Abstract
Purpose
To compare registration strategies to align arterial spin labeling (ASL) with 3D T1-weighted (T1w) images, with the goal of reducing the between-subject variability of cerebral blood flow (CBF) images.
Materials and Methods
Multi-center 3T ASL data were collected at 8 sites with 4 different sequences in the multi-centre GENetic Frontotemporal dementia Initiative (GENFI) study. In a total of 48 healthy controls, we compared the following image registration options: I) which images to use for registration (perfusion-weighted images to the segmented gray matter (GM) probability map (pGM) [CBF-pGM] or M0 to T1w [M0-T1w]); II) which transformation to use [rigid-body or non-rigid] and III) whether to mask or not [no masking, M0-based FSL BET masking]. In addition to visual comparison, we quantified image similarity using the Pearson correlation coefficient (CC), and used the Mann-Whitney U rank sum test.
Results
CBF-pGM outperformed M0-T1w (CC improvement 47.2% ± 22.0% (p<0.001), and the non-rigid transformation outperformed rigid-body (20.6% ± 5.3%, p<0.001). Masking only improved the M0-T1w rigid-body registration (14.5% ± 15.5%, p=0.007).
Conclusion
The choice of image registration strategy impacts ASL group analyses. The non-rigid transformation is promising but requires validation. CBF-pGM rigid-body registration without masking can be used as a default strategy. In patients with expansive perfusion deficits, M0-T1w may outperform CBF-pGM in sequences with high effective spatial resolution. BET-masking only improves M0-T1w registration when the M0 image has sufficient contrast.
Keywords: arterial spin labeling, cerebral blood flow, image registration
Introduction
Arterial spin labeling (ASL) is a non-invasive MRI perfusion technique with great potential to advance our understanding of the pathophysiology underlying neurodegenerative diseases such as frontotemporal dementia (FTD) (1, 2). Multi-center perfusion studies may help to establish ASL as an imaging biomarker with the ability to study brain physiology, to predict neurodegenerative disease onset and characteristics, as well as to monitor effects of potential disease-modifying drugs (1, 3, 4). An important step in the analysis of ASL studies is establishing standardized image processing methods (5).
A major challenge is the registration of ASL images to anatomical 3D T1-weighted (T1w) images, as there are inherent differences in image contrast, resolution and geometric distortion (2). Because of the relatively large cerebral blood flow (CBF) contrast between gray (GM) and white matter (WM) tissue, small alignment errors can have a large impact (6).
Once ASL data are aligned to the T1w images, the non-linear registration of T1w images to a common stereotactic space can be performed with relatively high precision, facilitating the identification of anatomical landmarks, creation of regions of interest, or performing group analyses (7, 8). In other words, the registration of ASL images to T1w images indirectly affects the alignment of CBF images between participants and consequently the ability of ASL to detect localized perfusion differences on a group level (8). Currently, there is no consensus on which registration strategies should be used for reliable and robust ASL image processing.
One obstacle in multi-center ASL studies is the heterogeneity in implementations, stemming both from the preferences of individual research groups and from the differences in commercially available ASL implementations from the major MRI vendors (9). As different centers employ scanners from different vendors, this contributes to the between-site variability and degrades statistical power for group inference (4, 10, 11). One major difference between ASL implementations is the design of the readout module. This leads to differences in effective spatial resolution, which is not only dependent on the reconstructed voxel-size but also on the acquisition point spread function (PSF), motion sensitivity and filtering procedures in the image reconstruction and post-processing (10, 12). These differences in image contrast may affect the registration performance and should be evaluated as part of the development of standardized ASL image processing methods (2).
The present study investigates three methodological components involved in subject-wise registration of ASL images to T1w images: I) which ASL and T1w images should be used for registration ‒ i.e. which image contrast results in an optimal registration (CBF to the GM probability map (pGM) [CBF-pGM] or M0 to T1w [M0-T1w]), II) which transformation algorithm should be used [rigid-body or non-rigid] and III) whether brain masking can improve registration [no masking or M0-based FSL BET masking]. A detailed motivation of these options can be found in the supplementary methods section.
Materials and Methods
Study design
Data for this study were drawn from the GENetic Frontotemporal dementia Initiative (GENFI) (13), a multi-center study aimed at identifying early brain changes in individuals who have a genetic risk of developing FTD. Inclusion and exclusion criteria are described elsewhere (13). Local ethical review boards approved the GENFI protocol and all participants provided written informed consent according to the declaration of Helsinki. For the first GENFI data freeze ‒ encompassing data collection from January 2012 to September 2013 ‒ 8 centers acquired ASL and T1w scans using the following 3T MR scanners and parameters: General Electric 3T MR750 with 3D spiral fast spin-echo pseudo-continuous ASL (PCASL) (3D spiral, 1 site), Philips Achieva 3T with 2D gradient-echo echo-planar imaging (EPI) PCASL (2D EPI, 3 sites) and 3T Siemens Trio with 3D gradient- and spin-echo pulsed ASL (PASL) (3D GRASE, 4 sites). One Philips site used different settings (background suppression, long post-label delay (PLD), referred to as 2D EPI Bsup) compared to two other sites (no background suppression, short PLD, referred to as 2D EPI noBsup). The four Siemens sites used an identical protocol (14), based on flow-sensitive alternating inversion recovery (FAIR) PASL with a defined bolus width (15). An M0 image was acquired for all ASL sequences except for 2D EPI noBsup (2). The T1w scan protocols were designed at the outset of GENFI to match them across scanners as much as possible. To avoid the confound of gene mutation effects on ASL perfusion, data for the current study were selected from healthy, unaffected participants who were mutation negative for one of three major FTD disease causing genes ‒ i.e. C9ORF72, GRN or MAPT. 12 healthy control subjects were randomly selected for each of the four ASL implementations (n=48 in total, 16 men/32 women, mean age 50.0 ± 16.1 years). Table 1 and Figure 1 provide an overview of the four different ASL implementations.
