Abstract
Although it has previously been shown that Neanderthals contributed DNA to modern humans1,2, not much is known about the genetic diversity of Neanderthals or the relationship between late Neanderthal populations at the time at which their last interactions with early modern humans occurred and before they eventually disappeared. Our ability to retrieve DNA from a larger number of Neanderthal individuals has been limited by poor preservation of endogenous DNA3 and contamination of Neanderthal skeletal remains by large amounts of microbial and present-day human DNA3–5. Here we use hypochlorite treatment6 of as little as 9 mg of bone or tooth powder to generate between 1- and 2.7-fold genomic coverage of five Neanderthals who lived around 39,000 to 47,000 years ago (that is, late Neanderthals), thereby doubling the number of Neanderthals for which genome sequences are available. Genetic similarity among late Neanderthals is well predicted by their geographical location, and comparison to the genome of an older Neanderthal from the Caucasus2,7 indicates that a population turnover is likely to have occurred, either in the Caucasus or throughout Europe, towards the end of Neanderthal history. We find that the bulk of Neanderthal gene flow into early modern humans originated from one or more source populations that diverged from the Neanderthals that were studied here at least 70,000 years ago, but after they split from a previously sequenced Neanderthal from Siberia2 around 150,000 years ago. Although four of the Neanderthals studied here post-date the putative arrival of early modern humans into Europe, we do not detect any recent gene flow from early modern humans in their ancestry.
The Middle to Upper Palaeolithic transition in Europe was characterized by major cultural and biological changes that coincided with the arrival of early modern humans and the disappearance of Neanderthals8,9. Analyses of the first Neanderthal genomes have provided evidence of gene flow from Neanderthals into modern humans between 50,000 and 60,000 years ago (ka), resulting in around 2% of Neanderthal DNA in the genomes of non-Africans today1,2,10. Additionally, genetic analyses of an approximately 39,000–42,000-year-old modern human from Romania (Oase 1) showed that interbreeding between Neanderthals and modern humans also happened in Europe at a later point in time11. However, little is known about the diversity of late Neanderthal populations across Europe and western Asia shortly before their disappearance, or about their relationship to the population that admixed with early modern humans. To date, only a handful of Neanderthal remains have been identified with a sufficiently high content of endogenous DNA and low enough levels of microbial and human DNA contamination3 to allow analysis of larger parts of their genomes1,2,7, limiting our ability to study their genetic history.
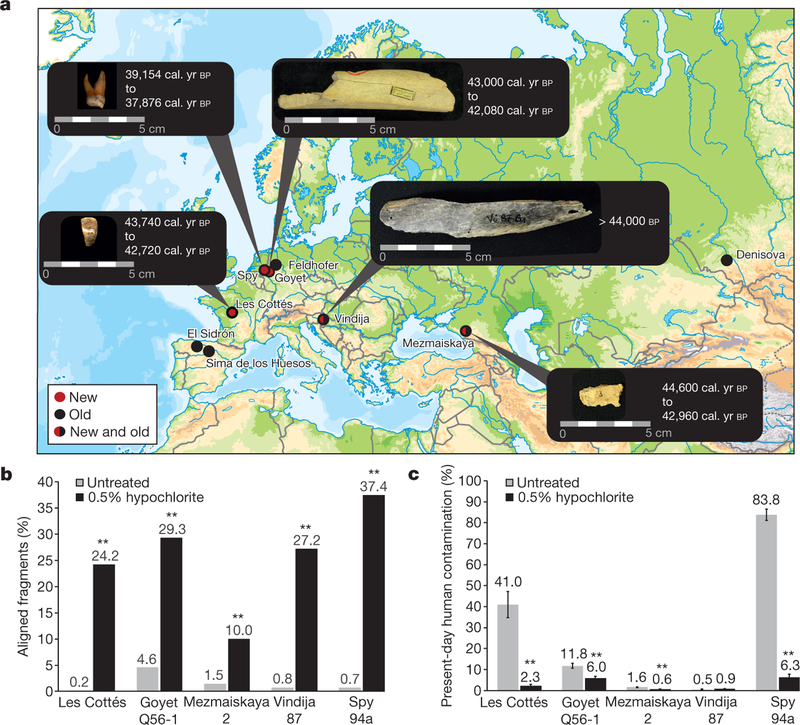
In an attempt to make more Neanderthal genomes available for population analyses, we identified five specimens with sufficient preservation of endogenous DNA to explore the possibility of nuclear genome sequencing (Fig. 1a and Supplementary Information 1): a fragment of a right femur (Goyet Q56–1) dated to 43,000–42,080 calibrated years (cal. yr) before present (bp; taken to be ad 1950) from the Troisième caverne of Goyet in Belgium12; an upper right molar (Spy 94a) that is associated with a maxillary fragment dated to 39,150–37,880 cal. yr bp from the neighbouring Spy cave in Belgium13; a tooth (Les Cottés Z4–1514) dated to 43,740–42,720 cal. yr bp from Les Cottés cave14 in France; an undiagnosed bone fragment found in Vindija cave in Croatia (Vindija 87) dated to be older than 44,000 uncalibrated years bp; and a skull fragment of an infant from Mezmaiskaya cave (Mezmaiskaya 2) in the Russian Caucasus dated to 44,600–42,960 cal. yr bp15.
Figure 1 |. Specimen information and the effects of 0.5% hypochlorite treatment.
a, Location and age of the five late Neanderthal specimens analysed in this study (new), and other sites for which genome-wide data of Neanderthal samples have previously been published (old). Map source: Vectormapcollection. b, Proportion of DNA fragments aligned to the human reference genome in untreated bone and tooth powder and in powder treated with 0.5% sodium hypochlorite (Supplementary Table 2.1). c, Proportion of present-day human contamination (with 95% binomial confidence intervals) inferred from mtDNA fragments in treated and untreated samples. Two-sided Pearson’s χ2 tests were used to calculate significant differences (**α ≪ 0.001) (Supplementary Table 2.5).
We extracted DNA from between 9 and 58 mg of bone or tooth powder. Approximately half of the powder from each specimen was treated with 0.5% hypochlorite solution before DNA extraction to remove present-day human and microbial DNA contamination6. Hypochlorite treatment increased the proportion of DNA fragments mapping to the human reference genome between 5.6- and 161-fold (Fig. 1b and Supplementary Information 2), and reduced present-day human contamination in four of the specimens between 2- and 18-fold (Fig. 1c and Supplementary Information 2). This substantial increase in the proportion of informative fragments made whole-genome sequencing of these previously inaccessible specimens feasible.
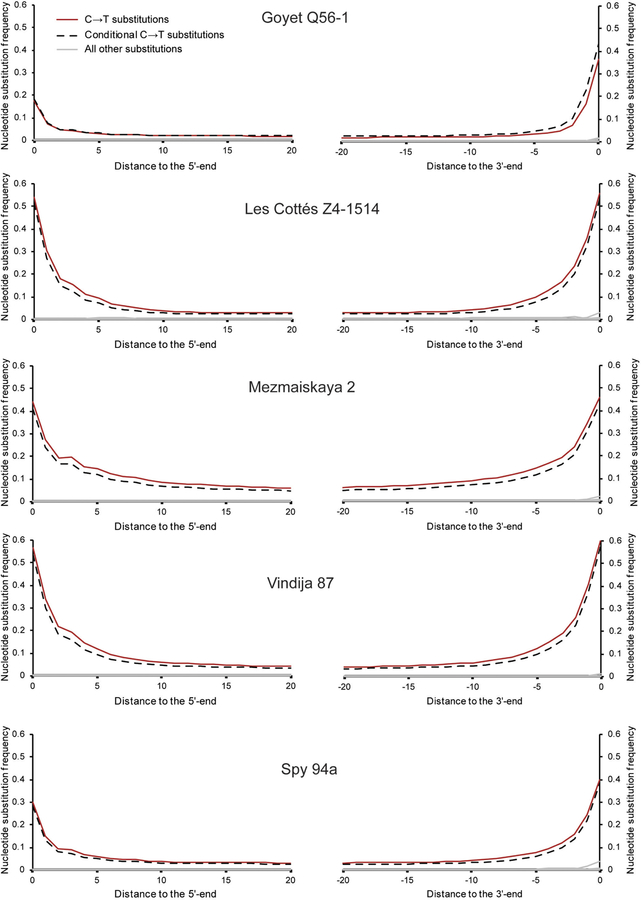
We generated additional single-stranded DNA libraries from selected extracts and sequenced them to an average genomic coverage of 2.7-fold for Les Cottés Z4–1514, 2.2-fold for Goyet Q56–1, 1.7-fold for Mezmaiskaya 2, 1.3-fold for Vindija 87 and 1-fold for Spy 94a (Extended Data Table 1 and Supplementary Information 3). Estimates of mitochondrial (mt) and nuclear DNA contamination ranged from 0.52% to 5.06% and from 0.18% to 1.75%, respectively (Supplementary Information 4). Therefore, we restricted analyses to fragments carrying cytosine (C) to thymine (T) substitutions at their ends as these derive from deamination of cytosine to uracil and indicate that DNA molecules are of ancient origin16,17 (Extended Data Figs 1, 2). This reduced the mtDNA contamination to 0.39–1.61% and the autosomal contamination to 0–0.81% (Supplementary Information 4). To mitigate the influence of deamination on genetic inferences, we further restricted the analyses to only transversion polymorphisms (see Supplementary Information 6).
