SUMMARY
Objectives:
Meningococcal conjugate vaccines (MenACWY) were licensed in the United States in 2005. We assessed the population structure of invasive Neisseria meningitidis (Nm) ten years after recommended use of MenACWY among adolescents.
Methods:
Meningococcal isolates obtained through Active Bacterial Core surveillance (ABCs) from 2000–05, 2006–10, and 2011–15 underwent whole genome or Sanger sequencing. Genome phylogenies were completed using maximum likelihood methods; and distribution of multilocus sequence typing (MLST) sequence type (ST) and clonal complex (CC), and PorA and FetA types were assessed.
Results:
Prevalent serogroups (B, C, Y and W), CCs, and PorA and FetA types were detected in all three time periods, but dynamic changes were observed. The proportion of serogroup W CC11 isolates increased in 2011–15 and were most related to South American strains. Changes in CC distribution were also observed in serogroup C and serogroup Y. Phylogenetic analysis showed that U.S. serogroup W CC11s are closely related to a subset of U.S. serogroup C isolates; combined global analysis demonstrated that some CCs, including CC11, exhibit regional clustering.
Conclusions
Overall, the Nm population structure has remained stable after MenACWY introduction. Dynamic changes in genotypes, unlikely related to vaccination, also occurred, highlighting the need for continued whole genome-based surveillance.
Keywords: Neisseria meningitides, MLST, Molecular typing, Molecular epidemiology
Background
Meningococcal disease is a life-threatening infection caused by Neisseria meningitidis (Nm). Since 2005, quadrivalent meningococcal conjugate vaccines (MenACWY), which target the capsule of serogroups A, C, W and Y (abbreviated NmA, NmC, NmW, and NmY), have been recommended for routine use in adolescents.1 A booster dose of MenACWY was recommended for adolescents in 2011 to address concerns about waning immunity.2 Men-ACWY helps provide protection to two of the three most common serogroups (NmB, NmC and NmY) that cause invasive disease in the United States.3 Vaccination coverage among adolescents has been steadily increasing, from 12% in 2006 to 81% in 2015.4,5 Since 2014, two protein-based NmB meningococcal vaccines (abbreviated MenB vaccines) have been licensed for use in persons aged 10–25 years. MenB vaccines are only recommended for routine use in persons at increased risk for serogroup B (NmB) meningococcal disease.6
Molecular surveillance has been implemented to monitor the vaccine impact on N. meningitidis genetic diversity and strain replacement. For example, this was essential for linking the NmW strain in Burkina Faso to the Hajj clone7 and detecting the emerging NmC strains from Niger and Nigeria8–10 that resulted in outbreaks after introduction of the NmA conjugate vaccine.11 Furthermore, molecular surveillance can detect capsule switching events that could lead to serogroup replacement, which was observed after implementation of PCV7, a conjugate vaccine for pneumococcal disease.12 Thus, continued monitoring of N. meningitidis genetic diversity is important and can be measured in multiple ways. Meningococcal genetic lineage is generally determined using multilocus sequence typing (MLST), which measures the nucleotide sequence diversity present in seven housekeeping genes, and is denoted as the sequence type (ST) or clonal complex (CC).13 The majority of invasive disease worldwide is caused by a limited number of CCs, which have been described as the hyperinvasive lineages.14 In addition, the highly variable peptide sequence of two outer membrane proteins, porin A (PorA) and ferric enterobactin transport (FetA), have been used for typing to provide additional characterization beyond ST and CC.15 More recently, whole genome sequencing has allowed analysis of all the core genes of meningococci to be simultaneously compared to measure strain diversity with higher resolution.16
In this report, we describe the population structure of invasive N. meningitidis isolates collected in the United States from 2011–2015, a time of high MenACWY coverage among adolescents. To address potential vaccine impacts, the 2011–15 population structure is compared to previously reported data from 2000–05 (no vaccine coverage) and 2006–10 (low vaccine coverage).17,18 We describe the diversity of meningococci using both classical typing methods (the distribution of CC/ST and the fine types for PorA and FetA) and phylogenetic analyses based on high resolution whole genome data. Finally, we compare the genetic similarity between U.S. and global isolates, with a specific analysis of the NmW CC11 lineages.
Methods
Meningococcal isolate collection
Isolates were collected through Active Bacterial Core surveil-lance (ABCs), a population- and laboratory-based active surveil-lance system that includes approximately 43.5 million U.S. residents.19 As of 2015, the catchment areas included California (CA - 3 bay area counties), Colorado (CO) (5 Denver counties), Connecticut (CT), Georgia (GA), Maryland (MD), Minnesota (MN), New Mexico (NM), New York (NY - 8 Albany and 7 Rochester counties), Oregon (OR), and Tennessee (TN - 20 urban counties). Not all ABCs sites were included between 2000 and 2015. NM was added in 2004 and 9 additional TN counties were added to in 2010.
From 2011–2015, there were 387 N. meningitidis cases reported through the ABCs and 350 (90%) isolates were available for molecular typing. Only one isolate per case was included in the analysis. The 2011–2015 isolates were compared to ABCs isolates collected in 2000–2005 (n = 1175) and 2006–10 (n = 638), which have been reported previously.17,18 An additional 28 previously unreported isolates from 2006–10 were included in this analysis.18 For this analysis, the 2000–05 isolates are considered to have been collected during a period of no vaccine coverage, the 2006–10 isolates represent low MenACWY coverage (12–63% in adolescents aged 13–17) and the 2011–15 isolates represent high MenACWY coverage (71–81% MenACWY in adolescents aged 13–17).4,5,20
Isolate identification and serogrouping were completed at the state public health laboratories. Serogroup was confirmed by slide agglutination and real-time PCR at CDC.21 Both cnl isolates and isolates with any mutations that impairs capsule synthesis would be detected by the combination of these methods and were considered as nongroupable (NmNG).