Table 1. Demographic and implementation characteristics for each sequence.
| 3D spiral (1 site, n=12) | 2D EPI Bsup (1 site, n=12) | 2D EPI noBsup (2 sites, n=12) | 3D GRASE (4 sites, n=12) | |
|---|---|---|---|---|
| Age (yrs) | 39.7 ± 18.2 | 52.6 ± 11.4 | 44.4 ± 18.1 | 51.2 ± 14.2 |
| Gender (M) | 5/12 | 4/12 | 3/12 | 4/12 |
| Scanner | 3T General Electric MR750 | 3T Philips Achieva | 3T Philips Achieva | 3T Siemens Trio |
| Labeling strategy | PCASL | PCASL | PCASL | FAIR Q2TIPS PASL |
| Labeling duration | 1450 ms | 1650 ms | 1650 ms | 800 ms (TI1) |
| Post-labeling delay (PLD) (range) | 1525 ms | 1525-2110 ms | 1200-1878 ms | 1200 ms (TI = 2000 ms) |
| PLD (mean) | 1525 ms | 1818 ms | 1540 ms | 1200 ms |
| Labeling plane planning | Fixed 22 mm below lower edge | 89 mm below, parallel to ACPC line | 89 mm below, parallel to ACPC line | Below lower edge of imaging slab (FAIR) |
| Readout module | 3D FSE interleaved stack-of-spirals | 2D gradient-echo single-shot EPI SENSE 2.5, CLEAR |
2D gradient-echo single-shot EPI SENSE 2.5, CLEAR |
3D GRASE |
| Acquisition matrix | 8 spirals, 512 sampling points | single shot, 80 x 80 | single-shot, 64 x 64 | 8 segments, 64 x 64 |
| Number of slices | 36 | 17 | 30 | 30 |
| Slice thickness | 4 mm | 7 mm | 4 mm | 4 mm |
| Acquisition voxel size (volume) | 3.75 x 3.75 x 4.0 mm (56 mm3) | 3.0 x 3.0 x 7.0 mm (63 mm3) | 3.75 x 3.75 x 4.0 mm (56 mm3) | 3.75 x 3.75 x 4.0 mm (56 mm3) |
| Reconstruction voxel size | 1.875 x 1.875 x 4.0 mm | 3.0 x 3.0 x 7.0 mm | 3.75 x 3.75 x 4.0 mm | 1.875 x 1.875 x 4.0 mm |
| Slice gap | n.a. | 0 mm | 0 mm | n.a. |
| TE/TR | 10.536/4632 ms | 13.8/4020 ms | 10-13/4000 ms | 14.86/5000 ms |
| M0 sequence | Yes (TR 4.6 s) | Yes (TR 10 s, 3.5 mm slices) | No (mean control image used) | Yes (TR 5 s) |
| Number of signal averages | 3 | 40 | 40 | 5 |
| Background suppression (n pulses) | yes (5) | yes (2) | No | Yes (2) |
| Acquisition duration | 4:19 min | 5:22 min | 5:20 min | 5:24 min |
ACPC = anterior-posterior commissure. ASL = arterial spin labeling, Bsup = background suppression, CLEAR = constant level appearance, EPI = echo-planar imaging, FAIR = flow-sensitive alternating inversion recovery, FSE = fast spin-echo, GRASE = gradient-echo and spin-echo, Q2TIPS = QUIPSS II with thin-slice TI1 periodic saturation, QUIPSS II = quantitative imaging of perfusion using a single subtraction, second version, PASL = pulsed ASL, PCASL = pseudo-continuous ASL, SENSE = sensitivity encoding, TE = echo time, TR = repetition time.
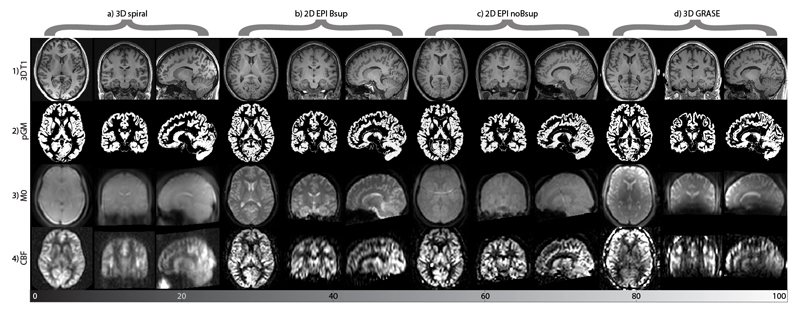
Figure 1.
Overview of different scans used for ASL image processing from a single representative subject from each ASL sequence (columns a-d). The rows show the images: 1) anatomical T1, 2) segmented gray matter probability map (pGM), 3) ASL reference image (M0), 4) perfusion-weighted image. For the 2D EPI sequence without background suppression (2D EPI noBsup) the mean control map is shown instead of an M0 image (3c). Note the fat shift aliasing artifact on this image. For illustrative purposes, all images were rigid-body registered, resampled to a 1.5x1.5x1.5 mm common space and scaled to the same median whole brain intensity.
Image processing
Image processing was performed with ExploreASL, an ASL toolbox developed to facilitate quality control and analyses for single- or multi-center ASL studies (9, 12). This toolbox is based on Matlab 7.12.0 (MathWorks, MA, USA), Statistical Parametric Mapping (SPM) 12 (Wellcome Trust Centre for Neuroimaging, University College London, UK) and Diffeomorphic Anatomical Registration analysis using Exponentiated Lie algebra (DARTEL) (4, 7). The processing is separated below into pre-registration parts containing ASL and T1w image processing to create intermediate images used for registration, the comparison of registration strategies and post-registration parts including the transformation to common space and CBF quantification (Figure 2).
