Abstract
Next-generation sequencing of the immunoglobulin gene repertoire (Ig-seq) produces large volumes of information at the nucleotide sequence level. Such data have improved our understanding of immune systems across numerous species and have already been successfully applied in vaccine development and drug discovery. However, the high-throughput nature of Ig-seq means that it is afflicted by high error rates. This has led to the development of error correction approaches. Computational error correction methods use sequence information alone, primarily designating sequences as likely to be correct if they are observed frequently. In this work, we describe an orthogonal method for filtering Ig-seq data, which considers the structural viability of each sequence. A typical natural antibody structure requires the presence of a disulfide bridge within each of its variable chains to maintain the fold. ABOSS, our AntiBOdy Sequence Selector uses the presence/absence of this bridge as a way of both identifying structurally viable sequences and estimating the sequencing error rate. On simulated Ig-seq datasets, ABOSS is able to identify more than 99% of structurally viable sequences. Applying our method to six independent Ig-seq datasets (1 mouse and 5 human), we show that our error calculations are in line with previous experimental and computational error estimates. We also show how ABOSS is able to identify structurally impossible sequences missed by other error correction methods.
1. Introduction
Effective recognition and elimination of noxious molecules from jawed vertebrates relies on the versatility of their immune systems. Antibodies, secreted products of B cells, play a key role in recognizing antigens – structural motifs on pathogenic molecules. Antibodies can be raised against potentially any antigen (1). As a result of this binding plasticity, antibodies are currently the most successful class of biotherapeutics (2, 3).
Next-generation sequencing of the immunoglobulin gene repertoire (Ig-seq) produces large volumes of information at the nucleotide sequence level, allowing interrogation of snapshots of antibody diversity. Such data have improved our understanding of immune systems across numerous species and have already been successfully applied in vaccine development and drug discovery e.g. (4, 5). However, the high-throughput nature of Ig-seq means that it is afflicted by high error rates, which makes it difficult to distinguish between Ig-seq artifacts and true nucleotide alterations introduced by the somatic hypermutation (SHM) machinery of B cells.
Several experimental Ig-seq error correction approaches have been proposed, however an agreed standard does not yet exist (6). Existing experimental approaches for error correction include taking invariant sequence portions as a proxy for estimating error or barcoding sequences that should be identical. For instance, Galson et al., (7) performed sequencing of the constant portions of the antibody heavy chain. As this region is typically sequence invariant, it offered an estimated error rate on the variable portions sequenced in the course of the same study. Khan et al., (8) barcoded individual antibody cDNA transcripts with unique molecular identifiers (UMI) prior to PCR. The resultant pool of genetic data was sequenced and identically barcoded sequences were put into separate clusters where a consensus sequence was devised. All other members of the cluster were corrected with respect to this consensus sequence. Error can be introduced even in this method in the early steps of sequencing sample preparation such as reverse transcription and PCR (9, 10). Devising a correct sequence within the clusters is heavily dependent on sequence redundancies, which precludes correction of singleton clusters using the barcode approach (9, 10).
Techniques such as barcoding or sequencing constant portions are time consuming and require specialized experimental setups. To address such issues, several computational error correction tools have been developed (6). These applications all operate by building consensus sequences using homology clustering. The majority of these tools work only in the remit of complementarity determining region 3 of the VH domain (CDR-H3) (11, 12), largely ignoring the rest of the sequence. MIXCR is the most commonly used Ig-seq error correction tool to date (13). It supports the analysis of entire VH or VL chains and performs sequencing error correction. MIXCR works by aligning sequences from an Ig-seq dataset to reference V, J and C genes followed by identifying “gene feature sequences”. This is a k-mer of residues identical across multiple sequences and is found in CDR-H3 by default. These “gene feature sequences” are then used to sort antibody sequences into sets of separate clonotypes. The number of unique clonotypes is always over-estimated due to PCR and sequencing errors. To overcome this, “correct” sequences are found by performing heuristic multilayer clustering on these clonotypes, where the most redundant clonotypes are treated as correct. A more recently developed antibody repertoire construction tool, IgReC (14), takes a different approach. It uses Hamming graphs to identify correct sequences. Benchmark analysis on barcoded Ig-seq data shows that the IgReC pipeline is as accurate as experimental error correction approaches (14). This suggests that advances in algorithm development can potentially alleviate the need for experimental Ig-seq correction. All currently available computational methods consider sequence information alone. In this paper, we consider how knowledge of an antibody structure may help to identify sequencing errors by finding sequences that are not structurally viable as structural viability is crucial for the correct functioning of an antibody. We then use this structural information to estimate sequencing error rates.
A typical antibody structure requires the presence of a disulfide bridge within each of the variable chains. This bridge helps to maintain the immunoglobulin fold. Cysteines at positions 23 and 104 (IMGT numbering (15)) must be present in structurally viable natural antibody sequences (16–19). There is evidence that some antibodies can still fold when the disulfide bond is ablated (20–22). However, such antibodies have been found via rational protein engineering where the conserved cysteines are mutated alongside further modifications to the rest of the antibody sequence that stabilize the overall structure (20–22). ABPC48 is the only example of an antibody that naturally lacks cysteine at position 104 (18). APBC48 is a mouse antibody derived from plasmacytoma (23). Although, ABPC48 antibody is able to fold, restoration of cysteine at position 104 significantly improves its stability (22).
Here we describe a novel computational tool, ABOSS (AntiBOdy Sequence Selector) that uses the presence/absence of the conserved cysteines in an antibody sequence to create a structure based estimate of the sequencing error rate in Ig-seq data. As opposed to other error correction tools which operate at the nucleotide sequence level, ABOSS uses amino acid sequences as they relate directly to protein structure. ABOSS both filters out amino acid sequences that are not structurally viable as well as those likely to contain erroneous residue/positions. Due to its use of structural information rather than homology clustering, ABOSS is orthogonal to all other computational methods for error estimation.
Examining ABOSS performance on simulated Ig-seq datasets indicated that ABOSS successfully isolates about 99% of structurally viable sequences, whilst preserving most of the SHM generated diversity. We tested ABOSS on six separate Ig-seq datasets and found that our error calculations based on structural viability were in line with error estimates declared in other recently published studies.
2. Materials and Methods
2.1. ANARCI Parsing
ABOSS supports several input formats. These can be amino acid sequences in the FASTA file or raw IgBlastn outputs (24).
The first step of ABOSS is to parse every sequence through ANARCI (25), an antibody numbering program. ANARCI parsing acts as a pre-filtering step removing sequences that 1) contain unusual insertions/deletions in the framework and canonical CDR regions 2) do not align to the respective species Hidden Markov Models (HMM) of the IMGT germline 3) have a J gene sequence identity of less than 50% to the IMGT germline (of the respective species) or truncated framework 4 region. Calculation of the J gene sequence identity allows us to remove sequences where indels have occurred in CDR-3 and framework 4. At this point, ABOSS also removes sequences in human and mouse datasets that have CDR-H3s longer than 37 amino acids. This cutoff is in place to remove sequences with erroneously long CDR-H3s (26, 27). These are chimeric sequences which arise as a result of PCR error. Sequences, which pass these initial tests, are numbered using the IMGT scheme (15). This provides a consistent frame of reference for sequences, and defines CDR and framework regions. We employ the IMGT numbering scheme in ABOSS since it assigns length mismatched CDRs located in roughly structurally equivalent space to identical residue numbers (15, 28).
