Abstract
Cancer cells acquire distinct metabolic adaptations to survive stress associated with tumour growth and to satisfy the anabolic demands of proliferation. The tumour suppressor protein p53 influences a range of cellular metabolic processes, including glycolysis1,2 oxidative phosphorylation3 (OXPHOS), glutaminolysis4,5 and anti-oxidant response6. In contrast to its role in promoting apoptosis during DNA damaging stress, p53 can promote cell survival during metabolic stress7, a function that may contribute not only to tumour suppression but also to non-cancer associated functions of p538. Here we show that cancer cells rapidly utilise exogenous serine and that serine deprivation triggered activation of the serine synthesis pathway (SSP) and rapidly suppressed aerobic glycolysis, resulting in increased flux to the TCA cycle. Transient p53-p21 activation and cell cycle arrest promoted cell survival efficiently channelling depleted serine stores to glutathione synthesis, preserving cellular anti-oxidant capacity. Cells lacking p53 failed to complete the response to serine depletion, resulting in oxidative stress, reduced viability and severely impaired proliferation. The role of p53 in supporting cancer cell proliferation under serine starvation was translated to an in vivo model, suggesting that serine depletion has a potential role in the treatment of p53-deficient tumours.
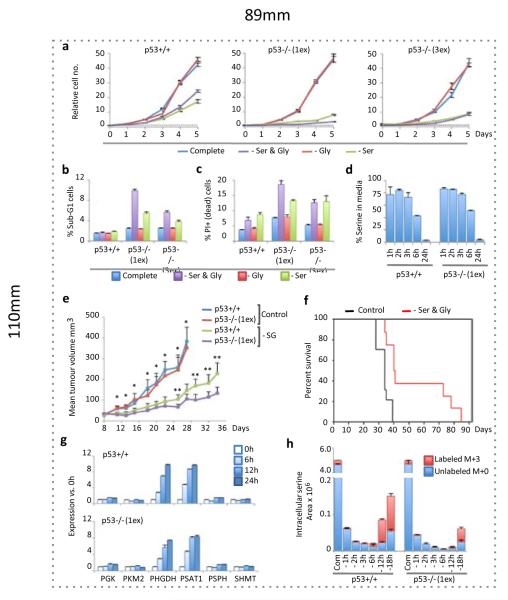
As p53 contributes to the survival of cells deprived of glucose7, we investigated whether removal of other nutrients found in normal media induced a differential response in p53+/+ and p53−/− HCT116 cells. While removal of the non-essential amino acids serine and glycine impaired proliferation of p53+/+ cells, p53−/− cells showed a more dramatic loss of proliferation (Fig. 1a) and substantial loss of viability (Fig. 1b&c). The contribution of p53 to growth and survival during serine and glycine depletion was also seen in RKO cells (Supp. Fig. 2a-c) and primary MEFs (Supp. Fig. 2d). By removing serine or glycine individually, we established that serine depletion was the major contributor to the starvation phenotype (Fig. 1a-c), as removal of glycine alone had no detrimental effect. While serine and glycine may be inter-converted by SHMT, serine to glycine conversion supports proliferation via methyl-tetrahydrofolate (THF) production (Supp. Fig. 1). Whereas, the reverse reaction (glycine to serine) depletes methyl-THF, which is presumably why excess glycine has been shown to inhibit proliferation9,10. As expected, removal of lysine (an essential amino acid) did not cause a differential response, being equally incompatible with proliferation in p53+/+ and p53−/− cells (Supp. Fig. 2e).
Figure 1. p53 promotes cell survival and proliferation during serine starvation in vitro and in vivo.
a, HCT116 cells were grown in complete media (containing serine and glycine) or equivalent media lacking these amino-acids (averages of triplicate wells). b, Viability of HCT116 cells was assessed by analysing sub-G1 DNA content (n=3) and c, PI exclusion (n=3). d, LC-MS was used to determine the relative consumption of serine by HCT116 cells fed complete media (averages of triplicate wells vs. fresh media). e, Nude mice were subcutaneously injected with HCT116 cells (p53+/+ right flank, p53−/− left flank); and fed diet with or without serine and glycine. Tumour volume is plotted until the first animal in each group reached the experimental end-point (*p<0.05 control diet group vs. –Ser & Gly group; **p<0.05 for p53+/+ vs. p53−/− within –Ser & Gly group). f, Kaplan Meier plot of survival until experimental end-point for diet groups (mean survival; Control = 33.3 days (n=10), -Ser & Gly = 53.2 days (n=8), Log rank p = 0.001, Wilcoxon p = 0.003). g, Expression of glycolytic and SSP genes (averages of triplicate qPCR). h, Intracellular serine levels in HCT116 cells fed serine and glycine deficient (−) media containing U-13C-glucose were measured by LC-MS (averages of triplicate wells). All error bars are SEM.
