Abstract
Background:
BRCA1 mutant breast tumors, in contrast to most sporadic tumors are typically Estrogen Receptor-alpha (ERα) negative. The purpose of this study was to provide a mechanism for the observed ERα deficiency in BRCA1 mutant tumors.
Methods:
Using a Breast Cancer Disease Specific Microarray, we identified differentially regulated transcripts between BRCA1 mutant and sporadic paraffin-embedded breast tumors. Using qPCR we specifically measured ERα mRNA expression in these tumor samples. Regulation of ERα transactivation, mRNA and protein expression in a BRCA1-dependent manner was also assessed in human breast cancer cell lines. Finally, we investigated cellular proliferation in response to anti-estrogen treatment in the presence and absence of functional BRCA1.
Results:
We show that ERα mRNA expression is reduced in the absence of functional BRCA1 in both BRCA1 deficient tumors and breast cancer cell lines. We demonstrate that the transcription factor Oct-1 recruits BRCA1 to the ERα promoter and that both BRCA1 and Oct-1 are required for ERα expression. We also provide evidence to demonstrate that the response of breast cancer cell lines to anti-estrogen treatment is dependent on functional BRCA1, an effect that is directly related to the ability of BRCA1 to modulate ERα expression levels.
Conclusions:
In this study we identify a novel mechanism to explain the link between mutation or aberration of BRCA1 expression and ERα deficiency. We demonstrate that BRCA1 can alter the response of breast cancer cells to anti-estrogen therapy by directly modulating ERα expression.
Germ-line mutations in the BRCA1 tumor suppressor gene confer a genetic predisposition to breast and ovarian cancer (1). In addition to the other reported roles for BRCA1 such as in the DNA damage response and ubiquitination pathway, a substantial body of evidence exists to suggest that BRCA1 is directly involved in transcriptional regulation. It was initially reported that the C-terminus of BRCA1 could act as a transcriptional activator when fused to a heterologous DNA binding domain (2,3). Subsequently it was shown that BRCA1 is a component of the RNA polymerase II holoenzyme complex, through an association with RNA Helicase A (4). BRCA1 has also been shown to act as either a co-activator or co-repressor through its ability to interact with a number of well characterized transcription factors including p53, STAT1, Oct-1, c-Myc and CtIP (5). Through these interactions BRCA1 can transcriptionally activate genes such as p21, GADD45 and 2′5′-OAS, and repress genes such as hTERT, ANG1 and S100A7 (5,6). The ability of BRCA1 to act as either a transcriptional co-activator or co-repressor may be related to its ability to recruit both the basal transcription machinery and proteins involved in chromatin remodelling (7,8).
BRCA1 mutant breast tumors exhibit a distinct pathological phenotype and are typically ERα negative (9,10). Furthermore, by gene expression profiling BRCA1 mutant tumors have been shown to cluster with the ERα negative basal subtype of breast cancer (11). The molecular basis for this ERα negative phenotype of BRCA1 mutant tumors has not been defined. However, evidence for a functional link between BRCA1 and estrogen signalling pathways has previously been revealed. BRCA1 has been shown to inhibit ERα signalling, an interaction which results in the regulation of downstream ERα target genes (12). BRCA1 has also been reported to regulate estrogen synthesis through transcriptional inhibition of the aromatase enzyme (13). In this study, we investigate further the relationship between BRCA1 and ERα and provide evidence to uncover the mechanism of ERα deficiency in BRCA1-linked breast tumors.
MATERIALS AND METHODS
The Breast Cancer DSA™ research tool
The Breast Cancer Disease Specific Microarray (DSA™) (Almac Diagnostics, Craigavon, United Kingdom) research tool contains 60,856 probesets and encodes approximately 60,000 transcripts confirmed as being expressed in breast cancer and normal tissue. The transcript information used to design the Breast Cancer DSA™ research tool was generated by a high throughput 3′-based sequencing approach to define the breast cancer transcriptome. Probes were generated at the 3′ end of each identified transcript and the breast cancer DSA™ research tool was custom designed by Affymetrix (Affymetrix, Santa Clara, CA). This combination of relevant disease specific content and 3′ based probe design allows robust profiling from Formalin Fixed Paraffin Embedded (FFPE) derived RNA. Comparing the Breast Cancer DSA™ research tool against the National Center for Biotechnology Information (NCBI) human Reference Sequence (RefSeq) RNA database (http://www.ncbi.nlm.nih.gov/RefSeq/) using BLAST analysis, 24,600 (41%) of transcripts are present and 30,600 (51%) of transcripts are absent from the human RefSeq database. Furthermore, 8% of the content represents expressed antisense transcripts to annotated genes.
Patients and Tumor Samples
Following ethical committee approval, FFPE specimens from 9 BRCA1 mutant and 7 sporadic breast tumors were received from Fergus Couch, Mayo Clinic, USA and in addition, specimens from 8 BRCA1 mutant and 7 sporadic breast tumors were received from Elaine Kay, Beaumont Hospital, Dublin, Ireland. All 31 samples were age and stage matched breast cancers according to the ‘optimal matching’ approach developed by Rosenbaum (1989).
Gene Expression Profiling from FFPE
Total RNA was extracted from one 10 micron section, with a minimum of 50% tumor content, for each of the 31 FFPE tissue specimens using the Qiagen RNeasy FFPE Kit (Qiagen, Crawley, United Kingdom) according to the manufacturer's instructions. RNA was converted into complementary deoxyribonucleic acid (cDNA) and then used in an in vitro transcription reaction to generate biotinylated cRNA. Biotinylated cRNA targets were prepared using the Two Cycle Target Labeling and Control Reagents (Affymetrix Santa Clara, CA). Using the Affymetrix GeneChip® Instrument System, biotinylated cRNA targets were fragmented and hybridized to Almac Diagnostics Breast Cancer DSA™ research tools. Hybridized cRNA was stained, washed and scanned using the Affymetrix GeneChip® Scanner 7G.
Data Analysis and Identification of Differentially Expressed Genes
After array scanning, the intensity for each probe was saved into Affymetrix CEL files. Gene expression indices were computed from these files with dChip using invariant set normalization and a perfect match (PM)-only model. To detect differential expression an unpaired t-test between mutant and wild-type samples was imposed using a p-value for differential expression of <0.05. Also, it was stipulated that the minimal fold-change between groups be at least ±2 between mutant and wild-type samples. To lessen the incidences of spurious differential expression it was required that any differential genes be classed “present” by the MAS5 algorithm on at least 20% of samples.
