Abstract
Lymphocyte functions triggered by antigen recognition and cosignals imply rapid and intense cell division, hence metabolism adaptation1. The cytidine nucleotide triphosphate (CTP) is a precursor required for the metabolism of DNA, RNA and phospholipids2-4. CTP originates from two sources: a salvage pathway and a de novo synthesis pathway that depends on two enzymes, the CTP synthase (or synthetase) 1 and 2 (CTPS1 and CTPS2), although their respective roles are not known5-7. CTP synthase activity is a potentially important step for DNA synthesis in lymphocytes8, 9. Here, we report the identification of a loss of function homozygous mutation (rs145092287) in CTPS1 in humans causing a novel and life threatening immunodeficiency characterized by an impaired capacity of activated T and B cells to proliferate in response to antigen receptor-mediated activation. In contrast, proximal and distal TCR signaling events and responses were only weakly affected by the absence of CTPS1. Activated CTPS1-deficient cells exhibited decreased levels of CTP. Normal T-cell proliferation was restored in CTPS1-deficient cells by expressing wild-type CTPS1 or by addition of exogenous CTP or its nucleoside precursor, cytidine. CTPS1 expression was found to be low in resting T cells, but rapidly upregulated following TCR activation. These results highlight a key and specific role of CTPS1 in the immune system by its capacity to sustain the proliferation of activated lymphocytes during the immune response. CTPS1 may therefore represent a therapeutic target of immunosuppressive drugs that could specifically dampen lymphocyte activation.
We initially studied two unrelated families (family 1 and 2) originating from the northwest region of England, whose four children suffered from severe and recurrent Epstein-Barr virus (EBV) infection, in whom known primary immunodeficiencies have been excluded10 (Fig. 1a and Table 1). Four additional patients (family 3 to 5) originating from the same geographical area were identified thereafter (Methods). All patients had early onset of severe chronic viral infections, mostly caused by herpes viruses, including EBV and Varicella Zooster Virus (VZV) and, also suffered from recurrent encapsulated bacterial infections, a spectrum of infections typical of a combined deficiency of adaptive immunity (CID)11 (Table 1 and data not shown). Overall, the clinical phenotype is severe with 3 patients have died. Six of 8 patients have undergone hematopoietic stem cell transplantation. Of note, none of the patients had extra-hematopoietic manifestations (Table 1).
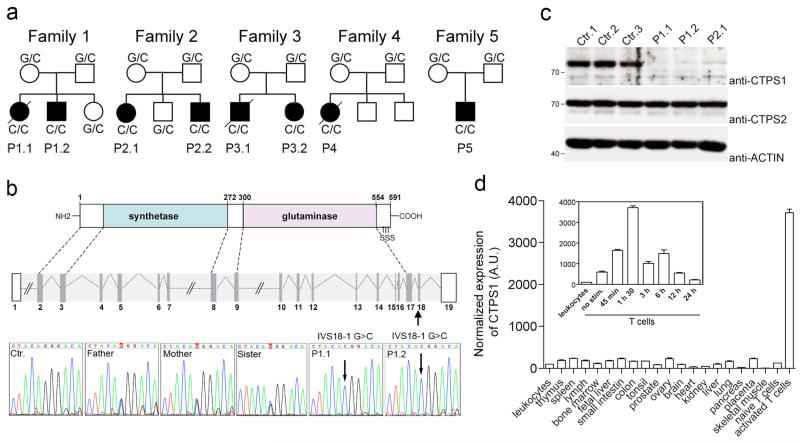
Figure 1. Identification of CTPS1 deficiency in patients with a combined immunodeficiency.
a, Pedigrees of the families in which an homozygous IVS18-1 G>C mutation in CTPS1 were identified. When known, the genotype of each individual is indicated. Black boxes represent affected individuals and diagonal bars indicate deceased individuals. Each patient (P) is identified by a number. b, Diagram of the CTPS1 intron-exon organization and protein domains with the serine phosphorylation sites (S) indicated and the coding exons in grey. DNA electropherograms showing the region containing the mutation in CTPS1 in family 1. The homozygous IVS18-1 G>C mutation is indicated by an arrow. c, Immunoblots for CTPS1 and CTPS2 expression in non stimulated EBV B-cell lines from healthy controls and CTPS1-mutated individuals (P1.1, P1.2 and P2.1). ACTIN serves as loading control. d, CTPS1 mRNA expression in normal tissues monitored by RT-qPCR in arbitrary units (A.U.). The inset shows the kinetic of CTPS1 mRNA expression following anti-CD3/CD28 beads stimulation.
Table 1. Clinical features of patients.
| Patient | Age at 1st symptoms | Viral infections | Bacterial infections | Extra-hematopoietic manifestations | Outcome (age in years) | ||
|---|---|---|---|---|---|---|---|
| EBV | VZV | Others | |||||
| P1.1 | 1 y. | SIM, chronic viremia | no | CMV, Novovirus, Rotavirus (gut) Parainfluenzae I (RTI) | H. influenzae (RTI) | no | HSCT (8 y.) died (GVHD) (8 y.) |
| P1.2 | 1 m. | SIM | no | Adenovirus, HHV-6, Novovirus (gut) | yes, n.k. (RTI) | no | alive (9 y.) |
| P2.1 | 5 y. | LPD (CNS) | yes | no | H. influenzae (RTI) | no | HSCT (9 y.) a.w. (17 y.) |
| P2.2 | 2 y. | chronic viremia | no | no | S. pneumoniae, H. influenzae (RTI) | no | HSCT (7 y.) a.w. (13 y.) |
| P3.1 | 1 y. | n.k. | yes (gastritis, pneumonitis) | no | S. pneumoniae (septis, meningitis) | no | died (disseminated VZV) (4 y.) |
| P3.2 | 3 m. | SIM, chronic viremia | yes | HHV-6 | no | no | HSCT (8 y.) a.w. (12 y.) |
| P4 | birth | LPD (CNS) | yes | CMV, Adenovirus, Rotavirus (gut) | no | no | HSCT (6 y.) died (LPD) (6 y.) |
| P5 | 3 m. | LPD (CNS, liver), chronic viremia | no | Novovirus (gut) Parainfluenzae III, Adenovirus, Rhinovirus (RTI) | N. meningitis B (meningitis) | no | HSCT (1 y.) alive (2 y.) |
y., year. m., month. SIM, severe infectious mononucleosis. CNS, central nervous system. EBV, Epstein-Barr virus. VZV, varicella zona virus. HHV-6, human herpes virus 6.
LPD, lymphoproliferative disease. RTI, respiratory tract infection. CMV, cytomegalovirus. HSCT, hematopoietic stem cell transplantation. GVHD, graft versus host disease. n.k., not known. a.w., alive and well.
Immunological investigations showed that most of patients had variable lymphopenia which was exacerbated during infection episodes with inversed CD4:CD8 T-cell ratio, while other blood cell counts were usually normal (Extended Data Table 1 and data not shown). Their immunoglobulin levels were normal or elevated with increased IgG but low IgG2 levels with low antibody titers to Streptococcus pneumoniae. Further analyses were performed in patient P1.2 showing naive CD4+ T-cell lymphopenia, increased numbers of effector memory T cells, low numbers of memory CD27+ B cells, a complete absence of both invariant T cell populations (CD3+Vα24+Vβ11+) iNKT and (CD3+CD161highVα7.2+) MAIT cells, as well as an impaired PHA- and antigen-induced proliferation of peripheral blood mononuclear cells (PBMCs) (Extended Data Table 2).
To identify the gene defect underlying the immunodeficiency in these patients, we performed whole exome sequencing (WES) in three patients (P1.1, P1.2 and P2.1). Intersection of the genetic variations found in the three patients pointed to an unique common homozygous G to C mutation in the CTPS1 gene encoding the CTP synthase 1 at position 41475832 in chromosome 1 with an assigned rsID (rs145092287) in the dbSNP database (Fig. 1b and Extended Data Fig. 1a,b). CTPS1 encodes a 67-kDa protein containing a CTP synthetase domain and a glutamine amide transfer domain promoting the formation of CTP from UTP and glutamine12. The identified mutation affects a splice donor site at the junction of intron 17-18 and exon 18 (IVS18-1 G>C) leading to the expression of an abnormal transcript lacking exon 18 (Extended Data Fig. 1b,c). This splice mutation was found to be deleterious since CTPS1 protein expression could not be detected in lysates of EBV-transformed B cells and T-cell blasts from patients (Fig. 1c, Fig. 2c and Extended Data Fig. 2). In contrast, CTPS2 was expressed normally in patient cell lysates. In the five affected families, all patients were homozygous for the IVS18-1 G>C mutation and all parents and tested healthy siblings were heterozygous carriers (Fig. 1a,b and data not shown). Sequencing of a cohort of 752 healthy individuals from the northwest of England gave an estimated frequency of homozygosity of 1:560,000. This represents more than a 10-fold increase compared to the frequency estimated from available exome databases. WES data and analysis of polymorphic microsatellite markers in all patients revealed a common region of homozygosity of 1.1 Mb surrounding the IVS18-1 G>C mutation (Supplementary information). All these data were indicative of a founder effect. These observations led us to conclude that the immunodeficiency resulting from the CTPS1 mutation in these patients could be primarly associated with a T-cell immunodeficiency.
