Summary
A barrage of information constantly assaults our senses, of which only a fraction is relevant at any given point in time. However, the neural circuitry supporting the suppression of irrelevant sensory distractors is not completely understood. The claustrum, a circuit hub with vast cortical connectivity, is an intriguing brain structure, whose restrictive anatomy, thin and elongated, has precluded functional investigation. Here, we describe the use of Egr2-CRE mice to access genetically-defined claustral neurons. Utilizing conditional viruses for anterograde axonal labeling, and retrograde trans-synaptic tracing, we validated this transgenic model for accessing the claustrum, and extended the known repertoire of claustral input/output connectivity. Addressing the function of the claustrum, we inactivated CLEgr2+ neurons, chronically as well as acutely, in mice performing an automated two-alternative forced-choice behavioral task. Strikingly, inhibition of CLEgr2+ neurons did not significantly impact task performance under varying delay times and cue durations, but revealed a selective role for the claustrum in supporting performance in the presence of an irrelevant auditory distractor. Further investigation of behavior, in the naturalistic maternal pup retrieval task, replicated the result of sensitization to an auditory distractor following inhibition of CLEgr2+ neurons. Initiating investigation into the underlying mechanism, we found that activation of CLEgr2+ neurons modulated cortical sensory processing, suppressing tone representation in auditory cortex. This functional investigation, utilizing selective genetic access, implicates the claustrum in supporting resilience to distraction, a fundamental aspect of attention.
Introduction
The claustrum is reported to be the most interconnected brain structure per regional volume [1], evoking curiosity regarding its function [2–5]. However, the anatomy of the claustrum, a thin elongated structure eponymously cloistered between the striatum and the cortex, has hindered its functional investigation. As a consequence, little is known about the role of this structure, despite an abundance of proposed theories. Most famously, it has been suggested to act as the ‘seat of consciousness’, binding sensory information across modalities to form a single unified conscious experience [5]. Other hypotheses have postulated that the claustrum functions to amplify and transmit cortical oscillations [6], coordinate active sensing [7], detect salience [8,9] and allocate attention [4]. However, functional experiments directly testing these hypotheses are largely lacking.
Multiple indications, including human functional imaging studies, suggest a role for the claustrum in sensory processing [10,11]. Furthermore, studies of executive function during attention-demanding tasks have revealed heightened activity of the insula and claustrum in participants with Attention Deficit Hyperactivity Disorder compared to controls [12,13]. In line with these findings are clinical observations from patients suffering temporary lesions of the claustrum, which suggest that the claustrum plays a role in supporting proper sensory processing and perception [14,15]. Additionally, the claustrum has been reported to respond to sensory stimuli [8,9,16–18], and electrical stimulation of the claustrum region has been shown to substantially impact the activity of large regions of cortex, primarily suppressing activity [19–23]. As eloquently advocated by Sir Francis Crick & Christof Koch, the next step in investigation of the claustrum, moving from correlation towards causation, requires genetic access to the claustrum, in order to perturb its function, and address the role of this structure in behavior [5].
In the present study, we describe the identification of such genetic access to claustral projection neurons through conditional transgene expression, enabled through viral targeting in Egr2-CRE knock-in mice [24]. Following a comprehensive characterization of the molecular composition and input-output connectivity of claustral Egr2-expressing neurons (CLEgr2+), we utilized this mode of genetic access to address the behavioral impact of chronic and acute suppression of CLEgr2+ neurons. Strikingly, in both an automated two-alternative forced-choice task, as well as in a naturalistic task of maternal pup retrieval, we found that inactivating CLEgr2+ neurons selectively impacted the performance of mice upon introduction of a distractor, suggesting a specific role for the claustrum in supporting resilience to distraction. Commencing the study of the underlying mechanism, we tested the impact of claustral activation on cortical sensory processing, and found that brief optogenetic activation of the claustrum was sufficient to significantly suppress the response to tone stimuli in auditory cortex. Taken together, our results provide functional evidence supporting the hypothesis that the claustrum enables resilience to distraction, potentially through its capacity to modulate cortical sensory responses.
Results
The Egr2-CRE line enables genetic access to the mouse claustrum
The need to identify and develop genetic tools which enable specific access to the claustrum has been emphasized for over a decade [5]. Even careful stereotactic targeting of non-conditional virus infection to the region of the claustrum is incapable of supporting selective access to claustral neurons due to the difficulty in precisely containing the spread of the virus to a thin and elongated structure. This motivated the identification of genes expressed in claustrum neurons, yielding a number of candidates with varying degrees of selectivity [25]. However, these genes have yet to be utilized for research into the function of this structure. Studying the spatial expression pattern of the Egr2 gene in the mouse brain, we observed that Egr2 is enriched in claustral neurons, delineating the structure of the claustrum (Figure 1A; Figure S1A). Stereotactic targeting of CRE-dependent virus expression to the claustrum of heterozygous Egr2-CRE knock-in mice enabled specific access to CLEgr2+ cells, largely covering the extent of the rostrocaudal and dorsoventral axes of the claustrum (Figures 1B, C). The localization of CLEgr2+ neurons within the anatomical boundaries of the claustrum was validated by their overlap with previously established prominent inputs from the anterior cingulate cortex and orbitofrontal cortex to the claustrum (Figure S1B, C) [7,25–28]. We next characterized the neurotransmitter identity of claustral neurons, including the CLEgr2+ subpopulation, using single molecule fluorescent in situ hybridization (smFISH), following conditional expression of eGFP in claustral neurons of Egr2-CRE mice (Figure 1D). Our analysis was consistent with the established composition of the claustrum, whereby excitatory projection neurons (identified by vGlut1 expression; Slc17a7) comprise the dominant cell type (83±2.8%), while GABAergic neurons (identified by GAD65 expression; Gad2) comprise a minority (4±1.4%) of cells in the claustrum (Figure 1E) [3,29–32]. Cells not labeled by these probes (16±2.6%) likely largely represent additional excitatory neurons expressing vGlut2 (Slc17a6) [29], as well as interneurons expressing GAD67 (Gad2) or non-neuronal cells. Interestingly, a majority of the GAD65-expressing neurons also expressed vGlut1 (73/84 cells) suggesting that a subpopulation of vGlut1-expressing claustral projection neurons may have the potential to co-release GABA, as has been reported for various projection neurons in other brain structures [33–35] (Figure S1D). CLEgr2+ neurons, as defined by eGFP-expression (comprising 33±7.1% of claustral cells), were, by and large, vGlut1+ positive (91±1.4% vGlut1; 6.8±3.26% GAD65; non-labeled 9±1.6%; Figure 1F). Virtually all eGFP+/GAD65+ neurons also expressed vGlut1 (49/51 cells), suggesting that Egr2 expression is largely selective to claustral projection neurons. Applying CLARITY [36] to image the intact claustrum and its efferents, we observed CLEgr2+ projections towards rostral cortical structures, as well as towards caudal cortical and subcortical structures. Axons extending towards the cortical structures immediately dorsal to the claustrum were also observed (Figure 1G & Supp. Video 1). Our results demonstrate the capacity of Egr2-CRE mice to provide selective genetic access to claustral projection neurons, enabling their detailed anatomical and functional investigation.
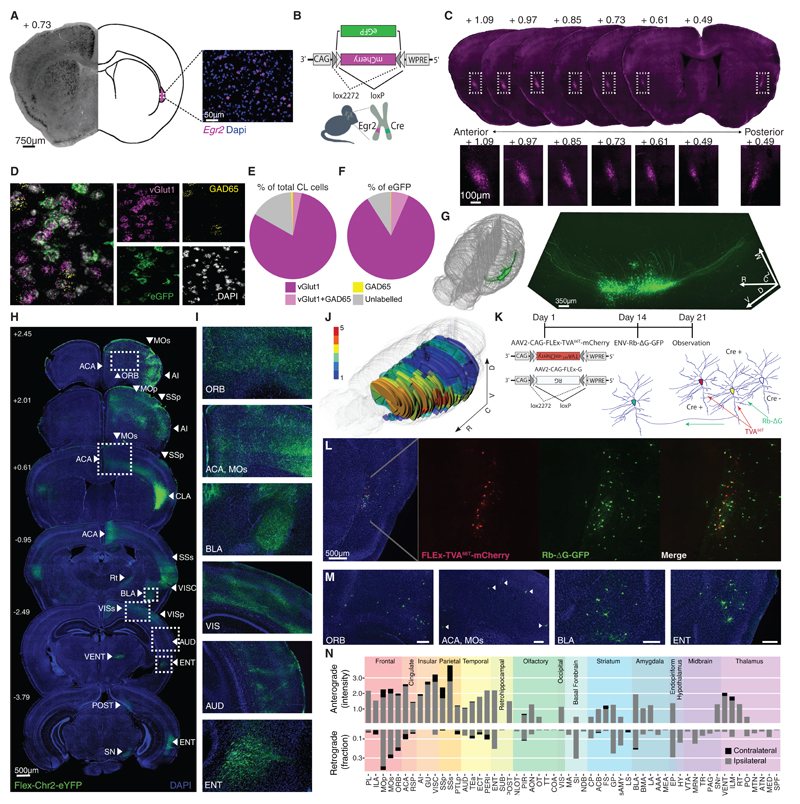
Figure 1. Genetic access to the claustrum in Egr2-CRE mice.
(A) In situ hybridization (left panel) and single molecule fluorescent in situ hybridization (smFISH; right panel) against Egr2 demonstrate robust expression of Egr2 in the claustrum (See also Figure S1A). (B) Illustration of the conditional approach of obtaining genetic access to the claustrum, using double-floxed inverse open reading frame (DIO) constructs to express transgenes in the claustrum of heterozygous Egr2-CRE knock-in mice. (C) Specific labeling of the claustrum along its anterior-posterior axis obtained by stereotactic targeting of an AAV-DIO-mCherry virus to the claustrum of Egr2-CRE mice. Bottom panels show increased magnification of the outlined frames (See also Figure S1B, C). (D) smFISH mRNA labeling of eGFP, vGlut1 and GAD65 in a section from an Egr2-CRE mouse injected with AAV-DIO-eGFP. Small panels show split channels: vGlut1 (Slc17a7), GAD65 (Gad2), eGFP, and DAPI (See also Figure S1D). (E-F) Quantification of smFISH labeling. Claustral neurons (n=1854 cells from two mice), and CLEgr2+ neurons (n=612/1854) in particular, are predominantly vGlut1+. (G) 3D-rendering of a CLARITY-processed hemisphere from an Egr2-CRE mouse in which the claustrum has been fluorescently labeled. Left: localization of the imaged claustrum within a volumetric scheme of the mouse brain. Right: reconstructed volume of the claustrum and its projections from stacked confocal images (see Supplemental Movie 1); R-rostral; C-caudal; L-lateral ; M-medial; D-dorsal ; V-ventral. (H) Brain-wide conditional anterograde tracing from CLEgr2+ neurons. Egr2-CRE mice were stereotactically infected with viruses encoding AAV-DIO-ChR2-eYFP, labeling axons of CLEgr2+ neurons. (I) Examples of the CLEgr2+ projections to cortical and subcortical structures. (ACA: anterior cingulate cortex; ORB: orbitofrontal cortex; MOs: motor cortex; BLA: basolateral amygdala; VIS: visual cortex; AUD: auditory cortex; ENT: entorhinal cortex). (J) Three-dimensional reconstruction of CLEgr2+ anterograde connectivity of an individual mouse. The density of CLEgr2+ projections is qualitatively represented on a numerical scale of 1 (sparse) to 5 (dense) (See also Figure S1E). (K) Experimental scheme for retrograde tracing of inputs to CLEgr2+ neurons, utilizing conditional pseudo-rabies virus (pRbV) in Egr2-CRE mice. In brief, the TVA receptor and pRbV glycoprotein (G) are conditionally expressed in CLEgr2+ neurons following infection with AAV viruses. pRbV, expressing GFP, is injected to the same site two weeks later, acquiring the RbV glycoprotein and enabling trans-synaptic labeling of presynaptic neurons. (L) Retrograde labeling of inputs to CLEgr2+ neurons. Red: expression of a mutant TVA receptor (TC66T); Green: input cells in the claustrum; Yellow: co-expressing starter cells. (M) Examples of cortical and subcortical input neurons, presynaptic to the claustrum. (N) Summary of CLEgr2+ connectivity. Top panel: numerical representation of anterogradely-labeled projection intensity; Bottom panel: retrograde input neurons (fraction of total 1501 neurons); Gray: ipsilateral; Black: contralateral.
