Abstract
Contractile injection systems mediate bacterial cell-cell interactions by a bacteriophage tail-like structure. In contrast to extracellular systems, the type 6 secretion system (T6SS) is defined by intracellular localization and attachment to the cytoplasmic membrane. Here we used cryo-focused ion beam milling, electron cryotomography, and functional assays to study a T6SS in Amoebophilus asiaticus. The in situ architecture revealed three modules including a contractile sheath-tube, a baseplate, and an anchor. All modules showed conformational changes upon firing. Lateral baseplate interactions coordinated T6SSs in hexagonal arrays. The system mediated interactions with host membranes and may participate in phagosome escape. Evolutionary sequence analyses predicted that T6SSs are more widespread than previously thought. Our insights form the basis for understanding T6SS key concepts and exploring T6SS diversity.
Contractile injection systems (CIS) deliver effectors to mediate bacterial cell-cell interactions. Their structural components are homologous to the contractile tails of phages (1). CIS consist of an inner tube surrounded by a contractile sheath, a spike capping the inner tube, and a baseplate complex at the base of the sheath. Sheath contraction propels the inner tube into the target. Two modes of action divide CIS into “extracellular CIS” (eCIS) and “type 6 secretion” (T6S) systems (T6SSs). eCIS resemble headless phages; they are released into the medium and bind to the target cell surface. Examples are antibacterial R-type bacteriocins (2), insecticidal antifeeding prophages (Afps) (3), and metamorphosis-inducing structures (MACs) (4). In contrast, the T6SS is defined by its cytoplasmic localization and anchoring to the inner membrane (5–9).
Amoebophilus asiaticus (hereafter referred to as Amoebophilus or amoebophili) is an obligate intracellular bacterial symbiont of amoebae (10). The Amoebophilus genome does not encode known secretion systems (11); however, it contains a gene cluster with similarities to Afps (12). We reasoned that the Afp-like gene cluster might encode a system that would give insight into T6SS structure, function and evolution.
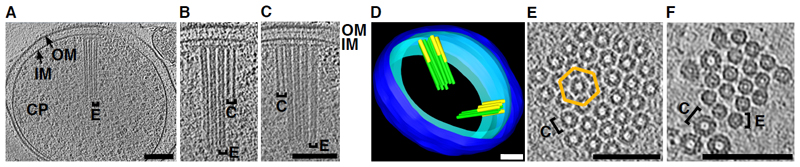
To investigate whether Amoebophilus produced any CIS, we imaged bacterial cells that were purified from infected amoeba cultures by electron cryotomography (ECT). 50 % of the imaged cells (n=92) contained phage tail-like assemblies. Like T6SSs and unlike eCIS, the structures were always located in the Amoebophilus cytoplasm and attached to the cytoplasmic membrane. The structures were always found in bundles of 2-34 parallel individual systems (8 on average; fig. 1A-C; S1A-D; movie S1). Cells contained either one or two (85 %) bundles (fig. 1D; S1E). Inside a bundle, the structures were arranged in ordered hexagonal arrays (fig. 1E/F). Extended and contracted conformational states could be distinguished by differences in length (242±7 nm, n = 254, and 122±6 nm, n = 153, respectively), diameter (14±2 nm and 19±2 nm, respectively), and surface properties of the sheath (helical ridges on the contracted structures) (fig. 1B/C; S1F-I). The narrow distribution of sheath lengths indicates a mechanism for length control. In addition, not all structures inside an array appeared to fire together (fig. 1B-D/F; S1E).
Figure 1. The Amoebophilus Afp-like gene cluster encodes for ordered arrays of T6S-like structures.
(A-C) Cryotomograms of purified Amoebophilus cells revealed bundles of cytoplasmic, membrane-bound, contractile structures in extended (“E”) and contracted (“C”) conformations. “CP”, cytoplasm; “IM”, inner membrane; “OM”, outer membrane. Shown are three different 14-nm slices through the same tomogram. (D) Bundles comprised 2-34 individual machines (green, extended; yellow, contracted) and were organized in 1-2 bundles per cell. Shown is a model of the tomogram shown in A-C. Blue, outer membrane; cyan, inner membrane. (E/F) Structures were arranged in hexagonal arrays (lattice indicated in orange). Shown are 15-nm (E) and 8-nm (F) cross-sectional slices. Bars: 100 nm.