Affymetrix control probesets were discarded. This imposition resulted in list of 636 genes defined as differentially expressed.
Reverse Transcription-PCR Amplification of ERα from FFPE Tumor Samples
Total RNA, extracted from 31 FFPE breast tumor samples, was reverse transcribed into cDNA using M-MLV Reverse Transcriptase (Invitrogen, Paisley, United Kingdom) using random primers according to the manufacturer's instructions. ERα primers were designed to the 3′ end of the ERα transcript for optimal detection of the cDNA sourced from FFPE RNA samples. ERα expression levels were normalised to GAPDH mRNA levels. ERα primers were 5′-GGTGCCTGAGACACAGACC-3′ and 5′-GTGAGAGAACAGAAACTGGC-3′ and GAPDH primers were 5′–AGGTGGTCTCCTCTGACTTCAA-3′and 5′-CACCCTGTTGCTGTAGCCAAATTC-3′. PCR was performed using the QuantiTect SYBR Green PCR Kit (Qiagen) according to the manufacturer's instructions. PCR reactions were carried out using the Opticon™ DNA Engine Therml Cycler (Bio-Rad, Hertfordshire, United Kingdom) and analysed using Opticon™ Monitor 3.1 software.
Cell lines
HCC-EV and HCC-BR cell lines were generated and maintained as previously described. T47D and MCF-7 cell lines were maintained as previously described (14).
Western blot analysis
Western blot assays were performed as previously described (15). Antibodies used were HC-20 ERα, D-9 BRCA1, C-21 Oct-1 (all from Santa Cruz Biotechnology, Santa Cruz, CA), and 6G5 GAPDH (Sigma, Dorset, United Kingdom).
Northern blot analysis
Total RNA was extracted from cells and analyzed by Northern blotting as previously described (14). The probe for ERα was generated by restriction digestion of an ERα pcDNA3.1/Hygro expression construct (A gift from Dr. G. Reid, The European Molecular Biology Laboratory (EMBL), Heidelberg, Germany). The GAPDH oligonucleotide probe was generated by RT-PCR.
Small Interfering RNA
Small interfering RNA (siRNA) transfections were performed using OligofectAMINE (Invitrogen) as instructed by the manufacturer. RNA and protein was extracted 48 hours following transfection. Each oligonucleotide was used at a concentration of 100nM. BRCA1 target sequence was 5′-GCGUGCAGCUGAGAGGCAU-3′, Oct-1 target sequence was 5′-CCAGCAGCUCACCUAUUAA-3′, ERα target sequence was 5′-UCAUCGCAUUCCUUGCAAA-3′ and scrambled control siRNA was 5′-CCUGGUAGCAGCGAGUCAG-3′.
Luciferase reporter assay
ProAB ERα promoter construct was a gift from Dr. R. Kiyama (AIST Central 6, Tsukuba, Ibaraki, Japan). Luciferase assay was performed as previously described (15).
Chromatin immunoprecipitation assays
Assays were performed as previously described (15). Antibodies used were CTD4H8 RNA Polymerase II (Upstate, Hampshire, UK), Ab-1 BRCA1 (Calbiochem, Nottingham, United Kingdom), C-21 Oct-1, HC-20 ERα (Santa Cruz Biotechnology). Specific primers were designed to amplify a 217 base pair promoter region directly upstream of the transcription start site of ESR1 from the extracted DNA (5′-AGGAGGGGGAATCAAACAGA-3′ and 5′-TTTACTTGTCGTCGCTGCTG-3′).
Anti-estrogen sensitivity studies
T47D and MCF-7 cells were transfected with scrambled or BRCA1-specific siRNA and after 24 hours seeded at 15,000 cells per well in 24 well culture dishes and at 150,000 cells per plate into 60mm plates. Following 24 hours, cells in the 24 well culture dishes were incubated in medium supplemented with the anti-estrogen drug ICI 182, 780 (Fulvestrant) at a concentration range from 1pM to 10μM and in parallel, 60mm plates were harvested for protein to measure the efficiency of the siRNA. After 72 hours of drug exposure, cells were detached with trypsin and counted using a Z2 particle and size analyzer (Coulter, Miami, FL). Each dose inhibition study was carried out three times. IC50 and IC40 values were calculated from respective sigmoidal dose response curves using Prism software. For ERα transient transfection studies, cells were transfected with BRCA1-specific siRNA and 24 hours later were seeded as before. Six hours later, cells were transfected with pcDNA3.1 empty expression vector or pcDNA3.1 ERα expression vector (0.25μg per well of 24 well plate, or 2.5μg per 60mm plate) using genejuice transfection reagent (Novagen, Nottingham, United Kingdom) according to the manufacturers instructions. After 24 hours cells were incubated with drug or extracted for protein and assayed as before.
RESULTS
ERα expression levels in tumor samples
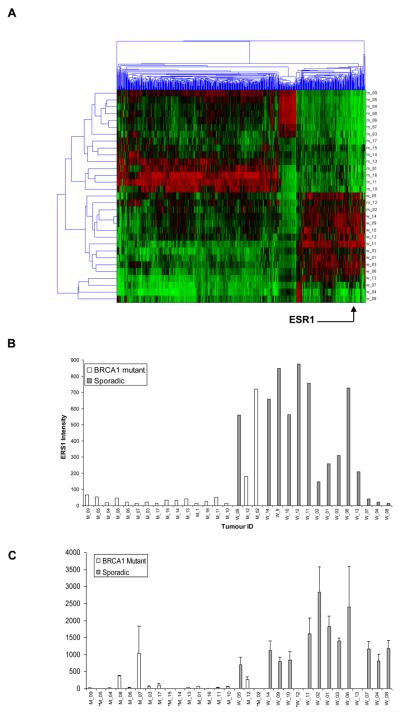
We utilised the Breast Cancer DSA™ research tool to identify genes differentially regulated between BRCA1 mutant and sporadic breast tumors. Our cohort consisted of 17 BRCA1 mutant and 14 sporadic tumors matched by stage and grade. Of the 636 genes defined as differentially expressed, ESR1, the gene which codes for ERα, was one of the most highly altered genes. ESR1 had a mean 5.4 fold downregulation (unpaired t-test, P-value = 0.0019) in BRCA1 mutant tumors compared to sporadic tumors (Fig. 1, A). The actual ERα intensity derived from the array data for each tumour sample was represented as a histogram to better visualise the difference in expression between the BRCA1 mutant and wild type tumours (Fig. 1,B). In order to confirm the loss of ERα mRNA expression in these tumours by an alternative approach quantitative real time PCR (qPCR) was also performed. qPCR confirmed the microarray finding, showing that BRCA1 mutant tumors display reduced expression of ERα mRNA in comparison to sporadic breast tumors (Fig. 1, C). Interestingly one of the BRCA1 tumours, MO2 displayed high levels of ERα mRNA expression and segregated with the wild type tumours by cluster analysis (Fig. 1, A).