The most recognized feature of the claustrum, its vast reciprocal connectivity, has been primarily investigated by studying individual claustral connections, or, at most, small subsets of claustral connectivity [7,31,32,37–39]. Broader studies focused on claustro-cortical connections, but did not account for subcortical connectivity [25–27]. Prominent reciprocal ipsilateral connections between the claustrum and the cortex have been described in detail [2,25,26]. Contralateral inputs to the claustrum are not as common, and emerge primarily from frontal and somatomotor structures [7,27,40]. Similarly, contralateral projections from the claustrum are sparse and predominantly limited to frontal and somatomotor regions [25]. Utilizing CLEgr2+ neurons as a seed for anterograde labeling, and retrograde trans-synaptic tracing, we characterized their connectivity with numerous cortical regions, as well as with subcortical structures (Figures 1H-N). Anterograde tracing was enabled by targeting conditional expression of an opsin-conjugated fluorophore to the claustrum, labeling the axonal projections of CLEgr2+ neurons throughout the brain (Figure 1H). CLEgr2+ projections were found along the entire ipsilateral cortical mantle, including frontal, motor, and sensory regions. Contralateral projections were scarce, primarily to frontal and somatosensory cortices. Notable subcortical nuclei innervated by the claustrum included sensorimotor thalamic nuclei, substania nigra pars reticulata, and basolateral amygdala (Figures 1I, N). The widespread propagation of claustral projections to the cortex, biased towards frontal, cingulate, and parietal cortices, was numerically represented by manual scoring of the projection density, and visualized with a 3-dimensional reconstruction (Figure 1J; Figure S1E).
Conducting the complementary experiment, we performed retrograde tracing by conditionally targeting pseudotyped rabies virus (pRbV) expression to CLEgr2+ starter cells, labeling their brain-wide monosynaptic inputs (Figures 1K, L). Labeled neurons were predominantly found in the hemisphere ipsilateral to the injection site. These were most prominent in frontal cortical regions, but also found throughout the cortical mantle, as well as in subcortical structures (Figures 1M, N). Numerical representation of the extent of anterograde and retrograde connectivity demonstrated the trend of reciprocal connectivity of CLEgr2+ neurons with cortical structures, consistent with cumulative reports in the literature regarding claustral connectivity. As our observations with the connectivity of the Egr2-Cre mice reliably replicate the sum of the literature regarding claustral connections, we view this as further validation that CLEgr2+ neurons faithfully represent claustro-cortical projection neurons. In addition, our observations regarding subcortical connections of the claustrum, which have received less attention to date, extend the principle of claustral reciprocal connectivity to its relation with sub-cortical structures (Figure 1N & Table 1).
Table 1. Glossary for Figure 1.
| Region: | Acronym: | Area: |
|---|---|---|
| Frontal | PL: | Prelimbic area |
| ILA: | Infralimbic area | |
| MOp: | Primary motor area | |
| MOs: | Secondary motor area | |
| ORB: | Orbital area | |
| Cingulate | ACA: | Anterior cingulate area |
| RSP: | Retrosplenial area | |
| Insular | AI: | Agranular insular area |
| GU: | Gustatory areas | |
| VISC: | Visceral area | |
| Parietal | SSp: | Primary somatosensory area |
| SSs | Secondary somatosensory area | |
| PTLp: | Posterior parietal association areas | |
| Temporal | AUD: | Auditory areas |
| TEa: | Temporal association areas | |
| ECT: | Ectorhinal area | |
| PERI: | Perirhinal area | |
| Retrohippocampal | ENT: | Entorhinal area |
| SUB: | Subiculum | |
| POST: | Postsubiculum | |
| Olfactory | NLOT: | Nucleus of the lateral olfactory tract |
| PIR: | Piriform area | |
| AON: | Anterior olfactory nucleus | |
| OT: | Olfactory tubercle | |
| TT: | Taenia tecta | |
| COA: | Cortical amygdalar area | |
| Occipital | VIS: | Visual areas |
| Basal Forebrain | MA: | Magnocellular nucleus |
| SI: | Substantia innominata | |
| NDB: | Diagonal band nucleus | |
| Striatum | CP: | Caudoputamen |
| ACB: | Nucleus accumbens | |
| FS: | Fundus of striatum | |
| GP: | Globus pallidus | |
| sAMY: | Striatum-like amygdalar nuclei | |
| LS: | Lateral septal nucleus | |
| Amydgala | BLA: | Basolateral amygdalar nucleus |
| BMA: | Basomedial amygdalar nucleus | |
| LA: | Lateral amygdalar nucleus | |
| AAA: | Anterior amygdalar area | |
| MEA: | Medial amygdalar nucleus | |
| Endopiriform | EP: | Endopiriform nucleus |
| Hypothalamus | HY: | Hypothalamus |
| Midbrain | VTA: | Ventral tegmental area |
| MRN: | Midbrain reticular nucleus | |
| TR: | Postpiriform transition area | |
| PAG: | Periaqueductal gray | |
| SNr: | Substantia nigra, reticular part | |
| Thalamus | VENT: | Ventral group of the dorsal thalamus |
| ILM | Intralaminar nuclei of the dorsal thalamus | |
| RT | Reticular nucleus of the thalamus | |
| PO | Posterior complex of the thalamus | |
| MTN | Midline group of the dorsal thalamus | |
| ATN | Anterior group of the dorsal thalamus | |
| MED | Medial group of the dorsal thalamus | |
| SPF | Subparafascicular nucleus | |
| Region: | Acronym: | Area: |
| Frontal | PL: | Prelimbic area |
| ILA: | Infralimbic area | |
| MOp: | Primary motor area | |
| MOs: | Secondary motor area | |
| ORB: | Orbital area | |
| Cingulate | ACA: | Anterior cingulate area |
| RSP: | Retrosplenial area | |
| Insular | AI: | Agranular insular area |
| GU: | Gustatory areas | |
| VISC: | Visceral area | |
| Parietal | SSp: | Primary somatosensory area |
| SSs | Secondary somatosensory area | |
| PTLp: | Posterior parietal association areas | |
| Temporal | AUD: | Auditory areas |
| TEa: | Temporal association areas | |
| ECT: | Ectorhinal area | |
| PERI: | Perirhinal area | |
| Retrohippocampal | ENT: | Entorhinal area |
| SUB: | Subiculum | |
| POST: | Postsubiculum | |
| Olfactory | NLOT: | Nucleus of the lateral olfactory tract |
| PIR: | Piriform area | |
| AON: | Anterior olfactory nucleus | |
| OT: | Olfactory tubercle | |
| TT: | Taenia tecta | |
| COA: | Cortical amygdalar area | |
| Occipital | VIS: | Visual areas |
| Basal Forebrain | MA: | Magnocellular nucleus |
| SI: | Substantia innominata | |
| NDB: | Diagonal band nucleus | |
| Striatum | CP: | Caudoputamen |
| ACB: | Nucleus accumbens | |
| FS: | Fundus of striatum | |
| GP: | Globus pallidus | |
| sAMY: | Striatum-like amygdalar nuclei | |
| LS: | Lateral septal nucleus | |
| Amydgala | BLA: | Basolateral amygdalar nucleus |
| BMA: | Basomedial amygdalar nucleus | |
| LA: | Lateral amygdalar nucleus | |
| AAA: | Anterior amygdalar area | |
| MEA: | Medial amygdalar nucleus | |
| Endopiriform | EP: | Endopiriform nucleus |
| Hypothalamus | HY: | Hypothalamus |
| Midbrain | VTA: | Ventral tegmental area |
| MRN: | Midbrain reticular nucleus | |
| TR: | Postpiriform transition area | |
| PAG: | Periaqueductal gray | |
| SNr: | Substantia nigra, reticular part | |
| Thalamus | VENT: | Ventral group of the dorsal thalamus |
| ILM | Intralaminar nuclei of the dorsal thalamus | |
| RT | Reticular nucleus of the thalamus | |
| PO | Posterior complex of the thalamus | |
| MTN | Midline group of the dorsal thalamus | |
| ATN | Anterior group of the dorsal thalamus | |
| MED | Medial group of the dorsal thalamus | |
| SPF | Subparafascicular nucleus |
CLEgr2+ inhibition impairs resilience to distraction
Genetic access to claustral neurons enables investigation of its role in supporting behavior. To this end, we constitutively inhibited CLEgr2+ neurons with conditional expression of the Kir2.1 potassium channel [41] (Figure 2A; for electrophysiological validation of efficacy see Figure S2). In order to initiate investigation into the role of the claustrum in cognition, we developed an automated version of the two-alternative forced choice task (2-AFC), simultaneously training co-housed control and experimental mice. The task was implemented in the ‘Intellicage’, an automated behavioral setup for multiple mice in their homecage environment (Figure 2B) [42]. The use of an automated behavioral setup allowed mice to perform the task in a self-paced fashion, motivated by the need to hydrate, initiating ±100 trials in each session with minimal experimenter intervention. Mice accessed the behavioral corner through a tube of limiting circumference connected to their homecage, initiating trials by breaking an infrared beam upon reaching the corner. Individual labeling of mice with unique radio-frequency identification (RFID) tags enabled precise recording of all individual visits, nose pokes, and licking events, providing detailed activity logs. Mice were trained to withhold their response for a progressively increasing delay period (0.2 - 3 seconds), in order to attend to a visual cue, randomly appearing above either the left or right port for a gradually decreasing duration (5 to 0.1 seconds). A nosepoke in the port corresponding to the visual cue was rewarded by its opening, allowing brief access to water (Figure 2C). In this design, there was no penalty for failure, other than a requirement to exit the behavioral corner to reinitiate a task. Required to perform a timed response to a brief cue presented following a substantial delay, mice achieved a suitable plateau performance rate of 70-80% successful trials (Figure 2D). While side errors were most common during the initial phase of learning, they were replaced by premature responses upon transition to longer delays, which diminished as the animals adjusted to the delays (Figures S2C-F).
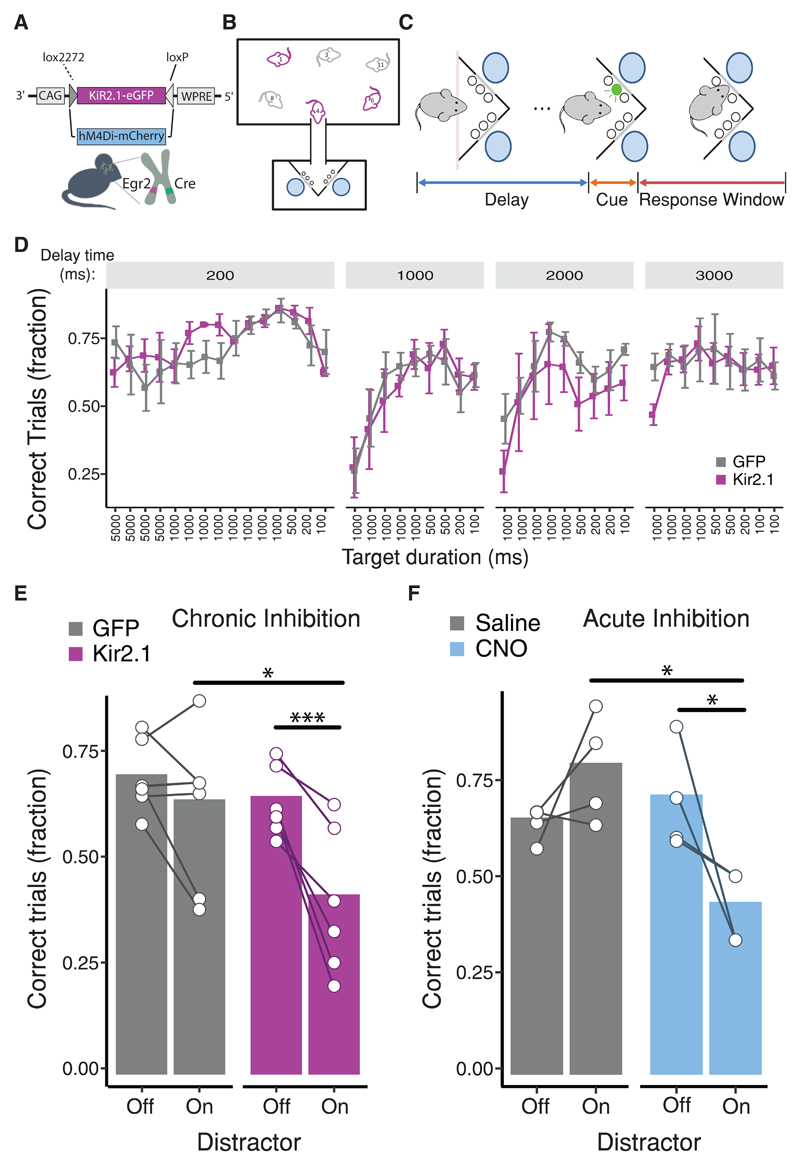
Figure 2. Inhibition of the claustrum confers susceptibility to distraction.