The arrays of contractile structures were encoded by the Amoebophilus Afp-like gene cluster. 12 of its components were detected in a sheath preparation (Table S1). Sheath structures were labeled by specific antibodies (fig. S1J/K). Furthermore, tomograms of purified sheath revealed contracted sheaths whose structure (fig. S1L/M) and dimensions (length 115±7 nm, diameter 19±2 nm, n = 51) were consistent with the structures observed in situ (fig. S1F-I).
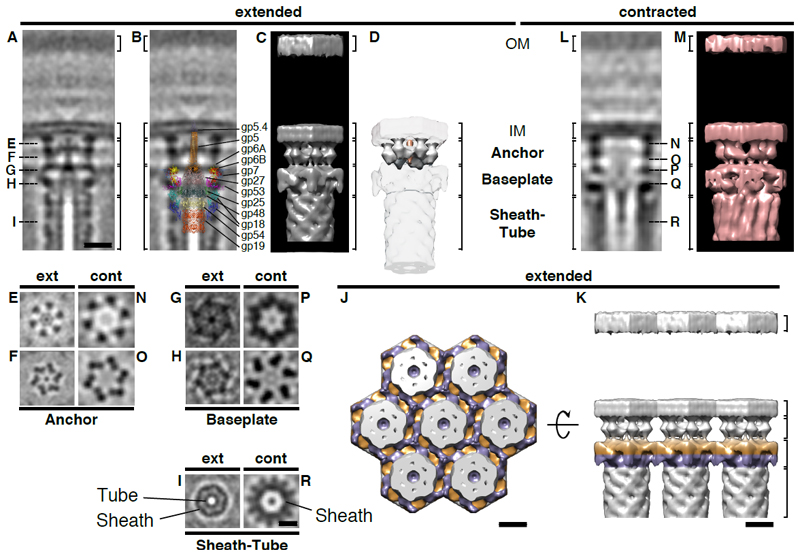
In order to observe macromolecular details, we averaged subvolumes of extended T6S-like machines (fig. 2A-K; movies S2, S3). The structure revealed three major modules: a sheath-tube assembly, a baseplate, and an anchoring complex (fig. 2A-I). This segmentation was supported by the comparison of the average with the structure of the minimal composition of a contractile injection system derived from the T4 phage tail (13) (fig. 2B). All three modules showed 6-fold rotationally symmetric features (fig. S2). Densities for the inner tube (7 nm diameter) and sheath (14 nm diameter) could be clearly discerned (fig. 2A/I). The baseplate was overall hexagonal (fig. 2G/H) and established connections with baseplates of neighboring structures, thereby coordinating multiple systems in hexagonal arrays (fig. 2J/K). The central baseplate region featured additional densities that reinforced in a 3-fold symmetrized average (fig. S2I-M). The anchoring complex consisted of a six-footed platform that attached the baseplate to the inner membrane (fig. 2A/D-F). Densities whose dimensions were consistent with a spike complex (14) were seen capping the inner tube and protruded through the centers of baseplate and anchor (fig. 2A/B/E-G). The averages lacked densities that would indicate the presence of an elaborate trans-envelope complex (fig. 2A; fig. S2A/E-G) such as TssJLM (9).
Figure 2. The T6SS in situ architecture reveals three major modules, conformational changes upon firing, and the structural basis of array formation.
Subtomogram averages of extended (A-K) and contracted (L-R) T6SSs revealed three major cytoplasmic modules: “Sheath-Tube”, “Baseplate”, and “Anchor” (indicated by brackets).
Shown are 0.81-nm (A/B, E-I) and 1.38-nm (L, N-R) longitudinal (A/B/L) and perpendicular (E-I, N-R) slices through 6-fold rotationally symmetrized averages, and their 3D models (C/D/J/K/M). The positions of perpendicular sections are indicated in (A/L). “OM”, outer membrane. “IM”, inner membrane. Bars: 10 nm (A-D/L/M to scale; E-I/N-R to scale).
The segmentation in three modules was supported by the overlay (B) with the structure of the minimal composition of a contractile injection system derived from the T4 phage tail [from (13)]. It allowed the putative assignment of densities corresponding to tube (gp19/gp48/gp54), sheath (gp18), spike complex (gp5/gp5.4/gp27), and baseplate wedge components (gp6/gp7/gp25/gp53). Densities that were not accounted for were assigned to the anchor module (segmented in D; white, anchor; orange, spike).