2.2. Residue Error Rate Estimation
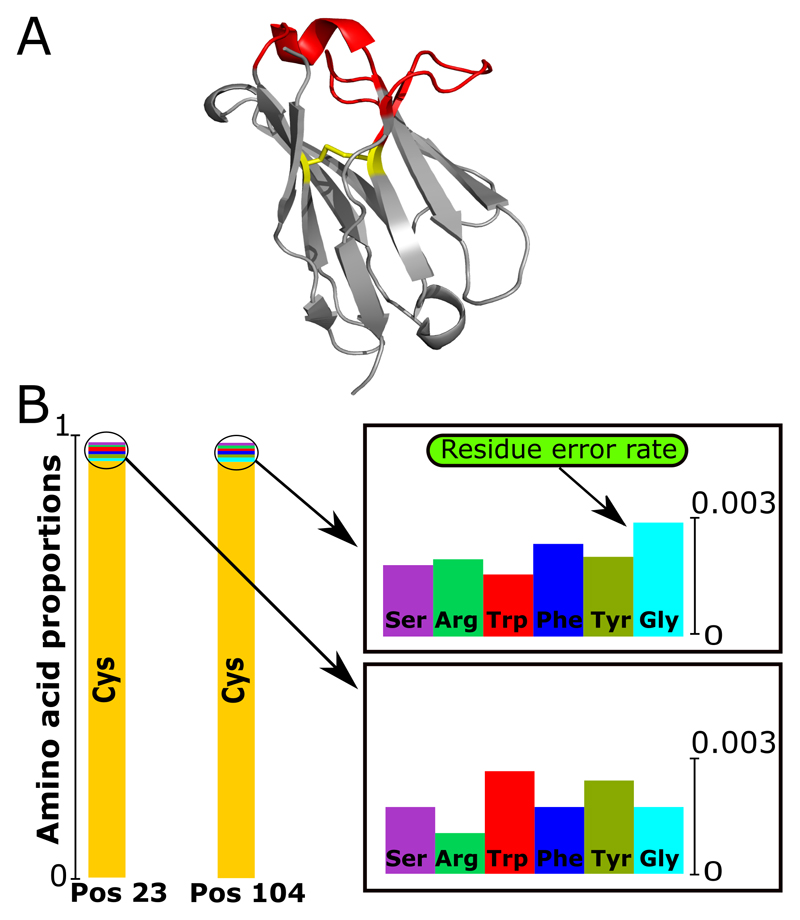
IMGT numbering enables the calculation of amino acid distributions by position. IMGT positions 23 and 104 are used to estimate the error rate in the data. In all naturally occurring antibodies both these positions are always a cysteine residue (16, 18). Some antibody pseudo V genes encode for a non-cysteine residue at position 23 and 104, but these antibodies are not structurally viable (17). Therefore, under a conservative model, any amino acid divergence from cysteine at these positions can be treated as error. We define the error to be equal to the largest non-cysteine amino acid proportion found at either of the two positions (Figure 1). The dataset used in Figure 1 would be ascribed an error rate of 0.0027 (the occurrence of glycine at position 104). Thus, all amino acids at a position that occur with proportion of less than 0.0027 across the dataset are considered erroneous. This will remove all the non-cysteine residue types at position 23 and 104 in the data as they all occur less than 0.0027 but will also indicate several other positions where residues may be erroneous. In this fashion, we provide an error estimate for individual residues, which can be extrapolated to the entire sequence.
Figure 1. Calculation of the residue error rate in terms of structural viability.
(A) Three-dimensional structure of the VH chain (PDB code: 5WUV) with the conserved disulfide bridge shown. Framework (grey) and CDRs regions (red), the cysteine bond between positions 23 and 104 in yellow. (B) The distribution of amino acid types found at positions 23 and 104 for an Ig-seq dataset. Since both positions in natural antibodies should be cysteines, the non-cysteine occurrence indicates possible sequencing error.
2.3. Structure based filtering of Ig-seq data
The next stage is ABOSS filtering. In this step if the proportion of an amino acid at a position is below the residue error rate, amino acids of that type at that position are flagged as potentially erroneous.
ABOSS creates a reference matrix, which contains the “allowed” amino acids at each IMGT position. The allowed amino acids are those whose proportion in the Ig-seq dataset are greater than the residues error rate at the respective position (see previous section). The reference matrix also contains the amino acids from IMGT germline sequences as they represent structurally viable antibodies. If less than 20 entries are used to calculate amino acid proportions at a position, this position is not included in the reference matrix.
Once the reference matrix is calculated, every sequence from the Ig-seq dataset is compared to it. For a given position in a sequence from the Ig-seq dataset a flag is placed if the amino acid in the sequence is not present in the reference matrix. ABOSS outputs a csv file of the ANARCI parsed sequences, their redundancies, CDR-H3 regions, flagged residue/positions, V and J genes, and query names of the original raw nucleotide sequences from the IgBlastn output. The ABOSS filtered dataset refers to the set of sequences with zero flagged residue/positions.
2.4. Data management
We have tested ABOSS on six Ig-seq datasets (Table I). Two datasets from Khan et al., (8), the raw sequences (Khan_R) and the error corrected sequences (Khan_C), each of these datasets comprised three immunized datasets of a single mouse that were pooled together (2.4m sequences). The Galson et al., (7) dataset (HEPB) consists of sequences from hepatitis B studies (29, 30) at a time point before the 11 participants were vaccinated (9.9m sequences). The third and fourth datasets are proprietary UCB Pharma Ltd datasets of 5.6m VH (UCB_H) and 9.3m VL (UCB_L) chain sequences (31). The UCB data were generated from the non-antigen challenged B cells of 494 pooled participants. The Vander Heiden et al., (32) datasets (Healthy_H, Healthy_L) include sequences from four healthy B cell donors. A mixture of VH and VL gene primers were used in sequencing material preparation, which produced pooled VH/VL Ig-seq datasets. Healthy_H and Healthy_L are the sorted heavy and light chain sequences respectively. This plethora of diversity of Ig-seq datasets was employed to test ABOSS across heterogeneous sequencing setups.
Table I. Summary of the datasets used.
The seven datasets (Khan_R, Khan_C, HEPB, UCB_H, UCB_L, Healthy_H and Healthy_L) were obtained from different sequencing methodologies, organisms and immunization protocols. The Khan_R and Khan_C datasets are the immunized mouse 1 dataset of the Khan et al., (8) study before and after the barcode correction approach. These datasets are from repeated Ig-seq of the same mouse. The majority of sequences in this Ig-seq dataset start at position 8. The Khan_R and Khan_C datasets consist of antibody amino acid and corresponding nucleotide sequences. The Khan_R dataset has the highest redundancy amongst the interrogated non-corrected datasets. We have removed the roughly 10% synthetic spike-ins in the Khan_R and Khan_C datasets. The HEPB dataset from Galson et al., (7) is from 11 participants. Standard Illumina Ig-seq was performed. The reads were gene-aligned and processed using IMGT/HighV-Quest. Due to selection of PCR primers, most of the sequences start at position 17. This dataset contains amino acid sequences only. The dataset’s redundancy is almost two times lower than the Khan_R data. The UCB proprietary Ig-seq datasets were obtained from 494 participants. The UCB_H and UCB_L datasets comprise 5.6m and 9.3m sequences respectively. The UCB_H and UCB_L datasets contain both antibody amino acid and corresponding nucleotide sequences. The UCB datasets were aligned with IgBlast (24), V and J genes identified, and pre-filtered for stop codons, they contain full-length variable chain sequences as described in Krawczyk et al., (31). The UCB_H and UCB_L datasets are the least redundant amongst the datasets. The Healthy_H and Healthy_L datasets come from four healthy human B cell donors from the Vander Heiden et al., (32) study. In this study, sequencing primers for both heavy and light chain genes were used at the same time forming pooled raw nucleotide samples. The raw nucleotide Ig-seq datasets were obtained from the OAS resource (36) followed by translating sequences into amino acids and antibody chain separation using IgBlastn (24).
| Dataset name | Study description | Total dataset size | Antibody chain | Dataset average redundancy | Participants |
|---|---|---|---|---|---|
| Khan_R | Raw sequences of Immunized mouse 1 from Khan et al., (8) | 2.4m | Heavy | 3.74 | 1 (mouse) |
| Khan_C | Barcode corrected sequences of immunized mouse 1 from Khan et al., (8) | 2.4m | Heavy | 45.3 | 1 (mouse) |
| HEPB | Human hepatitis B vaccination from Galson et al., (7) | 9.9m | Heavy | 1.93 | 11 |
| UCB_H | Proprietary UCB Ig-seq of the VH chain | 5.6m | Heavy | 1.15 | 494 |
| UCB_L | Proprietary UCB Ig-seq of the VL chain | 9.3m | Light | 1.12 | 494 |
| Healthy_H | VH chains from healthy human B cell donors from Vander Heiden et al., (32) | 1.4m | Heavy | 1.9 | 4 |
| Healthy_L | VL chains from healthy human B cell donors from Vander Heiden et al., (32) | 6.3m | Light | 2.96 | 4 |
2.5. In silico simulation
We performed in silico error simulation on two Ig-seq datasets, UCB_H and Khan_R. The simulations were performed at the nucleotide level. The nucleotide sequences that corresponded to the amino acid sequences that passed ABOSS with zero flagged residue/positions (see ABOSS filtered dataset in Table II) acted as starting points for our simulation. During the simulation each sequence in the starting dataset was subjected to randomized nucleotide mutations. The distribution of the number of nucleotide mutations was proportional to the distribution of flagged residue/positions in the respective redundant Ig-seq data determined by ABOSS analysis (see Section 3.2.), whilst mutation positions were stochastically selected along the VH chain. Only sequences where random mutations were introduced were added to the final simulation dataset. As UCB_H and Khan_R datasets were generated using Illumina sequencing technology, only nucleotide substitutions were considered in the error simulations.
Table II. ABOSS analysis of six Ig-seq datasets.
