Abstract
Largely owing to widespread deployment of microarray analysis, many of the transcriptional events associated with invasive cell migration are becoming clear. However, the transcriptional drives to invasive migration are likely modified by alternative splicing of pre-mRNAs to produce functionally distinct patterns of protein expression. Heterogenous nuclear ribonucleoprotein (hnRNP A2) is a known regulator of alternative splicing that is upregulated in a number of invasive cancer types. Here we report that, although siRNA of hnRNP A2 had little influence on the ability of cells to migrate on plastic surfaces, the splicing regulator was clearly required for cells to move effectively on 3D matrices and to invade into plugs of extracellular matrix (ECM) proteins. We used exon-tiling microarrays to determine that hnRNP A2 controlled approximately 6 individual splicing events in a 3D matrix-dependent fashion, one of which influenced invasive migration. Here we show that alternative splicing of an exon in the 5′UTR of a gene termed TP53INP2 is a key event downstream of hnRNP A2 that is necessary for cells to invade the ECM. Furthermore, we report that the consequences of altered TP53INP2 splicing on invasion are likely mediated via alterations in Golgi complex integrity during migration on 3D matrices.
Keywords: hnRNP A2, cell migration, alternative splicing, invasion, cancer, Golgi, TP53INP2
INTRODUCTION
One of the features of malignant cells, and one that makes cancer so difficult to treat, is their capacity to migrate invasively through the stroma to form metastases (1). Largely due to the use of microarrays to identify gene expression changes, many of the transcriptional events that drive cancer invasion are now clear. However, much of the complexity of gene expression is generated by mechanisms that act post-transcriptionally. One mechanism for deriving multiple proteins from a single gene is alternative splicing, and recent analyses have indicated that >90% of human genes are subjected to alternative splicing (2, 3). Alterations in mRNA splicing can alter gene expression in a way that contributes to acquisition of an invasive phenotype. A number of splicing-related mutations have been associated with malignancies, and some of these are within splice sites or splicing enhancers/silencers of cancer-related genes. For instance, ‘cis-acting’ mutations have been identified within splice sites of genes for DNA repair (BRCA1; (4)), oncogenic kinases (c-Kit; (5)) and cell-cell adhesion molecules (LI-cadherin; (6)), and these alter splicing in a way that likely contributes to malignant disease. Additionally, a number of altered splicing patterns that are not necessarily associated with mutations in the target gene itself are associated with cancer progression (reviewed in (7-9)). In some cases, these ‘trans-acting’ events may drive cancer progression by increasing cell migration and invasion. For instance, Ron is a receptor tyrosine kinase that can promote cell migration, and alterations in its splicing produces a constitutively active form of the kinase (ΔRon) that increases invasiveness (10). Furthermore Rac1b, an alternatively spliced form of the small GTPase Rac1 is generated by treatment of normal mammary epithelial cells with a matrix metalloprotease (MMP-3), and this may contribute to enhanced cell migration as these cells undergo epithelial to mesenchymal transition (EMT) (11). There are other examples of cancer-related splicing patterns of genes such as fibronectin, integrin β1, CD44 and uPAR, but the way in which these splicing events are regulated, and how they might drive cancer cell migration is currently unclear.
The way in which genes are spliced is dictated by the specific binding of proteins to regulatory elements within their mRNAs. Heterogeneous nuclear ribonucleoproteins (hnRNPs) are a family of proteins with central roles in processes such as telomere biogenesis, mRNA stability and turnover, cytoplasmic trafficking of mRNAs, and many hnRNP proteins participate in splicing control (12). Out of all the hnRNPs, a growing body of literature indicates that hnRNP A2 (and its splice variants hnRNP B1/A2b/B1b; hereafter referred to collectively as hnRNP A2) is up-regulated or mis-localised in human cancers (lung (13), colon (14), breast (15), pancreatic (16), and stomach (17) carcinoma) and tumour-derived cell lines, and that hnRNP A2 can be used as a marker of poor prognosis in lung cancer (13). hnRNP A2 has been implicated in the splicing of a number of genes (18) and these events may contribute to the progression of tumours in which hnRNP A2 is up-regulated. Here we describe a requirement for hnRNP A2 in cancer cell invasiveness, and have used genome-wide exon-tiling microarrays to identify hnRNP A2-dependent splicing events that occur only when cells are plated into 3D microenvironments. We have found that matrix-specific alternative splicing of an exon in the 5′ UTR of TP53INP2 (tumour protein p53 inducible nuclear protein 2) is a key event downstream of hnRNP A2 that is necessary for cancer cells to invade the ECM.
METHODS AND MATERIALS
Cell biological methods
The A2780 ovarian carcinoma cell line which was used for the exon array analysis, time lapse microscopy, immunofluorescence and invasion assays (as indicated) was cultured and transfected as described previously (19, 20). The BE colon carcinoma cells which were used for invasion assays as indicated were transfected using GeneFector (Venn Nova, USA). Inverted invasion assays were performed as described previously (21). Cell-derived matrix was generated as described previously (22, 23).
Time-lapse microscopy
A2780 cells were seeded onto cell-derived matrix-coated 6-well plates and incubated at 37°C until cells adhered and began migrating (8-10 hours). Cells were imaged with a 10x objective and an inverted microscope (Axiovert S100, Carl Zeiss MicroImaging, Inc.) in an atmosphere of 5% CO2 at 37 °C. Cells were imaged at 5 minute intervals for 500 minutes. Andor IQ and Tracker software were used to track cell nuclei.
