Abstract
An ability to mimic the boundaries of biological compartments would improve our understanding of self-assembly, and provide routes to new materials for the delivery of drugs and biologicals, and the development of protocells. We show that short designed peptides can be combined to form unilamellar spheres approximately 100 nanometers in diameter. The design comprises two, non-covalent, heterodimeric and homotrimeric coiled-coil bundles. These are joined back-to-back to render two complementary hubs, which when mixed form hexagonal networks that close to form cages. This design strategy offers control over chemistry, self-assembly, reversibility and size of such particles.
In viruses (1) and certain bacterial microcompartments (2), capsids and suprastructures are produced via the self-assembly of large folded proteins, usually in highly symmetric manners, and with exquisite positioning of non-covalent protein-protein interactions. Biomimetic assemblies have potential for creating simpler encapsulation systems, and for applications in controlled delivery and release, sensing, and the preparation of protocells for various aspects of synthetic biology (3-5). To these ends, others have produced macroscopic “sacs” from peptide amphiphiles (6); and engineered micelle-like structures (7, 8), small polyhedra (9, 10), extended protein arrays (11), and metal-directed assemblies (12, 13) using mainly natural peptides and proteins. Here we show that self-assembled cage-like particles, SAGEs, can be made from a set of short, de novo, α-helical, coiled-coil peptides by employing clear sequence-to-structure relationships and rational-design principles to direct stable and highly specific protein-protein interactions (14-16).
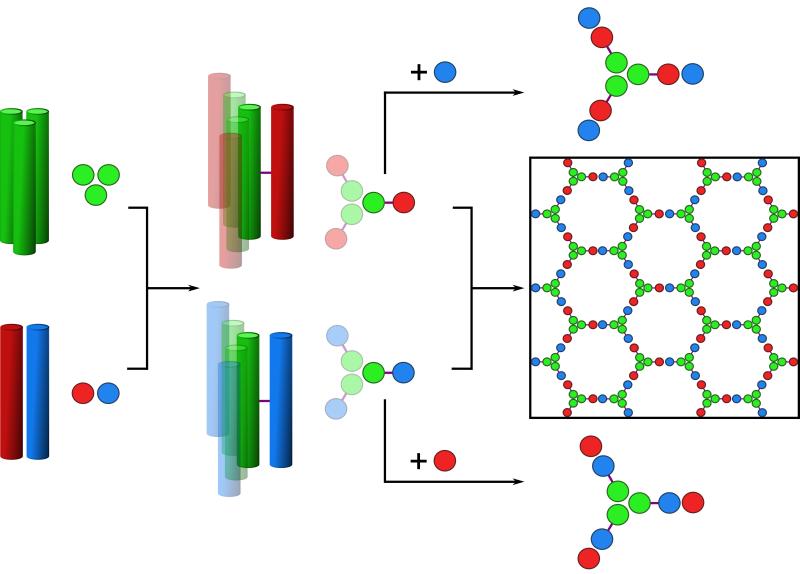
Previously, we developed a toolkit of coiled coils comprising homo-dimer, trimer and tetramers (17), and a number of heterodimers (18). These synthetic peptides, of ≈ 30 residues in length, assemble reversibly and form stable structures at micromolar to nanomolar concentrations. To expand this toolkit and to ease the construction of the building blocks for the SAGE design, we engineered two new coiled-coil modules: a shorter (~20 residues) homotrimer (CC-Tri3), and a similarly short obligate heterodimer (CC-Di-AB) comprising acidic (CC-Di-A) and basic (CC-Di-B) sequences (Fig. S1 and Table S1 (19)). We chose a heterodimer for the second module to give control in the following self-assembly process. Our goal was to link copies of CC-Tri3 and CC-Di-A or CC-Di-B through their external surfaces via disulfide bonds (Fig. 1). These covalent constructs, dubbed CC-Tri3—CC-Di-A and CC-Tri3—CCDi-B, should assemble into complementary trimeric hubs, hub A and hub B, respectively. Alone, these should be water-soluble, discrete, partly folded helical structures; i.e., CC-Tri3 should spontaneously assemble, leaving CC-Di-A and CC-Di-B orphaned on the outside of the assemblies. Upon mixing, however, the two hubs should co-assemble via association of the CC-Di-A and CC-Di-B modules to produce hexagonal networks with pores of ≈ 5 – 6 nm. Because the hubs are flexible and to maximize coiled-coil interactions, we argue that these networks should fold to form closed objects, i.e., SAGEs.