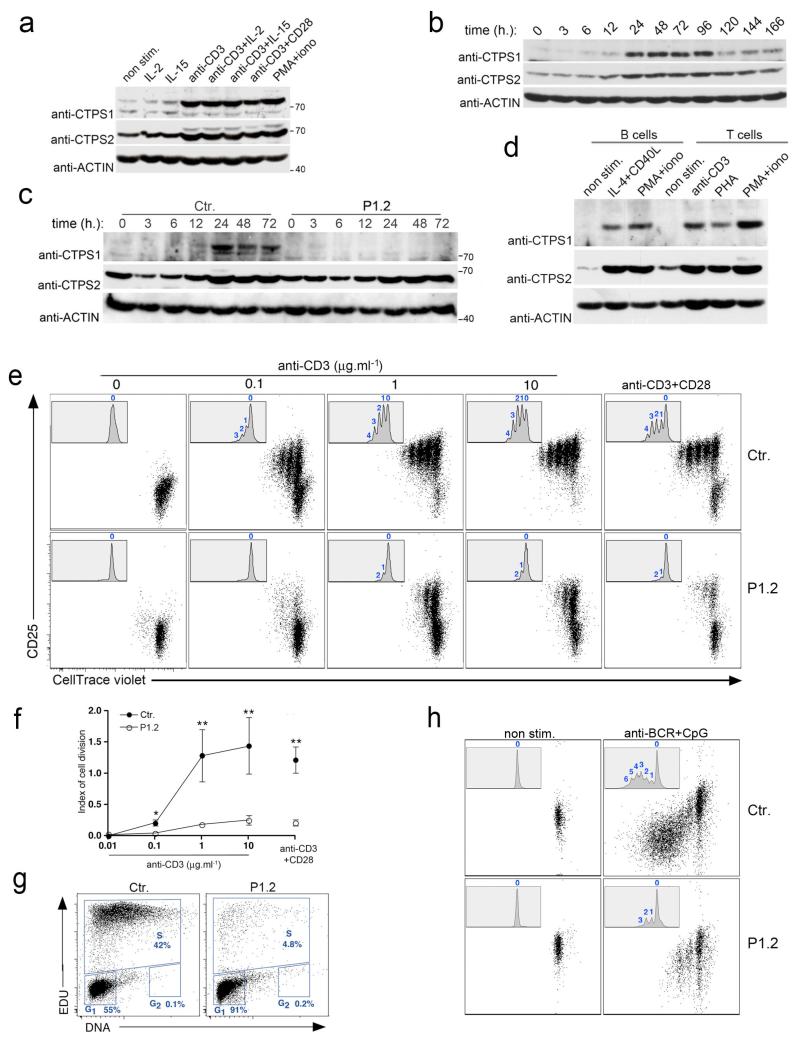
Figure 2. Induction of CTPS1 expression during T-cell activation and defective proliferation of activated CTPS1-deficient T-cells.
a-d, Immunoblots for CTPS1 and CTPS2 expression (a) in control T-cells (from an healthy donor) stimulated with various stimuli or (b) stimulated with anti-CD3 for different periods of time, (c) in control (Ctr.) or CTPS1-deficient cells from patient P1.2 stimulated with anti-CD3 for different periods of time and (d) in normal PBMCs sorted B- and T-cells stimulated with indicated stimuli. ACTIN serves as loading control. e, Representative dot plots showing cell divisions by dilution of the violet dye and expression of CD25 of control (Ctr.) or CTPS1-deficient T-cells (patient P1.2) stimulated with incremental doses of the anti-CD3 antibody or anti-CD3/CD28 coated beads. Inserts with histograms showing the violet dye dilution with the number cell divisions indicated at the top of each peak. Representative data from one of 4 independent experiments. f, Mean of index values of cell division of control T-cells (Ctr.) or CTPS1-deficient cells (P1.2) (n=4). Unpaired t-tests and **P<0.01. g, Representative dot plots of cell cycle progression of control (Ctr.) and CTPS1-deficient T-cells (patient P1.2) stimulated with anti-CD3 antibody. The percentages of cells in each stage are indicated. Data from one of 2 independent experiments. h, Proliferation of control (Ctr.) or CTPS1-deficient CD19+ B cells from PBMCs of healthy donor and patient P1.2. Cells were stimulated with anti-BCR plus CpG during 5 days. The proliferation was analyzed similarly as in (e). Representative data from one of 2 independent experiments.
We next examined CTPS1 expression in normal tissues. CTPS1 mRNA expression was comparable between the different tissues, except for T cells in which CTPS1 expression was strongly up regulated after cell activation in response to TCR-CD3 and CD28 co-stimulation (Fig. 1d). Interestingly, in lysates from T-cell blasts and T cells from PBMCs, CTPS1 protein was almost undetectable (Fig. 2a-d). In contrast, CTPS2 expression was readily detected. Activation of T cells by anti-CD3 antibody or phorbol 12-myristate 13-actate (PMA) and ionomycin stimulations induced CTPS1 protein expression while activation with IL-2 and/or IL-15 resulted in only weak effect. Under the same experimental conditions, CTPS2 expression was also induced but to a lesser extent. In TCR-CD3-stimulated T-cell blasts, CTPS1 protein expression was enhanced from 12 hours and persisted for up to 96 hours as a consequence of CTPS1 gene transcription activation (Fig.1d, inset and Fig. 2b). As expected, no expression of CTPS1 was detected in T-cell blasts from the CTPS1-deficient patient (P1.2) contrasting with detection of CTPS1 mRNA and suggesting protein instability (Fig. 2c and Extended Data Fig. 1c). These data indicate that T-cell activation through the TCR results in a rapid and sustained CTPS1 protein expression. Of note, in B cells activated by anti-BCR and CpG, IL-4 and CD40L or PMA and ionomycin, CTPS1 was also found to be upregulated (Fig. 2d and Extended Data Fig. 3a,b).
To further characterize the consequences of the CTPS1 deficiency in T cells, we investigated proximal T-cell activation signals as well as late responses. CTPS1-deficient T cells exhibited normal early and late responses with the exception of ERK1/2 phosphorylation and CD25 and CD69 upregulation which were found to be decreased (Extended Data Fig. 4). Basal and activation-induced cell death was also slightly increased (Extended Data Fig. 4g). These data suggest that CTPS1 deficiency had limited consequences in signaling downstream of TCR-CD3. Because the pool of CTP is potentially a limiting factor for DNA synthesis8, 13, we carefully analyzed proliferation of CTPS1-deficient T-cells. In response to activation by antigens, anti-CD3 antibody or co-stimulation by anti-CD3 and anti-CD28 antibodies, CTPS1-deficient cells from three patients (P1.1, P1.2 and P2.2) failed to sustain proliferative responses as measured by 3H-thymidine uptake and CFSE or violet cell tracer dye dilution (resulting in a weak index of cell proliferation) (Fig. 2e,f and Extended Data Table 1 and Extended Data Fig. 5, 6). Uptakes of 3H-Uridine and 3H-Cytidine were also found to be impaired in activated CTPS1-deficient T cells. This suggests that both RNA and DNA synthesis were affected (Extended Data Fig. 6). Defective proliferation of CTPS1-deficient T cells was associated with a lack of cell cycle progression since a majority of cells were arrested in the G1 phase (Fig. 2g). Proliferation of CTPS1-deficient B cells activated by anti-BCR and CpG was also found to be defective whereas that of IL-2-activated NK cells seemed to be less affected (Fig. 2h and Extended Data Fig. 5b).
Down-regulation of CTPS1 expression in control T cells, by lentiviral transduction of two distinct shRNA together with a GFP reporter gene, led to a specific decrease in the CD3-mediated proliferation of GFP-positive cells (Fig. 3a). No changes in proliferation were detected in non-targeted GFP-negative cells or in cells targeted with a scramble shRNA. The diminished proliferation resulting from the inhibition of CTPS1 expression led to a selective cell growth disadvantage with a decreased number of GFP targeted cells over time (middle panel). A similar decrease in proliferation rate was also observed in the Jurkat T-cell line in which CTPS1 expression was down-regulated (Extended Data Fig. 7).
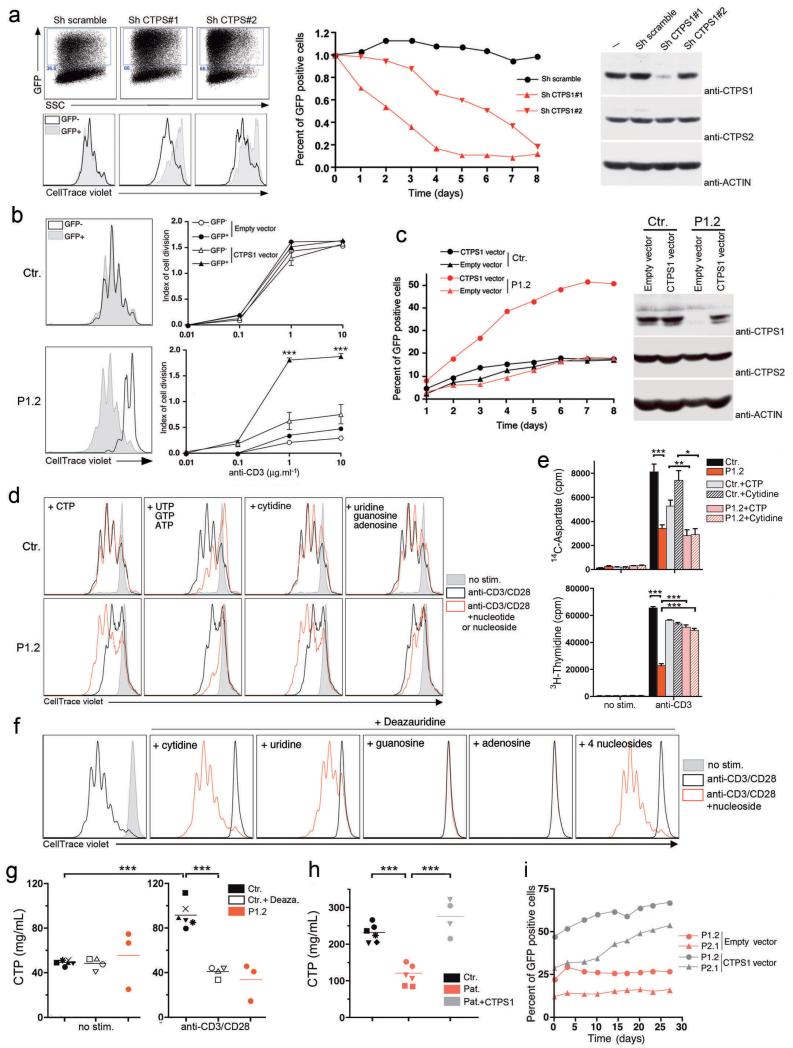
Figure 3. CTPS1 is required for proliferation of T-cells in response to TCR-CD3 activation.