Analysis of cell culture media by liquid chromatography – mass spectrometry (LC-MS) revealed rapid serine consumption by p53+/+ and p53−/− cells (Fig. 1d) while glycine uptake was low (Supp. Fig. 2f). A recent screen of NCI-60 cancer cell lines shows that elevated SHMT expression and glycine uptake are correlated with rapid proliferation11. Notably, all 60 lines consumed more serine than glycine, including seven lines with the shortest doubling times (<22h), which on average consumed 7.7-times more serine than glycine11. Furthermore, cells with the shortest doubling times and highest glycine uptake also consumed most of the available serine11, raising the possibility that these highly proliferative cells switch to glycine consumption because they have exhausted the available serine. Overall this demonstrates that cancer cells avidly consume serine, which may be converted through high SHMT expression, into methyl-THF and glycine.
Unlike essential amino acids, the chronic depletion of non-essential amino acids can be tolerated in vivo. Mice tolerated diets lacking serine and glycine well (Supp. Fig. 3a), and LC-MS confirmed a significant drop in serum levels of serine and glycine, but not other amino acids (Supp. Fig. 3b). HCT116 cells rapidly formed tumours in animals fed control diet, without a significant difference in tumour volume between p53+/+ and p53−/− tumours (Fig. 1e). However, animals fed matched diet lacking serine and glycine displayed significant reduction in the volume of tumours of both genotypes, and survived significantly longer before tumour size or ulceration endpoints (Fig. 1e&f). As with our in vitro studies, serine and glycine starvation had a more dramatic effect on p53−/− xenografts, which had significantly reduced volume compared to p53+/+ tumours in serine and glycine deprived animals (Fig. 1e).
Mammalian cells synthesise serine de novo by channelling the glycolytic intermediate 3-phosphoglycerate into the ‘phosphorylated pathway’ of synthesis12, flux through which is controlled primarily by the demand fro serine13 (Supp. Fig. 1). The SSP supports anabolism by providing precursors for biosynthesis of proteins, nucleotides, creatine, porphyrins, phospholipids and glutathione, and SSP up-regulation occurs in some breast cancers14,15,16. A recent study demonstrated that serine starvation activates the SSP17; we found that serine starvation induced strong p53-independent up-regulation of PHGDH and PSAT1, with a modest increase in PSPH (Fig. 1g, Supp. Fig. 4a&b). The failure of p53−/− cells to proliferate during serine starvation could not therefore be attributed to a deficiency in SSP enzyme expression. p53 has been shown to down-regulate PGAM18 – potentially allowing 3-phosphoglycerate to be channelled to the SSP. However, PGAM expression did not vary greatly during serine starvation (Supp. Fig. 4a&b). Consistent with their ability to activate the SSP, both p53+/+ and p53−/− cells achieved de novo serine synthesis, detected using U-13-C-glucose labeling (Fig. 1h). However, p53−/− cells had lower serine levels, suggesting some defect in the ability of these cells to adapt to de novo serine synthesis. We therefore sought to explore the mechanisms through which cells adapt to serine starvation.
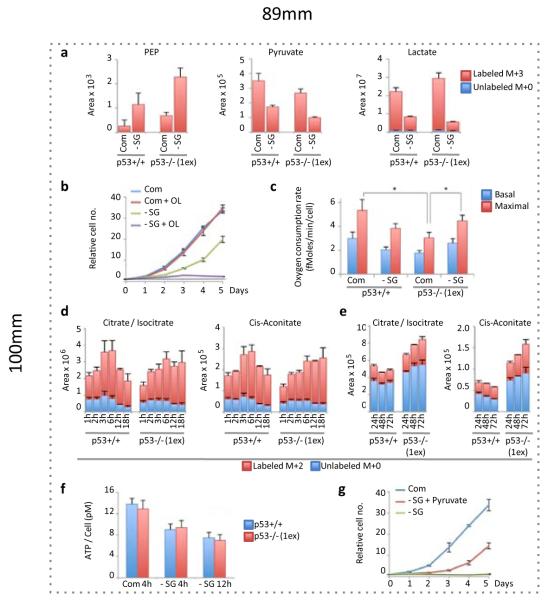
The mTOR pathway senses amino acid availability, and while mTORC1 activity was lowered by serine starvation, it was maintained at very similar levels in p53+/+ and p53−/− cells (Supp. Fig. 5). This demonstrates that the effect of serine starvation on mTORC1 was p53-independent and therefore unlikely to contribute to the enhanced sensitivity of p53−/− cells. A similar maintenance of mTORC1 activity in serine-starved cells has recently been shown, and is promoted by PKM2 expression17. Serine activates PKM219 and decreased PKM2 activity following serine starvation causes an accumulation of upstream glycolytic intermediates for diversion to the SSP20. To balance lower glycolysis following PKM2 inhibition, cells increase flux of pyruvate to the TCA cycle, requiring cells depleted of PKM to display increased O2 consumption to support elevated OXPHOS20. Both p53+/+ and p53−/− cells displayed elevated phosphoenopyruvate (PEP) levels and decreased pyruvate and lactate levels, evidence of low PKM2 activity following serine starvation (Fig. 2a). The importance of OXPHOS during serine starvation was demonstrated by treatment with the mitochondrial ATP synthase inhibitor Oligomycin (Fig. 2b), which completely inhibited the growth of serine-deprived p53+/+ cells. As p53 supports OXPHOS3,21,22, we considered the possibility that p53−/− cells would be unable to up-regulate OXPHOS in response to serine starvation.
