Abstract
Purpose
At the molecular level myeloma is characterised by copy number abnormalities and recurrent translocations into the immunoglobulin heavy chain (IGH) locus. Novel methodologies such as massively parallel sequencing have begun to describe the pattern of tumour acquired mutations but their clinical relevance has yet to be established.
Methods
We have performed whole exome sequencing on 463 presenting myeloma patients enrolled in the NCRI Myeloma XI trial (NCT01554852) for which complete molecular cytogenetic and clinical outcome data are available.
Results
We identified 15 significantly mutated genes comprising IRF4, KRAS, NRAS, MAX, HIST1H1E, RB1, EGR1, TP53, TRAF3, FAM46C, DIS3, BRAF, LTB, CYLD and FGFR3. The mutational spectrum is dominated by mutations in the RAS (43%) and NF-κB (17%) pathway, but while they are prognostically neutral they could be targeted therapeutically. Mutations in CCND1 and DNA repair pathway alterations (TP53, ATM, ATR and ZNFHX4 mutations) are associated with a negative impact on survival. In contrast, those in IRF4 and EGR1 are associated with a favourable overall survival. We have combined these novel mutation risk factors with the recurrent molecular adverse features and ISS to generate an ISS–MUT score that can identify a high-risk population that relapse and die prematurely.
Conclusion
We have refined our understanding of genetic events in myeloma and identified clinically relevant mutations that may be used to better stratify patients at presentation.
Introduction
Myeloma arises following the immortalisation of a plasma cell that subsequently acquires further genetic abnormalities leading to an increasingly malignant phenotype1. While the treatment of myeloma has improved with increasing numbers of survivors beyond ten years from diagnosis there are still a significant number of patients with both short progression-free survival (PFS) and overall survival (OS)2-4. It is important to identify these “high-risk” patients and to design trials aimed at improving their outcome. Traditionally, prognosis has been assessed by the use of clinical data such as the international staging system (ISS)3,5, but for individual patients this is insufficient to direct treatment. ISS can be improved by the addition of molecular cytogenetic data2,3 but this does not describe the full extent of the risk and could be improved by more comprehensively describing the genetic features of the disease and using it to define outcome.
The main clinically relevant molecular sub-groups of myeloma are defined by the balanced translocations into the IGH locus at 14q326-8 and copy number abnormalities (CNA). The translocation subgroups include the t(4;14) (13%) and the t(14;16)/t(14;20) translocated subgroups (5%) which are associated with poor prognosis with respective overall survival ranging from 22-60 months and 16-30 months, and the t(11;14) (13%) which is associated with a favourable prognosis (OS >5-10 years) 2,8-10.( More recently both the prevalence and adverse prognostic significance associated with MYC translocations (20%) has been realised (median OS 24 months) 11. A further set of clinically important prognostic abnormalities are defined by recurrent CNA the full spectrum of which have been defined by genome mapping experiments12. The clinically important copy number abnormalities include hyperdiploidy (HRD), associated with favourable prognosis, and gain(1q) (38%), del(1p) (8.4%), del(17p) (9.5%) and del(12p) (8.9%) being associated with adverse prognosis 2,9.
We have used FISH previously to show that patients with a greater number of adverse cytogenetic abnormalities have impaired clinical outcomes2. It has become clear that the value of these tests could be improved by identifying all of the prognostically important genetic abnormalities present and using all of the information to risk-stratify patients. A key technology with which to identify these abnormalities is whole exome sequencing plus a targeted pull down of regions of interest, which can identify structural variants as well as the spectrum of mutations13-15. In this study we have used such an approach on 463 presenting samples entered into a clinical trial and have used the data generated to define both the pattern of mutations and how they relate to survival. We have then combined this with the ISS to generate a predictive score to define high-risk clinical behaviour, which we term ISS-MUT.
Methods
Patient Samples
Samples were taken, following informed consent, from patients newly diagnosed with symptomatic myeloma and enrolled in the NCRI Myeloma XI trial (NCT01554852, CRUK/09/014). This is a phase III, open-label trial where patients were randomised between triplet immunomodulatory drug (IMiD) induction of either cyclophosphamide, thalidomide, dexamethasone (CTD) or cyclophosphamide, lenalidomide, dexamethasone (CRD). Patients with a suboptimal response (<very good partial response) were randomised to pre-transplant treatment with a proteasome inhibitor triplet (cyclophosphamide, bortezomib, dexamethasone, CVD). Older or less fit patients had appropriate dose reductions and did not receive an autologous stem cell transplant. All patients subsequently underwent further randomisation to either no maintenance, lenalidomide maintenance or lenalidomide and vorinostat maintenance, Overview of the Myeloma XI trial diagram. PFS and OS were measured from initial randomisation and the median (range) follow-up was 25 months (0.09, 42.97). The patient demographics are presented in Supplementary Table 1. The median PFS was 26.6 months (95%CI (23.6, 29.9)) and the 3-year overall survival rate was 66% (95%CI (60, 73)).
Exome Sequencing and mutation calling
Plasma cells were isolated from bone marrow samples using CD138+ MACSorting (Miltenyi Biotech, Bisley, UK). DNA from both tumour and peripheral blood were used in the exome capture protocol as previously described16. FastQC (v0.10.0) was used for basic quality control of Illumina paired-end sequencing data. Single nucleotide variants (SNVs) were called using MuTect (v1.1.4). Copy number across the exome was determined using Control-FREEC and Cancer Clonal fraction (CCF) calculated17.
Molecular cytogenetics
Translocations were called18 and manually curated. Simultaneously, translocations were determined by qRT-PCR19. When not concordant, the data were examined and a decision made. Copy number changes were assessed by Multiplex Ligation-Dependent Amplification using the SALSA P425-B1 probe mix (MRC-Holland, Amsterdam, The Netherlands)20. Copy number at each locus was estimated21 and determined as previously described22.
Correlation studies
Correlation between mutated genes and cytogenetic abnormalities using Bayesian inference was determined using the program “JAGS”23 and the R-interface Bayesmed 24,25. The probability of the observed data under the null hypothesis versus the alternative hypothesis or Bayes factor (BF) was computed. BF>1 was considered significant. BF 1-3, 3-20, 20-150 and >150 were considered weak, positive, strong and very strong associations respectively26. Correlation coefficients were plotted using corrplot27.
Survival analysis
Time-to-event analysis was performed in R28 using the survival29,30 and coin31,32 packages. Differences between survival functions were tested using the logrank test. Hazard ratios were estimated from Cox proportional hazard regression. All statistical tests were two-sided and evaluated at the 5% level. Power to detect prognostic associations was estimated using PowerSurvEpi33. Multivariable stepwise variable selection was performed using a standard backward elimination approach with p<0.05 taken as level of significance for variable retention to estimate an apparent predictive ability quantified using Harrell’s C-index. This predictive ability was then internally validated with a bootstrap resampling strategy using the package rms34 and the validate function where the stepwise selection strategy was also repeated. This analysis used 400 resamples and estimated an optimism-corrected C-index. Further details may be found in the Supplementary Methods.
Results
Significantly mutated genes and altered pathways
We identified 13 significantly mutated genes, Table 1, including KRAS, NRAS, TRAF3, TP53, FAM46C, DIS3, BRAF, LTB, CYLD, RB1, HIST1H1E, IRF4 and MAX. The location of the mutations may be found in Supplementary Figure 1.
Table 1.
Significantly mutated genes and their distributions in the main cytogenetic subgroups.
| Gene | Group | Samples with mutation in group (%) |
Non- synonymous Mutations (n=) |
Synonymous Mutations (n=) |
p-value | q-value |
|---|---|---|---|---|---|---|
| Overall | ||||||
| HIST1H1E | All | 2.8 | 15 | 1 | < 9×10−18 | < 9×10−18 |
| IRF4 | All | 3.2 | 15 | 1 | < 9×10−18 | < 9×10−18 |
| KRAS | All | 21.1 | 104 | 0 | < 9×10−18 | < 9×10−18 |
| MAX | All | 2.4 | 13 | 0 | < 9×10−18 | < 9×10−18 |
| NRAS | All | 19.4 | 91 | 0 | < 9×10−18 | < 9×10−18 |
| TP53 | All | 3.0 | 16 | 0 | < 9×10−18 | < 9×10−18 |
| TRAF3 | All | 3.7 | 19 | 0 | < 9×10−18 | < 9×10−18 |
| FAM46C | All | 5.6 | 26 | 1 | 5.00 ×10−15 | 1.05 ×10−11 |
| DIS3 | All | 8.6 | 48 | 1 | 5.88 ×10−15 | 1.11 ×10−11 |
| BRAF | All | 6.7 | 36 | 0 | 8.77 ×10−15 | 1.50 ×10−11 |
| LTB | All | 3.0 | 14 | 1 | 2.84 ×10−8 | 4.46 ×10−5 |
| CYLD | All | 2.4 | 14 | 0 | 6.65 ×10−6 | 9.66 ×10−3 |
| RB1 | All | 1.5 | 7 | 0 | 1.96 ×10−5 | 2.64 ×10−2 |
| t(4;14) | ||||||
| DIS3 | t(4;14) | 25.4 | 18 | 0 | 7.05 ×10−8 | 1.33 ×10−3 |
| FGFR3 | t(4;14) | 16.9 | 12 | 0 | 6.93 ×10−7 | 6.54 ×10−3 |
| t(11;14) | ||||||
| KRAS | t(11;14) | 33.7 | 31 | 0 | < 9×10−18 | < 9×10−18 |
| NRAS | t(11;14) | 25.6 | 22 | 0 | 1.44 ×10−15 | 1.36 ×10−11 |
| DIS3 | t(11;14) | 11.6 | 12 | 0 | 5.74 ×10−6 | 3.61 ×10−2 |
| IRF4 | t(11;14) | 10.5 | 9 | 0 | 1.18 ×10−5 | 5.57 ×10−2 |
| Hyperdiploidy | ||||||
| KRAS | HRD | 20.6 | 54 | 0 | < 9×10−18 | < 9×10−18 |
| NRAS | HRD | 24.4 | 53 | 0 | < 9×10−18 | < 9×10−18 |
| FAM46C | HRD | 7.1 | 15 | 1 | 6.99 ×10−9 | 4.40 ×10−5 |
| BRAF | HRD | 6.7 | 16 | 0 | 1.45 ×10−6 | 6.85 ×10−3 |
| EGR1 | HRD | 4.6 | 11 | 1 | 4.65 ×10−6 | 1.70 ×10−2 |
| CYLD | HRD | 2.9 | 11 | 0 | 1.89 ×10−5 | 5.09 ×10−2 |
The RAS/MAPK pathway is the most frequently mutated pathway (KRAS=21.2%, NRAS=19.4%, BRAF=6.7%) making up a total of 43.2% of patients with NRAS and KRAS tending to be mutually exclusive, but co-occurring in 2% of patients. They had no impact on survival, Supplementary Figure 2. The mean CCF for KRAS, NRAS and BRAF were 32%, 33% and 25% suggesting they are associated with progression. The hotspots of mutation in KRAS and NRAS included codons 12, 13 and 61 as well as codon 600 in BRAF, Supplementary Table 2-3.
Mutational activation of the NF-κB pathway genes35, Supplementary Figure 3, important in late B-cell development was seen. A total of 27 mutated genes falling within these pathways were identified, accounting for 17% of cases. These genes included TRAF3 (mutated in 3.7% and deleted in 13%), CYLD (mutated in 2.4% and deleted in 17%) and LTB (mutated in 3% of cases). The key plasma-cell survival gene and target of the IMiD drugs, IRF4, is mutated in 3.2% of cases, including eight cases with p.K123R. The pattern of these mutations and the down-regulation of MYC expression in these patients suggest they are activating. We identified mutations in another IMiD target gene36, EGR1 (n=17), all of which are located at the 5’ end of the gene.
We identified TP53 variants in 11% of cases (del(17p) in 9.5% and TP53 mutations in 3% of cases). Other mutated genes associated with the delivery of an apoptotic signal included ATM and ATR. Loss of ATM was seen in 1.3% and was mutated in 3% with ATR mutations and deletions seen in 1.5% of samples, resulting in combined TP53, ATM and ATR abnormalities present in 14.5% of patients. These abnormalities were seen in other datasets with similar frequencies (Supplementary Table 4, Supplementary Figures 4-5).
Association of Mutated Genes with Cytogenetic Sub-groups
Significantly mutated genes were seen within the cytogenetic sub-groups, Table 1. FGFR3, located on the der(14), is only mutated in the t(4;14) group where activating mutations are found in 17% of cases. CCND1 is significantly mutated in the t(11;14) subgroup (12% of cases) whereas the transcriptional regulator EGR1, is significantly mutated in the hyperdiploid samples. Integrating our data with previously published data13,15 established a series of 733 cases and identified 5 additional significantly mutated genes including PRDM1, BCL7A, ATRIP, NRM and PRKD2, Supplementary Table 5. PRKD2 mutations, which were present in 2.3% of cases, were correlated to t(4;14).
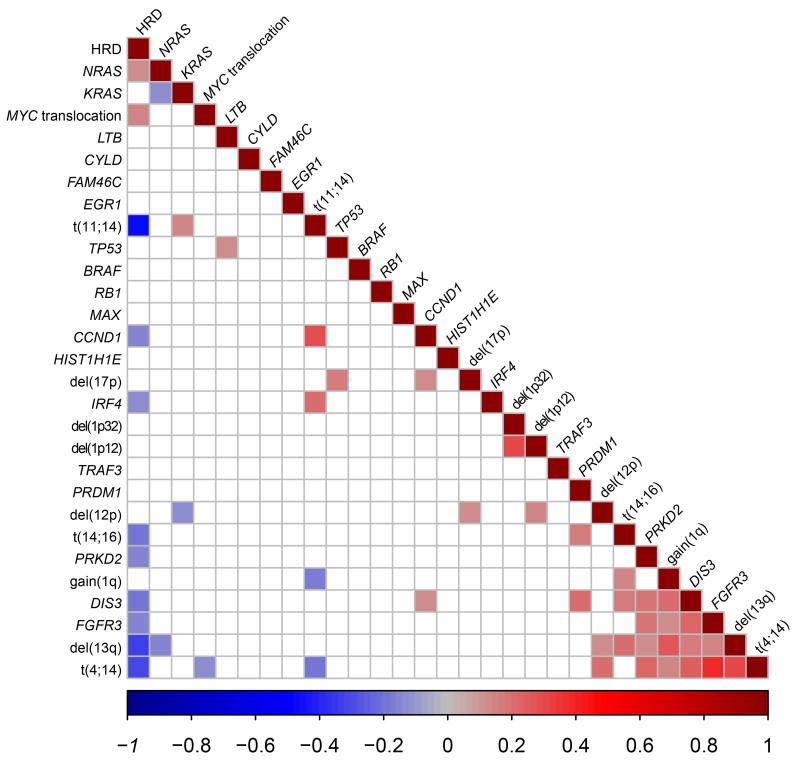
A number of adverse prognostic lesions were seen to co-segregate including t(4;14), del(13q), and gain(1q), Figure 1 and Supplementary Table 6. The t(11;14) was associated with KRAS and IRF4 mutations. Del(12p) was associated with t(4;14) and del(17p). MYC translocations were weakly correlated to HRD.
Figure 1.
Correlation between mutations and recurrent cytogenetic abnormalities. Intensity of colour shade represents the degree of correlation (blue=negative and red=positive) as per scale. Only significant correlations are represented on this plot with the insignificant correlations are in white.
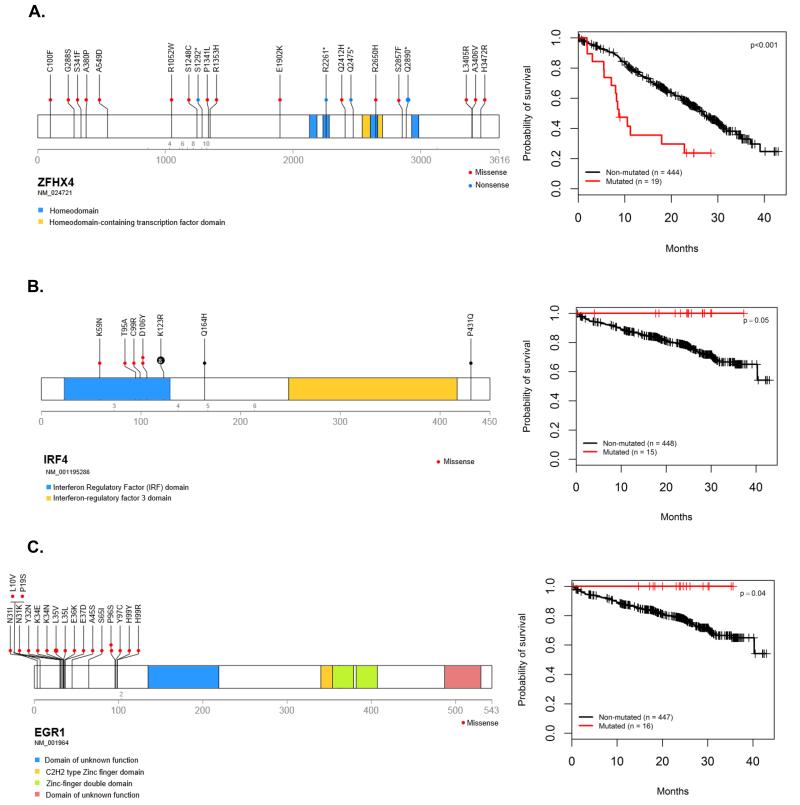
Within the region of del(12p) mutations in CHD4 were identified in 9 patients and deletions in 9% of patients. CHD4 interacts with ZFHX4 to modulate p53 function and mutations in ZFHX4 were seen in 20 cases, Supplementary Figure 6-Figure 3A.
Survival analysis
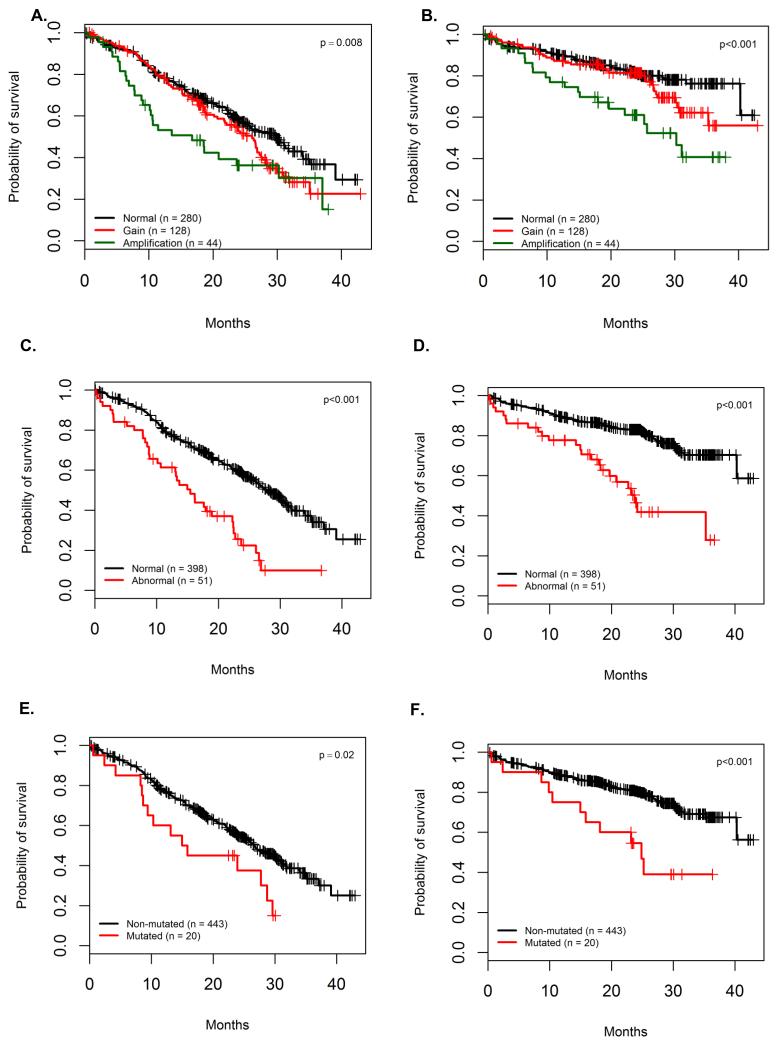
We carried out a time-to-event analysis of PFS and OS using the markers outlined above which included mutations occurring more than 10-times (excluding structural and housekeeping genes), copy number and structural abnormalities (CNSA) and mutated pathways with clinical features. This analysis had 80% power to identify features with a hazard ratio (HR) greater than 2.31 for PFS and 2.94 for OS, Supplementary Table 7. The dataset as a whole behaved as expected with negative associations being found for higher ISS stages, creatinine >150 μM, amp(1q), del(17p), MYC translocations, t(4;14), del(13q), del(1p32) and del(12p) and positive associations for hyperdiploidy, Table 2. Amp(1q) was prognostically relevant and seemed to drive the prognostic impact of gain(1q) (PFS: HR 1.8; 95%CI (1.2, 2.76); p=0.004 and OS HR 2.7; 95%CI (1.6, 4.5); p=0.0002), Figure 2A-B. Del(1p32) variants (including deletions and mutations) had a negative impact on OS (2y-OS 62%; 95%CI (48,79) versus 81%; 95%CI (77, 85); p=0.001), but not PFS (median 22.8 months; 95%CI (17.6, ∞) versus 26.7; 95%CI (24.0, 29.9); p=0.33).
Table 2.
Prognostic impact of the main variables in myeloma. Median survivals for mutations, CNSA and clinical variables are summarised in this table. NS stands for nonsignificant. NR for not reached.
| PFS present (months) |
PFS absent (months) |
p value | 2 year-OS present (%) |
2 year-OS absent (%) |
p value | Frequency (%) |
|
|---|---|---|---|---|---|---|---|
| Clinical features | |||||||
| Age>70 | 18.7 | 30.5 | <0.0001 | 76% | 81% | 0.06 | 40% |
| Creatinine >150 | 18.7 | 27.7 | 0.0003 | 67% | 81% | 0.0009 | 12% |
| ISS I | 39.1 | - | 87% | - | 30% | ||
| ISS II | 27.3 | 0.01 | 79% | 0.13 | 29% | ||
| ISS III | 22.0 | <0.0001 | 74% | 0.0004 | 34% | ||
| Mutations | |||||||
| ZFHX4 | 8.8 | 26.9 | <0.0001 | 72% | 80% | NS | 4% |
| TP53 | 13.7 | 26.9 | 0.0005 | 27% | 81% | 0.0001 | 3% |
| ATM/ATR mutations | 15.4 | 26.6 | 0.02 | 55% | 81% | 0.0008 | 4% |
| IRF4 | Not reached | 26.1 | 0.09 | 100% | 79% | 0.05 | 3.2% |
| EGR1 | 35.1 | 26.1 | NS | 100% | 79% | 0.04 | 3.5% |
| CCND1 | 10.7 | 26.6 | NS | 38% | 80% | 0.005 | 2.2% |
| NCKAP5 | 10.6 | 26.6 | NS | 53% | 80% | 0.04 | 2.2% |
| Translocations and copy number abnormalities | |||||||
| Del(17p)[TP53] | 15 | 27.5 | <0.0001 | 52% | 82% | <0.0001 | 9.5% |
| t(4;14) | 16.7 | 27.7 | 0.0001 | 71% | 80% | 0.08 | 12.7% |
| Del(13q) | 22.7 | 28.7 | 0.0008 | 75% | 82% | NS | 42.1% |
| Normal 1q [CKS1B] | 29.7 | - | 81% | - | 61% | ||
| Gain(1q) [CKS1B] | 26.1 | NS | 81% | NS | 28% | ||
| Amp(1q) [CKS1B] | 16.8 | 0.004 | 61% | 0.0002 | 9.5% | ||
| Hyperdiploidy | 29.6 | 23.7 | 0.009 | 81% | 77% | NS | 51.4% |
| MYC translocation | 21.7 | 27.5 | 0.008 | 68% | 82% | 0.006 | 18.4% |
| Del(12p) | 22.7 | 26.7 | 0.015 | 68% | 80% | 0.09 | 8.9% |
| MAF translocations | 17.6 | 26.6 | NS | 65% | 80% | NS | 4.5% |
| Del(1p)[FAF1 CDKN2C] | 21.7 | 26.7 | NS | 58% | 81% | 0.001 | 8.4% |
| t(14;16) | 17.6 | 26.6 | NS | 69% | 80% | NS | 3.8% |
| Del(1p) [FAM46C] | 26.6 | 26 | NS | 79% | 79% | NS | 24% |
| Del(16q) | 27 | 25.8 | NS | 77% | 81% | NS | 18.6% |
| t(11;14) | 26.6 | 26.1 | NS | 78% | 79% | NS | 18.6% |
| Del(14q32) [TRAF3] | 26.9 | 26.3 | NS | 80% | 79% | NS | 13.2% |
| Pathways | |||||||
| TP53 signal * | 15.4 | 28.4 | <0.0001 | 46% | 83% | <0.0001 | 11% |
| DIS3 signal * | 22.1 | 28.5 | 0.001 | 77% | 81% | NS | 40.2% |
| FAM46C signal * | 26.9 | 25.5 | NS | 79% | 79% | NS | 27% |
| RAS mutated | 27.5 | 23.9 | NS | 80% | 78% | NS | 43% |
| FAF1 signal * | 22.8 | 26.7 | NS | 62% | 81% | 0.001 | 9.9% |
| NF-κB signal * | 25.5 | 26.6 | NS | 78% | 81% | NS | 37% |
Signal refers to the combination of mutations and copy number changes
Figure 2.
Impact of DNA repair pathway alterations and CKS1B copy number changes in myeloma. The prognostic impact of the number of copies of amp(1q) is greater than gain(1q) for both PFS (A) and OS (B). TP53 mutations and deletions are also associated with a significant negative impact on PFS (C) and OS (D). ATM and ATR mutations are associated with a worse outcome on both PFS (E) and OS (F).
For the first time we are able to examine the impact of mutations discovered in a non-biased fashion on survival within a large clinical trial. We found that CCND1 mutations were associated with a negative impact on OS (2y-OS 38.1%; 95%CI (14, 100) versus 80% 95%CI (76, 84); p=0.005) (Supplementary Figure 7).
Inability to deliver an apoptotic signal was an important prognostic marker. Del(17p) and TP53 mutations have a significant negative impact on outcome (Figure 2C-D and Supplementary Figure 4). ATM mutations were associated with a trend towards impaired PFS (median 15.4 months; 95%CI (8.67, ∞) versus 26.6 months 95%CI (24.0, 30.0); p=0.05) and impaired OS (2y-OS 50%; 95%CI (30, 84) versus 80.3%; 95%CI (76, 84); p=0.01). ATR mutations were seen in 1.5% of cases and have a similar impact on prognosis in terms of PFS (median 23.9 months; 95%CI (10.35, ∞) versus 26.6 months; 95%CI (24.0, 29.9); p=0.3) and OS (2y-OS 67%; 95%CI (38, 100) versus 80%; 95%CI (76, 84); p=0.05). Combined together, ATM and ATR mutations, and TP53 mutations and del(17p) had a significant impact on both PFS and OS, Figure 2E-F and Supplementary Table 8. Mutations in ZFHX4, are associated with a negative impact on PFS (median 8.8 months; 95%CI (8.05, ∞) versus 26.9 months; 95%CI (25.0, 30.2); p<0.001), but not OS (2y-OS 72%; 95%CI (54, 96) versus 80%; 95%CI (76, 84); p=0.5). NCKAP5, coding for a Nck-adaptor protein, was associated with an adverse OS, Table 2 and Supplementary Figure 7-8.
Mutations in IRF4 had a positive impact on survival with a trend towards an improvement in PFS (2y-PFS 71%; 95%CI (50, 100) versus 54%; 95%CI (49, 60); p=0.09) and a significant impact on OS (2y-OS 100% versus 79%; 95%CI (75, 83); p=0.05), Figure 3B. Mutations in EGR1 had a positive impact on survival with a trend towards an improvement on PFS (median 35.1 months; 95% CI (26.1, ∞) versus 26.2 months; 95%CI (23.7, 28.7); p=0.14) and a significant impact on OS (2y-OS 100% versus 78%; 95%CI (75, 83); p=0.04), Figure 3C.
Figure 3.
Clinical impact and location of mutations in selected genes. Panel A: Location of the mutations and impact of ZFHX4 mutations on PFS. Panel B: Location of the mutations and impact of IRF4 mutations on OS. Panel C: Location of the mutations and impact of EGR1 mutations on OS
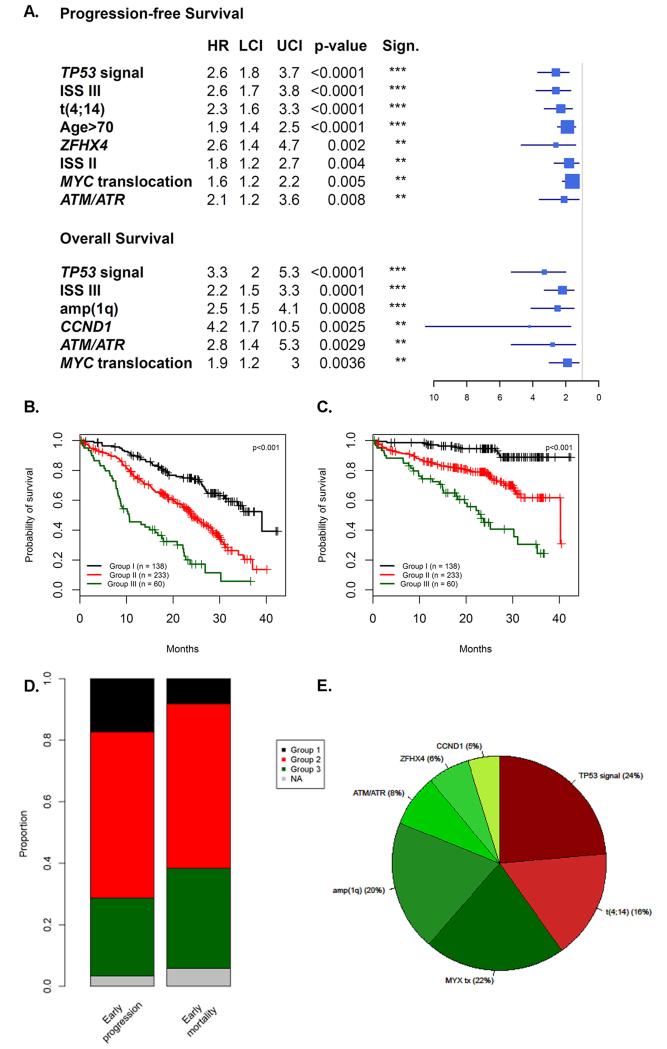
In a multivariable analysis for PFS, ISSIII, age>70, t(4;14), MYC translocations, TP53 variants, ATM/ATR mutations, ZFHX4 mutations, remained independent prognostic factors, Figure 4A. The apparent C-index was 0.67 with the bootstrap resampling strategy suggesting an optimism of 0.02, i.e. an internally validated C-index penalized for potential over-fitting of 0.65.
Figure 4.
Results of multivariate analysis (Panel A). The ISS-MUT identifies 3 prognostic groups (Group 1: ISS I/II with no CNSA or mutation, Group 2: ISS III with no CNSA or mutation or ISS I/II/III with one CNSA or mutation, Group 3: Two CNSA or mutation regardless of their ISS). It is an efficient tool to identify independent prognostic groups in terms of PFS (Panel B) and OS (Panel C). It also identified 81% and 90% of patients that both relapse and die prematurely (panel D) The adverse features that make up the HR group in the ISS-MUT score comprises not only the traditional ISS-FISH lesions, t(4;14) and del(17p) but also a variety of lesions previously not considered in the score that account for approximately 60% of the lesions (panel E).
In a multivariable analysis for OS, ISS III, TP53 variants, CCND1 mutations, ATM/ATR mutations, amp(1q) and MYC translocations, remained independent, Figure 4A. The apparent C-index was 0.65 with the bootstrap resampling strategy suggesting an optimism of 0.06, i.e. an internally validated C-index of 0.59.
Cumulative negative impact of mutations
We have previously shown that integrating ISS with CNSA is a key determinant of prognosis2. We further extended this to include poor prognosis mutations (MUT) including TP53, ZFHX4, CCND1 and ATM/ATR and novel CNSA such as MYC translocations and amp(1q). Using a CNSA-MUT score we were able to identify 4 different prognostic populations representing 50%, 33%, 11%, 2% of the population studied with increasingly poor PFS (one lesion: HR=1.7, p=0.0007; 2 lesions: HR=3.4, p<0.0001 and 3 lesions: HR=15.2, p<0.0001) and OS (one lesion: HR=2.0, p=0.002, 2 lesions: HR=4.8 p<0.0001, 3 lesions: HR=9.6, p<0.0001). A high-risk population can be identified comprising cases with 2 or more adverse features representing 13% of patients that both relapsed (median PFS 10.6 months; 95%CI (8.7, 17.9) versus 28.5 months; 95%CI (26.3, 31.3); p<0.0001) and died prematurely (2y-OS 45%; 95%CI (7, 33) versus 83%; (82, 89); p<0.0001). Nevertheless, using this acquired genetic lesion approach we show that 35% of patients that relapse before 18 months and 27% of patients that died before 24 months were not detected, Supplementary Figure 9.
In order to address this issue we incorporated the clinical information captured by the ISS, as has been done previously3. In this analysis combining CNSA and mutations with ISS, ISS-MUT, we identified 3 prognostic groups (Group 1: ISS I/II with no CNSA or mutation, Group 2: ISS III with no CNSA or mutation or ISS I/II/III with one CNSA or mutation, Group 3: Two CNSA or mutation regardless of their ISS). This approach adds sensitivity to the detection of early progression (EP) and early mortality (EM) (80.5% versus 65% for EP and 90% versus 75% for EM) (Supplementary Figure 9-10).
Discussion
We have identified the recurrent mutations characterising presentation myeloma and their impact on survival within this treatment setting. We show that there are a limited number of recurrent variants that are seen in a significant proportion of cases and that they co-segregate with the known recurrent CNSA typical of myeloma. Minimal differences in significantly mutated genes were seen between the major etiologic subtypes of myeloma and it seems likely that, once initiated, it is the same mutated pathways which push the disease forward. Based on this analysis and previous work, myeloma is clearly a disease driven by RAS pathway mutations and by MYC translocations11.
With the exception of NRAS and KRAS, all genes are mutated at a low percentage indicating the deregulation of key pathways, rather than mutations of single genes, could be important. We identified the central role of deregulation of the RAS/MAPK, the NF-κB pathway and apoptotic response, but interestingly mutation of the RAS/MAPK or NF-κB had no prognostic relevance in this clinical trial.
However, the inability to deliver an effective apoptotic response to DNA damage gave the most significantly prognostic mutational marker in this trial. Combining the known poor prognostic marker TP532,3,37 with additional mutations in ATM or ATR identifies 17% of presenting cases that have a significantly poor outcome. Mutations in ATM are known poor prognostic markers in CLL38,39, mantle cell40 and acute lymphoblastic T cell leukaemia41, but their prognostic impact has, until now, not been examined in myeloma42. ATR mutations, especially truncating mutations in exon 10, have been associated with a loss of function and an adverse prognosis in endometrial cancer43. Beyond ATM and ATR, we identified mutations in other members of the DNA-Damage-Repair (DDR) pathway including ZFHX4, member of the NuRD complex, involved in chromatin refolding 44,45,46. These data would suggest that patients with DNA repair pathway alterations may not benefit from alkylating agents, supporting the idea of therapy based on novel agents for these patients.
Myeloma XI is a trial based around IMiD drugs in which we have identified genes that are mutated and seem to have a positive impact on survival. In particular, we have identified mutations in IRF4 and EGR1. IRF4 is believed to be downstream of the IMiD target cereblon and other proteins downstream namely IKZF1 and KPNA2, that have been linked to survival differences in IMiD treated patients47. However, mutations in these genes are infrequent compared to IRF4.
In myeloma, EGR1 has recently been shown to be involved in recruitment of MYC to the promoters of NOXA and BIM inducing p53-independent apoptosis48,49. This is increased through bortezomib treatment where bortezomib enhances MYC and EGR1 expression49. Furthermore, EGR1 as a candidate gene for del(5q) in myelodysplastic syndromes has been associated with response to lenalidomide50. The role of mutations at the 5’ end of EGR1 remains to be ascertained.
The detection of mutations can improve our ability to detect high-risk patients that relapse and die early, but who may benefit from specific therapeutic interventions. We have previously shown that integration of ISS and cytogenetic data (ISS-FISH) can identify high-risk and ultra-high-risk patients2. The further integration of mutational prognostic data can improve this to a molecular level. We have shown that the greater the number of adverse CNSA (t(4;14), del(17p), MYC translocations and amp(1q)) present within an individual patient, the worse the outcome. Adding both ISS and mutations, in our ISS-MUT score, adds precision to early mortality and progression detection, Supplementary Figure 9. The number of molecular features required to identify this high-risk group is small (n=9) and could easily be incorporated into a molecular diagnostic test which could be readily translated into the modern molecular diagnostic laboratory. This predictive tool must be externally validated in further patient sample, but internal validation undertaken indicates robust predictive ability in bootstrap resamples.
The identification of actionable mutations within myeloma opens the way for targeted treatment. Key amongst these mutations in myeloma is the deregulation of the RAS/MAPK pathway with the most common being the recurrent mutations in NRAS and KRAS making it a major therapeutic target44,51-53. The other targetable pathway is NF-κB, which is consistently mutated in mature lymphoid malignancies, however, the spectrum of mutations seen in myeloma is different, Supplementary Figure 3D. Overall, we identified a set of potential actionable mutations comprising 309 targets applicable to 53% of patients. In the years to come we foresee this to increase to 440 targets applicable to 62% of patients, Table 3.
Table 3.
Actionable mutations. Tumour fractions and percentage of mutations are presented in this table with an example of agent and the phase of development that may be used
| Gene | Tumour fraction |
n | % patients |
Example | Phase (cancer) |
|---|---|---|---|---|---|
| KRAS | 32.3% | 98 | 21.17% | MEK inhibitor | Phase III |
| NRAS | 33.6% | 90 | 19.44% | MEK inhibitor | Phase III |
| BRAF | 25.2% | 31 | 6.70% | Vemurafenib | Phase III |
| CCND1 | 41.7% | 10 | 2.16% | Pablociclib | Phase I |
| FGFR3 | 37.1% | 10 | 2.16% | Masitinib | Phase II |
| MLL | 29.1% | 8 | 1.73% | EPZ-5676 | Phase I |
| ROS1 | 41.3% | 8 | 1.73% | Foretinib | Phase II |
| RET | 33.4% | 6 | 1.30% | Cabozantinib | Phase II |
| ERBB4 | 42.3% | 5 | 1.08% | Lapatinib | Phase II |
| FLT3 | 44.0% | 5 | 1.08% | Sunitinib | Phase III |
| ERBB2 | 11.9% | 3 | 0.65% | Herceptin | Phase III |
| FGFR2 | 66.7% | 3 | 0.65% | Masitinib | Phase II |
| KIT | 40.9% | 3 | 0.65% | Ponatinib | Phase III |
| MAP2K1 | 14.9% | 3 | 0.65% | MEK inh | Phase III |
| ABL1 | 53.3% | 2 | 0.43% | Gleevec | Phase III |
| BRCA2 | 52.2% | 2 | 0.43% | PPARP inhibitor (BMN673) | Phase I |
| DNMT3A | 39.2% | 2 | 0.43% | 5-azacytidin | Phase III |
| EGFR | 21.7% | 2 | 0.43% | Erlotinib | Phase III |
| EPHB2 | 37.8% | 2 | 0.43% | Herceptin | Phase III |
| PDGFRA | 17.4% | 2 | 0.43% | pazopanib | Phase I |
| BRCA1 | 6.4% | 1 | 0.22% | PPARP inhibitor (BMN673) | Phase I |
| DDR2 | 58.3% | 1 | 0.22% | Dasatinib | Phase III |
| FGFR4 | 28.6% | 1 | 0.22% | Masitinib | Phase II |
| HIF1A | 13.5% | 1 | 0.22% | PX-478 | Phase I |
| IDH2 | 17.9% | 1 | 0.22% | AG-221 | Phase I |
| KIF5B | 5.9% | 1 | 0.22% | Cabozantinib | Phase II |
| MET | 39.8% | 1 | 0.22% | MSC2156119J | Phase I |
| MPL | 51.9% | 1 | 0.22% | Eltrombopag | Phase III |
| PIK3CA | 45.1% | 1 | 0.22% | GDC-0941 | Phase I |
| RARA | 10.2% | 1 | 0.22% | ATRA | Phase III |
In summary, we have performed the first comprehensive molecular analysis of a clinical trial, in myeloma, that identifies key copy number and structural abnormalities and mutations that interact and identify high-risk patients who may benefit from alternative treatments.
Supplementary Material
Acknowledgements
The authors would like to thank all the patients and staff at centres throughout the UK whose participation made this study possible. The authors are grateful to the NCRI Haemato-oncology subgroup and to all principle investigators for their dedication and commitment to recruiting patients to the study. The principal investigators at the four top recruiting centres were Dr Don Milligan (Heart of England NHS Foundation Trust), Dr Jindriska Lindsay (Kent and Canterbury Hospital), Dr Nigel Russell (Nottingham University Hospital) and Dr Clare Chapman (Leicester Royal Infirmary). The support of the Clinical Trials Research Unit at The University of Leeds was essential to the successful running of the study and the authors would like to thank all the staff including Helen Howard, Corrine Collett, Jacqueline Ouzman and Alex Szubert. We also acknowledge The Institute of Cancer Research Tumour Profiling Unit for their support and technical expertise in this study.
Financial Support: This work was supported by a Myeloma UK program grant, Cancer Research UK CTAAC sample collection grants (C2470/A12136 and C2470/A17761) and a Cancer Research UK Biomarkers and Imaging Discovery and Development grant (C2470/A14261) as well as funds from the National Institute of Health Biomedical Research Centre at the Royal Marsden Hospital. EMB was supported by the Fédération Française de Recherche sur le myélome et les gammapathies grant.
References
- 1.Morgan GJ, Walker BA, Davies FE. The genetic architecture of multiple myeloma. Nat Rev Cancer. 2012;12:335–48. doi: 10.1038/nrc3257. [DOI] [PubMed] [Google Scholar]
- 2.Boyd KD, Ross FM, Chiecchio L, et al. A novel prognostic model in myeloma based on co-segregating adverse FISH lesions and the ISS: analysis of patients treated in the MRC Myeloma IX trial. Leukemia. 2012;26:349–55. doi: 10.1038/leu.2011.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avet-Loiseau H, Durie BG, Cavo M, et al. Combining fluorescent in situ hybridization data with ISS staging improves risk assessment in myeloma: an International Myeloma Working Group collaborative project. Leukemia. 2013;27:711–7. doi: 10.1038/leu.2012.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klein U, Jauch A, Hielscher T, et al. Chromosomal aberrations +1q21 and del(17p13) predict survival in patients with recurrent multiple myeloma treated with lenalidomide and dexamethasone. Cancer. 2011;117:2136–44. doi: 10.1002/cncr.25775. [DOI] [PubMed] [Google Scholar]
- 5.Greipp PR, San Miguel J, Durie BG, et al. International staging system for multiple myeloma. J Clin Oncol. 2005;23:3412–20. doi: 10.1200/JCO.2005.04.242. [DOI] [PubMed] [Google Scholar]
- 6.Walker BA, Wardell CP, Johnson DC, et al. Characterization of IGH locus breakpoints in multiple myeloma indicates a subset of translocations appear to occur in pregerminal center B cells. Blood. 2013;121:3413–9. doi: 10.1182/blood-2012-12-471888. [DOI] [PubMed] [Google Scholar]
- 7.Ross FM, Avet-Loiseau H, Ameye G, et al. Report from the European Myeloma Network on interphase FISH in multiple myeloma and related disorders. Haematologica. 2012;97:1272–7. doi: 10.3324/haematol.2011.056176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Avet-Loiseau H, Attal M, Moreau P, et al. Genetic abnormalities and survival in multiple myeloma: the experience of the Intergroupe Francophone du Myelome. Blood. 2007;109:3489–95. doi: 10.1182/blood-2006-08-040410. [DOI] [PubMed] [Google Scholar]
- 9.Avet-Loiseau H, Li C, Magrangeas F, et al. Prognostic significance of copy-number alterations in multiple myeloma. J Clin Oncol. 2009;27:4585–90. doi: 10.1200/JCO.2008.20.6136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fonseca R, Blood E, Rue M, et al. Clinical and biologic implications of recurrent genomic aberrations in myeloma. Blood. 2003;101:4569–4575. doi: 10.1182/blood-2002-10-3017. [DOI] [PubMed] [Google Scholar]
- 11.Walker BA, Wardell CP, Brioli A, et al. Translocations at 8q24 juxtapose MYC with genes that harbor superenhancers resulting in overexpression and poor prognosis in myeloma patients. Blood Cancer J. 2014;4:e191. doi: 10.1038/bcj.2014.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jenner MW, Leone PE, Walker BA, et al. Gene mapping and expression analysis of 16q loss of heterozygosity identifies WWOX and CYLD as being important in determining clinical outcome in multiple myeloma. Blood. 2007;110:3291–300. doi: 10.1182/blood-2007-02-075069. [DOI] [PubMed] [Google Scholar]
- 13.Lohr JG, Stojanov P, Carter SL, et al. Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer Cell. 2014;25:91–101. doi: 10.1016/j.ccr.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chapman MA, Lawrence MS, Keats JJ, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–72. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bolli N, Avet-Loiseau H, Wedge DC, et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat Commun. 2014;5:2997. doi: 10.1038/ncomms3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kozarewa I, Rosa-Rosa JM, Wardell CP, et al. A modified method for whole exome resequencing from minimal amounts of starting DNA. PLoS ONE. 2012;7:e32617. doi: 10.1371/journal.pone.0032617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boeva V, Popova T, Bleakley K, et al. Control-FREEC: a tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics. 2012;28:423–5. doi: 10.1093/bioinformatics/btr670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rausch T, Zichner T, Schlattl A, et al. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012;28:i333–i339. doi: 10.1093/bioinformatics/bts378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaiser MF, Walker BA, Hockley SL, et al. A TC classification-based predictor for multiple myeloma using multiplexed real-time quantitative PCR. Leukemia. 2013;27:1754–7. doi: 10.1038/leu.2013.12. [DOI] [PubMed] [Google Scholar]
- 20.Alpar D, de Jong D, Holczer-Nagy Z, et al. Multiplex ligation-dependent probe amplification and fluorescence in situ hybridization are complementary techniques to detect cytogenetic abnormalities in multiple myeloma. Genes Chromosomes Cancer. 2013;52:785–93. doi: 10.1002/gcc.22074. [DOI] [PubMed] [Google Scholar]
- 21.Schwab CJ, Jones LR, Morrison H, et al. Evaluation of multiplex ligation-dependent probe amplification as a method for the detection of copy number abnormalities in B-cell precursor acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2010;49:1104–13. doi: 10.1002/gcc.20818. [DOI] [PubMed] [Google Scholar]
- 22.Boyle EM, Proszek P, Kaiser M, et al. A molecular diagnostic approach able to detect the recurrent genetic prognostic factors typical of presenting myeloma. doi: 10.1002/gcc.22222. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.JAGS 3.4.0. Just Another Gibbs Sampler: (ed JAGS 3.4.0.) 2013. [Google Scholar]
