Abstract
RNA–protein complexes play pivotal roles in many central biological processes. While methods based on next-generation sequencing have profoundly advanced our ability to identify the specific RNAs bound by a particular protein, there is a dire need for precise and systematic ways to identify RNA interaction sites on proteins. We have developed an integrated experimental and computational workflow combining photo-induced cross-linking, high-resolution mass spectrometry, and automated analysis of the resulting mass spectra for the identification of cross-linked peptides and exact amino acids with their cross-linked RNA oligonucleotide moiety of such RNA-binding proteins. The generic workflow can be applied to any RNA–protein complex of interest. Application to human and yeast mRNA–protein complexes in vitro and in vivo demonstrates the powerful utility of the approach by identification of 257 cross-linking sites on 124 distinct RNA-binding proteins. The software pipeline developed for this purpose is available as open-source software as part of the OpenMS project.
Introduction
RNA molecules bind to proteins to form ribonucleoprotein complexes (RNPs). These are indispensable for the synthesis, stability, transport, and activity of mRNAs1 as well as of non-coding RNAs2, 3. RNA-binding proteins (RBPs) assume numerous functions in RNPs. RBPs can modulate or stabilize RNA structures, thereby making RNA catalytically active, e.g., during pre-mRNA splicing4. RNA can also guide a catalytically active RBP to its destination; examples of this are miRNA- or lncRNA-mediated translational control and epigenetic modulation5, 6. RBPs are also involved in splicing, can recruit or repel other proteins, induce hydrolysis of RNA, or protect it from degradation.
Consequently, much attention is devoted on the identification of proteins interacting directly with RNAs as well as to the identification of the corresponding RNA sites. Classical pull-down experiments can provide evidence that a certain RNA and protein interact with one another, directly or indirectly. UV-induced cross-linking studies and subsequent mass spectrometry (MS) have identified proteins in direct contact with RNA7-10. While the identification of RNAs bound by RBPs and their exact interaction sites benefits from next-generation sequencing techniques after cross-linking like PAR-CLIP11, so far no complementary approach has been described that allows the identification of the corresponding cross-link sites within the proteins.
Here, we report a novel methodology for biochemical purification of peptide–RNA oligonucleotide conjugates derived from in vitro or in vivo UV-irradiated RNP complexes that allows mass spectrometric sequencing of the cross-linked peptide and RNA moiety and the subsequent identification of the cross-linked peptides and RNA by automated database searches. The strategy was applied to three biological systems of human and yeast origin. Overall, we have identified 749 cross-links which were mapped to 257 unique amino acids or protein regions in 124 different proteins. The impartiality of the approach was demonstrated by the identification of proteins lacking classical RNA-binding motifs or annotated RNA-binding function.
Results
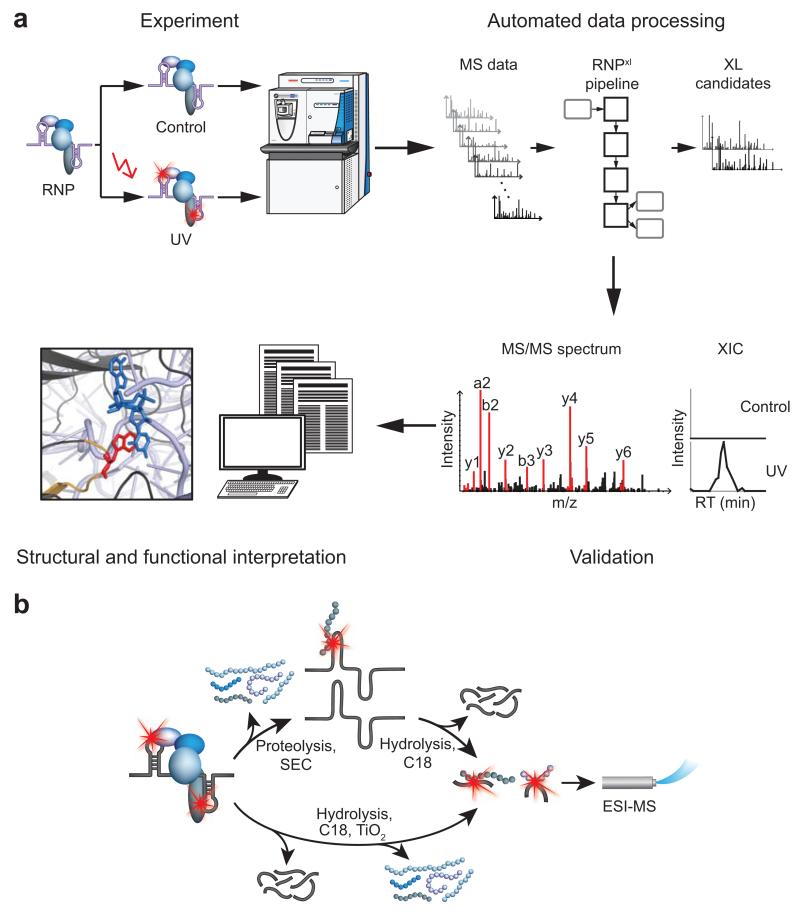
We designed an integrated experimental and computational strategy for the automated identification of protein–RNA cross-linking sites including the cross-linked amino acids and the cross-linked RNA moiety (Fig. 1a). Isolated and UV cross-linked RNPs are hydrolyzed with endoproteases and nucleases. The resulting peptide–RNA oligonucleotide heteroconjugates are enriched (Fig. 1b) and analyzed by electrospray-ionization (ESI) MS on Orbitrap instruments in data-dependent acquisition mode. MS data is submitted to a dedicated data-analysis workflow based on the OpenMS platform12, 13. In contrast to conventional MS search engines, the computational workflow introduced here removes all tandem mass spectra (MS/MS spectra) not corresponding to cross-linked species, calculates masses of possible peptide–RNA conjugates, and searches MS/MS spectra against a sequence database with OMSSA14. Fragment spectra assigned to peptide–RNA oligonucleotide cross-links are annotated in order to identify the cross-linked amino acid and nucleotides. We applied this approach independently to three biological systems: (1) RNPs derived from incubation of a transcribed pre-mRNA tagged with an MS2 aptamer and reconstituted in vitro with human nuclear extract, (2) purified (pre-) mRNA complexes from yeast, obtained by affinity capture of the nuclear cap-binding protein subunit 2 (Cpb20), and (3) entire yeast cells metabolically labeled with 4-thio-uridine (4SU) and cross-linked in vivo. Protein–RNA cross-linking in the first two systems was carried out at 254 nm, the protein moiety was digested with trypsin under denaturing conditions and noncross-linked peptides were removed by size-exclusion chromatography (SEC). The RNA pool was hydrolyzed with endonucleases, noncross-linked oligonucleotides were removed by reversed-phase chromatography (RPC), and remaining cross-linked peptide–oligonucleotide conjugates subjected to LC-MS/MS (Fig. 1b). In the third system, UV irradiation was performed at 365 nm in vivo, cells were lysed, and polyadenylated mRNA with its associated RBPs was recovered by oligo d(T) affinity selection. After digestion of the cross-linked protein and RNA moiety, noncross-linked oligonucleotides were removed by RPC and cross-linked conjugates were enriched with a TiO2 matrix prior to LC-MS/MS analysis (Fig. 1b, Online Methods).
Figure 1.