Upon firing, significant conformational changes were detected in all modules (movie S6 shows a morph between models shown in C and M). The sheath increased in diameter and formed surface ridges (A/C/I/L/M/R). Baseplate and anchor showed widening and loss of densities in the center (A/C/E-H/L/M/N-Q).
Ordered arrays were established by lateral interactions of the hexagonal baseplates (J/K). Shown are top (J) and side (K) views of a model that was assembled from masked averages. For viewing purposes, two different baseplate levels are colored in purple and orange.
The T4 phage baseplate triggers sheath contraction by a large-scale conformational change (13). We therefore calculated an average of contracted structures (fig. 2L-R; movies S4, S5). Again, sheath, baseplate, and anchor modules were identified (fig. 2L-R). All three modules revealed significant conformational changes as compared to the extended state (movie S6). Similar to the V. cholerae T6SS (5), the sheath diameter increased upon contraction, along with the appearance of helical surface ridges and the disappearance of the inner tube (fig. 2I/R). The baseplate showed a widening and a loss of densities in the center (fig. 2G/H/P/Q). Likewise, the anchoring platform showed lateral expansion (distance between opposing feet at the inner membrane increased from 16 nm to 19 nm) and a loss of the spike density in the center (fig. 2E/F/N/O). On a larger scale, the spacing between contractile structures increased from 22 nm between extended structures to 30 nm between contracted structures.
We then tested whether the Amoebophilus system secreted tube protein into extracellular space. Immunoblotting detected tube protein (Hcp), but no sheath, in the supernatant of a culture (fig. S3), indicating Hcp translocation. The system thus fulfills the functional hallmark of canonical T6SSs. Together with the structural data, this suggests that the Amoebophilus Afp-like gene cluster encodes for a T6SS rather than for an eCIS.
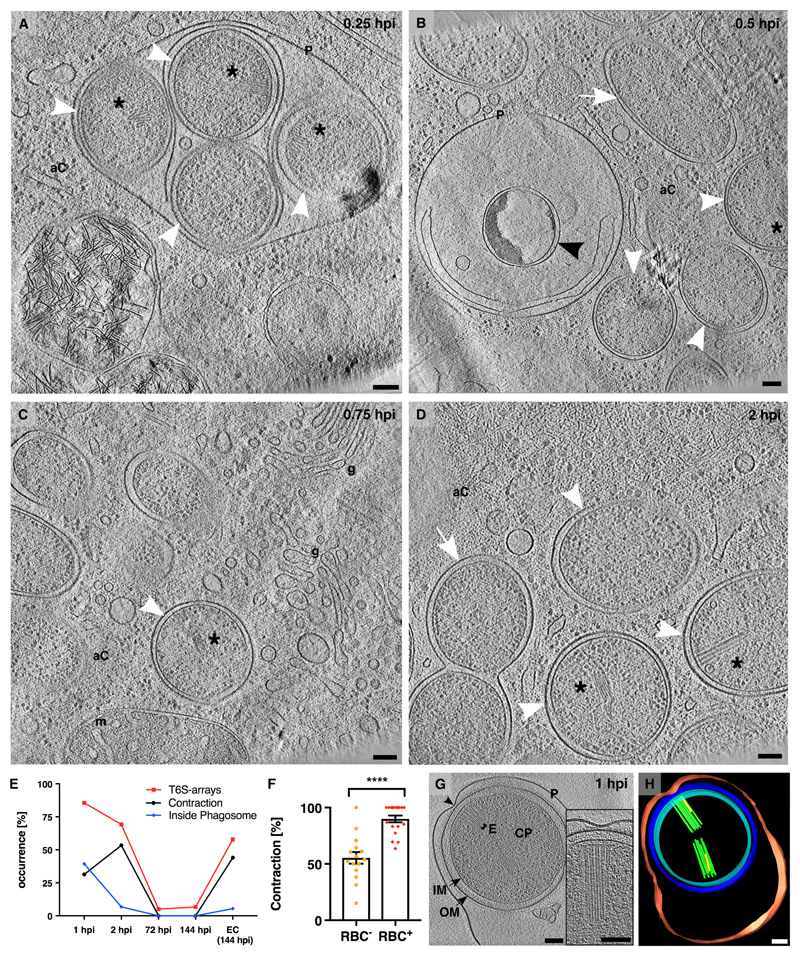
Next, we investigated the function of T6SS arrays. Amoebophili were internalized in the first two hours post infection (hpi) and exited the amoebae ~144 hpi (fig. S4). To observe intracellular amoebophili by ECT, we used cryo-focused ion beam milling to generate lamellae that were suitable for ECT (fig. S5) (15, 16). At 0.25 hpi, coccoid amoebophili were inside phagosomes (80 %, n=20, fig. 3A; movie S7). At later stages, most amoebophili had escaped into the cytosol (94 %, n=94), differentiated into rods, and replicated (fig. 3B-D; fig. S6A; movies S8-S10). The sheath mRNA level was 230-fold higher in extracellular amoebophili compared to the replicative stage (Table S2). Likewise, cryotomograms of amoebophili from different infection stages showed that T6SSs were most abundant in extracellular amoebophili (58 %, n=19) and at early infection stages (1 hpi, 96 %, n=25; 2 hpi, 69 %, n=13), while replicative amoebophili did usually not harbor structures (5 %, n=20) (fig. 3E; S6B-G). The process of exiting the phagosome during early infection (up to 2 hpi) correlated with increased fractions of contracted structures (fig. 3E). Experiments comparing