Molecular characterization
Molecular characterization of the 2011–15 isolates was completed by whole genome sequencing. DNA was extracted using the Gentra Puregene yeast/bacteria DNA extraction kit (Qiagen) or with a chemagic Prepito instrument (PerkinElmer) using the Cyto Pure Kit. Genomic libraries were generated with NEBNext Ultra DNA Library preparation kits according to manufacturer specifications. Libraries were sequenced at CDC using 250 bp paired-end reads on a HiSeq 2500 or a MiSeq (Illumina). Raw sequencing reads were trimmed to remove adapters and low quality bases. De novo genomic assembly was completed with SPAdes, version 3.7.0.22 Sequences for the MLST genes and the fine typing genes (porA and fetA) were identified by a BLAST search of the genome assembly using the PubMLST allele collection (www.pubmlst.org/Neisseria) to identify the sequence type (ST), clonal complex (CC), and PorA and FetA types. Sequencing data is deposited to GenBank (BioProject: SRP144480).
Whole genome phylogeny
Whole genome phylogenies were constructed using recombination-corrected maximum likelihood (ML) methods. Briefly, a whole genome core alignment was generated with Parsnp (Harvest suite, version 1.2), using the FAM18 reference genome (Accession: NC_008767).23 The whole genome core alignment was used to infer the ML phylogeny using RAxML with a general time reversible nucleotide model.24 Recombination corrections were completed with ClonalFrameML, which calculates ML estimates at every site on the alignment to identify the recombinant regions.25
To compare genomes of U.S. and global meningococcal strains, genomic assemblies from non-U.S. sources were obtained from PubML ST (PubMLST IDs listed in Supplemental Table 1).26 Only 2011–15 invasive isolates that belonged to the eight most common U.S. CCs (ST-103 complex, ST-11 complex, ST-23 complex, ST-167 complex, ST-22 complex, ST-32 complex, ST-4¼4 complex and the ST-35 complex) were included. Using this criterion, 1809 global isolate assemblies were identified and compared to all 2011–15 ABCs isolates. To compare U.S. and global NmW CC11 isolates, fifty-one previously published genomes collected between 1997 and 2015 were chosen. These genomes are representative of the NmW African lineages (including a Hajj clone isolate), the South American, and UK lineages described previously.7,27,28 Whole genome phylogenies were generated as described for the ABCs isolates alone.
Capsule switching events
Potential capsule switching events were detected by identification of an isolate with a different serogroup than the neighboring isolates within its phylogenetic clade. The presence or absence of the serogroup-specific capsule genes (csb for NmB or cssE/csc for NmC) were identified from the whole genome sequencing data to confirm the capsule switch event. The csb and csc genes are polysialytransferases and cssE is an O-acetyltransferase, essential for making the serogroup-specific capsule polysaccharides.29
Data analysis
Data were analyzed using SAS (version 9.4). Significant changes between time periods were calculated using χ2 statistics and exact methods (p values). Hyperendemic NmB disease has been reported in Oregon since 1993, with higher incidence of NmB within Oregon than the other ABCs sites between 2001 and 2010.30,31 To better estimate the distribution of NmB molecular types (ST, CC, PorA, FetA, PorA:FetA) circulating in the United States, NmB analyses were weighted based on the proportion of the ABCs population that is contributed by Oregon (10%). A weight of 0.1 was assigned to NmB isolates from Oregon and 0.9 for NmB isolates from non-Oregon ABCs sites, consistent with previous reports.17,18
Results
Serogroup distribution
During 2011–15, NmB, NmC, and NmY were the main causes of disease (Table 1). The serogroup distribution was NmC (28.5%), NmY (27.3%), NmB (25.7%), NmW (13.0%), and nongroupable (NmNG) (5.5%). The proportion of NmW isolates increased from 4.0% in 2006–10 to 13.0% in 2011–15 (p < .00 01). The proportion of NmY decreased (p < .003) from 37.2% (2006–10) to 27.4% (2011–15). All other serogroups exhibited no statistically significant changes.
Table 1.
Serogroup distribution of U.S. ABCs isolates characterized in this study.
| Serogroup | 2000–2005 No. of isolates (%) | 2006–2010 No. of isolates (%) | 2011–2015 No. of isolates (%) | P value 2006–10 vs. 2011–15 |
|---|---|---|---|---|
| A | 1 (0.10) | 0 (0) | 0 (0) | N/A |
| C | 277 (29.03) | 173 (31.11) | 88 (28.54) | 0.43 |
| Y | 319 (33.43) | 207 (37.22) | 84 (27.25) | <0.0001 |
| W | 27 (2.83) | 22 (3.96) | 40 (12.97) | 0.0029 |
| B* | 298 (31.26) | 129 (23.22) | 79 (25.72) | 0.41 |
| NG | 29 (3.04) | 19 (3.42) | 17 (5.51) | 0.14 |
| Other | 3 (0.31) | 6 (1.08) | 0 (0) | N/A |
| Total Isolates | 954 | 556 | 308 |
Results for 2000–05 [17] and 2006–10 [18] were reported previously. Nongroupable isolates are abbreviated NG.
Serogroup B isolates are represented as weighted values. Serogroup B isolates from Oregon are assigned a value of 0.10, while isolates from all other states are assigned 0.90. The uncorrected total number of isolates collected in each time period are 1175 in 2000–05, 638 in 2006–10, and 349 in 2011–15.