Fig. 1. ERα mRNA is downregulated in BRCA1 mutant tumors.
A) Gene tree showing clustering of 17 BRCA1 mutant (M_01-17) and 14 sporadic (W_01-14) samples) matched by stage and grade. Six hundred and thirty six genes were differentially expressed between the two groups, following an unpaired t-test analysis between mutant and sporadic samples with a p-value for differential expression of <0.05 and a minimum fold change of ±2. Highlighted is ESR1 (ERα) gene expression (p-value = 0.0019). B) Histogram showing intensities of ERα probe set in a cohort of 17 BRCA1 mutant (white) (M_01-17) and 14 sporadic (black) (W_01-14) tumors using the ordering of the samples from the clustering in A). C) qPCR analysis showing ERα mRNA levels in the cohort of 17 BRCA1 mutant tumors (white) and 14 sporadic tumors (black). Histogram illustrates ERα mRNA expression following normalisation of the data to GAPDH mRNA as a loading control. Data represents the average ± SD (n=3) of three experiments.
ERα expression is dependent on functional BRCA1
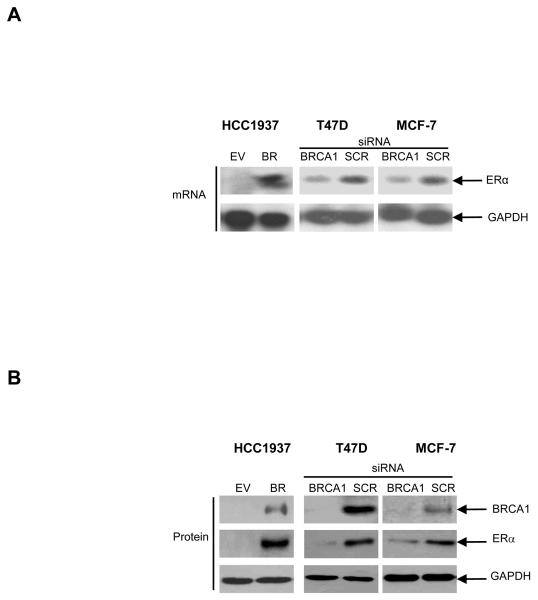
To investigate further the observed correlation between BRCA1 mutation and ERα negativity, we examined the effect on ERα expression levels of reconstituting exogenous BRCA1 in the BRCA1 deficient HCC1937 cell line (16). Northern blot analysis confirmed that ERα mRNA expression was substantially higher in the BRCA1 reconstituted HCC-BR cells compared to the vector transfected HCC-EV control cells (Fig. 2, A). Western blot analysis confirmed re-expression of ERα protein expression following reconstitution of exogenous BRCA1 into the HCC1937 cells demonstrating that loss of ERα protein in these cells was not an irreversible genetic effect (Fig. 2, B). To confirm this finding, we analysed ERα mRNA expression in the T47D and MCF-7 breast cancer cell lines following siRNA-mediated inhibition of endogenous BRCA1 – a model analogous to the downregulation of BRCA1 frequently seen in sporadic breast cancer (17). Consistent with the observations in BRCA1 mutated cells, siRNA-mediated inhibition of endogenous BRCA1 resulted in a marked reduction in expression of endogenous ERα mRNA and protein levels relative to scrambled controls (Fig. 2, A and B).
Fig. 2. Requirement for BRCA1 in ERα expression.
A) Northern blot analysis evaluating ERα mRNA expression in HCC-EV and HCC-BR cells; T47D and MCF-7 cells following siRNA-mediated inhibition of endogenous BRCA1 or following treatment with a scrambled control oligo (SCR). Reprobing for GAPDH confirmed equal RNA loading. B) Western blot analysis comparing levels of BRCA1, ERα and GAPDH protein in the breast cancer HCC-EV (BRCA1 mutant) and HCC-BR (BRCA1 wild-type) cells, and in the breast cancer T47D and MCF-7 cells following transfection of a scrambled control oligo (SCR) or following siRNA-mediated inhibition of endogenous BRCA1.
BRCA1 transcriptionally activates ERα through a mechanism dependent on Oct-1
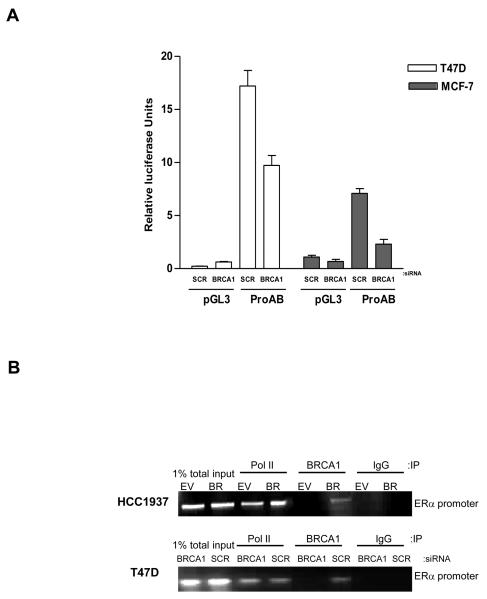
Having demonstrated a direct correlation between BRCA1 and ERα expression levels we wanted to establish if BRCA1 was directly regulating transactivation of ERα. ERα transactivation has been reported to be controlled from at least 7 regulatory promoter non-coding exons upstream of the translation start site (18). Previous studies have shown that in breast cell lines the predominant transcripts are generated from ERα promoter A and to a lesser extent promoter B (19). We obtained a construct that contained the ERα promoters A and B (ProAB) upstream of a luciferase reporter and used this construct to determine if BRCA1 could regulate expression from these promoters. Reporter assays revealed that siRNA-mediated inhibition of endogenous BRCA1 reduced luciferase activity by approximately 50% in both T47D and MCF-7 cell lines relative to scrambled controls (Fig. 3, A). This suggested BRCA1 was mediating transcriptional activation of ERα, at least in part, through ERα promoters A and B. To determine if BRCA1 was recruited to the ERα promoter, we performed chromatin immunoprecipitation (ChIP) assays and used primers specific to the ERα promoter ProAB region. ChIP patterns obtained on chromatin from the BRCA1 reconstituted HCC-BR cells showed that wild-type BRCA1 was recruited to the ERα promoter along with RNA polymerase II. In contrast, the pattern obtained from the BRCA1 mutant HCC-EV cells revealed recruitment of RNA Polymerase II but not BRCA1 (Fig. 3, B). Similarly, ChIP analysis demonstrated that BRCA1 and RNA Polymerase II were recruited to the ERα promoter in T47D cells (Fig. 3, B). These data confirmed that BRCA1 was associated with the ERα promoter and showed that full-length, wild-type BRCA1 was required for successful engagement with the promoter.