the potential of amoebophili from different infection stages to establish new infections showed that host infection rates positively correlated with T6SS expression (fig. S6H). Likewise, we tested amoebophili from different infection stages for hemolytic activity and found that red blood cell (RBC) lysis also positively correlated with T6SS expression (fig. S7). ECT imaging of amoebophili that had the possibility to interact with RBCs showed a 30 % increased fraction of contracted structures, compared to a control sample (fig. 3F; p < 0.0001). The analysis of tomograms of purified amoebophili that were found inside phagosomes (39 % at 1 hpi) revealed that any contact site between the phagosome membrane and the outer membrane of the bacterium correlated with the presence of T6SSs (with at least one contracted structure) (n=14, fig. 3G/H; S8; movie S11). Together, our data suggest that T6S arrays mediate interactions with host membranes and may participate in phagosome escape. It remains open whether phagosome rupturing is mediated by mechanical forces or membrane-targeting effectors.
Figure 3. T6S arrays are required during early infection stages and mediate interactions with host membranes.
(A-D) Bacteria inside their host were imaged by cryo-focused ion beam milling and ECT (fig. S5). At 0.25 hpi, most coccoid amoebophili (white arrowheads) were found inside phagosomes (“P”). At later time points (0.5-2 hpi), amoebophili had escaped into the amoeba cytosol (“aC”). Amoebophili differentiated into rods (white arrows) and divided. A small fraction did not escape, showing signs of degradation (black arrowhead). Shown are 15-nm slices through cryotomograms. Asterisk, T6S array; “g”, golgi apparatus; “m”, mitochondrion. Bars: 100 nm. (E) Abundance of T6S arrays was determined by ECT of amoebophili purified from synchronized cultures, and found to be highest in extracellular “EC (144 hpi)” and early intracellular infection stages (1 hpi, 2 hpi). The increase of the contraction rate between 1 and 2 hpi correlated with the escape from the phagosome. Shown are the percentages of cells with T6S arrays (red), percentages of T6S structures that were contracted (black), and percentages of cells found inside phagosomes (blue). T6S arrays, number of quantified amoebophili: n1hpi=25, n2hpi=13, n72hpi=20, n144hpi=15, nEC(144 hpi)=19; Contraction, number of quantified T6S structures: n1hpi=168, n2hpi=88, n72hpi=4, n144hpi=4, nEC(144 hpi)=59; Inside Phagosome, number of quantified amoebophili: n1hpi=121, n2hpi=118, n72hpi=218, n144hpi=337, nEC(144 hpi)=55). (F) Amoebophili showed hemolytic activity (fig. S7). Extracellular amoebophili that interacted with RBCs showed an increased T6S contraction rate (**** p < 0.0001; nRBC–=506; nRBC+=480). (G/H) Amoebophili residing in phagosomes revealed contact sites (black arrowhead) between the Amoebophilus outer membrane and the phagosome. Any such contact site correlated with a T6S array (n=14). Shown are a 15-nm tomographic slice (G) and the corresponding model (H). “P”/red, phagosome; “OM”/blue, outer membrane; “IM”/cyan, inner membrane; “CP”, cytoplasm; “E”/green, extended T6SS; yellow, contracted T6SS; Bars: 100 nm.