Fig. 1. Schematics for the design and self-assembly of peptide-based cages.
Left to right: Homotrimeric coiled coil (CC-Tri3, green) and heterodimeric coiled coils (CC-Di-AB); the latter comprises CC-Di-A and CC-Di-B, colored red and blue, respectively. CC-Tri and CC-Di-AB are linked via asymmetric disulfide bonds (purple lines) to render hub A (green-red) and hub B (green-blue). Mixing hub A with CC-Di-B, or hub B with CC-Di-A produces discrete 9-helix assemblies; whereas, mixing the hubs directly produces a hexagonal network, which should close.
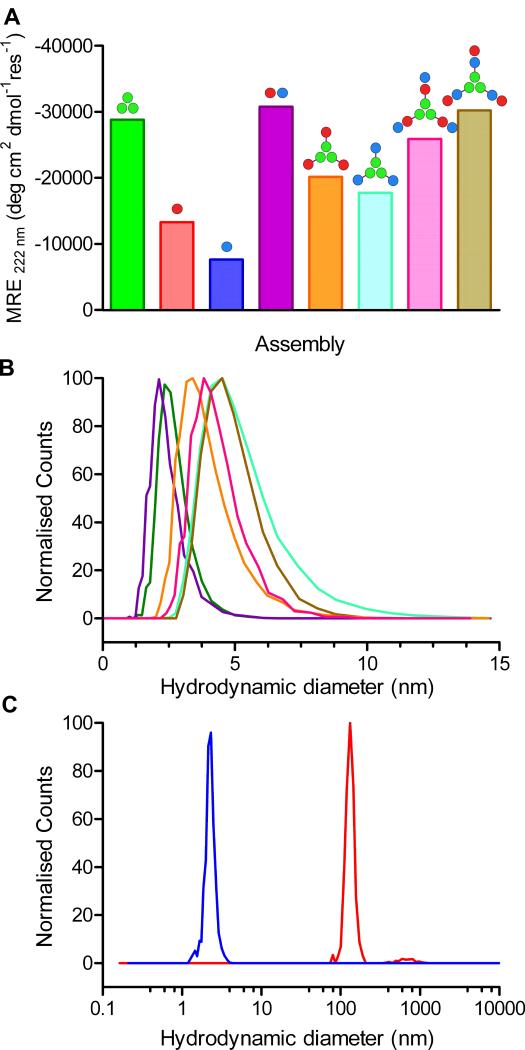
The two coiled coils were synthesized (Fig. S2) and characterized in solution using a combination of circular dichroism (CD) spectroscopy to measure secondary structure, stability, and dissociation constants (Kd values) (Figs. 2A, S3 and S4, Tables S2 and S3); dynamic light scattering (DLS, Figs. 2B & S5), and analytical ultracentrifugation (AUC, Fig. S6) to probe peptide association. These methods confirmed CC-Tri3 as a highly helical trimeric assembly, with concentration-dependent folding (Kd,20 °C = 3.99 × 10−14 M2), and a midpoint of thermal unfolding (TM) of 65 °C at 50 μM peptide. Similarly, CC-Di-A and CC-Di-B alone were unfolded in the micromolar range, but co-assembled when mixed to form a helical heterodimer, CC-Di-AB, (Kd, 20 °C = 5.83 × 10−8 M; TM = 51 °C) (Table 1). We verified that CC-Tri3 and CC-Di-AB did not form mixed species in the presence of each other by showing that the melting profile of the two coiled coils, when mixed, was the same as the average of the two independent profiles (Fig. S7).
Fig. 2. Solution-phase characterization of the designed peptide modules, hubs and discrete assemblies.