In the table, dataset sizes are given as the number of redundant sequences, the number of non-redundant sequences are shown in parentheses. Starting datasets are the inputs for ABOSS. ANARCI parsed datasets contain sequences that are successfully IMGT-numbered. ABOSS filtered datasets are the number of sequences that contain zero flagged residues. The percentage of sequences with zero flags are calculated as a percentage of redundant ABOSS passed sequences over the total number of starting redundant sequences. Residue error rates are calculated as described in Figure 1
| Data source | Starting dataset | ANARCI parsed dataset |
ABOSS filtered dataset | Sequence percentage with zero flags | ABOSS Residue Error Rate (%) |
|---|---|---|---|---|---|
| HEPB | 9,985,575 (5,175,036) |
9,700,893 (4,932,588) |
6,579,118 (3,226,473) |
65.9 % | 0.22 |
| Khan_R | 2,445,354 (653,520) |
2,247,761 (521,675) |
649,685 (47,593) |
26.6% | 1.5741 |
| UCB_L | 9,371,465 (8,380,540) |
8,021,407 (7,120,100) |
3,494,319 (2,983,103) |
37.3% | 0.4674 |
| UCB_H | 5,645,304 (4,925,532) |
5,277,305 (4,587,918) |
1,903,703 (1,561,082) |
33.7% | 0.5892 |
| Healthy_H | 1,422,405 (745,276) |
1,135,185 (558,171) |
486,437 (176,012) |
34.2% | 0.5427 |
| Healthy_L | 6,317,736 (2,135,745) |
4,860,389 (1,372,804) |
2,667,263 (386,165) |
42.2% | 0.4121 |
To assess the robustness of ABOSS, we varied both residue error rate and dataset size in our error simulations. To increase the residue error rate, every entry from the originally calculated distribution of flagged residue/positions was amplified by an error multiplier. Separate simulations were carried out for individual values of the error rate multiplier that ranged between one and eight. The simulation final dataset sizes were equivalent to the size of the respective ANARCI parsed Ig-seq dataset (see ANARCI parsed dataset in Table II). Separate simulations were also performed where the size of the simulation final dataset was varied to be between one and eight times smaller than the respective ANARCI parsed Ig-seq dataset.
2.6. Antibody SHM lineage tree simulation
We carried out two separate SHM simulations on the nucleotide sequences of the Healthy_H and UCB_H datasets. Nucleotide sequences, whose translated amino acids had zero ABOSS flagged residue/positions in Healthy_H, were assigned as the most recent common ancestors (MRCA) in the simulations. Two different clonal lineage trees (Lineage_A and Lineage_B) were employed for the number of progenitor sequences and SHM substitutions. We used the human HH_S5F targeting model (33) from the SHazaM package (http://shazam.readthedocs.io/en/version-0.1.9---baseline-fixes/) to perform SHM substitutions in the lineage trees. All MRCA and progeny sequences were added to the final SHM simulation datasets.
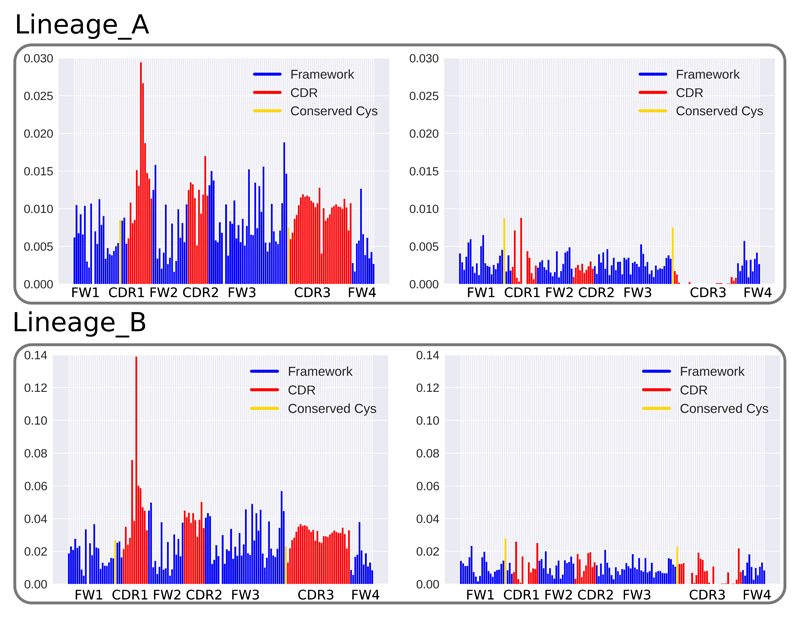
In the Lineage_A simulation experiment, two progenitor sequences originated from a single MRCA. Both of these sequences harbored two nucleotide SHM substitutions. In the Lineage_B SHM experiment, two progenitors were produced by MRCA with two and four nucleotide substitutions respectively. The former progenitor formed a further four offspring sequences with one, one, three and six SHM substitutions. Finally, the offspring with 3 SHM substitutions produced another progeny also with 3 nucleotide substitutions.
3. Results
3.1. The ABOSS Algorithm
ABOSS is a computational method that leverages structural antibody information to calculate the sequencing error rate and flag potentially erroneous residue/positions in Ig-seq sequences. Specifically, we exploit the knowledge of the conserved cysteines at positions 23 and 104, which shape and stabilize the conformation of the antibody variable chains. The presence of these conserved cysteines can be used as a way of both identifying structurally viable sequences and estimating the sequencing error rate.
ABPC48 is the only characterized natural antibody that lacks either cysteine at either position (20). A small number of structurally stable antibodies with pairwise substitutions of the conserved cysteines based on the ABPC48 antibody scaffold have been engineered (21, 22, 34). These pairwise substitutions require further stabilizing mutations to the antibody structure, often to the opposite variable chain (21, 22, 34). The known structurally viable non-cysteine pairs seen at positions 23 and 104 are summarized in (18). In our Ig-seq datasets, we rarely observe the pairwise substitution of cysteines. For instance, the total number of instances when the substitution of both cysteines was observed in the UCB_H data was 811 which corresponded to ~0.015% of UCB_H. Of these 811 pairwise substitutions, the potentially viable substitutions as described in (18) were serine – serine, serine – alanine, alanine – serine and tyrosine– valine which appeared 24, 2, 1 and 1 times respectively. The six amino acids that constitute the largest proportions of non-cysteine residue types at positions 23 and 104 in our six raw Ig-seq datasets are always the amino acids one nucleotide edit distance from the cysteine codons (Figure 1). The top non-cysteine residue type at positions 23 and 104 varies across our Ig-seq datasets, demonstrating the stochastic nature of this amino acid substitution. It was previously demonstrated that SHM substitutions are significantly reduced at positions 23 and 104 in gene-specific amino acid substitution profiles of SHM (35). This must be due to negative structural selection, as SHM substitution still takes place at these positions in passenger alleles and using the HH_S5F computational model (35). This evidence suggests that the substitutions in conserved cysteines seen in Ig-seq datasets are highly likely to be sequencing errors.
In the first step of the ABOSS protocol, all sequences are parsed using ANARCI (25), which IMGT numbers (15) the sequences. Antibody sequences with low sequence identities to ANARCI HMM profiles, unusual insertions/deletions along the antibody chain are discarded. Next, ABOSS calculates the residue error rate using the ANARCI parsed sequences. The residue error rate is taken as the largest non-cysteine amino acid proportion found at position 23 or 104 (Figure 1). The residue error rate is then used to flag specific residue/positions in individual sequences.
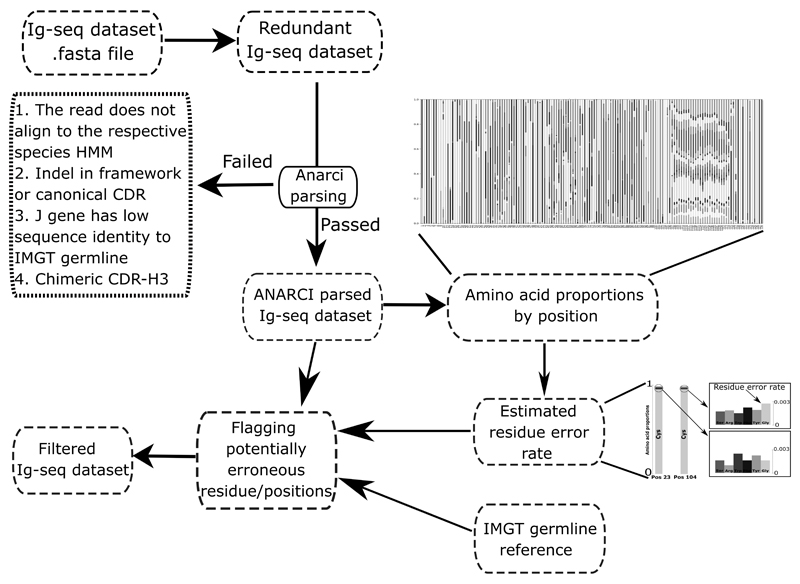
The workflow of the algorithm is summarized in Figure 2. ABOSS analysis takes less than 10h wall-clock time for 5m unique antibody amino acid sequences on a standard 8 core desktop computer (intel i7-6700). ABOSS is parallelized allowing for shorter run-times on more powerful machines. ABOSS is available via http://opig.stats.ox.ac.uk//resources.