a, Proliferation of T-cells in which CTPS1 expression was silenced with vectors containing shRNA for CTPS1 (Sh CTPS1#1 or Sh CTPS1#2) or containing a scramble shRNA (Sh scramble) with GFP gene reporter. Representative dot plots of GFP+ cells corresponding to transduced cells (left upper panels). Representative histograms of violet dye dilution showing the cell divisions after stimulation (left lower panels). Curves showing the percentage of GFP+ transduced cells in long-term expansions after repeated stimulation (middle panel). Immunoblots for CTPS1 and CTPS2 expression in transduced cells (right panels) and ACTIN as loading control. One representative of two experiments. b,c, Proliferation of control (Ctr.) and CTPS1-deficient T-cells (patient P1.2) transduced by empty or wild-type CTPS1-containing vector. Representative histograms of violet dye dilution (b, left panels). Means of indexes of cell division after stimulation (b, right panels) from triplicate of one representative of two experiments. Curves showing the percentage of GFP+ tranduced cells same as in (a) (c, left panel). Representative data from one of 2 independent experiments. Immunoblots same as in (a) (c, right panels). d, Representative histograms of violet dye dilution showing cell divisions of control (Ctr.) and CTPS-1-deficient T cells (P1.2) incubated with the indicated nucleotides or nucleosides before stimulation. Data from one of 3 independent experiments. e, Incorporation of 14C-aspartate, a tracer of the de novo pyrimidine nucleotide synthesis and 3H-thymidine as a control of proliferation/DNA synthesis. T cells were labelled during stimulation. Means of incorporated radioactivity (cpm) (n=6) f, Same as (d) excepted that control T-cells were incubated with deazauridine. Data from one representative of 3 independent experiments. g, Concentration of CTP in control T-cells incubated with deazauridine (Ctr.+Deaza., n=4) or not (Ctr., n=6) and CTPS1-deficient cells (P1.2, n=3) after stimulation with anti-CD3/CD28 coated beads. Data from 3 independent experiments. h, Concentration of CTP in cell extracts of EBV B-cell lines from healthy controls (Ctr.; n=6), and CTPS1-deficient patients transduced (Pat.+CTPS1, n=4) or not (Pat., n=6) with wild-type CTPS1-containing vector. P1.1 (squares), P1.2 (circles) and P2.1 (triangles). For controls, each symbol corresponds to cells of a different donor. Data from 2 independent experiments. i, Proliferation of CTPS1-deficient EBV B-cell lines (P1.2 and P2.1) transduced by empty or wild-type CTPS1-containing vector. Curves showing the percentage of GFP+-tranduced cells in culture. Unpaired t-tests and ***P<0.001, **P<0.01, *P<0.05 (b, e, g, h).
Together, these results indicate that CTPS1 deficiency causes a defect in T-cell proliferation in response to TCR-CD3 activation. To formally prove the causal relationship between CTPS1 deficiency and defective T-cell proliferation, we carried out reconstitution experiments with wild-type CTPS1 or by direct addition of CTP or its cytidine precursor that acts on CTP levels via the salvage pathway. Expression of ectopic CTPS1 in CTPS1-deficient T-cells fully restored proliferation upon CD3 stimulation (Fig. 3b) and enabled cells to expand selectively as shown by the accumulation of GFP-positive cells expressing CTPS1 (Fig. 3c, left panels). No such effect was detected in CTPS1-deficient cells transduced with an empty vector or in control cells transduced with the CTPS1-containing vector.
Proliferation and CD25 expression of CTPS1-deficient cells also recovered to a normal level by addition of CTP or cytidine (Fig. 3d and data not shown). In contrast, addition of a mix of UTP, GTP and ATP or uracil, guanine and adenosine did not result in increased proliferation of CTPS1-deficient cells. To determine the influence of the CTPS1 defect on the de novo pyrimidine synthesis pathway, we measured the incorporation of carbon from 14C-aspartate into nucleic acids of activated CTPS1-deficient T cells, which is a specific assay for de novo pyrimidine synthesis14 (Fig. 3e and Extended Data Fig. 6 b,c). Incorporation of 3H-thymidine and 3H-uridine was analyzed in parallel as control of the global RNA and DNA synthesis. TCR-CD3 activation-mediated incorporation of 14C-aspartate, 3H-thymidine and 3H-uridine was significantly decreased in CTPS1-deficient T cells. Addition of exogenous CTP or cytidine that bypassed the de novo synthesis pathway restored incorporation of 3H-thymidine but not of 14C-aspartate in CTPS1-deficient cells, thus demonstrating that the de novo CTP synthesis pathway is impaired in the absence of CTPS1. Deazauridine, an analogue of UTP and a known inhibitor of CTP synthetase activity15 completely blocked T-cell proliferation of control cells in response to CD3 activation without affecting proximal TCR-CD3-mediated responses, similar to results observed in CTPS1-deficient cells (Fig. 3f and data not shown). As expected, inhibition of T-cell proliferation by deazauridine was fully reverted by addition of CTP and partially by UTP, but not by ATP or GTP. Analysis of nucleotides pools in activated CTPS1-deficient T-cell blasts and CTPS1-deficient B/EBV cell lines revealed decreased levels of CTP, as also observed in activated normal cells treated with deazauridine (Fig. 3g,h). Defective CTPS1 expression or addition of deazauridine also led to reduced pools of ATP, GTP and UTP in activated T cells (Extended Data Fig. 8) suggesting interconnection in the nucleotide pools16. In contrast, CTP as well as ATP, GTP and UTP were found to be normal or increased in resting CTPS1-deficient T cells as the salvage pathway is predominent in quiescent cells17. Expression of wild-type CTPS1 in CTPS1-deficient B/EBV cell lines restored levels of CTP comparable to control cells and confered to cells a selective cell growth advantage (Fig. 3h,i).
This study reveals a critical role for CTPS1 in promoting the proliferation of T cells following their activation. However, proliferation of B cells was also found to be dependent of CTPS1. This may directly participate to the susceptibility to encapsulated bacterial infections seen in CTPS1-deficient patients and account for the low titers of S. pneumoniae antibodies as it is a T-independent B-cell response. The role of CTPS1 in B cells coud be different or/and less important than in T cells. Of note, CTPS1-deficient B cells preserve an intact capacity to expand upon transformation by EBV and patients had normal Ig levels and/or elevated IgG. Decreased expansion of NK cells and low numbers of iNKT and MAIT cells might also contribute to the CTPS1 immunodeficiency as these cells have been proposed to play a role in a broad range of immune responses including anti-EBV immunity18-21. The finding that CTPS1-deficiency causes no other significant clinical consequences favors a redundancy with CTPS2 activity in other cell lineages and tissues. Interestingly, pyrimidine pools including CTP have been previously shown to be strongly expanded in PHA-stimulated T cells via de novo pathways including increased CTPS activity8, 9. The induction of CTPS1 expression in activated T cells reported here thus appears as the major determinant of CTP pool increase. Indeed, proliferation of CTPS1-deficient T cells was restored to normal level by addition of CTP. The exact mechanism(s) by which TCR signaling induces a rapid expression and activation of CTPS1 in T cells remains to be determined, although we showed that the ERK pathway is required as well as tyrosine phosphorylation signals (Extended data Fig. 3c). It is interesting to note that T cell differentiation does not appear to be severely impaired by CTPS1 deficiency, suggesting that CTP pools in thymocytes may originate from the nucleoside salvage pathway and/or the CTPS2 activity8, 22-24. Notably though, CTPS1 activity is critical for the intense cell division induced by antigenic stimulation as exemplified by massive proliferation and expansion of CD8+ T cells during viral infections25, 26.
In the absence of CTPS1, we showed that de novo pyrimidine synthesis pathway is impaired but not totally abrogated. This residual activity is likely dependent of CTPS2. Recently, the de novo pyrimidine synthesis pathway was shown to be dependent on post-transcriptional regulation by mTORC1 and S6 protein (S6K) kinases that activate the first enzymatic steps required for pyrimidine synthesis14, 27, 28. Thus, distinct regulatory mechanisms control de novo pyrimidine synthesis. Based on the present study, CTPS1-mediated tuning of CTP synthesis in lymphocytes appears to be a key element in enabling adaptive immune responses. If CTPS1-specific inhibitors can be designed, they would potentially be highly specific immunosuppressive drugs able to inhibit auto- or allogenic-specific T and B cell responses without additional toxicity given the lymphocyte specificity of the CTPS1-deficiency phenotype. In conclusion, our results provide the first in vivo evidence of a role of the de novo pyrimidine synthesis pathway as a critical step for proliferation of T and B lymphocytes when activated by antigens.
METHODS
Cohorts of patients
Beside the four initially identified patients, four additional patients were identified by screening 10 patients (9 families) originating from the northwest of England with severe chronic viral infections, mostly caused by herpes viruses, including EBV and VZV. Furthermore, 24 patients (24 families) originating from different geographical areas with the same phenotype were also tested for all exons of CTPS1 in order to identify other mutations and none was found to be a carrier of CTPS1 mutations.
Exome sequencing and analysis
Exome capture was performed according to the manufacturer’s protocol using the Illumina TruSeq exome enrichment kit and sequencing of 100 bp paired end reads on an Illumina HiSeq. Approximately 10 Gb of sequence were obtained for each subject such that 90% of the coding bases of the exome defined by the consensus coding sequence (CCDS) project were covered by at least 10 reads. Adaptor sequences and quality trimmed reads were removed using the Fastx toolkit (http://hannonlab.cshl.edu/fastx_toolkit/) and a custom script was then used to ensure that only read pairs with both mates present were subsequently used. Reads were aligned to hg19 with BWA31, and duplicate reads were marked using Picard (http://picard.sourceforge.net/) and excluded from downstream analyses. Single nucleotide variants (SNVs) and short insertions and deletions (indels) were determined using samtools (http://samtools.sourceforge.net/) pileup and varFilter32 with the base alignment quality (BAQ) adjustment disabled, and they were then quality filtered to require at least 20% of reads supporting the variant call. Variants were annotated using both ANNOVAR33 and custom scripts to identify whether they affected protein coding sequences, and whether they had previously been seen in the dbSNP, the 1000 Genomes data set (1092 genomes), or in approximately 2073 exomes previously sequenced at our center. A variant detected in a patient was considered to be a candidate mutation if it had not been reported or had a frequency below 0.001% in the three databases indicated above. At the time the homozygous G>C mutation in CTPS1 (at position chr1:41475832) was identified by WES in the three patients (P1.1, P1.2 and P2.1), it was not described as a dbSNP or assigned to a rsID. Since, this mutation has been identified in the NHLBI GO Exome Sequencing Project (http://evs.gs.washington.edu/EVS/) with the assigned rsID: rs145092287. In the NHLBI GO Exome Sequencing Project, the rs145092287 is present three times in an heterozygous status among 4300 genomes from an European-American population and not found in the 2203 genomes of an African-American population. The rs145092287 was not found in an homozygous or heterozygous status in other available genome databases (NCBI, 1000 Genomes project and the 3519 genomes of our center). Homozygosity regions around the rs145092287 were determined in the exomes by looking at the homozygous variations. Between the positions chr1:40737516 (rs6677717) and chr1:42008077 (rs 63729761) a succession of 97 homozygous variations (without heterozygous variations) was found to be shared by the three patients.