Overview of the procedure and experimental workflow. (a) Schematic outline of the entire approach. UV irradiated protein–RNA complexes are processed for LC-ESI-MS/MS analysis on an Orbitrap mass spectrometer in parallel with a non-irradiated control. MS data is subjected to the data analysis workflow that yields potential cross-linked peptides. Results are validated by comparison of extracted ion chromatogram (XIC; RT: retention time) intensities in control versus UV-irradiated sample and evaluation of expected and observed fragmentation patterns in the MS/MS spectrum. Identified cross-links are compared to published RNA-binding functionality and structural data when available. (b) Isolation of cross-linked heteroconjugates. Cross-linked protein–RNA complexes are either enriched by size-exclusion (SEC) and reversed-phase C18 chromatography (upper workflow) or by C18 chromatography and titanium dioxide solid phase extraction (lower workflow; see Online Methods). Proteins are hydrolyzed before SEC, which separates RNA with or without cross-linked peptides from noncross-linked peptides. RNA-containing fractions are hydrolyzed and subsequently RNA oligonucleotides are removed from the cross-linked heteroconjugates by C18 chromatography. Alternatively, the sample is hydrolyzed completely. C18 chromatography removes the majority of noncross-linked RNA fragments, and subsequent enrichment with TiO2 retains the cross-linked heteroconjugates while noncross-linked peptides are eliminated. Enriched peptide–RNA oligonucleotide heteroconjugates from both experimental workflows are then directly analyzed by LC-ESI-MS/MS.
In all experiments with unlabeled RNA, UV-irradiated and non-irradiated samples were treated in parallel (Fig. 1a) for a reliable distinction between cross-linked peptide–RNA oligonucleotides and false positives (e.g., residual noncross-linked species). The experimental workflow resulted in two raw data files per experiment (cross-linked and control, except for yeast 4SU-labeled RNPs, Online Methods), which were then analyzed computationally. A typical data set from a cross-linked sample yields 5,000–10,000 MS/MS spectra.
MS data analysis
In previous studies with isolated proteins, cross-linked peptide–oligonucleotides have been shown to yield information-rich fragments of the cross-linked peptide moiety upon MS-based sequencing. These can be used to identify the cross-linked peptide15. While in principle the resulting spectra could be compared to theoretical spectra in a conventional database search approach (e.g., OMSSA14, MASCOT16), in the case of peptide–oligonucleotide cross-links such a search is complicated since database search engines require defined modification masses. The cross-linked RNA moiety is not known and can encompass different combinations of nucleotides including their derivatives (e.g., loss of water, terminal phosphate groups). Moreover, despite the biochemical purification procedures, the overall number of MS/MS spectra is quite large compared to the number of spectra containing cross-link information.
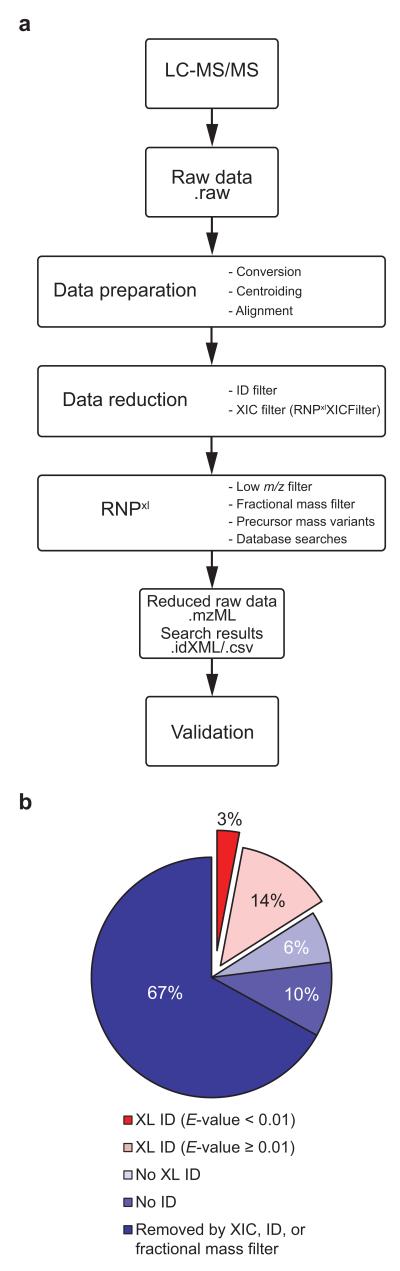
To evaluate larger MS datasets we designed a computational strategy to reduce data to the relevant spectra, search these spectra against entire proteomes, and annotate spectra of cross-linked peptide–RNA oligonucleotide conjugates (Fig. 2a; Online Methods, tutorial in Supplementary Note). Briefly, MS data are prepared for analysis (conversion to mzML format, centroiding, and alignment of chromatographic retention times of data from UV- and non-irradiated samples; Supplementary Fig. 1). Application of the different filter algorithms reduced the originally acquired mass spectra successively (Supplementary Fig. 2). These filters are: i) search against a target-decoy database to remove spectra corresponding to noncross-linked peptides with a false discovery rate of peptide-to-spectrum matches (PSM FDR) of 1 %; ii) an extracted ion chromatogram (XIC)-based filter to remove MS/MS spectra of precursors present in the UV- and non-irradiated control in a certain retention time window, iii) a fractional mass filter15, 17 eliminating pure RNA oligonucleotides. Finally, a precursor mass variant list is generated in which masses of all possible nucleotide combinations of putatively cross-linked oligonucleotides are subtracted from the experimental precursor mass. The resulting data are searched against the UniProt18 database containing the respective proteome (human or yeast; Supplementary Fig. 3) using OMSSA14.
Figure 2.
Data analysis workflow and achieved data reduction in representative datasets. (a) Outline of data analysis pipeline. Raw data from LC-ESI-MS/MS experiments is submitted to the data analysis pipeline. In preparation for subsequent steps, raw data is converted into an open mass spectrometry format (.mzML) and centroided; retention time alignment between the UV sample and the nonirradiated control is performed. Next, the overall amount of data is reduced by removing MS/MS spectra of confidently identified noncross-linked peptides (ID filter) and species appearing in both UV-irradiated sample and control with comparable intensities (XIC filter with RNPxlXIC). Lastly, the RNPxl tool removes MS/MS spectra with small precursors masses (low m/z filter) and residual short oligonucleotides (fractional mass filter). For the key steps in data analysis, RNPxl creates precursor mass variants, submits data into the search engine, and summarizes the search results. (b) Results of data analysis procedure for a single dataset of yeast RNA-binding proteins. XIC, ID, and fractional mass filter excluded approximately two thirds (67%) of the overall 9,728 fragment spectra. Additionally, 16% of the spectra could be disregarded as these did not yield any database search result for a cross-linked heteroconjugate. Of the remaining 17% potential cross link candidates, 14% had a low score (E-value ≥ 0.01) yielding a final list of 317 (3%) potential cross link candidates for manual validation.
In a single representative dataset of UV cross-linked RNPs from yeast (Fig. 2b), of an initial 9,728 spectra, two thirds were removed by the ID, XIC, and fractional mass filters. The ID filter removed 2,823 spectra (29%), the XIC filter 3,313 spectra (34%), and the fractional mass filter 335 spectra (3%) when applied in that order. Another 10% of the spectra remained completely unassigned while 6% were excluded because they matched noncross-linked peptides with medium or low confidence (FDR above 1%). 17% of the submitted MS/MS spectra represent potential peptide–RNA cross-links and among these, 3% (317 spectra) exhibit an E-value better than 0.01 and were kept as potential peptide-RNA cross-links (Supplementary Data).