Immunofluorescence
Cells were fixed briefly in ice-cold methanol, permeabilised with 0.2% (v/v) Triton X-100 in PBS for 5 min, and blocked with 1% BSA in PBS for 1-2 hours. Primary antibodies were; γ-tubulin (T5192; Sigma-Aldrich, UK) or gm130 (610822; BD, UK) in blocking solution at 4 °C overnight. Detection was with Alexa-fluor conjugated secondary antibodies (488 and 594; Invitrogen, UK), and confocal imaging performed (Fluoview FV1000, Olympus).
siRNA duplexes
Short RNA duplexes against hnRNP A2/B1 were A2#1: 5′ GGAUUAUUUAAUAACAUU A 3′ and A2#2: 5′ GGAGAGUAGUUGAGCCAAA 3′ respectively. The exon specific siRNAs were: TP53INP2 exon 2 (E2#1): 5′ UUGAAGUCCUAGAGUCC 3′; and TP53INP2 exon 2 (E2#2): 5′ GGAGAUUGGUUCACCUU 3′. MTA3 exon 5 (MTA3(E5)): 5′GGAUAGAAGAACUCAACAAUU 3′; and EPB41L4A exon 11 (EPB41L4A(E11)): 5′CCAAUUCACUGUCAAGAAAUU3′; MAP9 exon 4 (MAP9(E4)): 5′CCAAUAAAUCAAACGGUA3′. Dharmacon’s non-targetting smartpool was used as a negative control.
qRT-PCR
cDNA was prepared using Promega’s (WI, USA) ImpromII kit, and the oligo dT primer. Quantitative PCR was performed using SYBR green (Finnzymes, New England Biolabs, UK) and a Chromo4 DNA Engine (Biorad, UK). ΔΔC(t) was determined as described by Livak and Schmittgen (24), using β-actin as a reference point. Primers were: β-actin: Forward-5′AGCCATGTACGTAGCCATCC3′; Reverse-5′CTCTCAGCTGTGGTGGTGAA3′, amplicon 250 nt. TP53INP2 exon 5: Forward-5′CCTGTTCCCTTATTCTTCATTCC3′; Reverse-5′ATTCCCTCCATCTTCTCCCT3′, amplicon 253 nt. TP53INP2 Excluding exon 2: Forward-5′TCCCGCCCCCAGGTTTTT G3′; Reverse-5′CCAGCCGTCCACTTCATC3′, amplicon 166 nt. TP53INP2 Including exon 2: Forward-5′CCTCACTGTACCTTGAAGTC3′; Reverse-5′CTGAAGAAGAGGCTGGAGAG3′, amplicon 149 nt. MTA3 exon 5: Forward-5′ATTCCTCCAGCAACCCATACC3′; Reverse-5′TAGCATGCTTATCTGCGAGC3′, amplicon 144 nt. EPB41L4A exon 11: Forward-5′CAGCAAGTTTGGATCCATACG3′; Reverse-5′CTTAGGGTAAGTCTTGCTTCG3′, amplicon 136 nt. MAP9 exon 4: Forward-5′GGCACCTGATGGGTGTGAAG3′; Reverse-5′CTGTGTCAAGGCTGTTGTTTTC3′, amplicon 120 nt.
Exon-tiling microarray
Triplicate samples were derived from A2780 cells nucleofected with either the Con or A2#1 siRNA, which had been plated onto CDMs for 18 hours. RNA was extracted using Qiagen’s RNeasy kit. Biotinylated target cRNA was generated and hybridised onto Affymetrix exon arrays using the HuEx-1_0-st-v2 chips by the Cancer Research UK Paterson Institute Microarray Service. Data was normalised using the Robust Multi-Array Average algorithm, and non-paired t-tests carried out between triplicates to find probe sets with expression differences of +/- 2 (p-value 0.01). Splice indices were determined using the equation: splice index = (probe set intensity in sample 1/median gene intensity in sample 1)/(probe set intensity in sample 2/median gene intensity in sample 2), and probe sets with a splice index >1 or <-1 (which also fit the previous fold change and p-value criteria) identified. We have deposited the array data generated on the Affymetrix MIAMEVICE web page.
RESULTS
hnRNP A2 is required for invasive migration of tumour cells
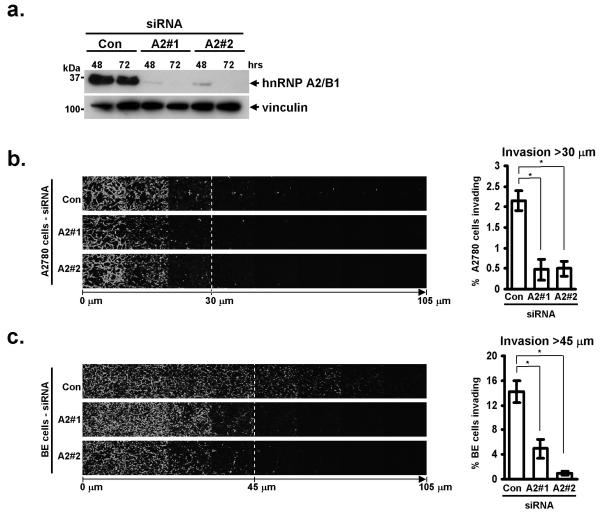
To suppress the cellular levels of hnRNP A2 we utilised two independent siRNA sequences (A2#1 and A2#2) which targetted all known isoforms of hnRNP A2. Both siRNAs suppressed the expression of hnRNP A2 in A2780 ovarian carcinoma (Fig. 1a) and BE colon cancer (not shown) cells, and the knockdown was stable for at least 72 hr (Fig. 1a). We did not detect differences in A2780 cell migration on plastic surfaces following knockdown of hnRNP A2 (not shown), indicating it was not required for the execution of basic processes of cell migration such as actin polymerisation. But there are key differences between the characteristics of cells migrating in 3D-versus on 2D-matrices, and experimental systems measuring migration across plastic surfaces may not accurately model the type of motility that would be deployed by a tumour cell to move away from the primary tumour and form metastases at distant sites. To elucidate the potential contribution that hnRNP A2 may make to the invasive phenotype of aggressive tumours, we employed an inverted invasion assay, in which cells migrate upward through Matrigel (predominantly a mixture of laminin and collagen IV) supplemented with fibronectin (FN) toward a gradient of serum and epithelial growth factor (EGF). Both A2780 (Fig. 1b) and BE cells (Fig. 1c) migrated efficiently into Matrigel/FN plugs, and this index of invasiveness was strongly opposed by knockdown of hnRNP A2 with two independent siRNA sequences.
Figure 1. siRNA knockdown of hnRNP A2 decreases invasiveness of A2780 and BE cells.
(a) A2780 cells were transfected with siRNA oligonucleotides targetting hnRNP A2 (A2#1, or A2#2), or a non-targeting siRNA (Con). Knockdown was confirmed by Western blotting for hnRNP A2, or vinculin as a loading control at 48 and 72 hours post transfection.