Genomic DNA sequencing
Genomic DNA from peripheral blood cells, EBV-B cell lines and/or fibroblasts of patients, their parents, and other family members was isolated according to standard methods. Genomic DNAs of 752 healthy control subjects born in the North West of England were obtained from the UK 1958 Birth Cohort (http://www2.le.ac.uk/projects/birthcohort/1958BC-About). The estimated frequency of the CTPS1 mutation in the populations was calculated according to the Hardy-Weinberg law. Oligonucleotide primers flanking the 3′ region of intron 17-18 and exon 18 of CTPS1 were used to amplify genomic DNA: Forward 5′-AGAGTTGGTGGTAGGGTGTGTGAC-3′ and reverse 5′-CTTGCAATCGCAGTGTGTTATCAC-3′. PCR products were amplified using high fidelity Platinum Taq DNA Polymerase (Invitrogen) according to the manufacturer’s recommendations, purified with the QIAquick gel extraction kit (Qiagen) and sequenced using the ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction Kit (PerkinElmer) according to the manufacturer’s recommendations. All collected sequences were analysed using 4peaks software (Version 1.7.2; A. Griekspoor and T. Groothuis, http://nucleobytes.com/index.php/4peaks).
Analysis of microsatellite markers
Microsatellite markers were genotyped using UniSTS sequences and mapping information available from the NCBI (http://www.ncbi.nlm.nih.gov). Genomic DNA from patients was used as templates to amplify by PCR with specific fluorescent labelled oligonucleotides, the polymorphic repeats corresponding to the microsatellite markers. PCR products were evaluated using an ABI 3100 DNA Fragment Analyzer (Applied Biosystems), and data were analyzed using Genescan and Genotyper software (Applied Biosystems).
Gene expression analysis
Total RNA was isolated from EBV-B cell and activated T cell blasts of P1.2, P2.1 patients and control donors using the RNeasy Mini kit (QIAGEN). The samples were depleted of genomic DNA and reverse transcription was performed using Superscript II First Strand Synthesis System (Invitrogen). cDNAs were used as a template to perform PCR amplifications of exon 15 to exon 19 of CTPS1 or exon 4 of ACTIN with the following primers using standard protocols: CTPS1 forward primer: 5′-GAGAGGCACCGCCACCGATTTG-3′, CTPS1 reverse primer: 5′-GCCAGTACACGTGATGGGACATGC-3′, ACTIN forward primer: 5′-CTCCTTAATGTCACGCACGAT-3′; ACTIN reverse primer: 5′-CTCCTTAATGTCACGCACGAT-3′. PCR to amplify full lengh CTPS1 cDNAs from control and patients cells were also performed using the following primers: forward primer: 5′-AGCTCTGTCGCTGACGGGAGGAT-3′ (exon1); reverse primer: 5′-GCCAGTACACGTGATGGGACATGC-3′ (exon 19). PCR products were verified by sequencing revealing the expression of an abnormal CTPS1 transcript lacking exon 18 in patients cells. Multiple tissue cDNA panels from Ozyme (Human MTC panel I, II and Human Immune Sytem MTC panel) were analyzed for CTPS1 and CTPS2 gene expression by RT-qPCR. Gene expression assays were performed with Assays-on-Demand probe and primer combinations (CTPS1, Hs01041858; CTPS2, Hs00219845; GAPDH, Hs027558991) from Applied Biosystem labeled with 6-carboxy-fluorescein (FAM) dye, and universal reaction mixture. Real time quantitative PCRs for GAPDH, CTPS1 and CTPS2 were performed in triplicate using a LightCycler VIIA7 System (Roche). Expression levels were determined by relative quantification using the comparative threshold cycle method 2ΔΔCt in which ΔΔCt is determined as followed: (Cttarget gene – Ctreference gene) target tissue – (Cttarget gene – Ctreference gene) calibrator tissue. The results shown in arbitrary units (A.U.) have been normalized to GAPDH gene expression and are presented as the relative change in gene expression normalized against the calibrator sample corresponding to leukocyte tissue.
Cell culture and stimulation
Peripheral blood mononuclear cells (PBMCs) collected from patients and healthy donors were isolated by Ficoll-Paque density gradient (Lymphoprep, Proteogenix) from blood samples using standard procedures. Expansion of T-cell blasts were obtained by incubating PBMCs for 72 h with phytohaemagglutinin (PHA) (2.5 μg ml−1, Sigma-Aldrich) in RPMI 1640 GlutaMax medium (Invitrogen) supplemented with 5% human AB serum (Etablissement Français du Sang), penicillin (100 U ml−1) and streptomycin (100 μg ml−1). After 3 days, dead cells were removed by Ficoll-Paque density gradient and blasts were maintained in culture with IL-2 (100 UI ml−1). For proliferation and cell cycle analyses, blasts were washed and cultured without IL-2 for 72 hours to synchronize the cells. Blasts or PBMCs were then cultured during 4 to 6 days in complete medium alone or in the presence of 0.1, 1 or 10 μg ml−1 coated anti-CD3 antibody (clone OKT3, eBiosciences), anti-CD3/CD28-coated beads (Invitrogen), PHA (2.5 μg ml−1, 10−5 M ionomycin (Sigma-Aldrich) plus 10−7 M Phorbol 12-myristate 13-acetate (PMA, Sigma-Aldrich), Candidin (5 μg ml−1, Bio-Rad), tetanus toxoid (1/8000 dilution, Statens Serum Institute) or tuberculosis antigen (50 μg ml−1, Statens Serum Institute). Proliferation and cell cycle were analysed at the indicated time points. 40 μM of 3-Deazauridine (DAZ, Sigma-Aldrich) was added for 12 hours before the stimulation. In complementation experiments, blasts were incubated with 100 μM of CTP, UTP, GTP or ATP (New England Biolabs) separately or in combination, or with 200 μM of Cytidine, Uridine, Guanosine or Adenosine (Sigma-Aldrich) separately or in combination 12 hours before the stimulation. For dosage of nucleotides, blasts were deprived of IL-2 for 72 hours before stimulation or not with anti-CD3/CD28-coated beads for 48 h and cell lysates were prepared. Jurkat cells29, 293-T cells and B EBV cell lines from patients were cultured in RPMI 1640 GlutaMax medium supplemented with 10 % heat-inactivated fetal calf serum (GIBCO), penicillin (100 U ml−1) and streptomycin (100 μg ml−1). Cells were free of mycoplasma contamination.
Proliferation and cell cycle assays
Cell proliferation was monitored by labelling cells with the cell trace violet dye (CellTrace violet proliferation kit, Invitrogen) or CFSE (5μM, Invitrogen) prior to stimulation, according to the manufacturer’s instructions. After 4 or 6 days of culture, cells were harvested and CellTrace violet or CSFE fluorescence dilution was assessed by flow cytometry. The division index of proliferation was calculated using Flowjo software (Tree Star) and corresponds to the average number of cell divisions per cell including the undivided peak. T-cell responses within total PBMCs were also measured by [3H]-thymidine incorporation after 6 days of stimulation. 0.074 MBq ml−1 of [3H]-thymidine was added during the last 18 h of stimulation. Cell proliferation was determined by cpm of [3H] thymidine incorporated in cells that were counted with TopCount NXT beta counter (PerkinElmer). Cell cycle analysis was determined by measuring the incorporation of the nucleoside analogue 5-ethynyl-2-deoxyuridine (EdU) into newly synthesized DNA, according to the manufacturer’s instructions (Click-iT EdU, Invitrogen) after 48 h of anti-CD3 stimulation. EdU incorporation in cells was measured following conjugation of EdU to azide-modified Alexa Fluor 647 dye. Cells were analyzed by flow cytometry with a FACS-Canto II flow cytometry system (BD Biosciences).
Nucleic acids and de novo pyrimidine synthesis assays
PBMCs were stimulated in the presence of 1 μg ml−1 coated anti-CD3 antibody (clone OKT3, eBiosciences) or 2.5 μg ml−1 PHA (Sigma-Aldrich) for 3 days or in the presence of candidin (5 μg ml−1, Bio-Rad), tetanus toxoid (1/8000 dilution, Statens Serum Institute) or PPD (tuberculin) (50 μg ml−1, Statens Serum Institute) for 6 days. 0.074 MBq ml−1 of [3H]-thymidine, [3H]-cytidine, [3H]-uridine or [3H]-leucine or 0.185 MBq ml−1 U-[14C]-aspartate were added during the last 18 h of stimulation. For [3H]-cytidine, this corresponds to 0.133 μM, which not allows to restore normal proliferation of CTPS1-deficient cells that required 50μM. Cells were harvested with a Filter Mate harvester (PerkinElmer) on filters for labelled cell assays (Unifilter™ plates, PerkinElmer) that retain nucleic acids and filters were then washed. Radioactivity (cpm) on filters (corresponding to radiolabelled compounds incorporated in nucleic acids) was measured by liquid scintillation counting with TopCount NXT beta counter (PerkinElmer).
Apoptosis assay
Apoptosis was determined by evaluating phosphatidylserine (PS) exposure in the outer leaflet of the cytoplasmic membrane with PE-conjugated Annexin-V in combination with 7-AAD Viaprobe (Apoptosis Detection Kit I, BD) 12 hours after stimulation with coated-OKT3 (0.1, 1 or 10 μg ml−1). Apoptosis was based on the percentage of Annexin V+/7AAD− cells to exclude necrotic cells. Cells were analyzed by flow cytometry.
Cytokine production and degranulation
For intracellular staining of cytokines, cells were stimulated overnight with PMA and ionomycin or anti CD3/CD28 beads in the presence of brefeldin A (GolgiPlug, BD). Cells were then fixed and permeabilized using the BD cytofix/cytoperm plus kit (BD Pharmingen) according to the manufacturer’s instructions. Cells were labelled with PE-anti-IL-2 (rat IgG2a, MHQ1-17H12), PE/Cy7-anti-TNF-α (mouse IgG1; MAb11), APC-anti-IFN-γ (mouse IgG1, 4S.B3) and isotype-matched mAbs purchased from Biolegend and analysed by flow cytometry. Degranulation was determined by analysis of the expression of CD107/LAMP, a marker of the exocytosis of lytic granules. Blasts were stimulated for 3 hours in the presence of 0.3, 3 or 30 μg ml−1 coated-OKT3 and simultaneously labelled with eFluor660-anti-CD107a (mouse IgG1; eBioH4A3), eFluor660-anti-CD107b (mouse IgG1; eBioABL-93) or isotype matched mAbs purchased from eBiosciences. Thereafter, cells were harvested, washed and stained with FITC-anti-CD3 and PE-anti-CD8 mAbs and analyzed by flow cytometry.