(A) Mean Residue Ellipticity (MRE) at 222 nm (MRE222) from CD spectroscopy in phosphate-buffered saline (PBS) at 20 °C for: CC-Tri3 (50 μM, green); CC-Di-A (50 μM, red); CC-Di-B (50 μM, blue); the mixture of CC-Di-A and CC-Di-B, i.e., CC-Di-AB, (50 μM + 50 μM, purple); hub A (CC-Tri3—CC-Di-A, 50 μM, orange); hub B (CC-Tri3—CC-Di-B, 50 μM, cyan); hub A plus CC-Di-B (50 μM + 50 μM, pink); hub B plus CC-Di-A (50 μM + 50 μM, brown). Full CD spectra and thermal denaturation curves are provided in Fig. S3 (19). The leucine-zipper peptide, GCN4-p1, gives a benchmark MRE222 for 100% α-helix of ≈ -36,000 deg cm2 dmol−1 res−1 (24). (B) Hydrodynamic diameters of the coiled-coil modules, hubs and 9-chain terminated assemblies as determined by dynamic light scattering; colors as per panel A. Expanded DLS data are given in Fig. S5. (C) DLS data for the assembled SAGE particles before (red) and after (blue) treatment with TCEP (tris(2-carboxyethyl)phosphine).
Table 1. Summary of solution-phase biophysical data.
| Assembly | MRE222† (deg cm2.dmol res−1) |
TM‡ (°C) |
Mass of Assembly by AUC [95% C.I.] (calculated mass) |
Hydrodynamic diameter by DLS (nm) |
|
|---|---|---|---|---|---|

|
CC-Tri3 | −28,780 | 65 | 7,944 Da. [7,858–8,009] (7,960 Da.) |
2.6 ± 0.5 |
|
|
CC-Di-A | −13,294 | ND | ND | ND |
|
|
CC-Di-B | −7,647 | ND | ND | ND |

|
CC-Di-AB | −30,746 | 51 | 5,350 Da. [5,289–5,442] (5,590 Da.) |
2.3 ± 0.6 |
|
(hub A)
CC-Tri3— CC-Di-A |
−20,168 | 68 | 16,910 Da. [16,720–17,110] (16,238 Da.) |
3.7 ± 0.6 |
|
(hub B)
CC-Tri3— CC-Di-B |
−17,764 | 56 | 15,810 Da. [15,662–15,963] (16,153 Da.) |
5.0 ± 0.3 |
|
hub A
+ CC-Di-B |
−25,894 | 54 | 24,980 Da. [24,795–25,143] (24,505 Da.) |
4.1 ± 0.6 |
|
hub B
+ CC-Di-A |
−30,218 | 57 | 31,690 Da. [31,574–31,793] (24,505 Da.) |
4.7 ± 0.2 |
Mean residue ellipticity (MRE) observed at 222 nm, 20 °C.
Midpoint, or point of inflection, in sigmoidal thermal unfolding curves recorded as the change in MRE222 whilst ramping the temperature from 5 to 90 °C at 40 °C/hour. All biophysical data were obtained from samples of 50 μM concentration of each peptide component listed, and in PBS at pH 7.4.
Building toward hubs A and B, the two-peptide constructs CC-Tri3—CC-Di-A and CC-Tri3—CC-Di-B had reduced mean residue ellipticities (MREs) compared with CC-Tri3 alone. Moreover, these values were close to averages of CC-Tri3 plus either CC-Di-A or CC-Di-B, respectively (Fig. 2A, S3, Table 1). In addition, the melting curves for the hubs were near simple averages of the component curves (Figs. S8 and 9). Next, we mixed three equivalents of CC-Di-A with hub B, and of CC-Di-B with hub A; i.e., equimolar amounts of the underlying peptide components CC-Di-A and CC-Tri3—CC-Di-B, and of CC-Di-B and CC-Tri3—CC-Di-A. In both cases, this should produce “terminated”, 9-helix assemblies (Fig. 1). Indeed, the increased MREs observed were indicative of near-complete folding of all of the modules (Figs. 2A and S3, Table 1). Moreover, the thermal denaturation curves for these assemblies were sigmoidal, and the apparent TM values measured were near the theoretical value for fully decoupled folding of the CC-Tri3 and CC-Di-AB components of 55 °C (Fig. S9, Tables 1 and S2). In all of these cases, DLS showed that the particle sizes of the peptide modules, hubs and terminated assemblies were ≈ 2 – 5 nm, consistent with discrete and appropriately sized objects (Fig. 2B). AUC (Fig. S6) gave solution molecular weights consistent with the compositions of each of the assemblies (Table 1); except for the terminated hub B, which had a mass higher than expected, but nonetheless was still a discrete assembly.