Protein–RNA cross-links
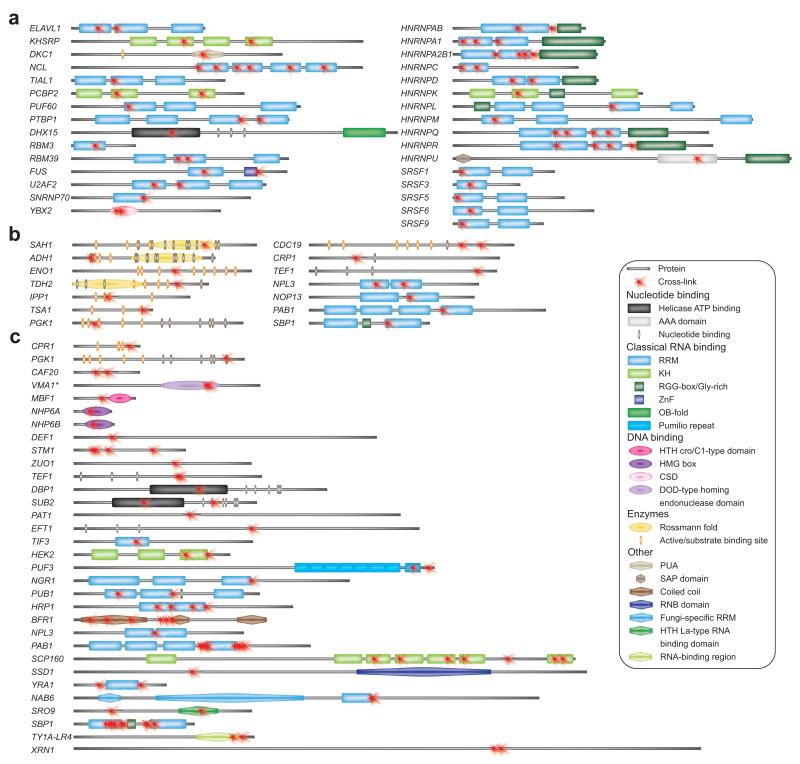
From the human protein–RNA complexes we identified 189 cross-links matching 60 tryptic peptides. In half of these peptides, the cross-linked amino acid was identified. The cross-linked peptides were mapped to 35 different proteins. A large majority of the cross-linked peptides (54) lie in known RNA-binding motifs such as RNA recognition motifs (RRMs) and K-homology (KH) domains (Fig. 3a, Supplementary Table 1, Supplementary Figs. 4-5, Supplementary Data spectra H01–H60).
Figure 3.
Distribution of cross-linking sites in identified RNA-binding proteins with annotated domain structure. The panels show (a) human proteins, (b) yeast proteins isolated with TAP tagged Cbp20, (c) yeast proteins cross-linked to 4-thio-U and isolated with oligo d(T). Ribosomal proteins are not included. Proteins are represented by their corresponding gene names to avoid ambiguity. (Putative) RNA-/DNA-binding domains, nucleotide binding sites, and active/substrate binding sites are given if annotated in protein databases. All appearing domains and sites are listed on the lower right with their assigned symbols. Information is derived from the UniProt database and supplemented with details from NCBI, Pfam, Superfamily, and CATH (Supplementary Tables 1–3). Yeast gene VMA1 encodes for V-type proton ATPase catalytic subunit A and Endonuclease PI-Scel, only the latter is shown since it contains the cross-linked regions.
In the UV-irradiated yeast RNPs (isolated by affinity purification of Cbp20), we identified 184 peptide–RNA oligonucleotide cross-links, which mapped to 64 tryptic peptides (Supplementary Table 2, Supplementary Fig. 6, Supplementary Data spectra Y01–Y64). In 39 of these, the cross-linked amino acid could be pinpointed. The cross-links involved 49 different proteins, the majority derived from the ribosome (137 cross-links in 34 ribosomal proteins). Non-ribosomal proteins included nucleolar proteins 3 and 13, polyadenylate-binding protein Pab1, and single-stranded nucleic acid-binding protein with cross-links located in the RRM (Fig. 3b). Of particular interest are three enzymes (adenosylhomocysteinase, alcohol dehydrogenase, and glyceraldehyde-3-phosphate dehydrogenase) containing a Rossmann fold19. These and five additional proteins (enolase, inorganic pyrophosphatase, peroxiredoxin Tsa1, phosphoglycerate kinase, and pyruvate kinase) are not classical RBPs (Fig. 3b).
The broadest spectrum of proteins cross-linked to RNA was obtained after isolation of polyadenylated mRNA from UV-irradiated yeast cells that had incorporated 4-thio-uridine into mRNA (Supplementary Table 3, Supplementary Figs. 7–8, Supplementary Data spectra Y65–Y104). Here, 376 cross-links were identified, which correspond to 133 unique cross-linking sites or regions in 57 different proteins. 161 cross-links were found in ribosomal and 215 in non-ribosomal proteins including metabolic enzymes (peptidyl-prolyl cis-trans isomerase and phosphoglycerate kinase), DNA binding proteins (e.g., non-histone chromosomal protein 6A and B, endonuclease PI-Scel, multiprotein-bridging factor 1), a nucleotide binding protein (elongation factor 1-alpha), and RBPs (Fig. 3c). Among the latter are RNA helicases (Dbp1, Sub2), proteins containing RRMs (e.g., Pub1), KH motifs (heterogeneous nuclear RNP K-like protein 2, Scp160), and Pumillio repeats (Puf3p), as well as proteins containing motifs that are not frequently associated with RNA binding, i.e., the coiled-coil domain (Bfr1), HTA-La-type RNA binding domain (Sro9), and RBPs with not yet well-defined motifs (Gag-p49 derived from TY1A-LR4).
Our data also reveals structural details of the protein–RNA interaction. Analyzing four cross-linked peptides, we could confirm that the cross-link locations agree with the known crystal structures of protein–RNA complexes (Fig. 4). U2AF65 is an RNA-binding protein that contains three RRMs and interacts with the polypyrimidine tract on pre-mRNAs20. The 3D structure of the first two RRMs in complex with poly(U) RNA has been described (PDB ID 2YH120). The MS/MS spectra narrowed down the cross-linking sites to Leu261 or Phe262 and Phe199, respectively (Fig. 4a). Phe262 corresponds to the conserved aromatic residue in the RNP2 consensus sequence and Phe199 to the second conserved aromatic residue in the RNP1 consensus sequence (Supplementary Fig. 4). The location of the cross-linked peptides agrees exactly with the available structure of RRM1 and RRM2 of U2AF65 together with an eight-mer poly(U) (Fig. 4b). Yeast ribosomal protein S1 was found cross-linked to uridine through a tryptophan residue (Fig. 4c). Within the crystal structure of the yeast ribosome21 (PDB IDs 3U5F, 3U5G, 3U5H, 3U5I), this tryptophan residue (Trp117) stacks perfectly with U1799 of the 18S rRNA (Fig. 4d). In mRNA-binding protein Puf3, residue Tyr825 was found cross-linked (Fig. 4e). In the structure of the protein’s RNA binding domain to one of its recognition sequences (PDB ID 3K4922), this residue stacks between U3 and G4 of the co-crystallized oligonucleotide (Fig. 4f).
Figure 4.
MS/MS fragment spectra of cross-linked heteroconjugates and structural interpretation. (a, c, e) Manually validated MS/MS spectra. Regular peptide fragments are shown in black, RNA fragments in blue, and specific fragment ions derrived from peptide–RNA cross-links in red (U0-p: U – H3PO ; U0 4 : U –H2O). (b, d, f) Structural interpretation of identified cross-links. Proteins are shown in gray, RNA (nucleotides) in blue, cross-linked peptides in orange, and amino acids in red. (a) Left: Fragment spectrum of peptide LFIGGLPNYLNDDQVK from human splicing factor U2AF 65kDa subunit (Leu261–Lys276) cross-linked to U via Leu261 or Phe262. Right: Fragment spectrum of U2AF peptide NFAFLEFR (N196-R203) cross-linked to a UU dinucleotide through F199. (b) Phe262 and Phe199 are found in close spatial proximity to uracil in the structure of RRM1 and RRM2 of U2AF with a poly-(U) oligonucleotide20. (c) MS/MS fragment spectrum of 40S ribosomal protein S1 peptide KWQTLIENANVTVK cross-linked to [U –H2O] via Trp117. (d) In the structure of the yeast ribosome21, S1 Trp117 is in close spatial proximity to U1799 of the 18S ribosomal RNA. (e) Fragment spectrum of peptide DQFANYVIQK from mRNA-binding protein Puf3 (Asp820-Lys829) cross-linked to [(4SU)U – H2S]. Shift of the y-series by a fragment of the cross-linked 4SU base identifies Tyr825 as the cross-linked amino acid. (f) Structure of the RNA-binding domain of Puf3 to a recognition sequence22.Tyr825 stacks between U3 and G4 of the co-crystallized oligonucleotide.
