Abstract
Faithful chromosome segregation depends on the ability of sister kinetochores to attach to spindle microtubules. The outer layer of kinetochores transiently expands in early mitosis to form a fibrous corona, and compacts upon microtubule capture. Here we show that the dynein adaptor Spindly and the RZZ (ROD-Zwilch-ZW10) complex drive kinetochore expansion in a dynein-independent manner. C-terminal farnesylation and MPS1 kinase activity cause conformational changes of Spindly that promote oligomerization of RZZ:Spindly complexes into a filamentous meshwork in cells and in vitro. Concurrent with kinetochore expansion, Spindly potentiates kinetochore compaction by recruiting dynein via three conserved short linear motifs. Expanded kinetochores unable to compact engage in extensive, long-lived lateral microtubule interactions that persist to metaphase, and result in merotelic attachments and chromosome segregation errors in anaphase. Thus, dynamic kinetochore size regulation in mitosis is coordinated by a single, Spindly-based mechanism that promotes initial microtubule capture and subsequent correct maturation of attachments.
Keywords: Mitosis, Chromosome segregation, Kinetochore, Fibrous corona, RZZ, Spindly, Dynein
Introduction
Fidelity of chromosome segregation in mitosis requires chromosomes to capture microtubules and achieve stable bioriented attachments before anaphase. To achieve this, chromosomes use an intricate kinetochore machinery1–3. The core of the kinetochore in animals and fungi is composed of the CCAN and the KMN networks, which associate with centromeric chromatin and microtubules, respectively1. The core kinetochore module is supplemented with proteins whose quantity at kinetochores changes during mitotic progression. These include proteins of the spindle assembly checkpoint (SAC), dynein recruiters (ROD-ZW10-Zwilch (RZZ) and Spindly), dynein regulators (CENP-F/Nude1/Nde1/CLIP-170), the kinesins CENP-E and Kif2b, and modifiers of microtubule dynamics such as CLASPs4,5.
The mammalian kinetochore exhibits great morphological plasticity during mitosis. While the core kinetochore module expands little, the more centromere-distal modules expand into large crescent shapes in early prometaphase and collapse into compact spherical structures in metaphase6–11. Recent computational modeling suggested that expanded kinetochores promote spindle assembly, and that correct rotation of the sister kinetochores followed by their compaction reduces the risk of erroneous attachments6. Electron micrographs of expanded kinetochores show a halo of low-density material referred to as the fibrous corona, which is absent from kinetochores that are bound by microtubules12–16. Although immuno electron microscopy (EM) studies have shown several proteins to reside at the corona17–20, it is unknown what proteins create the fibrous meshwork, impeding experimental interrogation of its functions. The dynamic behaviour of the fibrous corona correlates with that of the proteins that display early mitotic expansion and subsequent compaction. It is therefore likely that one or more of them can assemble into a fibrous meshwork and that regulation of the meshwork’s expansion/compaction will hinge on these components. A candidate is the RZZ complex. RZZ shows expected expansion/compaction behavior, is required for recruitment of several outer-kinetochore components, and has a molecular architecture resembling that of membrane coating proteins known to form polymeric states21–28. Here, we set out to identify the mechanisms of kinetochore expansion and compaction, and their functional relevance for chromosome segregation.
Results
Spindly recruits dynein to compact kinetochores upon microtubule attachment
In agreement with previous observations6,7,9, kinetochores expanded in nocodazole-treated cells shortly after NEB, as evidenced by the formation of ZW10-positive crescents (Figure 1a, examples 1 to 3). Crescents grew during mitosis, and those of sister kinetochores nearly connected to form large, ring-shaped structures (Figure 1a, example 4). Rings were absent from cells in which a spindle was allowed to form, and an attached kinetochore on a mono-oriented chromosome was more compact than its unattached sister kinetochore (Figure 1a, example 5). In Xenopus egg extracts, kinetochore size is regulated by mitotic phosphorylation11. However, small molecule inhibitors of Aurora B, PLK1, or MPS1, when added after expansion, did not cause compaction of kinetochores (Supplementary Figure 1).
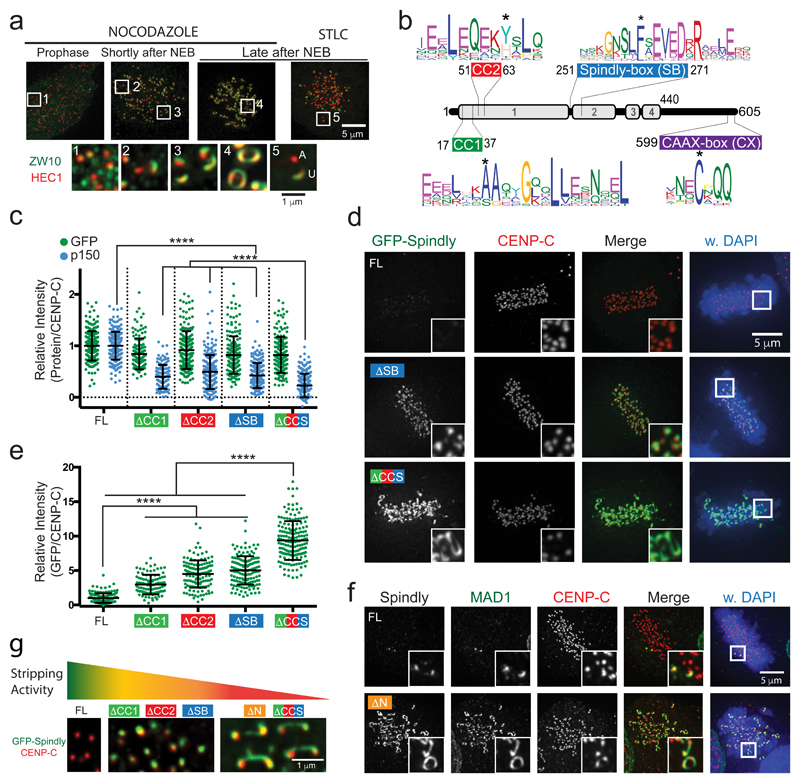
Figure 1. Spindly recruits dynein to compact kinetochores upon microtubule attachment.
(a) Immunofluorescence images of ZW10 and HEC1 in HeLa cells treated with nocodazole or STLC. (A, Attached; U, Unattached). The experiment was repeated at least three times with similar results. (b) Overview of the secondary structure of human Spindly with predicted coiled-coils (grey bars) and disordered regions, and with sequence logos of four conserved motifs. See also Supplementary Figure 2. (c) Quantification of the kinetochore levels of p150Glued in nocodazole-treated HeLa cells transfected with siRNAs to Spindly and expressing the indicated GFP-Spindly variants. The graph in shows the mean kinetochore intensity (±SD) normalized to the values of SpindlyFL. Each dot represents one cell: FL (n= 206 cells), ΔCC2 (n= 200 cells), ΔSB (n= 192 cells), ΔCCS (n= 194 cells) pooled from seven independent experiments. ΔCC1 (n= 122 cells) pooled from four independent experiments. Asterisks indicate significance (one-way ANOVA followed by Tukey's test; F (4, 909) = 238.5). ****P < 0.001. Representative images of cells are shown in Supplementary Figure 3c. (e,f) Representative images (e) and quantification (f) of metaphase HeLa cells transfected with siRNA to Spindly and expressing the indicated GFP-Spindly variants. The graph shows the mean fold change in kinetochore intensity (±SD) normalized to the values of SpindlyFL. Each dot represents one cell: FL (n= 139 cells), ΔCC2 (n= 136 cells), ΔSB (n= 141 cells), ΔCCS (n = 165 cells) pooled from five independent experiments. ΔCC1 (n= 97 cells) pooled from four independent experiments. Asterisks indicate significance (one-way ANOVA followed by Tukey's test; F (4, 673) = 362.5). ****P < 0.001. Representative images of all the mutants are shown in Supplementary Figure 3d. (g) Representative immunofluorescence images of HeLa cells transfected with siRNA to Spindly and expressing the indicated GFP-Spindly variants, and immuno-stained for the indicated antigens. The experiment was repeated at least three times with similar results. See siSpindly control in Supplementary Figure 3b. (h) Representative images of the morphology of Spindly and CENP-C in metaphase kinetochores expressing the indicated GFP-Spindly variants. Stripping activity is based on ability of Spindly variants to recruit dynein/dynactin. The experiment was repeated at least three times with similar results.
Dynein drives poleward transport of kinetochore proteins28–31. To investigate if dynein is required for kinetochore compaction, we examined its kinetochore adaptor Spindly32–35. Spindly localizes to kinetochores through interaction with the RZZ complex via its farnesylated C-terminal CAAX box21,26,36–38 (Figure 1b). Spindly in turn recruits dynein/dynactin through Spindly box (SB) and CC1 box motifs (Figure 1b and Supplementary Figure 2)21,26,38,39. Our ConFeaX pipeline40 additionally identified a Qxx[HY] motif close to the CC1 box, which we here named the CC2 box (Figure 1b). CC2-like boxes are also found in other dynein/dynactin adaptors (Supplementary Figure 2). Recruitment of the dynactin subunit p150glued to kinetochores was compromised in cells expressing GFP-Spindly mutated in any of the three motifs [A24V (SpindlyΔCC1), Y60A (SpindlyΔCC2), F258A (SpindlyΔSB)] (Figure 1c and Supplementary Figure S3c), and was nearly abolished when all three motifs were mutated (SpindlyΔCCS) (Figure 1c and Supplementary Figure S3c). Dynein removes Spindly from attached kinetochores26,32–35,38,39, and indeed the Spindly motif mutants defective in dynein recruitment were not removed from kinetochores of metaphase cells (Figure 1d,e and Supplementary Figure S3d). Strikingly, SpindlyΔCCS, which was the most compromised in recruiting dynein (Figure 1c) frequently appeared as expanded structures that apparently bridged the two sister kinetochores (Figure 1d). A similar phenotype was observed by deletion of the N-terminal 65 amino acids of Spindly (SpindlyΔN) that contain the CC1 and CC2 boxes (Figure 1f). These findings suggest that Spindly, via the recruitment of dynein, contributes to compaction of previously expanded kinetochores (Figure 1g).
Kinetochores expand by forming a structurally stable kinetochore sub-module
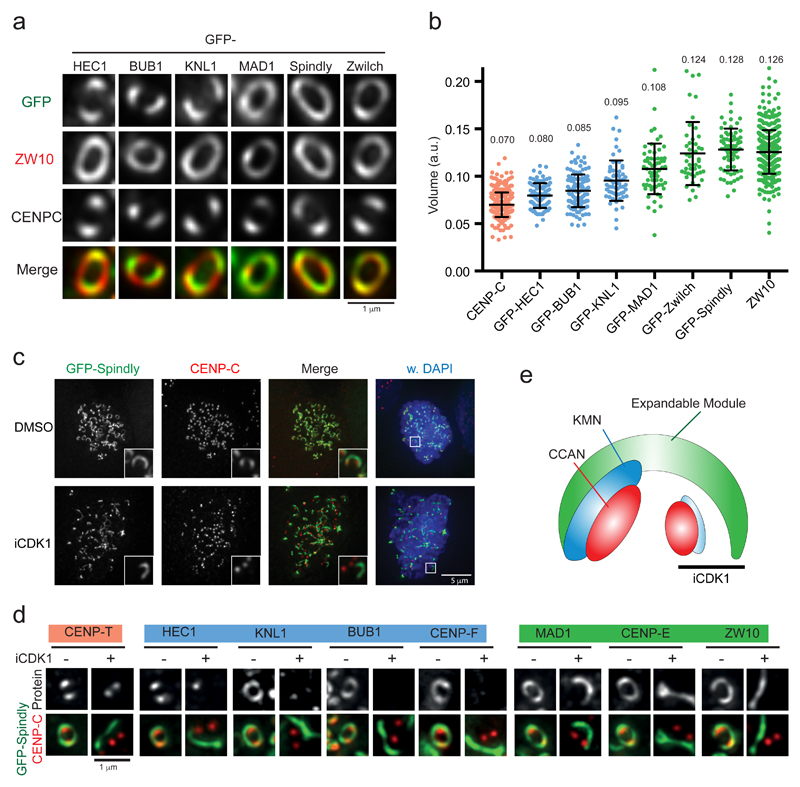
We next examined the molecular basis for kinetochore expansion. Quantitative immunofluorescence imaging of kinetochores in nocodazole-treated cells showed that the extent of expansion of different kinetochore modules inversely correlated with their proximity to the centromeric chromatin: RZZ, Spindly and MAD1 occupied the largest volumes, followed by the KMN network and the CCAN (Figure 2a,b). Interestingly, during analysis of expanded kinetochores under various experimental conditions, we noticed that brief (20 min.) CDK1 inhibition caused the disappearance of expanded outer kinetochores and the appearance, close to kinetochores, of crescent-shaped rods or extended filaments containing Spindly, ZW10, MAD1 and CENP-E (Figure 2c,d). Because these cells were still in mitosis (Figure 2c), we interpreted the rods to be expanded kinetochore modules that had fully or partially detached from the core kinetochore (examples of partial detachments: bottom-right images of the HEC1 and MAD1 examples in Figure 2d). These data suggest that the expanded kinetochore module is a relatively stable structure with a specific architecture, composition and regulation, distinct from other outer kinetochore modules such as the KMN network (Figure 3e).