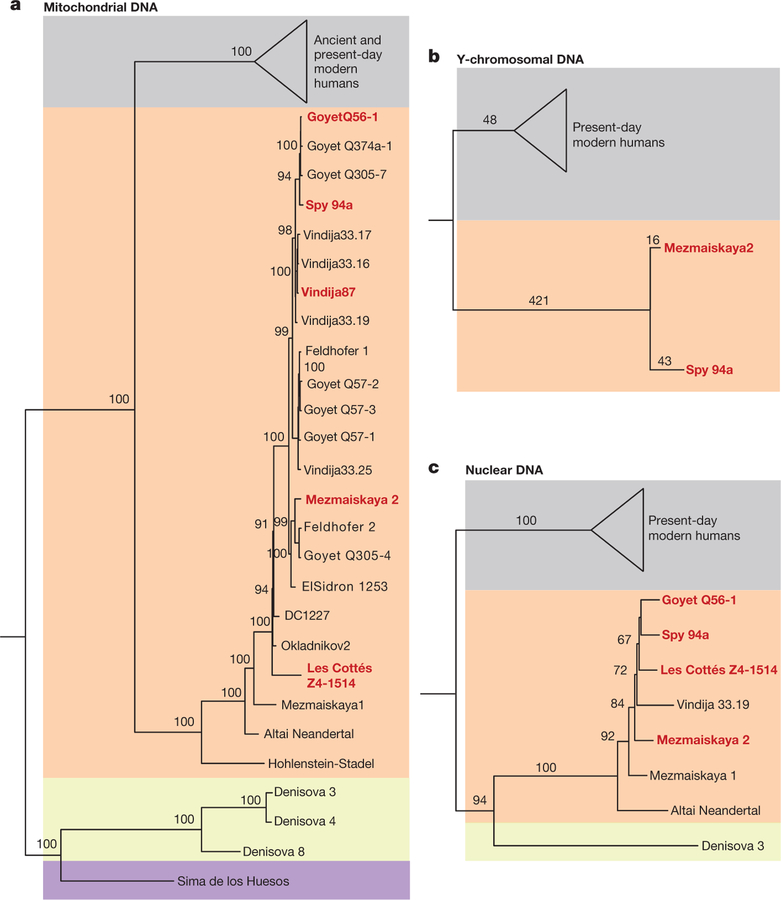
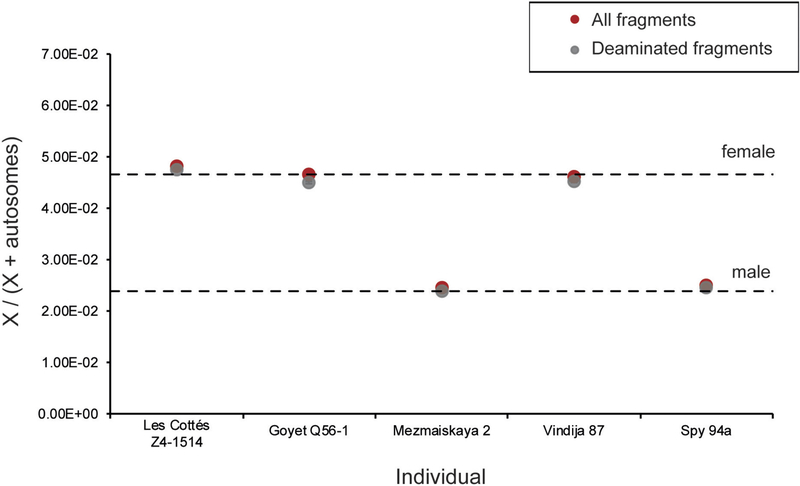
A phylogenetic tree (Fig. 2a) of the reconstructed complete mitochondrial genomes places these five specimens within the Neanderthal mtDNA variation. The relationship of the mtDNA of Les Cottés Z4–1514 to the mtDNAs of Neanderthals from across their entire geo-graphic range—including older Neanderthals from Okladnikov and Denisova cave (Supplementary Information 5)—together with the grouping of Mezmaiskaya 2 with late Neanderthals, including Feldhofer 2, challenges the previously proposed division between Eastern and Western mtDNAs in late-surviving Neanderthals18. As inferred from the sequence coverage of the X chromosome and the autosomes, Goyet Q56–1, Les Cottés Z4–1514 and Vindija 87 were females, whereas Mezmaiskaya 2 and Spy 94a were males (Extended Data Fig. 3). The Y chromosome sequences of both male individuals fall outside the known variation of present-day human Y chromosomes (Fig. 2b and Supplementary Information 5), as is the case for the Y chromosome of a Neanderthal from El Sidrón, Spain19.
Figure 2 |. Phylogenetic relationships of late Neanderthals.
a, Bayesian phylogenetic tree of mitochondrial genomes of 23 Neanderthals, 3 Denisovans, 64 modern humans and a hominin from Sima de los Huesos. The posterior probabilities for the branches are shown. b, Neighbour-joining tree of Y chromosome sequences of Mezmaiskaya 2, Spy 94a, 175 present-day humans21 and two present-day humans carrying the A00 haplogroup30. The number of substitutions is shown above the branches. c, Neighbour-joining tree of nuclear genomes based on autosomal transversions among late Neanderthals, Vindija 33.19, Mezmaiskaya 1, Altai Neanderthal, Denisovan and 12 present-day humans. Bootstrap support values after 1,000 replications are shown.
We analysed the genomes of the five late Neanderthals with the previously published high-quality genomes of an approximately 120,000-year-old Neanderthal from Siberia (Altai Neanderthal)2 and Neanderthal from Croatia (Vindija 33.19) that is more than 45,000 years old7, the low-coverage genome of a Neanderthal who lived approximately 60–70 ka from Mezmaiskaya cave (Mezmaiskaya 1)2,7, the composite low-coverage genome of three Neanderthals from Croatia1, the high-coverage genome of a Denisovan individual20 and a world-wide panel of present-day humans2,21. On the basis of derived alleles that are shared with the Vindija 33.19 genome7, we find that the Vindija 87 specimen originates from the same individual as Vindija 33.19. We therefore excluded Vindija 87 from subsequent analyses (Supplementary Information 7).
A neighbour-joining tree based on the number of pairwise trans-versions between individuals shows that all Neanderthals form a monophyletic clade relative to the Denisovan individual (Fig. 2c and Supplementary Information 7, 8). The tree reflects an apparent age-related division among the Neanderthals with the oldest specimen, the Altai Neanderthal branching off first, followed by Mezmaiskaya 1, whereas the late Neanderthals form a clade. To assess the relationship of the late Neanderthals to the high-coverage genomes of the Altai and Vindija 33.19 Neanderthals, we used two related statistics that measure the fraction of derived allele-sharing between the genomes. We find that all late Neanderthals and Mezmaiskaya 1 share significantly more derived alleles with Vindija 33.19 than with the Altai Neanderthal (−32.1 ≤ Z ≤ −58.1; Extended Data Table 2 and Supplementary Information 9) and that late Neanderthals share on average 49.0% (95% confidence interval, 44.2–54.2%) of the derived alleles seen in Vindija 33.19 and not in other high-coverage genomes in the analysis, whereas they share 17.2% (95% confidence interval, 15.9–18.3%) of the derived alleles in the Altai Neanderthal (Extended Data Table 3 and Supplementary Information 7).
Obtaining genome-wide data of multiple late Neanderthals from a broad geographical range enabled us to determine whether related-ness among Neanderthals is correlated to their geographical proximity, as is the case for present-day humans22. In support of this, we find that the two Neanderthals from Belgium share more derived alleles with each other than with any other Neanderthal (−3.65 ≤ Z ≤ −8.47; Supplementary Information 9), and in turn more derived alleles with Neanderthals from France and Croatia than with the late Neanderthal from the Caucasus. Similarly, the four Neanderthals from Vindija cave that come from a relatively narrow time range share more derived alleles with each other than with other Neanderthals (−2.2 ≤ Z ≤−14.5; Supplementary Information 9). Furthermore, specimens with a similar age and the largest geographical distance between them (Les Cottés Z4–1514 and Mezmaiskaya 2) shared the fewest derived alleles (Supplementary Information 9). By contrast, Mezmaiskaya 2 shared more derived alleles with the other late Neanderthals than with Mezmaiskaya 1 (−2.13 ≤ Z ≤ −9.56; Supplementary Information 9), suggesting that there was a population turnover towards the end of Neanderthal history. This turnover may have been the result of a population related to western Neanderthals replacing earlier Neanderthals in the Caucasus, or the replacement of Neanderthals in western Europe by a population related to Mezmaiskaya 2. The timing of this turnover coincides with pronounced climatic fluctuations during Marine Isotope Stage 3 between 60 and 24 ka23, when extreme cold periods in northern Europe may have triggered the local extinction of Neanderthal populations and subsequent re-colonization from refugia in southern Europe or western Asia24,25.