Discussion
We combined in vitro and in vivo UV cross-linking with MS and database search to identify RNA– protein contact sites at the peptide and amino acid as well as at the nucleotide level in the context – and against the background – of whole-cell extracts. Earlier studies established the specificity of combining UV cross-linking and MS in structural investigations of moderately complex protein–RNA complexes15, 23-27 and also through mutagenesis studies of identified cross-linked amino acids in single RNPs28, 29. More complex samples pose considerably greater challenges to MS data analysis.
Peptides cross-linked to RNA oligonucleotides cannot be comprehensively identified by conventional database search engines, as these would limit the identification of cross-linked peptides to a very restricted set of possible modifications (Supplementary Fig. 9). The purification procedure for peptide–RNA oligonucleotide cross-links applied here used various endonucleases to hydrolyze intact RNA. Consequently, neither the length distribution, nor the composition, nor possible modifications of the cross-linked RNA oligonucleotides could be predicted. The number of potential RNA adducts was too large to allow search strategies analogous to the identification of post-translational modifications with conventional database searches. Therefore, a comprehensive search with a dedicated search tool was called for. A conventional, limited database search for cross-linked species might nonetheless still be used as an initial survey in order to roughly anticipate the number of cross-links. Even then, a conventional database search (e.g., taking N-terminal uridinylation into account) only led to limited results, i.e., less than two thirds compared to the precursor variants approach (Supplementary Table 4).
We have therefore implemented a tool for a precursor variant search together with suitable filtering steps. It is available as part of the open-source package OpenMS. Integrating the method into a sustainable long-term software project will ensure the availability of the method, enable future algorithmic improvements, and enable vendor-dependent data processing. The tool allows the generation of freely defined precursor variants containing modified and non-modified nucleotides. In practice, the generation of precursor variants up to tetranucleotides will be sufficient for most analyses. Peptides cross-linked to longer RNA oligonucleotides are difficult to identify, as MS/MS spectra are dominated by RNA fragment ions, so that no (or very poor) sequence information on the cross-linked peptide can be obtained30. Nonetheless, cross-links to tri- or tetranucleotides can allow location of the cross-linking site on the RNA if the oligonucleotide composition identified matches a unique RNA sequence25.
The resulting datasets allowed the identification of the actual cross-link sites, i.e., the amino acid and the nucleotide. Detailed annotation of the spectra yielded several insights. (i) Nearly all amino-acid residues except Asp, Asn, Glu, and Gln were found to be cross-linked to nucleotides (Supplementary Tables 1–3). (ii) The cross-linked nucleotide was usually U or 4-thio-U. (iii) Cross-linking to 4-thio-U systematically showed a loss of H2S in the precursor mass, and fragment ions from the peptide moiety often showed an adduct mass of 94 Da, corresponding to a uracil derivative that has lost H2S15. (iv) Several cross-links, particularly from yeast ribosomal proteins co-purified with (pre-)mRNA by TAP-tagged Cbp20 protein, carried an additional mass of 151.9938 Da. Cross-links with an additional 152 Da were first reported in a cross-linking study of snurportin 1 to U1 snRNA26 and involve almost exclusively cysteine-containing peptides. We note that this adduct mass is associated with the presence of dithiothreitol (DTT) in the sample. DTT may promote formation of cross-links between cysteine residues and RNA bases under UV irradiation at 254 nm (personal communication; U. Zaman, MPI for Biophysical Chemistry).
Although we used different purification strategies for RNP isolation, the comparison of the cross-linked peptides derived from yeast RNP complexes irradiated at 254 nm (non-substituted RNA) and 365 nm (4-thio-U substituted RNA) demonstrates that the two cross-linking methods were complementary for numerous proteins, as has been observed before7. In Pgk1, elongation factor 1-alpha, nucleolar protein 3 and within several ribosomal proteins, different regions of the proteins were identified as being cross-linked to RNA, whereas in Pab1, Sbp1, and other ribosomal proteins the same amino acids or peptides have been identified as cross-linking site to RNA (Supplementary Table 5).
Our results highlight the utility of a UV-based irradiation approach to identify direct contact sites between proteins and RNA, and substantially extend the scope of previous studies where entire proteins – but not the specific cross-linking site – were identified by MS7-10. The majority of cross-linking sites identified reside in known RNA-binding domains, such as RRMs and KH motifs (Fig. 3, Supplementary Tables 1–3). Among the 20 hnRNP proteins in the human database, we found 13 (more than 60%) to be cross-linked. In the 44 canonical RNA-binding motifs (RRM and KH motifs) of these proteins we identified 25 distinct cross-linked peptides or amino acids; this accounts for more than 55% of the canonical RNA-binding motifs in these proteins. The cross-link sites also included less frequently described RNA-binding motifs such as Pumillo repeats, WD domains as well as other sequence motifs such as the AAA domain, RanBP-type zinc finger, PUA domain, coiled-coil domains, HMG box, and DOD-type homing endonuclease domain. Cross-linking sites within several metabolic enzymes underscore the enzymes’ ability to interact with RNA7. The RNA cross-links occured within the ATP- or substrate-binding sites, the Rossmann fold, and in other regions of the protein.
The comparison of the cross-linking sites with available 3D structures demonstrates the structural specificity of the cross-linking approach while at the same time proving useful for predicting (novel) RNA-binding sites. In the case of DNA-binding proteins the 3D structures of NHP6A in complex with dsDNA31 and of domain I of the S. cerevisiae homing endonuclease PI-Scel32 both show amino acids that were found to be cross-linked to RNA (Arg40 and Tyr328) while being located in the DNA-binding domain, thus suggesting a dual function of these proteins. Other examples for this are ribosomal and ribosome-associated proteins. In the 3D structure of the 80S yeast ribosome21 most ribosomal proteins – with their cross-linking sites – are found in close proximity to the 28S or 18S rRNA. The cross-linking sites identified in RACK1, and in the ribosome-associated proteins Stm121 and Zuo133 are not proximal to rRNA, suggesting alternative conformations of translating ribosomes on mRNA or involvement of these proteins in direct mRNA binding. In total, we have pinpointed 39 cross-linking sites in 26 different proteins (about one-third of yeast’s 79 ribosomal proteins).
In summary, the method reported here allows the systematic identification of cross-linked peptides and amino-acid residues in RBPs, irrespective of whether binding occurs in a classical RNA-binding domain or not. Although the quality of computational predictions for RNA–protein interactions has greatly improved, there are significant differences to our experimental approach. Computationally predicted RNA binding sites describe potential interactions, which are not necessarily realized in a specific protein–RNA complex. Since our approach is not biased towards known RNA-binding domains, it provides a basis for improving computational predictions of RNA-binding motifs, as in metabolic enzymes and transcription factors, in proteins that contain more than one RNA interaction site34, or in those proteins that form a composite RNA interaction site through protein–protein interactions35. It offers the counterpart to the now well-established RNA-sequencing approaches that identify cross-linked RNAs and nucleotides (e.g., PAR-CLIP11). In contrast to deep sequencing approaches, cross-linked peptides cannot be amplified, and the identification of corresponding cross-linking sites within proteins must rely on adequate enrichment procedures and the sensitivity of MS instruments. The number of cross-links identified and the nature of the cross-linked amino acids (e.g., proline and glycine residues) that had not been previously identified reflect the recent improvement in this area. The steady improvement of MS instrumentation can be expected to enable a more comprehensive identification of cross-linking sites by the method described here.