Figure 2. Workflow of ABOSS.
ABOSS input is antibody amino acid sequences in the FASTA format. Every sequence from the input file is IMGT-numbered with ANARCI (ANARCI parsing). The amino acid distribution by IMGT position is calculated for successfully ANARCI parsed sequences. The residue error rate is estimated based on the amino acid distributions at positions 23 and 104 (see Figure 1 for more details). The estimated residue error rate together with the ANARCI numbered IMGT germline genes are used to flag potentially erroneous residue/positions in individual Ig-seq sequences. Filtered Ig-seq dataset refers to a collection of sequences that pass ABOSS analysis with zero flagged residues/positions.
3.2. ABOSS analysis on raw Ig-seq data
We ran ABOSS on six Ig-seq datasets (Table I, Table II). We consider two sequences redundant if they have identical length and identical amino acid compositions. ANARCI parsing removed between 3-23% of sequences in the Ig-seq samples (Table II). The ANARCI parsing step removed the largest proportion of sequences from Healthy_L followed by the Healthy_H, UCB_L, Khan_R, UCB_H and HEPB datasets respectively. In the second step, ABOSS filtering, residue/position in the sequences are flagged as potential errors. In the Khan_R was the dataset with the smallest proportion of sequences with zero ABOSS flagged residue/positions (26.6%) (Table II). The HEPB dataset had the highest proportion of zero ABOSS flagged residue/positions (65.9%) followed by Health_L, UCB_L (37.3%), Healthy_H and UCB_H (33.7%).
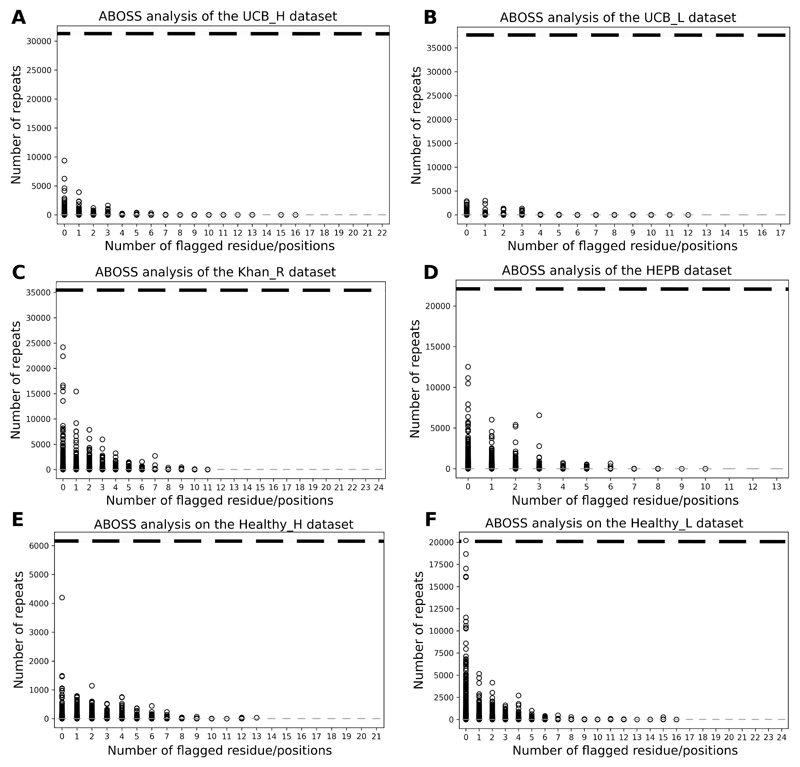
Current Ig-seq error correction pipelines assign greater confidence to highly redundant sequences and manipulate the nucleotide sequences of rare sequences (6, 8, 13, 14). In contrast, ABOSS does not have a direct link between sequence redundancy and “correct” sequences. To examine the performance overlap of ABOSS and redundancy based Ig-seq error correction tools, we compared the number of ABOSS flagged residue/positions to the sequence redundancy for our six datasets (Figure 3). In every dataset, sequences that are more redundant tend to have fewer ABOSS flagged residue/positions. This suggests that even though ABOSS is not a redundancy based technique, its results are still in line with the widely-adopted methodology based on sequence redundancy. ABOSS does flag residues as erroneous in a number of highly redundant clones which might be flagged as correct by redundancy-reliant methods. If a sequence was highly redundant it could in theory avoid any of its residues being flagged by ABOSS as every residue/position in this sequences would be present more times than the residue error rate. The horizontal dashed lines in Figure 3 shows the redundancy necessary to achieve this. Only a single sequence from the Healthy_L dataset reached such a level of redundancy (Figure 3F).
Figure 3. Sequence redundancy relative to the number of ABOSS flagged residue/positions in the sequences of our six datasets (see Table I for more details): UCB_H (A), UCB_L (B), Khan_R (C), HEPB (D), Healthy_H (E) and Healthy_L (F).
The ABOSS filtering step outputs the number of flagged residue/positions for every sequence in the ANARCI parsed Ig-seq dataset. Zero flagged residue/positions indicates that the sequence is structurally viable. The general trend in each Ig-seq dataset is that the more redundant the sequence the fewer ABOSS flagged residue/positions it has. The horizontal dashed line represents the residue error rate in terms of the number of entries required for a residue/position to be identified as structurally viable.
3.3. Ig-seq error simulation to estimate sequence volumes and error rates tolerated by ABOSS
In order to investigate the types of Ig-seq datasets that ABOSS can successfully analyze, we benchmarked ABOSS with respect to dataset redundancies, sequencing error rates and input sequence volumes. We tested ABOSS on two datasets with contrasting depth and breadth of coverage: the UCB_H and Khan_R datasets (see Table I). The starting datasets for the simulation consisted of sequences that passed ABOSS analysis with zero flagged residue/positions. The sizes of the simulation final datasets (UCB_H_Sim and Khan_R_Sim) were based on the number of sequences that passed the ANARCI parsing step (see Table II). We used the distribution of the number of flagged residue/positions in the UCB_H and Khan_R datasets as calculated by ABOSS (see Figure 3) to introduce erroneous residue/positions into our simulation starting datasets. The mutation substitution positions were stochastically selected along the VH chain. From these starting points, the simulations were performed as described in materials and methods (see section 2.5).
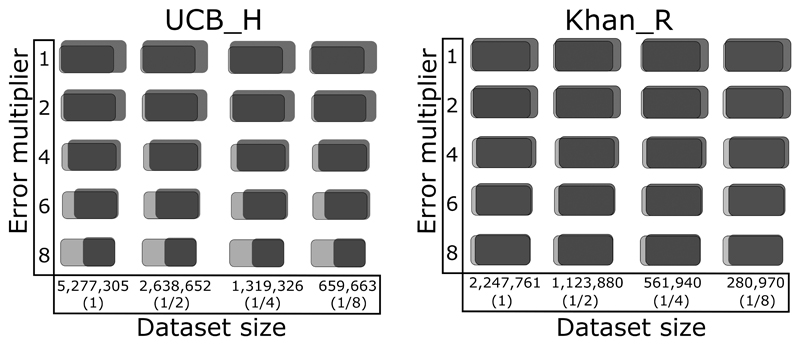
The simulation results are shown in Figure 4 and Supplemental Figure 1. Using sizes and error rates that match the original data ABOSS recovered 99.6% and 99% of the correct sequences incorporated into the UCB_H_Sim and Khan_R_Sim datasets respectively. Reducing the UCB_H_Sim and Khan_R_Sim dataset sizes does not appear to influence the percentage of correct sequences recovered by ABOSS analysis. Increasing the error rates has a minor effect on the recovered number of correct sequences from the Khan_R_Sim dataset and a much larger effect on the recovery of correct sequences from the UCB_H_Sim dataset. This difference is due to the far lower initial redundancy of the UCB_H data.
Figure 4. Examination of ABOSS performance on Ig-seq error simulation datasets.
Ig-seq data simulation was carried out based on the previously calculated numbers of ABOSS flagged residue/positions in two Ig-seq datsets, UCB_H and Khan_R (see Figure 3). The X-axis corresponds to UCB_H_Sim and Khan_R_Sim dataset sizes used for simulation (the percentages relative to the sizes of respective datasets that passed ANARCI are shown in parentheses). The Y-axis shows the multiplier of the original distribution of erroneous residue/positions in the Ig-seq datasets (see Figure 3). The total number of ABOSS filtered sequences (black) and the number of the correct sequences (grey) are pictured (The percentages are given in Supplemental Figure 1). The overlapping region indicates the proportion of the correct sequences that passed ABOSS relative to the total number of ABOSS filtered sequences.