We estimated the population split times between each of the low-coverage Neanderthal genomes and the two high-coverage Neanderthal genomes by determining the fraction of sites at which each of the low-coverage Neanderthal genomes shares a derived allele that occurs in the heterozygote state in one of the high-quality genomes (F(A|B) statistics1,20). This fraction was then used to estimate the population split times for each pair of Neanderthals using previous inferences of how Neanderthal population sizes changed over time2,7. Owing to the uncertainties in the mutation rate and generation times, we caution that although the times presented are likely to accurately reflect the relative ages of the population split times, the absolute estimates in years are approximate. We estimate that the late Neanderthals studied here separated from a common ancestor with the Altai Neanderthal around 150 ka (95% confidence interval, 142–186 ka), and from a common ancestor with Vindija 33.19 about 70 ka (95% confidence interval, 58–72 ka; Extended Data Table 4 and Supplementary Information 8). The estimates of the population split times from the common ancestors shared with the Denisovan and with modern humans are around 400 ka (95% confidence interval, 367–484 ka) and about 530 ka (95% confidence interval, 503–565 ka; Extended Data Table 4 and Supplementary Information 8), respectively, consistent with previous estimates using the Altai and Vindija 33.19 Neanderthal genomes1,2,20.
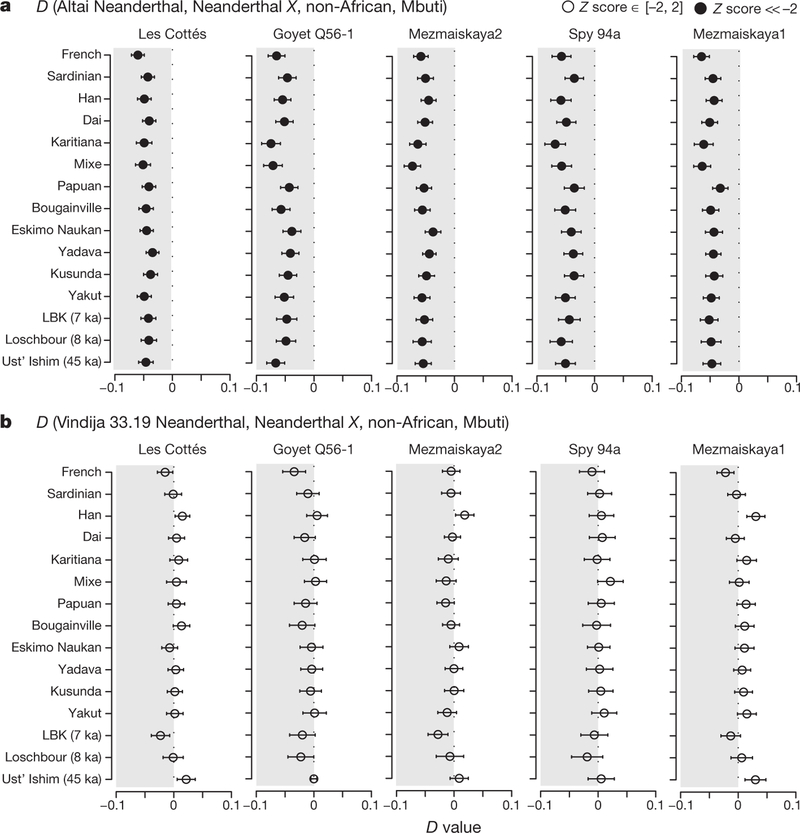
To investigate whether any of the Neanderthals sequenced to date is more closely related to the Neanderthal population that contributed genetic material to modern humans, we compared the Neanderthal genomes from this and previous studies to the genomes of 263 present-day humans21 as well as a number of early modern humans10,11,26,27. We find that all late Neanderthals and the older Mezmaiskaya 1 Neanderthal share significantly more derived alleles with the introgressing Neanderthals than the Altai Neanderthal does (−2.4 ≤ Z ≤−5.6; Fig. 3a and Supplementary Information 10), with no significant differences among them (−0.1 ≤ Z ≤ 1.8; Fig. 3b and Supplementary Information 10). Interestingly, this is also true for an approximately 45,000-year-old modern human from Siberia (Ust’-Ishim) (Fig. 3 and Supplementary Information 10), who was contemporaneous with late Neanderthals, but is not a direct ancestor of present-day humans10. Thus, the majority of gene flow into early modern humans appears to have originated from one or more Neanderthal populations that diverged from other late Neanderthals after their split from the Altai Neanderthal about 150 ka, but before the split from Mezmaiskaya 1 at least 90 ka (Extended Data Table 4). Owing to the scarcity of overlapping genetic data from Oase 1, whose genome revealed an unusually high percentage of Neanderthal ancestry11, we were unable to resolve whether one of these late Neanderthals was significantly closer than others to the introgressing Neanderthal in Oase 1.
Figure 3 |. Proximity to the introgressing Neanderthal populations in present-day and ancient humans calculated using D(Neanderthal1, Neanderthal2; non-African, African).
Three Mbuti individuals from the Simons Genome Diversity Project (SGDP)21 were used as an outgroup and standard errors were calculated using a weighted block jackknife (Supplementary Information 10). Shaded grey region corresponds to D < 0. a, All late Neanderthals and the older Mezmaiskaya 1 are significantly closer to the introgressing Neanderthal population(s) than the Altai Neanderthal. b, There is no significant difference between late Neanderthals, Mezmaiskaya 1 and Vindija 33.19 in their proximity to the introgressing Neanderthal population(s) in present-day and ancient humans.
Interbreeding between Neanderthals and early modern humans is likely to have occurred intermittently, presumably resulting in gene flow in both directions28. However, when we applied an approach that uses the extended length of haplotypes expected from recent introgression into the analysed late Neanderthals, we did not find any indications of recent gene flow from early modern humans to the late Neanderthals (Supplementary Information 11). We caution that given the small number of analysed Neanderthals we cannot exclude that such gene flow occurred. However, it is striking that Oase 1, one of two early modern humans that overlapped in time with late Neanderthals, showed evidence for recent additional Neanderthal introgression10,11 whereas none of the late Neanderthals analysed here do. This may indicate that gene flow affected the ancestry of modern human populations more than it did Neanderthals29. Further work is necessary to determine whether this was the case. Our work demonstrates that the generation of genome sequences from a large number of archaic human individuals is now technically feasible, and opens up the possibility to study Neanderthal populations across their temporal and geographical range.
METHODS
Data reporting.
No statistical methods were used to predetermine sample size. The experiments were not randomized and the investigators were not blinded to allocation during experiments and outcome assessment.
DNA extraction and library preparation.
All specimens were sampled in clean room facilities dedicated to the analysis of ancient DNA. Between 28 mg and 104 mg of tooth or bone powder was obtained by drilling once into the physically cleaned part of the specimen and split evenly (Supplementary Information 2; Supplementary Table 2.1). Approximately half of the powder was directly subjected to DNA extraction using a silica-based method31 as implemented in ref. 6 (the untreated sample), whereas the second half was treated with a 0.5% sodium hypochlorite solution6 before DNA extraction in an attempt to remove some of the microbial and present-day human DNA contamination3–5. Subsequently, 5 or 10 μl of each extract was converted into single-stranded DNA libraries32 with the modifications as in ref. 6. The initial libraries of Vindija 87, Goyet Q56–1 and Les Cottés Z4–1514 were treated with Escherichia coli uracil-DNA-glycosylase (UDG) and E. coli endonuclease VIII (Endo VIII)15,20 to excise uracils, while all other libraries were prepared without this enzymatic treatment (Supplementary Information 2). The libraries were amplified into plateau33 and tagged with two sample-specific indices6,34. An aliquot of each amplified library was additionally enriched for hominin mtDNA using a bead-based hybridization method and modern human mtDNA as a bait35–37. After analysing the first set of libraries, we selected the extracts with the highest proportion of endogenous DNA and the lowest levels of present-day human DNA contamination to produce the final set of single-stranded DNA libraries6,32 (Supplementary Information 2 and Supplementary Table 2.6).
Genome sequencing and data processing.
All libraries were initially sequenced together on Illumina’s MiSeq and HiSeq 2500 platforms to determine their suitability for whole-genome sequencing. Subsequently, 23 libraries from five Neanderthal specimens were selected and sequenced on 50 lanes of the Illumina HiSeq 2500 platform in rapid mode, using double index configuration (2× 76 bp)34 (Supplementary Information 2). Base calling was done using Bustard (Illumina) for the MiSeq runs and FreeIbis38 for the HiSeq runs. Adapters were trimmed and overlapping paired-end reads were merged into single sequences using leeHom39. The Burrows–Wheeler aligner (BWA, version 0.5.10-evan.9–1-g44db244; )40 was used to align the shotgun data to the modified human reference GRCh37 (ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/technical/reference/phase2_reference_assembly_sequence/) and to align the human mtDNA capture data to the revised Cambridge Reference Sequence (NC_01290) with parameters adjusted for ancient DNA (-n 0.01 –o 2 –l 16500)20. We developed a two-step algorithm called jivebunny (https://bioinf.eva.mpg.de/jivebunny) for de-multiplexing of the sequencing runs (Supplementary Information 3), retaining only fragments that were assigned to the correct library based on their index sequences for all of the downstream analyses. PCR duplicates were removed using bam-rmdup (version 0.6.3; ) and fragments were filtered for read length (≥35 bp) and mapping quality (MQ ≥ 25) using SAMtools (version 1.3.1)41.
The deamination of cytosine (C) to uracil (U) residues leaves characteristic C-to-T substitution patterns in ancient DNA molecules, which are particularly close to the alignment ends15 and thus provide evidence for the presence of authentic ancient DNA in specimens5,42 (Supplementary Information 2, 3 and Extended Data Fig. 1). We evaluated the frequency of these nucleotide substitution patterns characteristic of ancient DNA using an in-house Perl script. For determining the coverage of the nuclear genomes, we counted the number of bases with a base quality of at least 30 (BQ ≥ 30) in the fragments that overlapped highly mappable regions of the autosomes of the human genome (Map35_100% of ref. 2) and divided this number by the total length of those regions. We determined the sex of the Neanderthal individuals by counting the number of fragments that aligned to the X chromosome and the autosomes (Extended Data Fig. 3).