These findings all corroborate the modular design approach that underpins the SAGE concept.
When hub A and hub B were mixed in an equimolar ratio a fine white precipitate formed within minutes, accounting for the >90% of peptide initially in solution. Fresh samples diluted fivefold in PBS and analyzed by DLS indicated particles of hydrodynamic diameter 132 ± 42 nm (Fig. 2C). The role of the disulfide linkage in the assemblies was confirmed by adding the disulfide reducing agent TCEP to the suspension. This ruptured the particles producing smaller structures of diameter 2.3 ± 0.9 nm (Fig. 2C) similar to that observed for a mixture of CC-Tri3 and CC-Di-AB (2.5 ± 0.6 nm) (Fig. S10).
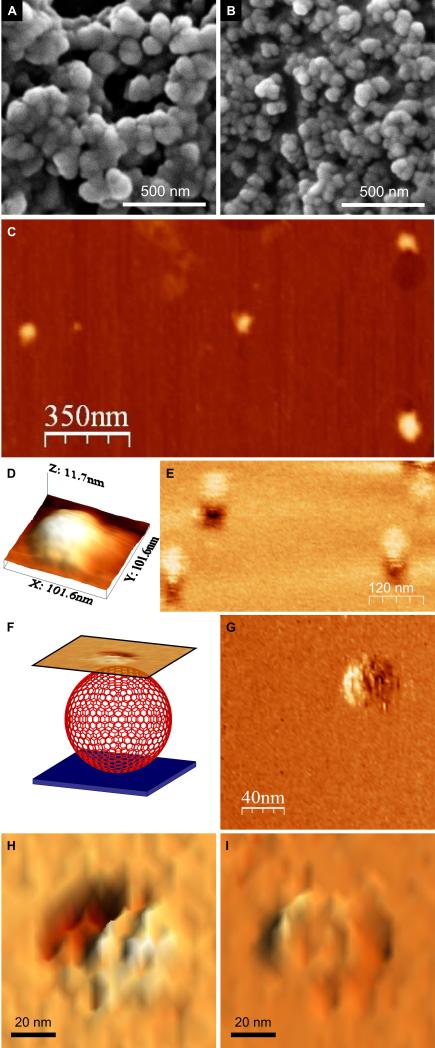
Scanning electron microscopy (SEM) revealed closed spherical objects of similar diameter (97 ± 19 nm, n = 135) (Fig. 3A, S11). Although the particles appear as aggregates in these particular micrographs, they dispersed in solution (Fig. 2C) and separated when deposited on porous membranes (Fig. S12). Tapping-mode atomic force microscopy (TM-AFM) was performed on particles deposited and dried onto mica. These particles were flattened disks 9.2 ± 1.0 nm thick (averaged from scans over 5 particles) with diameters of 95 ± 14 nm (from 4 measurements each on 5 particles) (Fig. 3, C and D and Fig. S13). As the coiled-coil modules are estimated to be ~3 nm in length, the observed thickness of these disks is strong evidence that, in solution, the spheres are hollow and unilamellar rather than being solid, multi-walled, or onion-like structures. That is, they collapse upon drying, presumably releasing water through pores in the assembly. This fits our concept for the SAGEs; i.e., a folded sheet comprising a hexagonal network of peptides (Fig. 1).
Fig. 3. (A and B) Scanning electron micrographs of the SAGE particles.
(A) Images recorded after mixing hub A and hub B in PBS, pH 7.4 (to final concentrations of 50 μM in each of the component peptides, CC-Tri3—CC-Di-A and CC-Tri3—CC-Di-B, i.e. 16.7 μM of each hub). After 1 h, resuspended material was transferred to a carbon-coated stub and sputter-coated with gold/palladium before imaging. (B) Smaller SAGE particles formed by mixing CC-Tri3—CC-Di-Ai and CC-Tri3—CC-Di-Bi. Full images are provided in Fig. S11. (C – I) Scanning probe microscopy of the SAGEs. (C) Tapping-mode AFM (TM-AFM) scan of 4 individual collapsed SAGEs dried onto a mica substrate. (D) 3D representation of the topography measured over a single collapsed SAGE via TM-AFM. (E) LMFM scan recorded in liquid in a noncontact regime with a constant separation of the tip from the glass substrate. (F) Schematic representation of the LMFM scanning regime. An optical feedback maintained the vertically oriented cantilever at a constant separation from the substrate. Mapping the shear-force interaction nanometers above the SAGE allowed an image to be collected in a non-contact mode. (G – I) LMFM images of the hexagonal ultra-structure on the surfaces of hydrated SAGEs.