Genetic diversity
Together, there were 372 STs detected during the three time periods with 177 STs unique to 2000–2005, 66 STs unique to 2006–10, and 58 STs unique to 2011–15. There were 23 STs present in all three time periods comprising 71.0% (2000–05), 75.2% (2006–10) or 69.1% (2011–15) of the population, respectively. Six of the 23 shared STs had more than 20 isolates in at least one time period including ST-11/CC11, ST-23/CC23, ST-32/CC32, ST-136/CC4¼4, ST-162/CC162 and ST-2006/CC103, which predominantly correspond to the main hyperinvasive lineages.14
Although there was some variance in CC composition, the common CCs were consistent over time. CCs unique to each time period were detected but only associated with a few isolates (6 unique CCs detected in 8 total 2000–5 isolates; 1 unique CC detected in 2 total 2006–10 isolates; 2 unique CCs detected in 2 total 2011–15 isolates) (Supplemental Table 2). Of the 29 distinct CCs detected between 2000 and 2015, 16 CCs were common to all three time periods comprising 95.5% of the isolate population. The four predominant CCs detected were CC11 (22.6%), CC23 (31.6%), CC32 (12.6%), and CC4¼4 (9.1%).
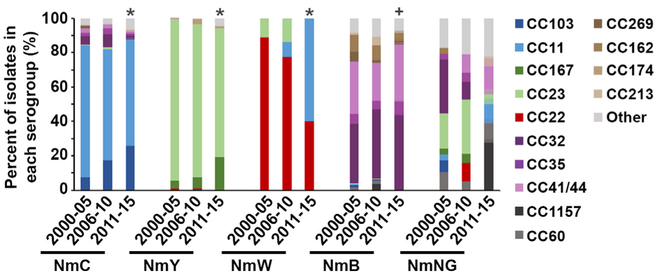
Consistent with previous reports, each serogroup mostly associated with distinct CCs in 2011–15 with the exception of CC11.17,18 NmB isolates had the highest diversity with CC32 (43.9%) and CC4¼4 (33.5%), comprising only 77.4% of the NmB isolate population (Fig. 1). In contrast > 88% of the vaccine-targeted serogroups ACWY were associated with two CCs each: NmC isolates were CC103 (26.1%) and CC11 (62.5%), NmY isolates were CC23 (75.0%) and CC167 (19.2%), and all NmW isolates were either CC22 (40.0%) or CC11 (60.0%). Among the NmW isolates, the proportion that were CC11 increased significantly, from 0% during 2000–05, 9.1% (n = 2) in 2006–10 and 60% (n = 24) in 2011–2015 (p < .0001) (Fig. 1). However, the majority of NmW isolates (17 of the 24 isolates) detected during 2011–2015 were collected from Georgia and may not be representative of nationwide trends. NmC and NmY also exhibited significant changes in CC distribution during 2011–15 (p < 0.0001 when compared to 2000–05). A single CC increased in each vaccine serogroup over time: CC103 increased from 7.2% (2000–05), 17.4% (2006–10), to 26.4% (2011–15) in NmC isolates and CC167 increased from 4.7% (2000–05), 6.3% (2006–10), to 19.2% (2011–15) in NmY isolates. One CC also increased in NmB with CC32 increasing from 34.5% (2000–05), 40.1% (2006–10), to 43.9% (2011–15), but the overall CC distribution in NmB did not significantly change (p = 0.18 for 2000–05 vs. 2011–15).
Fig. 1.
Graphical representation of the proportion of isolates associated with each clonal complex (CC) over time, analyzed by serogroup. CCs historically associated with NmC (CC103 and CC11) are shown in blue, with NmY (CC167 and CC23) are shown in green, with NmW (CC22) are shown in red and with NmB (CC32, CC35, CC4¼4) are depicted in purple. Other/ND represents isolates belonging to CCs that were detected in < 5 isolates or for isolates where a clonal complex was unassigned. NmB proportions reflect OR-corrected values. The “*” denotes a P < .0001 when comparing 2000–05 and 2011–15, while the “ + “ denotes a P > .18 when comparing 2000–05 and 2011–15. Results for 2000–05 (17) and 2006–10 (18) were reported previously.