(b, c) Invasive migration of A2780 (b) or BE (c) cells into plugs of Matrigel supplemented with fibronectin was determined using an inverted invasion assay. Invading cells were stained with Calcein-AM and visualised by confocal microscopy. Serial optical sections were taken at 15 μm intervals, and are presented as a sequence in which the individual optical sections are placed alongside one another with increasing depth from left to right, as indicated. Invasion assays were quantitated by measuring the fluorescence intensity of cells penetrating the Matrigel to depths of >30 μm (b) and >45 μm (c), this intensity is expressed as a percentage of the total fluorescence intensity of all cells within the plug. Data represent mean ± SEM, n>4. * indicates significant difference of p<0.003 (Mann-Whitney test for non-parametric data).
hnRNP A2 disrupts Golgi morphology and opposes cell migration on 3D matrices
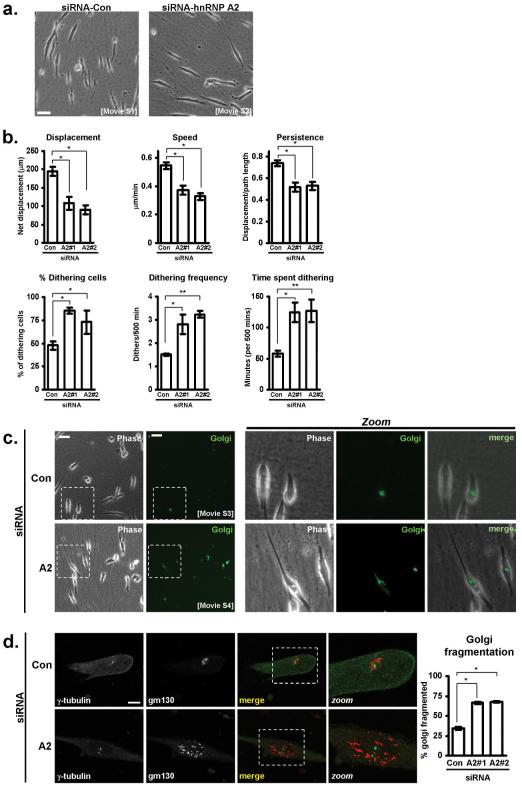
To study the role of hnRNP A2 on cell migration in the context of a 3D microenvironment, we used cell-derived matrix (CDM); a relatively thick, pliable matrix composed mainly of fibrillar collagen and fibronectin which recapitulates key aspects of the type of matrix found in connective tissues (23). When plated onto CDM, A2780 cells assumed a ‘slug-like’ morphology (20), and migrated with a single leading lamellipodium and a rounded rear end (Fig. 2a; Movie S1). However, following knockdown of hnRNP A2, A2780 cells had altered morphology (Fig. 2a) and reduced indices of migration (displacement, average speed, and persistence) (Fig. 2b). Indeed, hnRNP A2 knockdown cells lacked a rounded cell rear and frequently were seen to extend pseudopods at both ends (Fig. 2a; Movie S2). Furthermore, whereas control A2780 cells advanced consistently whilst maintaining front to back asymmetry, hnRNP A2 knockdown cells frequently stopped migrating and succumbed to prolonged episodes of hesitation or ‘dithering’ during which pseudopod dominance was suppressed (Fig. 1a; Movie S2). To obtain a quantitative index of hesitation, we defined a ‘dither’ as an episode of at least 30 minutes during which a cell moved less than 2 μm. This analysis revealed that knockdown of hnRNP A2 increased the proportion of cells that dithered during the time-lapse period (500 min) cells by approximately two-fold (Fig. 2b). Moreover, hnRNP A2 knockdown cells dithered more frequently and for longer than those transfected with control siRNAs (Fig. 2b).
Figure 2. hnRNP A2 knockdown disrupts Golgi morphology and migration of A2780 cells on cell-derived matrix.
(a) A2780 cells transfected with either a control (Con) siRNA, or one targeting hnRNP A2 (A2) were seeded onto cell-derived matrices, and allowed to adhere for approximately 10 hours prior to time lapse microscopy. Frames were captured every 5 minutes over a 500 minute period, and movies generated from these images (Movies S1 and S2). These images are representative stills from the movies. Bar, 50 μm.
(b) Cell tracking software was used to determine cell displacement, migrational persistence, and average speed of migration of A2780 cells on cell-derived matrix following either control (Con) siRNA, or knockdown of hnRNP A2 using two independent oligonucleotides (A2#1 and A2#2). By defining a ‘dither’ as a period of at least 30 minutes in which a cell moves less than 2 μm, the incidence and frequency of cells dithering within a 500 minute period was determined. Also shown is the amount of time during the 500 minute period that the cells spent dithering. Data represent mean ± SEM. * indicates significant difference of p<0.005 and ** indicates p<0.05 (Mann-Whitney test).
(c) A2780 cells were transfected with either a control (Con) or hnRNP A2-targetting siRNA (A2), in combination with a fluorescently-tagged Golgi marker and seeded onto cell-derived matrix. Approximately 10 hours after seeding, timelapse microscopy was used to capture phase-contrast (phase) and fluorescent (Golgi) frames every 5 minutes for a 500 minute time period, and movies were generated from these (Movies S3 and S4). The presented images are representative stills from the movies. Bar, 50 μm.
(d) A2780 cells were transfected with either a control (Con) or hnRNP A2-targetting (A2) siRNA, seeded onto cell-derived matrix, and allowed to adhere for 24 hours before being fixed and stained for γ-tubulin and the Golgi marker, gm130. Bar, 20 μm. The graph shows quantification of proportion of cells with scattered or fragmented Golgi complexes following transfection with either a control (Con) or two hnRNP A2-targetting (A2#1 and A2#2) siRNAs. Data represents mean ± SEM. * indicates statistical significance of p=<0.0001 (Mann-Whitney test).