(A) Illustration of conditional expression of Kir2.1 and hM4Di for chronic or acute inhibition of CLEgr2+ neurons, respectively (See also Figure S2A, B). (B) Illustration of the automated behavioral setup for multiple animals in the homecage environment. The behavioral corner, connected to the homecage by a slim tube, contains two (left and right) ports, located beneath the cue lights. (C) Behavioral paradigm: mice are trained to respond to a visual cue of gradually-decreasing duration, presented following a progressively-increasing delay. (D) Performance of mice following chronic silencing of CLEgr2+ neurons (Kir2.1) in comparison to control (GFP) mice. Each point represents a single day, in chronological order. The delay time from trial initiation to cue presentation, as well as the duration of cue presentation, were progressively made more difficult (See also Figure S2C-J). (E) Inhibition of CLEgr2+ neurons selectively sensitized mice to distraction by an auditory interference, with no effect on control mice [(interaction effect group:distractor: t=2.048, p<0.05; linear mixed effect model; n=6 per group). Contrasts: (Kir2.1.On - Kir2.1.Off), p<0.001, (Kir2.1.On - GFP.On), p=0.01, Tukey post hoc comparisons, with Bonferroni correction for multiple comparisons] (See also Figure S2K-N). (F) Acute chemogenetic inhibition of CLEgr2+ neurons. Mice expressing the DREADD hM4Di in CLEgr2+ neurons were assessed in the same task, in the presence or absence of auditory interference, following intraperitoneal injection of saline or CNO (clozapine N-oxide). Sensitivity to an auditory interference is selectively revealed upon acute inhibition of CLEgr2+ neurons (Interaction effect of the success rate “group:distractor”: t=4.316, p=0.005, linear mixed effect model). Bars represent group averages ± SEM. In panels E & F; connected white circles represent individual animals (See also Figure S3).
No significant impact of CLEgr2+ neuronal inhibition was observed on acquisition of the task. While not overall statistically significant, the observation that mice in which CLEgr2+ neurons were silenced displayed a trend towards poor performance on the first day following transitions to prolonged delay times is of potential interest (Figure 2D). Multiple other parameters of the task were not affected by CLEgr2+ inhibition, including visit count, lick count and reaction time (Figures S2G-J), suggesting that silencing CLEgr2+ neurons had no substantial impact on the motivation of the animals, their decision making, cognitive flexibility, or spatially-oriented light perception.
In contrast, upon introduction of an auditory distractor during the task, a significant decrease in the performance of the claustral-deficient group was observed. Addition of the distractor had no impact on the performance of control mice, yet resulted in a 40% reduction in the success rate of mice whose CLEgr2+ neurons were inhibited (Figure 2E). The distractor specifically impacted task performance, primarily manifested in an increase in premature responses (Figure S2K). Premature responses occurred following a significant delay from cue onset (±1.5 seconds), similar to the premature responses during training (Figure S2L), indicating that the mice were withholding their responses in anticipation of the cue. The inclusion of the distractor did not grossly impact the engagement with the task throughout the session, as control and experimental groups performed a similar number of trials and licks (Figures S2M, N).
In order to limit the inhibition of CLEgr2+ neurons to the duration of a single session, we repeated this experiment, using pharmacogenetics for acute inhibition of the claustrum. This was enabled by expression of the inhibitory DREADD (Designer Receptor Exclusively Activated by Designer Drug) hM4D(Gi), in a new cohort of Egr2-CRE mice [43] (Figure S3). Inhibition of CLEgr2+ neurons was induced selectively during the sessions in which an auditory distractor was interleaved. Similar to chronic inhibition, chemogenetic inhibition caused selective sensitization to an auditory distractor, resulting in a 40% reduction in performance (Figure 2F).
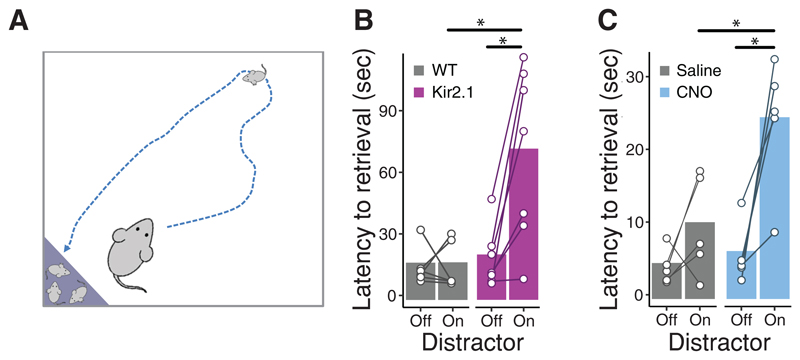
The ability to ignore irrelevant sensory stimuli is paramount for task performance in nature [44]. To assess the relevance of the claustrum in a naturalistic setting, we tested maternal pup retrieval, a well-established assay requiring multi-modal attention to sensory cues (Figure 3A) [45]. Neither chronic inhibition, by Kir2.1 expression, nor acute pharmacogenetic inhibition, by the DREADD hM4Di, affected pup retrieval in the absence of a distractor, while in its presence, chronic inhibition of CLEgr2+ neurons resulted in a 3.8-fold increase in latency to retrieval, and acute pharmacogenetic inhibition resulted in a 4.4-fold increase in latency to retrieval. This manifested in a trend towards longer and slower paths to reaching the pup, but no impact on the return path to the nest (Figure S4). Furthermore, dams spent more time in the nest in the presence of the distractor (Figure S4), primarily engaging with the pups and the nesting material. Thus, chronic inhibition of CLEgr2+ neurons, as well as acute pharmacogenetic inhibition, selectively sensitized dams to the introduction of an auditory interference.
Figure 3. CLEgr2+ inhibition sensitizes dams to auditory distraction during pup retrieval.
(A) Illustration of the maternal pup retrieval assay: female Egr2-CRE mice were injected with AAV viruses conditionally expressing the Kir2.1 channel (B) or the DREADD hM4Di (C) prior to exposure to male mice. Dams were tested 3-7 days after parturition in the pup retrieval paradigm, in the absence or presence of an auditory distractor. (B) Latency to pup retrieval of dams following chronic inactivation of CLEgr2+ neurons (Kir2.1, n=7) compared to controls (n=6), in the presence or absence of an auditory distractor (interaction effect group:distractor: t=-3.653, p<0.005, linear mixed effect model). (C) Latency to pup retrieval during chemogenetic inactivation of CLEgr2+ neurons (hM4Di + CNO, n=5) in comparison to within-animal controls (saline) in the presence or absence of an auditory distractor (interaction effect group:distractor: t=-2.758, p=0.015, linear mixed effect model). Bars represent group averages and lines represent individual mice (See also Figure S4).
Activation of CLEgr2+ neurons suppresses sensory representations in auditory cortex
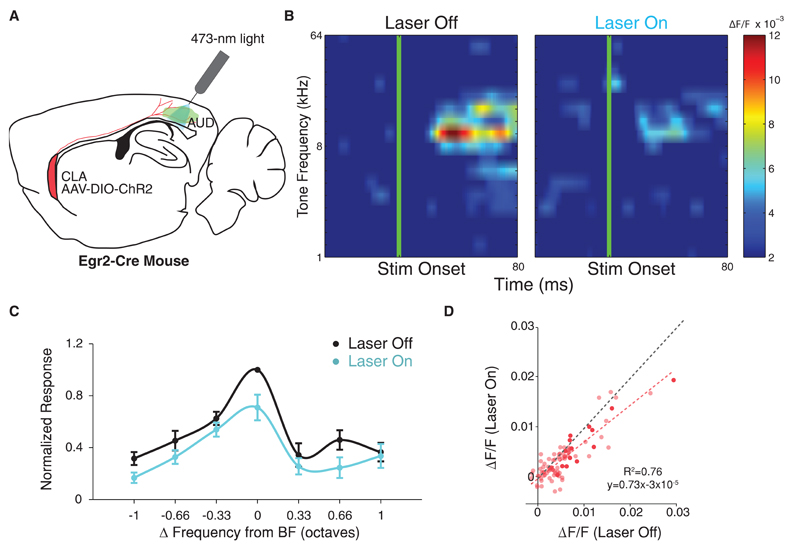
Our results demonstrate a role for the claustrum in supporting resilience to distraction both in a 2-AFC task, as well as in the naturalistic task of pup retrieval. In an effort to uncover a potential mechanism underlying this observation, we investigated the impact of claustral activation on sensory cortical processing. Early studies, in which the region of the claustrum was electrically stimulated, demonstrated a potential role for this structure in suppression of cortical activity [19–23]. Based on these reports, we hypothesized that a mechanism whereby the claustrum supports resilience to sensory distractors could be through limiting the cortical representation of task-irrelevant sensory information. Genetic access to CLEgr2+ neurons enables direct interrogation of claustral activation on cortical sensory processing by expression of optogenetic actuators in CLEgr2+ neurons. We chose to address the effect of claustral activation on auditory cortical processing of pure tones, since we implemented an auditory distractor in our behavioral analyses, and pure tone stimuli are well controlled in terms of content, temporal structure, intensity, and delivery. We performed fiber photometry recordings of a bolus-loaded calcium indicator, allowing detection of population calcium transients [46]. Experiments were performed in anesthetized Egr2-CRE mice, following conditional expression of the excitatory opsin channelrhodopsin (ChR2) in CLEgr2+ neurons, enabling optogenetic activation of claustral afferents to auditory cortex (Figure 4A). In each mouse, a number of sites were recorded. At each recording site, a protocol consisting of 19 pure tones (1 second inter-stimulus interval; each frequency repeated 20 times), logarithmically spaced between 1-64 kHz, was presented in a random sequence. In interleaved trials, immediately prior to tone onset, blue light intensity was briefly increased (5ms; from 1-5µW to 1-2mW, 473nm), inducing optogenetic stimulation of CLEgr2+ cortical afferents. Our analysis focused on the first 60 milliseconds after the stimulus onset, as early auditory cortical processing is represented in the fast initial peak of the calcium response [46]. We observed that activation of claustral afferents to the auditory cortex caused a reduction of the auditory population response (Figure 4B). To compare between multiple recording sites responding to different preferred frequencies, we normalized the tone-specific evoked responses at each site to the response at its best frequency (BF) with no laser stimulation, and aligned these values to produce a centered population tuning curve in the presence or absence of optogenetic stimulation of claustral terminals (Figure 4C). A linear mixed model, accounting for within and across animal variance, confirmed a primary effect of claustral activation (ANOVA, F1,172=14.07, p<0.0005) which, on average, suppressed the population response. Specifically, CLEgr2+ activation resulted in suppression of the auditory population responses by 25-30%, while maintaining the overall tuning, as evident by a linear fit of the data (Figure 4D). These results provide a potential mechanism whereby the claustrum could support resilience to distraction, by suppressing the cortical representation of sensory information not selected for attention.
Figure 4. Activation of CLEgr2+ neurons suppresses sensory representation in auditory cortex.