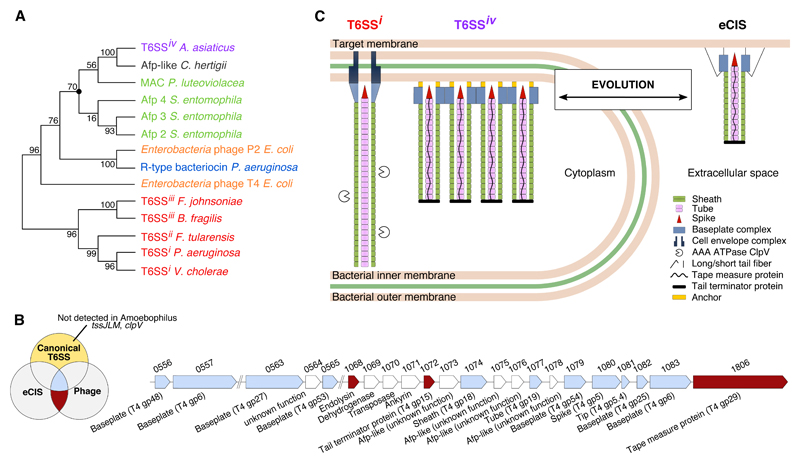
Next, we sought to understand the evolutionary history of the Amoebophilus Afp-like gene cluster. We compared three key components (sheath, tail tube, baseplate component gp25) to other CIS (Table S3). Similarities were highest with a gene cluster of unknown function in the related symbiont Cardinium hertigii (17). Moderate similarities were found for Afp and MAC, both mediating interactions with animal larvae (3, 4). Lowest (or no similarities at all) were detected for T6SSs [subtypes i, ii, iii (18)] and contractile phages. Likewise, phylogenetic analyses revealed that Amoebophilus (and Cardinium) sequences stably clustered in a monophyletic group with Afp and MAC, rather than with T6SSi-iii (fig. 4A; S9). With the exception of gp7, the analysis of gene content detected Amoebophilus homologues of all components that are conserved across CIS and phages (13). These include putative sheath (gp18/TssBC), inner tube (gp19/Hcp, gp48, gp54), spike (gp5/VgrG, gp5.4/PAAR, gp27) and three baseplate wedge components (gp6/TssF, gp25/TssE, gp53) (fig. 4B; Table S4). The lack of gp7 might be explained by the presence of two gp6 homologues, and the suggestion that gp6 and gp7 diverged from a common ancestor (13, 19). Components that are exclusively conserved in canonical T6SSs (and absent in eCIS/phages) were not found, including TssJLM [trans-envelope complex (9)], or ClpV [sheath recycling (20)]. In contrast, the Amoebophilus cluster encodes proteins that were thought to be specific for eCIS and contractile phages. The length of the Amoebophilus T6SSs, for instance, is likely controlled by Aasi_1072/1806, which are homologues of tail terminator and tape measure proteins in Afp and phages (21, 22). In fact, sheath length can be predicted from TmP sequence (22) and correlates well with the length of Amoebophilus sheaths (fig. S10). Another example is an Rz-like endopeptidase (Aasi_1068), which usually co-occurs with a holin to mediate the release of eCIS and phages from the bacterial cytoplasm (3, 23).
Figure 4. Amoebophilus T6SSiv is closely related to eCIS.
(A) Phylogenetic analyses of sheath protein sequences showed, that T6SSi-iii formed a monophyletic group with high support (bootstrap supports are indicated at nodes). Amoebophilus T6S sheath, however, stably clustered in a group with the sheath from a gene cluster in Cardinium (structure/function unknown), and with the eCIS sheaths of Afp and MAC. The marked node was stable in all calculated trees using different treeing methods and different components (fig. S9). (B) The Amoebophilus Afp-like gene cluster encodes for all components that are conserved (blue) among all contractile injection systems (canonical T6SS/eCIS/phages), while it lacks any homologues of components that are specific for canonical T6SSs (yellow). Instead, the cluster harbors genes that are typical of eCIS and phages (red). Shown are two Amoebophilus genomic regions, locus tags, and gene annotations. See also Table S4. (C) Schematic showing the major components of canonical T6SS, Amoebophilus T6SSiv, and eCIS (homologs in same colors). T6SSiv evolved either from eCIS, or alternatively, T6SSiv represents a primordial system that gave rise to eCIS/phages/T6SSi-iii. Both scenarios predict that T6SSs are more abundant than previously estimated (Table S5).