Figure 2. Kinetochores expand by forming a structurally stable kinetochore sub-module.
(a,b) Representative images (a) and volume quantification (b) of the indicated GFP-tagged proteins in expanded kinetochores of HeLa cells entering mitosis in the presence of nocodazole. Maximum expanded kinetochores were selected based on ZW10 staining and imaging acquisition was set to obtain similar mean intensity levels for the different proteins. The graph shows the mean kinetochore volume (±SD). Each dot represents a pair of sister kinetochores: CENP-C (n= 380 kinetochores pooled from 12 experiments), GFP-HEC1 (n=101 pooled from four experiments), GFP-BUB1 (n=124 pooled from five experiments), GFP-KNL1 (n=62 pooled from three experiments), GFP-MAD1 (n=61 pooled from three experiments), GFP-Zwilch (n=69 pooled from three experiments), GFP-Spindly (n=69, pooled from three experiments), and ZW10 (n=461 pooled from nine experiments). (c) Representative immunofluorescence images of HeLa cells transfected with siRNA to Spindly and expressing GFP-Spindly entering mitosis in the presence of nocodazole and subsequently treated with the CDK1 inhibitor RO-3306 for 20 minutes. The experiment was repeated at least three times with similar results. (d) Images of kinetochores of HeLa cells transfected with siRNA to Spindly and expressing GFP-Spindly entering mitosis in the presence of nocodazole and subsequently treated with RO-3306 for 20 minutes, and immunostained with the indicated antibodies. The experiment was repeated at least three times with similar results. Color codes in (b) and (c) are explained in panel (e). (e) The cartoon illustrates the different pools of kinetochore proteins and the observation that the expandable module of the kinetochore (green) appears as cytoplasmic rods detached from the core kinetochore after the KMN network is disassembled upon CDK1 inhibition.
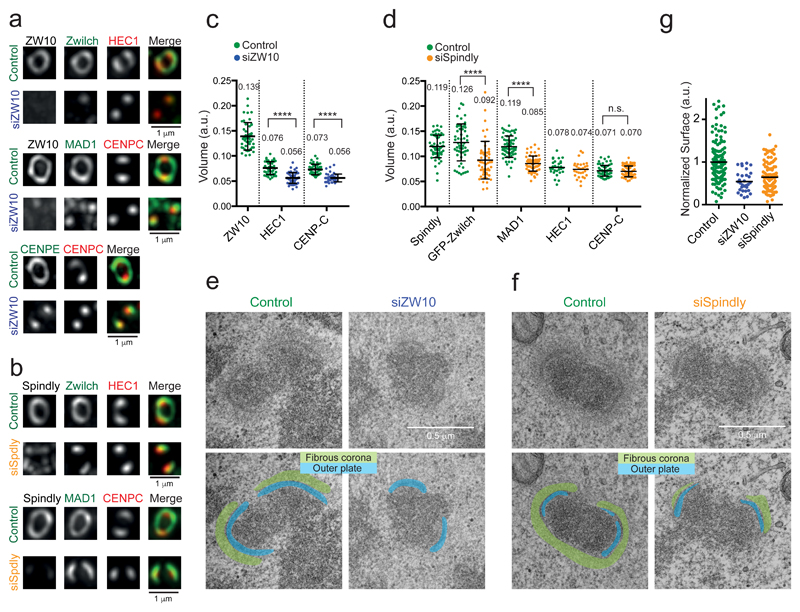
Figure 3. Spindly and RZZ are essential for kinetochore expansion.
(a-d) Representative images (a, b) and volume quantification (c, d) based on the indicated antigens of expanded kinetochores of nocodazole-treated HeLa cells transfected with siRNA to ZW10 (a,c) or Spindly (b,d). Imaging acquisition was set to obtain similar mean intensity levels for the different conditions. The graphs show the mean kinetochore volume (±SD). Each dot represents a pair of sister kinetochores. The sample size in (c) is: Control (n= 47 kinetochores), siZW10 (n= 55 kinetochores), pooled from three independent experiments. Asterisks indicate significance (Student’s t-test, two-tailed, unpaired; HEC1 (t=8.517 df=100); CENP-C: t=9.317 df=100). ****P < 0.0001. The sample size in (d) is: Spindly (n= 63 kinetochores pooled from three experiments), GFP-Zwilch (n= 55 kinetochores pooled from three three experiments), GFP-Zwilch in siSpindly (n= 52 kinetochores pooled from three three experiments), HEC1 (n= 30 kinetochores pooled from two experiments), HEC1 in siSpindly (n= 30 kinetochores pooled from two experiments, see Supplementary Table 3 for source data), MAD1 (n= 63 kinetochores pooled from three experiments), MAD1 in siSpindly (n= 63 kinetochores from three experiments), CENP-C (n= 63 kinetochores from three experiments), CENP-C in siSpindly (n= 63 kinetochores from three experiments). Asterisks indicate significance (Student’s t-test, two-tailed, unpaired; GFP-Zwilch (t=4.907 df=105); MAD1 (t=10.22 df=124); HEC1 (t=0.9724 df=58); CENP-C (t=0.5788 df=124). ****P < 0.0001; n.s., not significant. (e-g) Electron micrographs (e,f) and surface quantification (g) of the fibrous corona of nocodazole-treated cells depleted of ZW10 (e) or Spindly (f). The fibrous corona is highlighted in green and the outer plate in blue. Each dot represents one kinetochore: Control (n= 137 kinetochores), siSpindly (n= 122 kinetochores) pooled from two independent experiments. siZW10 (n=31 kinetochores from one experiment). Control is siGAPDH. See Supplementary Table 3 for source data. Quantification of the protein depletion efficiencies are shown in Supplementary Figure 4b,c.
Spindly and RZZ are essential for kinetochore expansion
Immunofluorescence imaging and EM showed absence of kinetochore expansion and absence of a fibrous corona, respectively, in ZW10 RNAi cells (Figure 3a,c,e,g and Supplementary Figure 4a-c). Interestingly, ZW10 RNAi also reduced the volumes occupied by the KMN network member HEC1 and the CCAN member CENP-C (Figure 3a,c). RZZ is thus not only important for full fibrous corona assembly but also for the (limited) expansion of inner- and more outer- kinetochore regions (Supplementary Figure 4a).
We next reasoned that if dynein recruitment were the predominant contribution of Spindly to kinetochore size dynamics, Spindly depletion should phenocopy expression of the Spindly motif mutants, i.e. persistently expanded kinetochores due to absence of the dynein-dependent kinetochore compaction mechanism (Supplementary Figure 3b). In contrast, volume measurements and EM revealed that cells depleted of Spindly were unable to expand kinetochores and had significantly compromised fibrous coronas (Figure 3b,d,f,g and Supplementary Figure 4a-c). Spindly is therefore essential not only for kinetochore compaction through dynein binding, but also for kinetochore expansion (Supplementary Figure 4a).
Spindly stimulates RZZ-Spindly polymerization in vitro and in vivo
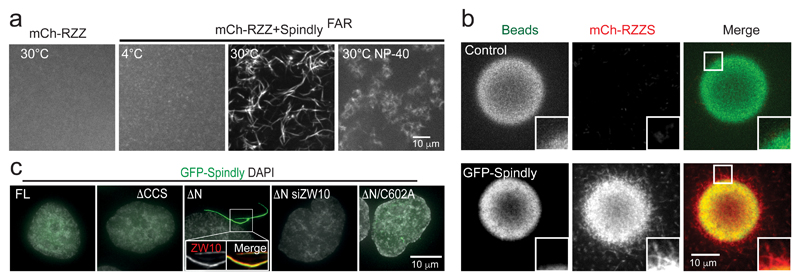
Dimeric RZZ structurally resembles coat scaffolds, which can self-assemble into polymeric states, and it was speculated that RZZ may be a structural fibrous corona precursor and driver of kinetochore expansion21. Purified recombinant RZZ, which assembles with 2:2:2 (ROD:ZW10:Zwilch) stoichiometry21, did not oligomerize, as assessed by direct visualization of mCherry-ROD (Figure 4a). Addition of purified farnesylated Spindly (SpindlyFAR), however, caused spontaneous oligomerization into filamentous structures at 30°C (Figure 4a, Supplementary Movie 1). In vitro filament formation of RZZ-Spindly (RZZS) complexes could be prevented by addition of detergent, suggesting that hydrophobic interactions underlie the assembly reaction (Figure 4a). Notably, RZZS oligomerization in the presence of GFP-Spindly-coated agarose beads resulted in association of a filamentous meshwork with the beads (Figure 4b).
Figure 4. Spindly stimulates RZZ-Spindly polymerization in vitro and in vivo.
(a) Fluorescence microscopy of purified mCherry-RZZ alone or in the presence of recombinant farnesylated Spindly (SpindlyFAR) and incubated as indicated. See also Supplementary Movie 1. The experiment was repeated with two independent RZZS preparations, with at least three replicates, and all samples gave similar results. (b) GFP-Spindly immobilized on beads and incubated in the presence of mCherry-RZZ and farnesylated Spindly (RZZS). Control is empty beads. The intensity level of the green channel in the control was enhanced to allow comparison with GFP-Spindly. The experiment was repeated with two independent RZZS preparations, with at least three replicates, and all samples gave similar results. (c) Immunofluorescence images of interphase HeLa cells expressing the indicated versions of GFP-Spindly and treated and stained as indicated. The experiment was repeated at least three times with similar results. See also Supplementary Movie 2.
Expression of SpindlyΔN but not SpindlyFL (full length) in interphase cells caused spontaneous formation of cytoplasmic filaments containing ZW10, Zwilch and ROD (Figure 4c, Supplementary Figure 5 and Supplementary Movie 2). Cytoplasmic filament formation was abolished upon ZW10 RNAi or mutation of the Spindly CAAX box (C602A), showing it relied on RZZ and its interaction with farnesylated Spindly (Figure 4c). SpindlyΔCCS, which like SpindlyΔN is unable to bind dynein/dynactin, did not induce filament formation (Figure 4c). Together, these results show that RZZS can polymerize in vivo and in vitro. This molecular behaviour is consistent with what is expected from proteins that are structural components of the expanded kinetochore.