Outer membrane protein fine typing
PorA
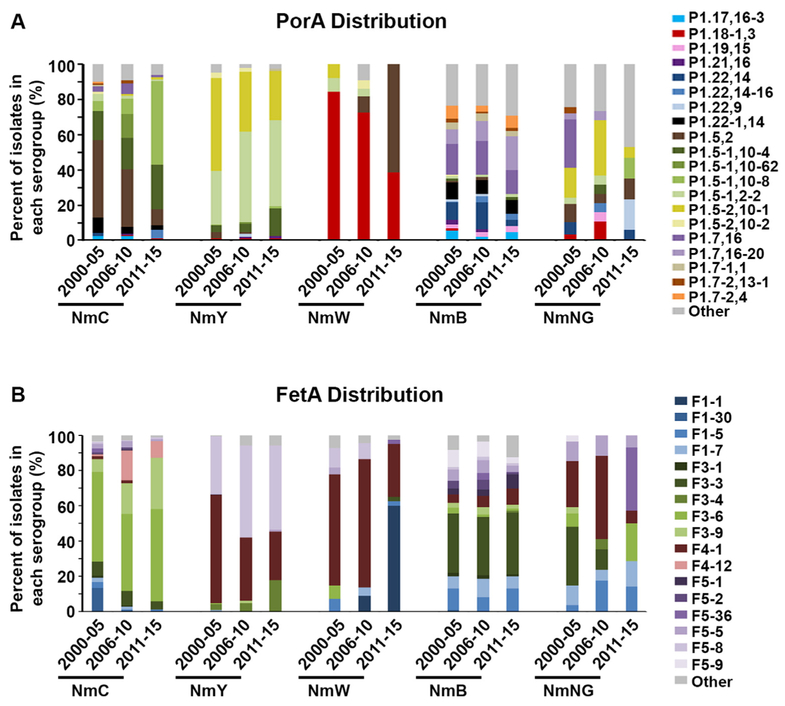
A total of 161 PorA types were detected in 2000–05, 2006–2010, or 2011–15. The majority of isolates (86.7%) were associated with the 23 PorA types detected in all three time periods, with less than 10% of the observed isolates having a PorA type unique to a single time period (73 unique PorA types in 2000–05; 26 unique types in 2006–10; 20 unique types detected in 2011–15). Most serogroups had a diverse array of PorA types in 2011–15 (Fig. 2A), and only 7 isolates (2.3%) were detected with a deletion in at least one of the PorA variable regions. NmB isolates were the most diverse with 35 PorA types detected (14 types in NmC, 2 types in NmW and 9 types in NmY). NmC, NmY and NmW exhibited statistically significant changes over time (p < .025), while NmB types were more stable (p = .12) (Fig. 2A). A large decrease in P1.5,2 was observed in NmC isolates (44.0% (2000–05), 32.4% (2006–10), and 9.1% (2011–15)), while a large increase in P1.5,2 was observed in NmW isolates (0.0% (2000–05), 9.1% (2006–10), and 60% (2011–15)). This is consistent with a reduction in NmC CC11s containing P1.5,2 over time and an increase in the NmW CC11s, which all contained P1.5,2 (Table 2). In 2011–2015, the majority of NmC CC11 isolates have P1.5–1,10–8, while the majority of NmW isolates have P1.5,2 (Table 2).
Fig. 2.
Graphical representation of the proportion of isolates associated with each PorA (A) or FetA (B) type. The Other category summarizes PorA or FetA types that were detected in < 10 isolates during 2000–2015 (representing 132 PorA types or 46 FetA types). In the PorA graph, types with the same VR1 region are depicted as multiple shades of the same color (Example: P1.5–1,10–4; P1.5–1,10–62; P1.5–1,10–8; and P1.5–1,2–2 are represented as shades of green). For the FetA graph, closely associated types are depicted as multiple shades of the same color (example: F1–1, F1–30, F1–5, and F1–7 are shown in hues of blue). NmB proportions are represented as OR-corrected values. Results for 2000–05 (17) and 2006–10 (18) were reported previously.
Table 2.
Strain genotypes with significant changes detected between 2000–05, 2006–10 and 2011–15.
| Serogroup:CC:PorA:FetA | 2000–05 No. of isolates (%) | 2006–10 No. of isolates (%) | 2011–15 No. of isolates (%) |
|---|---|---|---|
| B* Total Isolates | 298.3 | 129.1 | 79.3 |
| CC32 | |||
| P1.7,16–20:F3–3 | 24 (8.0) | 13 (9.8) | 15 (19.2) |
| CC41/44 | |||
| P1.7–2,4:F1–5 | 17 (5.8) | 2 (1.4) | 4 (4.8) |
| CC162 | |||
| P1.22,14:F5–9 | 23 (7.8) | 8 (6.4) | 1 (1.1) |
| C Total Isolates | 277 | 173 | 88 |
| CC103 | |||
| P1.5–1,10–4:F3–9 | 17 (6.1) | 29 (16.8) | 21 (23.9) |
| CC11 | |||
| P1.22–1,14:F3–6 | 23 (8.3) | 4 (2.3) | 0 (0.0) |
| P1.5–1,10–62:F3–6 | 0 (0.0) | 21 (12.1) | 0 (0.0) |
| P1.5–1,10–8:F3–6 | 12 (4.3) | 14 (8.1) | 39 (44.3) |
| P1.5,2:F3–6 | 57 (20.6) | 18 (10.4) | 0 (0.0) |
| P1.5,2:F4–12 | 3 (1.1) | 28 (16.2) | 6 (6.8) |
| P1.5,2:F1–30 | 32 (11.6) | 1 (0.6) | 0 (0.0) |
| W Total Isolates | 27 | 22 | 40 |
| CC22 | |||
| P1.18–1,3:F4–1 | 17 (63.0) | 14 (63.6) | 11 (27.5) |
| CC11 | |||
| P1.5,2:F1–1 | 0 (0.0) | 2(9.1) | 24 (60.0) |
| Y Total Isolates | 319 | 207 | 84 |
| CC23 | |||
| P1.5–2,10–1:F4–1 | 167 (52.4) | 64 (30.9) | 17 (20.2) |
| P1.5–1,2–2:F5–8 | 97 (30.4) | 103 (49.8) | 35 (41.7) |
| CC167 | |||
| P1.5–1,10–4:F3–4 | 11 (3.4) | 9 (4.3) | 10 (11.9) |
Results for 2000–05 [17] and 2006–10 [18] were reported previously.
NmB isolate counts are represented as OR-corrected values. Only the genotypes showing statistically significant changes over time are shown.
