Abstract
The great promise of comparative neuroscience is to understand why brains differ by investigating the relations between differences in the organization of different brains, their evolutionary history, and their current ecological niche. For this approach to be successful, the organization of different brains needs to be quantifiable. Here, we present an approach to formally comparing the connectivity of different cortical areas across different brains. We exploit the fact that cortical regions can be characterized by the unique pattern of connectivity, the so-called connectivity fingerprint. By comparing connectivity fingerprints between frontal cortical areas in the human and macaque brain we can identify between-species homologs, but also illustrate that is driving differences between species. We illustrate the approach by comparing the organization of the frontal cortex between humans and macaques, showing general similarities combined with some differences in the lateral frontal pole.
Keywords: Comparative neuroscience, brain architecture, connectivity, primate, connectivity fingerprint
Comparative neuroscience can provide crucial insights into why our brain is organized the way it is. Knowing about the organization of the brains of species related to us constrains how we interpret maps of our own neuroanatomy and theories on the function of particular brain areas. In recent years, a number of large-scale projects have been launched aimed at mapping the organization of entire brains across different species. These projects focus on different modalities, ranging from anatomical measures such as cytoarchitecure, receptor distribution, and the architecture of connections to functional activation profiles and genetic expression patterns, and they hold the potential to provide comparative maps across a wide phylogenetic range (Striedter et al. 2014). In primate comparative neuroscience, the increasing availability of magnetic resonance imaging-based techniques has the potential to substantially ease the acquisition of large volumes of data, allowing more diverse species to be studied at a higher rate than was previously possible (Mars et al. 2014). One of the main next challenges in this endeavour is to find formal methods to turn these diverse datasets into measures that can be meaningfully compared between brains.
The study of the architecture of connections between brain areas is one fruitful avenue for comparative neuroscience, especially in larger animals such as primates. This is mainly due to two reasons. First, connectivity has been proposed as one of the main mechanisms through which phenotypic diversity is realised (Krubitzer and Kaas 2005). Second, the availability of connectivity is increasingly widespread. Apart from established connectivity databases such as the CoCoMac for the macaque (Bakker et al. 2012) and the Allen atlas for the mouse (http://connectivity.brain-map.org), diffusion MRI tractography and other MRI-based techniques now allow the acquisition of whole-brain connectivity data in a short time period. Such approaches have become increasingly popular in primate comparative neuroscience, leading to a flurry of studies with sometimes quite different goals and approaches. For instance, a number of studies have qualitatively compared human MRI data to macaque tract tracing results (Thiebaut de Schotten et al. 2012, Margulies and Petrides 2013). