Abstract
WEE1 kinase regulates CDK1 and CDK2 activity to facilitate DNA replication during S-phase and to prevent unscheduled entry into mitosis. WEE1 inhibitors synergise with DNA damaging agents that arrest cells in S-phase by triggering direct mitotic entry without completing DNA synthesis, resulting in catastrophic chromosome fragmentation and apoptosis. Here, we investigated how WEE1 inhibition could be best exploited for cancer therapy by performing a functional genetic screen to identify novel determinants of sensitivity to WEE1 inhibition. Inhibition of kinases that regulate CDK activity, CHK1 and MYT1, synergised with WEE1 inhibition through both increased replication stress and forced mitotic entry of S-phase cells. Loss of multiple components of the Fanconi anaemia (FA) and homologous recombination (HR) pathways, in particular DNA helicases, sensitised to WEE1 inhibition. Silencing of FA/HR genes resulted in excessive replication stress and nucleotide depletion following WEE1 inhibition, which ultimately led to increased unscheduled mitotic entry. Our results suggest that cancers with defects in FA and HR pathways may be targeted by WEE1 inhibition, providing a basis for a novel synthetic lethal strategy for cancers harbouring FA/HR defects.
Keywords: WEE1, MK-1775, Fanconi anaemia, CHK1, mitosis, replication
Introduction
DNA replication and cell division are tightly controlled to maintain genome integrity. In response to DNA damage or replication stress, multiple checkpoints delay or block cell cycle progression to ensure that DNA synthesis and damage repair are completed before cells enter mitosis (1). Loss of normal cell cycle control is a hallmark of cancer, and this tumour specific loss of cell cycle control provides a potential therapeutic strategy for targeting cancer cells, either by blocking cell cycle progression or forcing cells inappropriately onwards through the cell cycle via disruption of the checkpoints (2).
WEE1 is a critical regulator of cell cycle progression that inhibits CDK1 and CDK2 activity through phosphorylation at Tyr15 (3, 4). Inhibition of WEE1 promotes mitotic entry through CDK1 activation (5). In addition, WEE1 inhibition results in elevated CDK activity during S-phase, which results in excessive replication origin firing and increased replication fork stalling (6, 7). WEE1 inhibitors have been used in combination with DNA damaging agents or radiation to override the G2/M or intra-S phase checkpoints to trigger mitotic entry from G2 (8, 9) or S-phase cells (10).
Although WEE1 inhibitors are currently in early phase and pre-clinical development, insights into the specific vulnerabilities that increase sensitivity to WEE1 inhibitors is limited. WEE1 inhibitors synergise with chemotherapeutic agents in a TP53 mutant background (11, 12). Combined inhibition of CHK1 and WEE1 has strong synergistic anti-tumour effects in the absence of chemotherapy (13-16), inducing cell death through DNA damage and collapsed replication forks during S-phase. Since p53 and CHK1 are both required to preserve genomic integrity and WEE1 overexpression correlates with increased genomic instability (17), it has been postulated that genetically unstable cancers in particular those with a strong oncogenic drive may be more sensitive to WEE1 inhibition.
Here, we investigate how WEE1 inhibition could be best exploited in the absence of DNA damaging agents by performing a functional genetic RNA interference (RNAi) screen. We show that loss of HR and Fanconi anaemia pathways sensitises to WEE1 inhibition, and investigate the mechanisms through which loss of these genes sensitises to WEE1 inhibition.
Materials and Methods
Cell lines and siRNA transfection
WiDr (December 2011), MCF7 (2008), SKBR3 (2009) and NCI-H508 (2012) cell lines were purchased from the ATCC; C2BBe1 and LS411N (May 2012) from the ECACC; CAL120 (2009) from DSMZ and SUM44PE (2008) from Asterand. Cells were maintained in MEM, DMEM or RPMI with 10% fetal bovine serum (FBS Gold, PAA Laboratories), and 2 mM L-glutamine (Sigma-Aldrich). All cell lines were banked in multiple aliquots upon receipt to reduce risk of phenotypic drift, and reauthenticated by STR profiling with the StemElite ID System (Promega) in 2012. Cells were routinely tested for mycoplasma contamination using the MycoAlert detection kit from Lonza.
For siRNA transfections, cells were reverse transfected with siRNA in 384-well plates or in 6-well plates (20 nM and 50 nM final siRNA concentration, respectively) using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturers’ instructions. Drug treatments were started 48 hours post transfection, and protein extracts were prepared 72 hours after transfection.
siRNAs were obtained from Dharmacon: CHEK1 siGENOME SMARTpool (siCHK1, M-003255-04) and individual siRNA duplexes (D-003255-06/07/08/26), MYT1 siGENOME SMARTpool (siMYT1, M-005026-02) and individual siRNA duplexes (D-005026-01/04/05/06), FANCM siGENOME SMARTpool (siFANCM, M-021955-01) and individual siRNA duplexes (D-021955-01/02/03/04), BRIP1 siGENOME SMARTpool (siBRIP1, M-010587-00) and individual siRNA duplexes (D-010587-01/02/03/04), MRE11A siGENOME SMARTpool (siMRE11, M-009271-01) and siGENOME Non-Targeting siRNA #1 (siCON1, D-001210-01).
Survival assays
For short-term survival assays, cells were reverse transfected in triplicate in 384-well plates at 300 cells per well. At 48 hours post transfection, cells were exposed to different concentrations of MK-1775 supplemented with nucleosides (EmbryoMax Nucleosides, 1:50; Millipore) where indicated. Cell viability was assessed after 96 hours exposure with the CellTiter-Glo Luminescent Cell Viability assay (Promega). For each experiment, average luminescence readings of drug-treated cells were normalised to the average readings of untreated control cells to calculate relative cell viability after drug and siRNA treatment.
For clonogenic survival assays, siRNA-transfected cells were seeded in triplicate at 500 cells/well in 6-well plates at 24 hours post-transfection. The next day, cells were exposed to MK-1775 and nucleosides for 72 hours, after which drugs were washed out. After 10 days, colonies were stained with sulforhodamine-B and counted using the GelCount colony counter platform (Oxford Optronics). Surviving fractions were calculated and normalised to untreated controls for each siRNA separately.
Chemical inhibitors and antibodies
The following chemical inhibitors were used at the indicated concentrations, unless stated otherwise, WEE1 inhibitor MK-1775 (250nM, Axon Medchem), CDK1 inhibitor RO-3306 (10μM; Tocris), hydroxyurea (HU; 3mM), mitomycin C (MMC, 100nM; both from Sigma-Aldrich).
Antibodies used were phospho-Histone H3-Ser10 (pH3; 06-570, Upstate), phospho-Histone-H2AX-Ser139 (05-636, Upstate; 9718, Cell Signaling Technology), β-actin (A5441), β-Tubulin (T4026, both from Sigma-Aldrich), CHK1 (sc-8408, Santa Cruz Biotechnology), MYT1 (4282), MRE11 (4895), Ezrin (3145; all from Cell Signalling Technology), BRIP1 (NB100-416), FANCD2 (NB100-182), 53BP1 (NB100-304; all from Novus Biologicals).
siRNA screening
Screening was performed in 384-well plates with a Dharmacon siGENOME SMARTpools library targeting all known protein kinases, phosphatases, tumour suppressor and DNA repair genes. The siRNA library was supplemented with non-targeting control siRNA, PLK1 siRNA as a viability control and CHK1 siRNA as a positive control. Briefly, cells were reverse transfected in triplicate at final siRNA concentration 20 nM. At 48 hours post transfection, cells were treated with 200 nM MK-1775 or vehicle, and survival was assessed after 96 hours exposure with CellTiter-Glo cell viability assay (Promega). CellTiter-Glo readings were first log2 transformed and then median centred. To assess the effect of siRNA on sensitivity to MK-1775 the log2 ratio between growth in MK-1775 plates and vehicle plates was assessed and expressed as a Z score, with the standard deviation estimated from the median absolute deviation (MAD). The MAD was calculated as the median of the absolute value of each value, xi, minus the median: median(∣xi - median(xi)∣). Z-score of siRNA X = (cell viability effect of siRNA X – median cell viability effect of all 1206 SMARTpool siRNAs of the library) / MAD of all 1206 SMARTpool siRNAs of the library). A Z score of <−2, approximately the 95% confidence intervals, was considered evidence of increased sensitivity to MK-1775.
Extraction and quantification of intracellular dTTPs
Deoxynucleotide availability was determined by a primer extension assay on a dTTP-specific template as described previously (18). Briefly, cells were plated in 100 mm dishes at 106 cells per dish and allowed to grow for 48 hours (or 72 hours post siRNA transfection) before treatment with MK-1775 as indicated. Extracts were prepared as described in Wilson et al. (18). Primer, template (dTTP-DT1) and detection probes were synthesised by Integrated DNA Technologies. Sequences, reaction mixtures and assay conditions were described previously (18). Thermal profiling and fluorescence detection was performed on an Applied Biosystems 7900HT Fast Real-Time PCR System. dTTP quantities were normalised based on cell counts and expressed relative to untreated untransfected or siCON1-transfected cells for each experiment separately.
Quantitative PCR
cDNA was synthesised from RNA using Superscript III and random hexamers (Invitrogen). Quantitative PCR was performed by absolute quantification with Taqman chemistry on an Applied Biosystems 7900HT Fast Real-Time PCR System with FANCM (FAM-labelled, Hs00326216) and GUSB control (VIC-labelled, Hs00939627) probes.
Flow cytometry
Cells were seeded in 60 mm dishes (3 × 105 cells per dish), and treated with MK-1775 after at least 48 hours post siRNA transfection for the indicated time points. Cells were fixed in ice-cold 70% ethanol, permeabilised with 0.25% Triton X-100 in phosphate-buffered saline (PBS), incubated with anti-γH2AX and anti-pH3 antibodies for 2 hours at 4°C, followed by a secondary antibody conjugated to AlexaFluor-488 and -633, respectively, for 1 hour at 4°C. DNA was stained with DAPI. γH2AX-positive cells were gated in a γH2AX-AlexaFluor488 (y-axis) versus DNA content (x-axis) plot. The γH2AXhigh and γH2AXlow cells were then back-gated in a pH3-AlexaFluor633 (y-axis) versus DNA content (x-axis) plot. Likewise, pH3-positive mitotic cells were colour back-gated in the γH2AX-AlexaFluor488 (y-axis) versus DNA content (x-axis) plot.
Western blot analysis
Cells were plated on 100 mm dishes, treated as indicated, and whole-cell extracts were prepared using NP-40 lysis buffer (10 mM Tris-HCl pH8.0, 150 mM NaCl, 1 mM EDTA pH8.0, 1% NP-40, 5 mM sodium pyrophosphate, 50 mM NaF, 1 mM Na3VO4, 5 mM DTT, Complete Protease Inhibitor Cocktail (Roche)). Lysates were run on precast 4-12% Bis-Tris gels (Invitrogen) and transferred onto nitrocellulose membranes (Bio-Rad). Signals were visualised using enhanced chemiluminescence (Amersham ECL Prime Western Blotting Detection Reagent, GE Healthcare).
Immunofluorescence
Cells were plated on coverslips and treated with MK-1775 as described. After drug treatment, cells were fixed in 4% paraformaldehyde for 1 hour, washed, permeabilised in 0.5% Triton X-100 in PBS for 10 minutes, washed and blocked in IFF (1% BSA, 2% FBS in PBS) followed by incubation with primary and secondary AlexaFluor-conjugated antibodies for 1-2 hours at RT each. DNA was stained with DAPI. Coverslips were mounted on glass slides using Vectashield. Images were acquired on a Leica confocal microscope (40x and 63x oil immersion objective). Intensity of γH2AX staining was quantified using Volocity 6.0.1 Software (PerkinElmer) of at least 3 images per condition for two independent experiments. Box and whisker plots show interquartile range, median (horizontal bar) and mean (+) fluorescence intensity. Whiskers denote data points within 1.5 times the interquartile range. Outliers are shown by black dots. 53BP1 foci were scored for at least 100 cells per condition for two independent experiments.
Statistical analysis
All statistical tests were performed with GraphPad Prism version 6.0. P values were two tailed and considered significant if P<0.05. Error bars represent SEM of three independent experiments unless stated otherwise.
Results
siRNA screen identifies novel determinants of WEE1 inhibitor sensitivity
To identify determinants of sensitivity to WEE1 inhibition, we screened an siRNA library consisting of the kinome, phosphatome, tumour suppressor genes and DNA repair genes (1206 genes in total) for modifiers of sensitivity to the WEE1 inhibitor MK-1775 (AZD1775). WiDr cells were transfected in triplicate with the siRNA library and at 48 hours post transfection cells were exposed to 200 nM of MK-1775 or vehicle control for 4 days after which cell viability was assessed using the CellTiter-Glo Cell Viability assay (Supplementary Fig. 1A and 1B). Vehicle control plates were used to examine for the effect of siRNA on cell survival/growth (Supplementary Fig. 1C), and the relative cell viability in the plates exposed to MK-1775 versus vehicle was used to identify siRNAs that altered sensitivity to MK-1775 (Fig. 1A).
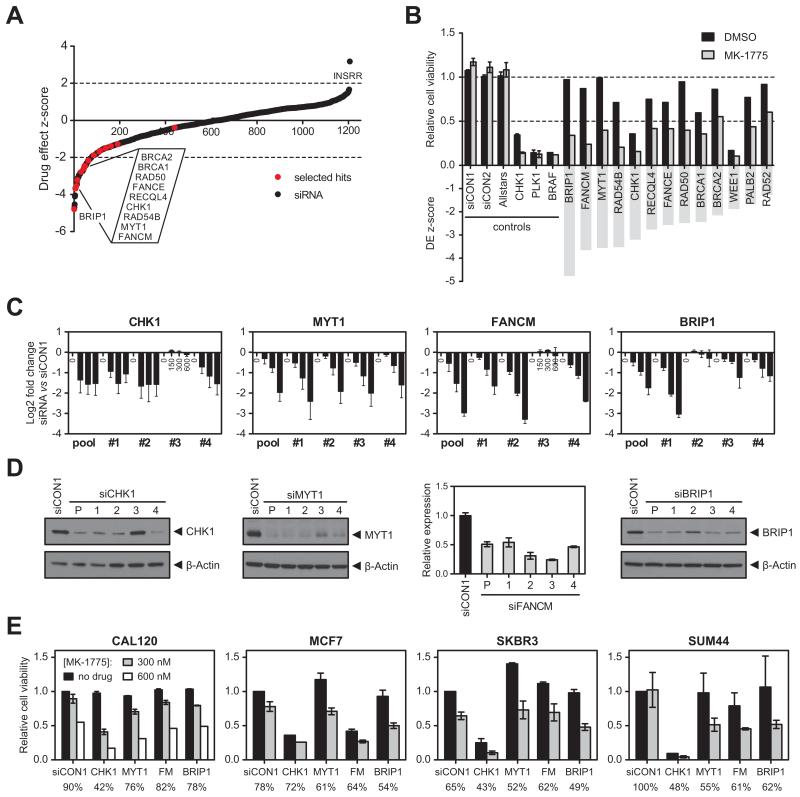
Figure 1. siRNA screen identifies novel determinants of WEE1 inhibitor sensitivity A.
Drug effect Z-scores for an siRNA/MK-1775 WEE1 inhibitor sensitivity screen in WiDr cells. Z-scores <−2 represent statistically significant sensitising effects to 200 nM MK-1775 (dotted line). Black dots, individual siRNA SMARTpools targeting 1206 genes; red dots, hits selected for further validation.
B. Relative cell viability (top) and drug effect (DE) Z-scores (bottom) for selected hits from the siRNA screen after treatment with DMSO (black bars) or MK-1775 (grey bars). siCON1, siCON2 and Allstars were included as non-targeting controls; siCHK1, siPLK1 and siBRAF were included as transfection controls.
C. Sensitisation to MK-1775 was validated with four individual siRNA duplexes targeting CHK1, MYT1, FANCM and BRIP1, or a pool of all four duplexes. At 48 hours post siRNA transfection, WiDr cells were exposed to 150, 300 or 600 nM MK-1775 for 4 days. Log2 fold change in cell viability is shown for each siRNA relative to siCON1.