FetA
Sixty four distinct FetA types were observed with 21 types in common between all three time periods (comprising 94.1% of isolates). In 2011–15, NmB exhibited the largest amount of diversity (24 types detected in NmB, 8 types detected in NmC and NmY, and 6 types detected in NmW). Two NmC isolates had FetA deletions. Analogous to PorA, the FetA types exhibited distinct profiles between serogroups (Fig. 2B). However, a few types overlapped multiple serogroups such as F4–1, which was found in NmY, NmW, NmB, and NmNG isolates. NmY and NmW had a significant change in FetA distribution between 2006–10 and 2011–15 (p < 0.005), while NmC isolates only had a significant change in FetA distribution when comparing 2000–05 and 2011–15 (p < .001), and no significant change in NmB was detected. The F4–1 decreased within NmY and NmW in 2011–15, consistent with the reduction of the NmY genotype CC23:P1.5–2,10–1:F4–1 and the NmW geno-type CC22:P1.18–1,3:F4–1 (Table 2). Furthermore, a large increase in F1–1 occurred in NmW (0.0% (2000–05), 9.1% (2006–10), and 60.0% (2011–15)) and was associated with the CC11:1.5,2:F1–1 genotype (Table 2 and Fig. 2B).
Genetic relatedness between U.S. and global strains
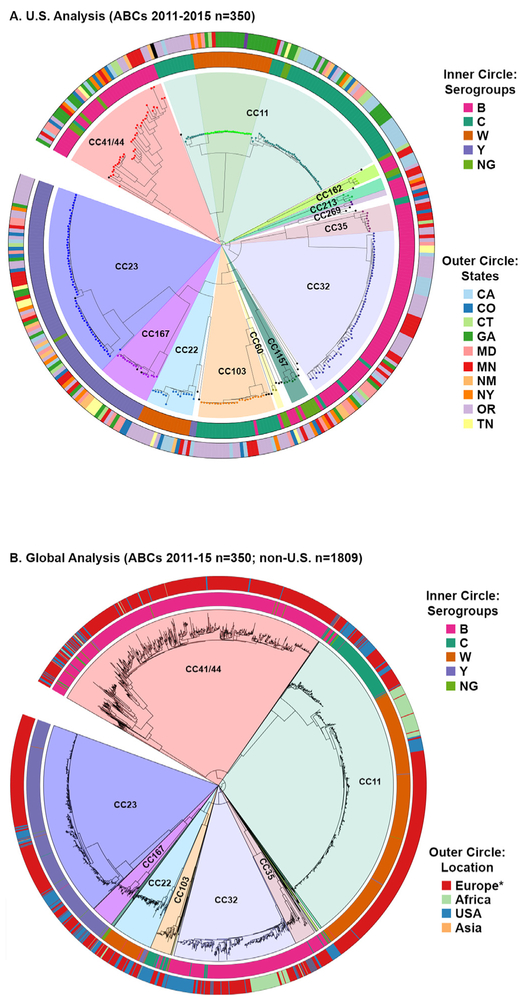
We assessed genetic relatedness between U.S. isolates by aligning the core genes present in all isolates and generating a maximum likelihood phylogeny (Fig. 3A). Isolates clustered by clonal complex, with each CC predominantly associated with a single serogroup (denoted by the inner circle). CC11 was comprised of two main branches. One contained predominantly NmC isolates and the second branch contained both NmC and NmW isolates. Most CCs showed no geographical association such as CC23 and CC4¼4, which were each detected in all 10 states (denoted by the outer circle). However, a few clonal complexes showed strong regional biases such as NmW CC11s, which were predominantly detected in eastern states (CT, GA, MD, MN, and NY) while the NmW CC22s were largely detected in the western states (CA, CO, MN, NM, OR, and TN).
Fig. 3.
Whole genome phylogenies for the (A) 2011–15 U.S. ABCs isolates alone or (B) the combined 2011–15 U.S. and global isolates associated with the main CCs circulating in the United States. Isolates from the same clonal complex cluster together and are highlighted by different colors. The inner circles depict the serogroup for each isolate and the outer circles represent the (A) state or (B) global region where the isolate was collected. *Most of European isolates (94%) are from the United Kingdom.
To better understand how U.S. isolates related to Nm circulating globally, we repeated the ML phylogeny analysis, adding non- U.S. isolates collected from 2011–15 and associated with the main U.S. CCs. The isolates clustered by CC, despite being collected from around the world (Fig. 3B). Notably, some CCs exhibited substantial intermixing of U.S. and global isolates such as CC4¼4 or CC22. Other CCs exhibited more distinct clustering of isolates based on their geographic origin such as CC23 or CC11.
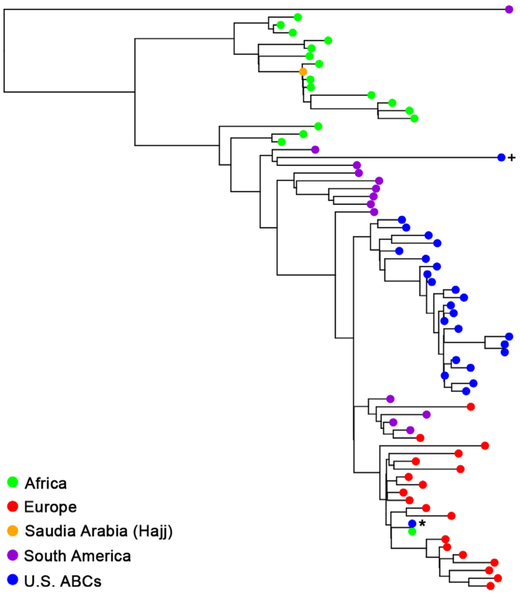
Finally, we completed a phylogenetic analysis specifically of NmW CC11 isolates to identify the most closely related lineage (Fig. 4). A total of 51 isolates representative of different NmW CC11 lineages were chosen including numerous African NmW CC11 lineages derived from the Hajj clone, the South American lineage, and the original and novel 2013 UK lineages depicted here as the European isolates.7,27,28 The Hajj clone and the closely related African lineages were not closely related to the 2011–15 ABCs NmW CC11 isolates. Nearly all (23/24) of the ABCs NmW CC11 isolates were most closely related to the South American lineage, clustering with isolates collected between 2008 and 2012 from Brazil, Chile and Argentina. Notably, one U.S. isolate, collected in 2012 from New York and denoted by the asterisks, was most closely related to a 2012 isolate from South Africa and present within the European clade.