Online Methods
Assembly and isolation of human protein–RNA complexes
In vitro transcribed MS2-tagged PM5 pre-mRNA37 was prebound with MS2-MBP fusion protein38and incubated for 30 min at 0°C with 10 ml HeLa nuclear extract39. Protein–RNA complexes were isolated by MS2 affinity selection with amylose beads (New England Biolabs) as previously reported38.
Preparation of yeast (pre-) mRNPs by TAP tag purification
Yeast strain
Amplification of TAP tag40 construct (CBP2 – PreScission protease cleavage site – 2 ProteinA) from pBS1539-Psc (URA3) plasmid was carried out under Phusion polymerase, with forward primer 5′-TCAGACCAGGTTTCGATGAAGAAAGAGAAGATGATAACTACGTACCTCAGTCCATGGAAAAGAGAAGAT-3′ and reverse primer 5′-TATATATATATCTGTGTGTAGAATCTTTCTCAGATATAAATTGATTGATTTACGACTCACTATAGGGCGA-3′. The PCR construct was transformed into yeast strain BJ2168. The positive yeast clone was confirmed by DNA sequencing and Western blotting.
Yeast cell extract
Yeast cells were grown in YPD (1% yeast extract, 2% peptone, 2% glucose) substituted with 50 mg/l ampicillin and 10 mg/l tetracycline, pelleted at 4 500 rpm for 10 min, suspended in 0.7 volumes (V/w) AGK buffer (10 mM Hepes pH 7.5, 1.5 mM MgCl2, 200 mM KCl, 10% glycerin) and flash frozen in liquid nitrogen. Cell beads were ground (ZM 200, Retsch). Cell debris were pelleted at 17 000 rpm for 30 min and optionally polysomes were separated in a second ultra-centrifugation at 37 000 rpm for 60 min.
TAP tag purification
Typically, 10 ml yeast cell extract (30-35 mg/ml protein) was incubated with 600 μl IgG beads suspension (IgG SepharoseTM 6 Fast Flow, GE Healthcare) for 2 h at 4°C. The supernatant was eluted by gravity and the beads were washed with 20 ml CBB buffer (calmodulin binding buffer; 25 mM Tris pH 7.9, 150 mM NaCl, 1 mM MgOAc2, 1 mM imidazole, 2 mM CaCl2, 2 mM DTT). The complex was released by incubation with 12 μl PreScission (10 mg/ml) in 2 ml CBB and 1 μl rRNasin (Promega) overnight.
The second purification step was carried out with 400 μl calmodulin beads suspension (Calmodulin Affinity Resin, Agilent). The sample was incubated with beads for 1 h at 4°C, washed with 20 ml CBB, and eluted twice by incubation with 1 ml CEB (calmodulin elution buffer; 25 mM Tris pH 7.9, 150 mM NaCl, 1 mM MgOAc2, 1 mM imidazole, 25 mM EGTA, 0.02% NP40, 2 mM DTT) for 5 min.
UV cross-linking (254 nm)
The cross-linking apparatus built in-house was equipped with four 8 W lamps (254 nm; G8T5, Sankyo Denki). Cross-linking was done on ice in custom made petri dishes in which either 10 or 1 ml sample solution had a depth of 1 mm. The petri dishes were placed on an ice-cold metal block at a distance of 1 cm under the lamps.
Cross-linking of yeast (pre-) mRNPs was typically carried out on the IgG eluate, in initial experiments also on cell extract and calmodulin eluate. For cross-linking of yeast cell extract, the extract was dialyzed against AGK without glycerin as glycerin is a radical scavenger. The sample was irradiated for 2 min and subsequently ethanol precipitated. Cross-links were typically isolated by size-exclusion and C18 chromatography (see below).
Human protein–RNA complexes were cross-linked after elution from amylose beads. Samples were UV irradiated for 10 min, followed by ethanol precipitation. Cross-links were isolated by size-exclusion and C18 chromatography followed by titanium dioxide solid phase extractions (see below).
Preparation of in vivo 4SU-labeled yeast mRNPs
Growth, in vivo labeling & cross-linking
Cells (strain BY4141) were grown in YPD (1% yeast extract, 2% peptone, 2% glucose) to OD600 0.5 and RNA was in vivo 4SU-labeled as described in 41. Cells were allowed to grow for another 3 hrs before cells were pelleted at 15,000 × g for 15 min. (m)RNPs were cross-linked by 365 nm UV light; cooled on ice using a XL-1500 Spectro Linker (Spectronics Corporation) as described41.
Purification of mRNPs
After pelleting cells at 2880 × g for 5 min, cells were resuspended in lysis buffer (20 mM Tris-HCl pH 7.5, 500 mM LiCl, 0.5% LiDS, 1 mM EDTA, 5 mM DTT, 1x Protease Inhibitor Cocktail EDTA-free, Roche). Cells were lysed using acid-washed glass beads in a FastPrep device (MPI) using 5 pulses at 6 m/s for 60 sec with 60 sec pausing in between. Lysates were then cleared by centrifugation at 9400 × g for 2.5 min in a table-top centrifuge. Purification of mRNPs was performed using magnetic oligo d(T) beads (NEB) as described in 7, 42. Finally, cross-links were isolated by C18 chromatography and titanium dioxide solid phase extraction (see below).
Enrichment and isolation of cross-links
To identify peptide-RNA oligonucleotide cross-links in MS analysis, the moiety of peptides and RNA oligonucleotides that are not-cross-linked has to be removed. This step is important, as residual noncross-linked peptides and RNA oligonucleotides will strongly interfere with the detection of peptide–RNA oligonucleotide cross-links in the mass spectrometer. Enrichment of cross-linked peptide RNA oligonucleotides and the removal of noncross-linked peptides using TiO2 has been established previously 15, 25. Size-exclusion chromatography (SEC) was performed for non-substituted yeast and human RNPs. Application of SEC or TiO2 solid phase extraction for purification of peptide– RNA oligonucleotides depends on the nature of the cross-linked RNPs and their RNA moieties. In principal, SEC and TiO2 enrichment are complementary as both techniques remove non-cross-linked peptides from the sample. SEC is generally beneficial for complexes isolated under native conditions, which might still contain “contaminating” noncross-linked proteins that do not interact with RNA directly. 4SU-substituted yeast RNPs were isolated under stringent conditions with oligo d(T) and contained almost exclusively cross-linked proteins; size-exclusion was omitted because the majority of proteins that are not cross-linked had been removed during this isolation. SEC is generally not applicable at all if peptides and RNA do not exhibit sufficient difference in size, e.g., for cross-linking experiments of proteins bound to short oligonucleotides (approximately 5–40 nucleotides). C18 reversed-phase (RP) chromatography is absolutely essential in order to remove the noncross-linked RNA oligonucleotides prior to MS analysis. Without this step, MS-based detection of peptide–RNA oligonucleotide heteroconjugates is severely hampered by residual noncross-linked RNA oligonucleotides.
For size-exclusion chromatography (SEC), the sample was digested with 1:50 (w/w) trypsin (sequencing grade modified trypsin, Promega) overnight in the presence of 0.1% SDS and 50 mM Tris pH 7.9. Protein amounts were determined by the Bradford assay. SEC was carried out on a SMART system (Pharmacia Biotech) equipped with a SuperdexTM 200 column (PC 3.2/30, 2.4 ml, Amersham Biosciences) at a flow rate of 40 μl/min with 20 mM Tris pH 7.5, 150 mM NaCl, 1.5 mM MgCl2 as running buffer. Fractions of 100 μl were collected. Fractions showing a strong absorbance at 254 nm over 280 nm (see Supplementary Fig. 10) were pooled and ethanol precipitated.