D. Western blot analysis of cell lysates harvested 72 hours after transfection with siRNAs targeting CHK1, MYT1 and BRIP1. β-Actin was used as loading control. For FANCM, cDNA was prepared from RNA isolated 48 hours after siRNA transfection. FANCM mRNA expression was normalised to GUSB mRNA expression. Error bars represent SEM of three technical replicates.
E. Effect of CHK1, MYT1, FANCM (FM) and BRIP1 siRNAs and siCON1 (non-targeting control) on sensitivity to MK-1775 in breast cell lines CAL120, MCF7, SKBR3 and SUM44. Cell viability (normalised to siCON1-transfected cells without MK-1775) is shown after exposure to 300 or 600 nM MK-1775 for 4 days as determined by CellTiter-Glo. Numbers below the graph indicate cell viability after 300 nM MK-1775 relative to no drug treatment for each siRNA separately. Error bars represent SEM of two independent experiments.
A number of siRNAs increased sensitivity to MK-1775, including siRNAs against checkpoint protein CHK1, CDK1 inhibitory kinase MYT1, Fanconi anaemia (FA) genes FANCM, BRIP1, FANCE, and PALB2, and genes involved in Homologous Recombination (HR) repair (RAD54B, RECQL4, RAD50, RAD52, BRCA1, BRCA2) (Fig. 1B). Multiple hits possessed DNA helicase or translocase activity, such as FANCM (19), BRIP1 (20), RAD54B (21) and RECQL4 (22, 23).
To validate the results of the screen, cell viability upon MK-1775 treatment was assessed for multiple siRNAs targeting each gene and expressed relative to siCON1-transfected cells (Fig. 1C). At least two independent siRNA duplexes for CHK1, MYT1, FANCM and BRIP1 conferred a two- to four-fold increase in sensitivity to MK-1775 in WiDr cells (Fig. 1C), and gene silencing was confirmed at the protein or mRNA level (Fig. 1D). Silencing WEE1 with siRNA recapitulated the sensitisation effects seen with MK-1775, confirming that the effects observed with MK-1775 were through inhibition of WEE1 (Supplementary Fig. 1D). Silencing of RAD54B, RECQL4, FANCE, PALB2, RAD50, RAD52, BRCA1, and BRCA2 sensitised to MK-1775 to a lesser extent (2-fold difference) or could be confirmed with only a single siRNA duplex (Supplementary Fig. 1E). Depletion of BRCA1, BRCA2, and PALB2 caused reduced cell viability in itself, which hampered validation of these siRNAs. However, there was no difference in sensitivity to MK-1775 in matched BRCA2-deficient and BRCA2-proficient DLD1 cell lines, in contrast to PARP-1 inhibitor olaparib (Supplementary Fig. 1F), suggesting that sensitisation to MK-1775 by BRCA2 depletion is context dependent.
Finally, we examined the effect of siRNAs against CHK1, MYT1, FANCM and BRIP1 on MK-1775 sensitivity across a panel of breast and colorectal cell lines (Fig. 1E, Supplementary Fig. 1G). Cell viability data assayed by CellTiter-Glo after 4 days of MK-1775 exposure, confirmed that across the panel silencing of these genes sensitised to MK-1775. This screen therefore identified two groups of genes that were required to tolerate WEE1 inhibition: other checkpoint kinases that regulate CDK activity (CHK1 and MYT1), and genes in the FA/HR pathways (FANCM and BRIP1).
WEE1 inhibition induces both replication stress and premature mitosis
To explore the mechanisms through which the identified hits sensitised to WEE1 inhibition, we first investigated the consequences of WEE1 inhibition alone. WEE1 kinase inhibits CDK1 and CDK2 activity and thereby plays an important role in regulating replication during S-phase (6) and in regulating mitotic entry (10). Consistent with the role of WEE1 in regulating replication, WEE1 inhibition induced pan-nuclear γH2AX staining in 23.4% of the cells at 24 hours, compared to 11.4% in untreated cells (Fig. 2A). BrdU pulse-chase experiments confirmed that the majority of γH2AX-positive cells were in S-phase prior to MK-1775 exposure (Supplementary Fig. 2A), suggesting that the pan-nuclear staining reflected replication stress. To investigate whether WEE1 inhibition induced replication fork stalling or collapse, we assessed 53BP1 foci formation as a marker of DNA double-strand breaks (DSBs). WEE1 inhibition did not induce 53BP1 foci formation (Fig. 2B, Supplementary Fig. 2B) and cells with pan-nuclear γH2AX staining were mutually exclusive to cells with 53BP1 foci, suggesting that WEE1 inhibition induced replication fork stalling but not collapse into DSBs. Together, this suggests that WEE1 inhibition results in an increase in cells with replication stress characterised by widespread replication fork stalling.
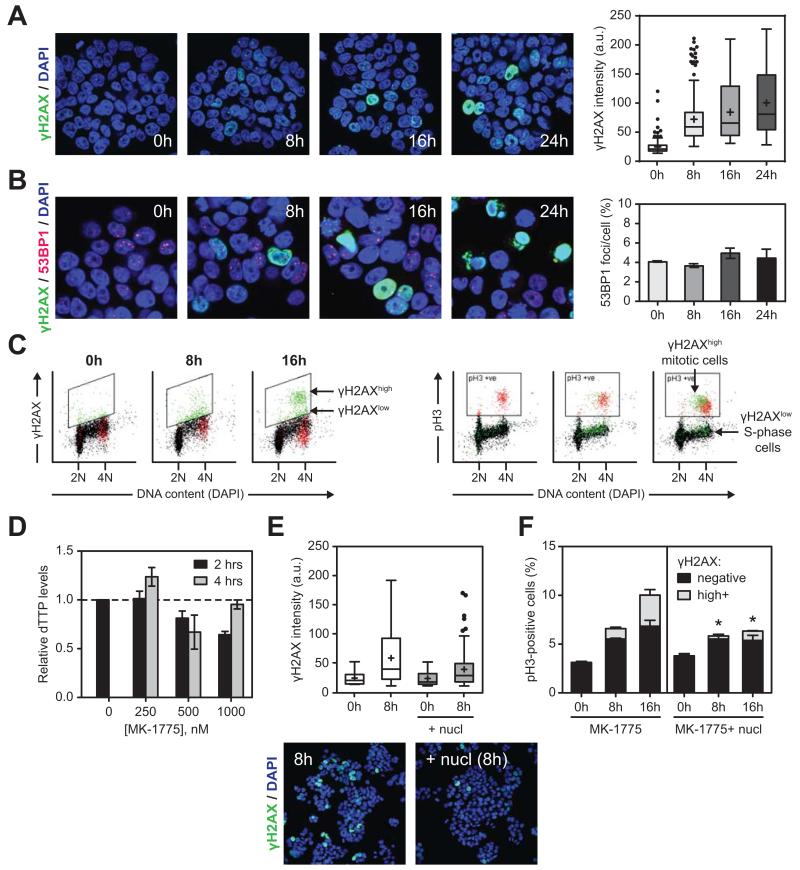
Figure 2. WEE1 inhibition induces replication stress and premature mitosis A.
Immunofluorescence of γH2AX (green) in WiDr cells treated with 250 nM MK-1775 for the indicated times. DNA was counterstained with DAPI (blue). Representative images and box and whisker plot of γH2AX intensity/cell are shown. Pan-nuclear γH2AX staining indicates widespread replication fork stalling.
B. Time course of 53BP1 (red) and γH2AX (green) immunofluorescence after WEE1 inhibition (250 nM). Representative images and average number of 53BP1 foci per cell of two independent experiments are shown.
C. Flow cytometry analysis of cells stained for γH2AX (left panel) and mitotic marker phospho-histone H3 (pH3; right panel) after treatment with 200 nM MK-1775 for the indicated times. Left panel illustrates two distinct populations of cells positive for γH2AX (green, in boxed area): γH2AXhigh and γH2AXlow; and a back-gated population of pH3-positive cells that is γH2AX-negative (in red). In the right panel, the γH2AXhigh and γH2AXlow cells are back-gated in a pH3 versus DNA content plot (pH3+ cells are gated in the boxed area). The γH2AXhighpH3+ double positive cells in these plots are premature mitotic (green cells in boxed area), while γH2AXlow cells were mainly in S or G2 phase (green cells outside boxed area). Normal mitotic cells positive for pH3 but not γH2AX are shown in red.
D. Intracellular dTTP levels were determined in cell extracts from WiDr cells treated with MK-1775 for 2 hours (black bars) or 4 hours (grey bars). Data was normalised to untreated control cells.
E. Nucleoside (nucl) supplementation reduced the number and intensity of γH2AX-positive cells after WEE1 inhibition. Representative immunofluorescence images and a box and whisker plot of γH2AX intensity/cell are shown of cells treated with 250 nM MK-1775 for 8 hours in the absence (white bars) or presence of nucleosides (grey bars).
F. Quantification of mitotic (pH3+) and premature mitotic (γH2AXhighpH3+) cells by flow cytometry analysis. Cells were treated as described in C. nucl indicates supplementation with nucleosides. For γH2AXhighpH3+ cells, * denotes P<0.04 versus MK-1775 alone (ratio paired Student’s t-test).
WEE1 inhibition induces increased entry into mitosis both from G2 cells (9, 24-26), as well as premature entry forced from S-phase (10). As anticipated, WEE1 inhibition resulted in an increase in pH3-positive mitotic cells as detected by flow cytometry (Fig. 2C). A fraction of the pH3-positive cells were also highly positive for γH2AX (γH2AXhigh). This γH2AXhigh phenotype was limited to cells that were also pH3-positive, identifying a cell population distinct from the pan-nuclear γH2AX cells that were observed by immunofluorescence. These γH2AXhighpH3+ double positive cells (green; Fig. 2C, right panels) had a lower DNA content compared to cells staining only for pH3+ (red), suggesting that these were cells forced into a premature mitosis directly from S-phase without completion of DNA synthesis. Such unscheduled mitosis has previously been shown to result in massive chromosome fragmentation (10), causing very high γH2AX expression. CDK1/2 inhibitor RO-3306 prevented both the induction of mitotic pH3-positive cells by WEE1 inhibition, and the induction of pan-nuclear γH2AX marking cells with stalled replication forks (Supplementary Fig. 2C).
Elevated CDK1/2 activity as a result of WEE1 inhibition has been shown to result in increased replication initiation and nucleotide shortage in S-phase (6). To assess whether nucleotide depletion could be the underlying cause of the observed replication stress, we determined the intracellular dTTP levels after WEE1 inhibition. Intracellular dTTP levels decreased after 2 hours of WEE1 inhibition, albeit only at relatively high doses of MK-1775 (Fig. 2D). Supplementation with nucleosides for 8 hours reduced the number (21.6% versus 11.2% with nucleosides) and fluorescence intensity of cells with pan-nuclear γH2AX detected by immunofluorescence (Fig. 2E), suggesting that nucleotide depletion in part contributes to replication stress. In addition, nucleotide supplementation decreased unscheduled mitosis, with the number of γH2AXhighpH3+ double positive cells decreasing 3-fold (3.2% after 16 hours of MK-1775 treatment versus 0.97% with nucleosides, Fig. 2F), suggesting that replication stress was a precursor to premature mitotic entry.
Together, these results show that WEE1 inhibition and unscheduled CDK activation cause two distinct but related phenotypes. In S-phase cells, nucleotide depletion results in replication stress and replication fork stalling, which is reflected by pan-nuclear γH2AX immunofluorescence staining. As a result of prolonged replication stress, cells stalled in late S-phase enter mitosis prior to completion of DNA synthesis, characterised by double γH2AXhighpH3+ staining by FACS analysis. Such unscheduled mitosis has been shown to result in chromosome fragmentation and apoptosis (10).
WEE1 inhibition promotes unscheduled mitosis after CHK1 and MYT1 depletion
Having established the assays for the phenotypes induced by WEE1 inhibition, we used these to investigate the underlying mechanism(s) by which the identified hits sensitised to WEE1 inhibition. We initially focused on the cell cycle kinases that regulate CDK activity. CHK1 is a key signalling kinase involved in the intra-S phase and G2/M checkpoints (27, 28). In response to replication stress or genotoxic insults, CHK1 phosphorylates and inactivates the CDC25 family of phosphatases, resulting in S or G2 arrest through inhibition of CDK activity. MYT1 kinase is functionally related to WEE1 and has been shown to similarly inhibit CDK1 and CDK2 through phosphorylation at Thr14 and/or Tyr15 (29). Thus, loss of CHK1 or MYT1 function cooperates with WEE1 inhibition by increasing CDK activity.
Depletion of CHK1 and MYT1 by siRNA indeed sensitised to WEE1 inhibitor MK-1775 in a clonogenic survival assay in WiDr cells (SF50 siCON1: 321 nM, siMYT1: 159 nM, siCHK1: 102 nM; Fig. 3A). We investigated whether sensitisation to WEE1 inhibition occurred as a consequence of increased replication stress, unscheduled mitosis, or both. Using FACS analysis, unscheduled mitotic γH2AXhighpH3+ cells were increased by siCHK1 and siMYT1 compared to siCON1 control cells after WEE1 inhibition for 16 hours (16.5% and 9.5% respectively versus 1.3%; Fig. 3B). CHK1 depletion alone induced unscheduled mitosis in 9.0% of cells, while MYT1 depletion did not (Fig. 3B and 3C). Dual inhibition of WEE1 and MYT1 with PD0166285 also triggered unscheduled mitosis (Supplementary Fig. 3). Silencing of MYT1 also induced replication stress as demonstrated by an increase in pan-nuclear γH2AX staining (22.9% in siMYT1 versus 12.8% in siCON1 after 8 hours; Fig. 3D). It was not possible to accurately assess replication stress after WEE1 inhibition in CHK1-depleted cells due to massive premature mitotic entry from S-phase (as demonstrated by a high number of γH2AXhighpH3+ double positive cells in Fig. 3B and 3C).
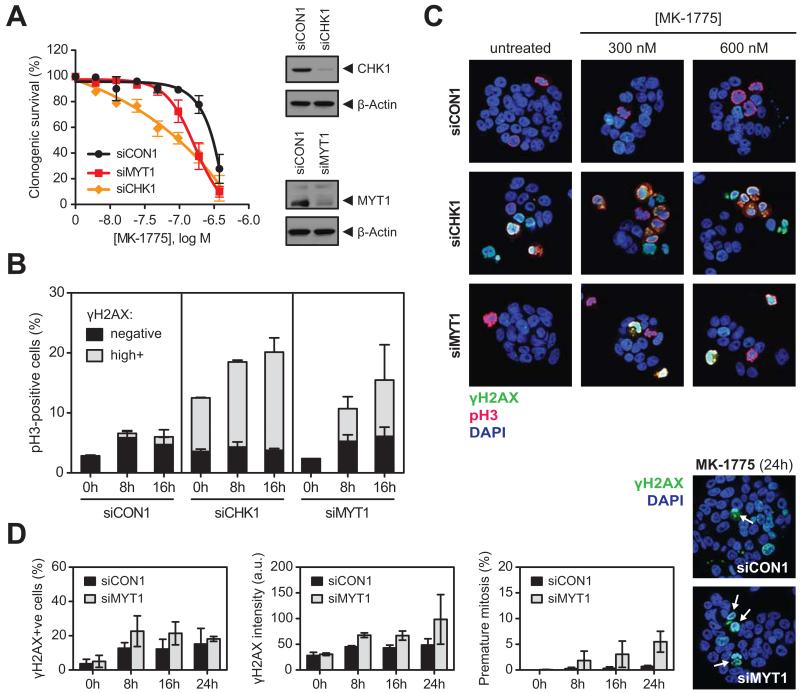
Figure 3. WEE1 inhibition promotes unscheduled mitosis after CHK1 and MYT1 depletion A.
Clonogenic survival of WiDr cells transfected with siCON1, siCHK1 or siMYT1, with colonies counted after 10 days of MK-1775 exposure. The number of colonies was normalised to untreated controls for each siRNA separately (clonogenic survival before normalisation in the absence of MK-1775: siCON1: 69%, siMYT1: 67% and siCHK1: 35%). Knockdown of CHK1 and MYT1 was confirmed by Western blot (β-Actin, loading control).
B. Flow cytometry analysis of γH2AX and pH3 after transfection with siCON1, siCHK1 and siMYT1. Cells were exposed to 200 nM MK-1775 for the indicated times. γH2AXhighpH3+ cells represent unscheduled mitosis. Error bars represent SEM of two independent experiments.
C. Immunofluorescence images showing induction of pan-nuclear γH2AX (green) and pH3 (red) staining in WiDr cells transfected with siCON1, siCHK1 or siMYT1 and exposed to 300 or 600 nM MK-1775 for 8 hours. Double γH2AX+pH3+ staining (yellow) indicates premature mitosis. DNA was counterstained with DAPI (blue).
D. WiDr cells transfected with siCON1 (black bars) or siMYT1 (grey bars) were exposed to 250 nM MK-1775 for the indicated times. Cells were stained for γH2AX and imaged by confocal microscopy. Representative immunofluorescence images show premature mitotic cells (indicated by arrows) after 24 hours of MK-1775 exposure. Bar charts show number of γH2AX-positive cells (left), γH2AX intensity (middle) and premature mitosis (right) in siMYT1-transfected cells compared to siCON1. Error bars are SEM of two independent experiments.
These results suggest that CHK1 depletion sensitised cells to WEE1 inhibition through unscheduled mitosis in S-phase cells, whilst in MYT1-depleted cells both replication stress and unscheduled mitotic entry contributed to sensitisation.
WEE1 inhibition in FANCM- or BRIP1-depleted cells causes replication stress and unscheduled mitosis
Next, we focused on the mechanism of sensitivity of FA and HR depleted cells, studying FANCM and BRIP1, two FA proteins involved in maintaining genome stability, as the most significant screen hits. The translocase activity of FANCM plays a critical role during DNA replication by stabilising stalled forks to prevent their collapse into DSBs (30). FANCM-deficient cells are defective in replication fork restart and complete DNA synthesis by firing dormant origins in response to replication stress (30, 31). BRIP1 (or FANCJ) helicase activity is required for facilitating replication past natural fork barriers caused by secondary DNA structures to ensure timely progression through S-phase (32). This function of BRIP1 is independent of a functional FA pathway.
Sensitisation to WEE1 inhibitor MK-1775 by FANCM and BRIP1 depletion was confirmed in a clonogenic survival assay in WiDr cells (Supplementary Fig. 4A). In FANCM- and BRIP1-depleted cells, WEE1 inhibition induced greater levels of pan-nuclear γH2AX immunofluorescence staining compared to siCON1 transfected cells, indicative of increased replication stress (Fig. 4A, Supplementary Fig. 4B). BrdU pulse-chase experiments confirmed that pan-nuclear γH2AX induction occurred predominantly in cells that were in S-phase at the time of MK-1775 addition (Supplementary Fig. 4C). In addition, WEE1 inhibition induced a greater level of unscheduled mitosis (assayed by double γH2AXhighpH3+ staining) demonstrated by FACS analysis (Fig. 4B and 4C, Supplementary Fig. 4D), although the level of induction of unscheduled mitosis was low compared to that seen after CHK1 depletion (Fig. 3B). Particularly in FANCM-depleted cells, the induction of γH2AXhighpH3+ double positive cells was more dramatic and occurred at lower DNA content compared to siCON1 (Fig. 4B), suggesting that unscheduled mitosis was triggered earlier in S-phase due to more widespread replication stress.
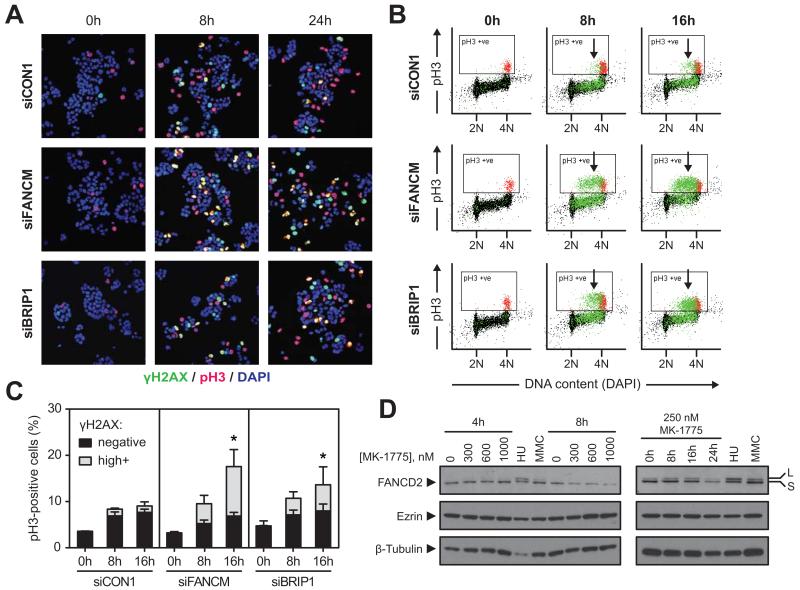
Figure 4. WEE1 inhibition in FANCM- or BRIP1-depleted cells causes replication stress and unscheduled mitosis A.
Immunofluorescence images showing induction of pan-nuclear γH2AX (green) and pH3 (red) staining in WiDr cells transfected with siCON1, siFANCM or siBRIP1 for 48 hours, and exposed to 250 nM MK-1775 for the indicated times. DNA was counterstained with DAPI (blue). Unscheduled mitosis is characterised by double γH2AX+pH3+ staining (yellow, quantified in Supplementary Fig. 4B).
B. Representative flow cytometry plots measuring pH3 (mitosis) and γH2AX (in green; events were back-gated from γH2AX versus DNA content plots shown in Supplementary Fig. 4D) in cells transfected with siCON1, siFANCM and siBRIP1 and exposed to 250 nM MK-1775 for the indicated times. Normal mitotic pH3+ cells are shown in red. Arrows indicate premature mitotic cells that are γH2AXhighpH3+ double positive. A back-gated population of S-phase cells that is γH2AXlow is shown in green. 2N DNA content indicates cells in G1 phase, 4N DNA content indicates cells in either G2 or M phase.
C. Quantification of mitosis (pH3+) and premature mitosis (γH2AXhighpH3+) by flow cytometry analysis of cells treated as described in B. For γH2AXhighpH3+ cells, * denotes P<0.05 versus siCON1 at 16h (ratio paired Student’s t-test).
D. Western blots of FANCD2 in cells exposed to 300, 600 and 1000 nM of MK-1775 for 4 or 8 hours (left) or to 250 nM MK-1775 for indicated times (right). Hydroxyurea (HU) or mitomycin C (MMC) treatment for 24 hours served as positive controls. Ezrin and β-Tubulin were used as loading controls. S, short non-ubiquitinated FANCD2 isoform. L mono-ubiquitinated long isoform.
WEE1 inhibition in cells with FA defects results in nucleotide depletion
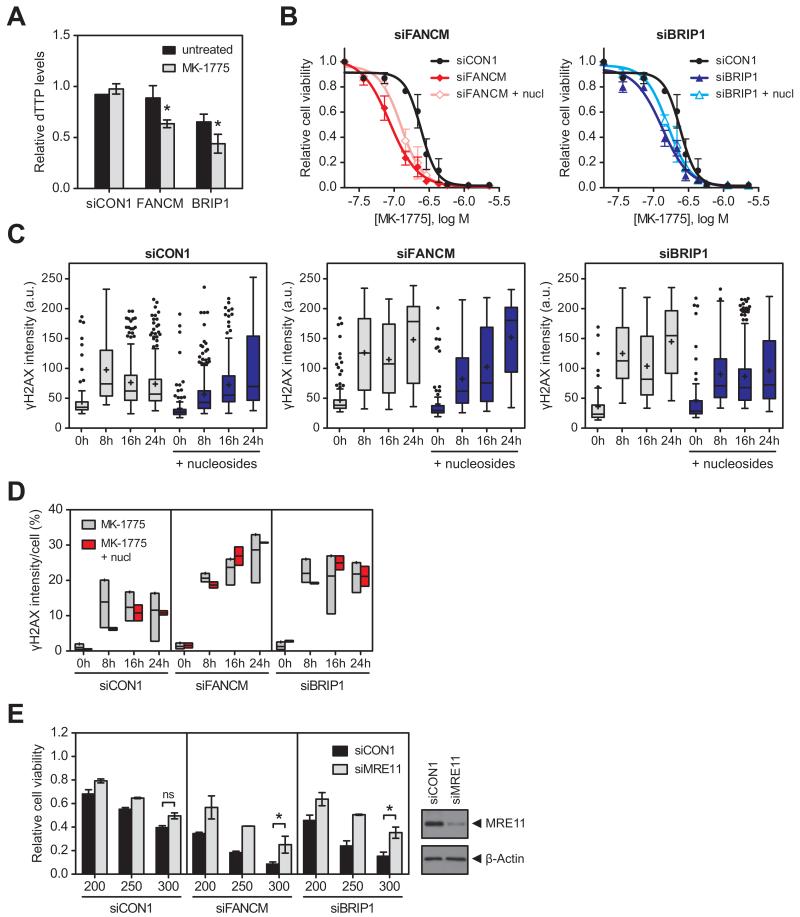
WEE1 inhibition did not induce FANCD2 mono-ubiquitination (Fig. 4D), suggesting that WEE1 inhibition did not lead to strong activation of the FA core complex, and we explored other mechanisms of synergy. FANCM-deficient cells have been shown to increase firing of replication origins to rescue DNA synthesis under conditions of replicative stress (31). In contrast, BRIP1-deficient cells recover from replication stress by an increase in repriming events downstream of the blockade (32). Because both events may increase nucleotide consumption, we determined the intracellular nucleotide levels following WEE1 inhibition in FANCM- or BRIP1-depleted cells. Indeed, intracellular dTTP levels were significantly reduced after 4 hours of WEE1 inhibition in siFANCM- or siBRIP1-transfected cells compared to siCON1 transfected cells (P=0.0015 and P=0.03, respectively; Fig. 5A). Nucleoside supplementation partially rescued the increased sensitivity to WEE1 inhibition after FANCM or BRIP1 knockdown in a short-term assay (Fig. 5B), deferring the onset of replication stress (Fig. 5C and 5D, Supplementary Fig. 5A) and unscheduled mitotic entry (γH2AXhighpH3+ double positive cells after 8 hours of MK-1775 alone versus supplemented with nucleosides: 1.5% versus 0.4% for siCON1, 4.4% versus 1.4% for siFANCM, 3.6% versus 0.5% for siBRIP1; compare Fig. 4C to Supplementary Fig. 5B).
Figure 5. WEE1 inhibition in cells with FA defects results in nucleotide depletion and nucleoside supplementation defers the onset of replication stress A.
WiDr cells were transfected with siCON1, siFANCM or siBRIP1, exposed to 250 nM MK-1775 for 4 hours, and processed for quantification of intracellular dTTPs. * denotes P=0.0015 for siFANCM and P=0.03 for siBRIP1 versus siCON1 + MK-1775 (paired Student’s t-test).
B. 48 hours post transfection with siCON1, siFANCM (left panel) or siBRIP1 (right panel), WiDr cells were treated with MK-1775 for 4 days in the presence (open symbols) or absence of nucleosides (nucl; closed symbols). Cell viability was determined using CellTiter-Glo and normalised to untreated cells for each siRNA separately. Error bars represent SEM of two independent experiments.
C. WiDr cells transfected with siCON1, siFANCM or siBRIP1 48 hours earlier were exposed to 250 nM MK-1775 for the indicated times in the absence (grey bars) or presence of nucleosides (blue bars). Cells were stained for γH2AX and imaged by confocal microscopy (representative images in Supplementary Fig. 5A). Box and whisker plots show representative quantification of γH2AX intensity/cell.
D. Quantification of γH2AX-positive cells treated as described in C. Floating bar chart shows the minimum, maximum and mean values for the average γH2AX intensity/cell after MK-1775 treatment in the absence (grey bars) or presence of nucleosides (red bars) of at least two independent experiments.
E. At 48 hours post transfection with siCON1, siFANCM or siBRIP1 in combination with siCON1 (black bars) or siMRE11 (grey bars), WiDr cells were exposed to 200, 250 or 300 nM MK-1775 for 4 days. Cell viability was determined using CellTiter-Glo and normalised to controls without MK-1775 for each siRNA combination separately. * denotes P<0.02; ns, not significant (ratio paired Student’s t-test). Knockdown of MRE11 was confirmed by Western blot analysis (β-Actin, loading control).
MRE11 resects stalled replication forks to generate intermediates for HR repair, which would involve both FANCM and BRIP1. Co-depletion of MRE11 significantly decreased the sensitivity of FANCM- and BRIP1-depleted cells to WEE1 inhibition (Fig. 5E), and attenuated the induction of pan-nuclear γH2AX (Supplementary Fig. 6A), and the level of unscheduled mitotic entry (Supplementary Fig. 6B). WEE1 has been shown to prevent DNA damage by controlling the MUS81-EME1 endonuclease during DNA replication (6, 7). Cleavage of stalled replication forks by endonucleases such as MUS81-EME1 and SLX1-SLX4 results in the formation of a DSB that can be repaired by HR. However, co-depletion of either MUS81 or SLX4 had no effect on cell survival (Supplementary Fig. 6C). The sensitivity of MYT1-depleted cells to WEE1 inhibition was not affected by co-depletion of MRE11 (Supplementary Fig. 6D), suggesting that the rescue by MRE11 siRNA was specific to genes required to repair stalled replication forks.
Taken together, these data suggest that WEE1 inhibition results in an increase in stalled replication forks that are processed in an MRE11-dependent manner to induce repair via HR/FA pathways. FANCM- and BRIP1-depleted cells fail to resolve these lesions, resulting in increased replication stalling and arrest, which is exacerbated by the resulting nucleotide depletion. Prolonged stalling in S-phase subsequently results in inappropriate mitotic entry of late S-phase cells and increased sensitivity to WEE1 inhibition.
Discussion
Successful clinical application of cell cycle kinase inhibitors or other targeted therapies depends on the identification of tumour specific vulnerabilities that can deliver a therapeutic window (2). Although WEE1 inhibitors are currently in early stage clinical trials in combination with chemotherapeutic agents, insight into the specific vulnerabilities that increase sensitivity to WEE1 inhibitors is limited. Here, through functional genetic screens we have identified novel determinants of sensitivity to WEE1 inhibition, and therapeutic partners to increase sensitivity. We demonstrate that WEE1 inhibition reduces cell survival through two related mechanisms, triggering both replication stress and unscheduled mitotic entry directly from S-phase. We identify two groups of kinases that modify sensitivity to WEE1 inhibition: the checkpoint kinases CHK1 and MYT1, inhibition of which likely sensitises to WEE1 inhibition through increasing CDK activity, and FA proteins FANCM and BRIP1, whose depletion sensitises to WEE1 inhibition likely by inhibiting repair of stalled replication forks generated by WEE1 inhibition.
CHK1, WEE1 and MYT1 all control genome integrity by restraining CDK activity. Several studies have reported the synergistic effects of combined CHK1 and WEE1 inhibition (13-16). In the absence of CHK1, cells are unable to maintain a functional intra-S phase checkpoint and accumulate DNA damage in S-phase (33). In combination with WEE1 inhibition this results in replication stress and DNA damage or rapid progression of S-phase cells into mitosis (10, 13). However, the synergy between CHK1 and WEE1 inhibition is observed in multiple cell lines and seems independent of p53 status (13, 15), suggesting that this combination may not be tumour-specific and could potentially induce excessive replication stress in normal cells. Silencing of MYT1 also resulted in increased sensitivity to MK-1775 and induction of DNA damage markers (Fig. 3) (34). Although MYT1 inhibitors are currently unavailable, combined inhibition of MYT1 and WEE1 could be promising, with potentially a greater therapeutic window than CHK1 and WEE1 inhibition. Alternatively, low MYT1 expression may serve as a biomarker for predicting MK-1775 sensitivity as suggested by others (34).
