neonatal mice. Nat Med. 2014;20(5):524–530. doi: 10.1038/nm.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khosravi A, et al. Gut microbiota promote hematopoiesis to control bacterial infection. Cell Host Microbe. 2014;15(3):374–381. doi: 10.1016/j.chom.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwartz DM, Kanno Y, Villarino A, Ward M, Gadina M, O’Shea JJ. JAK inhibition as a therapeutic strategy for immune and inflammatory diseases. Nat Rev Drug Discov. 2017;16(12):843–862. doi: 10.1038/nrd.2017.201. [DOI] [PubMed] [Google Scholar]
- 27.Das R, et al. Janus kinase inhibition lessens inflammation and ameliorates disease in murine models of hemophagocytic lymphohistiocytosis. Blood. 2016;127(13):1666–1675. doi: 10.1182/blood-2015-12-684399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Behrens EM, Beukelman T, Paessler M, Cron RQ. Occult macrophage activation syndrome in patients with systemic juvenile idiopathic arthritis. J Rheumatol. 2007;34(5):1133–1138. [PubMed] [Google Scholar]
- 29.Hinze CH, et al. Immature cell populations and an erythropoiesis gene-expression signature in systemic juvenile idiopathic arthritis: implications for pathogenesis. Arthritis Res Ther. 2010;12(3):R123. doi: 10.1186/ar3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Canna SW, et al. An activating NLRC4 inflammasome mutation causes autoinflammation with recurrent macrophage activation syndrome. Nat Genet. 2014;46(10):1140–1146. doi: 10.1038/ng.3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hall MW, et al. Immunoparalysis and nosocomial infection in children with multiple organ dysfunction syndrome. Intensive Care Med. 2011;37(3):525–532. doi: 10.1007/s00134-010-2088-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Munoz C, Carlet J, Fitting C, Misset B, Blériot JP, Cavaillon JM. Dysregulation of in vitro cytokine production by monocytes during sepsis. J Clin Invest. 1991;88(5):1747–1754. doi: 10.1172/JCI115493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scher JU, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife. 2013;2:e01202. doi: 10.7554/eLife.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jangi S, et al. Alterations of the human gut microbiome in multiple sclerosis. Nat Commun. 2016;7:12015. doi: 10.1038/ncomms12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kostic AD, et al. The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe. 2015;17(2):260–273. doi: 10.1016/j.chom.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lewis JD, et al. Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn’s disease. Cell Host Microbe. 2015;18(4):489–500. doi: 10.1016/j.chom.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu HJ, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32(6):815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci USA. 2011;108 Suppl 1:4615–4622. doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Silverman M, et al. Protective major histocompatibility complex allele prevents type 1 diabetes by shaping the intestinal microbiota early in ontogeny. Proc Natl Acad Sci USA. 2017;114(36):9671–9676. doi: 10.1073/pnas.1712280114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carvalho FA, et al. Transient inability to manage proteobacteria promotes chronic gut inflammation in TLR5-deficient mice. Cell Host Microbe. 2012;12(2):139–152. doi: 10.1016/j.chom.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Balmer ML, et al. Microbiota-derived compounds drive steady-state granulopoiesis via MyD88/TICAM signaling. J Immunol. 2014;193(10):5273–5283. doi: 10.4049/jimmunol.1400762. [DOI] [PubMed] [Google Scholar]
- 42.Trompette A, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. 2014;20(2):159–166. doi: 10.1038/nm.3444. [DOI] [PubMed] [Google Scholar]
- 43.Sigurdsson V, et al. Bile acids protect expanding hematopoietic stem cells from unfolded protein stress in fetal liver. Cell Stem Cell. 2016;18(4):522–532. doi: 10.1016/j.stem.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 44.Lv J, Wang L, Gao Y, Ding YQ, Liu F. 5-hydroxytryptamine synthesized in the aorta-gonad-mesonephros regulates hematopoietic stem and progenitor cell survival. J Exp Med. 2017;214(2):529–545. doi: 10.1084/jem.20150906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abt MC, et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity. 2012;37(1):158–170. doi: 10.1016/j.immuni.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Erny D, et al. Host microbiota constantly control maturation and function of microglia in the CNS. Nat Neurosci. 2015;18(7):965–977. doi: 10.1038/nn.4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McKenna P, et al. The macaque gut microbiome in health, lentiviral infection, and chronic enterocolitis. PLoS Pathog. 2008;4(2):e20. doi: 10.1371/journal.ppat.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu GD, et al. Sampling and pyrosequencing methods for characterizing bacterial communities in the human gut using 16S sequence tags. BMC Microbiol. 2010;10:206. doi: 10.1186/1471-2180-10-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 51.McDonald D, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6(3):610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Price MN, Dehal PS, Arkin AP. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5(3):e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lozupone CA, Hamady M, Kelley ST, Knight R. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol. 2007;73(5):1576–1585. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71(12):8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.