ABOSS also retained small numbers of sequences from UCB_H_Sim and Khan_R_Sim that were not present in the simulation starting datasets (Figure 4). These sequences are still structurally viable. The number of these sequences was larger in the UCB_H_Sim dataset (~30%) than in the Khan_R_Sim dataset (~17%) (Supplemental Figure 1). As the residue error rate was increased the simulation starting datasets constituted larger proportions of the ABOSS filtered UCB_H_Sim and Khan_R_Sim datasets (Figure 4, Supplemental Figure 1).
The outputs from these error simulations suggest that ABOSS performance becomes robust when either the Ig-seq data is redundant or more than ~600k of non-redundant sequences are available.
3.4. ABOSS analysis on SHM generated diversity
The SHM machinery of B cells increases antibody diversity by introducing nucleotide substitutions in the variable (V) region (37). SHM helps to fine tune an antibody to its cognate epitope (38). These substitutions are known to exhibit uneven frequencies along the V region (33, 35). We exploited the 5-mer nucleotide HH_S5F targeting model of SHM (33) to examine the ability of ABOSS to flag errors, whilst preserving SHM generated diversity in Ig-seq datasets.
The model requires a clonal tree reference to estimate rates of substitutions. We used two distinct architectures of antibody clonal lineage trees (Lineage_A and Lineage_B) to construct such substitutions matrixes. We used these two lineages to have coverage of the spectrum of SHM mutations as Lineage_A has a low substitution rate, whereas Lineage_B has a high one. Using the HH_S5F model combined with either of the Lineage_A or Lineage_B SHM references, the simulations were performed on Healthy_H and UCB_H. These two datasets were selected to test ABOSS performance on low (UCB_H) and high (Healthy_H) redundancy data. The sequences with zero ABOSS flagged residue/positions were used as the most recent common ancestor (MRCA) that were then employed as templates to which SHM mutations were introduced. The HH_S5F targeting model (33) introduced roughly the same SHM substitution ratios along the VH region in the two lineage trees (Figure 5, Supplemental Figure 2). There was a biased increase in SHM substitutions in framework 3, CDR regions and positions flanking the CDRs similar to previous results (33, 35). As the HH_S5F model does not consider structural selection pressure on the heavy chain positions, the conserved cysteines were mutated, which resulted in the residue error rates of 0.002567 (Lineage_A) and 0.008 (Lineage_B) in UCB_H, and 0.002513 (Lineage_A) and 0.0076 (Lineage_B) in Healthy_H simulation datasets respectively. The Lineage_A produced residue error rates were within the observed range of human Ig-seq data, whilst the Lineage_B generated residue error rates exceeded this range (Table II). ABOSS exhibited no preferential selection of unmutated germline V gene sequences over sequences that harbour SHM mutations in the Ig-seq data (Supplemental Figure 3).
Figure 5. ABOSS performance on SHM simulated Ig-seq data diversity.
Two antibody clonal lineage trees (Lineage_A and Lineage_B) were employed to provide the background mutational reference to introduce SHM substitutions into the ABOSS filtered UCB_H dataset using the human HH_S5F targeting model (33). The x-axis shows positions along the VH chain, and the y-axis shows the proportions of residue/positions in the simulation datasets. The figures on the left depict the proportion of SHM substitutions introduced at positions in the VH chain. The figures on the right represent the proportions of ABOSS flagged residue/positions in the simulation datasets.
Most of the HH_S5F generated diversity was preserved by ABOSS analysis (Figure 5). ABOSS flagged residue/positions uniformly along the VH chain, with the exception of CDR-H3, where fewer residue/positions were flagged. These proportions of ABOSS flags are unrelated to the pattern of generated SHM substitutions, which has a strong bias towards framework 3 and CDR loops.
These results demonstrate that ABOSS is able to flag structurally non-viable residue/positions, whilst preserving the majority of SHM substitutions. However, some true rare SHM substitutions may still be removed by ABOSS, as their positional presence in the V region is below the residue error rate. Therefore, highly SHM altered Ig-seq datasets may have a higher proportion of true mutations incorrectly flagged as erroneous.
3.5. ABOSS and IgReC, an Ig-seq computational error correction tool
We compared ABOSS to IgReC, a computational Ig-seq error correction tool. IgReC clusters and corrects PCR and sequencing errors in Ig-seq data based on sequence redundancy and homology. IgReC was recently benchmarked alongside other commonly used tools to error correct Ig-seq data (14). Its performance was considered comparable if not better than all other tools tested. IgReC relies on identification of clonotypes and sequence clustering.
We ran IgReC on the UCB_H and Healthy_H datasets as IgReC requires full-length VH or VL sequences and the HEPB and Khan_R datasets have truncated framework 1 regions. IgReC modifies sequences of Ig-seq datasets making it difficult to carry out an overlap comparison with ABOSS. IgReC removed approximately 1.5% of UCB_H and 8% of Healthy_H but modified nearly 50% and 30% of the sequences to ones not seen in the original UCB_H and Healthy_H datasets respectively (Table III).
Table III. Interrogation of IgReC performance on UCB_H and Healthy_H.
IgReC was run on the raw nucleotide UCB_H and Healthy_H datasets as well as the ABOSS filtered data. IgReC constructed datasets derived from the raw data contained roughly 50% and 30% of sequences that were different to ones found in the UCB_H and Healthy_H datasets respectively. When tested on the ABOSS filtered datasets, IgReC was unable to find V and J germline references for 8,843 sequences in ABOSS filtered UCB_H and 10,365 in ABOSS filtered Healthy_H. IgReC also generated ~42% and ~7% of sequences that were not present in the original UCB_H and Healthy_H datasets. The default parameters were used to run IgReC: ./igrec.py –s <reads.fasta> –l <IGH> –o <output_dir>. The number of non-redundant values are shown in parentheses.
| Outputs | % of sequences found in the original dataset | |
|---|---|---|
| ABOSS filtered UCB_H | 1,903,703 (1,561,082) | 100 (100) |
| IgReC-corrected UCB_H | 5,572,963 (4,069,318) | 51.4 (43.3) |
| IgReC on ABOSS filtered UCB_H | 1,894,860 (1,320,438) | 57.3 (47.9) |
| ABOSS on Healthy_H | 486,437 (176,012) | 100 (100) |
| IgReC-corrected Healthy_H | 1,303,128 (367,235) | 71.6 (60.4) |
| IgReC on ABOSS filtered Healthy_H | 476,072 (61,281) | 93.1 (87.9) |
For both datasets roughly 30% of the sequences in the IgReC-corrected set contained ABOSS flagged residue/positions. The redundancy of sequences that did not pass ABOSS but are found in the IgReC-corrected data is lower than the average of the IgReC-corrected data (Table S1).
As the data above suggests that IgReC and ABOSS remove different sequences ABOSS was run on the IgReC-corrected UCB_H and Healthy_H datasets. ABOSS filtered out 3,327,793 sequences (59.7%) from the IgReC-corrected UCB_H with a residue error rate of 0.0055. This error rate was very similar to that given by ABOSS for the original UCB_H dataset (see Table II). Among the IgReC-corrected UCB_H sequences filtered out by ABOSS, 37,671 (1.1%) sequences failed to pass ANARCI, while the rest contained ABOSS flagged residue/positions, of which 120,264 (3.6%) sequences lacked conserved cysteines. Applying ABOSS to the IgReC-corrected Healthy_H dataset yielded a residue error rate of 0.0041, which filtered 685,546 sequences (52%). Of these filtered sequences, 144,154 (21%) sequences failed ANARCI parsing, and of the rest with flagged residue/positions, 33,627 (4.9%) lacked cysteines at positions 23 and/or 104. IgReC analysis does not appear to correct stop codons, as at least one was identified in ~85.5% of the sequences that failed to pass ANARCI parsing from the IgRec-corrected Healthy_H dataset.
We then tested the reverse protocol running IgReC on the ABOSS filtered UCB_H and Healthy_H datasets. IgReC generates structurally incorrect sequences (~0.01%) when it is run on these ABOSS filtered datasets. Many of these sequences had a nucleotide indel introduced by IgReC. IgReC also altered the sequences of over 40% and 6% of the data to ones which were absent in the original UCB_H and Healthy_H datasets respectively (Table III).
Given this data, we would suggest that IgReC analysis can be enhanced by first using ABOSS to filter out structurally impossible Ig-seq data.