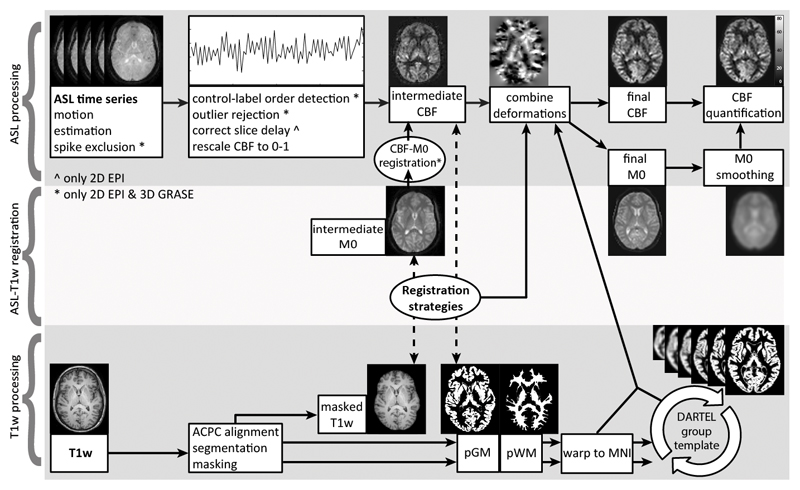
Figure 2.
Example images for the ExploreASL image processing pipeline from the first subject of the 2D EPI Bsup sequence. Note that some processing steps could be only applied to the 2D EPI or 3D GRASE sequences. The pipeline is subdivided into ASL (1st row) and T1w (3rd row) processing parts, which are connected by the registration strategies (2nd row) that are evaluated in the current study. Dashed lines connect the images used for the evaluation of registration strategies.
Image processing: T1w processing before registration
The T1w images were segmented into gray matter (pGM), white matter (pWM), cerebrospinal fluid and soft tissue probability maps after rigid-body realignment with the Montreal Neurological Institute (MNI) template. The segmentation was used to mask the skull out of the original T1w image. These T1w and pGM images were used as reference images for the M0-T1w and CBF-pGM registrations respectively.
Image processing: ASL processing before registration
3D rigid-body motion estimation was performed for the complete ASL time series, accounting for the signal intensity differences between control and label images as a zig-zag regressor (16). Afterwards, control-label pairs with the largest motion artifacts were discarded based on optimization of the mean GM temporal signal-to-noise ratio SNR (17). Motion correction was subsequently performed to create intermediate images used for registration. A voxel-wise outlier rejection was applied based on PWI signal intensities above or below the mean ± 3 temporal standard deviation, after which time series were averaged. These steps were conducted for the datasets in which ASL time series were available (2D EPI and 3D GRASE). For the 3D spiral data, the average PWI ‒ the CBF image ‒ was directly provided by the scanner. The M0 image was rigid-body registered to the mean control image for 2D EPI Bsup and 3D GRASE (2). The mean control image of 2D EPI noBsup was used as a surrogate M0 image, because this sequence did not have an M0 and did not use background suppression. The 3D spiral M0 image was not registered to the CBF image, because this did not improve its alignment on visual inspection.
Image processing: Transformation options
Before all registrations, the CBF image was clipped below zero and above the 95% non-zero quantile to remove potential macrovascular signal. For both the M0-T1w and CBF-pGM registration, a rigid-body SPM12 transformation was evaluated. An additional non-rigid transformation was only evaluated for the CBF-pGM approach. For the rigid-body transformation the 6-parameter SPM12 coregister method was used with default SPM12 settings, which optimizes a normalized mutual information objective function (18). For the non-rigid transformation, a DARTEL template was created separately for each participant from a CBF and a pGM image, using default SPM12 settings (7). The same rigid-body registration as described above was performed to provide a starting point for the non-rigid transformation. The non-rigid transformation was not tested for M0-T1w because it would require a mutual information or cross-correlation cost function and DARTEL only supports a sum-of-square cost function.
Image processing: Masking options
M0-based FSL BET masking was compared with no masking. In each instance, the same mask was applied to both the intermediate CBF and M0 images that were used for registration (not to the final images). The BET masks (Figure 3.2) were created by extracting the brain from the M0 image using BET2 with multiple iterations (option -r) (19).
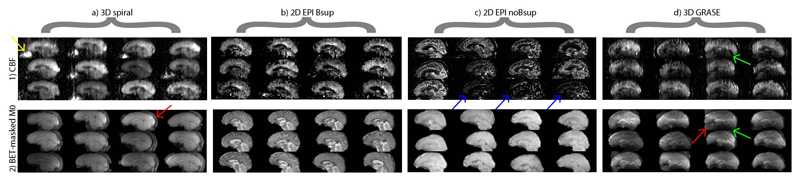
Figure 3.
Sagittal slices of perfusion-weighted images (PWI) (1) and M0 images (2) after being masked with the FSL Brain Extraction Tool (BET), for the 12 subjects and for each sequence (i.e. one image per subject). All images are shown in native space, only stretched to fit the Figure. Image intensities are scaled in such a way that the minimal and maximal intensities are the same for each image. Arrows denote the intensity bias field on 3D GRASE PWI and M0 images (green), vascular artifacts on some 2D EPI noBsup PWI (blue), high nose perfusion artifacts on 3D spiral PWI (yellow) and imperfect skull-stripped rims around the brain (red).
Image processing: Transformation to common space
The transformation obtained by the T1w segmentation was used to resample the pGM and pWM images into MNI space. These resampled images (i.e. 2 tissue type images x 48 participants) were used to create a group-wise DARTEL template. After all registrations were performed, these transformations were combined to transform all T1w and ASL images to common space for evaluation. In common space, a total GM mask was obtained by thresholding the pGM template per slice at 25%. For all intermediate and final images, the joint transformation from ASL native space to 1.5x1.5x1.5 mm3 MNI common space ‒ including motion correction and multiple registrations ‒ was applied in a single resampling step, to minimize the accumulation of interpolation artifacts.