Figure 2. Serine starvation differentially changes energy metabolism in p53+/+ and p53−/− cells.
a, HCT116 cells were fed complete (Com) or serine and glycine deficient (-SG) media for 24h, in the presence of U-13C-glucose for the final 2h. LC-MS was used to detect relative intracellular quantities of glycolytic intermediates (averages of triplicate wells). b, HCT116 cells were grown with or without serine, glycine and Oligomycin 1ng/ml (averages of triplicate wells). c, Oxygen consumption rate of HCT116 cells was measured after 48h serine and glycine starvation (n=3, *p<0.05). d, Relative intracellular levels of TCA cycle intermediates in HCT116 cells deprived of serine and glycine (in the constant presence of U-13C-glucose) were analysed by LC-MS (averages of triplicate wells), and e, after long term starvation, with U-13C-glucose added for the final hour (averages of triplicate wells). f, ATP levels were measured in HCT116 cells (n=3). g, HCT116 p53−/− (1ex) cells were grown in complete media or media lacking serine and glycine with or without pyruvate 5mM (averages of triplicate wells). All error bars are SEM.
As expected, p53−/− cells showed lower O2 consumption than p53+/+ cells under fed conditions. Surprisingly, p53−/− cells responded to serine starvation with increased O2 consumption, whereas p53+/+ cells showed lower O2 consumption (Fig. 2c). Closer analysis of metabolic flux into the TCA cycle revealed that while both p53+/+ and p53−/− cells showed an increase in glycolytic TCA cycle flux in the immediate response to serine starvation, this response was reversed over time in p53+/+ cells, but sustained in the p53−/− cells (Fig. 2d&e). This correlates with the changes in O2 consumption observed during serine starvation, suggesting the initial response of p53+/+ and p53−/− to serine depletion is similar, but p53−/− cells sustain a metabolic profile indicative of low PKM2 activity, including elevated O2 consumption.
We predicted that the disruption to glycolysis in the immediate response to serine starvation would impede ATP production, indeed ATP levels dropped in both p53+/+ and p53−/− cells (Fig. 2f). We considered that as glycolytic flux to pyruvate is lowered by serine starvation, increasing pyruvate levels (by adding exogenous pyruvate) would potentially alleviate the ATP shortage by further enhancing flux to the TCA cycle. We observed a significant recovery in proliferation of p53−/− cells after adding pyruvate (Fig. 2g). LC-MS analysis of serine-starved p53−/− cells fed unlabeled pyruvate (in the presence of U-13-C-glucose) confirmed increased TCA cycle flux (Supp. Fig. 6). However, this enhanced TCA cycle flux was not sufficient to fully restore proliferation (Fig. 2g).
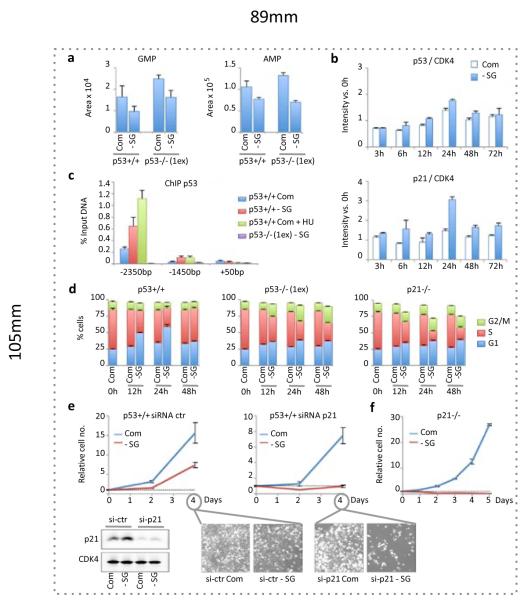
Serine contributes to the synthesis of purines via glycine, which is incorporated into GMP and AMP via IMP. Serine starvation resulted in lower GMP and AMP levels in p53+/+ and p53−/− cells (Fig. 3a). Previous studies show that GMP depletion can activate p53-dependent G1 arrest23,24. Consistently, we found that serine starvation led to a small elevation in p53 expression, accompanied by more marked, but transient, elevation of the p53 target protein p21 (Fig. 3b). Treatment of p53+/+ cells with low doses of mycophenolic acid (an inhibitor of GMP synthesis) replicated this subtle p53-p21 response (Supp. Fig. 7a&b). Recruitment of p53 to the p21 promoter during serine starvation was confirmed by chromatin-immunoprecipitation (Fig. 3c). As only a modest increase in p53 expression was observed, we speculate that p53 was activated via post-translational modification, rather than Mdm2 inhibition. While our data are consistent with p53 activation in response to nucleotide depletion, activation of AMPK (Supp. Fig. 7c) due to lower ATP levels could also contribute to the p53 response7.