Fig. 4.
Phylogenetic analysis of NmW CC11 isolates. The ABCs 2011–15 NmW CC11 isolates were compared to fifty-one isolates (collected 1997–2015) representative of the NmW African lineages, the South American, and UK lineages described previously (7, 27, 28). The ABCs isolate denoted by the (+) was collected in 2009 and was used as a positive control. The majority of U.S. isolates cluster together and are most similar to isolates from the South American lineage. One ABCs isolate, collected from NY in 2012 and denoted by the (*), clustered with the European isolates and was most closely related to a 2012 isolate from South Africa.
Capsule switching
We identified six isolates that switched from one serogroup to another (n = 5 B to C switches and n = 1 C to B switches). The B to C switch occurred within various clonal complexes (n = 1 CC4¼4, n = 1 C213, n = 1 CC35, n = 2 CC32) and the single C to B switch was CC103. WGS results verified these capsule switching events. The five NmC isolates that clustered with NmB contained the capsule locus genes cssE and csC but lacked csb, a serogroup B specific polymerase gene, while the NmB isolate that clustered with NmC had csb but lacked cssE and csC, The capsule switching events were detected in 1.7% of ABCs isolates 2011–15 (n = 5.1 of 308.33 total isolates).
Discussion
In this report, we provide a five year update characterizing the population structure of U.S. Nm isolates collected through the ABCs program during 2011–15. Although the U.S. meningococcal vaccination program targets adolescents, this analysis was completed on isolates collected from all ages because too few isolates were collected from the adolescent age group (n = 33). Results were compared to isolates collected in 2000–05 (no vaccine coverage) and in 2006–10 (low MenACWY coverage). Altogether, these results demonstrated that the U.S. N. meningitidis genetic structure has been relatively stable ten years after vaccine introduction. Dynamic changes in molecular profiles (ST, CC, and fine types) were observed in isolates of both vaccine targeted and non-targeted serogroups and thus, are likely representative of natural variation within the N. meningitidis population structure.
We used both classical molecular typing and high resolution phylogenetic comparisons. Using classical typing methods was essential to compare current data to historical isolates. Whole genome phylogenetic analyses allowed us to complete higher resolution genetic comparisons but notably, the phylogenetic analyses demonstrated that isolates clustered by CC, highlighting how accurately MLST predicts strain diversity.13 In the future, strain relatedness and population structure are expected to be assessed using WGS based methods.
NmB, NmC, and NmY remained the major cause of meningococcal disease during 2011–15, demonstrating that extensive serogroup replacement has not occurred after vaccine introduction. The proportion of capsule switching events detected in 2011–15 was lower than has been reported previously, which could at least partially be the result of the higher resolution phylogenetic methods used for detection.17,18 Widespread serogroup switching was not detected. Only one isolate exhibited capsule switching to the non-vaccine targeted NmB. Interestingly, we observed a statistically significant increase in the proportion of NmW isolates, which is targeted by MenACWY. However, 17 of the 40 isolates were collected in Georgia and may represent a localized increase and not a nationwide trend. This local increase is unlikely due to lack of vaccination because estimated MenACWY vaccine coverage within Georgia was high (87.0) and even better than the United States overall (81.3) in 2015.4 Other countries including Australia, the United Kingdom and the South American South Cone countries such as Chile and Argentina have reported an increase in NmW incidence in recent years, highlighting the need to assess NmW incidence using U.S. national data and these studies are currently ongoing.32–34
The population structure of N. meningitidis has been relatively stable after MenACWY introduction, with the majority of isolates from each time period belonging to common STs and CCs. However, dynamic trends in CC distribution within serogroups were detected. For example, an increase in CC103 within NmC, which is of particular interest because CC103 was recently associated with outbreaks and increased invasive disease in Brazil.35 The most notable change occurred within NmW, with a reduction of CC22 and a large increase in CC11 in 2011–15. The association of CC11 with NmW was highlighted by the 2000 Hajj outbreak and in the United States, CC11 caused a cluster of NmW disease in Florida in 2008–09.36,37 The NmW CC11 isolates detected in this study were most closely related to isolates from the South American lineage and were all the CC11:1.5,2:F1–1 genotype, indicative of a more clonal mode of expansion of CC11. The tight phylogenetic clustering of NmW CC11 isolates supports a possible clonal expansion, but distinct subclusters were detected, demonstrating that these isolates are still genetically distinct. Altogether, these results highlight that dynamic genetic variation continues to occur despite the overall stable Nm population structure.
We observed that some CCs exhibited distinct geographic distributions both within the United States and globally. In 2011–15, the U.S. NmW CC22s were associated with western states while the U.S. NmW CC11 isolates were detected in eastern states. In future years, it will be interesting to see if NmW CC11 spreads into the western states or remains geographically localized. To compare U.S. and global isolates, we analyzed isolate data from PubMLST, which provides comprehensive multi-country isolate genome data currently available but does not represent a population-based surveillance system. Globally, CCs such as CC11, CC23, and CC32 clustered more closely to isolates collected from the same continent, exhibiting regional associations. In contrast, CCs like CC4¼1 exhibited substantial genetic intermixing of isolates collected from different continents. The reason some CCs exhibit more clonal characteristics within a region is unclear. It is possible that some CCs are predisposed to undergo higher rates of horizontal gene transfer than others or more simply, this observation could be related to differences in transmission rates between various regions worldwide. For example, the United States predominantly experiences sporadic disease while many African countries experience extensive outbreaks, which could result in more clonal expansion of N. meningitidis.