A structural conformation of Spindly prevents RZZS oligomerization
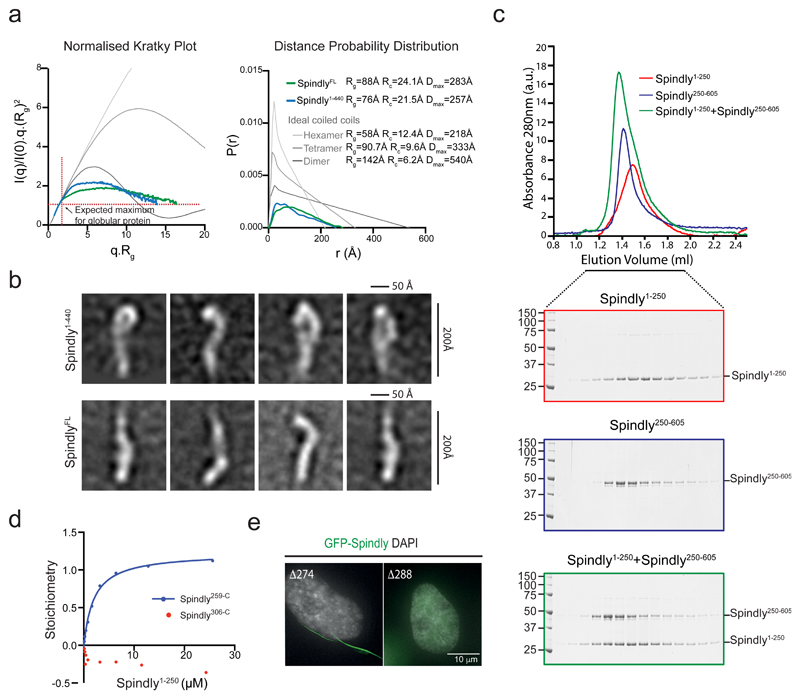
The ability of Spindly to induce formation of a RZZS meshwork was greatly enhanced by deletion of its N-terminal 65 amino acids (Figure 4c). To explain this, we needed to obtain a better understanding of Spindly’s structure and regulation. Residues 1-440 of Spindly are predicted to form a coiled-coil structure and the C-terminal region is likely disordered (Figure 1b)21,26,36. Intramolecular dimensions of recombinant Spindly1-440 as measured by Small Angle X-ray Scattering (SAXS) analysis were inconsistent with those of di-, tetra-, or hexameric coiled-coil models (Figure 5a). SpindlyFL showed an overall shape with dimensions similar to Spindly1-440 (Figure 5a). Negative stain EM on single Spindly1-440 and SpindlyFL particles showed that Spindly adopts elongated shapes with some characteristic “bends” along its length (Figure 5b). The EM analyses thus confirmed SAXS measurements, and suggested that purified Spindly may not form a typical coiled-coil. Finally, SAXS and SEC-MALLS experiments indicated that Spindly1-440 and SpindlyFL are dimeric and trimeric, respectively, in solution (Supplementary Figure 6b,c).
Figure 5. A structural conformation of Spindly prevents RZZS oligomerization.
(a) Normalized Kratky plot (left graph) and paired distance distribution (right graph) of purified SpindlyFL and Spindly1-440. The intersection of the red dotted lines represents the peak for a globular protein irrespective of its molecular weight. Comparison with dimeric, tetrameric and hexameric coiled-coil models of similar size are shown in shades of grey. Rg, radius of gyration; Rc, cross-sectional radius of gyration; Dmax, maximum inter-particle distance. See also Supplementary Figures 6a-b and Supplementary Table 3 for source data. (b) Representative class averages from the negative stain EM of indicated Spindly variant proteins. (c) Elution profiles and SDS-PAGE of SEC experiments on Spindly1-250 and Spindly250-605 and their stoichiometric combination. AU, arbitrary units. The experiment was repeated at least three times with similar results. See Supplementary Table 3 for source data. (d) Surface Plasmon Resonance (SPR) analyses of the interaction between immobilized Spindly259-C (blue) or Spindly306-C (red) and soluble Spindly1-250. The response (y-axis) was normalized to the molecular weight of the analyte to yield stoichiometry of binding. The experiment was repeated three times with similar results. See Supplementary Table 3 for source data. (e) Immunofluorescence images of Spindly in interphase HeLa cells expressing the indicated truncations of GFP-Spindly. The experiment was repeated at least three times with similar results.
Cross-link mass spectrometry of SpindlyFL supported the possibility of a more complicated Spindly fold, and revealed the existence of various long-range intramolecular interactions (Supplementary Figure 6d). Several of these interactions occurred around a cluster of cross-linked lysines at positions 276, 278, 283 and 284 (Supplementary Figure 6d). Size exclusion chromatography showed interaction between an N-terminal fragment of Spindly (Spindly1-250) and a Spindly fragment encompassing C-terminal sequences (Spindly250-605) (Figure 5c), which was verified by Surface Plasmon resonance (SPR) (Figure 5d). No interaction was observed between Spindly1-250 and Spindly306-605 (Figure 5d), suggesting amino acids 259-305 have a crucial role in long-range interactions with the N-terminal Spindly helices. This was consistent with the position of the lysine cluster in the cross-link mass spectrometry analysis (Supplementary Figure 6d). Notably, while a truncated Spindly lacking the N-terminal 274 amino acids (SpindlyΔ274) retained ability to form cytoplasmic filaments in cells, this was abolished by additional removal of 13 amino acids (SpindlyΔ287) (Figure 5e). Thus, this region that is crucial for Spindly intramolecular interactions is also important for in vivo filament formation.
Together, these data support the hypothesis that the Spindly N-terminal region imposes an auto-inhibitory configuration that precludes RZZ-Spindly oligomerization.
Release of Spindly autoinhibition promotes its interaction with RZZ
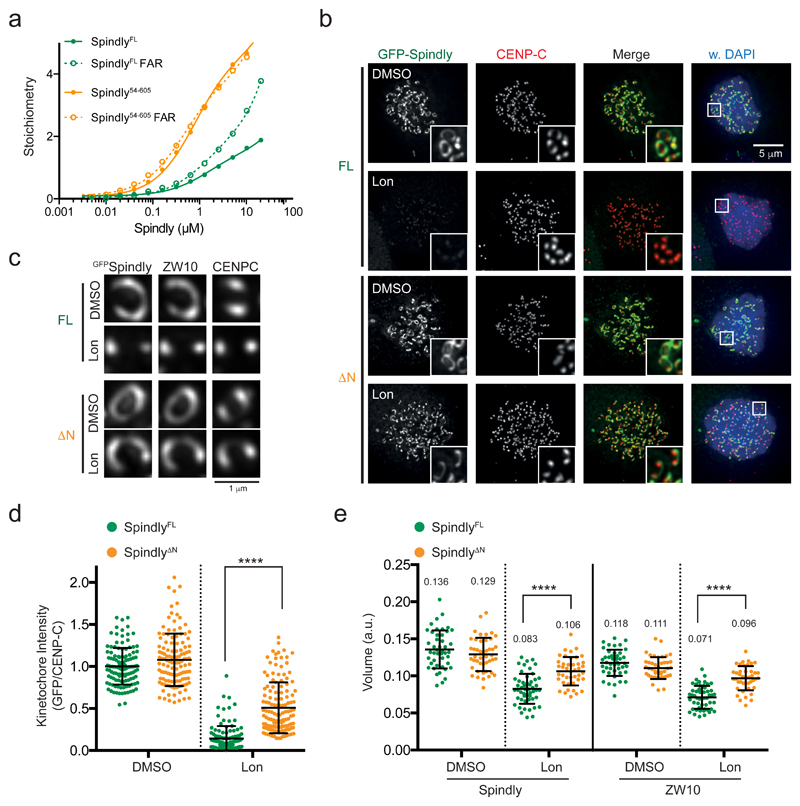
We next performed SPR analyses with immobilized, purified RZZ to examine interactions of recombinant Spindly versions with the RZZ scaffold. In the absence of C-terminal farnesylation, ˜2 molecules of SpindlyFL weakly bound one (dimeric) molecule of RZZ with a KD of ˜1 μM (Figure 6a). Farnesylation had little impact on overall interaction affinity in vitro but increased the number of Spindly molecules accumulating on RZZ. Similar observations were made with an alternative source of Spindly protein (Supplementary Figure 7a). The farnesyl group thus appeared to target Spindly to multiple sites on RZZ or to other Spindly molecules already on RZZ under these conditions. Spindly lacking the N-terminal helices (Spindly54-605) associated with RZZ with higher affinity (˜0.7 μM) and at higher stoichiometries: at least four molecules of Spindly could associate with RZZ. Notably, farnesylation no longer impacted interactions between Spindly54-605 and RZZ (Figure 6a).
Figure 6. Release of Spindly autoinhibition promotes its interaction with RZZ.
(a) SPR analyses of the indicated Spindly variant proteins. The response (y-axis) was normalized to the molecular weight of the analyte to yield stoichiometry of binding. The experiment was repeated three times with similar results. See Supplementary Table 3 for source data. (b-e) Immunofluorescence (b,c) and quantification of kinetochore levels (d) or volumes (e) of Spindly in HeLa expressing GFP-SpindlyFL or GFP-SpindlyΔN and treated with nocodazole and the farnesyl transferase inhibitor Lonafarnib (Lon). In (c), imaging acquisition was set to obtain similar mean intensity levels for the different conditions. The graph in (d) shows the mean kinetochore intensity (±SD) normalized to the values of SpindlyFL. Each dot represents one cell: FL in DMSO (n= 122 cells), FL in Lonafarnib (n= 123 cells), ΔN in DMSO (n= 117 cells), and ΔN in Lonafarnib (n= 136 cells), pooled from four independent experiments. Asterisks indicate significance (one-way ANOVA followed by Tukey's test, F (3, 494) = 354.4). ****P < 0.001. The graph in (e) shows the mean kinetochore volume (±SD). Each dot represents a pair of sister kinetochores: FL in DMSO (n= 45 kinetochores), FL in Lonafarnib (n= 51 kinetochores), ΔN in DMSO (n= 44 kinetochores), and ΔN in Lonafarnib (n= 42 kinetochores), pooled from three independent experiments. Asterisks indicate significance (one-way ANOVA followed by Tukey's test; GFP-Spindly: F (3, 179) = 57.70; ZW10: F (3, 177) = 78.50). ****P < 0.001.
Because farnesylation of SpindlyΔN was required for cytoplasmic filament formation (see Figure 4c) but was dispensable for RZZ-SpindlyΔN interactions when RZZ was on the SPR chip, we reasoned that farnesylation might facilitate initial RZZ-SpindlyΔN interactions that then promote additional, farnesyl-independent ones. To test this, we examined if RZZ complexes concentrated on mitotic kinetochores could recruit unfarnesylated Spindly molecules. As expected, mutation of the farnesylated cysteine (C602A) or treatment with the farnesyl transferase inhibitor Lonafarnib (SCH66336)41 prevented SpindlyFL localization and kinetochore expansion (Figure 6b-e and Supplementary Figure 7c,e). Removal of the N-terminal helices (SpindlyΔN) not only rescued localization of unfarnesylated Spindly but also rescued kinetochore expansion (Figure 6b-e). Rescue of kinetochore localization was dependent on the 275-287 region of Spindly and was independent of dynein (Supplementary Figure 7c-f).