Fig. 3. BRCA1 directly regulates ERα promoter activity.
A) Luciferase reporter assay of T47D (open bars) and MCF-7 (solid bars) cells transfected with a firefly luciferase construct driven by the ERα promoters A and B (ProAB). Cells were transfected with a BRCA1 specific or a scrambled control oligo (SCR) 48 hours prior to transfection with empty luciferase vector (pGL3) as a control or ProAB luciferase construct. Co-transfection of a Renilla luciferase plasmid served as an internal control. After incubation for 24 hours, luciferase activity was assayed and normalised to the internal Renilla control. The results are shown as the average ± SD (n=3) of three independent experiments. B) Chromatin Immunoprecipitation (ChIP) of HCC-EV and HCC-BR cells, and T47D cells treated with a scrambled control oligo (SCR) or BRCA1 specific siRNA. Recruitment of RNA Polymerase II (Pol II) and BRCA1 to the ERα promoter was analysed using primers specific to the ERα promoter. 1% of total input DNA was used as a loading control and isotype matched IgG was used as an internal control for the immunoprecipitation.
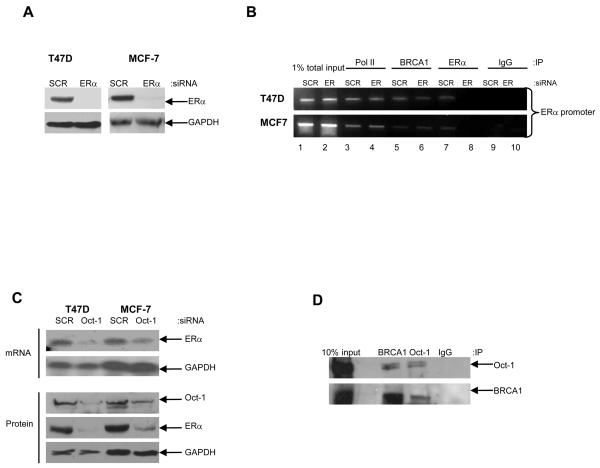
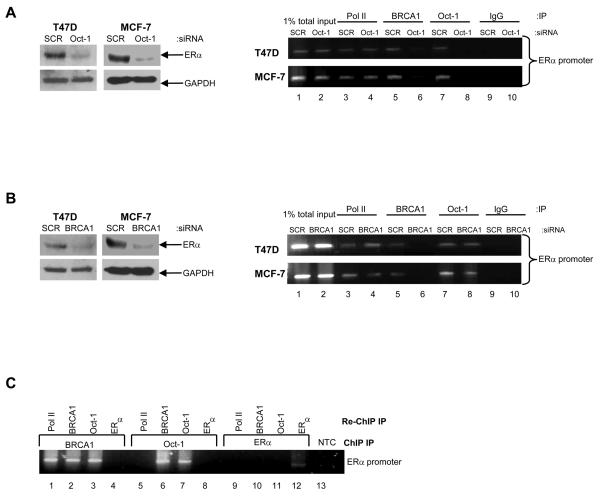
ERα is known to autoregulate its expression and activity (20). We investigated the hypothesis that BRCA1 may interact with ERα to induce expression of ERα by a positive feedback mechanism. To study this, we used ERα specific siRNA to inhibit endogenous ERα expression (Fig. 4, A). Our results demonstrate that ERα binds to it's own promoter (Fig. 4, B). However, BRCA1 can bind to the ERα promoter independent of functional ERα protein suggesting that the regulation occurs through a mechanism that is independent of ERα itself (Fig. 4, B, lanes 5 and 6). An investigation of putative transcription factor binding sites in the upstream promoter region of ERα revealed a number of potential octamer elements, which are the specific sequences recognised by the ubiquitous transcription factor Oct-1. To investigate if Oct-1 plays a functional role in the transcriptional regulation of ERα we utilised specific siRNA to inhibit endogenous Oct-1 in the BRCA1 wild-type T47D and MCF-7 cells. Compared to scrambled controls, depletion of endogenous Oct-1 protein using siRNA abrogated ERα mRNA and protein levels in both cell line models, as demonstrated by Northern blot and Western blot analysis, respectively (Fig. 4, C). These data imply a crucial role for Oct-1 in the transactivation of ERα in these cell line models. Interestingly, BRCA1 has previously been shown to bind to Oct-1 and regulate the expression of the genomic stability protein GADD45 and the checkpoint protein Mad2 (21,22). Consistent with these observations, we demonstrated a reciprocal interaction between endogenous BRCA1 and endogenous Oct-1 by co-immunoprecipitation experiments in T47D cells (Fig. 4, D). Furthermore, ChIP assays revealed that Oct-1, similar to BRCA1, was specifically recruited to the ERα promoter in both T47D and MCF-7 cells (Fig. 5, A, lane 7). The ability of Oct-1 to bind to the ERα promoter was shown to be independent of the presence of functional BRCA1, since Oct-1 was recruited to the ERα promoter following siRNA-mediated inhibition of endogenous BRCA1 (Fig. 5, B, lanes 7 and 8). Interestingly, the recruitment of BRCA1 to the ERα promoter was shown to be dependent on functional Oct-1, since this binding was abrogated following siRNA-mediated depletion of endogenous Oct-1 (Fig. 5, A, lanes 5 and 6). Furthermore, we demonstrated by re-ChIP assay that BRCA1 and Oct-1 form a complex on the ERα promoter, in contrast to BRCA1 and ERα, which did not interact on the ERα promoter (Fig. 5, C). These observations suggest that BRCA1 is acting as a co-activator for Oct-1 in the transcriptional regulation of ERα.