Flow cytometry
Cell staining and the flow-cytometry-based phenotypic analyses of PBMCs and cells were performed according to standard flow cytometry methods. The following mAbs were conjugated to fluorescein isothiocyanate (FITC), phycoerythrin (PE), phycoerythrin-Cyanine7 (PE/Cy7), phycoerythrin-Cyanine5, phycoerythrin-Cyanine5.5, allophycocyanin (APC), allophycocyanin-Vio7, View blue or View green: anti-CD25 (M-A251), anti-CD27 (M-T271), anti-CD31 (WM59), anti-CD45RA (HI100), anti-CD45RO (UCHL1), anti-CD197/CCR7 (3D12), anti-TCRαβ (T10B9), anti-TCRγδ(B1), anti-CD95 (DX2), anti-CD19 (HIB19), anti-CD21 (B-ly4), anti-IgM (G20-127), anti-IgD (IA6-2), anti-CD56 (B159) and anti-CD16 (3G8), all purchased from BD Biosciences and anti-CD3 (BW264/56), anti-CD4 (VIT4), anti-CD8 (BW135/80) and anti-CD69 (FN50) from Miltenyi Biotec. iNKT cells were detected by staining with anti-Vα24 (C15) and anti-Vβ11 (C21) (Beckman Coulter) and MAIT cells by staining with anti-Vα7.2 (3C10) and anti-CD161 (HP-3G10 (Biolegend). All data were collected on a FACS-Canto II cytometer (BD Biosciences) and analysed using FlowJo Version 9.3.2 software (TreeStar).
Immunoblotting and analysis CTPS1 protein expression
Cells (5. 106 cells/ml) were stimulated by anti-CD3 antibody (1 μg ml−1) cross-linking with a rabbit-anti-mouse IgG (2μg ml−1) or anti CD3/CD28 beads for the indicated time periods. Cells were then lysed in 1% NP40, 50 mM Tris pH 8, 150 mM NaCl, 2 0 mM EDTA, 1 mM Na3VO4, 1 mM NaF and complete protease inhibitor cocktail (Roche), as previously described30. Protein concentrations were quantitated by BCA assay (BIO-RAD). 80 μg of proteins were separated by SDS-PAGE and transferred on PVDF membranes (Millipore). Membranes were blocked with milk or BSA before incubation with antibodies. The following mAbs and rabbit polyclonal antibodies were used for immunoblotting: anti-PLC-γ1 (#2822S), anti-phosphorylated PLC-γ1 (#2821S), anti-phosphorylated ERK1/2 (clone E10, #9106S), anti-ERK1/2 (#4695S) anti-phosphorylated IκB—clone 5A5, #9246S), anti-phosphorylated PKCtheta (#9377S), NFκB (clone D14E12) anti-phosphorylated AKT (Serine 473, clone 587F11) and anti-phosphorylated tyrosine (4G10) purchased from Cell Signaling Technology and rabbit polyclonal antibodies anti-ACTIN (#A2066) and anti-CTPS1 raised against the residues 341 to 355 (#SAB111071) or 416-430 (#SAB111072) and anti-CTPS2 (#HPA017437) purchased from Sigma Aldrich. Anti-CTPS1 rabbit polyclonal antibodies (K-21) from Santa Cruz were also tested. Membranes were then washed and incubated with anti-mouse or anti-rabbit HRP-conjugated secondary antibodies from Cell Signaling and GE Healthcare, respectively. Pierce ECL western blotting substrate was used for detection. For inhibition assays of the signaling pathways after TCR-CD3 activation, cells have been stimulated with anti-CD3/CD28 beads for 48 h in the presence of 100 nM of the MAPK/ERK inhibitor PD0325901, 10μM of the Src family protein tyrosine kinase inhibitor PP1, 10μM of the Src family protein tyrosine kinase inhibitor PP2, 10μM of the selective Ca++ chelator 1,2-bis-(2-aminophenoxy) ethane-N,N,N′,N′-tetraacetic acid tetra(acetoxymethyl) ester (BAPTA/AM), 10μM of the IκBα phosphorylation inhibitor Bay 11-7085 or 10μM of PI3Kdelta inhibitor IC87114. All were from Sigma-Aldrich, excepted IC87114 from Calbiochem. The concentrations used were typical and previously reported. After 48 h incubation with the different inhibitors, cell viability was verified and was more than 90% in each condition. The activity and the selectivity of the inhibitors was verified in parallel by immunoblotting for phospho-tyrosine (for PP1 and PP2), IκBα phosphorylation (for Bay Bay 11-7085), ERK phosphorylation (for PD0325901) and AKT phosphorylation (for IC87114) (data not shown).
Calcium flux analysis
Ca2+ responses were assessed by flow cytometry, as previously described31. Briefly, cells were loaded with 5 μM Indo-1 AM (Molecular Probes), washed, incubated with anti-CD4-APC and anti-CD8-PE mAbs, stimulated by anti-CD3 antibody (0.125 μg ml−1) cross-linking with F(ab’)2 rabbit-anti-mouse IgG (10 μg ml−1) and then incubated with ionomycin (1 μM). Cells were analyzed with a FACS ARIA flow cytometer (BD Biosciences). Ca2+ flux data were obtained using kinetic analyses of FlowJo software package (TreeStar). Intracellular Ca2+ levels correspond to the normalized ratio of Ca2+-bound to Ca2+-free Indo-1 fluorescence and are plotted as a function of time.
Plasmid constructs, cell transfections and infections
A full length cDNA encoding wild-type CTPS1 and a full length cDNA encoding the mutant CTPS1delta18 were obtained by RT-PCR from control blasts and blasts from patient 1.2 respectively using the forward 5′-CGGGATCCCACCATGAAGTATATTCTGGTT-3′ and reverse 5′-CCGCTCGAGTCAGTCATGATTTAT TGA-3′ (for wild-type) and 5′-CCGCTCGAGTTAAAGAAAGTCTCCAAGC-3′ (for CTPS1delta18) specific primers. The cDNAs were verified by sequencing and inserted into a bicistronic lentiviral expression vector encoding the green fluorescent protein (GFP) as a reporter (pLenti7.3/V5-TOPO, Invitrogen). Viral particles for infection were obtained by coexpression of the lentiviral vector containing CTPS1 with third-generation lentiviral plasmids containing Gag-Pol, Rev and the G protein of the vesicular stomatitis virus (VSVG) into HEK 293T using calcium phosphate. Viral supernatants were collected every 12 h on 2 consecutive days, starting 48 h after transfection, and viral particles were concentrated by ultracentrifugation at 49,000 g for 1.5 h at 12°C. Cells were infected with viral particles at a minimal titer of 107 TU ml−1 and 48 h after infection, cells were deprived of IL-2 during 72 h for proliferation assays. To assess the selective advantage of GFP expression during long-term expansion, blasts were re-stimulated with antiCD3/CD28 beads (Invitrogen) every 48 h during 8 days. For CTPS1 gene knockdown, blasts or Jurkat cells were infected at day 3 of PHA stimulation with the pLKO.1 lentiviral vector containing a CTPS1-specific shRNA (Openbiosystems, n°TRCN0000045349 and n°TRCN0000045350) or a scrambled shRNA in which the puromycin resistance gene was replaced by the GFP gene. Proliferation was analyzed in GFP+ and GFP− blasts after 4 days of stimulation with anti-CD3/CD28 beads as previously described. For survey of loss of GFP expression in long-term expansions, blasts were repeatedly stimulated with anti-CD3/CD28 beads every 48 h during 8 days. Jurkat cells were maintained in culture after infection during 26 days. The proportions percentages of GFP+ cells were determined by flow cytometry.
Quantification of intracellular nucleotides
Intracellular pools of nucleotides were quantified based on previously described methods32,33. Briefly, 5 millions of cells were washed in 0.1 M phosphate buffer (pH=7.4) and lysed in 60 μL HClO4 1M, containing 2 μM 8-bromo-AMP (8-BrAMP) as an internal standard. After 12,000 g centrifugation for 5 min at 4°C, supernatants were transferred to a 384-wells plate and kept at 4°C in auto sampler before injection. Aliquots of 5 μL were injected onto a separation column (ACQUITY UPLC BEH300 C18, 1.7 μm, 2.1×100 mm reversed-phase column, Waters) with a flow rate of 0.5 mL/min and analyzed with a tandem mass spectrometry system consisting of an Acquity Ultra Performance Liquid Chromatography (UPLC Waters) interfaced with a xevo-TQ-S tandem quadrupole mass spectrometer (Waters). Mobile phase A was 0.1% formic acid in water and mobile phase B, 0.1% formic acid in acetonitrile. A programmed mobile phase-gradient was used during a 7-min run: 0 min, 1% B; 5 min, 10% B; 5.1 min, 100% B; 6 min, 100% B; 6.1 min, 1% B; 7 min, 1% B. The content of the 4 nucleotides ATP, GTP, UTP and CTP was quantified in the electrospray negative ion mode with multiple reaction monitoring (MRM). Transitions of m/z 505.9>408 and 505.9>272.9 were used for quantification and confirmation of ATP, respectively, and those of 521.9>158.9 and 521.9>177 for GTP, 482.8>158.9 and 482.8>79 for UTP, and 481.8>158.9 and 481.8>384 for CTP. Concentrations were determined by using calibration curves of the 4 nucleotides. The linearity, exactitude and variability were determined for the technical validation of this assay. The linearity gave a correlation coefficient of the linear regression curves greater than 0.99 for the 4 nucleotides. The minimum and maximum recovery of spiked samples with the 4 nucleotides at a concentration of 90 mg/L and 250 mg/L ranged from 72% to 123%. The maximum intra- and inter-assay variability was of 22% and 23%, respectively.