3.6. Comparison to experimental Ig-seq error correction methods
We also compared the results of ABOSS to two different experimental approaches. First to the work of Galson et al., (7). Their methodology of residue error estimation employs an analogous approach to ours. It is based on the proportion of nucleotide mismatches to the germline in the sequence invariant constant region, which was adjacent to the framework 4 region of the heavy chain (FW-H4). ABOSS analysis on their HEPB dataset estimated the residue error rate to be 0.22%. This is in the agreement with the residue error rates estimated by Galson et al., (7) which ranged between 0.19% and 0.79%.
Secondly, we contrasted ABOSS with the experimental/computational error correction protocol of Khan et al., (8). This method considers the entirety of the VH domain by applying barcodes to cDNA prior to sequencing, followed by clustering of identically barcoded sequences and error correction. The Khan_C dataset is the experimentally corrected version of the Khan_R dataset. In the process of this error correction protocol sequences are computationally modified (~33% of Khan_C sequences have been altered from the sequences experimentally determined in the Khan_R dataset). This maybe to another sequence present in the Khan_R dataset (increasing redundancy from 3.7 in Khan_R to 45.3 in Khan_C) or to a new sequence altogether. This modification means that the redundancy of sequences changes and that 0.5% of non-redundant (0.02% of redundant) sequences in the Khan_C dataset are not present in the original Khan_R dataset. These sequence changes make comparison with ABOSS difficult as within the ABOSS protocol no sequences are altered.
ABOSS analysis on Khan_R selects a similar number of non-redundant sequences to Khan_C (~50,000), but only ~6,000 of these sequences are directly observed in the Khan_C dataset (Table IV). In terms of redundant sequences, ABOSS selects a far smaller set. This reflects the fact that sequences have been modified to increase the redundancy of specific sequences in the Khan_C dataset. The redundant overlaps between the ABOSS filtered Khan_R dataset and the Khan_C dataset are 36.8% and 89.6% respectively (Table IV). Around 60% of Khan_C sequences are not seen in the ABOSS filtered Khan_R dataset of these about 1% fail the ANARCI parsing step (suggesting they would not produce viable antibodies), 0.04% are not found in Khan_R, the others contain residue/positions that are ABOSS flagged as below the residue error rate. Those flagged by ABOSS include ~0.2% redundant and ~8% non-redundant sequences that lack a cysteine at either position 23 or 104.
Table IV. Comparison analysis of ABOSS and the barcode approach of Khan et al., (8).
ABOSS was run on the Khan_R dataset. The ABOSS outputs were contrasted with the Khan_C dataset (see Table I for dataset information). (A) The overlap presents the percentage of total sequences that are shared between the Khan_C and ABOSS filtered Khan_R datasets. ABOSS appears to be more conservative than the barcode approach.
| Data source | Dataset size | Redundancy | Overlap |
|---|---|---|---|
| Khan_C | 2,385,080 (52,623) | 45.3 | 36.8% |
| ABOSS filtered Khan_R | 649,685 (47,593) | 13.7 | 89.6% |
ABOSS provides orthogonal functionality to Ig-seq data error correction and can be used to complement the UMI barcode approach, an increasingly common practice in Ig-seq data analysis (39). Performance of the barcode approach is heavily dependent on drawing a consensus sequence from a pool of identically barcoded sequences. Two common problems in the barcode approach are when a large number of the barcoded sequences are singletons or several identically barcoded sequences share the highest redundancies in a cluster. These problems hamper the ability of the approach to correct data efficiently. ABOSS can be used prior to clustering to prevent all structurally non-viable sequences from becoming consensus sequences.
UMI barcodes are also used for accurate detection of template amplification and quantification biases in Ig-seq datasets (8–10, 40). This allows for the precise calculation of the amount and diversity of sequencing templates (8). In this scenario, ABOSS should not be run prior to the barcode correction approach as it is a conservative tool that always reduces the dataset size and never alters antibody sequences.
3.7. The orthogonality of ABOSS
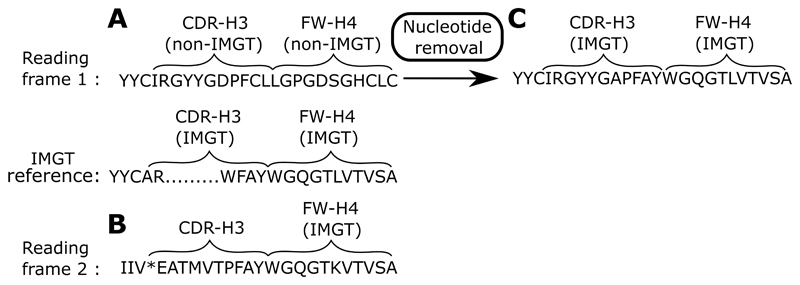
As an example of how ABOSS identifies potentially structurally not viable sequences that are not picked up by other techniques, Figure 6 shows an example of an antibody sequence from the Khan_C dataset (8). This sequence is translated into amino acids in the first reading frame. This sequence cannot be structurally viable as FW-H4 and the distal end of CDR-H3 do not align to the known IMGT amino acid germline. Translating this sequence into the second reading frame reveals a FW-H4 and a distal end of CDR-H3 that now align to the IMGT amino acid germline. This suggest that a single nucleotide insertion was introduced into CDR-H3.
Figure 6. ABOSS flags structurally non-viable antibody sequences.
(A) The distal part of the VH chain sequence of an antibody that is selected as correct in the Khan_C dataset. The closest germline matches of the V and J genes for this sequence are IGHV5-4*02 and IGHJ3*01 respectively. This sequence is shown in the first reading frame. The FW-H4 region and the distal end of CDR-H3 of this sequence do not align to an IMGT amino acid germline. (B) Translating this antibody sequence into the second reading frame creates FW-H4 and the distal end of CDR-H3 that align to the IMGT amino acid germline. (C) An arbitrary deletion of a nucleotide from the middle of the CDR-H3 region generates a structurally viable antibody sequence when it is translated in the first reading frame.
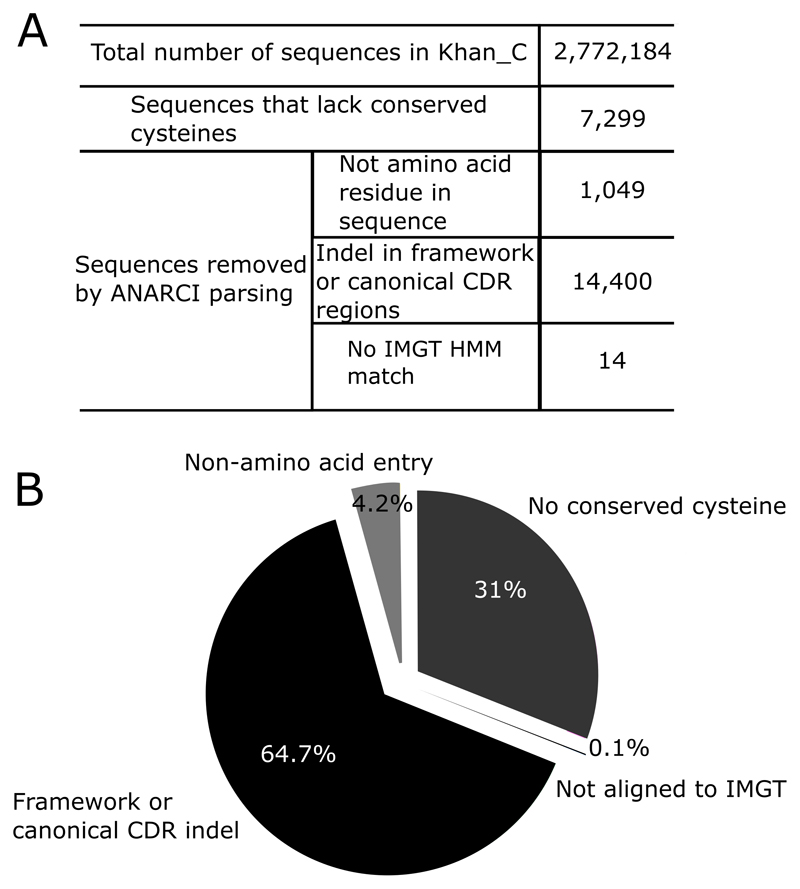
If we run ABOSS on Khan_C (the experimentally/computationally error corrected set of Khan_R), the ANARCI parsing step in conjunction with the check for conserved cysteines at positons 23 and 104 removed 11.6% of the unique sequences. These structurally impossible sequences correspond to 0.8% of the total redundant dataset (Figure 7). The inability of ANARCI to align the full-length FW-H4 to the IMGT germline was the main cause for sequences from the Khan_C dataset to fail ANARCI parsing as these sequences were considered to have a truncated FW-H4 region.
Figure 7. Identification of structurally non-viable antibody sequences using first steps of ABOSS on the Khan_C datasets.