The maintenance of replication fork stability is essential for faithful genome duplication. The FA-BRCA pathway is critical for DNA repair by HR and is needed for the repair of replication blocking lesions such as DNA interstrand cross-links (ICLs) (35). FANCM is targeted to stalled replication forks (36), where it loads the FA core complex (consisting of FANC A, B, C, E, F, G and L) onto the chromatin. This leads to monoubiquitination of FANCD2 and FANCI and activation of the downstream FA proteins FANCN (PALB2), FANCD1 (BRCA2) and FANCJ (BRIP1), and BRCA1 to repair the damage (37). Besides 5 members of the FA pathway (FANCM, BRIP1, FANCE, PALB2, and BRCA2), we identified four other genes (RAD54B, RECQL4, RAD50, and RAD52) whose depletion sensitised to WEE1 inhibition (Fig. 1B, Supplementary Fig. 1E). These genes are all involved in HR repair or preservation of replication fork integrity, suggesting that defects in these pathways present a general mechanism to increase sensitivity to WEE1 inhibition, and that tumours with inactivation of these pathways may show increased sensitivity to WEE1 inhibition.
While biallelic mutations in the FA-BRCA genes cause FA, monoallelic mutations in BRCA2, PALB2 and BRIP1 predispose to breast, ovarian and other cancers (38-40). Both BRIP1 and PALB2 directly interact with BRCA1 and BRCA2 and play critical roles in the recruitment of the BRCA complexes to sites of DNA damage to prevent DNA breakage and to promote DNA repair by HR. Consequently, mutations in these downstream FA genes are associated with perturbed DNA replication, an impaired DNA damage response, and increased genomic instability (41-43); features that make these cancers attractive targets for WEE1 inhibition. Interestingly, MK-1775 exhibited anti-tumour activity as a single agent in a patient with BRCA mutated head and neck cancer in a recent phase I clinical trial, confirming that our findings may translate into an effective synthetic lethal strategy in the clinic (44).
Our work corroborates previous findings that WEE1 inhibition results in nucleotide shortage, increased replication stress and unscheduled mitosis. However, we show that nucleotide shortage is only a minor component of the sensitivity of FANCM/BRIP1 depleted cells to WEE1 inhibition (Fig. 5A). WEE1 inhibition results in stalled replication forks that are processed by MRE11 to HR intermediates (Fig. 5E, Supplementary Fig. 6A and 6B). In contrast to previous reports (6, 7), MUS81/SLX4 appeared not to be involved in replication fork resection upon WEE1 inhibition in FA-depleted cells, since co-depletion of MUS81 or SLX4 did not prevent the induction of DNA damage nor improve cell survival after WEE1 inhibition in FA-depleted cells (Supplementary Fig. 6C). SLX4 has been identified as a novel FA protein (FANCP) (45), that coordinates DNA repair activities by binding to the structure-specific endonucleases XPF-ERCC1, MUS81-EME1 and SLX1, thereby stimulating their enzymatic activities (46). In our experiments, depletion of FANCM or BRIP1 may preclude MUS81/SLX4 activation and promote CtIP- and MRE11-dependent resection of stalled replication forks instead (20, 30). The ability to resolve these recombination intermediates results in prolonged replication stalling. In the absence of WEE1, such prolonged replication stalling may subsequently result in premature mitotic entry directly from S-phase, which is a lethal event (10).
Here, we conduct a functional genetic screen to identify determinants of sensitivity to the WEE1 inhibitor MK-1775. We identify potential therapeutic partners, including MYT1 kinase inhibitors, that synergise with WEE1 inhibition through increased replication stress and premature mitotic entry. We identify novel determinants of sensitivity to WEE1 inhibition, including cancers with defects in FA/HR pathways, suggesting potential novel cancer subtypes to be explored in future clinical studies of WEE1 inhibitors.
Supplementary Material
Acknowledgments
This work was supported by Cancer Research UK grant A10038, Susan G. Komen for the Cure, the Netherlands Organisation for Scientific Research (NWO; M. Aarts) and Breakthrough Breast Cancer Research. We acknowledge NHS funding to the NIHR Biomedical Research Centre at The Royal Marsden and the ICR.
Financial support: M. Aarts, A. Ashworth, N. C. Turner were supported by Cancer Research UK (A10038) and Susan G. Komen for the Cure. M. Aarts was supported by the Netherlands Organisation for Scientific Research (NWO). I. Bajrami, M. T. Herrera-Abreu, R. Elliott, R. Brough, and C. J. Lord were supported by Breakthrough Breast Cancer Research.
Footnotes
Conflict of interest: none declared
References
- 1.Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–23. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 2.Aarts M, Linardopoulos S, Turner NC. Tumour selective targeting of cell cycle kinases for cancer treatment. Curr Opin Pharmacol. 2013;13:529–35. doi: 10.1016/j.coph.2013.03.012. [DOI] [PubMed] [Google Scholar]
- 3.Parker LL, Piwnica-Worms H. Inactivation of the p34cdc2-cyclin B complex by the human WEE1 tyrosine kinase. Science. 1992;257:1955–7. doi: 10.1126/science.1384126. [DOI] [PubMed] [Google Scholar]
- 4.Watanabe N, Broome M, Hunter T. Regulation of the human WEE1Hu CDK tyrosine 15-kinase during the cell cycle. EMBO J. 1995;14:1878–91. doi: 10.1002/j.1460-2075.1995.tb07180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindqvist A, Rodriguez-Bravo V, Medema RH. The decision to enter mitosis: feedback and redundancy in the mitotic entry network. J Cell Biol. 2009;185:193–202. doi: 10.1083/jcb.200812045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beck H, Nahse-Kumpf V, Larsen MS, O’Hanlon KA, Patzke S, Holmberg C, et al. Cyclin-dependent kinase suppression by WEE1 kinase protects the genome through control of replication initiation and nucleotide consumption. Mol Cell Biol. 2012;32:4226–36. doi: 10.1128/MCB.00412-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dominguez-Kelly R, Martin Y, Koundrioukoff S, Tanenbaum ME, Smits VA, Medema RH, et al. Wee1 controls genomic stability during replication by regulating the Mus81-Eme1 endonuclease. J Cell Biol. 2011;194:567–79. doi: 10.1083/jcb.201101047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caretti V, Hiddingh L, Lagerweij T, Schellen P, Koken PW, Hulleman E, et al. WEE1 kinase inhibition enhances the radiation response of diffuse intrinsic pontine gliomas. Mol Cancer Ther. 2013;12:141–50. doi: 10.1158/1535-7163.MCT-12-0735. [DOI] [PubMed] [Google Scholar]
- 9.PosthumaDeBoer J, Wurdinger T, Graat HC, van Beusechem VW, Helder MN, van Royen BJ, et al. WEE1 inhibition sensitizes osteosarcoma to radiotherapy. BMC Cancer. 2011;11:156. doi: 10.1186/1471-2407-11-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aarts M, Sharpe R, Garcia-Murillas I, Gevensleben H, Hurd MS, Shumway SD, et al. Forced mitotic entry of S-phase cells as a therapeutic strategy induced by inhibition of WEE1. Cancer Discov. 2012;2:524–39. doi: 10.1158/2159-8290.CD-11-0320. [DOI] [PubMed] [Google Scholar]
- 11.Hirai H, Arai T, Okada M, Nishibata T, Kobayashi M, Sakai N, et al. MK-1775, a small molecule Wee1 inhibitor, enhances anti-tumor efficacy of various DNA-damaging agents, including 5-fluorouracil. Cancer Biol Ther. 2010;9:514–22. doi: 10.4161/cbt.9.7.11115. [DOI] [PubMed] [Google Scholar]
- 12.Hirai H, Iwasawa Y, Okada M, Arai T, Nishibata T, Kobayashi M, et al. Small-molecule inhibition of Wee1 kinase by MK-1775 selectively sensitizes p53-deficient tumor cells to DNA-damaging agents. Mol Cancer Ther. 2009;8:2992–3000. doi: 10.1158/1535-7163.MCT-09-0463. [DOI] [PubMed] [Google Scholar]
- 13.Carrassa L, Chila R, Lupi M, Ricci F, Celenza C, Mazzoletti M, et al. Combined inhibition of Chk1 and Wee1: in vitro synergistic effect translates to tumor growth inhibition in vivo. Cell Cycle. 2012;11:2507–17. doi: 10.4161/cc.20899. [DOI] [PubMed] [Google Scholar]