Statistical analysis
P values were calculated with the Students t-test using PRISM software (GraphPad Software), with a two-tailed distribution. The variance was similar between the groups that have been statistically compared.
Supplementary Material
Different immunological parameters of patients were tested from blood (numbers of cells), PBMCs (proliferation in response to PHA evaluated by incorporation of 3H-thymidine) and serum (immunoglobulin subclasses and specific antibodies).
Different T-cell subsets, B-cell subsets and NK cells from PBMCs were tested by flow cytometry. T-cell proliferation from PBMCs in response different stimuli including antigen-specific responses were analyzed.
a, Analysis of the single nucleotide variations (SNVs) detected by whole exome sequencing in the genome of P1.1, P1.2 and P2.1. The numbers of SNVs are indicated in the triangles. SNVs were filtered by removal of non-functional intronic and synonymous mutations, heterozygous variations and those present in dbSNPs, 1000 genomes databases. The intersection of the filtered SNVs in the three patients resulted in the identification of a single common splicing site variation in the CTPS1 gene. b, Exon-intron structure and sequences of exons 17, 18 and 19 of CTPS1. The position of the variation is indicated by an arrow. The boxed nucleotide corresponds to the alternative splice site which produces a shorter transcript lacking exon 18 detected in patient cells. The alternative stop codon is indicated by an asterisk. c, Expression of a CTPS1 transcript lacking exon 18 (CTPS1Δ18) in CTPS1-deficient patients. The relative expression of full length CTPS1, CTPS1Δ18 and ACTIN transcripts was examined by RT-PCR in EBV-B cell lines (patient P2.1) and T-cell blasts (patient P1.2) from CTPS1-deficient patients. RT-PCRs of ACTIN are shown as normalization controls of the cDNA samples. Three fold-serial dilutions of cDNAs (indicated as 1, 0.3 and 0.1) were used for amplification of each transcript. Base pair markers are shown on the left. PCR products were verified by sequencing showing the expression of an abnormal CTPS1 transcript lacking exon 18 in the cells of the patients.
a, Transient expression of CTPS1 and the mutant CTPS1Δ18 in 293-T cells transfected with vectors containing wild-type CTPS1 or the mutant CTPS1Δ18. Cell lysates were tested by immunoblotting for CTPS1 with different antibodies raised against CTPS1 and for ACTIN as a control for loading. The CTPS1Δ18 mutant protein is recognized by the rabbit polyclonal antibodies raised against the 341 to 355 (anti-341-355) or the 416 to 430 (anti-416-430) residues of CTPS1 but not by the rabbit polyclonal antibody K21. b, T-cell blasts from a healthy control (Ctr.) and the CTPS1-deficient patient P1.2 (P1.2) stimulated for 48 h with anti-CD3 were analyzed for CTPS1 expression with the rabbit polyclonal antibodies anti-416-430 and anti-341-355. ACTIN expression as control for loading. c, EBV B-cell lines from healthy controls (Ctr. 1 and Ctr.2) and CTPS1-mutated patients (P1.2 and P2.1) were analyzed for CTPS1 expression with the rabbit polyclonal antibody anti-416-430. ACTIN expression served as control for loading.
a, Immunoblots for CTPS1 expression in sorted CD19+B cells (from PBMCs of an healthy donor) stimulated with the indicated stimuli. ACTIN serves as loading control. b, Kinetic of CTPS1 mRNA expression monitored by RT-qPCR in sorted B cells that have been stimulated with anti-BCR+CpG. Expression is in arbitrary units (A.U.) normalized to the expression of the GADPH gene and leukocytes were used as calibrator. c, Immunoblots for CTPS1 expression in T-cell blasts (from an healthy donor) stimulated with anti-CD3/CD28 beads in the presence of selective inhibitors of NFκB, Src kinases, Ca++, ERK kinase and PI3Kdelta. ACTIN serves as loading control. The activity of the inhibitors was controlled in parallel (see methods and data not shown).
a, Immunoblots showing the phosphorylation of proximal signaling molecules in T-cell blasts from a control donor (Ctr.) and a CTPS1-deficient patient P1.2 (P1.2) stimulated with anti-CD3 antibodies for 0, 2, 5, 15, 30 and 60 minutes or PMA plus ionomycin (P+I). Cell lysates were immunoblotted with antibodies against tyrosine-phosphorylated residues (PY), phosphoPLCG1 (pPLCG1), PLCG1, NFAT2c, phosphoPKCtheta (pPKCtheta), IkBa, phosphoERK1/2 (pERK1/2) and ACTIN as a loading control. Molecular weights are on the left. Data correspond to one representative experiment of 2 or 3 independent experiments. b, Flow cytometry analyses of Ca2+-flux in T cells from PBMCs or T-cell blasts of a control donor (Ctr.) and a CTPS1-deficient patient P1.2 (P1.2) loaded with the Ca2+-sensitive fluorescent dye Indo-1. Cells were then stimulated with anti-CD3 antibodies (first arrow) crosslinked with rabbit anti-mouse antibodies (second arrow) and then incubated with ionomycin (third arrow) to induce a receptor-independent Ca2+ response. Intracellular Ca2+ levels are expressed in arbitrary units (A.U.). Data with the T-cell blasts correspond to one of three representative experiments. c, Analysis of the degranulation capacity of CD8+ T-cell blasts from two control donors (Ctr.1 and Ctr.2) and a CTPS1-deficient patient P1.2 (P1.2) stimulated with the indicated concentrations of anti-CD3 antibodies for 4 h. Cells were stained with antibodies against CD107a/b (LAMP1/2), a surface-exposed marker of the secretion of lytic granules, and then analyzed by FACS. Means with s.d of percentages of CD8+ CD107+ cells are presented. d, Flow cytometry analysis of intracellular IL-2 production in CD4+ and CD8+ T-cells from PBMCs of a control donor (Ctr.) and two CTPS1-deficient patients P1.2 and P2.2 (P1.2 and P2.2) stimulated for 36 hours with anti-CD28 and anti-CD3 antibodies. The percentages of CD4+IL-2+ and CD8+IL-2+ are shown. e, Flow cytometry analysis of intracellular IFN-γ and TNF-α production on gated CD3+ T-cell blasts of a control donor (Ctr.) and a CTPS1-deficient patient P1.2 (P1.2) stimulated for 12 hours with IL-2, anti-CD3 and anti-CD28 coated beads (anti-CD3/CD28), PMA plus Ionomycin or PHA. Data are representative of one of 3 independent experiments. Dot-plots in red correspond to the isotype control. f, Induction of CD25 and CD69 in CD3+ T-cell blasts from a control donor (Ctr.) and a CTPS1-deficient patient (P1.2) was assessed after 24 h of anti-CD3 stimulation for CD69 and 96 h for CD25. Expression was assessed by flow cytometry and the median fluorescence intensity (MFI) is presented. Data are means with s.d of four and eight independent experiments for CD69 and CD25, respectively. Unpaired t-test. ***P <0.001, *P <0.05. g, Analysis of activation-induced cell death (AICD) in CD3+ T-cell blasts from a control donor (Ctr.) and a CTPS1-deficient patient P1.2 (P1.2,) after stimulation with the indicated concentration of anti-CD3 antibodies for 12 h. Apoptotic cells were detected by annexin V and 7-AAD staining and the percentages of annexin V positive/7-AAD negative cells within the gated CD3 population are shown. Data are means with s.d. of four (P1.2, n=4) and eight (Ctr., n=8) independent experiments. Unpaired Student t-tests and *P <0.05.
a, Proliferation of CD3+ T cells from PBMCs of control donors (Ctr.) and CTPS1-deficient patients (P1.1, P1.2; left panels) or (P1.2, P2.2; right panels). Right panels and left panels correspond to 2 independent experiments. Cells were stimulated with immobilized anti-CD3 and soluble anti-CD28 antibodies during 6 days. The proliferation was determined by dilution of CFSE staining analyzed by flow cytometry. Histograms correspond to CFSE staining dilutions for which the number of cell divisions was indicated at the top of each peak. b, Proliferation of CD3+ T and CD16+CD56+ NK cells from PBMCs of a control donor (Ctr.) and a CTPS1-deficient patient (P1.2). Cells were stimulated with anti-CD3/CD28 coated beads for 3 days or IL-2 for 7 days. Representative dot plots showing cell divisions by dilution of the violet dye and expression of the activation marker CD69. Inserts with histograms showing the violet dye dilution with the number cell divisions indicated at the top of each peak.
a, Incorporation of 3H-thymidine, 3H-uridine, 3H-cytidine and 3H-leucine as tracers of DNA, RNA and protein synthesis in PBMCs from a control healthy donor (Ctr.) and a CTPS1-deficient patient (P1.2) stimulated or not (no stim.) for 3 days with anti-CD3 or PHA and for 6 days with tetanus toxoid, candidin or tuberculin. The concentration of 3H-cytidine used in these experiments is under the value allowing the restoration of normal proliferation in CTPS1-deficient cells (see also Methods). Data are means with s.d of two independent experiments with triplicates. Unpaired Student t-tests and ***P<0.001, **P<0.01, *P<0.05. b, c, Incorporation of 14C-aspartate and 3H-thymidine in PBMCs (b) or T-cell blasts (c) from a control healthy donor (Ctr.) and a CTPS1-deficient patient (P1.2) stimulated or not (no stim.) for 3 days with anti-CD3 or PHA. Data means with s.d of three independent samples.
a, Expression of CTPS1 in Jurkat cells transduced with lentiviral vectors containing two distinct CTPS1 shRNAs (Sh CTPS1 #1 or Sh CTPS1 #2) or a scrambled shRNA (Sh scramble). Cell lysates were analyzed by immunoblotting for CTPS1, CTPS2 and ACTIN protein expression. b, Proliferation of Jurkat cells, in which CTPS1 expression was silenced, was monitored as a function of the loss of GFP expression and compared with cells transduced with the scrambled shRNA. The percentages of GFP-positive cells were determined by flow cytometry at the indicated time points with “time 0” corresponding to 48 hours post transduction.
a, Concentration of ATP, GTP, UTP and CTP in cell extracts of T-cell blasts stimulated with anti-CD3/CD28 coated beads or not (no stim.) from control healthy donors (Ctr.) or from patient P1.2. Control cells were treated or not with deazauridine for 24 h before and during stimulation or not. Representative data from 3 independent experiments. b, same as in (a) with EBV B-cell lines from control healthy donors (Ctr.) and patients P1.1 (squares), P1.2 (circles) and P2.1 (triangles). For controls, each symbol corresponds to a different control cell line (from a different healthy donor). Representative data from two independent experiments with blinding during the measurements. Bars correspond to averages. Unpaired t-tests and *P<0.05, **P<0.01, ***P<0.001. CTP data are also shown in Figure 3g,h.