Image processing: CBF quantification
PWI were converted into CBF images using a single compartment quantification model, assuming that the label decays with the blood T1 relaxation rate (2). Before dividing the PWI by the M0 reference image, the M0 image was masked (i.e. clipped below 20% non-zero quantile) and subsequently smoothed with a 12 mm FWHM Gaussian kernel to reduce the M0 image to a smooth bias field and avoid the propagation of CBF-M0 registration effects into the CBF quantification (20). For each registration option, the resultant CBF images were scaled to a mean GM CBF of 50 mL/100g/min per ASL sequence (21), to reduce the potential confounding effects of other sequence-specific scaling factors such as background suppression, the effect of background suppression on labeling efficiency and incomplete longitudinal magnetization recovery.
Qualitative evaluation
All processing steps (intermediate images) and final images were visually inspected by HM and JP, with >5 years of image processing of multiple ASL sequences. Sequences were visually rated for their effective spatial resolution by looking at the GM-WM CBF contrast and the tissue contrast on the M0 images.
Quantitative evaluation: Pearson correlation coefficient
To quantify the similarity between two images, we used the Pearson correlation coefficient (CC). The CC ranges from 0 (completely dissimilar) to 100% (identical images). Assuming that a near perfect registration should still yield small differences between images due to physiological CBF variability, we regard Pearson CC > 50% as excellent agreement. The Pearson CC was computed per pair of CBF images, resulting in [n(n-1)]/2 = 66 unique pair-wise comparisons within one sequence (n=12) and 1128 unique pair-wise multi-sequence comparisons (n=48). The population distribution of the Pearson CC was summarized by the median ± mean absolute difference (MAD). Significance of differences was tested by a Mann-Whitney U rank sum test. Statistical significance was defined as p<0.05.
Results
Pre-registration sequence features and qualitative brain masking performance
Sequence features that were observed at a single participant level include large B1-field inhomogeneity in the 3D GRASE images (green arrows Figure 3), macro-vascular artifacts on some 2D EPI noBsup images (blue arrows Figure 3.1), a perceived nose perfusion on the 3D spiral images (yellow arrow Figure 3.1). The sequences visually differed in their effective spatial resolution (i.e. smoothness), from low to high effective resolution in the following order: 3D spiral, 3D GRASE, 2D EPI Bsup and 2D EPI noBsup. This was confirmed by their GM-WM CBF ratios: 1.84 ± 0.14 (3D spiral), 2.85 ± 0.50 (2D EPI Bsup), 5.62 ± 1.28 (2D EPI noBsup), 2.53 ± 0.45 (3D GRASE). FSL BET was visually able to mask the brain on the 2D sequences but was less robust for the 3D sequences, especially for 3D spiral (red arrows Figure 3.2).
Qualitative comparison of registration strategies
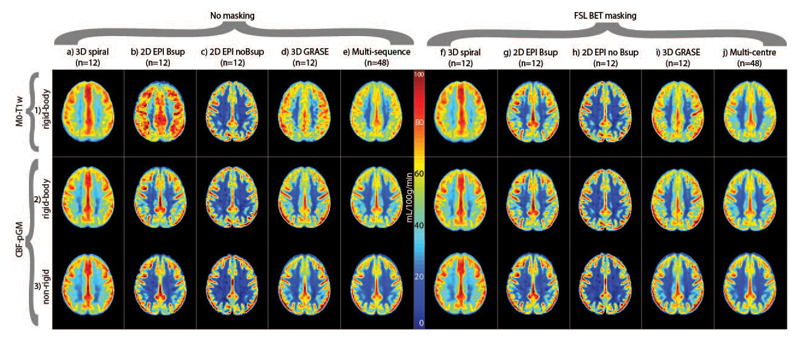
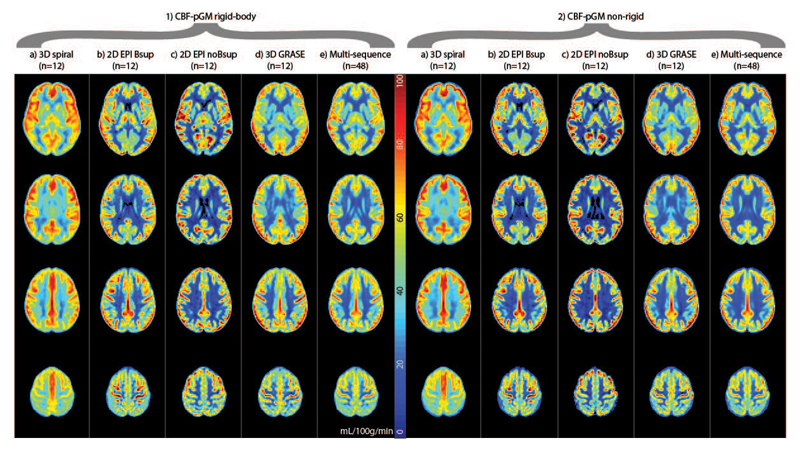
Figure 4 shows the mean CBF images for all registration strategies, for individual sequences and on a multi-sequence level. Differences between sequences and registration strategies can be appreciated by the CBF contrast between GM and WM, with higher GM-WM CBF contrast and a sharper GM-WM boundary reflecting improved alignment of CBF images between participants. This is visible as a sharper delineated GM region (yellow-red colors), a narrower light-blue color region of GM-WM transition and a larger dark-blue WM region (Figure 4). Irrespective of transformation or masking, the GM-WM CBF contrast was higher for the CBF-pGM registration (Figure 4.3 and Figure 4.4) than for M0-T1w (Figure 4.2) on a multi-sequence level, and mostly for 2D EPI Bsup and 3D GRASE, also for 3D spiral, but not for 2D EPI noBsup. The non-rigid transformation resulted in a higher image contrast than the rigid-body transformation on a group level, and mostly for single sequences 2D EPI Bsup and 3D GRASE. BET-masking visually improved the M0-T1w rigid-body registration on a multi-sequence level, and for single sequences 2D EPI Bsup and 3D GRASE (Figure 4.1).