Fig. 4. BRCA1 mediated transactivation of ERα requires Oct-1 but not ERα.
A) Western blot analysis demonstrating efficient siRNA-mediated inhibition of endogenous ERα in comparison to scrambled control oligo (SCR). Equal loading was confirmed by reprobing with GAPDH antibody. B) ChIP analysis, using primers specific to the ERα promoter, shows recruitment of RNA Polymerase II (Pol II), BRCA1 and ERα to the ERα promoter in T47D and MCF-7 cells. ERα is recruited to the ERα promoter in the scrambled control cells (SCR) (Lane 7) but absent with siRNA-mediated inhibition of endogenous ERα (Lane 8). BRCA1 is present on the ERα promoter in both the scrambled control cells (Lane 5) and with siRNA-mediated depletion of endogenous ERα (Lane 6). 1% of total input DNA was used as a loading control (lanes 1 and 2). RNA Pol II and isotype matched IgG were used as positive and negative controls respectively (lanes 3, 4, 9 and 10). C) Reduction of Oct-1 protein levels by siRNA reduces ERα mRNA and protein in T47D and MCF-7 cells. Cells were transfected with either a scrambled control oligo (SCR) or Oct-1 specific siRNA oligo. Reprobing for GAPDH confirmed equal loading. D) Co-immunoprecipitation analysis showing association of endogenous BRCA1 and Oct-1 in T47D cells. 1mg of protein lysate was immunoprecipitated with BRCA1 antibody or Oct-1 antibody and immunoblotted for Oct-1 (top panel) or BRCA1 (bottom panel). 10% input lysate was loaded as a positive control. IgG was used as a negative control for immunoprecipitation.
Fig. 5. Oct-1 recruits BRCA1 to the ERα promoter.
A) Western blot analysis confirming reduction of ERα protein expression with siRNA-mediated inhibition of endogenous Oct-1 in T47D and MCF-7 cells. From the identical experiment ChIP analysis shows recruitment of RNA Pol II, BRCA1 and Oct-1 to the ERα promoter in T47D cells. BRCA1 recruitment to the ERα promoter is impaired following siRNA-mediated depletion of endogenous Oct-1 (lane 6). 1% of total input DNA was used as a loading control (lanes 1 and 2). RNA Pol II and isotype matched IgG were used as a positive and negative controls respectively (lanes 3, 4, 9 and 10). B) Western blot analysis confirming the effect of siRNA-mediated inhibition of endogenous BRCA1 on ERα protein expression in T47D and MCF-7 cells. From the identical experiment, ChIP analysis demonstrates that recruitment of Oct-1 to the ERα promoter is independent of functional BRCA1 (lanes 7 and 8). 1% of total input DNA was used as a loading control (lanes 1 and 2). RNA Pol II and isotype matched IgG were used as a positive and negative controls respectively (lanes 3, 4, 9 and 10). C) Re-ChIP screening for interactions between factors recruited to the ERα promoter. Chromatin prepared from T47D cells was subjected to ChIP protocol, using antibodies for BRCA1 (lanes 1-4), Oct-1 (lanes 5-8) and ERα (lanes 9-12). Samples were then re-immunoprecipitated with antibodies for RNA Pol II, BRCA1, Oct-1 and ERα, respectively. NTC: no template control.
BRCA1 levels predict response to anti-estrogen treatment
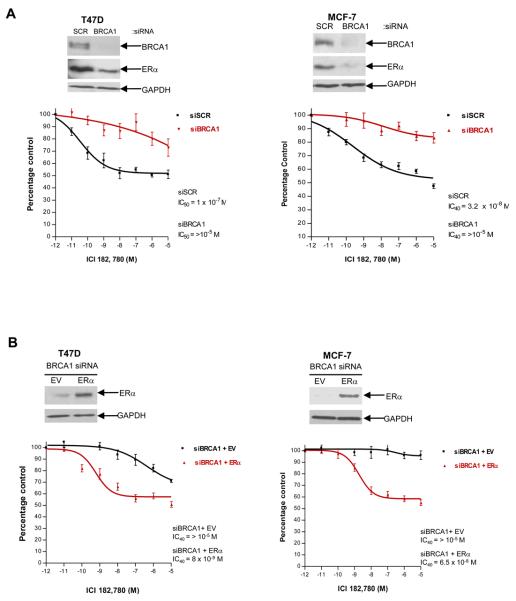
Given the importance of anti-estrogen therapy in both the preventative and adjuvant setting in breast cancer (23,24), we were interested in investigating whether BRCA1 could affect the response of breast cancer cells to anti-estrogen treatment. The anti-estrogen ICI 182, 780 (Fulvestrant) was selected for use in this study as it is a pure steroidal ERα antagonist and it does not exhibit the partial agonist properties frequently associated with Selective Estrogen Receptor Modulator (SERMs) (25,26). Fulvestrant also acts as an anti-estrogen by reducing the half-life of the ERα protein, which results in a downregulation of ERα. We transfected T47D and MCF-7 cells with scrambled control siRNA or BRCA1 specific siRNA oligonucleotides. Protein was harvested to determine the efficiency of the siRNA-mediated depletion of BRCA1 levels after 48 hours and in parallel, cells were treated with a range of concentrations of Fulvestrant. In both cell line models the scrambled control cells exhibited dose dependent growth inhibition following Fulvestrant treatment (T47D IC50 = 1 × 10−7 M, MCF-7 IC40 = 3.2 × 10−8M). However, depletion of functional BRCA1 rendered the cells substantially more resistant to anti-estrogen treatment (T47D IC50 >10−5 M, MCF-7 IC40 >10−5 M) (Fig. 6, A). These data suggest that functional BRCA1 is required for the response of tumor cells to Fulvestrant treatment. To examine if the insensitivity of BRCA1 depleted cells to anti-estrogen treatment is directly related to the observed downregulation of ERα, we carried out dose inhibition assays following expression of exogenous ERα in these cells. We demonstrated that compared to BRCA1 depleted cells transfected with empty vector (T47D and MCF-7 IC40 > 10−5 M), transient transfection of exogenous ERα restored the sensitivity of these cells to Fulvestrant (T47D IC40 = 8 × 10−9 M, MCF-7 IC40 = 6.5 × 10−8 M) (Fig. 6, B). Collectively, these data suggest that loss of BRCA1-mediated transcriptional activation of ERα expression results in increased resistance to an ERα antagonist, suggesting that ERα is a functionally important BRCA1 target.