Contamination estimates.
We used four methods to estimate the proportion of present-day human DNA contamination in the final dataset (Supplementary Information 4). We estimated the proportion of mtDNA contamination by present-day human DNA using two different sets of positions. First, we re-aligned the shotgun data of all HiSeq runs to the revised Cambridge Reference Sequence (rCRS, NC_012920) using BWA40. We then counted the number of fragments overlapping 63 positions at which 18 published Neanderthal mitochondrial genomes1,2,12,43–46 differ from those of 311 present-day humans43. In the second approach, we determined positions in the reconstructed mtDNA genomes of each of the five Neanderthals that are specific for that Neanderthal compared to 311 present-day humans. To mitigate the effect of deamination for both approaches, we ignored the alignments on the forward or reverse strands at positions where the informative base was a C or a G17.
We estimated the extent of present-day human DNA contamination in the autosomal sequences of the five late Neanderthals using a maximum likelihood approach described in ref. 1, based on all sites covering informative positions in the nuclear genome at which humans carry a fixed derived variant when compared to the great apes. Additionally, we estimated levels of present-day human DNA contamination using an ancestry model in which each low-coverage Neanderthal traces a portion of the genome either from a high-coverage uncontaminated Neanderthal, or from a modern human population related to present-day non-Africans. We built a two-source qpAdm47 model in which one part of the ancestry was modelled as being closely related to the high-coverage genome of Vindija 33.19 Neanderthal, and the other source of ancestry was modelled as being most closely related to the Dinka population of the Simons Genome Diversity Project (SGDP)21. We estimate the proportion of male contamination for the three female individuals, Les Cottés Z4–1514, Goyet Q56–1 and Vindija 87 by counting the number of fragments aligning to the unique regions of the Y chromosome and divided it with the number of fragments that would be expected if the individual was a male (Supplementary Information 4 and Supplementary Table 4.1).
mtDNA and Y chromosomes of Neanderthals.
We re-aligned the shotgun data of all HiSeq runs to the Vindija 33.16 mitochondrial genome (AM948965)43 using BWA40 to reconstruct complete mitochondrial genomes of five Neanderthals. A consensus base was called at each position covered by at least three fragments and where at least two-thirds of fragments had an identical base17 (Supplementary Information 5). We aligned the reconstructed mitochondrial genomes of the five late Neanderthals to the mtDNA genomes of 18 Neanderthals2,12,43–46,48,49, 311 present-day humans43, 10 ancient modern humans5,10,36,50–52, 3 Denisovans53–55, a hominin from Sima de los Huesos17 and a chimpanzee56 using MAFFT57 and calculated the number of pairwise differences among mitochondrial genomes using MEGA658. Bayesian phylogenetic analysis was performed using BEAST version 2.4.559 to infer the time to the most recent common ancestor of all Neanderthal mitochondrial genomes (Supplementary Information 5). Radiocarbon dates of Neanderthals and ancient modern humans were used as calibration points for the molecular clock and we used jModelTest260 and marginal likelihood estimation analysis61,62 to choose the best fitting substitution, clock and tree model (Supplementary Information 5).
For reconstructing the phylogeny of the Neanderthal Y chromosomes, we compared the two male Neanderthal individuals, Mezmaiskaya 2 and Spy 94a, to 175 present-day human males from the SGDP21 and two present-day humans with haplogroup A0030. For processing the Y chromosomal data of the presented Neanderthal individuals, we followed the processing of the previously published Neanderthal Y chromosome analysis19 and used the described parameters to call bases and infer genotypes (Supplementary Information 5).
Set of filters for nuclear data analyses.
As the data presented in this study were of low-coverage and contained elevated C-to-T substitutions, we investigated spurious correlations stemming from the properties of the data using D-statistics1,54,63 (Supplementary Information 6). All of the sampling schemes, except random read sampling and then restricting to transversion polymorphisms, as well as simulation of UDG treatment, resulted in significant spurious correlations between individuals (Supplementary Information 6 and Supplementary Table 6.1). Therefore, we applied random read sampling at each position in the genome that was covered for the low-coverage individuals. To diminish the effect of present-day human DNA contamination and enrich for the endogenous fragments17, we further selected the fragments that showed C-to-T substitutions relative to the human reference genome at the first three and/or the last three positions, that is, putatively deaminated fragments. The newly generated sequencing data of Mezmaiskaya 17, an approximately 60,000–70,000-year-old sample of a Neanderthal from Russia, were processed in the same way as the data of the other low-coverage individuals. For the high-coverage genomes of Altai2 and Vindija 33.197 Neanderthals, as well as the Denisovan20 individual, snpAD genotype calls (http://cdna.eva.mpg.de/neandertal/Vindija/VCF/) were used. For comparison to modern humans, we used hetfa files of 263 present-day human genomes of the SGDP21 and snpAD genotype calls of three high quality ancient modern humans, Ust’-Ishim, an approximately 45,000-year-old modern human from Siberia10; Loschbour, an around 8,000-year-old hunter-gatherer from Luxembourg26; and LBK, an approximately 7,000-year-old farmer from Stuttgart26. Variant sites across all genomes were extracted (https://bioinf.eva.mpg.de/heffalump) and converted into an input format for AdmixTools (version 4.1)63 or exported into combined VCF files. We further restricted all analyses to bi-allelic sites in the genome covered by at least one low-coverage Neanderthal genome and to transversion polymorphisms.
Principal component analysis.
We carried out principal component analysis64,65 using genomes of Vindija 33.197, the Altai Neanderthal2 and the Denisovan individual20 to estimate the eigenvectors of the genetic variation and then projected the low-coverage Neanderthal genomes onto the defined plane. This allowed us to explore the relationship of low-coverage Neanderthal genomes relative to the high-coverage Neanderthal genomes (Extended Data Fig. 4).
Lineage attribution and average sequence divergence of late Neanderthals.
We followed an approach described in ref. 66 based on the sharing of derived alleles with a certain hominin group to determine more precisely to which hominin group the nuclear genomes of Les Cottés Z4–1514, Goyet Q56–1, Spy 94a, Mezmaiskaya 2 and Vindija 87 are most closely related. We investigated the state of DNA fragments overlapping the positions at which the high-coverage genomes of the Altai Neanderthal2, the Vindija 33.19 Neanderthal7, the Denisovan individual20 and a present-day African (Mbuti, HGDP00982)2 differ from those of the great apes (chimpanzee, bonobo, gorilla and orangutan). We then determined the proportion of fragments for each of the low-coverage Neanderthal genomes that supported the derived state of each of the branches in the phylogenetic tree relating the four high-coverage genomes66 (Extended Data Table 3, Supplementary Information 7 and Supplementary Table 7.1).
We estimated the divergence of the five late Neanderthal genomes along the lineage from the ancestor shared with the chimpanzee and the high-coverage genomes of the Altai and Vindija 33.19 Neanderthals, the Denisovan individual, or a present-day human from the B-panel of ref. 2, using the triangulation method previously applied to a number of ancient genomes1,2,20,54,55. We calculated how many of the substitutions that were inferred to have occurred from the human– chimpanzee ancestor to the high-coverage genomes that occurred after the split from the low-coverage genome (Supplementary Information 7 and Supplementary Tables 7.2, S7.3). Standard errors were computed by a weighted block jackknife67 with a block size of 5 million base pairs (5 Mb) across all autosomes.
Split times of late Neanderthals and the neighbour-joining tree of nuclear genomes.
We conditioned on the heterozygous sites in the two high-coverage Neanderthals, and then computed the fraction of sites that show the same derived allele in randomly sampled fragments of the low-coverage individuals in order to estimate the split times between the low-coverage Neanderthals and the Altai2 and Vindija 33.197 Neanderthals (Extended Data Table 4 and Supplementary Information 8). In this F(A|B) statistic1,2,20, the expected value depends only on the demography of the high-coverage individuals from which heterozygous sites are determined. We performed these analyses on all randomly sampled fragments, and on deaminated fragments only (Extended Data Table 4 and Supplementary Information 8).
We used the low-coverage nuclear genomes of Les Cottés Z4–1514, Goyet Q56–1, Spy 94a, Mezmaiskaya 2 and Mezmaiskaya 1, the high-coverage genomes of Vindija 33.19, Altai Neanderthal, the Denisovan individual and 12 present-day humans from ref. 2 for the construction of a neighbour-joining tree. We counted the total number of transversions between all pairs of individuals and the human– chimpanzee common ancestor2,54. We constructed a neighbour-joining tree68 based on the pairwise number of transversions in windows of 5 Mb across all autosomes between all individuals with 1,000 bootstrap replications (Supplementary Information 8). The tree was constructed as implemented in the R-package phangorn69 and visualized with FigTree (version 1.4.2)(http://tree.bio.ed.ac.uk/software/figtree/).
Inferring the relationships between Neanderthals.