Acknowledgements
The authors thank the patients, their families and the healthy donors for cooperation. We thank S. Rigaud, S. Gérart, C. Synaeve and R. Rodriguez for help with experiments and P. Revy for discussion. This work was supported by grants from INSERM, ANR (ANR-08-MIEN-012-01, ANR-2010-MIDI-005-02 and ANR-10-IAHU-01), Fondation ARC (France), the European Research Council (ERC-2009-AdG_20090506 n°FP7-249816) and the Rare Diseases Fondation (France). S. L. is a senior scientist of CNRS (France). E. M. is supported by ANR (France) and Ligue contre le cancer (France). We are also grateful to the UK 1958 Birth Cohort (http://www2.le.ac.uk/projects/birthcohort/1958BC-About) for providing DNA from 752 individuals born in the North West of England. Access to these resources was enabled via the 58READIE Project funded by the Wellcome Trust and Medical Research Council (grant numbers WT095219MA and G1001799). A full list of the financial, institutional and personal contributions to the development of the 1958 Birth Cohort Biomedical resource is available at http://www2.le.ac.uk/projects/birthcohort.
Footnotes
The authors declare no competing financial interests.
References
- 1.MacIver NJ, Michalek RD, Rathmell JC. Metabolic regulation of T lymphocytes. Annu Rev Immunol. 2013;31:259–83. doi: 10.1146/annurev-immunol-032712-095956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Evans DR, Guy HI. Mammalian pyrimidine biosynthesis: fresh insights into an ancient pathway. J Biol Chem. 2004;279:33035–8. doi: 10.1074/jbc.R400007200. [DOI] [PubMed] [Google Scholar]
- 3.Higgins MJ, Graves PR, Graves LM. Regulation of human cytidine triphosphate synthetase 1 by glycogen synthase kinase 3. J Biol Chem. 2007;282:29493–503. doi: 10.1074/jbc.M703948200. [DOI] [PubMed] [Google Scholar]
- 4.Ostrander DB, O’Brien DJ, Gorman JA, Carman GM. Effect of CTP synthetase regulation by CTP on phospholipid synthesis in Saccharomyces cerevisiae. J Biol Chem. 1998;273:18992–9001. doi: 10.1074/jbc.273.30.18992. [DOI] [PubMed] [Google Scholar]
- 5.Kassel KM, Au da R, Higgins MJ, Hines M, Graves LM. Regulation of human cytidine triphosphate synthetase 2 by phosphorylation. J Biol Chem. 2010;285:33727–36. doi: 10.1074/jbc.M110.178566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nadkarni AK, et al. Differential biochemical regulation of the URA7- and URA8-encoded CTP synthetases from Saccharomyces cerevisiae. J Biol Chem. 1995;270:24982–8. doi: 10.1074/jbc.270.42.24982. [DOI] [PubMed] [Google Scholar]
- 7.van Kuilenburg AB, Meinsma R, Vreken P, Waterham HR, van Gennip AH. Identification of a cDNA encoding an isoform of human CTP synthetase. Biochim Biophys Acta. 2000;1492:548–52. doi: 10.1016/s0167-4781(00)00141-x. [DOI] [PubMed] [Google Scholar]
- 8.Fairbanks LD, Bofill M, Ruckemann K, Simmonds HA. Importance of ribonucleotide availability to proliferating T-lymphocytes from healthy humans. Disproportionate expansion of pyrimidine pools and contrasting effects of de novo synthesis inhibitors. J Biol Chem. 1995;270:29682–9. [PubMed] [Google Scholar]
- 9.van den Berg AA, et al. Cytidine triphosphate (CTP) synthetase activity during cell cycle progression in normal and malignant T-lymphocytic cells. Eur J Cancer. 1995;31A:108–12. doi: 10.1016/0959-8049(94)00442-8. [DOI] [PubMed] [Google Scholar]
- 10.Wynn RF, et al. Treatment of Epstein-Barr-virus-associated primary CNS B cell lymphoma with allogeneic T-cell immunotherapy and stem-cell transplantation. Lancet Oncol. 2005;6:344–6. doi: 10.1016/S1470-2045(05)70171-6. [DOI] [PubMed] [Google Scholar]
- 11.Notarangelo LD. Functional T cell immunodeficiencies (with T cells present) Annu Rev Immunol. 2013;31:195–225. doi: 10.1146/annurev-immunol-032712-095927. [DOI] [PubMed] [Google Scholar]
- 12.Kursula P, et al. Structure of the synthetase domain of human CTP synthetase, a target for anticancer therapy. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2006;62:613–7. doi: 10.1107/S1744309106018136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Traut TW. Physiological concentrations of purines and pyrimidines. Mol Cell Biochem. 1994;140:1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- 14.Ben-Sahra I, Howell JJ, Asara JM, Manning BD. Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science. 2013;339:1323–8. doi: 10.1126/science.1228792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McPartland RP, Wang MC, Bloch A, Weinfeld H. Cytidine 5′-triphosphate synthetase as a target for inhibition by the antitumor agent 3-deazauridine. Cancer Res. 1974;34:3107–11. [PubMed] [Google Scholar]
- 16.Qiu Y, et al. Mycophenolic acid-induced GTP depletion also affects ATP and pyrimidine synthesis in mitogen-stimulated primary human T-lymphocytes. Transplantation. 2000;69:890–7. doi: 10.1097/00007890-200003150-00038. [DOI] [PubMed] [Google Scholar]
- 17.van den Berg AA, et al. The roles of uridine-cytidine kinase and CTP synthetase in the synthesis of CTP in malignant human T-lymphocytic cells. Leukemia. 1994;8:1375–8. [PubMed] [Google Scholar]
- 18.Le Bourhis L, Mburu YK, Lantz O. MAIT cells, surveyors of a new class of antigen: development and functions. Curr Opin Immunol. 2013;25:174–80. doi: 10.1016/j.coi.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 19.Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. Functions of natural killer cells. Nat Immunol. 2008;9:503–10. doi: 10.1038/ni1582. [DOI] [PubMed] [Google Scholar]
- 20.Chung BK, et al. Innate immune control of EBV-infected B cells by invariant natural killer T cells. Blood. 2013;122:2600–8. doi: 10.1182/blood-2013-01-480665. [DOI] [PubMed] [Google Scholar]
- 21.Brennan PJ, Brigl M, Brenner MB. Invariant natural killer T cells: an innate activation scheme linked to diverse effector functions. Nat Rev Immunol. 2013;13:101–17. doi: 10.1038/nri3369. [DOI] [PubMed] [Google Scholar]
- 22.Toy G, et al. Requirement for deoxycytidine kinase in T and B lymphocyte development. Proc Natl Acad Sci U S A. 107:5551–6. doi: 10.1073/pnas.0913900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marijnen YM, et al. Studies on the incorporation of precursors into purine and pyrimidine nucleotides via ‘de novo’ and ‘salvage’ pathways in normal lymphocytes and lymphoblastic cell-line cells. Biochim Biophys Acta. 1989;1012:148–55. doi: 10.1016/0167-4889(89)90088-8. [DOI] [PubMed] [Google Scholar]
- 24.Austin WR, et al. Nucleoside salvage pathway kinases regulate hematopoiesis by linking nucleotide metabolism with replication stress. J Exp Med. 2012;209:2215–28. doi: 10.1084/jem.20121061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murali-Krishna K, et al. Counting antigen-specific CD8 T cells: a reevaluation of bystander activation during viral infection. Immunity. 1998;8:177–87. doi: 10.1016/s1074-7613(00)80470-7. [DOI] [PubMed] [Google Scholar]
- 26.Hislop AD, Taylor GS, Sauce D, Rickinson AB. Cellular responses to viral infection in humans: lessons from Epstein-Barr virus. Annu Rev Immunol. 2007;25:587–617. doi: 10.1146/annurev.immunol.25.022106.141553. [DOI] [PubMed] [Google Scholar]
- 27.Huang M, Graves LM. De novo synthesis of pyrimidine nucleotides; emerging interfaces with signal transduction pathways. Cell Mol Life Sci. 2003;60:321–36. doi: 10.1007/s000180300027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robitaille AM, et al. Quantitative phosphoproteomics reveal mTORC1 activates de novo pyrimidine synthesis. Science. 2013;339:1320–3. doi: 10.1126/science.1228771. [DOI] [PubMed] [Google Scholar]
- 29.Gülow K, Kaminski M, Darvas K, Süss D, Li-Weber M, Krammer PH. HIV-1 trans-activator of transcription substitutes for oxidative signaling in activation-induced T cell death. J Immunol. 2005 May 1;174(9):5249–60. doi: 10.4049/jimmunol.174.9.5249. [DOI] [PubMed] [Google Scholar]
- 30.Latour S, et al. Regulation of SLAM-mediated signal transduction by SAP, the X-linked lymphoproliferative gene product. Nat Immunol. 2001;2:681–90. doi: 10.1038/90615. [DOI] [PubMed] [Google Scholar]
- 31.Picard C, et al. Hypomorphic mutation of ZAP70 in human results in a late onset immunodeficiency and no autoimmunity. Eur J Immunol. 2009;39:1966–76. doi: 10.1002/eji.200939385. [DOI] [PubMed] [Google Scholar]
- 32.Luo B, Groenke K, Takors R, Wandrey C, Oldiges M. Simultaneous determination of multiple intracellular metabolites in glycolysis, pentose phosphate pathway and tricarboxylic acid cycle by liquid chromatography-mass spectrometry. J Chromatogr A. 2007;1147:153–64. doi: 10.1016/j.chroma.2007.02.034. [DOI] [PubMed] [Google Scholar]
- 33.Scavennec J, Maraninchi D, Gastaut JA, Carcassonne Y, Cailla HL. Purine and pyrimidine ribonucleoside monophosphate patterns of peripheral blood and bone marrow cells in human acute leukemias. Cancer Res. 1982;42:1326–30. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Different immunological parameters of patients were tested from blood (numbers of cells), PBMCs (proliferation in response to PHA evaluated by incorporation of 3H-thymidine) and serum (immunoglobulin subclasses and specific antibodies).