- 24.Nuijten M, Wetzels R, Matzke D, Dolan CV, Wagenmakers EJ. Default Bayesian hypothesis tests for correlation, partial correlation, and mediation, (ed 1.0) 2014. [DOI] [PubMed] [Google Scholar]
- 25.Wetzels R, Wagenmakers EJ. A default Bayesian hypothesis test for correlations and partial correlations. Psychon Bull Rev. 2012;19:1057–64. doi: 10.3758/s13423-012-0295-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kass R, Raferty A. Bayes Factors. Journal of the American Statistical Association. 1995;90:773–795. [Google Scholar]
- 27.Wei T. Corrplot: Visualization of a correlation matrix. 2013. [Google Scholar]
- 28.R development Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. R Foundation for statistical Computing; Vienna, Austria: 2013. [Google Scholar]
- 29.Therneau T, Grambsch P. Modeling Survival Data: Extending the Cox Model. Springer; New York: 2000. [Google Scholar]
- 30.Therneau T. A Package for Survival Analysis in S, (ed R package version 2.37-7) 2014. [Google Scholar]
- 31.Hothorn T, Hornik K, van de Wiel M, et al. Implementing a Class of Permutation Tests: The {coin} Package. Journal of Statistical Software. 2008;28:1–23. [Google Scholar]
- 32.Hothorn T, Hornik K, van de Wiel M, et al. A Lego System for Conditional Inference. The American Statistician. 2006;60:257–263. [Google Scholar]
- 33.Qiu Weiliang, C J, Lazarus Ross, Rosner Bernard, Jing Ma. powerSurvEpi: Power and sample size calculation for survival analysis of epidemiological studies, (ed 0.0.6) 2012. [Google Scholar]
- 34.Harrell FE. REGRESSION MODELING STRATEGIES with Applications to Linear Models, Logistic Regression, and Survival Analysis. Springer; 2001. [Google Scholar]
- 35.Annunziata CM, Davis RE, Demchenko Y, et al. Frequent engagement of the classical and alternative NF-kappaB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell. 2007;12:115–30. doi: 10.1016/j.ccr.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen L, Wang S, Zhou Y, et al. Identification of early growth response protein 1 (EGR-1) as a novel target for JUN-induced apoptosis in multiple myeloma. Blood. 2010;115:61–70. doi: 10.1182/blood-2009-03-210526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lode L, Eveillard M, Trichet V, et al. Mutations in TP53 are exclusively associated with del(17p) in multiple myeloma. Haematologica. 2010;95:1973–6. doi: 10.3324/haematol.2010.023697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skowronska A, Austen B, Powell JE, et al. ATM germline heterozygosity does not play a role in chronic lymphocytic leukemia initiation but influences rapid disease progression through loss of the remaining ATM allele. Haematologica. 2012;97:142–6. doi: 10.3324/haematol.2011.048827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guarini A, Marinelli M, Tavolaro S, et al. ATM gene alterations in chronic lymphocytic leukemia patients induce a distinct gene expression profile and predict disease progression. Haematologica. 2012;97:47–55. doi: 10.3324/haematol.2011.049270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fang NY, Greiner TC, Weisenburger DD, et al. Oligonucleotide microarrays demonstrate the highest frequency of ATM mutations in the mantle cell subtype of lymphoma. Proc Natl Acad Sci U S A. 2003;100:5372–7. doi: 10.1073/pnas.0831102100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meier M, den Boer ML, Hall AG, et al. Relation between genetic variants of the ataxia telangiectasia-mutated (ATM) gene, drug resistance, clinical outcome and predisposition to childhood T-lineage acute lymphoblastic leukaemia. Leukemia. 2005;19:1887–95. doi: 10.1038/sj.leu.2403943. [DOI] [PubMed] [Google Scholar]
- 42.Austen B, Barone G, Reiman A, et al. Pathogenic ATM mutations occur rarely in a subset of multiple myeloma patients. Br J Haematol. 2008;142:925–33. doi: 10.1111/j.1365-2141.2008.07281.x. [DOI] [PubMed] [Google Scholar]
- 43.Zighelboim I, Schmidt AP, Gao F, et al. ATR mutation in endometrioid endometrial cancer is associated with poor clinical outcomes. J Clin Oncol. 2009;27:3091–6. doi: 10.1200/JCO.2008.19.9802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsuoka S, Ballif BA, Smogorzewska A, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–6. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 45.Mu JJ, Wang Y, Luo H, et al. A proteomic analysis of ataxia telangiectasia-mutated (ATM)/ATM-Rad3-related (ATR) substrates identifies the ubiquitin-proteasome system as a regulator for DNA damage checkpoints. J Biol Chem. 2007;282:17330–4. doi: 10.1074/jbc.C700079200. [DOI] [PubMed] [Google Scholar]
- 46.Chudnovsky Y, Kim D, Zheng S, et al. ZFHX4 interacts with the NuRD core member CHD4 and regulates the glioblastoma tumor-initiating cell state. Cell Rep. 2014;6:313–24. doi: 10.1016/j.celrep.2013.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu YX, Braggio E, Shi CX, et al. Identification of cereblon-binding proteins and relationship with response and survival after IMiDs in multiple myeloma. Blood. 2014;124:536–45. doi: 10.1182/blood-2014-02-557819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boone DN, Qi Y, Li Z, et al. Egr1 mediates p53-independent c-Myc-induced apoptosis via a noncanonical ARF-dependent transcriptional mechanism. Proc Natl Acad Sci U S A. 2011;108:632–7. doi: 10.1073/pnas.1008848108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wirth M, Stojanovic N, Christian J, et al. MYC and EGR1 synergize to trigger tumor cell death by controlling NOXA and BIM transcription upon treatment with the proteasome inhibitor bortezomib. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Joslin JM, Fernald AA, Tennant TR, et al. Haploinsufficiency of EGR1, a candidate gene in the del(5q), leads to the development of myeloid disorders. Blood. 2007;110:719–26. doi: 10.1182/blood-2007-01-068809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mulligan G, Lichter DI, Di Bacco A, et al. Mutation of NRAS but not KRAS significantly reduces myeloma sensitivity to single-agent bortezomib therapy. Blood. 2014;123:632–9. doi: 10.1182/blood-2013-05-504340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takahashi N, Yamada Y, Taniguchi H, et al. Clinicopathological features and prognostic roles of KRAS, BRAF, PIK3CA and NRAS mutations in advanced gastric cancer. BMC Res Notes. 2014;7:271. doi: 10.1186/1756-0500-7-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bruce S, Leinonen R, Lindgren CM, et al. Global analysis of uniparental disomy using high density genotyping arrays. J.Med.Genet. 2005;42:847–851. doi: 10.1136/jmg.2005.032367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.