Each sequence from the Khan_C (Table I) dataset is examined for structural viability using ABOSS. (A) The tabulated outputs gives the total number sequences that did not pass ANARCI. (B) The pie chart shows the percentage of sequences that fail the ANARCI step and those that lack a cysteine at position 23 and 104. The reasons include: 1) A sequence lacks a cysteine at position 23 or 104 2) An indel is present in the framework or canonical CDR regions 3) A non-amino acid residue is present in a sequence. 4) A sequence does not align to the IMGT amino acid germlines of V or J genes.
These results demonstrate how leverage of our knowledge of immunoglobulin folding can help to filter data, even that, which has been generated by a barcoding approach.
We have examined the robustness of the ABOSS protocol by running it on a dataset parsed by either ANARCI or IgBlastn (24), a sequence-centered Ig-seq data processing tool. ANARCI parsing removed ~9% more sequences than IgBlastn (Table V). However, if ABOSS is run on the ANARCI parsed data or on data already passed by IgBlastn approximately, the same number of sequences are obtained. Examination of the sequences ANARCI removes and IgBlastn does not reveals that these sequences tend to not have a full-length framework 4 region or nothing at position 23, or had unusual indels in canonical CDR and framework regions. The ANARCI parsed Healthy_H and Healthy_L datasets contained almost all (>99.98%) sequences that IgBlastn called productive. ABOSS analysis generated almost identical outputs for Healthy_H and Healthy_H_IgBlastn, whereas there was an increase (~4%) in the number of Healthy_L_IgBlastn sequences compared to Healthy_L as a result of the slightly smaller residue error rate.
Table V. IgBlastn and ANARCI parsing.
Performance of IgBlastn and ANARCI parsing was investigated on two datasets, Healthy_H and Healthy_L (see Table I for more details). IgBlastn analysis was perform on the nucleotide sequences that were downloaded from the OAS resource (36). IgBlastn productive called sequences were put into Healthy_H_IgBlastn and Healthy_L_IgBlastn datasets respectively. ANARCI parsing was performed on the translated amino acid version of Healthy_H and Healthy_L (Table I). All four datasets were then subjected to ABOSS analysis.
| Dataset name | Starting dataset | ANARCI parsed dataset | ABOSS filtered dataset | ABOSS Residue Error Rate (%) |
|---|---|---|---|---|
| Healthy_H | 1,422,405 (745,276) |
1,135,185 (558,171) |
486,437 (176,012) |
0.5427 |
| Healthy_H_IgBlastn | 1,228,129 (597,976) |
1,135,116 (558,129) |
486,429 (176,008) |
0.5427 |
| Healthy_L | 6,317,736 (2,135,745) |
4,860,389 (1,372,804) |
2,667,263 (386,165) |
0.4121 |
| Healthy_L_IgBlastn | 5,361,955 (1,539,964) |
4,859,848 (1,372,519) |
2,776,176 (386,146) |
0.4118 |
4. Discussion
ABOSS is an orthogonal redundancy-neutral method that uses structural information to calculate sequencing error rate estimates for Ig-seq datasets. The novelty of our approach is founded in the application of current knowledge of immunoglobulin folding to identify and flag potential errors in Ig-seq sequences.
ABOSS has been tested on six different Ig-seq datasets ranging from 1,422,405 sequences to 9,985,575 sequences, which were generated by a variety of sequencing methods. The protocol is rapid and takes 10h to analyze 5m unique antibody sequences on a standard desktop computer. ABOSS calculated residue error rates agree well with experimental error rates where available as in Galson et al., (7).
ABOSS identified 99% of correct antibody sequences in the simulated Ig-seq data when using dataset sizes and error rates matching those in the experiments. Decreasing the size of the simulation Ig-seq data did not affect the percentage of correct sequences recovered. Our simulation results suggest that even at far higher error rates ABOSS performs well as long as either the redundancy is high or the dataset size is large enough (~600k unique sequences). The model selected to introduce in silico sequencing errors was based on Illumina technologies, where nucleotide substitutions can happen stochastically along the VH chain. For Roche 454 datasets (now one of the most common sources of Ig-seq data (36)) nucleotide indel introduction along the chain is the main origin of sequencing errors so further simulations of error might be necessary to understand the behaviour of ABOSS.
We also ran ABOSS on Ig-seq data with computationally simulated SHM diversity to assess its ability to preserve true mutations. These simulations indicated that ABOSS was able to spot structurally incorrect residue/positions, whilst preserving the SHM generated diversity. It is hard to assess the accuracy of SHM substitutions introduced by the HH_S5F model in the structural context. In the functional antibody repertoire there are a number of positions where SHM substitutions are not observed, in particular, positions 23 and 104, but are seen in the HH_S5F model (35). Therefore, SHM in functional genes has a negative reinforcement effect on the residue error rate, which will mean ABOSS is less likely to flag positions that harbour SHM substitutions.
The nature of ABOSS analysis is orthogonal to current Ig-seq correction techniques in particular it does not alter sequences but rather removes those it considers to contain impossible structural features. Comparison to leading experimental and computational Ig-seq error correction methods that do alter sequences shows that these approaches retain as well as create antibody sequences that are structurally non-viable (i.e. lack of cysteines at conserved position 23 and 104, or antibody regions that are out of the correct reading frame). These results suggest that ABOSS should be used alongside current state-of-art error-correction protocols to increase confidence of structural viability of Ig-seq sequences.
Supplementary Material
Acknowledgments
We would like to thank Dominic F. Kelly, Jacob D. Galson, Simon Friedensohn and Sai Reddy for proving the Ig-seq datasets as well as Jinwoo Leem from Oxford Protein Informatics Groups, whose comments have significantly improved the quality of our work. We also like to thank Michael Wright for generating UCB_H and UCB_L Ig-seq data.
Footnotes
This work was supported by funding from Biotechnology and Biological Sciences Research Council (BBSRC) [BB/M011224/1] and UCB Pharma Ltd awarded to AK.
References
- 1.Collis AVJ, Brouwer AP, Martin ACR. Analysis of the antigen combining site: Correlations between length and sequence composition of the hypervariable loops and the nature of the antigen. J Mol Biol. 2003;325:337–354. doi: 10.1016/s0022-2836(02)01222-6. [DOI] [PubMed] [Google Scholar]
- 2.Reichert JM. Antibodies to watch in 2017. MAbs. 2017;9:167–181. doi: 10.1080/19420862.2016.1269580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strohl WR. Current progress in innovative engineered antibodies. Protein Cell. 2017:1–35. doi: 10.1007/s13238-017-0457-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Georgiou G, Ippolito GC, Beausang J, Busse CE, Wardemann H, Quake SR. The promise and challenge of high-throughput sequencing of the antibody repertoire. Nat Biotech. 2014;32:158–168. doi: 10.1038/nbt.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parola C, Neumeier D, Reddy ST. Integrating high-throughput screening and sequencing for monoclonal antibody discovery and engineering. Immunology. 2017 doi: 10.1111/imm.12838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedensohn S, Khan TA, Reddy ST. Advanced Methodologies in High-Throughput Sequencing of Immune Repertoires. Trends Biotechnol. 2017;35:203–214. doi: 10.1016/j.tibtech.2016.09.010. [DOI] [PubMed] [Google Scholar]
- 7.Galson JD, Trück J, Fowler A, Münz M, Cerundolo V, Pollard AJ, Lunter G, Kelly DF. In-depth assessment of within-individual and inter-individual variation in the B cell receptor repertoire. Front Immunol. 2015;6 doi: 10.3389/fimmu.2015.00531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khan TA, Friedensohn S, de Vries ARG, Straszewski J, Ruscheweyh H-J, Reddy ST. Accurate and predictive antibody repertoire profiling by molecular amplification fingerprinting. Sci Adv. 2016;2:e1501371–e1501371. doi: 10.1126/sciadv.1501371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turchaninova MA, Davydov A, Britanova OV, Shugay M, Bikos V, Egorov ES, Kirgizova VI, Merzlyak EM, Staroverov DB, Bolotin DA, Mamedov IZ, et al. High-quality full-length immunoglobulin profiling with unique molecular barcoding. Nat Protoc. 2016;11:1599–1616. doi: 10.1038/nprot.2016.093. [DOI] [PubMed] [Google Scholar]