Lateral molecular-force microscopy (LMFM) with optical feedback was used in a non-contact regime to explore the assemblies in solution (20). Again, this showed approximately spherical objects (diameter 79 ± 12 nm (n = 19); height 82 ± 16 nm (averaged from scans over 6 particles; Fig. 3E). These dimensions are similar to those found by SEM, which should be ~10 nm larger because of the sputtered metal coating estimated from the manufacturer’s technical notes to be ~5 nm thick (19, 21). Moreover, and intriguingly, the LMFM revealed ultra-structure on the surfaces of the assemblies, notably clear hexagonal shapes, Figs. 4G - I. The edges of the hexagons averaged 7 ± 2 nm (n = 22); although such x- and y-dimensions in scanning probe microscopies are tip dependent and are not as reliable as measurements made in z.
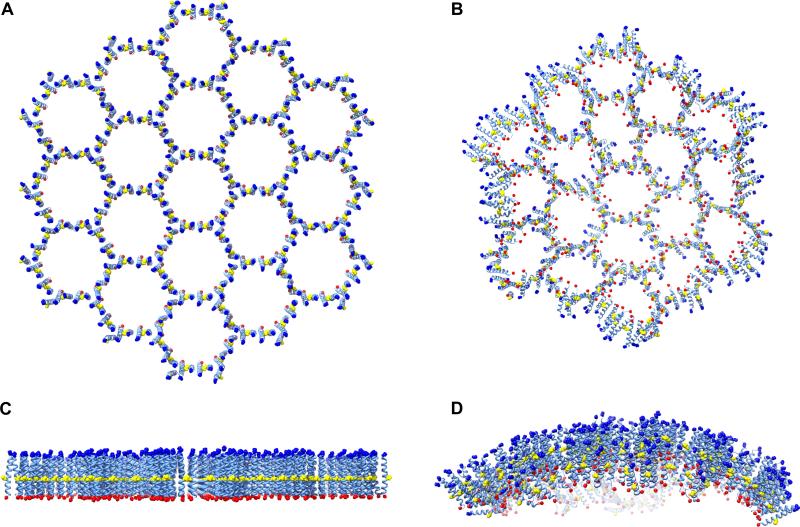
Fig. 4. Molecular dynamics of a model for the SAGEs.
(A and C) Atomistic molecular model of 19 tessellated hexagons comprising hub A and hub B across three perspectives. These were generated from models for CC-Tri3 and CC-Di-AB aligned “back-to-back” along the 3- and 2-fold symmetry axes and linked through disulfide bonds. As for the synthesized peptides, the termini of the modeled peptides were capped and are represented in blue (acylated N-termini) and red (amidated C-termini), with the Cys-Cys disulfide linkers colored yellow. (B and D) Curvature observed following 5 ns molecular dynamics simulations. These were performed under periodic boundary conditions using explicit water (TIP3P) at pH 7 and with 150 mM NaCl present (full details provided as supplementary material (19), along with a movie S1).
Our observations of closed spheres with a tight size distribution, confirmed by three independent methods, is intriguing. It raises two immediate questions: How do the hexagonal networks fold and close, and why are the resulting closed structures so uniform in size?
The first question arises because rigid hexagonal networks should form flat assembles (like a graphite sheet); and closing a sphere (as illustrated by a football) cannot be achieved with hexagons alone and requires, for example, 12 pentagons. However, the coiled-coil modules and hubs of the SAGEs are more flexible, and the assemblies that they produce may tolerate imperfections required to close. Such imperfections, which are inevitable when closing such structures, could include a few mismatched hub pairings, rather than the perfect hexagonal array shown in Fig. 1.