Figure 4.
Mean CBF images for each ASL implementation (columns a-d, f-i) and all combined (columns e and j). Rows correspond to the different ASL registration options. Bsup = background suppression. Data are shown without and with FSL Brain Extraction Tool (BET) masking. The CBF images were scaled to a mean GM CBF of 50 mL/100g/min per ASL sequence.
Quantitative comparison of registration strategies
Table 2 shows the median total GM Pearson CC values for all registration strategies, for individual sequences (n=12) and on a multi-sequence level (n=44). The CC variability between participants was relatively high for 2D EPI noBsup (22.0-30.7%), intermediate for 3D spiral (6.3-15.8%) and 3D GRASE (4.5-22.9%) and relatively low for 2D EPI Bsup (4.5-12.8%).
Table 2. Pearson correlation coefficients.
| Registration approach | 3D spiral (n=12) |
2D EPI Bsup (n=12) |
2D EPI noBsup (n=12) |
3D GRASE (n=12) |
Multi-sequence (n=48) |
|---|---|---|---|---|---|
|
M0-T1w no masking Rigid-body |
0.57 ± 0.09 | 0.39 ± 0.05 | 0.38 ± 0.10 | 0.35 ± 0.08 | 0.28 ± 0.11 |
|
M0-T1w BET masking Rigid-body |
0.57 ± 0.09 | 0.52 ± 0.04 | 0.38 ± 0.11 | 0.43 ± 0.05 | 0.36 ± 0.11 |
|
CBF-pGM no masking Rigid-body |
0.74 ± 0.06 | 0.56 ± 0.03 | 0.40 ± 0.12 | 0.56 ± 0.03 | 0.49 ± 0.10 |
| Non-rigid | 0.79 ± 0.05 | 0.66 ± 0.03 | 0.50 ± 0.11 | 0.67 ± 0.03 | 0.60 ± 0.09 |
|
CBF-pGM BET masking Rigid-body |
0.74 ± 0.06 | 0.56 ± 0.03 | 0.39 ± 0.12 | 0.54 ± 0.05 | 0.47 ± 0.11 |
| Non-rigid | 0.79 ± 0.05 | 0.66 ± 0.03 | 0.49 ± 0.11 | 0.65 ± 0.04 | 0.59 ± 0.09 |
Pearson correlation coefficients illustrating the between-subject cerebral blood flow (CBF) similarity for individual sequences (n=12) and for all multi-center ASL data combined (n=48). The Pearson correlation coefficient was calculated for each image comparison, and of all unique pair-wise CBF image comparisons that were performed the median (± mean absolute difference from the mean) are shown here.
With a rigid-body transformation and without masking, CBF-pGM provided 47.2% ± 22.0% (p<0.001) higher Pearson CC than the M0-T1w registration, when considering the multi-sequence (i.e. all data) comparison. On a single sequence level this CC difference was statistically significant for all sequences (CC improvement for 3D spiral was 15.9% ± 10.4%, p=0.003; 2D EPI Bsup 36.1% ± 6.4%, p<0.001; 3D GRASE 48.8% ± 16.5%, p<0.001) except for 2D EPI noBsup (4.5% ± 7.0%, p=0.396). For CBF-pGM without masking, the non-rigid transformation outperformed the rigid-body transformation on the multi-sequence comparison (CC improvement 20.6% ± 5.3%, p<0.001) and on a single sequence level for 3D spiral (6.9% ± 1.8%, p=0.029), 2D EPI Bsup (16.3% ± 1.9%, p<0.001), 2D EPI noBsup (22.2% ± 7.8%, p=0.041) and 3D GRASE (17.1% ± 1.7%, p<0.001).
FSL BET improved the M0-T1w CC on a multi-sequence level (14.5% ± 15.5%, p=0.007) and on a single sequence level improved CC for 2D EPI Bsup (30.0% ± 8.7%, p<0.001) and 3D GRASE (19.9% ± 20.1%, p=0.016) but not for 3D spiral (0.4% ± 1.6%, p=0.481) or 2D EPI noBsup (-0.4% ± 2.4%, p=0.487).
FSL BET did not improve the CBF-pGM registrations on a multi-sequence level (-0.6% ± 1.0%, p=0.722) or for the individual sequences (3D spiral 0.0% ± 0.0%, p=0.487, 2D EPI Bsup 0.0% ± 0.0%, p=0.534, 2D EPI noBsup -0.9% ± 1.3%, p=0.578, 3D GRASE -4.0% ± 1.8%, p=0.951).
Optimal registration strategies
Figure 5 shows the CBF images for the two registration strategies with the visually highest GM-WM CBF contrast and highest Pearson CC. Visual differences between sequences can still be appreciated, with 3D spiral showing a homogeneous CC image and the 2D sequences showing highest GM-WM contrast. The 3D GRASE images appear in between the 3D spiral and 2D EPI images in terms of image homogeneity and GM-WM CBF contrast. The non-rigid transformation (Figure 5.2) visually showed a higher GM-WM CBF contrast compared with the rigid-body transformation (Figure 5.1). This is visible in Figure 5 as a deeper red color in a sharper delineated GM region, as well as a narrower light-blue color region. This difference was observed for all sequences but was visually largest for the 2D sequences and on a multi-sequence level.
Figure 5.
Multiple slices of mean cerebral blood flow images for the two best registration strategies, which were CBF-pGM rigid-body only (1) or with an additional non-rigid transformation (2). The CBF images were scaled to a mean GM CBF of 50 mL/100g/min per ASL sequence.