In conclusion, the population structure of U.S. meningococcal disease has remained relatively stable with dynamic changes continuing to persist after high rates of vaccine coverage have been achieved 10 years after MenACWY introduction. Localized emergence of new strains such as NmW CC11 highlight the continued need for monitoring the molecular characteristics of N. meningitidis. WGS based methods enabled the discrimination between isolates of identical genotypes, like the NmW CC11:1.5,2:F1–1 isolates, and are expected to refine and improve our understanding of the Nm population structure in the future.
Supplementary Material
Acknowledgments
We would like to acknowledge the members of the ABCs team for their support and for providing the N. meningitidis isolates used in this study. Specifically, we would like to thank Arthur Reingold (California Emerging Infections Program), Lisa Miller (Colorado Emerging Infections Program), Sue Petit (Connecticut Emerging Infections Program), Monica Farley and Melissa Tobin-D’Angelo (Georgia Emerging Infections Program–Emory University and Atlanta Veterans Administration Medical Center), Ruth Lynfield (Minnesota Emerging Infections Program), Brooke Doman (New Mexico Emerging Infections Program), Shelley Zansky (New York Emerging Infections Program), Ann Thomas (Oregon Emerging Infections Program), and William Schaffner (Tennessee Emerging Infections Program). We would also like to thank the Office of Advanced Molecular Detection and the Biotechnology Core Facility Genome Sequencing Laboratory for their help and support. We thank the Laboratory Leadership Service Fellowship for their support of C.C. Potts during this study. And finally, we would like to thank our colleagues in the Bacterial Meningitis Laboratory and the MVPDB epidemiology team for their input and feedback related to this project.
Funding
This work was supported by the Centers for Disease Control and Prevention.
Footnotes
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Conflict of interest
Dr. Harrison has served on a scientific advisory board for GlaxoSmithKline.
Supplementary materials
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jinf.2018.06.008.
References
- 1.Bilukha OO. Rosenstein NNational Center for Infectious Diseases CfDC, Prevention Prevention and control of meningococcal disease. Recommendations of the Advisory Committee on Immunization Practices (ACIP). MMWR Recomm Rep 2005;54(RR-7):1–21. [PubMed] [Google Scholar]
- 2.Cohn AC. MacNeil JR. Clark TA. Ortega-Sanchez IR. Briere EZ. Meissner HC. et al. Prevention and control of meningococcal disease: recommendations of the Advisory Committee on Immunization Practices (ACIP). MMWR Recomm Rep 2013;62(RR-2):1–28. [PubMed] [Google Scholar]
- 3.Cohn A, MacNeil J. The changing epidemiology of meningococcal disease. Infect Dis Clin North Am 2015;29(4):667–77. [DOI] [PubMed] [Google Scholar]
- 4.Reagan-Steiner S, Yankey D, Jeyarajah J, Elam-Evans LD, Curtis CR, MacNeil J, et al. National, regional, state, and selected local area vaccination coverage among adolescents aged 13–17 years - United States, 2015. MMWR Morb Mortal Wkly Rep 2016;65(33):850–8. [DOI] [PubMed] [Google Scholar]
- 5.(CDC) CfDCaP National vaccination coverage among adolescents aged 13–17 years-United States, 2006. MMWR Morb Mortal Wkly Rep 2007;56(34):885–8. [PubMed] [Google Scholar]
- 6.Folaranmi T, Rubin L, Martin SW, Patel M, MacNeil JRCenters for Disease C Use of serogroup B meningococcal vaccines in persons aged >/=10 years at increased risk for serogroup B meningococcal disease: recommendations of the advisory committee on immunization practices, 2015. MMWR Morb Mortal Wkly Rep 2015;64(22):608–12. [PMC free article] [PubMed] [Google Scholar]
- 7.Retchless AC, Hu F, Ouedraogo AS, Diarra S, Knipe K, Sheth M, et al. The establishment and diversification of epidemic-associated serogroup W meningococcus in the African meningitis belt, 1994 to 2012. mSphere. 2016;1 (6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sidikou F, Zaneidou M, Alkassoum I, Schwartz S, Issaka B, Obama R, et al. Emergence of epidemic Neisseria meningitidis serogroup C in Niger, 2015: an analysis of national surveillance data. Lancet Infect Dis 2016;16(11):1288–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kretz CB, Retchless AC, Sidikou F, Issaka B, Ousmane S, Schwartz S, et al. Whole-Genome characterization of epidemic Neisseria meningitidis Serogroup C and resurgence of serogroup W, Niger, 2015. Emerg Infect Dis 2016;22(10):1762–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chow J, Uadiale K, Bestman A, Kamau C, Caugant DA, Shehu A, et al. Invasive meningococcal meningitis serogroup C outbreak in Northwest Nigeria, 2015 -Third consecutive outbreak of a new strain. PLoS Curr 2016:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trotter CL, Lingani C, Fernandez K, Cooper LV, Bita A, Tevi-Benissan C, et al. Impact of MenAfriVac in nine countries of the African meningitis belt, 2010–15: an analysis of surveillance data. Lancet Infect Dis 2017. [DOI] [PubMed] [Google Scholar]
- 12.Weinberger DM, Malley R, Lipsitch M. Serotype replacement in disease after pneumococcal vaccination. Lancet 2011;378(9807):1962–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maiden MC, Bygraves JA, Feil E. Morelli G, Russell JE, Urwin R. et al. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci USA 1998;95(6):3140–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caugant DA.. Genetics and evolution of Neisseria meningitidis: importance for the epidemiology of meningococcal disease. Infect Genet Evol 2008;8(5):558–65. [DOI] [PubMed] [Google Scholar]