Figure 3. Serine starvation causes recruitment of p53 to the p21 promoter and activation of a transient p21-dependent G1 arrest.
a, LC-MS was used to quantify total relative amounts of intracellular purine nucleotides GMP and AMP in serine and glycine fed (Com) and starved (-SG) HCT116 cells (averages of triplicate wells). b, p53 and p21 protein expression was quantified in p53+/+ HCT116 cells via western blot and detection with infra-red conjugated secondary antibodies (n=3). c, Chromatin-immunoprecipitation (ChIP) was performed for p53 with qPCR for two p53-response elements (-2350bp and -1450bp) and the transcription initiation region (+50bp) of the p21 promoter (n=3). d, BrdU labeling and PI staining followed by flow cytometry were used to asses cell cycle (n=3). e, p21 was transiently knocked down in p53+/+ HCT116 cells using si-RNA (averages of triplicate wells). f, p21−/− HCT116 cells (retaining wild-type p53+/+) were grown in media with or without serine and glycine (averages of triplicate wells). All error bars are SEM.
As expected following p53 activation, p53+/+ cells starved of serine and glycine initially showed an accumulation of cells in G1 and reduced S-phase, correlating with the increased expression of p21 (Fig. 3d). By 48 hours, these cells resumed a normal cell cycle. By comparison, p53−/− cells failed to establish a strong G1 arrest, showing a more gradual decrease in S-phase (corresponding to an increase in sub-G1 cells – Fig. 1b) and did not recover normal cell cycle (Fig. 3d). Since p21 is a major mediator of p53-induced G1 arrest25, we examined the effect of p21 loss in p53+/+ cells. p21−/− cells showed a similar cell cycle response as p53−/− cells (Fig. 3d), suggesting that induction of p21 may be critical for adaptation to serine starvation. Indeed, p53+/+ cells depleted of p21 by siRNA were unable to increase in number without serine (similar to p53−/− cells, Fig. 3e), a response even more dramatic in cells genetically deleted of p21 (Fig. 3f).
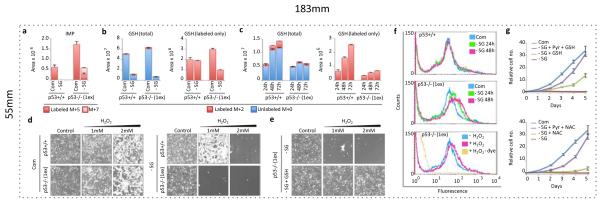
We next considered whether p53+/+ and p53−/− cells utilise the low levels of intracellular serine available under conditions of starvation differently. Specifically we analysed the balance between purine-nucleotide and glutathione synthesis, both of which require serine/glycine. LC-MS showed that after 24 hours, p53−/− cells retained flux of de novo serine/glycine into IMP (M+7), but this was inhibited in p53+/+ cells (Fig. 4a). Re-feeding serine-starved cells with U-13C,15N-L-Serine confirmed sustained serine flux into IMP, GMP and AMP in p53−/− cells (Supp. Fig. 8a). However, in serine-starved p53+/+ cells, flux of the replenished labeled-serine to nucleotides was blocked despite plentiful intracellular serine. This demonstrates that a feature of p53-p21 induced cell cycle arrest in response to serine starvation is inhibition of nucleotide synthesis, a known function of p2126,27. Reduced glutathione (GSH) is the principal cellular anti-oxidant, and we found that levels of GSH dropped significantly in p53+/+ and p53−/− serine-starved cells (Fig. 4b). However, in contrast to nucleotides, p53+/+ cells maintained and, over time, enhanced flux to GSH synthesis during starvation. Strikingly, this maintenance of flux to glutathione was not seen in p53−/− and p21−/− cells (Fig. 4b & Supp. Fig. 8b). Consequently, total GSH levels showed recovery in p53+/+ but not p53−/− or p21−/− cells (Fig. 4c & Supp. Fig. 8c).
Figure 4. p53-p21 activation allows serine deprived cells to synthesise GSH in preference to nucleotides.
a, LC-MS was used to detect relative intracellular quantities of IMP and b, GSH in HCT116 cells fed complete media (Com) or media lacking serine and glycine (-SG) for 24h, in the presence of U-13C-glucose for the final 2h, and c, fed –SG media for the indicated times in the presence of U-13C-glucose for the final hour (averages of triplicate wells). d, HCT116 cells were grown with or without serine and glycine, or e, media lacking serine and glycine with 5mM glutathione (GSH) for 48h in the presence of hydrogen peroxide (H2O2) for the final 24h. f, HCT116 cells were treated with an oxidation-activated fluorescent dye and analysed by flow cytometry. g, HCT116 p53−/− (1ex) cells were grown with or without serine, glycine, pyruvate 5mM (Pyr) and / or GSH 5mM, or N-acetyl cysteine 0.2mM (NAC) (averages of triplicate wells). All error bars are SEM.