Discussion
The main results of this study were threefold: I) the registration of CBF-pGM images outperformed M0-T1w, II) the non-rigid transformation outperformed the rigid-body transformation and III) FSL BET-masking improved the M0-T1w registration for 2D EPI Bsup and 3D GRASE. These results were similar for the qualitative and quantitative analysis. Although it may be less important for analyses within large region-of-interests, these findings highlight the importance of adequate registration procedures for voxel-based analyses or accurate partial volume correction.
Both the M0-T1w registration strategy and M0-based BET performed poorly for the 3D sequences, which we attribute to the relatively low contrast of these smooth M0 images. Apparently, for acquisitions with relatively low effective resolution, such as 3D spiral, the smooth M0 contrast is challenging for registration whereas the CBF contrast is preserved even when the PSF is significantly larger than the nominal voxel resolution. Another factor that may explain the poorer performance of the M0-T1w approach compared to the CBF-pGM approach, is the requirement of two registration steps to be optimized, both the CBF-M0 and the M0-T1w registration. This can be problematic when background suppression reduces the contrast of the control image that is used for the CBF-M0 registration. For the CBF-pGM registration, any CBF-M0 misalignment is much less problematic, as the M0 image is only used for quantification, for which the M0 image can be spatially smoothed to reduce the effect of misalignment (2, 20). This may also explain why the 2D EPI noBsup sequence was the only sequence for which the CBF-pGM did not outperform the M0-T1w strategy, as for this sequence the mean control image was used as the M0, in which case the CBF-M0 alignment is perfect by design.
Increased registration performance with the non-rigid transformation has previously also been reported for diffusion tensor imaging (5). For ASL, this registration optimization may especially be of value to improve the correction of partial volume effects (22). However, we believe that the assumption that the pGM is a high-resolution CBF image is not completely valid, as it assumes both WM and cerebrospinal fluid (CSF) CBF to be 0, whereas the WM CBF may vary from 0.2 to 0.5 of the GM CBF value, depending on the ASL sequence in addition to physiology (6, 12). This potential mismatch between CBF and pGM images can explain the visually apparent cortical thinning and the apparent remodeling of the subcortical perfusion pattern by the non-rigid transformation in comparison to the rigid-body transformation. Future research should investigate to what extent the reduced variance between CBF images by the non-rigid transformation is the result of improved alignment, or whether this transformation also reduced meaningful perfusion variance between participants, and how this will affect group analyses. Furthermore, it is unclear how the non-rigid transformation will perform in the presence of perfusion deficits, although non-rigid transformations have been designed to be robust to pathophysiological differences between images to a certain extent (7). For these reasons, we are cautiously optimistic that non-rigid registration is promising but future studies that directly address how the non-rigid transformation reduces perfusion variance between participants ‒ also in the presence of pathological perfusion deficits ‒ is encouraged as subject of future research.
The fact that masking improved M0-T1w but not the CBF-pGM registration was surprising, considering the presence of artifacts in some CBF images, such as frontal in the 3D spiral images. Perhaps, the CBF-pGM contrast and/or image similarity is already sufficient such that it is not impacted by the CBF artifacts, whereas the M0-T1w contrast and/or image similarity is lower and still benefits from improvement by masking (23). Higher Pearson CC were obtained with the M0-T1w registration for 2D EPI Bsup than with 2D EPI noBsup. These higher CC may be due to the larger contrast in the M0 image compared to the mean control image. This could also contribute to improved skull-stripping, which can explain why the CC improved significantly with BET skull-stripping for 2D EPI Bsup but not for 2D EPI noBsup.
Our ranking of sequences based on their effective spatial resolution, deduced by visual inspection and the GM-WM CBF ratios, was in agreement with previous studies (10, 12). The two 2D EPI readouts showed very different GM-WM CBF ratios, which is probably not only explained by the differences in voxel size. The different noise properties because of differences in background suppression and post-label delay may also have contributed to the GM-WM CBF ratio. Interestingly, the effective spatial resolution of each sequence was associated with its Pearson CC. For instance, the 3D spiral sequence had the lowest effective spatial resolution and the highest Pearson CC, whereas the sequence with the highest effective resolution ‒ 2D EPI noBsup ‒ showed the lowest CC. It can be envisioned that the smooth images of a high SNR but low effective resolution sequence have a lower between-subject voxel-wise CBF variability, resulting in higher correlations. In other words, the comparison of Pearson CC between sequences is complicated by differences in effective resolution.
These observations highlight the importance of further readout design development for 3D ASL sequences, as the loss of effective spatial resolution due to blurring associated with long echo trains may detract when it comes to group level analyses, where accurate registration is an important component. Reducing the echo train length by increasing the number of shots could improve the PSF of the 3D sequences but this would increase the acquisition duration and make them more prone to smoothing caused by head motion. Our data encourage the development of approaches to increase the effective spatial resolution of 3D acquisitions, such as incorporation of parallel imaging or sparse data sampling.
Furthermore, these effective resolution differences between sequences were still visible after optimal registration. Therefore, different ASL sequences can be expected to contribute differently to group analyses, with respect to their effective spatial resolution and consequential partial volume errors. Although these observations also encourage to further adapt image processing for these sequence differences in directions such as adaptive smoothing, deblurring or partial volume correction (22, 24), they emphasize the importance of ASL sequence harmonization.