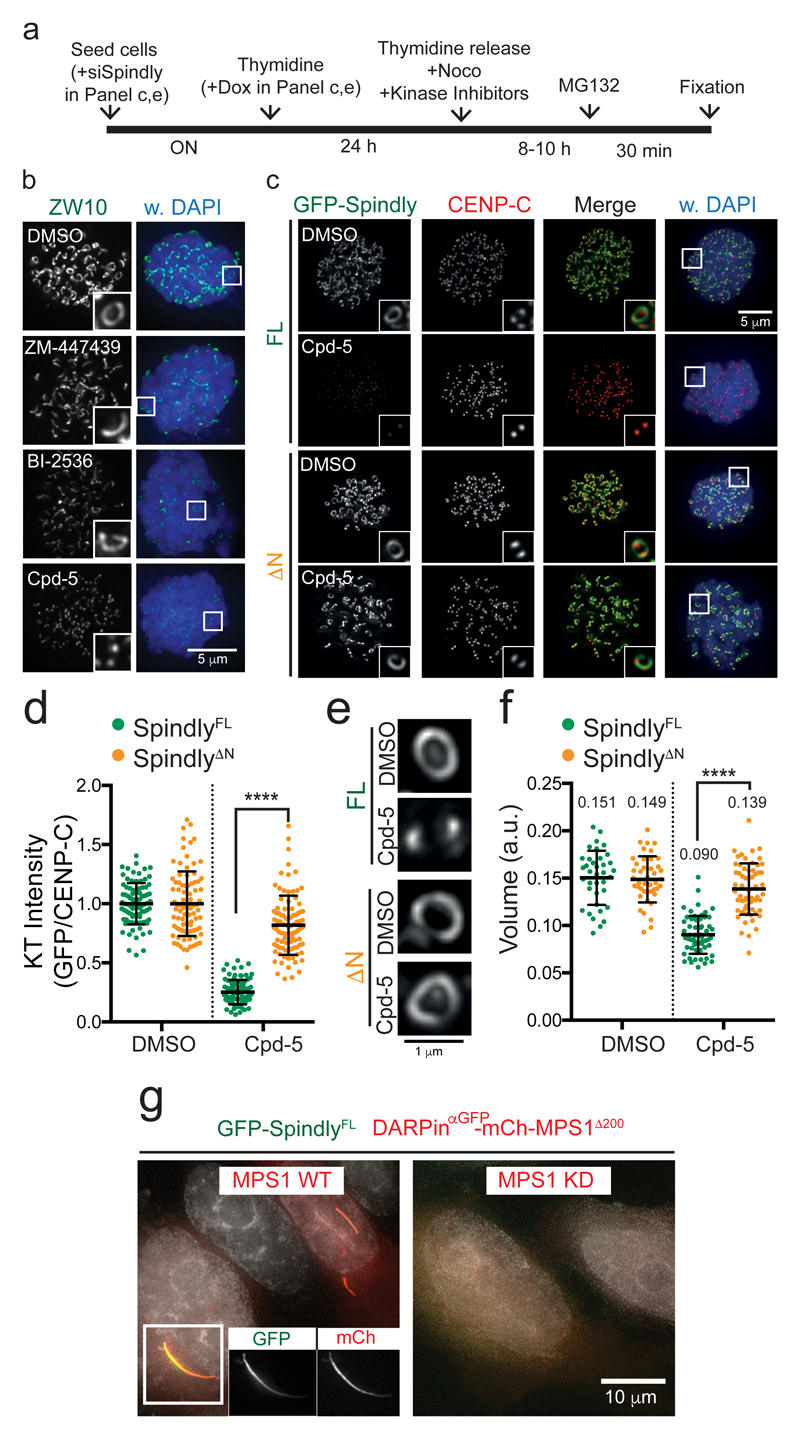
MPS1 promotes RZZS meshwork formation and kinetochore expansion
Our data thus far suggest a mechanism for kinetochore expansion in which release of Spindly auto-inhibition enables direct interactions with RZZ and other Spindly molecules to form the RZZS meshwork. Such a release is expected to occur at or near kinetochores in mitosis, and we reasoned that kinetochore-localized kinases may cause the triggering event. Unlike inhibition of Aurora B or PLK1 (Figure 7a,b), inhibition of MPS1 prior to mitotic entry substantially affected Spindly localization and kinetochore expansion to an extent similar to Spindly depletion (Figure 7b,c-f). Importantly, Spindly kinetochore levels and kinetochore expansion were rescued in MPS1-inhibited cells by deletion of the N-terminal helices of Spindly (Figure 7c-f). To examine if MPS1 is sufficient to trigger Spindly-dependent RZZ oligomerization, we targeted MPS1 to cytoplasmic Spindly by co-expressing GFP-SpindlyFL and DARPinαGFP-MPS1Δ200. This MPS1 variant is unable to localize to kinetochores42 but can bind Spindly’s GFP tag by virtue of a GFP-binding DARPin moiety43. Strikingly, when bound by active but not inactive MPS1, GFP-SpindlyFL was able to induce interphasic filament formation (Figure 7g), just like SpindlyΔN could in the absence of MPS1 activity (Figure 4a). We conclude that MPS1 kinase activity triggers RZZ-Spindly oligomerization by impacting the mechanism that releases inhibitory intra-molecular Spindly interactions.
Figure 7. MPS1 promotes RZZS meshwork formation and kinetochore expansion.
(a) Timeline of the treatments with kinase inhibitors and nocodazole (Noco) of the experiments shown in b-f. (b) Representative images of ZW10 immunostainings of cells treated as indicated in (a). The intensity levels of the zoom ins (on right) were equalized to facilitate the direct comparison of the size of the kinetochores. The experiment was repeated at least three times with similar results. (c,d) Representative images (c) and quantification (d) of kinetochore localization of the indicated GFP-Spindly variants in HeLa cells treated with nocodazole and Cpd-5 as indicated in (a). The graph shows the mean kinetochore intensity (±SD) normalized to the values of SpindlyFL. Each dot represents one cell: FL in DMSO (n= 87 cells), FL in Cpd-5 (n = 92 cells), ΔN in DMSO (n= 87 cells), and ΔN in Cpd-5 (n= 93 cells), pooled from three independent experiments. Asterisks indicate significance (one-way ANOVA followed by Tukey's test; F (3, 355) = 255.6). ****P < 0.001. (e,f) Representative images (e) and volume quantification (f) of immunostained kinetochores of HeLa cells expressing the indicated versions of GFP-Spindly and treated as indicated in (a). In (e), imaging acquisition was set to obtain similar mean intensity levels for the different conditions. The graph shows the mean kinetochore volume (±SD). Each dot represents a pair of sister kinetochores: FL in DMSO (n= 37 kinetochores), FL in Cpd-5 (n= 48 kinetochores), ΔN in DMSO (n= 43 kinetochores), and ΔN in Cpd-5 (n= 60 kinetochores), pooled from three independent experiments. Asterisks indicate significance (one-way ANOVA followed by Tukey's test; F (3, 194) = 68.18). ****P < 0.001. (g) Representative images of HeLa cells overexpressing GFP-SpindlyFL and an active (WT) or kinase dead (KD) version of mCherry-MPS1Δ200 targeted to GFP-Spindly by DARPinαGFP. The experiment was repeated two times with similar results.
The expanded kinetochore module interacts with microtubule lattices
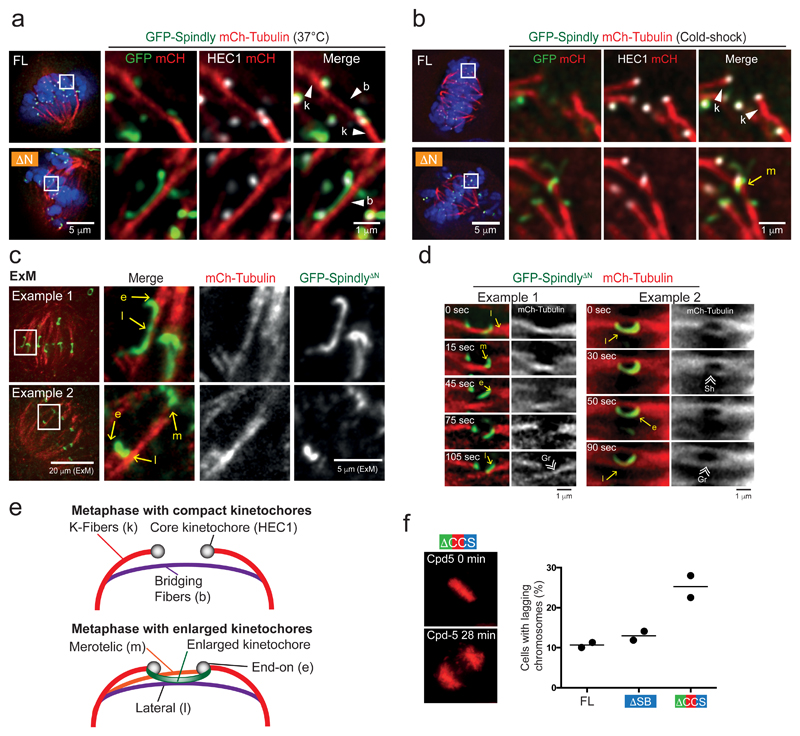
Elegant ultrastructural work had suggested that fibrous coronas facilitate lateral microtubule capture44,45 and mathematical modeling had predicted the same for expanded kinetochores6. The persistent presence of an expanded kinetochore in cells expressing SpindlyΔCCS and SpindlyΔN provided a means to examine the functional relevance of kinetochore expansion and subsequent compaction. Imaging of congressed chromosomes in SpindlyΔN-expressing cells showed that the expanded kinetochores of sister chromatids engaged with the sides of microtubules (Figure 8a) and had lower Astrin levels, indicative of fewer mature end-on kinetochore-microtubule interactions (Supplementary Figure 8a,b)46. The microtubules that engaged expanded kinetochores were positive for PRC1 (Supplementary Figure 8c) and thus likely were anti-parallel bridging fibers (‘b’ in Figure 8a and Supplementary Figure 8c)47. Cold treatment caused depolymerization of PRC1-positive bundles (Supplementary Figure 8c) and revealed the additional presence of stable, end-on microtubule interactions (Figure 8b and Supplementary Figure 8c). Super-resolution imaging by Expansion Microscopy (ExM)48 confirmed the co-occurrence of lateral and end-on attachments on the same kinetochore (‘l’ and ‘e’ in Figure 8c) and showed an extensive kinetochore surface capable of lateral microtubule interactions (Example 1 in Figure 8c). Live imaging of mCherry-Tubulin and GFP-SpindlyΔN further showed that the expanded kinetochore maintained interactions with the lattices of microtubules that go through rounds of growth and shrinkage (‘Gr’ and ‘Sh’ in Example 2 of Figure 8d and Supplementary Figure 8d, and Supplementary Movies 4-5), and with the lattices of microtubules that move away from the sister kinetochores (Example 1 in Figure 8d and Supplementary Movie 3). In line with a proposed role for the fibrous corona6,15, the expanded kinetochore thus efficiently captures and maintains interactions with the lattices of dynamic microtubules (Figure 8e).
Figure 8. The expanded kinetochore module interacts with microtubule lattices and prevent bi-orientation.
(a,b) Representative single z-plane immunofluorescence images of HeLa cells in metaphase, transfected with siRNA to Spindly and expressing the indicated GFP-Spindly variants and mCherry-tubulin. Cells were fixed at 37°C (a) or after cold-shock (b). (‘b’, bridging fibers; ‘k’, k-fibers; ‘m’ merotelic attachment). The experiment was repeated at least three times with similar results. (c) Two examples of ExM (expansion microscopy) images of HeLa cells transfected with siRNA to Spindly and expressing GFP-SpindlyΔN and mCherry-tubulin. Single z-plane images show pairs of enlarged kinetochores with simultaneous end-on (e) and lateral (l) attachments (example 1) or end-on (e), lateral (l), and merotelic (m) attachments (example 2). The experiment was repeated at least three times with similar results. (d) Live-cell imaging of HeLa cells transfected with siRNA to Spindly and expressing GFP-SpindlyΔN and mCherry-tubulin. (‘Gr’, growing microtubules; ‘Sh’ shrinking microtubules; ‘e', end-on attachment; ‘l’, lateral attachment; ‘m’, merotelic attachment). Maximum projections of several z-planes are shown. See also Supplementary Movies 3 (Example 1) and 4 (Example 2). The experiment was repeated at least three times with similar results. (e) Cartoon summarizing the interaction of compacted or enlarged kinetochores with microtubule fibers. (f) Quantification of chromosome segregation errors by live-cell imaging of metaphase HeLa cells transfected with siRNA to Spindly, expressing the indicated GFP-Spindly variants, and treated with Cpd-5. The bar graph shows the mean percentage of cells showing lagging chromosomes of two independent experiments. (FL n= 222 cells; ΔSB n= 179 cells; ΔCCS n= 167). See Supplementary Table 3 for source data.
Persistently expanded kinetochores cause attachment errors and chromosome missegregations
We noticed that the expanded kinetochores of sister chromatids in SpindlyΔN cells frequently formed merotelic attachments (‘m’ in Figure 8b and Example 2 of Figure 8c). Live imaging captured these events: During successive cycles of capture-release, we observed short-lived events in which expanded kinetochores simultaneously bound to dynamic microtubules and stable k-fibers from opposite orientations, briefly causing merotelic attachments (see second 15 in Example 1 of Figure 8d and Supplementary Movie 3). Merotely is an important cause of chromosome segregation errors in both healthy and cancerous cells49. To examine if inability to compact kinetochores increases the frequency of chromosome segregation errors, we live imaged Spindly mutant cells undergoing anaphase, induced by treatment with an MPS1 inhibitor to bypass the SAC silencing defect resulting from the absence of kinetochore dynein38,39. We analyzed only cells that achieved full chromosome alignment so as not to bias for alignment problems associated with persistently expanded kinetochores (Figure 8f). In contrast to cells expressing Spindly variants that allowed kinetochore compaction (SpindlyFL and SpindlyΔSB), cells expressing SpindlyΔCCS showed a high rate of lagging chromosomes in anaphase, indicative of persistent merotelic attachments. Kinetochore compaction is therefore important to establish proper amphitelic attachments of sister chromatids.