The positioning of the Golgi complex along with the microtubule organising centre (MTOC) is known to be associated with generation of cell polarity during cell migration (25). The asymmetric bipolar phenotype and reduction of pseudopod dominance induced by hnRNP A2 knockdown likely reflected a defect in establishment of cellular polarity. We therefore investigated the requirement for hnRNP A2 in Golgi complex and MTOC positioning during cell migration. To visualise the Golgi complex in migrating cells we expressed a fluorescent version of galactosyl N-acetyl transferase; an enzyme that is resident to the trans-Golgi stacks (26). In control cells, the Golgi complex was seen as a single condensed structure located in the perinuclear region anterior to the direction of migration (Fig. 2c; Movie S3). However, knockdown of hnRNP A2 led to scattering of Golgi membranes with the fragmentation being most apparent when cells were dithering (Fig. 2c, d; Movie S4). The positioning of the Golgi complex is thought to be dependent on the integrity of the MTOC. However, even in cells that displayed extensive disruption and scattering of Golgi membranes, the MTOC (Fig. 2d) and overall microtubular organisation (not shown) was not noticeably altered. Taken together these data indicate that hnRNP A2 plays a key role in generating appropriate organisation of the Golgi complex (and not the MTOC), and that this is required for polarised cell migration on cell-derived matrices and for tumour cell invasion into 3D microenvironments.
hnRNP A2 regulates 3D matrix-dependent alternative splicing of TP53INP2
hnRNP A2 is a multi-functional protein, and one of its key roles is to control alternative splicing via both exon skipping and exon inclusion events (12). A recent study characterising the requirement for a range of hnRNP proteins in alternative splicing of apoptotic genes has indicated that the splicing events controlled by hnRNP proteins varies between cell lines (27). These indications that hnRNP-regulated splicing is context dependent, in combination with our findings that hnRNP A2 affects cell migration in a 3D matrix-dependent fashion prompted us to screen for hnRNP A2-regulated alternative splicing events in both 2D and 3D environments. To do this we plated control or hnRNP A2 knockdown cells onto either plastic or CDMs and compared their alternative splicing profiles using Affymetrix HuEx-1_0-st-v2 exon-tiling chips. These arrays comprise approx. 5 million probes grouped into 1.4 million probesets, interrogating over 1 million exon clusters (28). Using expression differences of +/- 2 (for individual probe sets), and splice indices of >1 or <-1 (relative to probe sets in different exons within the same gene), six genes were identified as having altered splicing patterns between control and hnRNP A2 knockdown cells plated onto cell-derived matrix. Genes that displayed significantly altered splicing indices following hnRNP A2 knockdown are listed in table 1 and splice maps corresponding to these data are displayed in supplementary figure S1. No alterations in splicing of these genes was detected when cells were plated onto plastic (not shown).
Table 1.
A2780 cells transfected with either a control or hnRNP A2-targetting siRNAs were seeded onto cell-derived matrices, and allowed to adhere for 18 hours before RNA was harvested. RNA was labelled and hybridised to Affymetrix HuEx-1_0-st-v2 chips. Genes containing probesets with ± 2-fold change between the two conditions, and with splice indices of >1 or <-1 were mapped to known exons, generating a list of six genes whose splicing is altered by the loss of hnRNP A2/B1
| Probe set ID |
Gene ID | Gene name | Effect of hnRNPA2 knockdown |
Fold change |
Splice index |
p-value |
|---|---|---|---|---|---|---|
| 2478948 | NM_020744 | Metastasis associated gene 3 (MTA3) |
Skipping of exon 5 |
↓ 3.6 | -1.77 | <0.01 |
| 2790849 | NM_001039580 | Microtubule- associated protein 9 (MAP9) |
Skipping of exon 4 |
↓ 3.4 | -1.48 | <0.005 |
| 2871029 | NM_022140 | Erythrocyte membrane protein band 4.1 like 4A (EPB41L4A) |
Skipping of exon 11 |
↓ 2.4 | -1.01 | <0.002 |
| 3883017 | NM_021202 | Tumor protein p53 inducible nuclear protein 2 (TP53INP2) |
Skipping of exon 2 |
↓ 2.2 | -1.10 | <0.01 |
| 2749508 | NM_021634 | Relaxin receptor 1 (RXFP1) |
Inclusion of exon 1 |
↑ 2.7 | +1.50 | <0.005 |
| 2990076 | NM_001007157 | PHD finger protein 14 (PHF14) |
Inclusion of exon 3 |
↑ 4.0 | +2.00 | <0.01 |
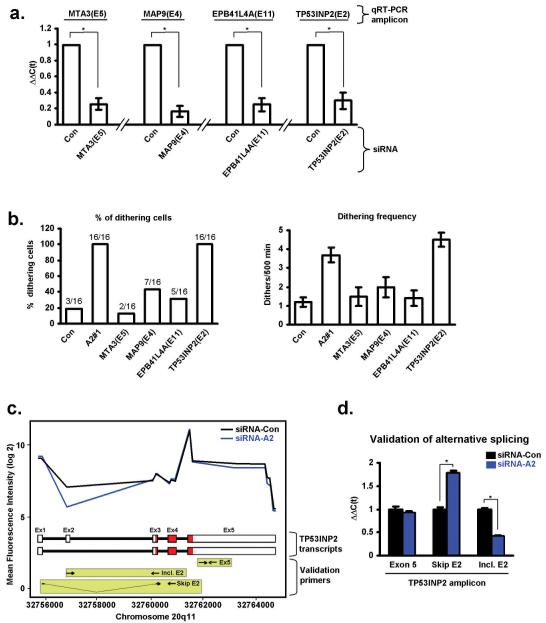
As can be seen from the data in table 1 and their corresponding splice maps (supplementary fig. S1), knockdown of hnRNP A2 promoted 2 exon inclusion and 4 exon skipping events. To determine whether any of these four exon skipping events were responsible for hnRNP A2’s influence on cell migration, we designed siRNA oligonucleotides to target the differentially spliced exons of MTA3 (exon 3), MAP9 (exon 4), EBP41L4A (exon 11) and TP53INP2 (exon 2) and transfected them into A2780 cells. Then, having confirmed that these siRNA duplexes were effective to reduce expression of the appropriate targetted exon (Fig. 3a), we assessed migration on cell-derived matrix. Of these 4 validated siRNA oligonucleotides, only the one targetting exon 2 of TP53INP2 recapitulated the migratory phenotype (high incidence of dithering) seen following knockdown of hnRNP A2 (Fig. 3b).