We used D-statistics1,54,63 to investigate the population relationships among Neanderthal individuals. We co-analysed all low-coverage Neanderthal genomes with the high-coverage genomes of the Altai2 and Vindija 33.197 Neanderthals. We used the whole-genome alignments of the chimpanzee, orangutan and rhesus macaque to the human reference genome70–72 to infer the ancestral states for the analyses of D(Altai, Vindija 33.19; Neanderthal, outgroup). Furthermore, we used the genomes of the Dinka and Mbuti individuals from the SGDP21 as outgroups for the statistics of D(Neanderthal1, Neanderthal2; Neanderthal3, outgroup). The standard errors were computed using a weighted block jackknife63,67 with equally sized blocks of 5 Mb over all autosomes. We further restricted these analyses to bi-allelic sites in the genome covered by at least one low-coverage Neanderthal and transversion polymorphisms.
Inferring the relationship to the introgressed Neanderthals in present-day and ancient modern humans.
We analysed the low-coverage late Neanderthal genomes together with the high-coverage genomes of the Altai2 and Vindija 33.19 Neanderthals7, the high-coverage genomes of the Denisovan individual20 and 263 present-day humans of the SGDP21 (Supplementary Information 10). We included Ust’-Ishim, an approximately 45,000-year-old modern human from Siberia10; Loschbour, an around 8,000-year-old hunter-gatherer from Luxembourg26; and LBK, an around 7,000-year-old farmer from Stuttgart26 to study the differences among Neanderthals in their proximity to the introgressed Neanderthal DNA detected in ancient modern humans. Analyses were restricted to transversion polymorphisms and to bi-allelic sites in the genome covered by at least one low-coverage Neanderthal. As above, D-statistics were used to infer the relationships between individuals1,54,63 and standard errors were computed using a weighted block jackknife63,67 over all autosomes (block size: 5 Mb).
Early modern human gene flow into late Neanderthals.
We modelled admixture from modern humans into Neanderthals with an ascertainment scheme in which both the Denisovan individual and Altai Neanderthal were fixed for the ancestral allele and at least half of the alleles in present-day African populations are derived. We applied the method as previously described73 and estimated the date of recent early modern human admixture into Neanderthals to be around 10–100 generations ago. At each single-nucleotide polymorphism in the genome, we considered data from all Yoruba individuals from the 1000 Genomes Project74 covered by at least three fragments that passed a pre-defined set of filters. Furthermore, we restricted the analysis to sites in the genome at which ≥24 Yoruba individuals as well as the Altai Neanderthal and Denisovan individual had allele calls (Map35_50% filter from ref. 2). The ancestral states were taken from the inferred ancestor of humans and chimpanzees (Ensembl Compara version 64)75,76. We introduced a more complex demographic history that is loosely based on the model described in ref. 77 (details in Supplementary Information 11).
Extended Data
Extended Data Figure 1 |. Frequency of nucleotide substitutions at the beginning and the end of nuclear alignments for the final dataset of Les Cottés Z4–1514, Goyet Q56–1, Mezmaiskaya 2, Vindija 87 and Spy 94a.
Only fragments of at least 35 bp that mapped to the human reference genome with a mapping quality of at least 25 (MQ ≥ 25) were used for this analysis. Solid lines depict all fragments and dashed lines the fragments that have a C-to-T substitution at the opposing end (‘conditional’ C-to-T substitutions). All other types of substitutions are marked in grey.
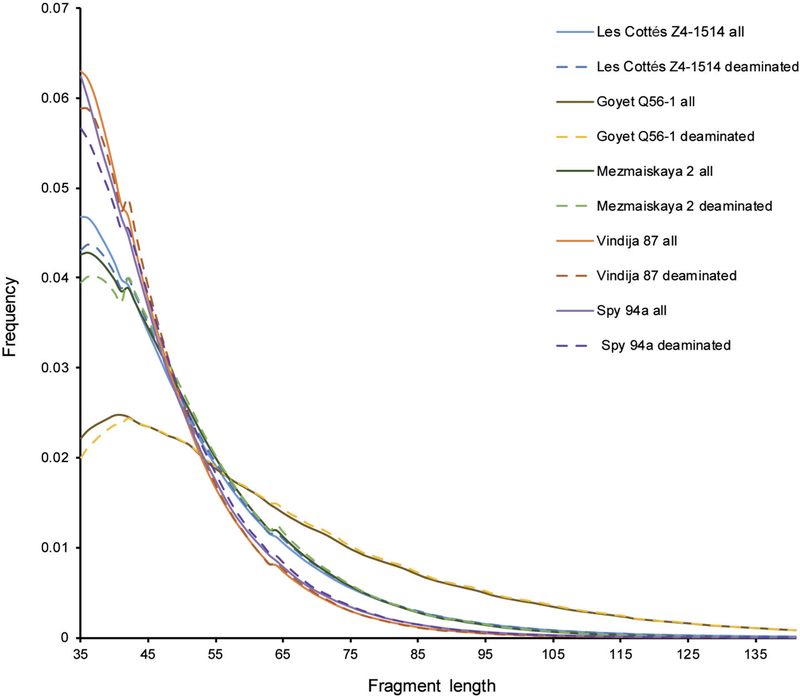
Extended Data Figure 2 |. Fragment size distribution of fragments longer than 35 bp mapped to the human reference genome with MQ ≥ 25 for each of the five late Neanderthals.
All fragments are depicted in solid lines and fragments with C-to-T substitutions to the reference genome (putatively deaminated fragments) are depicted with dashed lines.
Extended Data Figure 3 |. Sex determination based on the number of fragments aligning to the X chromosome and the autosomes.
The expected ratios of X to (X + autosomal) fragments for a female and a male individual are depicted as dashed lines. The results were concordant for all fragments (in red) and for deaminated fragments only (in grey).
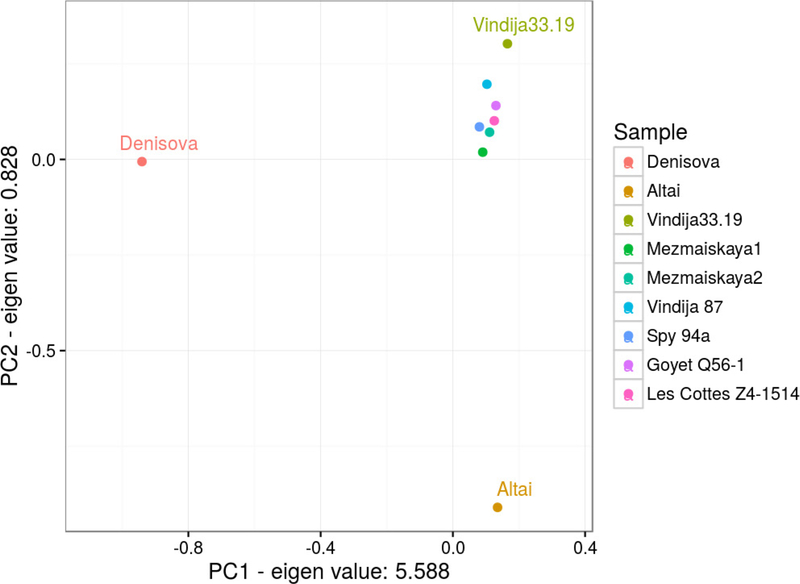
Extended Data Figure 4 |. Principal component analysis of the genomes of Vindija 33.19, Altai, the Denisovan individual, five late Neanderthals and Mezmaiskaya 1.
Genomes of the high-coverage archaics were used to estimate the eigenvectors of the genetic variation and low-coverage Neanderthals were projected onto the plane. Only transversion polymorphisms and bi-allelic sites were considered for the analysis, to a total of 1,010,417 sites as defined by the high-coverage genomes. PC, principal component.
Extended data table 1 |.
Amount of data generated for Les Cottés Z4–1514, Goyet Q56–1, Mezmaiskaya 2, Vindija 87 and Spy 94a
| Specimen | All fragments | Fragments with terminal C-to-T substitutions to the reference genome | ||||||
|---|---|---|---|---|---|---|---|---|
| Number of sequenced fragments | Number of fragments ≥ 35 bp | Number of mapped fragments ≥ 35 bp, MQ ≥ 25, Map35_100% | Number of unique fragments ≥ 35 bp, MQ ≥ 25, Map35_100% | Obtained nuclear coverage | Number of mapped fragments ≥ 35 bp, MQ ≥ 25, Map35_100% | Number of unique fragments ≥ 35 bp, MQ ≥ 25, Map35_100% | Obtained nuclear coverage | |
| Les Cottés Z4–1514 | 1,776,108,129 | 978,981,412 | 253,134,472 | 121,336,498 | 2.71 | 59,777,476 | 41,804,332 | 1.00 |
| Goyet Q56–1 | 917,798,995 | 575,597,226 | 124,582,445 | 80,515,019 | 2.18 | 13,585,646 | 9,859,280 | 0.27 |
| Mezmaikaya 2 | 2,571,128,011 | 1,356,302,134 | 125,815,832 | 74,407,074 | 1.74 | 32,838,281 | 23,521,855 | 0.56 |
| Vindija 87 | 1,353,546,508 | 678,005,556 | 110,385,510 | 60,034,976 | 1.27 | 32,597,516 | 22,828,056 | 0.49 |
| Spy 94a | 733,212,063 | 278,595,622 | 72,962,863 | 46,836,553 | 1.00 | 14,301,192 | 10,003,560 | 0.22 |
The number of fragments after merging all of the sequencing libraries together is shown. The coverage of the nuclear genomes is determined by counting the number of bases with a base quality of at least 30 in fragments longer than 35 bp with MQ ≥ 25 that overlapped highly mappable regions of autosomes of the human genome, and dividing that number by the total length of these regions.