By design this study investigated registration approaches among healthy adults. Thus, results may vary in clinical populations since local perfusion deficits or severe atrophy could contribute to differences between the CBF and pGM image and consequently lower registration performance ‒ especially for non-rigid transformations. However, cortical thinning ‒ the most prevalent structural deficit leading to focal reduced apparent perfusion due to partial volume effects ‒ is expected to have a similar effect on CBF as on pGM (22). Furthermore, discrepancies between perfusion and anatomy that we expect to observe in dementias are often subtle, for which the CBF-pGM option will be sufficiently robust. A specific disparity between CBF and pGM is the presence of macro-vascular artifacts, which were visible in the 2D EPI noBsup sequence, because of the short PLD used in this sequence. Some CBF images of this sequence were more angiography-weighted, which we expect to have degraded the CBF-pGM performance. Although these cases are often excluded from analyses and sequences with longer PLDs or vascular crushing are expected to have less macro-vascular artifacts (2), the CBF-pGM registration should be used with caution in vascular compromised patients. The main advantage of CBF-pGM over M0-T1w is the similarity of image contrast, especially in smooth 3D sequences. Which extent of perfusion deficits or vascular artifacts will reduce this similarity of image contrast to the point where the CBF-pGM performs worse than M0-T1w, cannot be predicted with these data. For sequences with relatively high effective spatial resolution, the M0-T1w registration is a good alternative for CBF images with vascular artifacts, provided that adequate masking is performed.
Another limitation is that we did not compare all available registration strategies or settings but rather a selection of those that are most commonly used in the literature, deviating from SPM12 default settings as little as possible. From a practical perspective for the standardization of ASL image processing, we aimed to compare registration strategies that are readily accessible through commonly used software packages such as AFNI, FSL or SPM. We did not evaluate registration options with images of different contrast ‒ i.e. CBF-T1w or M0-pGM ‒ because these options would not be beneficial in terms of registration quality but would only work optimally under the assumption of the absence of a bias field, and homogeneous GM and WM intensities. Additionally, these combinations require a mutual information cost function which is not practical for non-linear registrations with high degrees of freedom.
The choice of registration strategy from the ASL image to the higher resolution anatomical reference image has an impact on single- or multi-center ASL perfusion studies. The CBF-pGM rigid-body registration without masking can be used for all ASL sequences as a default strategy. In patients with expansive perfusion deficits, M0-T1w rigid-body registration may outperform CBF-pGM and should be attempted as second-choice alternative for sequences with high effective spatial resolution. BET-masking only improves M0-T1w registration when the M0 image has sufficient contrast and effective spatial resolution. Better results can be achieved with the non-rigid transformation but this requires further validation. We anticipate that the standardization of ASL image processing will facilitate the development of ASL as a biomarker for diseases such as FTD.
Supplementary Material
Acknowledgments
The authors are grateful to D Marcus and R Herrick of Radiologics for their support in setting up of the GENFI XNAT database and AD Robertson, PhD for providing the BET masks. The authors express their gratitude to the participants and their families for taking part in the GENFI, and thank the GENFI investigators: Martin Rossor, Nick Fox, Jason Warren, Martina Bocchetta, Katrina Dick, Michela Pievani, Roberta Ghidoni, Luisa Benussi, Alessandro Padovani, Maura Cosseddu, Alexandre Mendonça, Giovanni Frisoni, Enrico Premi, Silvana Archetti, Elio Scarpini, Giorgio Fumagalli, Andrea Arighi, Chiara Fenoglio, Sara Prioni, Veronica Redaelii, Marina Grisoli, Pietro Tiraboschi, Sandra Black, Ekaterina Rogaeva, Morris Freedman, Maria Carmela Tartaglia, David Tang-Wai, Ron Keren, Jessica Panman, Lieke Meeter, Lize Jiskoot, Rick van Minkelen, Gemma Lombardi, Cristina Polito, Benedetta Nacmias, Vesna Jelic, Christin Andersson, Linn Öijerstedt, Marie Fallström, Hakan Thonberg, Ana Verdelho, Carolina Maruta.
Funding
This study was carried out within the context of the GENFI, which was supported by the UK Medical Research Council, the Italian Ministry of Health, and the Canadian Institutes of Health Research as part of a Centres of Excellence in Neurodegeneration grant. JDR and MR acknowledge the support of the National Institute for Health Research Queen Square Dementia Biomedical Research Unit, Leonard Wolfson Experimental Neurology Centre, the Brain Research Trust, and the University College London Hospitals NHS Trust Biomedical Research Centre. CG acknowledges the support of Swedish Brain Power. HM & MM acknowledge the support of the Weston Brain Institute. BM and HM acknowledge the support of the Canadian Partnership for Stroke Recovery. DT, EV, DC, XG, MR and JR acknowledge the support of the UK Department of Health’s NIHR Biomedical Research Centres funding scheme. JBR is supported by the Wellcome Trust (103838). MM acknowledges the support from the Department of Medicine at Sunnybrook Health Sciences Centre and University of Toronto, and from the Sunnybrook Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviation key
- ASL
arterial spin labeling
- BET
brain extraction tool
- CBF
cerebral blood flow
- CSF
cerebrospinal fluid
- DARTEL
Diffeomorphic Anatomical Registration analysis using Exponentiated Lie algebra
- EPI
echo-planar imaging
- FSL
FMRIB software library
- FTD
frontotemporal dementia
- FWHM
full width at half maximum
- GENFI
GENetic Frontotemporal dementia Initiative
- GM
gray matter
- GRASE
gradient-echo and spin-echo
- MNI
Montreal neurological institute
- pGM