Figure 3. Knockdown of hnRNP A2 leads to altered mRNA splicing of TP53INP2.
(a) siRNAs were designed to target the following exons: MTA3(E5), MAP9(E4), EPB41L4A(E11) and TP53INP2(E2). siRNAs were transfected into A2780 cells and the cells plated onto cell-derived matrix. The efficacy of the siRNA knockdowns was confirmed using qRT-PCR to amplify the appropriate exons (qRT-PCR amplicon). Values are mean ±SEM, * indicates significant difference of p<0.01 (Mann-Whitney test).
(b) siRNAs targetting MTA3(E5), MAP9(E4), EPB41L4A(E11) and TP53INP2(E2) were transfected into A2780 cells. These were then seeded onto cell-derived matrices and time-lapse microscopy was used as before to measure the incidence of dithering.
(c) Fluorescence intensity of labelled RNA derived from Con (black line) and hnRNP A2 (blue line) knockdown A2780 cells following hybridisation to probe sets representing exons from the TP53INP2 gene. The schematic beneath indicates the exon/intron structure of two known TP53INP2 transcripts (ENST00000374810 (exon 2 inclusive; top) and ENST00000374809 (exon 2 skipping; bottom)). Primers used for qRT-PCR to report on exon 2 inclusive (Incl. E2), exon 2 skipping (Skip E2) and exon 5 containing (Ex. 5) are indicated in the light green boxes. The coding regions of the transcripts are in red.
(d) qRT-PCR validation of the exon-tiling microarray. RNA was extracted from Con (black bars) and hnRNP A2 (blue bars) knockdown A2780 cells and analysed forTP53INP2 transcripts using qRT-PCR. The primers indicated in the green boxes in (c) were used to amplify portions of exon 5 (common to both known TP53INP2 transcripts), and exon 2 skipping (Skip E2) or exon 2 inclusive (Incl. E2) transcripts. Values are mean ±SEM, * indicates significant difference of p<0.01 (Mann-Whitney test).
Examination of the fluorescence intensity of the exon array probesets with reference to the genomic structure of TP53INP2 (Fig. 3c) indicated that siRNA knockdown of hnRNP A2/B1 (blue line) decreased the number of exon 2-containing transcripts. To confirm this, we designed primers (schematically indicated in the light green boxes in Fig. 3c) to amplify either a portion of exon 5 (Ex 5, common to both transcripts), exon 2-skipping (Skip E2) transcripts, or exon 2-including (Incl. E2) transcripts, and performed qRT-PCR. This quantitative analysis confirmed that knockdown of hnRNP A2 significantly reduced the number of TP53INP2 transcripts containing exon 2, and correspondingly increased the level of exon 2-excluding transcripts (Fig. 3d). Furthermore, as indicated by the use of PCR primers reporting on exon 5 (Ex 5), the total level of TP53INP2 transcripts (exon 2 inclusive plus exon 2 exclusive) was unaltered by hnRNP A2 knockdown (Fig. 3d).
The influence of hnRNP A2 on cell migration and invasion is mediated via alternative splicing of TP53INP2’s exon 2
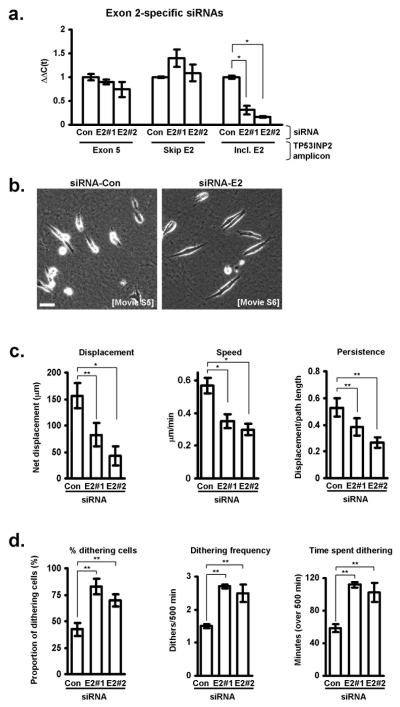
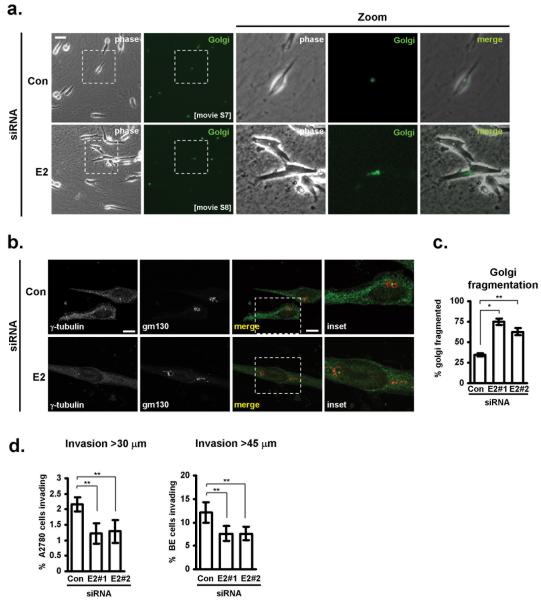
To fully investigate the role of TP53INP2’s alternative splicing in cell migration, we needed to selectively suppress the expression of exon 2-containing TP53INP2 transcripts. We designed two siRNA oligonucleotides to target TP53INP2’s exon 2 (E2#1 and E2#2) and assessed their efficacy and selectivity using qRT-PCR. Both of these siRNAs significantly reduced the level of exon 2-containing TP53INP2 transcripts (Incl. E2), without affecting expression of those excluding exon 2 (Skip E2 & Exon 5) (Fig. 4a). Using these two validated exon-specific siRNAs, we proceeded to investigate the role that exon 2-containing TP53INP2 transcripts had on cell migration and invasion. Interestingly, we found that selective suppression of exon 2-containing TP53INP2 transcripts recapitulated the same cellular phenotype as was seen following knockdown of hnRNP A2. Indeed, in cells with exon 2-specific TP53INP2 knockdowns we consistently observed: a) reduced migration speed and persistence on cell-derived matrices (Fig. 4c; Movies S5&S6); b) increased dithering and dilatory migratory behaviour comprising episodes of reduced back/front polarity (Fig. 4b,d; Movies S5&S6); c) scattered Golgi membranes particularly in cells that were dithering (Fig. 5a,b,c; Movies S7&S8); and d) reduced invasion into Matrigel for both the A2780 and BE tumour cell lines (Fig. 5d).