In conclusion, our structural and functional data showed that the Amoebophilus Afp-like gene cluster encodes for a T6SS, while sequence analyses indicated a close relationship to eCIS. We therefore introduce the term “T6SS subtype 4” (T6SSiv). In contrast to the distant relationships of T6SSi-iii to eCIS and phages that obstruct the reconstruction of an evolutionary path (1, 24), we can hypothesize that T6SSiv evolved from an Afp/MAC-like eCIS (independently of T6SSi-iii) by the loss of tail fibers, loss of holin, and the establishment of interactions with the cytoplasmic membrane. Alternatively, T6SSiv represents a primordial system from which eCIS, phages, and T6SSi-iii have evolved (fig. 4C). Both scenarios predict that T6SSs are more abundant than previously thought. In fact, T6SSiv-like gene clusters were detected in six diverse bacterial phyla (Table S5). The finding that diverse T6SS subtypes do not share a conserved gene set that would distinguish them from eCIS/phages, emphasizes the necessity of an integrative approach to discover and characterize new systems.
Supplementary Material
One Sentence Summary.
A type 6 secretion model system reveals the structure, dynamics and evolution of apparatus attachment to the membrane.
Acknowledgements
We thank F. Bosia, A. Harreither, P. Gasser, S. Rutz, P. Szwedziak, P. Tittmann, R. Wepf, and C. Zaubitzer for technical support. ScopeM is acknowledged for instrument access at ETH Zürich. We thank O. Medalia for microscope access at the University of Zürich. T. Ishikawa, R. Wepf, and Pilhofer Lab members are acknowledged for discussions. Rok Kostanjsek is acknowledged for generating preliminary data. W.-D. Hardt, H. Hilbi, R. Kooger, V. Panse, N. Shikuma, M. Swulius, and E. Tocheva are acknowledged for comments on the manuscript. We thank N. Taylor for providing us with the T4 model shown in fig. 2B. Preliminary observations were made in the lab of G. J. Jensen. MP is supported by the European Research Council, the Swiss National Science Foundation, and the Helmut Horten Foundation. MH is supported by the European Research Council (ERC StG, No. 281633) and Austrian Science Fund (I 291-B09). Subtomogram averages were deposited in the Electron Microscopy Data Bank (accession numbers EMD-3791 and EMD-3793 to EMD-3801).
References
- 1.Leiman PG, Shneider MM. Contractile tail machines of bacteriophages. Adv Exp Med Biol. 2012;726:93–114. doi: 10.1007/978-1-4614-0980-9_5. [DOI] [PubMed] [Google Scholar]
- 2.Uratani Y, Hoshino T. Pyocin R1 inhibits active transport in Pseudomonas aeruginosa and depolarizes membrane potential. J Bacteriol. 1984;157:632–636. doi: 10.1128/jb.157.2.632-636.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hurst MRH, Glare TR, Jackson TA. Cloning Serratia entomophila antifeeding genes--a putative defective prophage active against the grass grub Costelytra zealandica. J Bacteriol. 2004;186:5116–5128. doi: 10.1128/JB.186.15.5116-5128.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shikuma NJ, Pilhofer M, Weiss GL, Hadfield MG, Jensen GJ, Newman DK. Marine Tubeworm Metamorphosis Induced by Arrays of Bacterial Phage Tail–Like Structures. Science. 2014;343:529–533. doi: 10.1126/science.1246794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature. 2012;483:182–186. doi: 10.1038/nature10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hachani A, Wood TE, Filloux A. Type VI secretion and anti-host effectors. Curr Opin Microbiol. 2016;29:81–93. doi: 10.1016/j.mib.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 7.Chang Y-W, Rettberg LA, Ortega DR, Jensen GJ. In vivo structures of an intact type VI secretion system revealed by electron cryotomography. EMBO Rep. 2017;18:1090–1099. doi: 10.15252/embr.201744072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Basler M. Type VI secretion system: secretion by a contractile nanomachine. Philos Trans R Soc Lond B Biol Sci. 2015;370 doi: 10.1098/rstb.2015.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Durand E, Nguyen VS, Zoued A, Logger L, Péhau-Arnaudet G, Aschtgen MS, Sinelli S, Desmyter A, Bardiaux B, Dujeancourt A, Roussel A, et al. Biogenesis and structure of a type VI secretion membrane core complex. Nature. 2015;523:555–560. doi: 10.1038/nature14667. [DOI] [PubMed] [Google Scholar]