Closing the particles may be driven by thermodynamic and geometric constraints: Regarding thermodynamics, the hubs are designed to associate with their complementary partners, which has two consequences: (1) hubs from solution co-assemble to grow the network; and (2) these expanding edges have unsatisfied coiled coils, which drive the sheets to close and satisfy as many coiled-coil interactions as possible. In terms of geometry, it is likely there is some intrinsic tendency for the hubs to prefer tripod-like structures, with arms arranged at less than 120° creating curvature. We tested these ideas computationally and experimentally as follows.
Complete SAGEs are too large for atomistic simulations, so we modeled smaller fragments of the hexagonal network. From x-ray crystal structures (17) and standard coiled-coil parameters (22), we generated an array of 19 tessellated hexagons built from CC-Tri3 and CC-Di-AB modules, and with 306 chains in total (Figs. 4A, C). After 5 ns of molecular dynamics (MD) in water, uniform curvature was evident in both the x and y directions (Figs. 4B, D and movie S1). This was reproducible: in this, and multiple MD simulations for smaller 7-hexagon networks, the CC-Tri3 modules remained perpendicular to the curved surface with their N-termini always facing “out”.
A sphere of diameter 100 nm has a girth of ≈ 314 nm, corresponding to ≈ 40 equatorial hexagons. Thus, each hexagon is required to be wedge-shaped subtending an angle of ≈ 10° at the center of the sphere. Further examination of the MD trajectories, and retrospective inspection of the designed sequences suggest a molecular interpretation for this wedging: The disulphide bridges linking the coiled coils are slightly offset towards the C-termini; and each peptide has a positively charged lysine residue at the f site between these bridges and the N-termini (Fig. S1). As borne out by the MD, the positively charged lysine residues repel each other, while the disulfide bonds act as a tether. The overall effect is to splay the collective N-termini of each coiled-coil unit apart resulting in wedge-shaped hubs (Fig. S14), producing local and then global curvature.
The question regarding the tight size distribution of the SAGEs is more difficult to rationalize, though this is likely to involve elements such as hub rigidity, the proportion of imperfections required to close a sphere, and entropic factors. To examine how hub rigidity and any preferred local curvature may vary, we analyzed multiple MD simulations of 7-hexagon tessellates from different starting conditions. After 10 ns simulations, the hub-hub angle approached equilibrium settling to 33.9 ± 17.2° (Fig. S15). Clearly, the simulations overestimate the local, and therefore, global curvature. Nonetheless, the 10° angle estimated from the experiments is sampled in the simulations (Fig. S15).
To exploit this apparent flexibility, and to test the importance of burying unsatisfied edges en route to closure, we attempted to engineer smaller SAGE particles. We prepared an additional heterodimer module, CC-Di-AiBi (Table S1). In these peptides, Asn→Ile mutations were made at complementary a sites in the hydrophobic face to give a variant with more than two orders of magnitude higher affinity than the CC-Di-AB parent (17); otherwise, we do not expect this change to alter coiled-coil or hub structure or geometry. Thus, the free-energy penalty associated with unsatisfied edges, and proposed to drive closure, should be higher for the variant. When compared by SEM (Fig. 3, A and B), the parent SAGE particles had diameters of 97 ± 19 nm (n = 135), whereas those incorporating the variant had diameters of 68 ± 12 nm (n = 97) (p < 0.001). This translates to the latter having about half the surface area, and provides strong evidence that satisfying coiled-coil interactions on the edge of a growing disk is a key driving force in closing assemblies. Moreover, it illustrates another advantage of our modular design strategy; namely, that altering the Kd of the individual coiled coils can be used to control SAGE size.
The SAGE concept, though inspired by natural examples, offers routes to closed systems of reduced complexity with the potential for encapsulation. Because the components are modular, interchangeable, and bear termini and side chains that could be derivatized, it should be possible to tune their properties for applications such as vehicles for drug and biomolecule delivery, cages for trapping functional enzyme cascades that allow flux of starting materials and products, components of sensing systems, and as new frameworks for the development of protocells (23).
Supplementary Material
Acknowledgements
We thank the Woolfson group for valuable discussions; the BBSRC for funding to DNW and PJB (BB/G008833/1) and for a studentship to ALB; and the EPSRC for studentships to RLH and THS. JM is supported by a Wellcome University Award. We are grateful to the University of Bristol Advanced Computing Research Centre and the eInfraStructureSouth Consortium for high performance computing; and Jonathan Jones and Electron Microscopy Unit (EMU) School of Chemistry University of Bristol for EM access. The peptides described have been added to the Pcomp database for synthetic biology (http://coiledcoils.chm.bris.ac.uk/pcomp/index.php).