(A) Schematic of the experimental setup for fiber photometry recording of population calcium responses in the auditory cortex during optogenetic activation of CLEgr2+ axons. The claustrum and the auditory cortex are presented on the same sagittal plane for visualization. An optic fiber is used for recording the emission of a bolus-loaded calcium-sensitive dye (green), and for optogenetic excitation of CLEgr2+ afferents (red). (B) Example of tone-evoked responses from a single recording site without (left) and with (right) CLEgr2+ activation. (C) Average auditory tuning curves (n=14 recording sites from 4 mice) centered and normalized according to the response at the BF without laser stimulation (black), compared to following laser stimulation (blue). Activation of CLEgr2+ axons suppressed the population response without affecting tuning (p<0.001, two-sided Wilcoxon signed-rank test). Error bars represent SEM. (D) Trial-averaged evoked calcium response with laser (Y axis) vs no laser (X axis). Dashed red line indicates linear fit (Slope=0.73; Intercept<0.001, R2=0.76). Dark circles indicate responses at the BF, light circles indicate responses at frequencies within one octave of the BF. Unity line in gray.
Discussion
In this manuscript we describe a functional investigation of the claustrum, enabled by specific genetic access to claustral projections neurons in the Egr2-CRE mouse. The Egr2/KROX-20 gene encodes a zinc-finger transcription regulatory factor, the sustained expression of which is important for the development of peripheral & central nervous systems, as well as adaptive immunity [47,48]. In some experimental systems, Egr2 expression is inducible [49], however we find no evidence for significant dynamic regulation of Egr2 expression in the claustrum. Our analyses of the molecular composition and connectivity of CLEgr2+ neurons validate the specificity and penetrance of the Egr2-CRE line as a tool for functional investigation of a significant population of claustrum neurons. Anterograde and retrograde tracing of genetically-defined CLEgr2+ connectivity corroborates the validity of our transgenic access to the claustrum, as it replicates and extends the reported connectivity of the claustrum [2,4,5,50]. The vast ipsilateral claustro–cortical projections we identify (to frontal, association, motor, and sensory cortices) are consistent with the majority of previous reports, including findings summarized in recent comprehensive investigations of claustral connectivity [25,26,51]. We further extend the description of claustral afferents, identifying substantial projections to subcortical structures, notably nuclei in the midbrain, thalamus, striatum, amygdala, and basal forebrain. Our results of retrograde mapping of cortical inputs are consistent with the observations found in the literature, as we observe claustral inputs originating from different sensory and motor cortical regions, as well as association regions of the cortex [25–28].
An additional important aspect of claustral connectivity is its reciprocity, as cortical regions targeting the claustrum are often recipients of claustral efferents [2,4,5,50]. Our results corroborate these findings, and further extend the principle of reciprocity to the connections between the claustrum and subcortical structures. The subcortical connectivity of the claustrum remains largely uncharacterized, and is worthy of attention in future studies. With regard to functional implications, the presence of reciprocal connections between the claustrum and midbrain dopaminergic, as well as basal forebrain cholinergic centers, are of special interest for future investigation. The claustrum is rich in neuromodulatory receptors [52], and our current observations suggest that it may additionally impact the activity of subcortical neuromodulatory structures, which are highly implicated in regulating attention [53], potentially tying into our current observations regarding a role for the claustrum in supporting resilience to distraction.
Addressing the function of the claustrum, we implemented an automated version of a two-alternative forced choice task (similar in design to [54,55]). To test a naturalistic behavior of multi-sensory nature, we turned to maternal pup-retrieval [45]. The impact of inhibition of CLEgr2+ neurons was observed only upon introducing an auditory distractor during the tasks. The role of the claustrum in maintaining goal-directed behavior in the presence of distraction could be interpreted in the context of top-down attention; a deficit of which is expected to selectively sensitize an affected animal to a distractor [56]. A recent study demonstrating a role for frontal inputs to the claustrum in top-down control [28] is consistent with our proposal that the claustrum acts to support resilience to distraction, a fundamental cognitive aspect of attention and sensory gating.
Addressing potential mechanisms whereby the claustrum may act to support resilience to distraction, we examined the effects of claustral stimulation on cortical sensory representation. Utilizing fiber photometry in the auditory cortex, while interleaving optogenetic stimulation of CLEgr2+ afferents, we found that activation of the claustrum suppressed auditory cortical responses to pure tone presentations. We interpret our observations as supporting the hypothesis that the claustrum acts to diminish the representation of sensory input currently not selected for attention. Indeed, claustral inhibition rendered mice susceptible to an auditory distractor in our behavioral experiments. Possibly, the claustrum acts to globally suppress sensory representation within non-selected modalities, suppressing distractors as well as potentially relevant sensory information. However, we favor the hypothesis that the claustrum modulates cortical processing in a refined manner, selectively diminishing the representation of sensory distractors, while retaining task-relevant sensory information, even within the same modality. This function would support effective behavior in multi-sensory environments, a simple example of which is the pup retrieval task. Determining the precise nature of the impact of the claustrum on cortical processing is a topic for future investigation.
Given that CLEgr2+ afferents are excitatory, a mechanistic question arises regarding how activation of the claustrum could drive net inhibition of sensory cortex. Electrical stimulation of the claustrum area has been shown to drive prolonged inhibition of cortical responses [20,21], and claustral projections have been previously reported to impinge on spineless dendrites of putative inhibitory interneurons [39,57]. Recently, two reports describe powerful, widespread, and long-lasting feedforward inhibition of cortical activity driven by claustral neurons [58,59]. Both groups, studying the impact of the claustrum on activity in frontal, association and sensory cortical structures, describe an underlying mechanism whereby claustral neurons drive firing of cortical inhibitory interneurons, driving net inhibition of primary cortical neurons. Interestingly, we report co-expression of the GABA-biosynthetic enzyme, GAD65, within vGlut1-expressing claustral neurons, suggesting that a subset of claustral projection neurons may harbor the potential to co-release GABA. A variety of potential functional consequences could be mediated by direct release of GABA, ranging from gain control to increased temporal precision of target neuron firing [33]. Future work, requiring the development of experimental setups for the recording and modulation of claustral activity in behaving mice, will further address whether in addition to suppression of sensory responses, the claustrum can facilitate responses to task-relevant stimuli.
STAR methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Ami Citri (ami.citri@mail.huji.ac.il).
Experimental Model and Subject Details
All mice described in this study were heterozygous Egr2-CRE knock-in mice [24], crossed onto a C57BL/6JOLAHSD strain. All mice were kept on a 12-hour light-dark cycle in a specific pathogen-free (SPF) animal facility with free access to food and water. Mice were housed in a groups of same-sex littermates. Littermates of the same sex were randomly assigned to experimental groups. All experimental procedures, handling, surgeries and care of laboratory animals used in this study were approved by the Hebrew University Institutional Animal Care and Use Committee (IACUC).
Sex and ages
Connectivity experiments: Females, 8-12 weeks old at the time of virus injections.
Behavior in the 'Intellicage': Males, 6-10 weeks old
Behavior in the pup retrieval assay: Females, 8 weeks old at the injection (prior to conception and delivery), pups were 3-7 days postnatal.
Calcium fiber photometry: Females, 8-12 weeks old.
Each experiment was conducted on homogenous groups. No sex-specific analysis was performed.
Method Details
Histology
Mice were sacrificed by anesthesia with 5% isoflurane, followed by cervical dislocation and rapid decapitation. Brains were harvested, placed in disposable histology molds with OCT embedding medium (Tissue-Tek®), snap-frozen in liquid nitrogen and stored in -80ºC until further processing. Brains were sectioned to 14 μm thickness using a cryostat (Leica CM1860) at -20°C and collected onto slides (SuperFrost® Plus). Slides were kept at -80°C until further processing.
Probe preparation
Egr2 ISH Probe plasmid [60] was digested for 2 hours at 37°C with XbaI to create a DNA template for an RNA probe. Restriction was validated via 1% Agarose gel electrophoresis. Linearized plasmids were extracted using Phenol Chloroform Isoamyl alcohol and precipitated using 3M Na-Acetate and 100% Ethanol at -20°C overnight. On the following day, tubes were centrifuged at 4°C for 1 hour and the pellet was air-dried and dissolved in DDW for 10 minutes at 60°C. In vitro transcription was performed at 37°C for 2 hours using T3 RNA polymerase and Digoxigenin (DIG)-labeled Uracil Ribonucleotide (DIG RNA Labeling Mix, Roche). RNA probes were hydrolyzed to ~250 bp fragments at 60°C for 12 minutes and precipitated using 4M LiCl and Ethanol at -20°C overnight. On the following day, tubes were centrifuged at 4°C for 1 hour and pellet was air-dried and dissolved in DDW for 15 minutes at 60°C. A dot-blot was performed in order to quantify the hydrolyzed DIG-labeled RNA probes; serially diluted probe solutions were crosslinked to a hybridization transfer membrane (GeneScreen) by UV. The membrane was washed according to manufacturer’s instructions and incubated with a chromogenic reaction eliciting substrate (BCIP+NBT ready mix, Roche). After washing with DDW, the membrane was dried and compared to a serially-diluted positive control probe, of known concentration.
In Situ Hybridization
Our protocol followed previously described work [61]. Briefly, slides were fixed in 4% PFA in PBS (for 10 minutes at 4°C), washed in PBS, acetylated using Triethanolamine (TEA at 1.2% v/v) and acetic anhydride (0.3% v/v) and washed in PBS. Sections were circled using a hydrophobic barrier pen (ImmEdge), placed in a humid chamber and mounted with pre-hybridization buffer overnight at room temperature. On the following day, for Egr2 DIG-labeled probe hybridization, pre-hybridization buffer was replaced with hybridization buffer containing 200 ng Egr2 DIG-labeled RNA probe, slides were covered with hybridization cover (Bio-Labs HybriSlip™), placed in a humidified chamber and incubated at 71.5°C overnight. On the following day, slides were washed in 5X concentrated (0.75M NaCl, 0.075M) sodium chloride and sodium citrate solution (5X SSC) at 65°C for removing of the coverslips followed by a wash in 0.2X SSC. Slides were mounted with wash buffer, followed by blocking buffer and immunolabeled with sheep Anti-Digoxigenin-AP Fab fragments (Roche; catalog No. 11093274910; final dilution to 1:5000 in blocking buffer). Slides were then washed with wash buffer and mounted with a chromogenic reaction eliciting substrate (BCIP+NBT ready mix, Roche), covered with coverslips to prevent contact with air and incubated in the dark, overnight at room temperature. On the following day the chromogenic reaction was stopped by 10mM Tris-HCl, 1mM EDTA, pH 8, slides were mounted with coverslips and air dried prior to image acquisition.
Single molecule fluorescence in-situ hybridization (smFISH)
Egr2-CRE mice were stereotactically injected with AAVdj-DIO-eGFP (see Stereotactic surgery and virus injections). Four weeks after the injection mice were deeply anesthetized with isoflurane, decapitated, and their brains were rapidly removed and briefly washed in PBS. Brains were placed in molds containing O.C.T embedding medium (Scigen Scientific Gardena, CA 90248 U.S.A) and snap-frozen on dry ice. Embedded brains were sectioned on a Leica CM1850 cryostat into 14µm sections, mounted onto SuperFrost Plus slides and kept at -80°C (see Histology). Slides were processed according to ACD RNAscope fresh frozen tissue pretreatment and fluorescent multiplex assay manuals. All probes were purchased from Advanced Cell Diagnostics (RNAscope Probe – EGFP: Cat No. 400281, RNAscope Probe- Mm-Slc17a7-C2: Cat No. 416631-C2, RNAscope Probe- Mm-Gad2-C3: Cat No. 439371-C3). Slides were counterstained with DAPI for 30s, coverslipped with mounting medium (PermaFluor Aqueous Mounting Media, Thermo Scientific, Cat No. 94-TA-030-FM) and imaged using a high-speed fully-motorized multi-channel light microscope (Olympus IX-81) at 40x magnification. Data was analyzed using the CellProfiler v.2.1.1 (www.cellprofiler.org) speckle counting pipeline with minor modifications [62,63].