- 15.Jolley KA, Brehony C, Maiden MC. Molecular typing of meningococci: recommendations for target choice and nomenclature. FEMS Microbiol Rev 2007;31(1):89–96. [DOI] [PubMed] [Google Scholar]
- 16.Maiden MC, Jansen van Rensburg MJ, Bray JE, Earle SG, Ford SA, Jolley KA, et al. MLST revisited: the gene-by-gene approach to bacterial genomics. Nat Rev Microbiol 2013;11(10):728–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harrison LH, Shutt KA, Schmink SE, Marsh JW, Harcourt BH, Wang X. et al. Population structure and capsular switching of invasive Neisseria meningitidis isolates in the pre-meningococcal conjugate vaccine era-United States, 2000–2005. J Infect Dis 2010;201(8):1208–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X, Shutt KA, Vuong JT, Cohn A, MacNeil J, Schmink S, et al. Changes in the population structure of invasive Neisseria meningitidis in the United States After quadrivalent meningococcal conjugate vaccine licensure. J Infect Dis 2015;211(12):1887–94. [DOI] [PubMed] [Google Scholar]
- 19.Langley G, Schaffner W, Farley MM, Lynfield R, Bennett NM, Reingold A, et al. Twenty years of active bacterial core surveillance. Emerg Infect Dis 2015;21(9):1520–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.(CDC) CfDCaP. National and state vaccination coverage among adolescents aged 13–17 years – United States, 2011. MMWR Morb Mortal Wkly Rep. 2012;61(34):671–7. [PubMed] [Google Scholar]
- 21.Mothershed EA, Sacchi CT, Whitney AM, Barnett GA, Ajello GW, Schmink S, et al. Use of real-time PCR to resolve slide agglutination discrepancies in serogroup identification of Neisseria meningitidis. J Clin Microbiol 2004;42(1):320–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 2012;19(5):455–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Treangen TJ, Ondov BD, Treangen S, Phillippy AM. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol 2014;15(11):524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stamatakis A, Aberer AJ, Goll C, Smith SA, Berger SA, Izquierdo-Carrasco F. RAxML-Light: a tool for computing terabyte phylogenies. Bioinformatics 2012;28(15):2064–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Didelot X, Wilson DJ. ClonalFrameML: efficient inference of recombination in whole bacterial genomes. PLoS Comput Biol 2015;11(2):e1004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jolley KA, Maiden MC. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinform 2010;11: 595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lucidarme J, Scott KJ, Ure R, Smith A, Lindsay D, Stenmark B. et al. An international invasive meningococcal disease outbreak due to a novel and rapidly expanding serogroup W strain, Scotland and Sweden, July to August 2015. Euro Surveill. 2016;21(45). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mustapha MM, Marsh JW, Krauland MG, Fernandez JO, de Lemos AP, Dunning Hotopp JC. et al. Genomic epidemiology of hypervirulent serogroup W, ST-11 Neisseria meningitidis. EBioMedicine 2015;2(10):1447–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harrison OB, Claus H, Jiang Y, Bennett JS, Bratcher HB, Jolley KA. et al. Description and nomenclature of Neisseria meningitidis capsule locus. Emerg Infect Dis 2013;19(4):566–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cohn AC, MacNeil JR, Harrison LH, Hatcher C, Theodore J, Schmidt M, et al. Changes in Neisseria meningitidis disease epidemiology in the United States, 1998–2007: implications for prevention of meningococcal disease. Clin Infect Dis 2010;50(2):184–91. [DOI] [PubMed] [Google Scholar]
- 31.Diermayer M, Hedberg K, Hoesly F, Fischer M, Perkins B, Reeves M. et al. Epidemic serogroup B meningococcal disease in Oregon: the evolving epidemiology of the ET-5 strain.JAMA 1999;281(16):1493–7. [DOI] [PubMed] [Google Scholar]
- 32.Carville KS, Stevens K, Sohail A, Franklin LJ, Bond KA, Brahmi A. et al. Increase in meningococcal serogroup W disease, Victoria, Australia, 2013–2015. Emerg Infect Dis 2016;22(10):1785–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abad R, Lopez EL, Debbag R, Vazquez JA. Serogroup W meningococcal disease: global spread and current affect on the southern Cone in Latin America. Epidemiol Infect 2014;142(12):2461–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ladhani SN, Beebeejaun K, Lucidarme J, Campbell H, Gray S, Kaczmarski E. et al. Increase in endemic Neisseria meningitidis capsular group W sequence type 11 complex associated with severe invasive disease in England and Wales. Clin Infect Dis 2015;60(4):578–5. [DOI] [PubMed] [Google Scholar]
- 35.Sardinha G, Cordeiro S, Gomes E, Romanelli C, Andrade C, Reis J, et al. Replacement of Neisseria meningitidis C cc11/ET-15 variant by a cc103 hypervirulent clone, Brazil 2005–2011. Diagn Microbiol Infect Dis 2013;76(4):524–5. [DOI] [PubMed] [Google Scholar]
- 36.Taha MK, Achtman M, Alonso JM, Greenwood B, Ramsay M, Fox A, et al. Serogroup W135 meningococcal disease in Hajj pilgrims. Lancet 2000;356(9248):2159. [DOI] [PubMed] [Google Scholar]
- 37.Doyle TJ, Mejia-Echeverry A, Fiorella P, Leguen F, Livengood J, Kay R. et al. Cluster of serogroup W135 meningococci, southeastern Florida, 2008–2009. Emerg Infect Dis 2010;16(1):113–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.