Figure 4. Knockdown of exon 2 inclusive TP53INP2 transcripts affects migration on cell-derived matrices.
(a) Two siRNAs (E2#1 & E2#2) were designed to target exon 2 of TP53INP2 and their influence on expression of exon 2 inclusive (Incl. E2), exon 2 skipping (Skip E2) transcripts and total expression of TP53INP2 (Exon 5; common exon) was determined using quantitative RT-PCR. Values are mean ±SEM, * indicates significant difference of p<0.03 (Mann-Whitney test).
(b-d) A2780 cells transfected with either a control (Con) siRNA, or one targeting exon 2 inclusive transcripts of TP53INP2 (E2) were seeded onto cell-derived matrices, and their migratory characteristic analysed and quantified as for figure 2 b-c. Bar in b, 50 μM. Data represent mean ± SEM. * indicates significant difference of p<0.001 and ** indicates p<0.05 (Mann-Whitney test).
Figure 5. Exon 2-containing TP53INP2 transcripts are required to support Golgi morphology and invasive migration of tumour cells.
(a-c) A2780 cells were transfected with either control (Con) siRNAs or those targetting exon 2-containing TP53INP2 transcripts (E2), in combination with a fluorescently-tagged Golgi marker and seeded onto cell-derived matrix, and their Golgi morphology analysed and quantified as for figure 2 c&d. Bar in a, 50 μm. Bar in b, 20μm. Data represents mean ± SEM. * indicates statistical significance of p=<0.0001 and ** indicates p<0.05 (Mann-Whitney test).
(d) A2780 or BE cells were transfected with either control (Con) siRNAs or those targetting exon 2-containing TP53INP2 transcripts (E2) and their invasion into plugs of Matrigel supplemented with fibronectin was determined using an inverted invasion assay. Invasion assays were quantitated by measuring the proportion of cells within the plug that penetrate the Matrigel to depths of >30 μm (for A2780 cells) and >45 μm (for BE cells). Data represent mean ± SEM, n>4. ** indicates significant difference of p<0.05 (Mann-Whitney test for non-parametric data).
DISCUSSION
The role of hnRNP A2 in alternative splicing of TP53INP2 transcripts
Although there is well-documented redundancy between the splicing activities of hnRNPs A1 and A2 (18), our data is consistent with a recent report indicating that the function of these two proteins do not completely overlap (27). Indeed, we find that suppression of hnRNP A2 alone is sufficient to generate a migratory phenotype without altering the levels or splicing of hnRNP A1 or by regulating splicing of established hnRNP A1 targets such as caspase-2 or c-Src (29, 30). hnRNP A2 promotes inclusion of the TP53INP2 exon 2 which is located in the 5′ UTR. Furthermore this event was completely dependent on cells being within a flexible 3D microenvironment, and was not detected when cells were plated onto a rigid substrate. It is now clear that alternative splicing is influenced by extracellular signals via the activation of various signalling pathways (31). Although examples of direct links between signalling kinases and the splicing machinery are somewhat rare, it has recently been reported that hnRNP A2 is phosphorylated by the Src-family kinase, Fyn (32). Src signalling is strongly promoted on CDMs (33), and it will be interesting to determine whether matrix-dependent phosphorylation of hnRNP A2 by Src family kinases and other pathways influenced by the ECM environment, alters its capacity to promote exon 2-inclusive splicing of TP53INP2.
As 5′ UTRs are known to direct the efficiency of translation, it is likely that hnRNP A2 splicing will dictate the levels of TP53INP2 protein within the cell. Indeed, inspection of the TP53INP2 5′ UTR sequence indicates the presence of a CCUCCC motif which could suggest that a functional IRES (34) may reside within exon 2. We have found that siRNAs targetting sequences within the coding region and 3′ UTR of TP53INP2 affect cell migration in a way that is similar to exon 2-specific knockdowns (not shown). Such striking similarity between the consequences of transfecting siRNAs that specifically target exon 2-specific and those that target a coding exon (that would be expected to suppress overall TP53INP2 protein levels) suggests that exon 2-containing transcripts are more efficiently translated than those lacking this exon.
How does alternative splicing of TP53INP2 influence Golgi integrity and cell migration?
TP53INP2 was originally described as a gene involved in mammalian craniofacial development (35). Subsequent independent studies have found TP53INP2 to be differentially expressed in diabetic rats, and possibly to influence the progression of diabetes by binding to the thyroid hormone receptor and functioning as a transcriptional co-activator within the nucleus (36). More recently, TP53INP2 has been found to have a role outwith the nucleus. Following induction of autophagy, TP53INP2 has been seen to relocate from the nucleus to autophagic vacuoles, and thereupon promote recruitment of the autophagy mediator, LC3, to these structures (37). Although this study highlights a potential role for TP53INP2 outwith the nucleus, the overexpression approaches employed by these workers will have masked the influence of alternative splicing on its function. Our results highlight the need to determine the influence of regulated inclusion/skipping of exon 2 on TP53INP2 protein synthesis and localisation with respect to cytoplasmic structures such as autophagosomes and the Golgi complex.
Although mislocalisation of the Golgi complex often accompanies defects in polarised cell migration, it has long been debated whether this is a cause or a consequence of aberrant cell polarity. More recent studies have shown that manipulations designed specifically to target Golgi structural proteins are themselves sufficient to disturb cell migration, most likely by disrupting polarised secretion to regions such as the leading edge of migrating cells (38-40). The scattering and fragmentation of Golgi membranes that we observe following knockdown of hnRNP A2 or TP53INP2 exon 2-containing transcripts is similar to that which has been reported following interference with Golgi structural proteins such as golgin-160 (40), GRASP65 (38) or GM130 (39). This indicates that TP53INP2 may play a direct role in regulating Golgi integrity, and that its influence on cell migration is a consequence of this. It is interesting to note that VMP1, which was identified by Nowak et al. (37), as a binding partner for TP53INP2, has been reported to be localised to the Golgi (41) and has been independently identified in an siRNA screen as a novel regulator of Golgi function (42). VMP1 has also been proposed to play a role in cell-cell adhesion and metastasis (43). We are, therefore, considering the possibility that loss of exon 2-inclusive TP53INP2 transcripts leads to reduced levels and/or mis-localisation of TP53INP2 and that this alters Golgi function and thereby cell migration via its interaction with VMP1. Indeed, we have found that knockdown of hnRNP A2 reduces levels of VMP1 (not shown), consistent with a mechanism whereby exon 2 of TP53INP2 is required for efficient translation of the TP53INP2 which, in turn, acts to stabilise VMP1 to support Golgi function during cell migration.