Stereotactic surgery and virus injections
Mice were anesthetized by IP injection of ketamine (75 mg/kg) and medetomidine (1 mg/kg). Following validation of the depth of anesthesia, mice were secured in a stereotaxic apparatus (David KOPF instruments, Tujunga CA). Following incision of the scalp, a small hole was made in the skull using a fine drill burr (model 78001, RWD Life Science, San Diego CA). A microsyringe (33GA Hamilton syringe, Reno, NV) loaded with the virus was lowered into the target structure. The viral tracer was injected at 50-100nl/min via an UltraMicroPump (World Precision Instruments, Saratosa, FL), following which the microsyringe was left in the tissue for 5-10 minutes after the termination of the injection before being slowly retracted. Finally the incision was closed with bioadhesive and the animals were injected with saline (for hydration), antisedan (to negate the anesthesia) and rimadyl (analgesic) and recovered under gentle heating. Coordinates for stereotactic injection were based on the Paxinos and Franklin mouse brain atlas [64]. The coordinates used for claustral injections were lateral-medial (LM): -2.8, rostral-caudal (RC): 1, dorsal-ventral (DV): -3.68, relative to Bregma. Where noted, an additional injection was made at LM: -3.25, RC: 0, DV: -4.15. Unless noted otherwise, viruses were prepared at the vector core facility of the Edmond and Lily Safra Center for Brain Sciences, as described previously [27]. Each batch of viruses used in the study was titrated so as to ensure specific infection of the claustrum, limiting spillover to adjacent insular, piriform and striatal structures.
Frontal projections
To identify whether the frontal projections from the ACA overlap with the Egr2+ population in the claustrum, Egr2-CRE mice were stereotactically injected with 200nl AAVdj-CMV-eGFP to the ACA (LM: 0.25, RC: 1.1, DV: -1.9), or to the ORB (LM: 1, RC: 2.55, DV: -2.4) and 250nl AAV2-hSyn-DIO-mCherry (purchased from the UNC viral core facility) to the claustrum. Two weeks later, animals were sacrificed for histology and immunohistochemistry.
Connectivity
For anterograde tracing, 80-100nl of AAV9-CBA-DIO-ARCH-eGFP-WPRE-SV40 (purchased from the UPENN viral core facility, # AV-9-PV2432) or AAVdj-DIO-ChR2-eYFP (purchased from the Stanford viral core facility) were stereotactically injected unilaterally into the claustrum of Egr2-CRE mice. Analysis presented in the manuscript is based on the analysis of one mouse injected with each virus. A number of additional mice, demonstrating essentially overlapping projection maps, were excluded from analysis due to virus infection extending somewhat beyond the boundaries of the claustrum. Mice were sacrificed for histology and immunohistochemistry 2-4 weeks later. For retrograde mono-trans-synaptic labeling, we made use of two AAV viruses, combined with a pseudotyped rabies virus (pRbV). This system (graphically represented in Figure 1K) utilizes a mutant RbV, which lacks the rabies gene encoding the envelope glycoprotein (RG) required for viral spread. This mutant RbV is pseudotyped (pRbV) with the avian sarcoma leucosis virus EnvA envelope protein, restricting the cells the virus can transduce to those that express the EnvA receptor TVA. To reconstitute infectious pRbV particles in vivo, which will selectively label the neurons presynaptic to the target population (the ‘starter cell’ population), neurons are first transduced with conditional AAV vectors that express TVA and RG in a CRE-dependent manner [65,66]. To this end, we targeted the claustrum with a combination of two AAV viruses: 100nl of 1:1 mixture of AAV2-CAG-DIO-TC66T-mCherry and AAV2-CAG-DIO-G were stereotactically injected unilaterally into the claustrum of three Egr2-CRE mice. The first virus expresses (in a CRE-dependent fashion) a modified receptor for avian virus (TVA66T) and mCherry – labeling infected cells fluorescently red. The TC66T mutation reduces EnvA-enveloped viral transduction by ±10 fold, and has been reported to eliminate off-target (CRE-independent) transduction events by EnvA-pseudotyped RbV, at the expense of a reduced number of starter cells [66]. The second AAV virus conditionally expresses the RbV glycoprotein, enabling the complementation of pRbV for trans-synaptic tracing. Two weeks following injection of the AAV viruses, we stereotactically targeted the claustrum with a modified monosynaptic rabies virus, from which the glycoprotein was deleted and replaced with EnvA and GFP (500nl). This pRbV can only infect cells expressing TVA. Since the Cre-dependent AAV provides Cre+ cells with a source of rabies glycoprotein, newly formed rabies virus particles can spread retrogradely from these Cre+ cells to their directly connected inputs. These input cells do not express TVA or rabies glycoprotein, preventing the rabies virus from spreading further, effectively restricting rabies virus infection to Cre+ cells and their direct, monosynaptic inputs [65]. Thus, starter cells express both red and green fluorophores and appear yellow after merging channels, while presynaptic neurons are labeled in green. Following 6 days allowing for trans-synaptic delivery and expression of pRbV-driven fluorescence, mice were sacrificed for histology, brains were fixed and prepared for imaging. Data presented in Figure 1N is an average of analysis of two mice for anterograde tracing and two mice for retrograde tracing. Mice in which the infection was not precisely limited to the claustrum were eliminated from analysis, although, these mice still largely replicated the observations we report herein. It is important to note that it is likely that the inputs we identify under-represent the magnitude of cortical projections to the claustrum. Under-representation appears to be pervasive in studies applying conditional pRbV monosynaptic labeling [65,67–69], and may be exacerbated by the use of the highly specific TVA66T mutant in our study [66,70]. Preferring to err on the side of caution, we opted for specificity, minimizing the danger of infecting starter cells outside the claustrum. Potentially, newer versions of pRbV, such as CVS-N2cΔG [71] or RVΔGL [72] may be helpful in maintaining specificity while increasing the efficacy of presynaptic labeling.
Histology and immunohistochemistry
Mice were sacrificed by anesthesia with 5% isoflurane, followed by rapid decapitation. Brains were removed and fixed in 4% PFA overnight at 4°C. On the following day, brains were thoroughly rinsed in a 0.9% NaCl Phosphate buffered saline (PBS) solution and sectioned on a Vibratome (7000 smz-2) at 60µM thickness in the coronal plane. Two series of sections were collected from each brain, resulting in two copies of brain slices at 120µm apart, corresponding to the division of the mouse brain atlas (as in [73]). In order to enhance the GFP signal in terminal endings for anterograde tracing experiments, floating section immunohistochemistry was performed. Sectioned brain slices were washed twice in PBS, followed by blocking in 3% normal horse serum and 0.3% Triton X-100 in PBS for 1 hour. Sections were then incubated overnight at 4ºC in a rabbit anti-GFP primary antibody (life technologies; catalog No. A-6455; http://1degreebio.org/company/life-technologies---novex/, final dilution to 1:500 in 3% normal horse serum, see table 2). 16 hrs later the sections were washed three times in PBS. Washes were followed by 2 hours of incubation at room temperature with donkey anti-rabbit IgG H&L Alexa Fluor 488 (Abcam; catalog No. ab150065; final dilution to 1:500) in 3% normal horse serum. Finally the sections were washed three times in PBS and then counterstained with DAPI (Roche; catalog No. 10-236-276; final dilution 1:1000 in PBS) to detect cell nuclei and then quickly washed twice, mounted onto slides and covered.
Clarity
Brains were cleared based on previously published protocols [74]. Briefly, mice were transcardially perfused with ice cold PBS followed by 4% PFA, brains were removed and kept in 4%PFA overnight at 4°C. Brains were then transferred to a hydrogel solution (PBS with: 1% acrylamide, bio-rad #161-0140; 0.125% Bisacrylamide, bio-rad #161-0142; 0.25% VA-044 initiator, Wako, 011-19365; 4% PFA) for 2 days. The samples were then degassed with N2 for 45min and polymerized in 37°C for 4 hours. The samples were then washed overnight in 200mM NaOH-Boric buffer (sigma, #B7901) containing 8% SDS (sigma, #L3771), to remove PFA residuals. Samples were then stirred in a clearing solution (100mM Tris-Boric buffer, bio-lab, #002009239100 with 8% SDS) at 37°C for 16 days. After the samples became transparent, they were washed with PBST (PBS with 0.2% tritonX100; ChemCruz, #sc-29112A) for 24 hours at 37°C with mild shaking and for another 24 hours with fresh PBST 0.2% at RT. Finally the samples were incubated in the refractive index matched solution Rapiclear (RI=1.47; SunJin lab, #RC147002) for 6 hours at 37°C and 2 days at room temperature before imaging.
Image acquisition
Slides were scanned on a high-speed fully-motorized multi-channel light microscope (Olympus IX-81) in the microscopy unit of the Alexander Silberman Institute of Life Sciences. All slices were imaged at 10X magnification (NA=0.3), green and red channels exposure times were selected for optimal clarity and were kept constant within each brain series. DAPI was acquired using excitation filters of 350±50nm, emission 455±50nm; eGFP excitation 490±20nm, emission 525±36; mCherry excitation 555±25nm, emission 605±52nm. Figures were prepared using Photoshop, Illustrator, and InDesign (Adobe, San Jose, CA). Figures showing stained brain tissue were adjusted using a uniform brightness/contrast mask created in linear-mode in Photoshop, and applied consistently to all slices within a single brain. Images were then scaled or cropped to improve data presentation and increase signal visibility. Digitization was performed on raw, non-adjusted images. CLARITY Scanning was performed using the Olympus FV1200/IX83 Bio-Imaging confocal microscope (Bio-Imaging Unit, the Alexander Silberman Institute of Life Sciences). A 20X/0.45 objective was used, excitation 561nm and emission 570-620nm, Z step 4mm. 36 fields were tiled.
Intellicage experiment
Animals:
Seven Egr2-CRE male mice, 6-8 weeks old, were injected with 100-120nl AAVdj-EF1a-DIO-Kir2.1-t2A-zsGreen (Kir2.1) to the claustrum in two injection sites. As a control group, six Egr2-CRE male mice, 6-8 weeks old, were injected with 100-120nl AAV9-CAG-DIO-eGFP (purchased from the UPENN viral core facility, # AV-9-ALL854) to the same coordinates. Mice were randomly assigned to either the experimental or the control groups. Two weeks after injection, mice were introduced to the behavior cage (the ‘Intellicage’) for behavioral training. While the automated setup has the advantage of enabling self-paced experiments without experimenter involvement, it is limited in the number of mice that can be included in each experiment. Sample size was maximized under the limitation of the setup (the setup can support up to 8 mice in each group, as they are co-housed and share a window of 5 hours in which they are permitted to engage with the task), therefore the experiment was performed in two repetitions. The first experiment consisted of five mice (3 injected with DIO-Kir2.1, and 2 with DIO-GFP), the second experiment consisted of eight mice (4 DIO-Kir2.1 and 4 DIO-GFP). The two batches did not differ in performance. (The success rate of the two Kir2.1 groups: p=0.9372, two sided Mann-Whitney U Test, for the two GFP groups: p=0.9333, two sided Mann-Whitney U Test). One DIO-Kir2.1 injected mouse from the second batch failed to engage with the task in early stages, and was removed from the analysis. For pharmacogenetic inhibition of the claustrum, four Egr2-CRE male mice (6 weeks old), were injected with AAV2-hSyn-DIO-hM4D(Gi)-mCherry (purchased from the UNC viral core facility, # AV-4-500a) to the claustrum. An IP injection of CNO (10 mg/kg) preceded the sessions in which a distractor was interleaved. The automated nature of the Intellicage setup supported blinding of experimenters to the experimental groups throughout training and test sessions, as well as during data analysis.
Behavioral setup:
The experiment was conducted using the ‘intellicage’, an automated behavioral setup for multiple animals in the homecage environment (New Behaviour/TSE, Germany) [75,42] The intellicage consists of two compartments connected by a tube corridor. One compartment, a normal mouse cage, serves as the homecage, where the animals have access to food. The second compartment of the intellicage is the behavior ‘corner’, which consists of two ports, one at each side of the corner, with controllable doors and nose poke detectors, behind the doors are positioned two drinking bottles as the sole source of water. The doors to the ports can be opened or closed depending on the demands of the experiment. The learning corner incorporates light (LED positioned above the two doors) and sound (loudspeaker positioned directly behind the corner) stimuli and is programmable as a customized operant conditioning chamber. Mice are implanted with RFID tags, enabling documentation of all visits, nose pokes and licking events of individual mice. Entrance into the corner (a ‘visit’) is detected by both an antenna located at its opening that reads the implanted transponder, and the activation of a heat sensor within the corner.