Extended data table 2 |.
relationship of the late Neanderthals and Mezmaiskaya 1 to the Altai and Vindija 33.19 Neanderthals calculated as D(Altai, Vindija 33.19, Neanderthal, outgroup) for all fragments and deaminated fragments, restricted to transversions
| D (Altai, Vindija 33.19, Neanderthal, outgroup) | All fragments | Deaminated fragments | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| W | X | Y | Z | % D | Z-score | BABA | ABBA | % D | Z-score | BABA | ABBA |
| Altai | Vindija33.19 | Les Cottés Z4–1514 | Dinka | 48.87 | −55.31 | 30,276 | 10,399 | 49.08 | −52.83 | 19,048 | 6,506 |
| Altai | Vindija33.19 | Les Cottés Z4–1514 | Mbuti | 49.08 | −57.83 | 30,340 | 10,362 | 49.34 | −55.25 | 19,077 | 6,471 |
| Altai | Vindija33.19 | Les Cottés Z4–1514 | Chimp | 49.16 | −59.83 | 29,427 | 10,028 | 49.38 | −56.69 | 18,597 | 6,302 |
| Altai | Vindija33.19 | Les Cottés Z4–1514 | Orang | 48.88 | −60.25 | 28,149 | 9,664 | 49.15 | −57.21 | 17,775 | 6,060 |
| Altai | Vindija33.19 | Les Cottés Z4–1514 | Rhesus | 48.35 | −58.96 | 25,856 | 9,003 | 48.38 | −55.77 | 16,281 | 5,664 |
| Altai | Vindija33.19 | Goyet Q56–1 | Dinka | 55.87 | −66.92 | 33,208 | 9,403 | 56.32 | −56.88 | 8,673 | 2,423 |
| Altai | Vindija33.19 | Goyet Q56–1 | Mbuti | 56.00 | −68.27 | 33,269 | 9,383 | 56.32 | −58.37 | 8,689 | 2,428 |
| Altai | Vindija33.19 | Goyet Q56–1 | Chimp | 55.96 | −68.97 | 32,188 | 9,090 | 56.76 | −59.25 | 8,485 | 2,340 |
| Altai | Vindija33.19 | Goyet Q56–1 | Orang | 55.68 | −68.11 | 30,735 | 8,749 | 56.79 | −59.39 | 8,125 | 2,239 |
| Altai | Vindija33.19 | Goyet Q56–1 | Rhesus | 55.40 | −67.04 | 28,312 | 8,125 | 56.31 | −58.09 | 7,483 | 2,092 |
| Altai | Vindija33.19 | Mezmaiskaya1 | Dinka | 36.58 | −36.63 | 20,392 | 9,468 | 37.09 | −32.69 | 8,379 | 3,845 |
| Altai | Vindija33.19 | Mezmaiskaya1 | Mbuti | 36.80 | −37.33 | 20,516 | 9,479 | 37.43 | −33.41 | 8,438 | 3,842 |
| Altai | Vindija33.19 | Mezmaiskaya1 | Chimp | 36.70 | −38.66 | 19,860 | 9,197 | 37.00 | −33.45 | 8,126 | 3,736 |
| Altai | Vindija33.19 | Mezmaiskaya1 | Orang | 36.18 | −37.38 | 18,904 | 8,860 | 36.77 | −32.27 | 7,758 | 3,586 |
| Altai | Vindija33.19 | Mezmaiskaya1 | Rhesus | 36.00 | −37.77 | 17,433 | 8,204 | 36.36 | −32.05 | 7,129 | 3,327 |
| Altai | Vindija33.19 | Mezmaiskaya2 | Dinka | 47.44 | −42.97 | 26,802 | 9,553 | 46.84 | −40.59 | 12,477 | 4,516 |
| Altai | Vindija33.19 | Mezmaiskaya2 | Mbuti | 47.83 | −47.39 | 26,781 | 9,451 | 47.13 | −44.43 | 12,471 | 4,481 |
| Altai | Vindija33.19 | Mezmaiskaya2 | Chimp | 47.92 | −51.11 | 26,019 | 9,160 | 47.37 | −47.06 | 12,233 | 4,369 |
| Altai | Vindija33.19 | Mezmaiskaya2 | Orang | 47.86 | −53.48 | 24,921 | 8,788 | 47.07 | −47.53 | 11,700 | 4,210 |
| Altai | Vindija33.19 | Mezmaiskaya2 | Rhesus | 47.33 | −49.32 | 22,908 | 8,189 | 46.65 | −44.74 | 10,799 | 3,929 |
| Altai | Vindija33.19 | Spy 94a | Dinka | 54.86 | −56.62 | 21,416 | 6,242 | 55.17 | −50.39 | 6,558 | 1,895 |
| Altai | Vindija33.19 | Spy 94a | Mbuti | 55.18 | −59.76 | 21,453 | 6,196 | 55.53 | −52.89 | 6,567 | 1,878 |
| Altai | Vindija33.19 | Spy 94a | Chimp | 55.32 | −60.58 | 20,814 | 5,988 | 55.39 | −50.67 | 6,354 | 1,824 |
| Altai | Vindija33.19 | Spy 94a | Orang | 55.01 | −60.26 | 19,793 | 5,746 | 55.33 | −50.10 | 6,052 | 1,740 |
| Altai | Vindija33.19 | Spy 94a | Rhesus | 54.46 | −58.94 | 18,179 | 5,360 | 54.93 | −48.52 | 5,584 | 1,624 |
All late Neanderthals and Mezmaiskaya 1 are significantly closer to Vindija 33.19 than to the Altai Neanderthal, irrespective of the outgroup used. Blue denotes Z << − 2. A total of 1,567,449 sites that are transversions among Neanderthals and for which at least one late Neanderthal has coverage were used. Chimp, chimpanzee; Orang, orangutan; Rhesus, rhesus macaque.
Extended data table 3 |.
the fraction of derived alleles among putatively deaminated fragments that each of the low-coverage individuals shares with the Altai Neanderthal, Vindija 33.19, the denisovan individual and a present-day human genome
| Human (%) | Neanderthal (%) | Altai Neanderthal (%) | Vindija 33.19 (%) | Denisovan (%) | Neanderthal-Denisova (%) | Human-Neanderthal (%) | Human-Denisova (%) | |
|---|---|---|---|---|---|---|---|---|
| Les Cottés Z4–1514 | 0.67 [0.65–0.70] | 93.01 [92.89–93.13] | 18.00 [17.69–18.32] | 46.41 [46.06–46.77] | 0.86 [0.82–0.89] | 97.90 [97.83–97.98] | 97.30 [97.17–97.43] | 3.22 [3.07–3.38] |
| Goyet Q56–1 | 0.60 [0.56–0.65] | 94.38 [94.19–94.57] | 16.47 [15.93–17.02] | 53.55 [52.91–54.18] | 0.80 [0.74–0.86] | 98.17 [98.04–98.29] | 97.72 [97.49–97.92] | 2.52 [2.29–2.78] |
| Spy 94a | 0.71 [0.66–0.77] | 93.89 [93.65–94.12] | 16.52 [15.90–17.17] | 51.45 [50.70–52.21] | 0.80 [0.73–0.87] | 97.83 [97.66–97.99] | 97.62 [97.35–97.87] | 3.06 [2.75–3.39] |
| Vindija 87 | 0.36 [0.33–0.38] | 97.08 [96.97–97.19] | 6.81 [6.53–7.09] | 75.93 [75.49–76.36] | 0.37 [0.34–0.40] | 99.05 [98.98–99.12] | 98.91 [98.79–99.02] | 1.38 [1.24–1.53] |
| Mezmaiskaya 2 | 0.65 [0.62–0.69] | 92.78 [92.62–92.93] | 17.91 [17.51–18.31] | 44.65 [44.19–45.12] | 0.83 [0.79–0.88] | 97.74 [97.63–97.84] | 97.53 [97.36–97.68] | 3.38 [3.18–3.59] |
| Mezmaiskaya 1 | 0.76 [0.72–0.80] | 91.87 [91.67–92.06] | 20.30 [19.81–20.80] | 38.38 [37.85–38.91] | 0.97 [0.91–1.03] | 97.61 [97.48–97.73] | 96.80 [96.58–97.00] | 3.73 [3.49–4.00] |
| Vindija 33.16 | 3.60 [3.50–3.71] | 93.39 [93.18–93.59] | 17.09 [16.55–17.65] | 58.99 [58.36–59.63] | 3.94 [3.81–4.08] | 96.14 [95.95–96.32] | 95.46 [95.15–95.74] | 4.89 [4.56–5.24] |
| Vindija 33.25 | 3.04 [2.94–3.15] | 94.17 [93.95–94.37] | 16.69 [16.10–17.30] | 60.81 [60.11–61.49] | 3.41 [3.28–3.55] | 96.53 [96.33–96.72] | 95.79 [95.46–96.09] | 3.77 [3.46–4.12] |
| Vindija 33.26 | 3.49 [3.38–3.62] | 93.54 [93.31–93.77] | 17.04 [16.43–17.67] | 59.65 [58.94–60.36] | 3.70 [3.55–3.85] | 96.25 [96.04–96.45] | 95.40 [95.04–95.72] | 4.56 [4.21–4.95] |
| Feldhofer 1 | 4.71 [2.40–9.01] | 94.38 [87.51–97.58] | 20 [8.86–39.13] | 50.00 [34.07–65.93] | 1.80 [0.50–6.33] | 90.63 [81.02–95.63] | 96.00 [80.46–99.29] | 0 [0–12.87] |
| El Sidron 1253 | 1.29 [0.44–3.72] | 90.99 [84.21–95.03] | 17.50 [8.75–31.95] | 42.86 [30.02–56.73] | 4.35 [2.22–8.34] | 95.24 [88.39–98.13] | 97.44 [86.82–99.55] | 2.33 [0.41–12.06] |
| Denisova 4 | 2.63 [1.51–4.54] | 2.31 [1.06–4.94] | 2.13 [0.59–7.43] | 2.04 [0.56–7.14] | 71.43 [65.87–76.40] | 96.15 [91.86–98.23] | 10.00 [5.15–18.51] | 90.32 [80.45–95.49] |
| Denisova 8 | 1.60 [1.39–1.85] | 6.75 [6.13–7.43] | 1.75 [1.23–2.48] | 1.83 [1.37–2.45] | 60.02 [58.92–61.12] | 92.53 [91.66–93.32] | 15.32 [13.72–17.06] | 87.72 [86.01–89.25] |
The state of DNA fragments overlapping the positions at which the high-coverage genomes of the Altai Neanderthal, the Vindija 33.19 Neanderthal, the Denisovan individual and a present-day African (Mbuti, HGDP00982) differ from those of the great apes were investigated. Fragments longer than 35 bp with MQ ≥ 25 and within the highly mappable regions of the genome that had terminal C-to-T substitutions reported in the Supplementary Table 3.2 were used. 95% binomial confidence intervals are provided in brackets.