- 10.Shugay M, Britanova OV, Merzlyak EM, Turchaninova Ma, Mamedov IZ, Tuganbaev TR, Bolotin Da, Staroverov DB, Putintseva EV, Plevova K, Linnemann C, et al. Towards error-free profiling of immune repertoires. Nat Methods. 2014;11:653–5. doi: 10.1038/nmeth.2960. [DOI] [PubMed] [Google Scholar]
- 11.Kuchenbecker L, Nienen M, Hecht J, Neumann AU, Babel N, Reinert K, Robinson PN. IMSEQ-A fast and error aware approach to immunogenetic sequence analysis. Bioinformatics. 2015;31:2963–2971. doi: 10.1093/bioinformatics/btv309. [DOI] [PubMed] [Google Scholar]
- 12.Yu Y, Ceredig R, Seoighe C. LymAnalyzer: A tool for comprehensive analysis of next generation sequencing data of T cell receptors and immunoglobulins. Nucleic Acids Res. 2015;44 doi: 10.1093/nar/gkv1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bolotin DA, Poslavsky S, Mitrophanov I, Shugay M, Mamedov IZ, Putintseva EV, Chudakov DM. MiXCR: software for comprehensive adaptive immunity profiling. Nat Methods. 2015;12:380–381. doi: 10.1038/nmeth.3364. [DOI] [PubMed] [Google Scholar]
- 14.Shlemov A, Bankevich S, Bzikadze A, Turchaninova MA, Safonova Y, Pevzner PA. Reconstructing Antibody Repertoires from Error-Prone Immunosequencing Reads. J Immunol. 2017;199:3369–3380. doi: 10.4049/jimmunol.1700485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lefranc M-P, Pommié C, Ruiz M, Giuducelli V, Foulquier E, Truong L, Thouvenin-Contet V, Lefranc G. IMGT unique numbering fro immunoglobulin and T cell receptor variable domains and Ig superfamily V-like domains. Dev Comp Immunol. 2003;27:55–77. doi: 10.1016/s0145-305x(02)00039-3. [DOI] [PubMed] [Google Scholar]
- 16.Glockshuber R, Schmidt T, Plückthun A. The disulfide bonds in antibody variable domains: effects on stability, folding in vitro, and functional expression in Escherichia coli. Biochemistry. 1992;31:1270–1279. doi: 10.1021/bi00120a002. [DOI] [PubMed] [Google Scholar]
- 17.Lefranc MP. IMGT, the international ImMunoGeneTics database®. Nucleic Acids Res. 2003;31:307–310. doi: 10.1093/nar/gkg085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hagihara Y, Saerens D. Engineering disulfide bonds within an antibody. Biochim Biophys Acta - Proteins Proteomics. 2014;1844:2016–2023. doi: 10.1016/j.bbapap.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 19.Koenig P, Lee CV, Walters BT, Janakiraman V, Stinson J, Patapoff TW, Fuh G. Mutational landscape of antibody variable domains reveals a switch modulating the interdomain conformational dynamics and antigen binding. Proc Natl Acad Sci. 2017;114:E486–E495. doi: 10.1073/pnas.1613231114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rudikoff S, Pumphrey JG. Functional antibody lacking a variable-region disulfide bridge. Proc Natl Acad Sci U S A. 1986;83:7875–8. doi: 10.1073/pnas.83.20.7875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wörn A, Plückthun A. Mutual stabilization of V(L) and V(H) in single-chain antibody fragments, investigated with mutants engineered for stability. Biochemistry. 1998;37:13120–13127. doi: 10.1021/bi980712q. [DOI] [PubMed] [Google Scholar]
- 22.Proba K, Honegger A, Plückthun A. A natural antibody missing a cysteine in V(H): Consequences for thermodynamic stability and folding. J Mol Biol. 1997;265:161–172. doi: 10.1006/jmbi.1996.0726. [DOI] [PubMed] [Google Scholar]
- 23.Auffray C, Sikorav JL, Ollo R, Rougeon F. Correlation between D region structure and antigen-binding specificity: evidences from the comparison of closely related immunoglobulin VH sequences. Ann Immunol (Paris) 132D:77–88. [PubMed] [Google Scholar]
- 24.Ye J, Ma N, Madden TL, Ostell JM. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 2013;41:W34–W40. doi: 10.1093/nar/gkt382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dunbar J, Deane CM. ANARCI: Antigen receptor numbering and receptor classification. Bioinformatics. 2015;32:298–300. doi: 10.1093/bioinformatics/btv552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He L, Sok D, Azadnia P, Hsueh J, Landais E, Simek M, Koff WC, Poignard P, Burton DR, Zhu J. Toward a more accurate view of human B-cell repertoire by next-generation sequencing, unbiased repertoire capture and single-molecule barcoding. Sci Rep. 2014;4:6778. doi: 10.1038/srep06778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zemlin M, Klinger M, Link J, Zemlin C, Bauer K, Engler JA, Schroeder HW, Kirkham PM. Expressed murine and human CDR-H3 intervals of equal length exhibit distinct repertoires that differ in their amino acid composition and predicted range of structures. J Mol Biol. 2003;334:733–749. doi: 10.1016/j.jmb.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 28.Kovaltsuk A, Krawczyk K, Galson JD, Kelly DF, Deane CM, Trück J. How B-Cell Receptor Repertoire Sequencing Can Be Enriched with Structural Antibody Data. Front Immunol. 2017;8:1753. doi: 10.3389/fimmu.2017.01753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galson JD, Trück J, Fowler A, Clutterbuck EA, Münz M, Cerundolo V, Reinhard C, van der Most R, Pollard AJ, Lunter G, Kelly DF. Analysis of B Cell Repertoire Dynamics Following Hepatitis B Vaccination in Humans, and Enrichment of Vaccine-specific Antibody Sequences. EBioMedicine. 2015;2:2070–2079. doi: 10.1016/j.ebiom.2015.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galson JD, Trück J, Clutterbuck EA, Fowler A, Cerundolo V, Pollard AJ, Lunter G, Kelly DF. B-cell repertoire dynamics after sequential hepatitis B vaccination and evidence for cross-reactive B-cell activation. Genome Med. 2016;8:68. doi: 10.1186/s13073-016-0322-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krawczyk Konrad, Kelm Sebastian, Kovaltsuk Aleksandr, Galson Jacob D, Kelly Dominic, Trück Johannes, Regep Cristian, Leem Jinwoo, Wong Wing Ki, Nowak Jaroslaw, Snowden James, et al. Structurally Mapping Antibody Repertoires. Front Immunol. 2018 doi: 10.3389/fimmu.2018.01698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vander Heiden JA, Stathopoulos P, Zhou JQ, Chen L, Gilbert TJ, Bolen CR, Barohn RJ, Dimachkie MM, Ciafaloni E, Broering TJ, Vigneault F, et al. Dysregulation of B Cell Repertoire Formation in Myasthenia Gravis Patients Revealed through Deep Sequencing. J Immunol. 2017;198:1460–1473. doi: 10.4049/jimmunol.1601415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yaari G, Vander Heiden JA, Uduman M, Gadala-Maria D, Gupta N, Joel JN, O’Connor KC, Hafler DA, Laserson U, Vigneault F, Kleinstein SH. Models of somatic hypermutation targeting and substitution based on synonymous mutations from high-throughput immunoglobulin sequencing data. Front Immunol. 2013 doi: 10.3389/fimmu.2013.00358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Proba K, Wörn A, Honegger A, Plückthun A. Antibody scFv fragments without disulfide bonds made by molecular evolution. J Mol Biol. 1998;275:245–253. doi: 10.1006/jmbi.1997.1457. [DOI] [PubMed] [Google Scholar]
- 35.Sheng Z, Schramm CA, Kong R, Mullikin JC, Mascola JR, Kwong PD, Shapiro L. Gene-specific substitution profiles describe the types and frequencies of amino acid changes during antibody somatic hypermutation. Front Immunol. 2017;8 doi: 10.3389/fimmu.2017.00537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kovaltsuk A, Leem J, Kelm S, Snowden J, Deane CM, Krawczyk K. Observed Antibody Space: a resource for data mining next generation sequencing antibody repertoires. bioRxiv. 2018 doi: 10.4049/jimmunol.1800708. 316026. [DOI] [PubMed] [Google Scholar]
- 37.Peled JU, Kuang FL, Iglesias-Ussel MD, Roa S, Kalis SL, Goodman MF, Scharff MD. The Biochemistry of Somatic Hypermutation. Annu Rev Immunol. 2008;26:481–511. doi: 10.1146/annurev.immunol.26.021607.090236. [DOI] [PubMed] [Google Scholar]
- 38.Di Noia JM, Neuberger MS. Molecular Mechanisms of Antibody Somatic Hypermutation. Annu Rev Biochem. 2007 doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- 39.Miho E, Yermanos A, Weber CR, Berger CT, Reddy ST, Greiff V. Computational Strategies for Dissecting the High-Dimensional Complexity of Adaptive Immune Repertoires. Front Immunol. 2018;9:224. doi: 10.3389/fimmu.2018.00224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Friedensohn S, Lindner JM, Cornacchione V, Iazeolla M, Miho E, Zingg A, Meng S, Traggiai E, Reddy ST. Synthetic Standards Combined With Error and Bias Correction Improve the Accuracy and Quantitative Resolution of Antibody Repertoire Sequencing in Human Naïve and Memory B Cells. Front Immunol. 2018;9:1401. doi: 10.3389/fimmu.2018.01401. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.