The failure of p53−/− cells to recover GSH levels combined with their elevated oxygen consumption suggested that these cells would accumulate increased intracellular ROS levels. p53 has well established anti-oxidant functions1,6, but hydrogen peroxide treatment demonstrated that p53−/− cells were only slightly more susceptible than p53+/+ to oxidative stress under normal conditions (Fig. 4d). While serine and glycine starvation increased the sensitivity of both genotypes to peroxide treatment, this effect was more marked in p53−/− cells (Fig. 4d), and rescued by adding GSH (Fig. 4e). Staining with an oxidation-activated fluorescent dye confirmed increased intracellular ROS in p53−/− cells during serine starvation (Fig. 4f & Supp. Fig. 9a). To assess the importance of ROS in limiting proliferation, we tested the effect exogenous GSH or N-acetyl cysteine. While anti-oxidant treatment alone modestly improved proliferation of serine-starved p53−/− cells, either of the ROS-limiting treatments almost completely rescued the proliferation of serine-starved p53−/− cells in combination with pyruvate (Fig. 4g). We confirmed that the exogenous GSH did not lead to accumulation of intracellular glycine or serine, demonstrating that rescue was achieved by increasing the intracellular GSH pool (Supp. Fig. 9b). Adding GSH to serine-starved p53+/+ cells did not greatly enhance proliferation, supporting the theory that p53+/+ cells are able to maintain GSH pools independently (Supp. Fig. 9c).
Our data therefore shows that the sensitivity of p53−/− cells to serine depletion is due to a combination of impaired glycolysis and elevated ROS. While the initial response of cells with or without p53 to serine depletion was similar, p53+/+ cells underwent p21-dependent G1 arrest, allowing the limited levels of de novo serine to be channelled to GSH production to counter oxidative stress. Depletion of p21 (while retaining p53) caused severe sensitivity to serine depletion (Fig. 3e&f), suggesting that activation of other arms of the p53 response may contribute to the death of these cells. Our data also demonstrates that the ability of p53-deficient cells to engage higher rates of OXPHOS is not entirely defective, but that OXPHOS in these cells is likely limited by a requirement to prevent ROS generation. This observation ties in with the suggestion that cancer cells adopt aerobic glycolysis (the Warburg effect) to avoid generation of metabolic ROS28. It has recently been shown that increased ROS levels inhibit PKM229, providing an explanation for why p53−/− cells show evidence of sustained PKM2 inhibition during serine starvation.
In conclusion, our study underlines the importance of p53 in coordinating metabolic remodelling in response to metabolic stress. We demonstrate that serine uptake supports the Warburg effect, indicating that many cancer cells may show some sensitivity to serine depletion, particularly those lacking p53. However it is likely that other genetic alterations (such as PHGDH amplification14-16) may circumvent serine-dependence in other cancer cells. Taken together, our work suggests the therapeutic utility of serine depletion – either by removal from the diet, enzymatic depletion in vivo, or other means – is worthy of further investigation.
Methods
Cell culture
Unless otherwise stated, chemicals were obtained from Sigma-Aldrich, and cell culture reagents from Gibco (Invitrogen). HCT116 p53+/+/ p21+/+ [parental], p53−/− (1ex) [deletion of p53 exon 2], p53−/− (3ex) [deletion of p53 exons 2, 3 & 4], p21−/− and RKO p53+/+ and p53−/− cells were a gift of Prof. B. Vogelstein (Johns Hopkins University)30. Litter-matched wild-type and p53−/− MEFs (passage <4) were prepared from E14.5 embryos derived from mating p53+/− mice31 (C57Bl6/J background, >20 gen). Cells were kept at 37°C in humidified 5% CO2 in air, stock HCT116 and RKO cells were maintained in McCoys 5A medium (26600) containing 2mM L-glutamine and 10% FBS (PAA Laboratories). For experiments cells were seeded in DMEM (21969) with 10% FBS and 2mM L-glutamine. Complete (Com) media was formulated to closely match the nutrient composition of DMEM (which contains 0.4mM serine and 0.4mM glycine); complete media consisted of MEM (21090) supplemented with additional 1×MEM vitamins (11120), 10% dialysed-FBS (Hyclone, Thermo Scientific), L-glutamine 2mM, additional D-glucose (to 25mM), serine 0.4mM and glycine 0.4mM. For starvation experiments cells were fed the same media formulation without serine and glycine (-SG media).
Proliferation assays
HCT116 and RKO cells were seeded in 24-well plates (8 × 104/well), MEFs in 12-well plates (p53+/+ 2 × 104/well, p53−/− 1 × 104/well) in DMEM and allowed to grow for 16-24h. Cells were then washed with PBS and received complete or –SG media supplemented with the stated nutrients and/or drugs. To maintain constant nutrient levels and remove nutrients liberated from dead cells, media was replaced every 24h. Cells counts were performed with a CASY TT cell counter (Innovatis, Roche Applied Science). siRNA for p21 (sc-29427, Santa Cruz Biotechnology, UGUCAGAACCGGCUGGGGA & UCCCCAGCCGGUUCUGACA) or non-targeting control (sictr, ON-TARGETplus, Dharmacon D-001810-10-20, UGGUUUACAUGUCGACUAA, UGGUUUACAUGUUGUGUGA, UGGUUUACAUGUUUUCUGA & UGGUUUACAUGUUUUCCUA) was transfected with Metafectene SI (Biontex) in DMEM for 24h before washing and adding complete or –SG media.