Extended data table 4 |.
time of separation of late Neanderthals and Mezmaiskaya 1 (A) from the ancestor with the high-coverage genomes of Altai and Vindija 33.19 Neanderthals, denisovan individual and a present-day human (B), when measured in terms of time of split from the B individual (split A–B), or time from present (split-time + branch shortening)
| A | B | All fragments |
Deaminated fragments |
||||||
|---|---|---|---|---|---|---|---|---|---|
| % F(A|B) | Split A-B (ky) | Split time + bs (kya) | 95% CI (kya) | % F(A|B) | Split A-B (ky) | Split time + bs (kya) | 95% CI (kya) | ||
| Les Cottés Z4–1514 | Altai | 35.2 | 35.0 | 157.5 | 144.6–160.7 | 34.7 | 37.8 | 160.2 | 156.4–163.5 |
| Goyet Q56–1 | Altai | 35.8 | 22.3 | 144.7 | 142.8–157.6 | 35.9 | 22.2 | 144.6 | 141.5–159.3 |
| Mezmaiskaya 2 | Altai | 35.4 | 33.8 | 156.2 | 143.9–159.7 | 34.2 | 40.2 | 162.6 | 158.7–166.5 |
| Spy 94a | Altai | 34.4 | 39.2 | 161.7 | 158.2–165.2 | 33.6 | 43.1 | 165.5 | 160.4–186.2 |
| Vindija 87 | Altai | 35.2 | 35.4 | 157.8 | 144.7–161.1 | 34.6 | 38.3 | 160.8 | 156.2–164.8 |
| Mezmaiskaya 1 | Altai | 34.8 | 37.4 | 159.9 | 156.1–163.0 | 34.4 | 42.9 | 165.3 | 161.1–178.6 |
| Les Cottés Z4–1514 | Vindija 33.19 | 35.3 | 15.1 | 66.9 | 66.1–68.7 | 34.3 | 18.3 | 70.0 | 67.9–72.4 |
| Goyet Q56–1 | Vindija 33.19 | 37.8 | 8.3 | 60.1 | 58.8–61.7 | 38.0 | 8.1 | 59.9 | 57.8–61.9 |
| Mezmaiskaya 2 | Vindija 33.19 | 35.6 | 14.3 | 66.1 | 64.3–67.7 | 34.3 | 18.3 | 70.0 | 67.4–74.5 |
| Spy 94a | Vindija 33.19 | 36.9 | 10.4 | 62.2 | 60.6–64.3 | 37.2 | 9.9 | 62.7 | 59.1–64.8 |
| Vindija 87 | Vindija 33.19 | 46.8 | 0.0 | 51.8 | 51.8–51.8 | 45.0 | 0.0 | 61.7 | 51.8–51.8 |
| Mezmaiskaya 1 | Vindija 33.19 | 31.6 | 42.0 | 93.8 | 81.5–96.9 | 30.6 | 46.9 | 98.7 | 94.5–102.9 |
| Les Cottés Z4–1514 | Denisovan | 12.6 | 333.1 | 405.1 | 381.9–448.2 | 12.2 | 346.7 | 418.7 | 390.4–470.8 |
| Goyet Q56–1 | Denisovan | 12.8 | 328.6 | 400.6 | 377.3–438.0 | 13.1 | 322.4 | 394.3 | 367.1–434.0 |
| Mezmaiskaya 2 | Denisovan | 12.4 | 338.2 | 410.2 | 387.0–458.4 | 11.9 | 362.0 | 434.0 | 398.9–483.8 |
| Spy 94a | Denisovan | 12.5 | 336.0 | 407.9 | 384.7–451.6 | 12.4 | 339.4 | 411.3 | 382.4–470.8 |
| Vindija 87 | Denisovan | 12.5 | 337.1 | 409.1 | 385.8–455.0 | 12.2 | 346.7 | 418.7 | 390.9–470.2 |
| Mezmaiskaya 1 | Denisovan | 12.3 | 341.6 | 413.6 | 389.2–463.5 | 12.3 | 345.0 | 417.0 | 388.7–471.4 |
| Les Cottés Z4–1514 | Mbuti | 17.5 | 525.9 | 525.9 | 511.3–539.8 | 17.2 | 549.3 | 549.3 | 533.2–565.4 |
| Goyet Q56–1 | Mbuti | 17.7 | 515.0 | 515.0 | 501.8–528.9 | 17.6 | 520.8 | 520.8 | 502.6–540.6 |
| Mezmaiskaya 2 | Mbuti | 17.6 | 521.6 | 521.6 | 506.9–536.2 | 17.4 | 535.4 | 535.4 | 517.9–553.0 |
| Spy 94a | Mbuti | 18.0 | 502.6 | 502.6 | 488.7–517.2 | 17.3 | 542.7 | 542.7 | 518.6–566.1 |
| Vindija 87 | Mbuti | 17.6 | 523.0 | 523.0 | 509.1–537.6 | 17.3 | 542.0 | 542.0 | 524.5–558.8 |
| Mezmaiskaya 1 | Mbuti | 17.8 | 512.8 | 512.8 | 498.9–528.1 | 17.1 | 552.2 | 552.2 | 534.7–569.8 |
Results reported for all fragments and for deaminated fragments only (Supplementary Tables 3.1, 3.2). 95% confidence intervals for F(A|B) values are estimated via weighted block jackknife, using blocks of 5 Mb. In order to obtain estimates of split times from present, we correct ‘split A–B’ by adding the age of the high-coverage B-individual estimated from branch-shortening (column: split time + branch shortening (ka)).
Supplementary Material
Acknowledgements
We thank A. Weihmann and B. Höber for their help with DNA sequencing, U. Stenzel for computational support and advice for data analysis, R. Barr for the help with the graphics, V. Slon for helpful discussions and comments on the manuscript. Q.F. is funded in part by NSFC (91731303, 41672021, 41630102), CAS (QYZDB-SSW-DQC003, XDB13000000, XDA19050102, XDPB05) and the Howard Hughes Medical Institute (grant number 55008731). D.R. is supported by the US National Science Foundation (grant BCS-1032255) and by an Allen Discovery Center of the Paul Allen Foundation and is an investigator of the Howard Hughes Medical Institute. This study was funded by the Max Planck Society and the European Research Council (grant agreement number 694707 to S.P.). M.So. thanks the owner of Les Cottés, and the French Ministry of Culture for financial support and excavation permits.
Footnotes
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of the paper.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Code availability. All software packages used for analysis are cited and are publicly available. All custom developed software is freely available at .
Data availability. The aligned sequences have been deposited in the European Nucleotide Archive under accession numbers PRJEB21870 (Goyet Q56–1), PRJEB21875 (Les Cottés Z4–1514), PRJEB21881 (Mezmaiskaya 2), PRJEB21882 (Vindija 87) and PRJEB21883 (Spy 94a). The mitochondrial consensus sequences reported in this paper are available in GenBank with accession numbers MG025536, MG025537, MG025538, MG025539 and MG025540.
Reviewer Information Nature thanks C. Lalueza-Fox, C. Stringer and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Online Content Methods, along with any additional Extended Data display items and Source Data, are available in the online version of the paper; references unique to these sections appear only in the online paper.