Discussion
While our understanding of the assembly mechanisms of the core kinetochore has seen great progress in recent years50, the molecular mechanisms and functions of kinetochore size dynamics have remained enigmatic. Our data are consistent with a model in which kinetochore expansion, driven by Spindly and RZZ, enables efficient capture of microtubule lattices. We propose (Supplementary Figure 8e) that before localizing to kinetochores, Spindly exists in an auto-inhibited state that masks a surface required for polymerization of RZZ. Farnesylation-dependent targeting of Spindly to kinetochores by virtue of an initial interaction with ROD then enables kinetochore-localized MPS1 activity to (directly or indirectly) release Spindly auto-inhibition and stimulate RZZ-Spindly polymerization. Inhibitory intramolecular interactions have also been observed in the Spindly-related dynein adaptor BICD251. Meanwhile, Spindly-driven kinetochore expansion has set the stage for future compaction by dynein once the expanded kinetochore module interacts with microtubules. Removal of the module may then promote exposure of the KMN network, facilitating end-on microtubule interactions and chromosome biorientation (Supplementary Figure 8e).
The expanded kinetochore can remain relatively intact when disconnected from the underlying outer kinetochore (Figure 2), and therefore appears to be a stable kinetochore sub-module like the CCAN and the KMN network, albeit more transient. We speculate that the RZZS meshwork is the underlying scaffold of the expanded kinetochore module. The properties of this meshwork resemble those of other cellular polymers that assemble through a ‘collaborative’ mechanism52. Collaborative filaments form on a supporting matrix, such as DNA or membranes, and frequently use hydrophobic interactions as a driving force for their assembly. Analogously, RZZ assembles on the core kinetochore in a manner dependent on BUB1 and KNL153,54, and subsequently forms a filamentous structure involving hydrophobic interactions, which is maintained even when its association to the core kinetochore is lost after brief CDK1 inhibition.
Pioneering ultrastructural studies during the second half of the 20th century had observed that outer-plates of expanded kinetochores have a fibrous corona (original works reviewed in12) and that at least CENP-E, dynein and CLASP1 are part of its constituents17–20. Because of their similar localization to expanded kinetochores, CENP-F, RZZ, Spindly, and MAD1 are also likely fibrous corona constituents6,21,25,26,28,29,55 (Figures 2 and 3). We now show that fibrous corona formation and kinetochore expansion both rely on RZZS (Figure 3), implying they are mechanistically similar if not identical. This is consistent with absence of RZZS from metaphase kinetochores, which are compact and devoid of a corona12. It will be important to examine how the RZZS meshwork interacts with other corona constituents and which of these, if any, additionally contribute to kinetochore expansion and corona formation. Having such insights will greatly facilitate understanding of how fibrous coronas capture the lateral sides of microtubules, how they integrate with SAC signaling, and how their disassembly enables correct interactions with microtubule plus ends.
Methods
Cell Culture and generation of stable cell lines
HeLa Flp-In cells were grown in Dulbecco’s modified Eagle medium (DMEM; Sigma D6429) supplemented with 9% Tetracycline-free Fetal Bovine Serum (FBS), penicillin-Streptomycin (50 μg/ml; Sigma P0781) and Ala-Gln (2 mM; Sigma G8541) at 37°C and 5% CO2. Live-cell imaging was performed in DMEM without Phenol red (Sigma; D1145) supplemented with 9% FBS, penicillin-streptomycin (50 μg/ml; Sigma P0781) and Ala-Gln (2 mM; Sigma G8541). Constructs were expressed by addition of 1 μg/ml doxycycline.
Plasmids were transfected into Flp-in HeLa cells using Fugene HD (Promega) according to the manufacturer’s instructions. To generate stably integrated HeLa FlpIN, with LAP -tagged genes stably integrated in the FRT site and TetR inducible, pCDNA5-constructs were co-transfected with pOG44 recombinase in a 1:9 and kept in Hygromycin (Roche, 10843555001) selection for three weeks. To generate stably expressing mCherry-Tubulin and DARPInGFP-mCherry-MPS1Δ200 Hela FlpIn cell lines, pmCherry-α-tubulin-IRES-puro2 56 (Addgene #21043) or pCDNA4-DARPInGFP-mCherry-Mps1Δ200 variants were transfected and selected in Puromycin (Sigma, P7255) for three weeks. For generation of stably iCas9 LAP-MAD1GR cell lines, Hela Flp-In cells were first infected with lentiviral iCas9 and cultured in puromycin selection for three weeks. Cells were then seeded to clonal density in 96-well plates and grown in Puromycin. A colony from a single well was expanded, and validated for inducible expression of Cas9 upon doxycyclin addition. This monoclonal cell line was transfected with pCDNA5-LAP-MAD1 U6-GuideA and cultured in Hygromycin and Puromycin for three weeks. Cas9 and LAP-MAD1GR were induced with doxycyclin addition and cells were then seeded to clonal density in 96-well plates and kept in the presence of puromycin, hygromycin and doxycycline. A colony from a single well was expanded, and validated for presence of LAP-MAD1 and reduced levels of endogenous MAD1. Protein levels of the different Spindly constructs are shown in Supplementary Figures 3a and 7b.
Plasmids, cloning and virus production
Cloning was performed by Gibson assembly and restriction-ligation strategies. The list of plasmids and primers used in this study can be found in Supplementary Table 1. Spindly mutants derived from pCDNA5-LAP-Spindly (a gift from Reto Gassmann39) were created by assembling two PCR fragments generated using primers 33/34 in combination with the Spindly primers 1-12. pCDNA5-LAP-Zwilch encodes full-length, N-terminally LAP-tagged and siRNA-resistant Zwilch. Zwilch ORF was cloned into pCDNA5-LAP and mutated with primers 19-22. pCDNA5-LAP-MAD1GR U6-GuideA expresses a sgRNA that targets MAD1 (CCTCCCCTTCAGTGCGTTGA) and additionally encodes an N-terminally LAP-tagged MAD1 version resistant to the sgRNA (LAP-MAD1GR). MAD1 ORF was cloned into pCDNA5-LAP and mutated using primers 30-32. The U6 promoter-sgRNA scaffold containing the Guide A was introduced in pCDNA5 with primers 13-16. pCDNA4-DARPinαGFP-mCherry-Δ200-MPS1 and pCDNA4- DARPinαGFP-mCherry-Δ200-MPS1KD encode a resistant version of MPS1 to Cpd-5 (C604Y) and kinase dead (D664A), respectively, both lacking the first 200 amino acids and tagged with mCherry and a DARPin that recognizes GFP (DARPin 3G86.32)43. The DARPinαGFP fragment was synthesized by GenScript and cloned into the BamHI/NotI sites of pCDNA5-LAP-Δ200-MPS142. Subsequently, mCherry amplified with primers 23/24 was introduced into the NotI site, and MPS1 was mutated with primers 25-28. Finally, DARPinαGFP-mCherry-Δ200-MPS1 constructs were subcloned into the KpnI and ApaI sites of pCDNA4.
E. coli constructs were generated as follows. The full length Spindly and the 306-605 constructs were cloned in NKI LIC 1.2 and NKI LIC 1.1 vectors respectively which resulted in the introduction of a cleavable N-terminal 6x His-tag to these constructs. Spindly 1-440, 54-605 and 259-605 constructs were cloned in NKI LIC 1.10 vector which resulted in the introduction of a cleavable N-terminal 6x His-SUMO tag to these constructs. The genes for these constructs were PCR amplified from pCDNA5-LAP-Spindly using primers 35-44.
Virions were first generated by transient transfection of HEK 293T cells with the pCW-9 and separate plasmids that express Gag-Pol, Rev, Tat and VSV-G. Supernatants were clarified by filtration.
Cell treatments, transfections and reconstitution
For knockdown experiments, siRNAs (see Supplementary Table 1 for sequences and concentrations) were transfected using Hiperfect (Qiagen) or RNAi Max (Thermo Fisher Scientific) according to manufacturer’s instructions. After 16 hours of siRNA treatment, cells were arrested in S-phase by addition of thymidine (2 mM; Sigma-Aldrich Cat#T1895). For the rescue experiments, doxycycline (1 μg/ml; Sigma-Aldrich Cat#D9891) was also added at this point to induce the expression of the constructs. In the experiments in which farnesyl transferase activity was inhibited, Lonafarnib (5 μM; Selleckchem Cat#: S2797) was added together with thymidine, and the induction of the constructs with doxycycline was performed 8 hours later. After 24 hours of thymidine addition, cells were released and treated with the indicated drugs [ZM-447439 (2 μM; Tocris Bioscience, Cat#2458); BI-2536 (100 nM; Advanced ChemBlocks Cat#10293); Cpd-5 (250 nM; gift from R.H. Medema); RO-3306 (10 μM; Tocris Bioscience Cat#4181); Nocodazole (3.3 μM; Sigma-Aldrich Cat#M1404); MG-132 (5 mM), Cat#C2211]. Cells were used for experiments between 6-10 hours after thymidine release.
Immunofluorescence and Expansion Microscopy
For immunofluorescence, HeLa Flp-In cells grown on 12-mm coverslip (No. 1.5) were permeabilized for 1 minute with warm 0.2% Triton in PHEM buffer, followed by fixation for 10 minutes with 4% PFA in PBS. For tubulin visualization, mCherry-αTubulin expressing cells were additionally stained with anti-α-tubulin antibody. For analysis of cold-stable microtubules, cells were placed 15 minutes on ice prior pre-extraction and fixation. For staining of p150glued, cells were fixed for 20 minutes in methanol at -20°C. After fixation, coverslips were washed three times with PBS and blocked with 3% BSA in PBS for 1 hour at RT. Primary antibodies diluted in 3% BSA were added to the coverslips and incubated for 16 hours at 4°C. (A list with the primary and secondary antibodies can be found in Supplementary Table 2). Subsequently, cells were washed three times with 0.1% Triton in PBS and incubated with DAPI and secondary antibodies in 3% BSA for another hour at RT. Finally, coverslips were washed three times with 0.1% Triton in PBS and mounted onto glass slides using Prolong Gold antifade. All images were acquired on a deconvolution system (DeltaVision Elite Applied Precision/GE Healthcare) with a x100/1.40 NA UPlanSApo objective (Olympus) using SoftWorx 6.0 software (Applied Precision/GE Healthcare). Images were acquired as z-stacks at 0.2-μm intervals and deconvolved using SoftWoRx.
Expansion Microscopy (ExM) was performed following the protocol described in 48. Cells were seeded in 12-mm coverslips, pre-extracted with 0.5% Triton in PHEM buffer, and fixed for 10 minutes with 3.2% PFA and 0.1% Glutaraldehyde in PHEM. Coverslips were briefly washed with PBS, quenched in 10 mM sodium Borohydrate for 5 minutes, washed three times with PBS, and then stained with a 5-fold excess of primary and secondary antibodies. Samples were then treated with 0.25% Glutaraldehyde in PBS for 10 minutes, washed three times with PBS and incubated for 1 hour in monomer solution [1X PBS, 2 M NaCl, 2,5% (wt/wt) acrylamide 0,15% (wt/wt) N,N’-methylenebisacrylamide 8,625% (wt/wt) sodium acrylate]. Coverslips were placed on top of a drop of 70 μl of freshly prepared gelation solution [monomer solution supplemented with 0,2% (wt/wt) TEMED and 0,2% (wt/wt) APS] and incubated for 30 minutes at RT. Gels were then incubated in digestion solution (8 units/mL Proteinase K, 1x TAE, 0,5% TX-100, 0,8 M guanidine HCl) for 30 minutes at 37°C, and expanded in a 10-cm plate by several washings of 30 minutes with excess volume of Milli-Q water. Expanded samples were imaged at 500 nm z-slices on an Andor CSU-W1 spinning disk (50 µm disk) with 60x 1.3 NA water objective lens (Nikon) and 1.5 zoom.