Different T-cell subsets, B-cell subsets and NK cells from PBMCs were tested by flow cytometry. T-cell proliferation from PBMCs in response different stimuli including antigen-specific responses were analyzed.
a, Analysis of the single nucleotide variations (SNVs) detected by whole exome sequencing in the genome of P1.1, P1.2 and P2.1. The numbers of SNVs are indicated in the triangles. SNVs were filtered by removal of non-functional intronic and synonymous mutations, heterozygous variations and those present in dbSNPs, 1000 genomes databases. The intersection of the filtered SNVs in the three patients resulted in the identification of a single common splicing site variation in the CTPS1 gene. b, Exon-intron structure and sequences of exons 17, 18 and 19 of CTPS1. The position of the variation is indicated by an arrow. The boxed nucleotide corresponds to the alternative splice site which produces a shorter transcript lacking exon 18 detected in patient cells. The alternative stop codon is indicated by an asterisk. c, Expression of a CTPS1 transcript lacking exon 18 (CTPS1Δ18) in CTPS1-deficient patients. The relative expression of full length CTPS1, CTPS1Δ18 and ACTIN transcripts was examined by RT-PCR in EBV-B cell lines (patient P2.1) and T-cell blasts (patient P1.2) from CTPS1-deficient patients. RT-PCRs of ACTIN are shown as normalization controls of the cDNA samples. Three fold-serial dilutions of cDNAs (indicated as 1, 0.3 and 0.1) were used for amplification of each transcript. Base pair markers are shown on the left. PCR products were verified by sequencing showing the expression of an abnormal CTPS1 transcript lacking exon 18 in the cells of the patients.
a, Transient expression of CTPS1 and the mutant CTPS1Δ18 in 293-T cells transfected with vectors containing wild-type CTPS1 or the mutant CTPS1Δ18. Cell lysates were tested by immunoblotting for CTPS1 with different antibodies raised against CTPS1 and for ACTIN as a control for loading. The CTPS1Δ18 mutant protein is recognized by the rabbit polyclonal antibodies raised against the 341 to 355 (anti-341-355) or the 416 to 430 (anti-416-430) residues of CTPS1 but not by the rabbit polyclonal antibody K21. b, T-cell blasts from a healthy control (Ctr.) and the CTPS1-deficient patient P1.2 (P1.2) stimulated for 48 h with anti-CD3 were analyzed for CTPS1 expression with the rabbit polyclonal antibodies anti-416-430 and anti-341-355. ACTIN expression as control for loading. c, EBV B-cell lines from healthy controls (Ctr. 1 and Ctr.2) and CTPS1-mutated patients (P1.2 and P2.1) were analyzed for CTPS1 expression with the rabbit polyclonal antibody anti-416-430. ACTIN expression served as control for loading.
a, Immunoblots for CTPS1 expression in sorted CD19+B cells (from PBMCs of an healthy donor) stimulated with the indicated stimuli. ACTIN serves as loading control. b, Kinetic of CTPS1 mRNA expression monitored by RT-qPCR in sorted B cells that have been stimulated with anti-BCR+CpG. Expression is in arbitrary units (A.U.) normalized to the expression of the GADPH gene and leukocytes were used as calibrator. c, Immunoblots for CTPS1 expression in T-cell blasts (from an healthy donor) stimulated with anti-CD3/CD28 beads in the presence of selective inhibitors of NFκB, Src kinases, Ca++, ERK kinase and PI3Kdelta. ACTIN serves as loading control. The activity of the inhibitors was controlled in parallel (see methods and data not shown).
a, Immunoblots showing the phosphorylation of proximal signaling molecules in T-cell blasts from a control donor (Ctr.) and a CTPS1-deficient patient P1.2 (P1.2) stimulated with anti-CD3 antibodies for 0, 2, 5, 15, 30 and 60 minutes or PMA plus ionomycin (P+I). Cell lysates were immunoblotted with antibodies against tyrosine-phosphorylated residues (PY), phosphoPLCG1 (pPLCG1), PLCG1, NFAT2c, phosphoPKCtheta (pPKCtheta), IkBa, phosphoERK1/2 (pERK1/2) and ACTIN as a loading control. Molecular weights are on the left. Data correspond to one representative experiment of 2 or 3 independent experiments. b, Flow cytometry analyses of Ca2+-flux in T cells from PBMCs or T-cell blasts of a control donor (Ctr.) and a CTPS1-deficient patient P1.2 (P1.2) loaded with the Ca2+-sensitive fluorescent dye Indo-1. Cells were then stimulated with anti-CD3 antibodies (first arrow) crosslinked with rabbit anti-mouse antibodies (second arrow) and then incubated with ionomycin (third arrow) to induce a receptor-independent Ca2+ response. Intracellular Ca2+ levels are expressed in arbitrary units (A.U.). Data with the T-cell blasts correspond to one of three representative experiments. c, Analysis of the degranulation capacity of CD8+ T-cell blasts from two control donors (Ctr.1 and Ctr.2) and a CTPS1-deficient patient P1.2 (P1.2) stimulated with the indicated concentrations of anti-CD3 antibodies for 4 h. Cells were stained with antibodies against CD107a/b (LAMP1/2), a surface-exposed marker of the secretion of lytic granules, and then analyzed by FACS. Means with s.d of percentages of CD8+ CD107+ cells are presented. d, Flow cytometry analysis of intracellular IL-2 production in CD4+ and CD8+ T-cells from PBMCs of a control donor (Ctr.) and two CTPS1-deficient patients P1.2 and P2.2 (P1.2 and P2.2) stimulated for 36 hours with anti-CD28 and anti-CD3 antibodies. The percentages of CD4+IL-2+ and CD8+IL-2+ are shown. e, Flow cytometry analysis of intracellular IFN-γ and TNF-α production on gated CD3+ T-cell blasts of a control donor (Ctr.) and a CTPS1-deficient patient P1.2 (P1.2) stimulated for 12 hours with IL-2, anti-CD3 and anti-CD28 coated beads (anti-CD3/CD28), PMA plus Ionomycin or PHA. Data are representative of one of 3 independent experiments. Dot-plots in red correspond to the isotype control. f, Induction of CD25 and CD69 in CD3+ T-cell blasts from a control donor (Ctr.) and a CTPS1-deficient patient (P1.2) was assessed after 24 h of anti-CD3 stimulation for CD69 and 96 h for CD25. Expression was assessed by flow cytometry and the median fluorescence intensity (MFI) is presented. Data are means with s.d of four and eight independent experiments for CD69 and CD25, respectively. Unpaired t-test. ***P <0.001, *P <0.05. g, Analysis of activation-induced cell death (AICD) in CD3+ T-cell blasts from a control donor (Ctr.) and a CTPS1-deficient patient P1.2 (P1.2,) after stimulation with the indicated concentration of anti-CD3 antibodies for 12 h. Apoptotic cells were detected by annexin V and 7-AAD staining and the percentages of annexin V positive/7-AAD negative cells within the gated CD3 population are shown. Data are means with s.d. of four (P1.2, n=4) and eight (Ctr., n=8) independent experiments. Unpaired Student t-tests and *P <0.05.
a, Proliferation of CD3+ T cells from PBMCs of control donors (Ctr.) and CTPS1-deficient patients (P1.1, P1.2; left panels) or (P1.2, P2.2; right panels). Right panels and left panels correspond to 2 independent experiments. Cells were stimulated with immobilized anti-CD3 and soluble anti-CD28 antibodies during 6 days. The proliferation was determined by dilution of CFSE staining analyzed by flow cytometry. Histograms correspond to CFSE staining dilutions for which the number of cell divisions was indicated at the top of each peak. b, Proliferation of CD3+ T and CD16+CD56+ NK cells from PBMCs of a control donor (Ctr.) and a CTPS1-deficient patient (P1.2). Cells were stimulated with anti-CD3/CD28 coated beads for 3 days or IL-2 for 7 days. Representative dot plots showing cell divisions by dilution of the violet dye and expression of the activation marker CD69. Inserts with histograms showing the violet dye dilution with the number cell divisions indicated at the top of each peak.
a, Incorporation of 3H-thymidine, 3H-uridine, 3H-cytidine and 3H-leucine as tracers of DNA, RNA and protein synthesis in PBMCs from a control healthy donor (Ctr.) and a CTPS1-deficient patient (P1.2) stimulated or not (no stim.) for 3 days with anti-CD3 or PHA and for 6 days with tetanus toxoid, candidin or tuberculin. The concentration of 3H-cytidine used in these experiments is under the value allowing the restoration of normal proliferation in CTPS1-deficient cells (see also Methods). Data are means with s.d of two independent experiments with triplicates. Unpaired Student t-tests and ***P<0.001, **P<0.01, *P<0.05. b, c, Incorporation of 14C-aspartate and 3H-thymidine in PBMCs (b) or T-cell blasts (c) from a control healthy donor (Ctr.) and a CTPS1-deficient patient (P1.2) stimulated or not (no stim.) for 3 days with anti-CD3 or PHA. Data means with s.d of three independent samples.
a, Expression of CTPS1 in Jurkat cells transduced with lentiviral vectors containing two distinct CTPS1 shRNAs (Sh CTPS1 #1 or Sh CTPS1 #2) or a scrambled shRNA (Sh scramble). Cell lysates were analyzed by immunoblotting for CTPS1, CTPS2 and ACTIN protein expression. b, Proliferation of Jurkat cells, in which CTPS1 expression was silenced, was monitored as a function of the loss of GFP expression and compared with cells transduced with the scrambled shRNA. The percentages of GFP-positive cells were determined by flow cytometry at the indicated time points with “time 0” corresponding to 48 hours post transduction.
a, Concentration of ATP, GTP, UTP and CTP in cell extracts of T-cell blasts stimulated with anti-CD3/CD28 coated beads or not (no stim.) from control healthy donors (Ctr.) or from patient P1.2. Control cells were treated or not with deazauridine for 24 h before and during stimulation or not. Representative data from 3 independent experiments. b, same as in (a) with EBV B-cell lines from control healthy donors (Ctr.) and patients P1.1 (squares), P1.2 (circles) and P2.1 (triangles). For controls, each symbol corresponds to a different control cell line (from a different healthy donor). Representative data from two independent experiments with blinding during the measurements. Bars correspond to averages. Unpaired t-tests and *P<0.05, **P<0.01, ***P<0.001. CTP data are also shown in Figure 3g,h.