To conclude, hnRNP A2 isoforms have been documented as both up-regulated and mis-localised in a number of cancer types, but it has hitherto been unclear how this might contribute to the malignant phenotype. The metastatic potential of tumour cells is dictated as much by their migratory potential as by their proliferative and/or apoptotic indices. Our description of the alternative splicing of TP53INP2 and its influence on cell migration provides the first evidence that a hnRNP-mediated splicing event contributes to tumour cell invasiveness, and we propose that this mechanism may account for the correlation between high levels of hnRNP A2 and poor cancer outcomes.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by Cancer Research UK and by the BBSRC. We would like to thank Sian Dibben, Yvonne Hey and Stuart Pepper of Cancer Research UK’s Paterson Institute microarray facility for their invaluable assistance with the analysis of the exon-tiling array data. We would like to thank Margaret O’Prey, Tom Gilbey and David Strachen of the Beatson Institute Advanced Imaging Facility for their assistance with the microscopy and image analysis.
REFERENCES
- 1.Sahai E. Mechanisms of cancer cell invasion. Curr Opin Genet Dev. 2005;15:87–96. doi: 10.1016/j.gde.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 2.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–5. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 3.Wang ET, Sandberg R, Luo S, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–6. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mazoyer S, Puget N, Perrin-Vidoz L, Lynch HT, Serova-Sinilnikova OM, Lenoir GM. A BRCA1 nonsense mutation causes exon skipping. Am J Hum Genet. 1998;62:713–5. doi: 10.1086/301768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen LL, Sabripour M, Wu EF, Prieto VG, Fuller GN, Frazier ML. A mutation-created novel intra-exonic pre-mRNA splice site causes constitutive activation of KIT in human gastrointestinal stromal tumors. Oncogene. 2005;24:4271–80. doi: 10.1038/sj.onc.1208587. [DOI] [PubMed] [Google Scholar]
- 6.Wang XQ, Luk JM, Leung PP, Wong BW, Stanbridge EJ, Fan ST. Alternative mRNA splicing of liver intestine-cadherin in hepatocellular carcinoma. Clin Cancer Res. 2005;11:483–9. [PubMed] [Google Scholar]
- 7.Srebrow A, Kornblihtt AR. The connection between splicing and cancer. J Cell Sci. 2006;119:2635–41. doi: 10.1242/jcs.03053. [DOI] [PubMed] [Google Scholar]
- 8.Venables JP. Aberrant and alternative splicing in cancer. Cancer Res. 2004;64:7647–54. doi: 10.1158/0008-5472.CAN-04-1910. [DOI] [PubMed] [Google Scholar]
- 9.Venables JP. Unbalanced alternative splicing and its significance in cancer. Bioessays. 2006;28:378–86. doi: 10.1002/bies.20390. [DOI] [PubMed] [Google Scholar]
- 10.Ghigna C, Giordano S, Shen H, et al. Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol Cell. 2005;20:881–90. doi: 10.1016/j.molcel.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 11.Radisky DC, Levy DD, Littlepage LE, et al. Rac1b and reactive oxygen species mediate MMP-3-induced EMT and genomic instability. Nature. 2005;436:123–7. doi: 10.1038/nature03688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martinez-Contreras R, Cloutier P, Shkreta L, Fisette JF, Revil T, Chabot B. hnRNP proteins and splicing control. Adv Exp Med Biol. 2007;623:123–47. doi: 10.1007/978-0-387-77374-2_8. [DOI] [PubMed] [Google Scholar]
- 13.Wu S, Sato M, Endo C, et al. hnRNP B1 protein may be a possible prognostic factor in squamous cell carcinoma of the lung. Lung Cancer. 2003;41:179–86. doi: 10.1016/s0169-5002(03)00226-5. [DOI] [PubMed] [Google Scholar]
- 14.Ushigome M, Ubagai T, Fukuda H, et al. Up-regulation of hnRNP A1 gene in sporadic human colorectal cancers. Int J Oncol. 2005;26:635–40. [PubMed] [Google Scholar]
- 15.Zhou J, Allred DC, Avis I, et al. Differential expression of the early lung cancer detection marker, heterogeneous nuclear ribonucleoprotein-A2/B1 (hnRNP-A2/B1) in normal breast and neoplastic breast cancer. Breast Cancer Res Treat. 2001;66:217–24. doi: 10.1023/a:1010631915831. [DOI] [PubMed] [Google Scholar]
- 16.Yan-Sanders Y, Hammons GJ, Lyn-Cook BD. Increased expression of heterogeneous nuclear ribonucleoprotein A2/B1 (hnRNP) in pancreatic tissue from smokers and pancreatic tumor cells. Cancer Lett. 2002;183:215–20. doi: 10.1016/s0304-3835(02)00168-4. [DOI] [PubMed] [Google Scholar]
- 17.Lee CH, Lum JH, Cheung BP, et al. Identification of the heterogeneous nuclear ribonucleoprotein A2/B1 as the antigen for the gastrointestinal cancer specific monoclonal antibody MG7. Proteomics. 2005;5:1160–6. doi: 10.1002/pmic.200401159. [DOI] [PubMed] [Google Scholar]
- 18.Mayeda A, Munroe SH, Caceres JF, Krainer AR. Function of conserved domains of hnRNP A1 and other hnRNP A/B proteins. Embo J. 1994;13:5483–95. doi: 10.1002/j.1460-2075.1994.tb06883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caswell PT, Chan M, Lindsay AJ, McCaffrey MW, Boettiger D, Norman JC. Rab-coupling protein coordinates recycling of alpha5beta1 integrin and EGFR1 to promote cell migration in 3D microenvironments. J Cell Biol. 2008;183:143–55. doi: 10.1083/jcb.200804140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caswell PT, Spence HJ, Parsons M, et al. Rab25 associates with alpha5beta1 integrin to promote invasive migration in 3D microenvironments. Dev Cell. 2007;13:496–510. doi: 10.1016/j.devcel.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 21.Hennigan RF, Hawker KL, Ozanne BW. Fos-transformation activates genes associated with invasion. Oncogene. 1994;9:3591–600. [PubMed] [Google Scholar]