Behavioral training:
Training comprised several stages. First, mice were habituated to the behavioral corner, they were allowed free access to water, with the two doors in the corner open for 24 hours. Next, mice were taught to associate the visual cue with water availability during a defined time window (19:00-24:00). During inactive hours any nose poke resulted in a negative reinforcer tone (4 kHz, 100ms). During active hours, 200ms from entry to the corner one door was open (randomly) associated with a green LED. The cue and the opening of the door lasted for 5 seconds. Third, mice had to learn to nose poke in order to receive the reward (water access). 200ms after entry to the corner the LED on top of one of the ports turned on, nose poke in the correct side triggered opening of the door and access to water for 3 seconds. Wrong nose poke (before light on, after light off, or at the wrong side) triggered the negative reinforcer, closing of the door, and deactivation of the LED cue. In order to receive a reward after a wrong choice, mice had to leave to corner and re-enter to initiate a new trial. Following training, mice were tested on variations of cue duration, as well as variable duration of delay to cue presentation. When the stimulus presentation was shorter than one second, the mice had a full second from cue onset to respond correctly. The parameters were fixed during a 5-hour experimental session, but varied between sessions as a function of performance (a threshold of 70% success was defined before further modifying any parameter). For any fixed delay duration (200/1000/2000/3000ms), cue presentation was shortened (1000/500/200/100ms) in consecutive sessions. In the pharmacogenetic experiment a similar training paradigm was employed, in the absence of a negative reinforcer tone, and during the session with the distractor, the delay period was randomized between 2-4 seconds, and the cue was presented for 500ms.
Distractor session:
The session with the distractor was identical to previous sessions with 2 sec delay and 100ms cue duration, with the introduction of an auditory interference (5 second clip of the song ‘Pluto’ by the artist Bjork) randomly interleaved in half of the trials. The distractor initiated 200ms after entry to the corner and lasted for 5 second, or until the animal left the corner. The negative reinforcer was disabled during the test session.
Data analysis:
A linear mixed effect model, ‘successRate~injection*distractor+(1|animal)’, was applied for statistical analysis, as described below.
Pup retrieval experiment
Chemogenetic inhibition using DREADDs:
6 females, 8 weeks old, were injected with 250nl AAV2-hSyn-DIO-hM4D(Gi)-mCherry (purchased from the UNC viral core facility, #AV-4-500a) targeting the claustrum. 7 days after surgery, mice were placed in 3 breeding cages that contained 1 male & 2 injected females and cotton for nesting. Each female conceived and delivered 3-10 pups. The experiment was performed at 3-7 days postnatal (P3-P7). On the first day of the experiments the mothers were intraperitoneally (i.p.) injected with saline, while on the second day of the experiment the mothers were injected with CNO (10 mg/kg, i.p). After the injection, the injected mother was placed back in the homecage, while the other mother was placed in a new cage with the pups. After the first mother completed the experiment the second mother underwent i.p. injection and was placed in the homecage for habituation. The experiment started with 40 minutes habituation to the new environment in the absence of pups, following which single pups were returned to the homecage, and placed on the opposite side of the nest from the mother. 5 pups were added without any interference and 5 pups were added in the presence of a distractor (a scrambled version of the song ‘Pluto’ by the artist Bjork) with 10 seconds of silence between each pup. The retrieval of the first pup was regarded as ‘training’ and was removed from the analysis.
Chronic inhibition using Kir2.1:
Female Egr2-CRE mice were injected with 300nl AAVdj-EF1a-DIO-Kir2.1-t2A-zsGreen (Kir2.1, purchased from the Stanford viral core facility [41]) to the claustrum. 7 days after the surgery mice were put in 2 breeding cages that contained a pregnant mother, a virally-injected female and cotton for nesting. The experiment was performed at 3-7 days postnatal (P3-P7). The experiment started with 1 hour habituation to the new environment in the absence of pups, following which single pups were returned to the homecage, and placed on the opposite side of the nest from the mother. 4 pups were added without any interference and 3 pups were in the presence of a distractor (the song ‘Pluto’ by the artist Bjork, played continuously)
Data analysis:
We measured the time it took the mother to collect the pups, the measurement started when the pup was put inside the cage and stopped when the mother picked up the pup. A linear mixed effect model, 'duration~injection*distractor+(1| animal)', was used for statistical analysis of the data, as described below.
Ex vivo slice physiology
Slice preparation:
Acute coronal brain slices (260um thick) were generated for in vitro recording and optical stimulation. After anesthetizing the mice with isoflurane, brains were isolated in an ice-cold sucrose solution composed of the following (in mM): 26 NaHCO3, 10 glucose, 204 sucrose, 2.5 KCl, 1 NaH2PO4, 1 CaCl2, 4 MgSO4, pH 7.3, 300 mOsm. Coronal slices were sectioned on a vibratome (7000 SMZ, Campden Instruments) in the same ice-cold sucrose solution. Slices were then transferred to warm (32°C) solution of artificial CSF (aCSF) composed of the following (in mM): 124 NaCl, 26 NaHCO3, 4.4 KCl, 1 NaH2PO4, 1.3 MgSO4-7H2O, 11 glucose, 2.5 CaCl2-2H2O, pH 7.3, ~300 mOsm. The solution was then allowed to cool (27°C). This same aCSF solution was also used for all subsequent recordings. All solutions were continuously bubbled with 95% O2/5% CO2. For electrophysiological recordings, slices were transferred to a submersion chamber on an upright microscope (Olympus BX51WI) and were continuously superfused (2-4 ml/min) with warm (32-34°C), oxygenated aCSF. Fluorescently-labeled CLEgr2+ neurons were visualized with a digital camera (QImaging 2000R) using transmitted light with infrared differential interference contrast optics and epifluorescence. Neurons were targeted within the labeled region using glass recording electrodes (3–7 MΩ) filled with a solution containing the following (in mM): 2.8 KCl, 145 K-Gluconate, 10 HEPES, 1 EGTA, 2.4 MgATP, 0.4 NaGTP, 0.1 CaCl2, 2 NaCl, pH 7.3, 295 mOsm. Whole-cell patch-clamp recordings were obtained from claustral neurons using a Multiclamp 700B amplifier (Molecular Devices) in current-clamp modes controlled by custom-written routines in Clampex 10.4 (Molecular devices). All signals were low-pass filtered at 10 kHz and digitized at 20-100 kHz (Digidata 1440A, Molecular devices). Data acquisition was performed using Clampex 10.4 (Molecular devices).
Calcium fiber photometry:
Animals:
4 Egr2-CRE female mice were used for in vivo fiber photometry. At age 8-12 weeks mice were injected with 100-150nl of either AAV9-CAGGS-DIO-ChR2-tdTomato (purchased from the UPENN viral core facility, # AV-9-18917P) or AAVdj-Ef1α-DIO-ChR2-EYFP (purchased from the Stanford viral core facility) to the left claustrum, in two coordinates. Following virus injection, a custom metal bar was placed on the skull and fixed to the bone using dental cement. After letting the virus express in claustral axon terminals in the cortex (16.25±2.7 weeks post injection), animals were anesthetized with Sevoflurane (~4%). Anesthesia depth was assessed by monitoring the pinch withdrawal reflex and respiration rate and adjusted accordingly. Rectal temperature was monitored continuously and maintained at 36 ± 1°C. The head of the mice was positioned relative to the speaker (facing the right ear) and held in place by connecting the head bar to a custom stage. To access the brain, the muscle overlying the left auditory cortex was removed, and a craniotomy (~1 × 1mm) was performed over A1 (coordinates, 2.3mm posterior and 4.2mm lateral to bregma) as described previously [76,77]
Recording:
A membrane-permeant acetoxymethyl (AM)-ester of the calcium-sensitive dye Oregon green 488 BAPTA-1 (Molecular Probes, Eugene, Oregon) was dissolved in DMSO containing 20% Pluronic F-127 acid to a final concentration of 10 mM. The stocks were diluted in a solution containing (in mM): 150 NaCl, 2.5 KCl, 10 HEPES, yielding a final dye concentration of 0.5 mM, which was bolus loaded into the cortex with a glass micropipette as described previously [46]. For optical recordings multi-mode fibers (FT-200-URT, Thorlabs, Grünberg, Germany) with a diameter of 200 μm and a numerical aperture (NA) of 0.48 were used. After removing the shielding from its tip the fiber was glued into a metal tube (0.9mm o.d., about 10mm length) with cyanoacryl glue (UHU, Buhl-Baden, Germany). The fiber tip protruded by about 1mm from the metal tube. 30min after dye application the fiber tip was inserted with a micromanipulator into the stained region and lowered ventrally into the cortex, recording from sites separated by 100-200µm (recording sites were located at cortical depths of between 50-800µm). Low intensity stimulation (typically <0.1mW at the tip of the optical fiber) was applied for excitation of the calcium-sensitive dye and recording of calcium-dependent fluorescence. Brief transitions (5ms) to high intensity stimulation (2mW at the tip of the optical fiber) were randomly interleaved for activation of ChR2, terminating coincidently with sound onset. The conditions for optogenetic activation through the fiber used for recording were previously established [46,78]. The auditory protocol comprised of 15 or 19 pure tones (100ms duration, 900ms inter-stimulus interval) logarithmically spaced between 1-64 kHz, and presented at 3db attenuation. Each stimulus-condition pair was repeated 10 times and stimuli were presented in random order. Photometry was performed using a device developed and built in the lab of Prof. Arthur Konnerth (Technical University Munich, Germany). An optical fiber was used for delivering the excitation light from a 20 mW solid state laser (Sapphire, 488 nm, Coherent, Dieburg, Germany) to the stained brain region and for transmitting the fluorescence to the detector, in our case an avalanche photodiode (APD, S5343, Hamamatsu Photonics, Herrsching, Germany). Switching between two laser light intensities was enabled by an acousto-optic modulator (AOM 3080-125, Crystal Technology, Palo Alto, California, USA): high intensities (1-2 mW at the tip of the optical fiber) for activation of ChR2 and low intensities (typically 0.01-0.05 mW at the tip of the optical fiber) for excitation of the calcium sensitive dye and recording of the calcium-dependent fluorescence. The resulting voltage signal was sampled at 2.4 kHz (Alpha-Omega SNR data acquisition system) and stored, together with stimulus time markers, in a file for offline analysis.
Quantification and Statistical Analysis
Cell counting and quantification
Anterograde projections from CLEgr2+ neurons were manually analyzed. For each slide we identified the best matching image from the Allen mouse brain atlas. The density of the CLEgr2+ projections was scored between 0 and 5, such that 0 indicates the absence of observed projections, and 1-5 provide a scale for the range of relative density of the projections (see also Figure S1E). In order to quantify the inputs to the CLEgr2+ neurons, slides from retrograde labeling were manually analyzed. After finding the best matching image from the Allen mouse brain atlas, eGFP-labeled cells, representing inputs to CLEgr2+ neurons, were counted. Input neurons in the vicinity of the virus injection sites were subtracted from the analysis.
Statistical analysis of behavioral data
For each statistical analysis provided in the manuscript, an appropriate statistical comparison was performed. For small sample sizes (n < 5) non-parametric tests were used by default. For larger sample size we applied a linear mixed effect model in order to avoid the assumptions of normality and variance homogeneity. After applying the linear model, the normality of the residuals was assessed using the Shapiro-Wilk test of normality test. In several cases where repeated measures ANOVA was applied upon the linear model Bartlett's K-squared test for homogeneity of the variances was performed. Statistical analysis of the behavioral results was done in R (Ver. 3.3.1) and RStudio (Ver. 1.0.44).