- 10.Horn M, Harzenetter MD, Linner T, Schmid EN, Müller KD, Michel R, Wagner M. Members of the Cytophaga–Flavobacterium–Bacteroides phylum as intracellular bacteria of acanthamoebae: proposal of “Candidatus Amoebophilus asiaticus”. Environ Microbiol. 2001;3:440–449. doi: 10.1046/j.1462-2920.2001.00210.x. [DOI] [PubMed] [Google Scholar]
- 11.Schmitz-Esser S, Tischler P, Arnold R, Montanaro J, Wagner M, Rattei T, Horn M. The genome of the amoeba symbiont “Candidatus Amoebophilus asiaticus” reveals common mechanisms for host cell interaction among amoeba-associated bacteria. J Bacteriol. 2010;192:1045–1057. doi: 10.1128/JB.01379-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Penz T, Horn M, Schmitz-Esser S. The genome of the amoeba symbiont “Candidatus Amoebophilus asiaticus” encodes an afp-like prophage possibly used for protein secretion. Virulence. 2010;1:541–545. doi: 10.4161/viru.1.6.13800. [DOI] [PubMed] [Google Scholar]
- 13.Taylor NMI, Prokhorov NS, Guerrero-Ferreira RC, Shneider MM, Browning C, Goldie KN, Stahlberg H, Leiman PG. Structure of the T4 baseplate and its function in triggering sheath contraction. Nature. 2016;533:346–352. doi: 10.1038/nature17971. [DOI] [PubMed] [Google Scholar]
- 14.Shneider MM, Buth SA, Ho BT, Basler M, Mekalanos JJ, Leiman PG. PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature. 2013;500:350–353. doi: 10.1038/nature12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marko M, Hsieh C, Schalek R, Frank J, Mannella C. Focused-ion-beam thinning of frozen-hydrated biological specimens for cryo-electron microscopy. Nat Methods. 2007;4:215–217. doi: 10.1038/nmeth1014. [DOI] [PubMed] [Google Scholar]
- 16.Rigort A, Bäuerlein FJ, Leis A, Gruska M, Hoffmann C, Laugks T, Böhm U, Eibauer M, Gnaegi H, Baumeister W, Plitzko JM. Micromachining tools and correlative approaches for cellular cryo-electron tomography. J Struct Biol. 2010;172:169–179. doi: 10.1016/j.jsb.2010.02.011. [DOI] [PubMed] [Google Scholar]
- 17.Penz T, Schmitz-Esser S, Kelly SE, Cass BN, Müller A, Woyke T, Malfatti SA, Hunter MS, Horn M. Comparative Genomics Suggests an Independent Origin of Cytoplasmic Incompatibility in Cardinium hertigii. PLoS Genet. 2012;8:e1003012. doi: 10.1371/journal.pgen.1003012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Russell AB, Wexler AG, Harding BN, Whitney JC, Bohn AJ, Goo YA, Tran BQ, Barry NA, Zheng H, Peterson SB, Chou S, et al. A Type VI Secretion-Related Pathway in Bacteroidetes Mediates Interbacterial Antagonism. Cell Host & Microbe. 2014;16:227–236. doi: 10.1016/j.chom.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Büttner CR, Wu Y, Maxwell KL, Davidson AR. Baseplate assembly of phage Mu: Defining the conserved core components of contractile-tailed phages and related bacterial systems. Proc Natl Acad Sci USA. 2016;113:10174–10179. doi: 10.1073/pnas.1607966113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bönemann G, Pietrosiuk A, Diemand A, Zentgraf H, Mogk A. Remodelling of VipA/VipB tubules by ClpV-mediated threading is crucial for type VI protein secretion. EMBO J. 2009;28:315–325. doi: 10.1038/emboj.2008.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rybakova D, Radjainia M, Turner A, Sen A, Mitra AK, Hurst MR. Role of antifeeding prophage (Afp) protein Afp16 in terminating the length of the Afp tailocin and stabilizing its sheath. Mol Microbiol. 2013;89:702–714. doi: 10.1111/mmi.12305. [DOI] [PubMed] [Google Scholar]
- 22.Rybakova D, Schramm P, Mitra AK, Hurst MRH. Afp14 is involved in regulating the length of Anti-feeding prophage (Afp) Mol Microbiol. 2015;96:815–826. doi: 10.1111/mmi.12974. [DOI] [PubMed] [Google Scholar]
- 23.Wang IN, Smith DL, Young R. Holins: the protein clocks of bacteriophage infections. Annu Rev Microbiol. 2000;54:799–825. doi: 10.1146/annurev.micro.54.1.799. [DOI] [PubMed] [Google Scholar]