Fig. 6. BRCA1 levels predict response to the ERα antagonist, Fulvestrant.
A) Dose response curves demonstrate that siRNA-mediated inhibition of endogenous BRCA1 results in increased resistance to the ERα antagonist Fulvestrant (ICI 182, 780) compared to scrambled controls (SCR) in T47D and MCF-7 cells. Western blot analysis confirms the reduction in ERα protein expression levels with siRNA-mediated inhibition of endogenous BRCA1 in T47D and MCF-7 cells (top). Reprobing for GAPDH confirmed equal loading. In parallel, cells were treated with a range of concentrations of Fulvestrant. IC50 and IC40 values were calculated from the respective sigmoidal dose response curves. The results are shown as the average ± SD (n=3) of three independent experiments. B) Dose response curves showing that transient reconstitution of BRCA1-depleted T47D and MCF-7 cells with exogenous ERα re-sensitises cells to Fulvestrant (ICI 182, 780). Western blot analysis demonstrates ERα expression levels in BRCA1 siRNA treated cells following transfection of pcDNA3.1 EV or pcDNA3.1 ERα expression vectors. Reprobing for GAPDH confirmed equal loading. In parallel, cells were treated with a range of concentrations of Fulvestrant. IC40 values were calculated from the respective sigmoidal dose response curves. The results are shown as the average ± SD (n=3) of three independent experiments.
DISCUSSION
One of the unexplained features of BRCA1 mutant tumors is the observed ERα negativity. BRCA1 mutant tumors exhibit loss of ERα expression in up to 90% of cases (9,10). In this study we identify a novel transcriptional mechanism to explain the link between BRCA1 mutation and ERα negativity. The results presented here suggest that BRCA1 mutant tumors fail to express ERα due to a loss of BRCA1-mediated transcriptional activation. In sporadic breast cancers BRCA1 expression is frequently reduced during the transition from carcinoma in situ to invasive cancer (17). The reduction/absence of BRCA1 in sporadic cancers occurs through mechanisms involving loss of heterozygosity (LOH), hypermethylation of the BRCA1 promoter, and loss of transcriptional regulation of BRCA1 (27-29). Importantly, reduced BRCA1 expression in sporadic cancers is associated with ERα negativity (28,29). In agreement with our findings, a recent study demonstrated a positive association between BRCA1 and ERα mRNA expression levels in sporadic breast cancers (30). Interestingly, in support of a role for BRCA1 as a positive regulator of ERα, this study also revealed an inverse association between tumor levels of ERα and ID4, a transcription factor which has previously been identified as a negative regulator of BRCA1 expression (31).
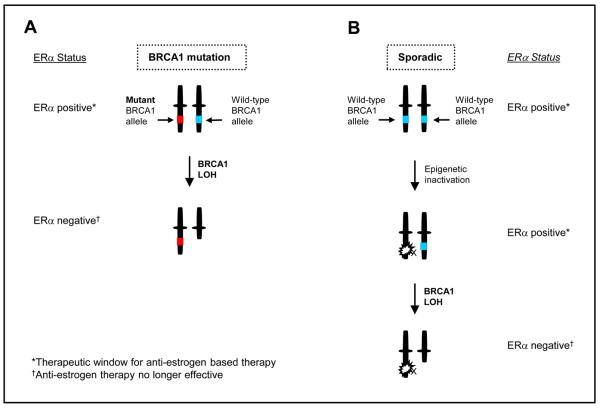
We suggest that loss of the wild-type BRCA1 allele as occurs during neoplastic development in BRCA1 mutation carriers has a direct impact on ERα transactivation which can result in the loss of ERα mRNA and protein expression. We therefore propose a model whereby the observed loss of ERα expression in BRCA1 mutant tumors is likely to occur after loss of the wild-type BRCA1 allele. The results also imply that the reduction in BRCA1 levels that frequently occurs in sporadic breast cancers may directly impact on ERα transactivation and expression (Fig. 7).
Fig. 7. Schematic highlighting a possible sequence of events leading to ERα deficiency in BRCA1 mutated tumors and sporadic tumors.
A) With loss of the wild-type allele in BRCA1 mutant breast cells, transactivation of ERα is lost and the cells revert to an ERα negative phenotype. B) In sporadic tumors, reduction of BRCA1 expression by mechanisms involving LOH and epigenetic inactivation may result in a decrease in ERα expression. A complete loss of BRCA1 function in sporadic cancers would result in ERα negativity of the tumor.
While it is clear that the majority of BRCA1 mutant tumors lack ERα expression, there are a small number of these tumors that retain ERα positivity. In this study, 2 of 17 (12%) of BRCA1 mutant samples (M_02 and M_12) had microarray intensity values above 100 (Fig 1, A and B) and segregated with the wild type tumours The mechanism of retention of ERα is unknown in these cases, however it could be due to an increase in expression of a positive regulator of ERα through a pathway that is independent of functional BRCA1. An alternative mechanism could be that in certain circumstances, mutation does not abrogate the transcriptional activity of the BRCA1 protein. This effect could be mutation specific, where certain mutant proteins retain some ability to transcriptionally activate ERα. Further research is required to investigate the mechanism for ERα positivity in these tumors.
Gene expression profiling has allowed the classification of breast cancers into at least three distinct subtypes - Luminal, Basal and HER2 positive. Interestingly, BRCA1 mutant tumor expression profiles have been shown to cluster with the basal subtype of breast cancer (11), and further studies have characterised BRCA1 mutant tumors as associated with this distinct breast cancer phenotype (9,32). The basal subtype of breast cancer is correlated with a lack of luminal epithelial cell markers such as ERα, while exhibiting a concomitant over-expression of basal epithelial cell markers such as keratin-5 and keratin-17 (33). In future studies it would be intriguing to investigate if the loss of BRCA1 transcriptional activity may play a direct role in driving the basal-like breast cancer phenotype.