- 14.Davies KD, Cable PL, Garrus JE, Sullivan FX, von Carlowitz I, Huerou YL, et al. Chk1 inhibition and Wee1 inhibition combine synergistically to impede cellular proliferation. Cancer Biol Ther. 2011;12:788–96. doi: 10.4161/cbt.12.9.17673. [DOI] [PubMed] [Google Scholar]
- 15.Guertin AD, Martin MM, Roberts B, Hurd M, Qu X, Miselis NR, et al. Unique functions of CHK1 and WEE1 underlie synergistic anti-tumor activity upon pharmacologic inhibition. Cancer Cell Int. 2012;12:45. doi: 10.1186/1475-2867-12-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Russell MR, Levin K, Rader J, Belcastro L, Li Y, Martinez D, et al. Combination therapy targeting the Chk1 and Wee1 kinases shows therapeutic efficacy in neuroblastoma. Cancer Res. 2013;73:776–84. doi: 10.1158/0008-5472.CAN-12-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vriend LE, De Witt Hamer PC, Van Noorden CJ, Wurdinger T. WEE1 inhibition and genomic instability in cancer. Biochim Biophys Acta. 2013;1836:227–35. doi: 10.1016/j.bbcan.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 18.Wilson PM, Labonte MJ, Russell J, Louie S, Ghobrial AA, Ladner RD. A novel fluorescence-based assay for the rapid detection and quantification of cellular deoxyribonucleoside triphosphates. Nucleic Acids Res. 2011;39:e112. doi: 10.1093/nar/gkr350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whitby MC. The FANCM family of DNA helicases/translocases. DNA Repair (Amst) 2010;9:224–36. doi: 10.1016/j.dnarep.2009.12.012. [DOI] [PubMed] [Google Scholar]
- 20.Suhasini AN, Sommers JA, Muniandy PA, Coulombe Y, Cantor SB, Masson JY, et al. Fanconi anemia group J helicase and MRE11 nuclease interact to facilitate the DNA damage response. Mol Cell Biol. 2013;33:2212–27. doi: 10.1128/MCB.01256-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wesoly J, Agarwal S, Sigurdsson S, Bussen W, Van Komen S, Qin J, et al. Differential contributions of mammalian Rad54 paralogs to recombination, DNA damage repair, and meiosis. Mol Cell Biol. 2006;26:976–89. doi: 10.1128/MCB.26.3.976-989.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thangavel S, Mendoza-Maldonado R, Tissino E, Sidorova JM, Yin J, Wang W, et al. Human RECQ1 and RECQ4 helicases play distinct roles in DNA replication initiation. Mol Cell Biol. 2010;30:1382–96. doi: 10.1128/MCB.01290-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu X, Rochette PJ, Feyissa EA, Su TV, Liu Y. MCM10 mediates RECQ4 association with MCM2-7 helicase complex during DNA replication. EMBO J. 2009;28:3005–14. doi: 10.1038/emboj.2009.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bridges KA, Hirai H, Buser CA, Brooks C, Liu H, Buchholz TA, et al. MK-1775, a novel Wee1 kinase inhibitor, radiosensitizes p53-defective human tumor cells. Clin Cancer Res. 2011;17:5638–48. doi: 10.1158/1078-0432.CCR-11-0650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mir SE, De Witt Hamer PC, Krawczyk PM, Balaj L, Claes A, Niers JM, et al. In silico analysis of kinase expression identifies WEE1 as a gatekeeper against mitotic catastrophe in glioblastoma. Cancer Cell. 2010;18:244–57. doi: 10.1016/j.ccr.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarcar B, Kahali S, Prabhu AH, Shumway SD, Xu Y, Demuth T, et al. Targeting radiation-induced G(2) checkpoint activation with the Wee-1 inhibitor MK-1775 in glioblastoma cell lines. Mol Cancer Ther. 2011;10:2405–14. doi: 10.1158/1535-7163.MCT-11-0469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dai Y, Grant S. Targeting Chk1 in the replicative stress response. Cell Cycle. 2010;9:1025. doi: 10.4161/cc.9.6.11155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sorensen CS, Syljuasen RG. Safeguarding genome integrity: the checkpoint kinases ATR, CHK1 and WEE1 restrain CDK activity during normal DNA replication. Nucleic Acids Res. 2012;40:477–86. doi: 10.1093/nar/gkr697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mueller PR, Coleman TR, Kumagai A, Dunphy WG. Myt1: a membrane-associated inhibitory kinase that phosphorylates Cdc2 on both threonine-14 and tyrosine-15. Science. 1995;270:86–90. doi: 10.1126/science.270.5233.86. [DOI] [PubMed] [Google Scholar]
- 30.Blackford AN, Schwab RA, Nieminuszczy J, Deans AJ, West SC, Niedzwiedz W. The DNA translocase activity of FANCM protects stalled replication forks. Hum Mol Genet. 2012;21:2005–16. doi: 10.1093/hmg/dds013. [DOI] [PubMed] [Google Scholar]
- 31.Schwab RA, Blackford AN, Niedzwiedz W. ATR activation and replication fork restart are defective in FANCM-deficient cells. EMBO J. 2010;29:806–18. doi: 10.1038/emboj.2009.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwab RA, Nieminuszczy J, Shin-ya K, Niedzwiedz W. FANCJ couples replication past natural fork barriers with maintenance of chromatin structure. J Cell Biol. 2013;201:33–48. doi: 10.1083/jcb.201208009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beck H, Nahse V, Larsen MS, Groth P, Clancy T, Lees M, et al. Regulators of cyclin-dependent kinases are crucial for maintaining genome integrity in S phase. J Cell Biol. 2010;188:629–38. doi: 10.1083/jcb.200905059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guertin AD, Li J, Liu Y, Hurd MS, Schuller AG, Long B, et al. Preclinical evaluation of the WEE1 inhibitor MK-1775 as single-agent anticancer therapy. Mol Cancer Ther. 2013;12:1442–52. doi: 10.1158/1535-7163.MCT-13-0025. [DOI] [PubMed] [Google Scholar]
- 35.D’Andrea AD. Susceptibility pathways in Fanconi’s anemia and breast cancer. N Engl J Med. 2010;362:1909–19. doi: 10.1056/NEJMra0809889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singh TR, Saro D, Ali AM, Zheng XF, Du CH, Killen MW, et al. MHF1-MHF2, a histone-fold-containing protein complex, participates in the Fanconi anemia pathway via FANCM. Mol Cell. 2010;37:879–86. doi: 10.1016/j.molcel.2010.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deans AJ, West SC. DNA interstrand crosslink repair and cancer. Nat Rev Cancer. 2011;11:467–80. doi: 10.1038/nrc3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rahman N, Seal S, Thompson D, Kelly P, Renwick A, Elliott A, et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2007;39:165–7. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seal S, Thompson D, Renwick A, Elliott A, Kelly P, Barfoot R, et al. Truncating mutations in the Fanconi anemia J gene BRIP1 are low-penetrance breast cancer susceptibility alleles. Nat Genet. 2006;38:1239–41. doi: 10.1038/ng1902. [DOI] [PubMed] [Google Scholar]
- 40.Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, et al. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789–92. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- 41.Kumaraswamy E, Shiekhattar R. Activation of BRCA1/BRCA2-associated helicase BACH1 is required for timely progression through S phase. Mol Cell Biol. 2007;27:6733–41. doi: 10.1128/MCB.00961-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nikkila J, Parplys AC, Pylkas K, Bose M, Huo Y, Borgmann K, et al. Heterozygous mutations in PALB2 cause DNA replication and damage response defects. Nat Commun. 2013;4:2578. doi: 10.1038/ncomms3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roy R, Chun J, Powell SN. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat Rev Cancer. 2012;12:68–78. doi: 10.1038/nrc3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tu Do K, Wilsker D, Balasubramanian P, Zlott J, Jeong W, Lawrence SM, et al. Phase I trial of AZD1775 (MK1775), a wee1 kinase inhibitor, in patients with refractory solid tumors. J Clin Oncol. 2014;32:5s. doi: 10.1200/JCO.2014.60.4009. (suppl; abstr 2503) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim Y, Lach FP, Desetty R, Hanenberg H, Auerbach AD, Smogorzewska A. Mutations of the SLX4 gene in Fanconi anemia. Nat Genet. 2011;43:142–6. doi: 10.1038/ng.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cybulski KE, Howlett NG. FANCP/SLX4: a Swiss army knife of DNA interstrand crosslink repair. Cell Cycle. 2011;10:1757–63. doi: 10.4161/cc.10.11.15818. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.