Xenografts
Bilateral subcutaneous injections of 3 × 106 HCT116 cells were carried out on 8 week CD-1-Foxn1nu female mice (Charles River); p53+/+ on right flank and p53−/− (1ex) on the left. Immediately following injection mice were placed either on control diet (n=10) (containing serine and glycine as part of the amino-acid mix) or diet deficient in serine & glycine (n=10) (Test Diet, International Product Supplies) – formulations available on request. The diets had equal calorific value and equal total amino acid content. Animals were housed in sterile IVC cages, monitored thrice weekly and humanely sacrificed when tumours reached clinical endpoint of predetermined size (volume = (length × width2)/2) or ulceration. All animal work was approved by the Ethical Review Process (University of Glasgow) and undertaken in line with the UK Animals (Scientific Procedures) Act of 1986 (PPL 60/4181) and the EU directive 2010.
Western blot
Cells were seeded in 6-well plates and grown in DMEM for ~40h. Cells were washed with PBS then received complete or –SG media. Cell numbers were titrated at the time of seeding to be ~90% confluent at protein isolation. Cell lysates were prepared in RIPA buffer with complete protease inhibitors (Roche), resolved via PAGE and transferred to nitrocellulose. Primary antibodies: Phosho-p70S6K (9206) & total-p70S6K (2708) Phospho-S6 (2215) & total-S6 (2217), AMPKα (2532) & Phospho-AMPKα (2535) all from Cell Signaling Technology, PHGDH (Sigma Life Science, HPA021241), PSAT1 (Novus Biologicals, 21020002), PSPH (sc-98683), CDK4 (sc-260), p53 DO-1 (sc-126), p21 (sc-397) and PGAM1 (sc-130334) all from Santa Cruz Biotechnology. Secondary antibodies were IRDye800CW or IRDye6980LT conjugated (LiCor Biosciences), and were detected using an Odyssey infrared scanner (LiCor Biosciences) and quantified with Odyssey software.
Oxygen consumption Rates (OCR)
An XF24 Extracellular Flux Analyser (SeaHorse Bioscience) recorded OCR. HCT116 cells were grown in XF-plates for 48h in complete or –SG media (90-100% confluent at measurement). OCR was recorded after equilibration and after CCCP (0.5uM) and antimycin (1.5uM) treatments, these values were used to calculate basal and maximal OCR. All OCR measurements were normalized with well-by-well haemocytometer cell counts.
Peroxide sensitivity assay
HCT116 cells were seeded in 12-well plates in DMEM. After 16-20h cells were washed with PBS and received complete or serine & glycine deficient media. After 24h the media was replaced with matching media containing the stated concentrations of hydrogen peroxide, after a further 24h cells were washed and images captured using a light microscope. We noted that cell number had a large impact on peroxide sensitivity; therefore cell seeding was carefully titrated so that cell number was equal across the different experimental conditions at the time of peroxide addition.
ROS detection
CellROX deep red reagent (Invitrogen) was added to cell culture media for 30 minutes. For flow cytometry, cells were detached by washing with PBS-EDTA followed by treatment with PBS-EDTA with trypsin (0.025%) and analysed on a FACSCalibur cytometer (BD Bioscience). For confocal microscopy, live cells were imaged using an inverted confocal fluorescence microscope (Fluoview FV1000, Olympus).
Liquid Chromatography-Mass spectrometry (LC-MS)
HCT116 cells (0.5–1.5 × 106) were seeded in triplicate wells of 6-well plates in DMEM, duplicate plates were seeded for cell counts. After 16-24h cells were washed with PBS and received complete or –SG media for the stated times. For glucose flux experiments, media was replaced with HEPES buffered Krebs-Ringer solution with 25mM U-13C-D-Glucose (Cambridge Isotopes), 10% dialysed FBS, 1×MEM amino acids (11130), 2×MEM vitamins and 2mM L-glutamine. For Serine flux experiments, medium was replaced with complete media, with serine substituted for 0.4mM U-13C,15N-L-Serine (Cambridge Isotopes). Cells were washed with PBS and metabolites extracted using Methanol/Acetonitrile/dH2O (5:3:2) (2–4 × 106 cells/ml). Samples were snap frozen, thawed at 4°C, spun at 16000g for 15 minutes and supernatants collected and filtered through 0.45um PTFE membranes (Millipore). Serum samples were collected at sacrifice; 20ul of serum was added to 980ul of extraction buffer and prepared as above. LC-MS analyses were performed on an Orbitrap Exactive (Thermo Scientific) in line with an Accela autosampler and an Accela 600 pump (Thermo Scientific). The Exactive operated in the polarity-switching mode with positive voltage 4.5kV and negative voltage 3.5kV. Column hardware consisted of a Sequant ZIC-pHILIC column (2.1×150 mm, 5um) coupled to a Sequant ZIC-pHILIC guard column (2.1×20 mm, 5um) (Merck). Flow rate was 100uL/min, buffers consisted of Acetonitrile (ACN) for A, and 20 mM (NH4)2CO3, 0.1% NH4OH in H2O for B. Gradient ran from 80% to 40% ACN in 20min, followed by a wash at 20% ACN and re-equilibration at 80% ACN. Metabolites were identified and quantified using LCquan software (Thermo Scientific). Metabolites were positively identified on the basis of exact mass within 5ppm, and further validated by concordance with standard retention times.