Live-cell Imaging
For live-cell imaging, cells were seeded in 8-well plates (μ-Slide 8 well, Ibidi). Imaging of mCherry-Tubulin and GFP-Spindly was performed over 7 Z-slices separated by 200 nm every 5-15 seconds at 1x1 binning on an Andor CSU-W1 spinning disk (50 µm disk) with 100x 1.45 NA oil objective lens (Nikon). 488 and 561 nm laser were used for sample excitation and images were acquired using Andor iXon-888 EMCCD camera. Emission filters were 525nm-50 bandpass for GFP and 655nm-150 bandpass for mCherry. Cells were kept at 37°C and 5% CO2 using a cage incubator and Boldline temperature/CO2 controller (OKO-Lab).
For mitotic progression experiments, chromatin was visualized with siR-DNA (Spirochrome). Eight hours later after thymidine release, Cpd-5 was added and filming was immediately started, taking 8 z-slices separated by 2 μm every 4 minutes at 2x2 binning on a Nikon Ti-E motorized microscope equipped with a Zyla 4.2Mpx sCMOS camera (Andor) and 40x 1.3 NA objective lens (Nikon). Fluorescence excitation was done using Spectra X LED illumination system (Lumencor) and Chroma-ET filtersets. Cells were kept at 37°C and 5% CO2 using a cage incubator and Boldline temperature/CO2 controller (OKO-Lab). Maximal intensity projections were performed and scored for chromosome missegregations. Only cells showing all chromosomes aligned before Cpd-5 addition were selected for analysis.
Settings of image acquisition and quantification
All images used for quantification were deconvolved using SoftWoRx and analysed using Image J (https://imagej.nih.gov/ij/)/Fiji (http://fiji.sc/#). For quantification of kinetochore levels, all images of similarly stained experiments were acquired with identical illumination settings. For analysis of the localization of the different Spindly variants, cells with maximum GFP levels on kinetochores were selected for imaging. Kinetochore levels were determined on maximum projections of z-stacks using an Image J macro which thresholds the CENP-C signal within the DAPI area as described in 57.
For kinetochore volume measurements, illumination settings on the DeltaVision Elite microscope were set to obtain a mean intensity of 3000 in the middle region of the cell. Z-stacks of single kinetochores showing maximal expansion were selected for analysis. Volume was measured using the 3D object counter macro of Fiji with the threshold set at the mean intensity of the kinetochore. The mean intensity of the kinetochore was determined as the mean intensity within the region selected by the auto-threshold function of image J applied on the maximal intensity projection of the Z-stack of the kinetochore.
Electron microscopy and quantification
Cells were seeded in 6-well culture plates, treated with the indicated siRNAs, synchronized in Thymidine and released in nocodazole. Mitotic cells were chemically fixed by adding PHEM buffer (pH=6.9) containing 2.5% glutaraldehyde (Merck) and 2% paraformaldehyde (Sigma) to the culture medium for 10 minutes. This was replaced by fresh fixative for 2 hours at room temperature after which cells were scraped and pelleted in 2% low-melting point agarose in PHEM. Cells were postfixed in PHEM buffer containing 1% osmium tetroxide and 1.5% potassium ferrocyanide for 2 hours at 4°C, followed by 0.5% uranyl acetate in dH2O for 1 hour at 4°C. Cells were dehydrated using a graded acetone series and embedded in epon epoxy resin (Polysciences).
Ultrathin sections of 70 nm were collected on formvar and carbon coated, 50 mesh copper grids. The sections were contrasted using 1% uranyl acetate and Reynold’s lead citrate using an AC20 automated grid stainer (Leica). Kinetochores were imaged using a Tecnai 12 Spirit transmission electron microscope (Thermo Scientific) equipped with a Veleta 2k x 2k CCD camera (EMSIS) at a pixel size of 1 nm. All photos were blinded and independently analyzed using Fiji. The surface area of the fibrous corona was measured for all conditions using the polygonal area selection tool. Per experiment, at least 50 kinetochores were imaged from random sections. In 21 out of 50 images of siZW10 condition, the surface of the fibrous corona could not be determined, like in the example shown in Figure 3e. In the control and siSpindly samples these numbers were 21 out of 162 and 16 out of 138, respectively.
Conserved Feature Extraction (ConFeaX)
We utilised our previously established workflow40 to establish the orthologs of Spindly (Figure 1b) and other dynein-dynactin adapters (Supplementary Figure 2) in a set of 100 eukaryotic genomes (see suppl. sequence file 1 and 2). The orthologous sequences were masked using IUpred (disorder/order threshold = 0,4) 58 and MARCOIL (coiled-coil threshold = 90)59. ConFeaX starts with a probablistic search for short conserved regions using the MEME algorithm (option:any number of repeats) 60. Significant motif hits are extended on both sides by five residues to compensate for the strict treatment of alignment information by the MEME algorithm and aligned using MAFFT-LINSI 61 to introduce gaps. The alignments were modelled using the HMMER package62 and sensitive profile HMM searches were iterated (jackhmmer-like approach; E-value=1) until convergence. For the multiple sequence alignment of the Spindly box in various dynein-dynactin adapters (Supplementary Figure 2) we followed the coordinates previously established by 26.
Western Blotting
Cells were collected by mitotic shake-off and lysed in 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 2% NP-40, 1 mM EDTA and protease inhibitor cocktail (Roche). Lysates were sonicated and centrifuged for 10’at 20.000 x g at 4°C. Supernatants were collected, supplied with Laemmli buffer and boiled for 5 minutes at 96°C. Electrophoresis in SDS-acrylamide gel, transfer to nitrocellulose membranes and immunoblotting were performed using standard protocols. Western blot signals were detected by chemiluminescence using an ImageQuant LAS 4000 (GE Healthcare) imager.
Expression and purification of mCherry-RZZ
mCherry-RZZ was produced using the biGBac system for baculovirus expression. Specically, the coding sequence of ROD, Zwilch and ZW10 were subcloned into the multiple cloning site of pLIB. The mCherry coding sequence was then inserted by PCR at the 5′ end of ROD. Bacmid was produced from EMBacY cells and subsequently used to transfect Sf9 cells and produce baculovirus. The latter was amplifed through three rounds of amplifcation and used to infect TnaO38 cells. Cells infected with the mCherryRZZ virus were cultured for 72 h before harvesting. Cells were resuspended in lysis buffer (50 mM Hepes, pH 8, 250 mM NaCl, 10% glycerol, 20 mM imidazole, and 2 mM tris(2-carboxyethyl) phosphine [TCEP]). Resuspended cells were lysed by sonication in the presence of the protease inhibitor mix HP Plus (Serva), DNaseI (Roche), and 1 mM PMSF before clearance at 100,000 g at 4°C for 45 min. The cleared lysate was applied to a 5ml NiNTA Fast Flow column (GE Healthcare) preequilibrated in lysis buffer. The column was washed with 100 column volumes of lysis buffer, and the bound protein was eluted with lysis buffer supplemented with 300 mM imidazole. The eluate was diluted five times with dilution buffer (50 mM Hepes, pH 8, and 2 mM TCEP) to a final NaCl concentration of 50 mM and applied to a 6 ml Resource Q anion exchange column (GE Healthcare) pre-equilibrated in buffer A (50 mM Hepes, pH 8, 50 mM NaCl, and 2 mM TCEP). Elution of bound protein was achieved by a linear gradient from 50–400 mM NaCl in 20 column volumes. Relevant fractions were concentrated in 30kD molecular mass cutoff Amicon concentrators (EMD Millipore) and applied to a Superose6 10/300 column (GE Healthcare) equilibrated in SEC buffer (50 mM Hepes, pH 8, 250 mM NaCl, and 2 mM TCEP). SEC was performed under isocratic conditions at a flow rate of 0.4 ml/min, and the relevant fractions were pooled, concentrated, flash frozen in liquid nitrogen, and stored at −80°C.
Reconstitution of mCherryRZZ/Spindly
Spindly was incubated with mCherryRZZ, farnesyl transferase and 3M excess of farnesyl di-phosphate for 90 min at 25°C in the reaction buffer (50 mM Hepes, pH 8.0, 250 mM NaCl, 10 mM MgCl2, and 2 mM TCEP). The sample was then loaded onto a Superose 6 5/150 column (GE Healthcare) pre-equilibrated in the reaction buffer and the relevant fractions were pooled, concentrated, flash frozen in liquid nitrogen, and stored at −80°C.
Protein expression and purification of Spindly 1-605, 1-440, 1-250, 259-605 and 306-605 in E. coli
Protein expression in E. coli BL21 DE3 cells was induced by adding 0.3mM IPTG to the culture at OD600 of 0.6. Post induction, the cultures were grown overnight at 16°C, harvested and stored at -20°C. The cells were lysed by sonication in 40 mM HEPES pH 7.5, 500 mM NaCl, 2 mM TCEP and supplemented with protease inhibitor tablets (ThermoFisher). For all the constructs, a three-step purification was followed–immobilized metal affinity chromatography followed by ion-exchange and gel filtration. Except for the 259-C construct, the affinity tags for all the other constructs were removed. The proteins were stored in 40 mM HEPES pH 7.5, 100 mM NaCl and 2 mM TCEP. SDS-PAGE gels and Coomassie staining of the constructs can be found in Supplementary Figure 6a.
Expression and purification of Spindly1-605, Spindly76-605, Spindly250-605 and Spindly1-250 in insect cells
Expression of Spindly constructs was performed with the biGBac system 63 in TnaO38 cells. Specically, the coding sequence of Spindly constructs were subcloned into the multiple cloning site of pLIB with a 6-histidine tag at the 5’end. Lysis of a pellet from 500 ml expression culture was performed by sonication in 100 ml lysis buffer (50 mM Hepes, pH 8.5, 200 mM NaCl, 10% glycerol, 20 mM imidazole, 2mM TCEP, 1 mM PMSF, and 1 mM protease inhibitor cocktail [Serva]). The cleared lysate was loaded onto an equilibrated 5-ml HisTrap Fast Flow column (GE Healthcare) using a peristaltic pump (2 ml/min ow rate). The column was washed with 500 ml wash buffer (50 mM Hepes, pH 8.5, 200 mM NaCl, 10% glycerol, 20 mM imidazole, and 2mM TCEP). Elution was performed with wash buffer supplemented with 250 mM imidazole. The 2-ml fractions were analyzed by SDS- PAGE, and those containing Spindly were concentrated up to a volume of 10 ml. The protein solution was diluted five times with buffer A and subsequently purified on a Resource Q anion exchange column. Peak fractions were analyzed by SDS-PAGE, and those containing Spindly constructs were concentrated up to 500 μl and applied to a Superose 6 10/300 column. The peak fractions were analyzed by SDS-PAGE, and those containing pure Spindly proteins were concentrated up to 20 mg/ml, flash frozen in liquid nitrogen, and stored at −80°C.
In vitro farnesylation of Spindly
Spindly was incubated with farnesyl transferase and 3M excess of farnesyl di-phosphate for 90 min at 25°C in the reaction buffer (50 mM Hepes, pH 8.0, 250 mM NaCl, 10 mM MgCl2, and 2 mM TCEP). The sample was then loaded onto a Superose 6 5/150 column (GE Healthcare) pre-equilibrated in the reaction buffer and the relevant fractions were pooled, concentrated, flash frozen in liquid nitrogen, and stored at −80°C.
In vitro filamentation assays
Filamentation was performed in 10 μl of 50 mM Hepes, pH 7.5, 10 mM KCl, 1 mM MgCl2 containing 2 μM of mCherry-RZZ and 8 μM of farnesylated SpindlyFL isolated from insect cells. Reaction was incubated at the indicated conditions for 1 hour and filaments were imaged in #1.5 glass bottom 96-well plates (Cellvis) on an Andor CSU-W1 spinning disk (50 µm disk) with 100x 1.45 NA oil objective lens (Nikon). Where indicated, filament reaction was induced in the presence of GFP-Spindly beads or empty beads at 30°C. Before imaging, beads were diluted in 100 μl of buffer. To immobilize GFP-Spindly in agarose beads, HeLa cells expressing GFP-Spindly were arrested in nocodazole and collected by mitotic shake-off. Cells were lysed in 50 mM Hepes, pH 7.5, 10 mM KCl, 1 mM MgCl2 supplemented with 1% NP-40 and protease inhibitor cocktail (Roche). Lysates were sonicated and centrifuged for 10’ at 20.000 x g at 4°C. Supernatants were collected and incubated with GFP-Trap beads (Chromotek) for 2 hours at 4°C and washed 5 times in the same buffer without NP-40.