- 24.Leiman PG, Basler M, Ramagopal UA, Bonanno JB, Sauder JM, Pukatzki S, Burley SK, Almo SC, Mekalanos JJ. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc Natl Acad Sci USA. 2009;106:4154–4159. doi: 10.1073/pnas.0813360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wagner M, Haider S. New trends in fluorescence in situ hybridization for identification and functional analyses of microbes. Curr Opin Biotechnol. 2012;23:96–102. doi: 10.1016/j.copbio.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 26.Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–25. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Müller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 28.Weiss GL, Medeiros JM, Pilhofer M. In Situ Imaging of Bacterial Secretion Systems by Electron Cryotomography. Methods Mol Biol. 2017;1615:353–375. doi: 10.1007/978-1-4939-7033-9_27. [DOI] [PubMed] [Google Scholar]
- 29.Hayles MF, Stokes DJ, Phifer D, Findlay KC. A technique for improved focused ion beam milling of cryo-prepared life science specimens. J Microsc. 2007;226:263–269. doi: 10.1111/j.1365-2818.2007.01775.x. [DOI] [PubMed] [Google Scholar]
- 30.Zheng SQ, Keszthelyi B, Branlund E, Lyle JM, Braunfeld MB, Sedat JW, Agard DA. UCSF tomography: an integrated software suite for real-time electron microscopic tomographic data collection, alignment, and reconstruction. J Struct Biol. 2007;157:138–147. doi: 10.1016/j.jsb.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 31.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 32.Mastronarde DN. Correction for non-perpendicularity of beam and tilt axis in tomographic reconstructions with the IMOD package. J Microsc. 2008;230:212–217. doi: 10.1111/j.1365-2818.2008.01977.x. [DOI] [PubMed] [Google Scholar]
- 33.Nicastro D, Schwartz C, Pierson J, Gaudette R, Porter ME, McIntosh JR. The Molecular Architecture of Axonemes Revealed by Cryoelectron Tomography. Science. 2006;313:944–948. doi: 10.1126/science.1128618. [DOI] [PubMed] [Google Scholar]
- 34.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 35.Evans BC, Nelson CE, Yu SS, Beavers KR, Kim AJ, Li H, Nelson HM, Giorgio TD, Duvall CL. Ex Vivo Red Blood Cell Hemolysis Assay for the Evaluation of pH-responsive Endosomolytic Agents for Cytosolic Delivery of Biomacromolecular Drugs. J Vis Exp. 2013;73:50166. doi: 10.3791/50166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bleumink-Pluym NMC, van Alphen LB, Bouwman LI, Wösten MMSM, van Putten JPM. Identification of a functional type VI secretion system in Campylobacter jejuni conferring capsule polysaccharide sensitive cytotoxicity. PLoS Pathog. 2013;9:e1003393. doi: 10.1371/journal.ppat.1003393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Söding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 38.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Y, Schmidt B, Maskell DL. MSAProbs: multiple sequence alignment based on pair hidden Markov models and partition function posterior probabilities. Bioinformatics. 2010;26:1958–1964. doi: 10.1093/bioinformatics/btq338. [DOI] [PubMed] [Google Scholar]
- 40.Felsenstein J. PHYLIP - Phylogenetic Interference Package (Version 3.2) Cladistics. 1989;5:163–166. [Google Scholar]
- 41.Kudryashev M, Wang RY, Brackmann M, Scherer S, Maier T, Baker D, Dimaio F, Stahlberg H, Egelman EH, Basler M. Structure of the Type VI Secretion System Contractile Sheath. Cell. 2015;160:952–962. doi: 10.1016/j.cell.2015.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leiman PG, Arisaka F, van Raaij MJ, Kostyuchenko VA, Aksyuk AA, Kanamaru S, Rossmann MG. Morphogenesis of the T4 tail and tail fibers. Virol J. 2010;7:355. doi: 10.1186/1743-422X-7-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Planamente S, Salih O, Manoli E, Albesa-Jové D, Freemont PS, Filloux A. TssA forms a gp6-like ring attached to the type VI secretion sheath. EMBO J. 2016;35:1613–1627. doi: 10.15252/embj.201694024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.