Protein and RNA digestion as well as C18 desalting and titanium dioxide enrichment was done according to established protocols15, 25.
For hydrolysis of proteins and RNA, precipitated samples from cross-linking or size-exclusion chromatography were dissolved in the presence of 50 μl 4 M urea, 50 mM Tris pH 7.9, 1.5 mM MgCl2 and diluted with 150 μl 50 mM Tris pH 7.9, 1.5 mM MgCl2 to a final concentration of 1 M urea, 50 mM Tris pH 7.9, 1.5 mM MgCl2. RNA was hydrolyzed with 1 μl benzonase (25 U, Novagen) for 30 min at 37°C and subsequently with 1 μl each of RNases A and T1 (both Ambion; RNase A 1 μg/μl, RNase T1 1 U/μl) for 60 min at 52°C. Protein digestion was carried out with 1:20 (w/w) trypsin (sequencing grade modified trypsin, Promega) at 37°C overnight. Protein amounts were determined by the Bradford assay. For samples completely proteolyzed prior to size exclusion chromatography, 1 μg trypsin was added instead for hydrolysis of nucleases and residual longer protein fragments.
Desalting was done immediately after digestion for all samples. 10 μl acetonitrile (ACN) and 2 μl 10% formic acid (FA) (v/v) were added to the sample (volume 200 μl) prior to loading on C18 columns (in-house; C18 AQ 120 Å 5 μm, Dr. Maisch GmbH). Samples were washed with 120 μl 0.1% FA (v/v) and eluted stepwise with 120 μl 50% ACN (v/v), 0.1% FA and 60 μl 80% ACN (v/v), 0.1% FA (v/v). The combined eluate was dried in a centrifugal evaporator. Samples were either enriched further with titanium dioxide or directly subjected to LC-MS/MS analysis.
For titanium dioxide enrichment, samples were dissolved in 60 μl buffer A (200 mg/ml dihydroxy benzoic acid, 80% ACN, 5% trifluoroacetic acid (TFA)) and loaded on TiO2 micro columns (in-house; titansphere, 5 μm, GL Science). The sample was washed with buffer A (180 μl) and extensively with buffer B (80% ACN, 5% TFA; 300 μl) to remove any residual DHB and finally eluted with 120 μl ammonia. The eluate was dried in a centrifugal evaporator.
Mass spectrometry
Pellets from C18 desalting or TiO2 enrichment were dissolved with 50% ACN, 0.1% FA and diluted to a final concentration of 10% ACN and 0.1% FA. Each sample was injected twice either on the same or different instruments.
Most LC-MS/MS analyses were carried out with an LTQ Orbitrap Velos (Thermo) directly coupled to a nanoflow-liquid chromatography system (Agilent 1100 series, Agilent). Samples were loaded on a C18 trapping column at a flow rate of 10 μl/min in 3% buffer B (buffer A: 0.1% FA; buffer B: 95% ACN, 0.1% FA) and washed for 5 min. A linear gradient of 3 to 36% buffer B (flow rate 300 nl/min) flushed the analytes onto the analytical column and separated them in an elution time of 37 min (60 min overall run time) or 97 min (120 min overall run time). Residual analytes were eluted by raising buffer B to 95% for 7.5 min. The columns were built in house (trapping column: inner diameter 150 μm, length 2 cm; analytical column: inner diameter 75 μm, length 15 cm; C18 material see above). The instrument was run in data-dependent acquisition mode. MS1 was acquired from 350 to 1600 m/z (or Thomson, Th) at a resolution setting of 30 000 FWHM (Full Width Half Maximum). The ten most intense precursors were chosen for fragmentation by higher-energy collision induced dissociation (HCD). For MS/MS, the following parameters were set: minimal signal required 5000, isolation width 2 Th, normalized collision energy 45, dynamic exclusion 20 s, resolution setting 7 500 FWHM.
Alternatively, LC-MS/MS experiments were conducted with a Q Exactive instrument (Thermo) coupled to an EASY-nLC II (Thermo). The sample was loaded on a C18 trapping column (inner diameter 100 μm, length 4 cm, C18 material see above) and washed with 25 μl buffer A. A linear gradient of 4 to 36% buffer B within 92 min at a flow rate of 250 nl/min separated the analytes on the C18 analytical column (inner diameter 50 μm, length 10 cm; C18 AQ 120 Å 3 μm, Dr. Maisch GmbH). A final elution step at 95% buffer B for 8 min removed any residual analytes. The instrument was set to a TOP12 method in data-dependent acquisition mode. MS1 was recorded from 350 to 1600 m/z at a resolution setting of 70 000 FWHM. MS/MS fragmentation was done on the 12 most intense precursors with HCD fragmentation. MS/MS parameter were chosen as follows: minimal signal required 10 000, isolation with 2 Th, normalized collision energy 30, dynamic exclusion 20 sec, resolution setting 17 500 FWHM.
Data analysis with RNPxl
The experimental workflow resulted in two raw data files per experiment (cross-linked and control, except for yeast 4SU-labeled RNPs), which were then subjected to the computational workflow (see below). For samples derived from yeast 4SU-labeled RNPs, no non-UV irradiated control was performed. In initial MS analyses we confirmed that noncross-linked proteins were completely removed through very stringent washing steps. Accordingly, the control MS data did not provide any additional benefit for the automated data analysis
MS raw data in Thermo’s .raw format was converted into the .mzML-format43 with msconvert of the ProteoWizard software package (http://proteowizard.sourceforge.net; 44).
All subsequent steps were performed with tools from the OpenMS software library (see detailed instructions as well as links to sample pipelines and a test dataset in Supplementary Tutorial).
Data in profile mode was centroided. The non-irradiated control was aligned relative to the UV irradiated sample based on high intense features to correct for retention time shifts45.
Next, a search was performed against a target-decoy version of the UniProt yeast database, also including contaminant sequences as distributed with the MaxQuant46 software package. Oxidation of methionine, carbamylation of lysines and N-termini, as well as phosphorylation of tyrosine, serine, and threonine were considered as variable modifications. All MS/MS spectra with a confident peptide-to-spectrum match (FDR < 1%) were filtered from the sample’s dataset.
Of all remaining MS/MS spectra, differential analysis was performed on precursor intensities calculated from extracted ion chromatograms of UV irradiated sample and non-irradiated control. If the same precursor was observed in the control at a comparable intensity (fold change less than 2), the corresponding MS/MS spectrum was removed from the UV sample data file.
The reduced sample data file was subjected to our novel data analysis tool, RNPxl. Prior to generation of precursor variants, spectra were filtered if their precursor corresponded to a small RNA oligonucleotide according to fractional mass (M < 1750 Da and fractional mass < .2) or if the precursor was too small for an identifiable cross-link (M < 600 Da).
Precursor variants for unlabeled RNA were generated with all calculated masses of RNAs meeting the following criteria: maximum length of four nucleotides, at least one U in sequence, RNA modifications: none, (-H2O), (-HPO3), (-H3PO4), (-H2O +152), (-HPO3 +152), (-H3PO4 +152). In addition, (+152) was considered as a modification without an additional nucleotide. This lead to a maximum number of 281 precursor variants per MS/MS spectrum.