Footnotes
Supplementary Materials www.sciencemag.org/…. Materials and Methods Figs. S1 to S15 Tables S1 to S3 References (25 – 41) Movie S1
References and Notes
- 1.Lodish HF. Molecular cell biology. ed. 6th W.H. Freeman; New York: 2008. [Google Scholar]
- 2.Tanaka S, et al. Science. 2008;319:1083. doi: 10.1126/science.1151458. [DOI] [PubMed] [Google Scholar]
- 3.Agapakis CM, Boyle PM, Silver PA. Nat. Chem. Biol. 2012;8:527. doi: 10.1038/nchembio.975. [DOI] [PubMed] [Google Scholar]
- 4.Hammer DA, Kamat NP. FEBS Lett. 2012;586:2882. doi: 10.1016/j.febslet.2012.07.044. [DOI] [PubMed] [Google Scholar]
- 5.Uchida M, et al. Adv. Mater. 2007;19:1025. [Google Scholar]
- 6.Capito RM, Azevedo HS, Velichko YS, Mata A, Stupp SI. Science. 2008;319:1812. doi: 10.1126/science.1154586. [DOI] [PubMed] [Google Scholar]
- 7.Boato F, et al. Angew. Chem. Int. Ed. Engl. 2007;46:9015. doi: 10.1002/anie.200702805. [DOI] [PubMed] [Google Scholar]
- 8.Raman S, Machaidze G, Lustig A, Aebi U, Burkhard P. Nanomedicine. 2006;2:95. doi: 10.1016/j.nano.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 9.King NP, et al. Science. 2012;336:1171. doi: 10.1126/science.1219364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lai YT, Cascio D, Yeates TO. Science. 2012;336:1129. doi: 10.1126/science.1219351. [DOI] [PubMed] [Google Scholar]
- 11.Sinclair JC, Davies KM, Venien-Bryan C, Noble MEM. Nat. Nanotechnol. 2011;6:558. doi: 10.1038/nnano.2011.122. [DOI] [PubMed] [Google Scholar]
- 12.Brodin JD, et al. Nat. Chem. 2012;4:375. doi: 10.1038/nchem.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pires MM, Lee J, Ernenwein D, Chmielewski J. Langmuir. 2012;28:1993. doi: 10.1021/la203848r. [DOI] [PubMed] [Google Scholar]
- 14.Bromley EHC, Channon K, Moutevelis E, Woolfson DN. ACS Chem. Biol. 2008;3:38. doi: 10.1021/cb700249v. [DOI] [PubMed] [Google Scholar]
- 15.Lupas AN, Gruber M. Adv. Prot. Chem. 2005;70:37. doi: 10.1016/S0065-3233(05)70003-6. [DOI] [PubMed] [Google Scholar]
- 16.Woolfson DN. Adv. Prot. Chem. 2005;70:79. doi: 10.1016/S0065-3233(05)70004-8. [DOI] [PubMed] [Google Scholar]
- 17.Fletcher JM, et al. ACS Synth. Biol. 2012;1:240. doi: 10.1021/sb300028q. [DOI] [PubMed] [Google Scholar]
- 18.Bromley EHC, Sessions RB, Thomson AR, Woolfson DN. J. Am. Chem. Soc. 2009;131:928. doi: 10.1021/ja804231a. [DOI] [PubMed] [Google Scholar]
- 19.See supplementary materials on Science Online.
- 20.Harniman RL, et al. Nanotechnology. 2012;23 doi: 10.1088/0957-4484/23/8/085703. [DOI] [PubMed] [Google Scholar]
- 21. http://www.quorumtech.com/pdf/SputterCoatingTechniques/Sputter_coating_technica l_brief.pdf.
- 22.Offer G, Sessions R. J. Mol. Biol. 1995;249:967. doi: 10.1006/jmbi.1995.0352. [DOI] [PubMed] [Google Scholar]
- 23.Mansy SS, et al. Nature. 2008;454:122. doi: 10.1038/nature07018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O’Shea EK, Rutkowski R, Kim PS. Science. 1989;243:538. doi: 10.1126/science.2911757. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.