Negative stain EM
Concentrated protein samples were diluted to a concentration of ~30 μg/mL. Samples were applied to glow-discharged carbon-coated copper grids, washed quickly with distilled water and negatively stained with 2% (w/v) uranyl acetate and observed using a JEOL-1230 operated at 100 kV. Images of single molecules were obtained automatically using a TVIPS F416 CMOS and a final magnification of 54,926.
All image processing was performed using the Scipion platform (http://scipion.cnb.csic.es), which is an image processing framework that integrates several software packages into a unified interface 64. For each sample (FL, 1-440), a total of (10767, 13400) particles were extracted from (58, 64) micrographs respectively using the particle picking tool of Xmipp 65. The particles were classified in 2D using Xmipp 66.
Cross-linking and Mass Spectrometry
86 μM of recombinant SpindlyFL purified from E. coli was cross-linked with 860 μM of freshly prepared bis(sulfosuccinimidyl)suberate (BS3) (ThermoFisher Scientific) for 30 minutes at RT. Reaction was quenched by adding Tris to a final concentration of 50 mM for 10 min at RT. The sample was partially dried down and denatured in 8 M urea. Disulfide bonds were reduced using 10 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP) (Sigma), and the free cysteines alkylated using 50 mM 2-chloroacetamide (Sigma). The sample was diluted to 2 M urea using 500 mM ammonium bicarbonate (Fluka) containing 3 μg of trypsine/lys-c (Promega). Protein was digested overnight at 37 °C, desalted on a C-18 zip-tip and washed with 0.1% formic acid in water. SCX fractionation was then performed using the Thermo Ultimate 3000 HPLC system equipped with a PolyLC Polysulfoethyl A column (100 x 1.0mm, 3 micron, cat. no. 101SE0303). Fractions were dried down and again desalted on C-18 zip-tips. Fractions were loaded on a 30 cm in-house prepared column containing 1.9 micron C-18 beads and measured on the Thermo Orbitrap Fusion Tribrid mass spectrometer using a Proxeon 1000 bar nano-LC system and a 150 min gradient. MS1 resolution was set at 120K, HCD (35%) MS2 at 30K also in the orbitrap with a max injection time of 100 ms and AGC target of 50000. MS2 was triggered for peptides with at least 3+ charges. The resulting data was analysed using the Xlinkx workflow in Thermo Proteome Discoverer 2.2.
SEC-SAXS data collection and analysis
Synchrotron X-ray data for SpindlyFL and Spindly1-440 constructs were collected on a Pilatus 1M detector at the ESRF beamline BM29 67. 50 μl of each sample, at a concentration 8 mg ml-1 were loaded onto a Superose 6 Increase 3.2/300 column. The flow rate for SAXS data collection was 0.2 ml min-1 and a scattering profile was integrated every second. Frames for each dataset were selected based on the examination of the Size Exclusion profile together with the calculated Rg and Dmax values. At least 20 frames for each dataset were selected, scaled and averaged using PRIMUS 68 following the standard procedures. SAXS data analysis was performed using the ScÅtter software package 69. The forward scattering I(0) was evaluated using the Guinier approximation70 assuming the formula I(q) = I(0)exp(-(qRg)2/3) for a very small range of momentum transfer values (qRg< 1.3). Calculation of the pair distribution function and maximum distance Dmax was performed using ScÅtter which uses modification of the Moore function for transforming the data to real space. The Rg was estimated by Guinier approximation. The molecular mass was calculated using the Porod volume, and the QR method 69.
SEC-MALLS data collection and analysis
Size exclusion chromatography-multi angle laser light scattering (SEC-MALLS) was performed using an AKTA HPLC system connected to miniDAWN Tristar detector (Wyatt Technologies). 100 µl of purified Spindly1-440 or SpindlyFL at 5 mg/ml was loaded onto a Superose 6 Increase 10/300 column at a flow rate of 0.4 ml/min in a buffer containing 40 mM HEPES pH 7.5, 100 mM NaCl and 2 mM TCEP. The scattering data were analysed by ASTRA (Wyatt Technologies) software and estimation of molecular weight was done by using the refractive index signal as measure of the concentration.
Surface Plasmon Resonance (SPR)
For binding affinity determination by SPR, a Streptavidin sensor chip (GE) was used to immobilize the Spindly constructs or RZZ. Before immobilization, the non-specific binding of the Spindly constructs or RZZ to the Streptavidin chip was determined by flowing over these constructs over the sensor chip and monitoring the response in resonance units (RUs). Constructs which had little or no non-specific binding were selected as analytes. Based on these results, Spindly 259-C, 306-C and RZZ were selected as ligands and biotinylated via lysines by adding 1mg/ml of NHS-Biotin (Thermofisher) to each of these ligands in a 2:1 molar ratio (Protein: NHS-Biotin) to ensure that preferably a single Lysine per molecule is cross-linked. The biotinylation reaction was carried at room temperature for 30 minutes.~80 RUs of Spindly 259-C and 306-C were immobilized on two different channels of the Streptavidin sensor chip and Spindly 1-250 flown over in subsequently increasing concentration (5 nM to 20.48 µM).
For RZZ, ~235 RUs were immobilized on the Streptavidin sensor chip and SpindlyFL, SpindlyΔN or their farnesylated forms were flown over in increasing concentrations (5 nM to 20.48 µM). The binding buffer composed of 40 mM HEPES pH 7.5, 100mM NaCl, 2mM TCEP supplemented with 0.05% Tween-20, 1 mg/ml BSA and 1 mg/ml CM-Dextran to reduce non-specific binding. The experiments were performed using Biacore T200 (GE Healthcare).
Equilibrium binding responses were plotted as a function of the concentration in GraphPad PRISM v. 7.0c (GraphPad Software Inc.) and fitted using the equation for non-linear regression: Y=Bmax*X/(Kd+X) where, Bmax is the maximum specific binding in the same units as Y. Kd is the equilibrium binding constant, in the same units as X. The response plotted on the y-axis was normalized for the molecular weight of the analyte to yield the stoichiometry of binding by using the formula: Y = Y / K where, K = (RUs of Ligand immobilized / Mol. weight of Ligand) * Mol. weight of Analyte.
Statistics and reproducibility
Representative results are displayed or all data were reported, as specified in individual figure legends. Results from immunofluorescence images from different experiments were pooled and no difference was observed between different experimental sets. The comparisons most pertinent for the conclusions and number of independent experiments are specified in the figures and legends. Two-tailed, unpaired t-tests or one-way ANOVA followed by Tukey's test were performed to compare experimental groups in immunofluorescence quantifications when n≥3. In those cases in which n<3, no statistical analysis was performed and the source data is provided in Supplemental Table S3. Data are presented as mean ± SD, and P < 0.001 was considered statistically significant. The source data of the experiments performed with purified proteins is provided in Supplementary Table 3. All replicates showed similar results and a representative experiment was reported.
Supplementary Material
Acknowledgments
We thank all lab members for suggestions and discussions. We are grateful to Andrea Murachelli for help with EM data figure preparation, to Eleonore von Castelmur, Tatjana Heidebrecht and Yoshitaka Hiruma for help with Spindly structure experiments, to Joshua Vaughan for help with ExM, to Reto Gassmann for sharing unpublished results and Spindly constructs, to Iain Cheeseman, Susanne Lens and René Medema for reagents, and to Anko de Graaf of the Hubrecht Imaging Center. The Horizon 2020 iNEXT project (653706) provided financial support and access to EM infrastructures. This work is part of the Oncode Institute which is partly financed by the Dutch Cancer Society. This work was further supported by the Netherlands Organisation for Scientific Research (NWO) (gravitation program CancerGenomiCs.nl; VICI grant (865.12.004 to GJPLK)), the Dutch Cancer Society (KWF/HUBR-11080 to GJPLK), and the ERC (675737 to AM). V. Groenewold is supported by the Proteins@Work initiative of the Netherlands Proteomics Centre.
Footnotes
Code availability
Custom Image J macros used in this study are available upon request.
Data availability
Sequences analysed here can be found in Supplementary Tables 4 and 5. Source data for Figures 3d (HEC1), 3g, 5a,c,d, 6a, 8f and Supplementary Figures 4c, 6b,c,d 7a, 7e, 7f and 8b have been provided as Supplementary Table 3. All other data supporting the findings of this study are available from the corresponding author upon request.
Author Contributions
C.S. and G.J.P.L.K. conceived the project. C.S., G.J.P.L.K., M.A. A.P., J.K. and A.M designed experiments and interpreted data. C.S. performed the cell biology experiments. M.A. and J.K. performed the in vitro experiments with the help of A.F.. J.F. and J.K. performed and analysed the electron microscopy experiments. V.G. performed the cross-linking experiments. E.T. performed the comparative sequence analysis. R.M. and J.M.C performed the electron microscopy of Spindly. C.S. and G.J.P.L.K. wrote the manuscript with the help of A.P. and A.M. and the input of the rest of authors.
Declaration of Interests
The authors declare no competing interests.