We have also demonstrated that BRCA1 expression levels affect response to the ERα antagonist, Fulvestrant. This suggests that there is a functionally relevant link between BRCA1-mediated regulation of ERα expression and response to Fulvestrant therapy. Hence, ERα deficiency in these tumors may result in reduced response to an anti-estrogen drug that function as an ERα antagonist (e.g. a pure ERα antagonist such as Fulvestrant and a partial antagonist SERM such as Tamoxifen). These data add further support to the suggestion that adjuvant anti-estrogen therapy may be ineffective for the treatment of BRCA1 mutant tumors. However, it is plausible that anti-estrogen therapy may have a role to play in the prevention of breast cancer in these women, prior to the loss of the wild-type allele. In support of this concept, bilateral prophylactic oophorectomy has been shown to significantly reduce the risk of breast cancer in BRCA1 mutation carriers (34) and furthermore, treatment with the anti-estrogen Tamoxifen has been reported to reduce the risk of contralateral breast cancers after the initial diagnosis of breast cancers in BRCA1 mutation carriers (35).
Previous studies have demonstrated that BRCA1 can inhibit ERα signalling (12). In addition, BRCA1 has been implicated in the regulation of estradiol production (13), which is the biological ligand for ERα; while on another level, estrogens are known to upregulate BRCA1 expression (36). These observations are likely to have important implications for the development of breast cancers in BRCA1 mutation carriers prior to the loss of the wild-type allele. The current study adds further insight into the complex interaction between BRCA1 and ERα and provides a simple mechanism to explain the high frequency of ERα negativity observed in BRCA1 deficient tumors.
Acknowledgments
We thank Linda Wadum, study co-ordinator, for collection of BRCA1 tumor specimens; Dr. R Kiyama for the gift of the ERα luciferase construct; and Dr. G Reid for the gift of the ERα expression construct. This work was supported by funding from Cancer Research UK, The Breast Cancer Campaign, Action Cancer, The R&D Office Northern Ireland.
REFERENCES
- 1.Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 2.Chapman MS, Verma IM. Transcriptional activation by BRCA1. Nature. 1996;382:678–9. doi: 10.1038/382678a0. [DOI] [PubMed] [Google Scholar]
- 3.Monteiro AN, August A, Hanafusa H. Evidence for a transcriptional activation function of BRCA1 C-terminal region. Proc Natl Acad Sci U S A. 1996;93:13595–9. doi: 10.1073/pnas.93.24.13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson SF, Schlegel BP, Nakajima T, Wolpin ES, Parvin JD. BRCA1 protein is linked to the RNA polymerase II holoenzyme complex via RNA helicase A. Nat Genet. 1998;19:254–6. doi: 10.1038/930. [DOI] [PubMed] [Google Scholar]
- 5.Mullan PB, Quinn JE, Harkin DP. The role of BRCA1 in transcriptional regulation and cell cycle control. Oncogene. 2006;25:5854–63. doi: 10.1038/sj.onc.1209872. [DOI] [PubMed] [Google Scholar]
- 6.Furuta S, Wang JM, Wei S, Jeng YM, Jiang X, Gu B, et al. Removal of BRCA1/CtIP/ZBRK1 repressor complex on ANG1 promoter leads to accelerated mammary tumor growth contributed by prominent vasculature. Cancer Cell. 2006;10:13–24. doi: 10.1016/j.ccr.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 7.Bochar DA, Wang L, Beniya H, Kinev A, Xue Y, Lane WS, et al. BRCA1 is associated with a human SWI/SNF-related complex: linking chromatin remodeling to breast cancer. Cell. 2000;102:257–65. doi: 10.1016/s0092-8674(00)00030-1. [DOI] [PubMed] [Google Scholar]
- 8.Yarden RI, Brody LC. BRCA1 interacts with components of the histone deacetylase complex. Proc Natl Acad Sci U S A. 1999;96:4983–8. doi: 10.1073/pnas.96.9.4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lakhani SR, Reis-Filho JS, Fulford L, Penault-Llorca F, van der Vijver M, Parry S, et al. Prediction of BRCA1 status in patients with breast cancer using estrogen receptor and basal phenotype. Clin Cancer Res. 2005;11:5175–80. doi: 10.1158/1078-0432.CCR-04-2424. [DOI] [PubMed] [Google Scholar]
- 10.Foulkes WD, Metcalfe K, Sun P, Hanna WM, Lynch HT, Ghadirian P, et al. Estrogen receptor status in BRCA1- and BRCA2-related breast cancer: the influence of age, grade, and histological type. Clin Cancer Res. 2004;10:2029–34. doi: 10.1158/1078-0432.ccr-03-1061. [DOI] [PubMed] [Google Scholar]
- 11.Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100:8418–23. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosen EM, Fan S, Isaacs C. BRCA1 in hormonal carcinogenesis: basic and clinical research. Endocr Relat Cancer. 2005;12:533–48. doi: 10.1677/erc.1.00972. [DOI] [PubMed] [Google Scholar]
- 13.Hu Y, Ghosh S, Amleh A, Yue W, Lu Y, Katz A, et al. Modulation of aromatase expression by BRCA1: a possible link to tissue-specific tumor suppression. Oncogene. 2005;24:8343–8. doi: 10.1038/sj.onc.1208985. [DOI] [PubMed] [Google Scholar]
- 14.Andrews HN, Mullan PB, McWilliams S, Sebelova S, Quinn JE, Gilmore PM, et al. BRCA1 regulates the interferon gamma-mediated apoptotic response. J Biol Chem. 2002;277:26225–32. doi: 10.1074/jbc.M201316200. [DOI] [PubMed] [Google Scholar]
- 15.Kennedy RD, Gorski JJ, Quinn JE, Stewart GE, James CR, Moore S, et al. BRCA1 and c-Myc associate to transcriptionally repress psoriasin, a DNA damage-inducible gene. Cancer Res. 2005;65:10265–72. doi: 10.1158/0008-5472.CAN-05-1841. [DOI] [PubMed] [Google Scholar]