qPCR
Primers: PGK-F:CTGTGGCTTCTGGCATACCT, PGK-R:CGAGTGACAGCCTCAGCATA, PHGDH-F:ATCTCTCACGGGGGTTGTG, PHGDH-R:AGGCTCGCATCAGTGTCC, PSAT1-F:CGGTCCTGGAATACAAGGTG, PSAT-R:AACCAAGCCCATGACGTAGA, PSPH-F: GAGCGGACTCCCTTTTAAGC, PSPH-R:CAGGGAGGTGAGCTGTGC, SHMT-F: CCCTCCCCATTTGAACACT, SHMT-R:GGGATCCACACTTTTCACTCC, PKM2-F: CTCGGGCTGAAGGCAGT, PKM2-R:AATTGCAAGTGGTAGATGGCA, Actin-F:TCC ATCATGAAGTGTGACG, Actin-R:TACTCCTGCTTGCTGATCCAC. Reactions used SYBR Green master-mix on a 7500 Fast Real-Time PCR System (both Applied Biosystems).
Chromatin-Immunoprecipitation
Assays were performed as described previously32. Cells (6–8 × 105) were seeded in 6-well plates in DMEM, allowed to grow for ~40h, then washed with PBS and received complete or –SG media for 15 hours, hydroxyurea (HU, 0.4mM) was added as a positive control.
Cell cycle analysis
For sub-G1 analysis, cells were grown and detached from plates as described above, then fixed and stained with propidium iodide (PI, 50ug/ml) for 30min. For cell cycle analysis BrdU (Invitrogen) was added to live cells for 100 minutes and detected as described previously32. Flow cytometry was performed on a FACSCalibur cytometer.
PI exclusion
PI was added to cell culture media (1ug/ml) for 5min. Non-adherent and adherent cells were harvested and analysed by Flow cytometry on a FACSCalibur cytomter.
ATP assay
Aliquots of cell suspension were added to TE buffer (pH 7.75) heated to 99°C and assayed with an ATP Determination kit (Invitrogen) on a Veritas Microplate luminometer (Turner Biosystems). Cell suspensions were counted to normalize for cell number.
Statistics
Survival was assessed by non-parametric distribution analysis (right censoring), using log rank and Wilcoxon tests calculated on Minitab 16 (Minitab Ltd). The following t-test comparisons were performed with Microsoft Excel (v12.3.4): Tumour volume between diet groups; unpaired, one tail. Tumour volume within diet groups; paired, one tail. Serum amino acids between diet groups; unpaired, one tail. Oxygen consumption rate within genotype; paired, one tail. Oxygen consumption rate between genotypes; unpaired, one tail.
Supplementary Material
Acknowledgements
This work was funded by Cancer Research UK. C.R.B. is a recipient of a Rubicon Fellowship from the Netherlands Organisation for Scientific Research. The authors thank Arnaud Vigneron, Barbara Chaneton, Margaret O’Prey, Eric Cheung, Dimitris Athineos, Gabriela Kalna, Gillian Mackay and Bob Ludwig for advice and technical assistance.
Footnotes
This manuscript is accompanied by supplementary figures.