- 22.Bass MD, Roach KA, Morgan MR, et al. Syndecan-4-dependent Rac1 regulation determines directional migration in response to the extracellular matrix. J Cell Biol. 2007;177:527–38. doi: 10.1083/jcb.200610076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cukierman E, Pankov R, Stevens DR, Yamada KM. Taking cell-matrix adhesions to the third dimension. Science. 2001;294:1708–12. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Kupfer A, Louvard D, Singer SJ. Polarization of the Golgi apparatus and the microtubule-organizing center in cultured fibroblasts at the edge of an experimental wound. Proc Natl Acad Sci U S A. 1982;79:2603–7. doi: 10.1073/pnas.79.8.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giraudo CG, Daniotti JL, Maccioni HJ. Physical and functional association of glycolipid N-acetyl-galactosaminyl and galactosyl transferases in the Golgi apparatus. Proc Natl Acad Sci U S A. 2001;98:1625–30. doi: 10.1073/pnas.031458398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Venables JP, Koh CS, Froehlich U, et al. Multiple and specific mRNA processing targets for the major human hnRNP proteins. Mol Cell Biol. 2008;28:6033–43. doi: 10.1128/MCB.00726-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yates T, Okoniewski MJ, Miller CJ. X:Map: annotation and visualization of genome structure for Affymetrix exon array analysis. Nucleic Acids Res. 2008;36:D780–6. doi: 10.1093/nar/gkm779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang ZH, Zhang WJ, Rao Y, Wu JY. Regulation of Ich-1 pre-mRNA alternative splicing and apoptosis by mammalian splicing factors. Proc Natl Acad Sci U S A. 1998;95:9155–60. doi: 10.1073/pnas.95.16.9155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rooke N, Markovtsov V, Cagavi E, Black DL. Roles for SR proteins and hnRNP A1 in the regulation of c-src exon N1. Mol Cell Biol. 2003;23:1874–84. doi: 10.1128/MCB.23.6.1874-1884.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shin C, Manley JL. Cell signalling and the control of pre-mRNA splicing. Nat Rev Mol Cell Biol. 2004;5:727–38. doi: 10.1038/nrm1467. [DOI] [PubMed] [Google Scholar]
- 32.White R, Gonsior C, Kramer-Albers EM, Stohr N, Huttelmaier S, Trotter J. Activation of oligodendroglial Fyn kinase enhances translation of mRNAs transported in hnRNP A2-dependent RNA granules. J Cell Biol. 2008;181:579–86. doi: 10.1083/jcb.200706164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Damianova R, Stefanova N, Cukierman E, Momchilova A, Pankov R. Three-dimensional matrix induces sustained activation of ERK1/2 via Src/Ras/Raf signaling pathway. Cell Biol Int. 2008;32:229–34. doi: 10.1016/j.cellbi.2007.08.029. [DOI] [PubMed] [Google Scholar]
- 34.Mitchell SA, Spriggs KA, Bushell M, et al. Identification of a motif that mediates polypyrimidine tract-binding protein-dependent internal ribosome entry. Genes Dev. 2005;19:1556–71. doi: 10.1101/gad.339105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bennetts JS, Rendtorff ND, Simpson F, Tranebjaerg L, Wicking C. The coding region of TP53INP2, a gene expressed in the developing nervous system, is not altered in a family with autosomal recessive non-progressive infantile ataxia on chromosome 20q11-q13. Dev Dyn. 2007;236:843–52. doi: 10.1002/dvdy.21064. [DOI] [PubMed] [Google Scholar]
- 36.Baumgartner BG, Orpinell M, Duran J, et al. Identification of a novel modulator of thyroid hormone receptor-mediated action. PLoS ONE. 2007;2:e1183. doi: 10.1371/journal.pone.0001183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nowak J, Archange C, Tardivel-Lacombe J, et al. The TP53INP2 protein is required for autophagy in mammalian cells. Mol Biol Cell. 2009;20:870–81. doi: 10.1091/mbc.E08-07-0671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bisel B, Wang Y, Wei JH, et al. ERK regulates Golgi and centrosome orientation towards the leading edge through GRASP65. J Cell Biol. 2008;182:837–43. doi: 10.1083/jcb.200805045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Preisinger C, Short B, De Corte V, et al. YSK1 is activated by the Golgi matrix protein GM130 and plays a role in cell migration through its substrate 14-3-3zeta. J Cell Biol. 2004;164:1009–20. doi: 10.1083/jcb.200310061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yadav S, Puri S, Linstedt AD. A primary role for Golgi positioning in directed secretion, cell polarity, and wound healing. Mol Biol Cell. 2009;20:1728–36. doi: 10.1091/mbc.E08-10-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dusetti NJ, Jiang Y, Vaccaro MI, et al. Cloning and expression of the rat vacuole membrane protein 1 (VMP1), a new gene activated in pancreas with acute pancreatitis, which promotes vacuole formation. Biochem Biophys Res Commun. 2002;290:641–9. doi: 10.1006/bbrc.2001.6244. [DOI] [PubMed] [Google Scholar]
- 42.Simpson JC, Wellenreuther R, Poustka A, Pepperkok R, Wiemann S. Systematic subcellular localization of novel proteins identified by large-scale cDNA sequencing. EMBO Rep. 2000;1:287–92. doi: 10.1093/embo-reports/kvd058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sauermann M, Sahin O, Sultmann H, et al. Reduced expression of vacuole membrane protein 1 affects the invasion capacity of tumor cells. Oncogene. 2008;27:1320–6. doi: 10.1038/sj.onc.1210743. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.