- 16.Tomlinson GE, Chen TT, Stastny VA, Virmani AK, Spillman MA, Tonk V, et al. Characterization of a breast cancer cell line derived from a germ-line BRCA1 mutation carrier. Cancer Res. 1998;58:3237–42. [PubMed] [Google Scholar]
- 17.Thompson ME, Jensen RA, Obermiller PS, Page DL, Holt JT. Decreased expression of BRCA1 accelerates growth and is often present during sporadic breast cancer progression. Nat Genet. 1995;9:444–50. doi: 10.1038/ng0495-444. [DOI] [PubMed] [Google Scholar]
- 18.Kos M, Reid G, Denger S, Gannon F. Minireview: genomic organization of the human ERalpha gene promoter region. Mol Endocrinol. 2001;15:2057–63. doi: 10.1210/mend.15.12.0731. [DOI] [PubMed] [Google Scholar]
- 19.Flouriot G, Griffin C, Kenealy M, Sonntag-Buck V, Gannon F. Differentially expressed messenger RNA isoforms of the human estrogen receptor-alpha gene are generated by alternative splicing and promoter usage. Mol Endocrinol. 1998;12:1939–54. doi: 10.1210/mend.12.12.0209. [DOI] [PubMed] [Google Scholar]
- 20.Castles CG, Oesterreich S, Hansen R, Fuqua SA. Auto-regulation of the estrogen receptor promoter. J Steroid Biochem Mol Biol. 1997;62:155–63. doi: 10.1016/s0960-0760(97)00023-x. [DOI] [PubMed] [Google Scholar]
- 21.Fan W, Jin S, Tong T, Zhao H, Fan F, Antinore MJ, et al. BRCA1 regulates GADD45 through its interactions with the OCT-1 and CAAT motifs. J Biol Chem. 2002;277:8061–7. doi: 10.1074/jbc.M110225200. [DOI] [PubMed] [Google Scholar]
- 22.Wang RH, Yu H, Deng CX. A requirement for breast-cancer-associated gene 1 (BRCA1) in the spindle checkpoint. Proc Natl Acad Sci U S A. 2004;101:17108–13. doi: 10.1073/pnas.0407585101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.EBCTCG Tamoxifen for early breast cancer: an overview of the randomised trials. Early Breast Cancer Trialists' Collaborative Group. Lancet. 1998;351:1451–67. [PubMed] [Google Scholar]
- 24.Fisher B, Costantino JP, Wickerham DL, Redmond CK, Kavanah M, Cronin WM, et al. Tamoxifen for prevention of breast cancer: report of the National Surgical Adjuvant Breast and Bowel Project P-1 Study. J Natl Cancer Inst. 1998;90:1371–88. doi: 10.1093/jnci/90.18.1371. [DOI] [PubMed] [Google Scholar]
- 25.Vendrell JA, Magnino F, Danis E, Duchesne MJ, Pinloche S, Pons M, et al. Estrogen regulation in human breast cancer cells of new downstream gene targets involved in estrogen metabolism, cell proliferation and cell transformation. J Mol Endocrinol. 2004;32:397–414. doi: 10.1677/jme.0.0320397. [DOI] [PubMed] [Google Scholar]
- 26.Howell A, Osborne CK, Morris C, Wakeling AE. ICI 182,780 (Faslodex): development of a novel, “pure” antiestrogen. Cancer. 2000;89:817–25. doi: 10.1002/1097-0142(20000815)89:4<817::aid-cncr14>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 27.Baldassarre G, Battista S, Belletti B, Thakur S, Pentimalli F, Trapasso F, et al. Negative regulation of BRCA1 gene expression by HMGA1 proteins accounts for the reduced BRCA1 protein levels in sporadic breast carcinoma. Mol Cell Biol. 2003;23:2225–38. doi: 10.1128/MCB.23.7.2225-2238.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Catteau A, Harris WH, Xu CF, Solomon E. Methylation of the BRCA1 promoter region in sporadic breast and ovarian cancer: correlation with disease characteristics. Oncogene. 1999;18:1957–65. doi: 10.1038/sj.onc.1202509. [DOI] [PubMed] [Google Scholar]
- 29.Schmutzler RK, Bierhoff E, Werkhausen T, Fimmers R, Speiser P, Kubista E, et al. Genomic deletions in the BRCA1, BRCA2 and TP53 regions associate with low expression of the estrogen receptor in sporadic breast carcinoma. Int J Cancer. 1997;74:322–5. doi: 10.1002/(sici)1097-0215(19970620)74:3<322::aid-ijc15>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 30.Roldan G, Delgado L, Muse IM. Tumoral expression of BRCA1, estrogen receptor alpha and ID4 protein in patients with sporadic breast cancer. Cancer Biol Ther. 2006;5:505–10. doi: 10.4161/cbt.5.5.2597. [DOI] [PubMed] [Google Scholar]
- 31.Beger C, Pierce LN, Kruger M, Marcusson EG, Robbins JM, Welcsh P, et al. Identification of Id4 as a regulator of BRCA1 expression by using a ribozyme-library-based inverse genomics approach. Proc Natl Acad Sci U S A. 2001;98:130–5. doi: 10.1073/pnas.98.1.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Foulkes WD, Stefansson IM, Chappuis PO, Begin LR, Goffin JR, Wong N, et al. Germline BRCA1 mutations and a basal epithelial phenotype in breast cancer. J Natl Cancer Inst. 2003;95:1482–5. doi: 10.1093/jnci/djg050. [DOI] [PubMed] [Google Scholar]
- 33.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 34.Rebbeck TR, Levin AM, Eisen A, Snyder C, Watson P, Cannon-Albright L, et al. Breast cancer risk after bilateral prophylactic oophorectomy in BRCA1 mutation carriers. J Natl Cancer Inst. 1999;91:1475–9. doi: 10.1093/jnci/91.17.1475. [DOI] [PubMed] [Google Scholar]
- 35.Narod SA, Brunet JS, Ghadirian P, Robson M, Heimdal K, Neuhausen SL, et al. Tamoxifen and risk of contralateral breast cancer in BRCA1 and BRCA2 mutation carriers: a case-control study. Hereditary Breast Cancer Clinical Study Group. Lancet. 2000;356:1876–81. doi: 10.1016/s0140-6736(00)03258-x. [DOI] [PubMed] [Google Scholar]
- 36.Spillman MA, Bowcock AM. BRCA1 and BRCA2 mRNA levels are coordinately elevated in human breast cancer cells in response to estrogen. Oncogene. 1996;13:1639–45. [PubMed] [Google Scholar]