gray matter probability
- PASL
pulsed ASL
- PCASL
pseudo-continuous ASL
- PLD
post-label delay
- PSF
point spread function
- PWI
perfusion-weighted image
- SC
similarity coefficient
- SNR
signal-to-noise ratio
- SPM
statistical parametric mapping
- T1w
T1-weighted
- WM
white matter
Reference List
- 1.Steketee RM, Bron EE, Meijboom R, et al. Early-stage differentiation between presenile Alzheimer's disease and frontotemporal dementia using arterial spin labeling MRI. Eur Radiol. 2015 doi: 10.1007/s00330-015-3789-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alsop DC, Detre JA, Golay X, et al. Recommended implementation of arterial spin-labeled perfusion MRI for clinical applications: A consensus of the ISMRM perfusion study group and the European consortium for ASL in dementia. Magn Reson Med. 2014 doi: 10.1002/mrm.25197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bateman RJ, Xiong C, Benzinger TL, et al. Clinical and biomarker changes in dominantly inherited Alzheimer's disease. N Engl J Med. 2012;367:795–804. doi: 10.1056/NEJMoa1202753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stonnington CM, Tan G, Kloppel S, et al. Interpreting scan data acquired from multiple scanners: a study with Alzheimer's disease. Neuroimage. 2008;39:1180–1185. doi: 10.1016/j.neuroimage.2007.09.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vollmar C, O'Muircheartaigh J, Barker GJ, et al. Identical, but not the same: intra-site and inter-site reproducibility of fractional anisotropy measures on two 3.0T scanners. Neuroimage. 2010;51:1384–1394. doi: 10.1016/j.neuroimage.2010.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pohmann R. Accurate, localized quantification of white matter perfusion with single-voxel ASL. Magn Reson Med. 2010;64:1109–1113. doi: 10.1002/mrm.22476. [DOI] [PubMed] [Google Scholar]
- 7.Ashburner J. A fast diffeomorphic image registration algorithm. Neuroimage. 2007;38:95–113. doi: 10.1016/j.neuroimage.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 8.Gonzalez-Castillo J, Duthie KN, Saad ZS, Chu C, Bandettini PA, Luh WM. Effects of image contrast on functional MRI image registration. Neuroimage. 2013;67:163–174. doi: 10.1016/j.neuroimage.2012.10.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mutsaerts HJ, van Osch MJ, Zelaya FO, et al. Multi-vendor reliability of arterial spin labeling perfusion MRI using a near-identical sequence: implications for multi-center studies. Neuroimage. 2015;113:143–152. doi: 10.1016/j.neuroimage.2015.03.043. [DOI] [PubMed] [Google Scholar]
- 10.Vidorreta M, Wang Z, Rodriguez I, Pastor MA, Detre JA, Fernandez-Seara MA. Comparison of 2D and 3D single-shot ASL perfusion fMRI sequences. Neuroimage. 2012;66C:662–671. doi: 10.1016/j.neuroimage.2012.10.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mutsaerts HJ, Steketee RM, Heijtel DF, et al. Reproducibility of pharmacological ASL using sequences from different vendors: implications for multicenter drug studies. MAGMA. 2015 doi: 10.1007/s10334-014-0480-1. [DOI] [PubMed] [Google Scholar]
- 12.Mutsaerts HJ, Steketee RM, Heijtel DF, et al. Inter-vendor reproducibility of pseudo-continuous arterial spin labeling at 3 tesla. PLoS One. 2014;9:e104108. doi: 10.1371/journal.pone.0104108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rohrer JD, Nicholas JM, Cash DM, et al. Presymptomatic cognitive and neuroanatomical changes in genetic frontotemporal dementia in the Genetic Frontotemporal dementia Initiative (GENFI) study: a cross-sectional analysis. Lancet Neurol. 2015;14:253–262. doi: 10.1016/S1474-4422(14)70324-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Vita E, Gunther M, Golay X, Thomas DL. Magnetisation transfer effects of Q2TIPS pulses in ASL. MAGMA. 2012;25:113–126. doi: 10.1007/s10334-011-0298-z. [DOI] [PubMed] [Google Scholar]
- 15.Wong EC, Buxton RB, Frank LR. Quantitative imaging of perfusion using a single subtraction (QUIPSS and QUIPSS II) Magn Reson Med. 1998;39:702–708. doi: 10.1002/mrm.1910390506. [DOI] [PubMed] [Google Scholar]
- 16.Wang Z, Aguirre GK, Rao H, et al. Empirical optimization of ASL data analysis using an ASL data processing toolbox: ASLtbx. Magn Reson Imaging. 2008;26:261–269. doi: 10.1016/j.mri.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shirzadi Z, Crane DE, Robertson AD, et al. Automated removal of spurious intermediate cerebral blood flow volumes improves image quality among older patients: A clinical arterial spin labeling investigation. J Magn Reson Imaging. 2015;42:1377–1385. doi: 10.1002/jmri.24918. [DOI] [PubMed] [Google Scholar]
- 18.Collignon A, Maes F, Delaere D, Vandermeulen D, Suetens P, Marchal G. Proc. Information Processing in Medical Imaging. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. Automated multi-modality image registration based on information theory; pp. 263–274. [Google Scholar]
- 19.Smith SM. Fast robust automated brain extraction. Hum Brain Mapp. 2002;17:143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beaumont H. Multimodal Magnetic Resonance Imaging of Frontotemporal Lobar Degeneration. University of Manchester, Faculty of Medical and Human Sciences; 2015. pp. 102–126. [Google Scholar]
- 21.Heijtel DF, Mutsaerts HJ, Bakker E, et al. Accuracy and precision of pseudo-continuous arterial spin labeling perfusion during baseline and hypercapnia: a head-to-head comparison with (1)(5)O H(2)O positron emission tomography. Neuroimage. 2014;92:182–192. doi: 10.1016/j.neuroimage.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 22.Asllani I, Borogovac A, Brown TR. Regression algorithm correcting for partial volume effects in arterial spin labeling MRI. Magn Reson Med. 2008;60:1362–1371. doi: 10.1002/mrm.21670. [DOI] [PubMed] [Google Scholar]
- 23.Fischmeister FP, Hollinger I, Klinger N, et al. The benefits of skull stripping in the normalization of clinical fMRI data. Neuroimage Clin. 2013;3:369–380. doi: 10.1016/j.nicl.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oliver RA. Improved quantification of arterial spin labeling images using partial volume correction techniques. London, UK: University College London; 2015. pp. 160–184. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.