For 4SU substituted RNA, the following parameters were chosen for precursor mass variant generation: maximum RNA length four nucleotides; at least one 4SU in sequence; RNA modifications: none, (-H2O), (-H2S), (-HPO3), (-H2S –HPO3), (-H3PO4). Alternatively, a mass of a post-translational modification was defined on all 20 amino acid, resembling 4SU cross-linked under loss of H2S with a stable adduct of 94.0167 Da (C4H2N2O) after fragmentation15: PTM mass 306.0253 (C9H11N2O8P), neutral loss 212.0086 Da (C5H9O7P). For the corresponding searches, RNA masses for precursor mass variant generation were calculated with the following criteria: maximum length three nucleotides, RNA modification (-H2O). The resulting precursor mass variants were searched with OMSSA in four separate searches allowing the described PTM on sets of five amino acids: K, F, H, R, and Y; S, G, P, W, and M; A, V, T, C, and L; or I, N, D, Q, and E. In these searches, only oxidation of methionine was considered as an additional variable modification.
Precursor mass variants with a mass-to-charge ratio below 250 m/z were disregarded for further processing. Parameters for OMSSA searches were chosen as follows: Precursor mass tolerance 10 ppm; fragment mass tolerance 0.01 Da; variable modifications (unless noted otherwise above): oxidation of methionines and carbamylation of N-termini and lysines. Typically, all results with an OMSSA score better than 1×10−5 (native RNA) or 1×10−8 (4SU-labeled RNA) were considered for manual validation.
In general, the computational workflow runs on standard personal computers and is integrated into the OpenMS environment12, 13 (see Supplementary Tutorial for details). The search results are reported in two formats: a tabular comma-separated values (CSV) file, which can be imported into spreadsheet applications (e.g., Microsoft Excel), and an idXML file, used to annotate the raw data with the search results in mzML format in TOPPView47 (a graphical MS data visualization tool that is part of OpenMS).
Validation of search results
First, correct assignment of monoisotopic peak and charge state was confirmed. For experiments with unlabeled RNA, extracted ion chromatograms of non-irradiated control and UV-irradiated sample were compared with Xcalibur. Cross-link candidates were rejected if the same precursor was observed in the control measurement with an extracted ion chromatogram area of more than half of the area in the UV-irradiated sample.
The assignment of peptide fragments was verified by annotating the raw data in mzML format with the search result output of the RNPxl tool in idXML format using TOPPView. Unassigned peaks, especially those of high intensity, were annotated manually, either as internal peptide fragments, RNA marker ions, or peptide sequence ions shifted by the mass of the cross-linked RNA or fragments thereof. In the majority of fragment spectra, y- and/or b-type fragment ion series were observed that unambiguously identified the cross-linked peptide sequence (see Supplementary Spectra). The fragment spectra of the putatively cross-linked species were omitted if i) RNA marker ions did not match to the computed RNA composition, i.e., an adenosine predicted but not visible as a strong marker ion in the lower m/z regime; ii) a large number of high intensity signals, especially in the higher m/z range was observed but could not be annotated, and iii) a peptide sequence tag was manually assigned that did not correspond to the computed candidate sequence and also could not be explained by an overlapping precursor and its corresponding fragment spectra. Special focus was on the following peptide and RNA ions: the a2/b2-ion pair usually well observable in HCD fragment spectra; high intense immonium ions; fragments resulting from cleavage N- and C-terminal to proline, where the N-terminal ion should have high intensity and the C-terminal ion should be barely or not observable, and RNA marker ions of the nucleic acid bases when more than one nucleotide was cross-linked. Localization of the cross-linking site on the peptide was done by comparing automatically annotated regular peptide fragments and manually annotated signals corresponding to peptide fragments shifted by the mass of the cross-linked RNA or its fragments. Only if the last regular peptide fragment and the first shifted fragment clearly pointed to a single amino acid or if an immonium ion bound to RNA was observed, localization to a single amino acid was possible. More details are given in the tutorial that is part of the Supplementary Material. For peptide fragmentation characteristics with HCD and a collection of further literature about collision induced fragmentation see 48.
Comparison to standard database search
Comparative database searches with OMSSA14 and Mascot16 were performed on the MS dataset of cross-linked human RNPs. A novel post-translational modification was defined on arginine and lysine with a mass of 324.0359 Da (C9H13N2O9P) and a neutral loss with the same mass. At least one arginine or lysine are present in tryptic peptides (with the possible exception of the protein C-terminus).
For OMSSA, data was processed as described above (conversion, peak picking). Next, the ID filter pipeline was modified by only taking the newly defined modification and oxidation of methionine into account as variable modifications. All other parameters remained unchanged. Consequently, results up to an FDR of 1% were reported and are listed in Supplementary Table 4.
For Mascot (version 2.3.02), data was converted into the text-based .msm file format with Raw2MSMS version 1.1049. Variable PTMs were the same as for OMSSA. Two missed cleavages were allowed for trypsin. Peptide tolerance was set to 10 ppm, MS/MS tolerance to 20 mmu, Instrument to ESI-TRAP. The same version of the human Uniprot database with the MaxQuant contaminant database was used as for OMSSA but without reverse sequences. A Mascot score of 29 (p < 0.05, determined by Mascot) was applied for the comparison, the complete results are also listed in Supplementary Table 4.
Data Deposition and Software Availability
The mass spectrometry data described in this work have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository36 with the dataset identifier PXD000513 and can be viewed with the PRIDEInspector Tool. The software described above is available in source code for all major platforms as well as precompiled binaries for Windows and OS X as part of the OpenMS software suite at www.OpenMS.de. The software is distributed as open-source software under a three-clause BSD license. Detailed documentation and a tutorial on the use of the software is part of the supplementary material.
Supplementary Material
Acknowledgements
We dedicate this article to the memory of our colleague and friend Andreas Bertsch.
The authors wish to acknowledge M. Raabe, U. Pleßmann and T. Conrad for excellent technical assistance, U. Zaman for providing unpublished data, R. Hofele for helpful discussions, and R. Lührmann for support and providing infrastructure (all are colleagues at the MPI for Biophysical Chemistry). We are also grateful to S. Klinge (ETH Zurich) for help with the yeast ribosome structure in PyMol. We would like to thank S. Aiche (FU Berlin) and C. Bielow (MDC Berlin) for helpful discussions on workflow implementation. This work was supported by a DFG grant to H.U. (SFB860, INST 186/859-1), a HEC/DAAD stipend to S.Q., funding from the European Research Council under the Union’s Seventh Framework Programme (FP7/2007-2013) / ERC-2011-ADG_20110310 to M.W.H., and a Marie Curie Fellowship (FP7/2007-2013) / MC-IEF-301031 to B.M.B..
Footnotes
Accession Codes The mass spectrometry data described in this work have been deposited in PRIDE36 with the dataset identifier PXD000513; the software is available as part of the OpenMS software suite at www.OpenMS.de (further details see Online Methods).
Competing Financial Interest The authors declare no competing financial interests.