References
- 1.Green RE et al. A draft sequence of the Neandertal genome. Science 328, 710–722 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prüfer K et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505, 43–49 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pääbo S et al. Genetic analyses from ancient DNA. Annu. Rev. Genet 38, 645–679 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Gilbert MT, Bandelt HJ, Hofreiter M & Barnes I Assessing ancient DNA studies. Trends Ecol. Evol 20, 541–544 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Krause J et al. A complete mtDNA genome of an early modern human from Kostenki, Russia. Curr. Biol 20, 231–236 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Korlević P et al. Reducing microbial and human contamination in DNA extractions from ancient bones and teeth. Biotechniques 59, 87–93 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Prüfer K et al. A high-coverage Neandertal genome from Vindija Cave in Croatia. Science 358, 655–658 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hublin J-J The modern human colonization of western Eurasia: when and where? Quat. Sci. Rev 118, 194–210 (2015). [Google Scholar]
- 9.Higham T et al. The timing and spatiotemporal patterning of Neanderthal disappearance. Nature 512, 306–309 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Fu Q et al. Genome sequence of a 45,000-year-old modern human from western Siberia. Nature 514, 445–449 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu Q et al. An early modern human from Romania with a recent Neanderthal ancestor. Nature 524, 216–219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rougier H et al. Neandertal cannibalism and Neandertal bones used as tools in Northern Europe. Sci. Rep 6, 29005 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Semal P et al. New data on the late Neandertals: direct dating of the Belgian Spy fossils. Am. J. Phys. Anthropol 138, 421–428 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Soressi M et al. in Préhistoire entre Vienne et Charente — Hommes et sociétés du Paléolithique mémoire 38 (eds Buisson-Catil J & Primault J) 221–234 (Association des Publications Chauvinoises, 2010). [Google Scholar]
- 15.Pinhasi R, Higham TF, Golovanova LV & Doronichev VB Revised age of late Neanderthal occupation and the end of the Middle Paleolithic in the northern Caucasus. Proc. Natl Acad. Sci. USA 108, 8611–8616 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Briggs AW et al. Patterns of damage in genomic DNA sequences from a Neandertal. Proc. Natl Acad. Sci. USA 104, 14616–14621 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyer M et al. A mitochondrial genome sequence of a hominin from Sima de los Huesos. Nature 505, 403–406 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Dalén L et al. Partial genetic turnover in Neandertals: continuity in the East and population replacement in the West. Mol. Biol. Evol 29, 1893–1897 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Mendez FL, Poznik GD, Castellano S & Bustamante CD The divergence of Neandertal and modern human Y chromosomes. Am. J. Hum. Genet 98, 728–734 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meyer M et al. A high-coverage genome sequence from an archaic Denisovan individual. Science 338, 222–226 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mallick S et al. The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature 538, 201–206 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cavalli-Sforza LL, Menozzi P & Piazza A The History and Geography of Human Genes (Princeton Univ. Press, 1994). [Google Scholar]
- 23.Svensson A et al. A 60 000 year Greenland stratigraphic ice core chronology. Clim. Past 4, 47–57 (2008). [Google Scholar]
- 24.Hublin J-J & Roebroeks W Ebb and flow or regional extinctions? On the character of Neandertal occupation of northern environments. C. R. Palevol 8, 503–509 (2009). [Google Scholar]
- 25.Müller UC et al. The role of climate in the spread of modern humans into Europe. Quat. Sci. Rev 30, 273–279 (2011). [Google Scholar]
- 26.Lazaridis I et al. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature 513, 409–413 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fu Q et al. The genetic history of Ice Age Europe. Nature 534, 200–205 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuhlwilm M et al. Ancient gene flow from early modern humans into Eastern Neanderthals. Nature 530, 429–433 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Overmann K & Coolidge F Human species and mating systems: Neandertal– Homo sapiens reproductive isolation and the archaeological and fossil records. J. Anthropol. Sci 91, 91–110 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Karmin M et al. A recent bottleneck of Y chromosome diversity coincides with a global change in culture. Genome Res 25, 459–466 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dabney J et al. Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proc. Natl Acad. Sci. USA 110, 15758–15763 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gansauge MT & Meyer M Single-stranded DNA library preparation for the sequencing of ancient or damaged DNA. Nat. Protoc 8, 737–748 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Dabney J & Meyer M Length and GC-biases during sequencing library amplification: a comparison of various polymerase-buffer systems with ancient and modern DNA sequencing libraries. Biotechniques 52, 87–94 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Kircher M, Sawyer S & Meyer M Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res 40, e3 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maricic T, Whitten M & Pääbo S Multiplexed DNA sequence capture of mitochondrial genomes using PCR products. PLoS ONE 5, e14004 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fu Q et al. DNA analysis of an early modern human from Tianyuan Cave, China. Proc. Natl Acad. Sci. USA 110, 2223–2227 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Welker F et al. Palaeoproteomic evidence identifies archaic hominins associated with the Châtelperronian at the Grotte du Renne. Proc. Natl Acad. Sci. USA 113, 11162–11167 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Renaud G, Kircher M, Stenzel U & Kelso J freeIbis: an efficient basecaller with calibrated quality scores for Illumina sequencers. Bioinformatics 29, 1208–1209 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Renaud G, Stenzel U & Kelso J leeHom: adaptor trimming and merging for Illumina sequencing reads. Nucleic Acids Res 42, e141 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H & Durbin R Fast and accurate long-read alignment with Burrows– Wheeler transform. Bioinformatics 26, 589–595 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sawyer S, Krause J, Guschanski K, Savolainen V & Pääbo S Temporal patterns of nucleotide misincorporations and DNA fragmentation in ancient DNA. PLoS ONE 7, e34131 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Green RE et al. A complete Neandertal mitochondrial genome sequence determined by high-throughput sequencing. Cell 134, 416–426 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Briggs AW et al. Targeted retrieval and analysis of five Neandertal mtDNA genomes. Science 325, 318–321 (2009). [DOI] [PubMed] [Google Scholar]
- 45.Skoglund P et al. Separating endogenous ancient DNA from modern day contamination in a Siberian Neandertal. Proc. Natl Acad. Sci. USA 111, 2229–2234 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brown S et al. Identification of a new hominin bone from Denisova Cave, Siberia using collagen fingerprinting and mitochondrial DNA analysis. Sci. Rep 6, 23559 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haak W et al. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature 522, 207–211 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gansauge MT & Meyer M Selective enrichment of damaged DNA molecules for ancient genome sequencing. Genome Res 24, 1543–1549 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Posth C et al. Deeply divergent archaic mitochondrial genome provides lower time boundary for African gene flow into Neanderthals. Nat. Commun 8, 16046 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ermini L et al. Complete mitochondrial genome sequence of the Tyrolean Iceman. Curr. Biol 18, 1687–1693 (2008). [DOI] [PubMed] [Google Scholar]
- 51.Gilbert MTP et al. Paleo-Eskimo mtDNA genome reveals matrilineal discontinuity in Greenland. Science 320, 1787–1789 (2008). [DOI] [PubMed] [Google Scholar]
- 52.Fu Q et al. A revised timescale for human evolution based on ancient mitochondrial genomes. Curr. Biol 23, 553–559 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Krause J et al. The complete mitochondrial DNA genome of an unknown hominin from southern Siberia. Nature 464, 894–897 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reich D et al. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature 468, 1053–1060 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sawyer S et al. Nuclear and mitochondrial DNA sequences from two Denisovan individuals. Proc. Natl Acad. Sci. USA 112, 15696–15700 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Horai S et al. Man’s place in Hominoidea revealed by mitochondrial DNA genealogy. J. Mol. Evol 35, 32–43 (1992). [DOI] [PubMed] [Google Scholar]
- 57.Katoh K & Standley DM MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tamura K, Stecher G, Peterson D, Filipski A & Kumar S MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol 30, 2725–2729 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bouckaert R et al. BEAST 2: a software platform for Bayesian evolutionary analysis. PLOS Comput. Biol 10, e1003537 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Darriba D, Taboada GL, Doallo R & Posada D jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9, 772 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Baele G et al. Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty. Mol. Biol. Evol 29, 2157–2167 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baele G, Li WLS, Drummond AJ, Suchard MA & Lemey P Accurate model selection of relaxed molecular clocks in Bayesian phylogenetics. Mol. Biol. Evol 30, 239–243 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Patterson N et al. Ancient admixture in human history. Genetics 192, 1065–1093 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Patterson N, Price AL & Reich D Population structure and eigenanalysis. PLoS Genet 2, e190 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Price AL et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet 38, 904–909 (2006). [DOI] [PubMed] [Google Scholar]
- 66.Meyer M et al. Nuclear DNA sequences from the Middle Pleistocene Sima de los Huesos hominins. Nature 531, 504–507 (2016). [DOI] [PubMed] [Google Scholar]
- 67.Busing FMTA, Meijer E & Van Der Leeden R Delete-m jackknife for unequal m. Stat. Comput 9, 3–8 (1999). [Google Scholar]
- 68.Saitou N & Nei M The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol 4, 406–425 (1987). [DOI] [PubMed] [Google Scholar]
- 69.Schliep KP phangorn: phylogenetic analysis in R. Bioinformatics 27, 592–593 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.The Chimpanzee Sequencing and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437, 69–87 (2005). [DOI] [PubMed] [Google Scholar]
- 71.Locke DP et al. Comparative and demographic analysis of orang-utan genomes. Nature 469, 529–533 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gibbs RA et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science 316, 222–234 (2007). [DOI] [PubMed] [Google Scholar]
- 73.Moorjani P et al. A genetic method for dating ancient genomes provides a direct estimate of human generation interval in the last 45,000 years. Proc. Natl Acad. Sci. USA 113, 5652–5657 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.The 1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Paten B, Herrero J, Beal K, Fitzgerald S & Birney E Enredo and Pecan: genome-wide mammalian consistency-based multiple alignment with paralogs. Genome Res 18, 1814–1828 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Paten B et al. Genome-wide nucleotide-level mammalian ancestor reconstruction. Genome Res 18, 1829–1843 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gravel S et al. Demographic history and rare allele sharing among human populations. Proc. Natl Acad. Sci. USA 108, 11983–11988 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.