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
References
- 1.Bensaad Karim, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126(1):107. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 2.Jiang Peng, et al. p53 regulates biosynthesis through direct inactivation of glucose-6-phosphate dehydrogenase. Nat Cell Biol. 13(3):310. doi: 10.1038/ncb2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matoba Satoaki, et al. p53 regulates mitochondrial respiration. Science. 2006;312(5780):1650. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki Sawako, et al. Phosphate-activated glutaminase (GLS2), a p53-inducible regulator of glutamine metabolism and reactive oxygen species. Proc Natl Acad Sci U S A. 107(16):7461. doi: 10.1073/pnas.1002459107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu Wenwei, et al. Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc Natl Acad Sci U S A. 107(16):7455. doi: 10.1073/pnas.1001006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Budanov Andrei V., et al. Regeneration of peroxiredoxins by p53-regulated sestrins, homologs of bacterial AhpD. Science. 2004;304(5670):596. doi: 10.1126/science.1095569. [DOI] [PubMed] [Google Scholar]
- 7.Jones Russell G., et al. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Molecular cell. 2005;18(3):283. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 8.Maddocks Oliver D. K., Vousden Karen H. Metabolic regulation by p53. J Mol Med (Berl) 89(3):237. doi: 10.1007/s00109-011-0735-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rose ML, et al. Dietary glycine prevents the development of liver tumors caused by the peroxisome proliferator WY-14,643. Carcinogenesis. 1999;20(11):2075. doi: 10.1093/carcin/20.11.2075. [DOI] [PubMed] [Google Scholar]
- 10.Rose ML, Madren J, Bunzendahl H, Thurman RG. Dietary glycine inhibits the growth of B16 melanoma tumors in mice. Carcinogenesis. 1999;20(5):793. doi: 10.1093/carcin/20.5.793. [DOI] [PubMed] [Google Scholar]
- 11.Jain M, et al. Metabolite Profiling Identifies a Key Role for Glycine in Rapid Cancer Cell Proliferation. Science. 2012;336:1040. doi: 10.1126/science.1218595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Snell Keith. The duality of pathways for serine biosynthesis is a fallacy. Trends in Biochemical Sciences. 1986;11(6):241. [Google Scholar]
- 13.Snell K, Fell DA. Metabolic control analysis of mammalian serine metabolism. Adv Enzyme Regul. 1990;30:13. doi: 10.1016/0065-2571(90)90006-n. [DOI] [PubMed] [Google Scholar]
- 14.Pollari Sirkku, et al. Enhanced serine production by bone metastatic breast cancer cells stimulates osteoclastogenesis. Breast Cancer Res Treat. 125(2):421. doi: 10.1007/s10549-010-0848-5. [DOI] [PubMed] [Google Scholar]
- 15.Possemato Richard, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 476(7360):346. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Locasale Jason W., Cantley Lewis C. Genetic selection for enhanced serine metabolism in cancer development. Cell Cycle. 10(22):3812. doi: 10.4161/cc.10.22.18224. [DOI] [PubMed] [Google Scholar]
- 17.Ye Jiangbin, et al. Pyruvate kinase M2 promotes de novo serine synthesis to sustain mTORC1 activity and cell proliferation. Proc Natl Acad Sci U S A. 109(18):6904. doi: 10.1073/pnas.1204176109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kondoh Hiroshi, et al. Glycolytic enzymes can modulate cellular life span. Cancer research. 2005;65(1):177. [PubMed] [Google Scholar]
- 19.Mazurek Sybille. Pyruvate kinase type M2: a key regulator of the metabolic budget system in tumor cells. Int J Biochem Cell Biol. 43(7):969. doi: 10.1016/j.biocel.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 20.Chaneton B, et al. Serine is a natural ligand and allosteric activator of pyruvate kinase. Nature. 2012 doi: 10.1038/nature11540. (In Press) DOI 10.1038/nature11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okamura S, et al. Identification of seven genes regulated by wild-type p53 in a colon cancer cell line carrying a well-controlled wild-type p53 expression system. Oncology Research. 1999;11(6):281. [PubMed] [Google Scholar]
- 22.Stambolsky P, et al. Regulation of AIF expression by p53. Cell death and differentiation. 2006;13(12):2140. doi: 10.1038/sj.cdd.4401965. [DOI] [PubMed] [Google Scholar]
- 23.Linke SP, et al. A reversible, p53-dependent G0/G1 cell cycle arrest induced by ribonucleotide depletion in the absence of detectable DNA damage. Genes Dev. 1996;10(8):934. doi: 10.1101/gad.10.8.934. [DOI] [PubMed] [Google Scholar]
- 24.Messina Elisa, et al. Guanine nucleotide depletion triggers cell cycle arrest and apoptosis in human neuroblastoma cell lines. Int J Cancer. 2004;108(6):812. doi: 10.1002/ijc.11642. [DOI] [PubMed] [Google Scholar]
- 25.Deng C, et al. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82(4):675. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 26.Almasan A, et al. Deficiency of retinoblastoma protein leads to inappropriate S-phase entry, activation of E2F-responsive genes, and apoptosis. Proc Natl Acad Sci U S A. 1995;92(12):5436. doi: 10.1073/pnas.92.12.5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dimri GP, et al. Inhibition of E2F activity by the cyclin-dependent protein kinase inhibitor p21 in cells expressing or lacking a functional retinoblastoma protein. Mol Cell Biol. 1996;16(6):2987. doi: 10.1128/mcb.16.6.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gruning Nana-Maria, Ralser Markus. Cancer: Sacrifice for survival. Nature. 480(7376):190. doi: 10.1038/480190a. [DOI] [PubMed] [Google Scholar]
- 29.Anastasiou Dimitrios, et al. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science. 334(6060):1278. doi: 10.1126/science.1211485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bunz F, et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282(5393):1497. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 31.Donehower LA, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356(6366):215. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 32.Vigneron Arnaud M., Ludwig Robert L., Vousden Karen H. Cytoplasmic ASPP1 inhibits apoptosis through the control of YAP. Genes Dev. 24(21):2430. doi: 10.1101/gad.1954310. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.