Calcium fiber photometry Data Analysis
Raw traces were low-pass filtered at 50 Hz. To correct for slow drifts of the baseline, the fluctuations of the fluorescence traces were first fitted by a 3rd degree polynomial. The fit was subtracted from the trace, and the baseline was set at the 1st percentile of the subtracted waveform (so that 1% of the values were smaller from the baseline). The fluorescence was transformed into ΔF/F units. Due to the high temporal resolution of OGB, baseline activity was then subtracted on a trial-by-trial bases to align the responses at stimulus onset. Calcium transients were averaged over repetitions, and the peak fluorescence occurring in a 60ms time window following stimulus onset was considered as the early responses [46]. For each responsive recording site (n=14), best frequency (BF) was determined as the frequency which elicited the maximal early response with no stimulation of CLEgr2+ neurons, averaged over repetitions. In order to quantify the effect of CLEgr2+ neurons stimulation on the neuronal response to frequency sweeps, while accounting for multiple comparisons and repeated measures, we employed a linear mixed effects model (implemented by the fitlme function in MATLAB) with one regressor (ABF) accounting for the absolute distance of a tone from the BF, and one for condition (Laser On/Off), as well as a quadratic term allowing for random variation in the population tuning curves bewteen recording sites (DF2). An indicator function (IBF) was used to assess the interaction between laser activation and the response at the BF, for a final formula of: 'response ~ ABF + condition+ IBF : condition + (DF2 | site') Finally, a one-way ANOVA was run on the results of the model, followed by a post-hoc Wilcoxon signed-rank test.
Data and Software Availability
All the data that support the findings of this study are available from the corresponding author upon reasonable request. Computer codes used for analysis in this study were implemented in MATLAB (MathWorks), R and RStudio software. Codes will be made freely available upon request. Requests should be directed to the corresponding author.
Supplementary Material
Acknowledgments
The authors thank our friends and colleagues for their comments, especially Dr. Mickey London. We are indebted to Dr. Naomi Book-Melamed, head of the microscopy unit at the Life Science Institute for technical support and access to software. Work in the Citri lab is funded by the Edmond and Lily Safra Center for Brain Sciences (ELSC), the Brain and Behavior Foundation (NARSAD #18795); the German-Israel Foundation (GIF # 2299-2291.1/2011); the Binational Israel-USA Foundation (BSF #2011266); the EU Marie Curie Career Integration Grant (CIG # PCIG13-GA-2013-618201); the Israel Anti-Drug Authority; the Israel Science Foundation (ISF #393/12), the National Institute for Psychobiology in Israel – Founded by the Charles E. Smith Family (109-15-16), the Adelis Award for Advances in Neuroscience, the European Research Council (ERC-CoG #770951), a seed grant from the Eric Roland Fund for interdisciplinary research administered by ELSC, and contributions from anonymous philanthropists in Los Angeles and Mexico City. AC is a supported member of the Israeli Center for Research Excellence (iCORE) Program of the Israel Science Foundation (#1796/12) and the Canadian Institutes for Advanced Research program in Child and Brain Development.
Footnotes
Author Contributions:
Study concept and design: GA, AT, NR & AC. ISH: BJG. Claustral targeting and anterograde connectivity: AT, GA & NR. Connectivity reconstruction: NR, OZ, AT & GA. Retrograde connectivity: GA, AT & GT (with support of AM, initial experiments supported by BL). CLARITY: RR & AT (with support of IG). Behavioral analysis: AT, NR & GP. Ex vivo slice physiology: GP & YG. Calcium photometry: GA & KS (with support of IN). Virus preparation: MG. Drafting of the manuscript: GA, AT, NR & AC. Obtained funding: AC. Study supervision: AC.
Declaration of Interests:
The authors declare no competing interests.
References
- 1.Torgerson CM, Irimia A, Goh SYM, Van Horn JD. The DTI connectivity of the human claustrum. Hum Brain Mapp. 2015;36:827–838. doi: 10.1002/hbm.22667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Edelstein LR, Denaro FJ. The claustrum: a historical review of its anatomy, physiology, cytochemistry and functional significance. Cell Mol Biol. 2004;50:675–702. [PubMed] [Google Scholar]
- 3.Druga R. Chapter 2 - The Structure and Connections of the Claustrum. In: Smythies JR, Edelstein LR, Ramachandran VS, editors. The Claustrum. San Diego: Academic Press; 2014. pp. 29–84. [Google Scholar]
- 4.Goll Y, Atlan G, Citri A. Attention: The claustrum. Trends Neurosci. 2015;38:486–495. doi: 10.1016/j.tins.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 5.Crick FC, Koch C. What is the function of the claustrum? Philos Trans R Soc L B Biol Sci. 2005;360:1271–1279. doi: 10.1098/rstb.2005.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smythies J, Edelstein L, Ramachandran V. Hypotheses relating to the function of the claustrum. Front Integr Neurosci. 2012;6 doi: 10.3389/fnint.2012.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith JB, Alloway KD. Interhemispheric claustral circuits coordinate sensory and motor cortical areas that regulate exploratory behaviors. Front Syst Neurosci. 2014;8:93. doi: 10.3389/fnsys.2014.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Remedios R, Logothetis NKK, Kayser C. A role of the claustrum in auditory scene analysis by reflecting sensory change. Front Syst Neurosci. 2014;8:44. doi: 10.3389/fnsys.2014.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Remedios R, Logothetis NK, Kayser C. Unimodal responses prevail within the multisensory claustrum. J Neurosci. 2010;30:12902–12907. doi: 10.1523/JNEUROSCI.2937-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naghavi HRR, Eriksson J, Larsson A, Nyberg L. The claustrum/insula region integrates conceptually related sounds and pictures. Neurosci Lett. 2007;422:77–80. doi: 10.1016/j.neulet.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 11.Vohn R, Fimm B, Weber J, Schnitker R, Thron A, Spijkers W, Willmes K, Sturm W. Management of attentional resources in within-modal and cross-modal divided attention tasks: an fMRI study. Hum Brain Mapp. 2007;28:1267–1275. doi: 10.1002/hbm.20350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castellanos FX, Margulies DS, Kelly C, Uddin LQ, Ghaffari M, Kirsch A, Shaw D, Shehzad Z, Di Martino A, Biswal B, et al. Cingulate-precuneus interactions: a new locus of dysfunction in adult attention-deficit/hyperactivity disorder. Biol Psychiatry. 2008;63:332–337. doi: 10.1016/j.biopsych.2007.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dickstein SG, Bannon K, Castellanos FX, Milham MP. The neural correlates of attention deficit hyperactivity disorder: an ALE meta-analysis. J Child Psychol Psychiatry. 2006;47:1051–1062. doi: 10.1111/j.1469-7610.2006.01671.x. [DOI] [PubMed] [Google Scholar]
- 14.Koubeissi MZ, Bartolomei F, Beltagy A, Picard F. Electrical stimulation of a small brain area reversibly disrupts consciousness. Epilepsy Behav. 2014;37:32–35. doi: 10.1016/j.yebeh.2014.05.027. [DOI] [PubMed] [Google Scholar]
- 15.Sperner J, Sander B, Lau S, Krude H, Scheffner D. Severe transitory encephalopathy with reversible lesions of the claustrum. Pediatr Radiol. 1996;26:769–771. doi: 10.1007/BF01396197. [DOI] [PubMed] [Google Scholar]
- 16.Olson CR, Graybiel AM. Sensory maps in the claustrum of the cat. Nature. 1980;288:479–481. doi: 10.1038/288479a0. [DOI] [PubMed] [Google Scholar]
- 17.Sherk H, LeVay S. Visual claustrum: topography and receptive field properties in the cat. Science (80-.) 1981;212:87–89. doi: 10.1126/science.7209525. [DOI] [PubMed] [Google Scholar]
- 18.Spector I, Hassmannova J, Albe-Fessard D. Sensory properties of single neurons of cat’s claustrum. Brain Res. 1974;66:39–65. [Google Scholar]
- 19.Cortimiglia R, Crescimanno G, Salerno MT, Amato G. The role of the claustrum in the bilateral control of frontal oculomotor neurons in the cat. Exp Brain Res. 1991;84:471–477. doi: 10.1007/BF00230958. [DOI] [PubMed] [Google Scholar]
- 20.Ptito M, Lassonde MC. Effects of claustral stimulation on the properties of visual cortex neurons in the cat. Exp Neurol. 1981;73:315–320. doi: 10.1016/0014-4886(81)90066-2. [DOI] [PubMed] [Google Scholar]
- 21.Tsumoto T, Suda K. Effects of stimulation of the dorsocaudal claustrum on activities of striate cortex neurons in the cat. Brain Res. 1982;240:345–349. doi: 10.1016/0006-8993(82)90233-5. [DOI] [PubMed] [Google Scholar]
- 22.Salerno MT, Cortimiglia R, Crescimanno G, Amato G, Infantellina F. Effects of claustrum stimulation on spontaneous bioelectrical activity of motor cortex neurons in the cat. Exp Neurol. 1984;86:227–239. doi: 10.1016/0014-4886(84)90183-3. [DOI] [PubMed] [Google Scholar]
- 23.Salerno MT, Cortimiglia R, Crescimanno G, Amato G. Effect of claustrum activation on the spontaneous unitary activity of frontal eye field neurons in the cat. Neurosci Lett. 1989;98:299–304. doi: 10.1016/0304-3940(89)90418-7. [DOI] [PubMed] [Google Scholar]
- 24.Voiculescu O, Charnay P, Schneider-Maunoury S. Expression pattern of a Krox-20/Cre knock-in allele in the developing hindbrain, bones, and peripheral nervous system. Genesis. 2000;26:123–126. doi: 10.1002/(sici)1526-968x(200002)26:2<123::aid-gene7>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 25.Wang Q, Ng L, Harris JA, Feng D, Li Y, Royall JJ, Oh SW, Bernard A, Sunkin SM, Koch C, et al. Organization of the connections between claustrum and cortex in the mouse. J Comp Neurol. 2017;525:1317–1346. doi: 10.1002/cne.24047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zingg B, Hintiryan H, Gou L, Song MY, Bay M, Bienkowski MS, Foster NN, Yamashita S, Bowman I, Toga AW, et al. Neural networks of the mouse neocortex. Cell. 2014;156:1096–1111. doi: 10.1016/j.cell.2014.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Atlan G, Terem A, Peretz-Rivlin N, Groysman M, Citri A. Mapping synaptic cortico-claustral connectivity in the mouse. J Comp Neurol. 2016;520:633–655. doi: 10.1002/cne.23997. [DOI] [PubMed] [Google Scholar]
- 28.White MG, Cody PA, Bubser M, Deutch AY, Deutch AY, Mathur BN. Cortical hierarchy governs rat claustrocortical circuit organization. J Comp Neurol. 2017;525:1347–1362. doi: 10.1002/cne.23970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hur EE, Zaborszky L. Vglut2 afferents to the medial prefrontal and primary somatosensory cortices: a combined retrograde tracing in situ hybridization study [corrected] J Comp Neurol. 2005;483:351–373. doi: 10.1002/cne.20444. [DOI] [PubMed] [Google Scholar]
- 30.Rahman FE, Baizer JS. Neurochemically defined cell types in the claustrum of the cat. Brain Res. 2007;1159:94–111. doi: 10.1016/j.brainres.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 31.Riche D, Lanoir J. Some claustro-cortical connections in the cat and baboon as studied by retrograde horseradish peroxidase transport. J Comp Neurol. 1978;177:435–444. doi: 10.1002/cne.901770306. [DOI] [PubMed] [Google Scholar]
- 32.Markowitsch HJ, Irle E, Bang-Olsen R, Flindt-Egebak P. Claustral efferents to the cat’s limbic cortex studied with retrograde and anterograde tracing techniques. Neuroscience. 1984;12:409–425. doi: 10.1016/0306-4522(84)90062-9. [DOI] [PubMed] [Google Scholar]