References
- 1.Glisovic T, Bachorik JL, Yong J, Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008;582:1977–1986. doi: 10.1016/j.febslet.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matera AG, Terns RM, Terns MP. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- 3.Yates LA, Norbury CJ, Gilbert RJ. The long and short of microRNA. Cell. 2013;153:516–519. doi: 10.1016/j.cell.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 4.van der Feltz C, Anthony K, Brilot A, Pomeranz Krummel DA. Architecture of the spliceosome. Biochemistry. 2012;51:3321–3333. doi: 10.1021/bi201215r. [DOI] [PubMed] [Google Scholar]
- 5.Sabin LR, Delas MJ, Hannon GJ. Dogma derailed: the many influences of RNA on the genome. Mol Cell. 2013;49:783–794. doi: 10.1016/j.molcel.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20:300–307. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- 7.Castello A, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 8.Baltz AG, et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol Cell. 2012;46:674–690. doi: 10.1016/j.molcel.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 9.Mitchell SF, Jain S, She M, Parker R. Global analysis of yeast mRNPs. Nat Struct Mol Biol. 2013;20:127–133. doi: 10.1038/nsmb.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klass DM, et al. Quantitative proteomic analysis reveals concurrent RNA-protein interactions and identifies new RNA-binding proteins in Saccharomyces cerevisiae. Genome Res. 2013;23:1028–1038. doi: 10.1101/gr.153031.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hafner M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kohlbacher O, et al. TOPP--the OpenMS proteomics pipeline. Bioinformatics. 2007;23:e191–197. doi: 10.1093/bioinformatics/btl299. [DOI] [PubMed] [Google Scholar]
- 13.Sturm M, et al. OpenMS - an open-source software framework for mass spectrometry. BMC Bioinformatics. 2008;9:163. doi: 10.1186/1471-2105-9-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geer LY, et al. Open mass spectrometry search algorithm. J Proteome Res. 2004;3:958–964. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- 15.Kramer K, et al. Mass-spectrometric analysis of proteins cross-linked to 4-thio-uracil- and 5-bromo-uracil-substituted RNA. Int J Mass Spectrom. 2011;304:184–194. [Google Scholar]
- 16.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 17.Pourshahian S, Limbach PA. Application of fractional mass for the identification of peptide-oligonucleotide cross-links by mass spectrometry. J Mass Spectrom. 2008;43:1081–1088. doi: 10.1002/jms.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reorganizing the protein space at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2012;40:D71–75. doi: 10.1093/nar/gkr981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hentze MW. Enzymes as RNA-binding proteins: a role for (di)nucleotide-binding domains? Trends in biochemical sciences. 1994;19:101–103. doi: 10.1016/0968-0004(94)90198-8. [DOI] [PubMed] [Google Scholar]
- 20.Mackereth CD, et al. Multi-domain conformational selection underlies pre-mRNA splicing regulation by U2AF. Nature. 2011;475:408–411. doi: 10.1038/nature10171. [DOI] [PubMed] [Google Scholar]
- 21.Ben-Shem A, et al. The structure of the eukaryotic ribosome at 3. Science. 2011;334:1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 22.Zhu D, Stumpf CR, Krahn JM, Wickens M, Hall TM. A 5′ cytosine binding pocket in Puf3p specifies regulation of mitochondrial mRNAs. Proc Natl Acad Sci U S A. 2009;106:20192–20197. doi: 10.1073/pnas.0812079106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Urlaub H, Thiede B, Muller EC, Brimacombe R, Wittmann-Liebold B. Identification and sequence analysis of contact sites between ribosomal proteins and rRNA in Escherichia coli 30 S subunits by a new approach using matrix-assisted laser desorption/ionization-mass spectrometry combined with N-terminal microsequencing. J Biol Chem. 1997;272:14547–14555. doi: 10.1074/jbc.272.23.14547. [DOI] [PubMed] [Google Scholar]
- 24.Kuhn-Holsken E, Dybkov O, Sander B, Luhrmann R, Urlaub H. Improved identification of enriched peptide RNA cross-links from ribonucleoprotein particles (RNPs) by mass spectrometry. Nucleic Acids Res. 2007;35:e95. doi: 10.1093/nar/gkm540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo X, et al. Structural and Functional Analysis of the E. Mol Cell. 2008;32:791–802. doi: 10.1016/j.molcel.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuhn-Holsken E, et al. Mapping the binding site of snurportin 1 on native U1 snRNP by cross-linking and mass spectrometry. Nucleic Acids Res. 2010;38:5581–5593. doi: 10.1093/nar/gkq272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mozaffari-Jovin S, et al. The Prp8 RNase H-like domain inhibits Brr2-mediated U4/U6 snRNA unwinding by blocking Brr2 loading onto the U4 snRNA. Genes Dev. 2012;26:2422–2434. doi: 10.1101/gad.200949.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghalei H, Hsiao HH, Urlaub H, Wahl MC, Watkins NJ. A novel Nop5-sRNA interaction that is required for efficient archaeal box C/D sRNP formation. RNA. 2010;16:2341–2348. doi: 10.1261/rna.2380410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muller M, et al. A cytoplasmic complex mediates specific mRNA recognition and localization in yeast. PLoS Biol. 2011;9:e1000611. doi: 10.1371/journal.pbio.1000611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmidt C, Kramer K, Urlaub H. Investigation of protein-RNA interactions by mass spectrometry--Techniques and applications. J Proteomics. 2012;75:3478–3494. doi: 10.1016/j.jprot.2012.04.030. [DOI] [PubMed] [Google Scholar]
- 31.Allain FH, et al. Solution structure of the HMG protein NHP6A and its interaction with DNA reveals the structural determinants for non-sequence-specific binding. EMBO J. 1999;18:2563–2579. doi: 10.1093/emboj/18.9.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Werner E, Wende W, Pingoud A, Heinemann U. High resolution crystal structure of domain I of the Saccharomyces cerevisiae homing endonuclease PI-SceI. Nucleic Acids Res. 2002;30:3962–3971. doi: 10.1093/nar/gkf523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leidig C, et al. Structural characterization of a eukaryotic chaperone--the ribosome-associated complex. Nat Struct Mol Biol. 2013;20:23–28. doi: 10.1038/nsmb.2447. [DOI] [PubMed] [Google Scholar]
- 34.Schmitzova J, et al. Crystal structure of Cwc2 reveals a novel architecture of a multipartite RNA-binding protein. EMBO J. 2012;31:2222–2234. doi: 10.1038/emboj.2012.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Urlaub H, Raker VA, Kostka S, Luhrmann R. Sm protein-Sm site RNA interactions within the inner ring of the spliceosomal snRNP core structure. EMBO J. 2001;20:187–196. doi: 10.1093/emboj/20.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vizcaino JA, et al. The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 2013;41:D1063–1069. doi: 10.1093/nar/gks1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
References (Methods-only)
- 37.Bessonov S, Anokhina M, Will C, Urlaub H, Lührmann R. Isolation of an active step I spliceosome and composition of its RNP core. Nature. 2008;452:846–850. doi: 10.1038/nature06842. [DOI] [PubMed] [Google Scholar]
- 38.Deckert J, et al. Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Mol Cell Biol. 2006;26:5528–5543. doi: 10.1128/MCB.00582-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rigaut G, et al. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 41.Creamer TJ, et al. Transcriptome-wide binding sites for components of the Saccharomyces cerevisiae non-poly(A) termination pathway: Nrd1, Nab3, and Sen1. PLoS genetics. 2011;7:e1002329. doi: 10.1371/journal.pgen.1002329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Castello A, et al. System-wide identification of RNA-binding proteins by interactome capture. Nature protocols. 2013;8:491–500. doi: 10.1038/nprot.2013.020. [DOI] [PubMed] [Google Scholar]
- 43.Martens L, et al. mzML--a community standard for mass spectrometry data. Mol Cell Proteomics. 2011;10:R110 000133. doi: 10.1074/mcp.R110.000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chambers MC, et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat Biotechnol. 2012;30:918–920. doi: 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lange E, et al. A geometric approach for the alignment of liquid chromatography-mass spectrometry data. Bioinformatics. 2007;23:i273–281. doi: 10.1093/bioinformatics/btm209. [DOI] [PubMed] [Google Scholar]
- 46.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 47.Sturm M, Kohlbacher O. TOPPView: an open-source viewer for mass spectrometry data. J Proteome Res. 2009;8:3760–3763. doi: 10.1021/pr900171m. [DOI] [PubMed] [Google Scholar]
- 48.Michalski A, Neuhauser N, Cox J, Mann M. A systematic investigation into the nature of tryptic HCD spectra. J Proteome Res. 2012;11:5479–5491. doi: 10.1021/pr3007045. [DOI] [PubMed] [Google Scholar]
- 49.Olsen JV, et al. Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol Cell Proteomics. 2005;4:2010–2021. doi: 10.1074/mcp.T500030-MCP200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.