References
- 1.Musacchio A, Desai A. A Molecular View of Kinetochore Assembly and Function. Biology (Basel) 2017;6:5. doi: 10.3390/biology6010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Joglekar AP, Kukreja AA. How Kinetochore Architecture Shapes the Mechanisms of Its Function. Current Biology. 2017;27:R816–R824. doi: 10.1016/j.cub.2017.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Etemad B, Kops GJ. Attachment issues: Kinetochore transformations and spindle checkpoint silencing. Current Opinion in Cell Biology. 2016;39:101–108. doi: 10.1016/j.ceb.2016.02.016. [DOI] [PubMed] [Google Scholar]
- 4.Maiato H. The dynamic kinetochore-microtubule interface. J Cell Sci. 2004;117:5461–5477. doi: 10.1242/jcs.01536. [DOI] [PubMed] [Google Scholar]
- 5.Maiato H, Gomes A, Sousa F, Barisic M. Mechanisms of Chromosome Congression during Mitosis. Biology (Basel) 2017;6:13. doi: 10.3390/biology6010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Magidson V, et al. Adaptive changes in the kinetochore architecture facilitate proper spindle assembly. Nat Cell Biol. 2015;17:1134–1144. doi: 10.1038/ncb3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wynne DJ, Funabiki H. Heterogeneous architecture of vertebrate kinetochores revealed by three-dimensional superresolution fluorescence microscopy. Mol Biol Cell. 2016;27:3395–3404. doi: 10.1091/mbc.E16-02-0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thrower DA, Jordan MA, Wilson L. Modulation of CENP-E organization at kinetochores by spindle microtubule attachment. Cell Motil Cytoskeleton. 1996;35:121–133. doi: 10.1002/(SICI)1097-0169(1996)35:2<121::AID-CM5>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 9.Hoffman DB, Pearson CG, Yen TJ, Howell BJ, Salmon ED. Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol Biol Cell. 2001;12:1995–2009. doi: 10.1091/mbc.12.7.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Magidson V, et al. Unattached kinetochores rather than intrakinetochore tension arrest mitosis in taxol-treated cells. J Cell Biol. 2016;212:307–319. doi: 10.1083/jcb.201412139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wynne DJ, Funabiki H. Kinetochore function is controlled by a phosphodependent coexpansion of inner and outer components. J Cell Biol. 2015;210:899–916. doi: 10.1083/jcb.201506020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rieder CL. The Formation, Structure, and Composition of the Mammalian Kinetochore and Kinetochore Fiber. Int Rev Cytol. 1982;79:1–58. doi: 10.1016/s0074-7696(08)61672-1. [DOI] [PubMed] [Google Scholar]
- 13.McEwen BF, Hsieh C-E, Mattheyses AL, Rieder CL. A new look at kineochore structure in vertebrate somatic cells using high-pressure freezing an freeze substitution. Chromosoma. 1998;107:366–375. doi: 10.1007/s004120050320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jokelainen PT. The ultrastructure and spatial organization of the metaphase kinetochore in mitotic rat cells. J Ultrastruct Res. 1967;19:19–44. doi: 10.1016/s0022-5320(67)80058-3. [DOI] [PubMed] [Google Scholar]
- 15.Cassimeris L, Rieder CL, Rupp G, Salmon ED. Stability of microtubule attachment to metaphase kinetochores in PtK1 cells. J Cell Sci. 1990;96(Pt 1):9–15. doi: 10.1242/jcs.96.1.9. [DOI] [PubMed] [Google Scholar]
- 16.McEwen BF, Arena JT, Frank J, Rieder CL. Structure of the Colcemid-treated PtK1 kinetochore outer plate as determined by high voltage electron microscopic tomography. J Cell Biol. 1993;120:301–312. doi: 10.1083/jcb.120.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pereira ALALJ, et al. Mammalian CLASP1 and CLASP2 cooperate to ensure mitotic fidelity by regulating spindle and kinetochore function. Mol Biol Cell. 2006;17:4526–42. doi: 10.1091/mbc.E06-07-0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wordeman L, Steuer ER, Sheetz MP, Mitchison T. Chemical subdomains within the kinetochore domain of isolated CHO mitotic chromosomes. J Cell Biol. 1991;114:285–94. doi: 10.1083/jcb.114.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yao X, Anderson KL, Cleveland DW. The microtubule-dependent motor centromere-associated protein E (CENP- E) is an integral component of kinetochore corona fibers that link centromeres to spindle microtubules. J Cell Biol. 1997;139:435–447. doi: 10.1083/jcb.139.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cooke CA, Schaar B, Yen TJ, Earnshaw WC. Localization of CENP-E in the fibrous corona and outer plate of mammalian kinetochores from prometaphase through anaphase. Chromosoma. 1997;106:446–455. doi: 10.1007/s004120050266. [DOI] [PubMed] [Google Scholar]
- 21.Mosalaganti S, et al. Structure of the RZZ complex and molecular basis of its interaction with Spindly. J Cell Biol. 2017;216:961–981. doi: 10.1083/jcb.201611060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kops GJPL, et al. ZW10 links mitotic checkpoint signaling to the structural kinetochore. J Cell Biol. 2005;169:49–60. doi: 10.1083/jcb.200411118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buffin E, Lefebvre C, Huang J, Gagou ME, Karess RE. Recruitment of Mad2 to the kinetochore requires the Rod/Zw10 complex. Curr Biol. 2005;15:856–861. doi: 10.1016/j.cub.2005.03.052. [DOI] [PubMed] [Google Scholar]
- 24.Défachelles L, et al. RZZ and Mad1 dynamics in Drosophila mitosis. Chromosom Res. 2015;23:333–342. doi: 10.1007/s10577-015-9472-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Starr DA, et al. Conservation of the centromere/kinetochore protein ZW10. J Cell Biol. 1997;138:1289–1301. doi: 10.1083/jcb.138.6.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gama JB, et al. Molecular mechanism of dynein recruitment to kinetochores by the Rod-Zw10-Zwilch complex and Spindly. J Cell Biol. 2017;216:943–960. doi: 10.1083/jcb.201610108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Çivril F, et al. Structural analysis of the RZZ complex reveals common ancestry with multisubunit vesicle tethering machinery. Structure. 2010;18:616–626. doi: 10.1016/j.str.2010.02.014. [DOI] [PubMed] [Google Scholar]
- 28.Basto R, et al. In Vivo Dynamics of the Rough Deal Checkpoint Protein during Drosophila Mitosis. Curr Biol. 2004;14:56–61. doi: 10.1016/j.cub.2003.12.025. [DOI] [PubMed] [Google Scholar]
- 29.Howell BJ, et al. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J Cell Biol. 2001;155:1159–1172. doi: 10.1083/jcb.200105093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wojcik E, et al. Kinetochore dynein: Its dynamics and role in the transport of the Rough deal checkpoint protein. Nat Cell Biol. 2001;3:1001–1007. doi: 10.1038/ncb1101-1001. [DOI] [PubMed] [Google Scholar]
- 31.Famulski JK, Vos LJ, Rattner JB, Chan GK. Dynein/dynactin-mediated transport of kinetochore components off kinetochores and onto spindle poles induced by Nordihydroguaiaretic acid. PLoS One. 2011;6 doi: 10.1371/journal.pone.0016494. e16494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Griffis ER, Stuurman N, Vale RD. Spindly, a novel protein essential for silencing the spindle assembly checkpoint, recruits dynein to the kinetochore. J Cell Biol. 2007;177:1005–1015. doi: 10.1083/jcb.200702062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gassmann R, et al. A new mechanism controlling kinetochore-microtubule interactions revealed by comparison of two dynein-targeting components: SPDL-1 and the Rod/Zwilch/Zw10 complex. Genes Dev. 2008;22:2385–2399. doi: 10.1101/gad.1687508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ying WC, et al. Mitotic control of kinetochore-associated dynein and spindle orientation by human Spindly. J Cell Biol. 2009;185:859–874. doi: 10.1083/jcb.200812167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McKenney RJ, Huynh W, Tanenbaum ME, Bhabha G, Vale RD. Activation of cytoplasmic dynein motility by dynactin-cargo adapter complexes. Science. 2014;345:337–341. doi: 10.1126/science.1254198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moudgil DK, et al. A novel role of farnesylation in targeting a mitotic checkpoint protein, human spindly, to kinetochores. J Cell Biol. 2015;208:881–896. doi: 10.1083/jcb.201412085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holland AJ, et al. Preventing farnesylation of the dynein adaptor Spindly contributes to the mitotic defects caused by farnesyltransferase inhibitors. Mol Biol Cell. 2015;26:1845–1856. doi: 10.1091/mbc.E14-11-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barisic M, et al. Spindly/CCDC99 is required for efficient chromosome congression and mitotic checkpoint regulation. Mol Biol Cell. 2010;21:1968–1981. doi: 10.1091/mbc.E09-04-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gassmann R, et al. Removal of Spindly from microtubule-attached kinetochores controls spindle checkpoint silencing in human cells. Genes Dev. 2010;24:957–971. doi: 10.1101/gad.1886810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tromer E, Bade D, Snel B, Kops GJPL. Phylogenomics-guided discovery of a novel conserved cassette of short linear motifs in BubR1 essential for the spindle checkpoint. Open Biol. 2016;6:1–11. doi: 10.1098/rsob.160315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Njoroge FG, et al. (+)-4-[2-[4-(8-Chloro-3,10-dibromo-6,11-dihydro-5H-benzo[5,6]cyclohepta[1,2-b]- pyridin-11(R)-yl)-1-piperidinyl]-2-oxo-ethyl]-1-piperidinecarboxamide (SCH-66336): A Very Potent Farnesyl Protein Transferase Inhibitor as a Novel Antitumor Agent. J Med Chem. 1998;41:4890–4902. doi: 10.1021/jm980462b. [DOI] [PubMed] [Google Scholar]
- 42.Nijenhuis W, et al. A TPR domain-containing N-terminal module of MPS1 is required for its kinetochore localization by Aurora B. J Cell Biol. 2013;201:217–231. doi: 10.1083/jcb.201210033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brauchle M, et al. Protein interference applications in cellular and developmental biology using DARPins that recognize GFP and mCherry. Biol Open. 2014;3:1252–1261. doi: 10.1242/bio.201410041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rieder CL, Alexander SP. Kinetochores Are Transported Poleward along a Single Astral Microtubule during Chromosome Attachment to the Spindle in Newt Lung Cells. J Cell Biol. 1990;110:81–95. doi: 10.1083/jcb.110.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kapoor TM, et al. Chromosomes can congress to the metaphase plate before biorientation. Science. 2006;311:388–91. doi: 10.1126/science.1122142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shrestha RL, Draviam VM. Lateral to end-on conversion of chromosome-microtubule attachment requires kinesins cenp-e and MCAK. Curr Biol. 2013;23:1514–1526. doi: 10.1016/j.cub.2013.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kajtez J, et al. Overlap microtubules link sister k-fibres and balance the forces on bi-oriented kinetochores. Nat Commun. 2016;7 doi: 10.1038/ncomms10298. 10298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chozinski TJ, et al. Expansion microscopy with conventional antibodies and fluorescent proteins. Nat Methods. 2016;13:485–488. doi: 10.1038/nmeth.3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gregan J, Polakova S, Zhang L, Tolić-Nørrelykke IM, Cimini D. Merotelic kinetochore attachment: Causes and effects. Trends in Cell Biology. 2011;21:374–381. doi: 10.1016/j.tcb.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pesenti ME, Weir JR, Musacchio A. Progress in the structural and functional characterization of kinetochores. Current Opinion in Structural Biology. 2016;37:152–163. doi: 10.1016/j.sbi.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 51.Hoogenraad CC, Akhmanova A. Bicaudal D Family of Motor Adaptors: Linking Dynein Motility to Cargo Binding. Trends Cell Biol. 2016;26:327–340. doi: 10.1016/j.tcb.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 52.Ghosal D, et al. Collaborative protein filaments. EMBO J. 2015;34:2312–2320. doi: 10.15252/embj.201591756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang G, Lischetti T, Hayward DG, Nilsson J. Distinct domains in Bub1 localize RZZ and BubR1 to kinetochores to regulate the checkpoint. Nat Commun. 2015;6 doi: 10.1038/ncomms8162. 7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Caldas GV, et al. The RZZ complex requires the N-terminus of KNL1 to mediate optimal Mad1 kinetochore localization in human cells. Open Biol. 2015;5 doi: 10.1098/rsob.150160. 150160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Karess R. Rod-Zw10-Zwilch: A key player in the spindle checkpoint. Trends Cell Biol. 2005;15:386–392. doi: 10.1016/j.tcb.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 56.Steigemann P, et al. Aurora B-Mediated Abscission Checkpoint Protects against Tetraploidization. Cell. 2009;136:473–484. doi: 10.1016/j.cell.2008.12.020. [DOI] [PubMed] [Google Scholar]
- 57.Saurin AT, van der Waal MS, Medema RH, Lens SMA, Kops GJPL. Aurora B potentiates Mps1 activation to ensure rapid checkpoint establishment at the onset of mitosis. Nat Commun. 2011;2 doi: 10.1038/ncomms1319. 316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dosztanyi Z, Csizmok V, Tompa P, Simon I. The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J Mol Biol. 2005;347:827–839. doi: 10.1016/j.jmb.2005.01.071. [DOI] [PubMed] [Google Scholar]
- 59.Delorenzi M, Speed T. An HMM model for coiled-coil domains and a comparison with PSSM-based predictions. Bioinformatics. 2002;18:617–625. doi: 10.1093/bioinformatics/18.4.617. [DOI] [PubMed] [Google Scholar]
- 60.Bailey TL, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–8. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eddy SR. Accelerated Profile HMM Searches. PLoS Comput Biol. 2011;7 doi: 10.1371/journal.pcbi.1002195. e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weissmann F, et al. biGBac enables rapid gene assembly for the expression of large multisubunit protein complexes. Proc Natl Acad Sci USA. 2016;113:E2564–9. doi: 10.1073/pnas.1604935113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.de la Rosa-Trevín JM, et al. Scipion: A software framework toward integration, reproducibility and validation in 3D electron microscopy. J Struct Biol. 2016;195:93–99. doi: 10.1016/j.jsb.2016.04.010. [DOI] [PubMed] [Google Scholar]
- 65.Abrishami V, et al. A pattern matching approach to the automatic selection of particles from low-contrast electron micrographs. Bioinformatics. 2013;29:2460–8. doi: 10.1093/bioinformatics/btt429. [DOI] [PubMed] [Google Scholar]
- 66.Sorzano COS, et al. A clustering approach to multireference alignment of single-particle projections in electron microscopy. J Struct Biol. 2010;171:197–206. doi: 10.1016/j.jsb.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pernot P, et al. Upgraded ESRF BM29 beamline for SAXS on macromolecules in solution. J Synchrotron Radiat. 2013;20:660–664. doi: 10.1107/S0909049513010431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Petoukhov MV, et al. New developments in the ATSAS program package for small-angle scattering data analysis. J Appl Crystallogr. 2012;45:342–350. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rambo RP, Tainer JA. Accurate assessment of mass, models and resolution by small-angle scattering. Nature. 2013;496:477–481. doi: 10.1038/nature12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Iyer LM, Zhang D, Maxwell Burroughs A, Aravind L. Computational identification of novel biochemical systems involved in oxidation, glycosylation and other complex modifications of bases in DNA. Nucleic Acids Res. 2013;41:7635–7655. doi: 10.1093/nar/gkt573. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.