- 33.Tritsch NX, Granger AJ, Sabatini BL. Mechanisms and functions of GABA co-release. Nat Rev Neurosci. 2016;17:139–145. doi: 10.1038/nrn.2015.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Root DH, Mejias-Aponte CA, Zhang S, Wang H-L, Hoffman AF, Lupica CR, Morales M. Single rodent mesohabenular axons release glutamate and GABA. Nat Neurosci. 2014;17:1543–1551. doi: 10.1038/nn.3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shabel SJ, Proulx CD, Piriz J, Malinow R. Mood regulation. GABA/glutamate co-release controls habenula output and is modified by antidepressant treatment. Science. 2014;345:1494–8. doi: 10.1126/science.1250469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ye L, Allen WE, Kimberly R, Luo L, Jennifer A, Deisseroth K. Wiring and Molecular Features of Prefrontal Article Wiring and Molecular Features of Prefrontal Ensembles Representing Distinct Experiences. 2016:1–13. doi: 10.1016/j.cell.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kitanishi T, Matsuo N. Organization of the claustrum-to-entorhinal cortical connection in mice. J Neurosci. 2016;37:269–280. doi: 10.1523/JNEUROSCI.1360-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carey RG, Neal TL. Reciprocal connections between the claustrum and visual thalamus in the tree shrew (Tupaia glis) Brain Res. 1986;386:155–168. doi: 10.1016/0006-8993(86)90152-6. [DOI] [PubMed] [Google Scholar]
- 39.LeVay S. Synaptic organization of claustral and geniculate afferents to the visual cortex of the cat. J Neurosci. 1986;6:3564–3575. doi: 10.1523/JNEUROSCI.06-12-03564.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith JB, Alloway KD. Functional specificity of claustrum connections in the rat: interhemispheric communication between specific parts of motor cortex. J Neurosci. 2010;30:16832–16844. doi: 10.1523/JNEUROSCI.4438-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rothwell PE, Fuccillo MV, Maxeiner S, Hayton SJ, Gokce O, Lim BK, Fowler SC, Malenka RC, Südhof TC. Autism-associated neuroligin-3 mutations commonly impair striatal circuits to boost repetitive behaviors. Cell. 2014;158:198–212. doi: 10.1016/j.cell.2014.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Hoz L, Nelken I. Frequency tuning in the behaving mouse: Different bandwidths for discrimination and generalization. PLoS One. 2014;9 doi: 10.1371/journal.pone.0091676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roth BL. DREADDs for Neuroscientists. Neuron. 2016;89:683–694. doi: 10.1016/j.neuron.2016.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Forster S, Lavie N. Failures to ignore entirely irrelevant distractors: the role of load. J Exp Psychol Appl. 2008;14:73–83. doi: 10.1037/1076-898X.14.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cohen L, Rothschild G, Mizrahi A. Multisensory integration of natural odors and sounds in the auditory cortex. Neuron. 2011;72:357–369. doi: 10.1016/j.neuron.2011.08.019. [DOI] [PubMed] [Google Scholar]
- 46.Grienberger C, Adelsberger H, Stroh A, Milos R-I, Garaschuk O, Schierloh A, Nelken I, Konnerth A. Sound-evoked network calcium transients in mouse auditory cortex in vivo. J Physiol. 2012;590:899–918. doi: 10.1113/jphysiol.2011.222513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schneider-Maunoury S, Topilko P, Seitanidou T, Levi G, Cohen-Tannoudji M, Poumin S, Babinet C, Chamay P. Disruption of Krox-20 Results in Alteration of Rhombomeres 3 and 5 in the Developing Hindbrain. Cell. 1993;75:1199–1214. doi: 10.1016/0092-8674(93)90329-o. [DOI] [PubMed] [Google Scholar]
- 48.Miao T, Symonds ALJ, Singh R, Symonds JD, Ogbe A, Omodho B, Zhu B, Li S, Wang P. Egr2 and 3 control adaptive immune responses by temporally uncoupling expansion from T cell differentiation. J Exp Med. 2017;214:1787–1808. doi: 10.1084/jem.20160553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mukherjee D, Ignatowska-Jankowska BM, Itskovits E, Gonzales BJ, Turm H, Izakson L, Haritan D, Bleistein N, Cohen C, Amit I, et al. Salient experiences are represented by unique transcriptional signatures in the mouse brain. Elife. 2018;7:e31220. doi: 10.7554/eLife.31220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baizer JS, Sherwood CC, Noonan M, Hof PR. Comparative organization of the claustrum: what does structure tell us about function? Front Syst Neurosci. 2014;8:117. doi: 10.3389/fnsys.2014.00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Milardi D, Bramanti P, Milazzo C, Finocchio G, Arrigo A, Santoro G, Trimarchi F, Quartarone A, Anastasi G, Gaeta M. Cortical and subcortical connections of the human claustrum revealed in vivo by constrained spherical deconvolution tractography. Cereb Cortex. 2015;25:406–414. doi: 10.1093/cercor/bht231. [DOI] [PubMed] [Google Scholar]
- 52.Baizer JS. The Claustrum: Structural, Functional, and Clinical Neuroscience. Elsevier; 2014. The Neurochemical Organization of the Claustrum; pp. 85–118. [Google Scholar]
- 53.Noudoost B, Moore T. The role of neuromodulators in selective attention. Trends Cogn Sci. 2011;15:585–591. doi: 10.1016/j.tics.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wimmer RD, Schmitt LI, Davidson TJ, Nakajima M, Deisseroth K, Halassa MM. Thalamic control of sensory selection in divided attention. Nature. 2015;526:705–9. doi: 10.1038/nature15398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Terreros G, Jorratt P, Aedo C, Elgoyhen AB, Delano PH. Selective Attention to Visual Stimuli Using Auditory Distractors Is Altered in Alpha-9 Nicotinic Receptor Subunit Knock-Out Mice. J Neurosci. 2016;36:7198–7209. doi: 10.1523/JNEUROSCI.4031-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsuchiya N, Koch C. The Neurology of Conciousness. Second Edition. San Diego: Academic Press; 2016. Chapter 5 - The Relationship Between Consciousness and Top-Down Attention BT - The Neurology of Conciousness; pp. 71–91. [Google Scholar]
- 57.da Costa NM, Fursinger D, Martin KAC, Fürsinger D, Martin KAC. The synaptic organization of the claustral projection to the cat’s visual cortex. J Neurosci. 2010;30:13166–13170. doi: 10.1523/JNEUROSCI.3122-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jackson JC, Karnani MM, Burdakov D, AKL Investigating the mechanisms of claustrum-cortical connectivity. Program No. 291.12. 2017 Neuroscience Meeting Planner; Washington, DC: Society for Neuroscience; 2017. Online. [Google Scholar]
- 59.Narikiyo K, Mizuguchi R, Ajima A, Mitsui S, Shiozaki M, Hamanaka H, Johansen JP, Mori K, Yoshihara Y. The Claustrum Coordinates Cortical Slow-Wave Activity. 2018 doi: 10.1038/s41593-020-0625-7. [DOI] [PubMed] [Google Scholar]
- 60.Maro GS, Vermeren M, Voiculescu O, Melton L, Cohen J, Charnay P, Topilko P. Neural crest boundary cap cells constitute a source of neuronal and glial cells of the PNS. Nat Neurosci. 2004;7:930–938. doi: 10.1038/nn1299. [DOI] [PubMed] [Google Scholar]
- 61.Schaerenwiemers N, Gerfinmoser a. A Single Protocol to Detect Transcripts of Various Types and Expression Levels in Neural Tissue and Cultured-Cells - in-Situ Hybridization Using Digoxigenin-Labeled Crna Probes. Histochemistry. 1993;100:431–440. doi: 10.1007/BF00267823. [DOI] [PubMed] [Google Scholar]
- 62.Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kamentsky L, Jones TR, Fraser A, Bray M-A, Logan DJ, Madden KL, Ljosa V, Rueden C, Eliceiri KW, Carpenter AE. Improved structure, function and compatibility for CellProfiler: modular high-throughput image analysis software. Bioinformatics. 2011;27:1179–1180. doi: 10.1093/bioinformatics/btr095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Paxinos G, Franklin KBJ. Paxinos and Franklin’s the mouse brain in stereotaxic coordinates. Elsevier; 2013. Elsevier. [Google Scholar]
- 65.Wall NR, Wickersham IR, Cetin A, De La Parra M, Callaway EM. Monosynaptic circuit tracing in vivo through Cre-dependent targeting and complementation of modified rabies virus. Proc Natl Acad Sci U S A. 2010;107:21848–21853. doi: 10.1073/pnas.1011756107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Miyamichi K, Shlomai-Fuchs Y, Shu M, Weissbourd BC, Luo L, Mizrahi A. Dissecting local circuits: Parvalbumin interneurons underlie broad feedback control of olfactory bulb output. Neuron. 2013;80:1232–1245. doi: 10.1016/j.neuron.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wall N, DeLaParra M, Callaway E, Kreitzer A. Differential innervation of direct- and indirect-pathway striatal projection neurons. Neuron. 2013;79:347–360. doi: 10.1016/j.neuron.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Beier KTT, Steinberg EEE, DeLoach KEE, Xie S, Miyamichi K, Schwarz L, Gao XJJ, Kremer EJJ, Malenka RCC, Luo L. Circuit Architecture of VTA Dopamine Neurons Revealed by Systematic Input-Output Mapping. Cell. 2015;162:622–634. doi: 10.1016/j.cell.2015.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schwarz LA, Miyamichi K, Gao XJ, Beier KT, Weissbourd B, DeLoach KE, Ren J, Ibanes S, Malenka RC, Kremer EJ, et al. Viral-genetic tracing of the input–output organization of a central noradrenaline circuit. Nature. 2015;524:88–92. doi: 10.1038/nature14600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Callaway EM, Luo L. Monosynaptic Circuit Tracing with Glycoprotein-Deleted Rabies Viruses. J Neurosci. 2015;35:8979–8985. doi: 10.1523/JNEUROSCI.0409-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Reardon TR, Murray AJ, Turi GF, Schnell MJ, Jessell TM, Losonczy A, Wirblich C, Croce KR. Rabies Virus CVS-N2c Strain Enhances Retrograde Synaptic Transfer and Neuronal Viability Highlights d A new rabies virus strain for monosynaptic circuit mapping and manipulation d Enhanced retrograde transsynaptic transfer d Reduction in neurotoxicity permits optogenetic access to neural circuits Neuron NeuroResource Rabies Virus CVS-N2c Strain Enhances Retrograde Synaptic Transfer and Neuronal Viability. 2016 doi: 10.1016/j.neuron.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chatterjee S, Sullivan HA, MacLennan BJ, Xu R, Hou Y, Lavin TK, Lea NE, Michalski JE, Babcock KR, Dietrich S, et al. Nontoxic, double-deletion-mutant rabies viral vectors for retrograde targeting of projection neurons. Nat Neurosci. doi: 10.1038/s41593-018-0091-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pollak Dorocic I, Fürth D, Xuan Y, Johansson Y, Pozzi L, Silberberg G, Carlén M, Meletis K, Furth D, Xuan Y, et al. A whole-brain atlas of inputs to serotonergic neurons of the dorsal and median raphe nuclei. Neuron. 2014;83:663–678. doi: 10.1016/j.neuron.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 74.Ye L, Allen WE, Thompson KR, Tian Q, Hsueh B, Ramakrishnan C, Wang AC, Jennings JH, Adhikari A, Halpern CH, et al. Wiring and Molecular Features of Prefrontal Ensembles Representing Distinct Experiences. Cell. 2016;165:1776–1788. doi: 10.1016/j.cell.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Endo T, Maekawa F, Voikar V, Haijima A, Uemura Y, Zhang Y, Miyazaki W, Suyama S, Shimazaki K, Wolfer DP, et al. Automated test of behavioral flexibility in mice using a behavioral sequencing task in IntelliCage. Behav Brain Res. 2011;221:172–181. doi: 10.1016/j.bbr.2011.02.037. [DOI] [PubMed] [Google Scholar]
- 76.Stiebler I, Neulist R, Fichtel I, Ehret G. The auditory cortex of the house mouse: left-right differences, tonotopic organization and quantitative analysis of frequency representation. J Comp Physiol A. 1997;181:559–71. doi: 10.1007/s003590050140. [DOI] [PubMed] [Google Scholar]
- 77.Maor I, Shalev A, Mizrahi A. Distinct Spatiotemporal Response Properties of Excitatory Versus Inhibitory Neurons in the Mouse Auditory Cortex. Cereb Cortex. 2016;26:4242–4252. doi: 10.1093/cercor/bhw266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim CK, Yang SJ, Pichamoorthy N, Young NP, Kauvar I, Jennings JH, Lerner TN, Berndt A, Lee SY, Ramakrishnan C, et al. Simultaneous fast measurement of circuit dynamics at multiple sites across the mammalian brain. Nat Methods. 2016;13:325–328. doi: 10.1038/nmeth.3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data that support the findings of this study are available from the corresponding author upon reasonable request. Computer codes used for analysis in this study were implemented in MATLAB (MathWorks), R and RStudio software. Codes will be made freely available upon request. Requests should be directed to the corresponding author.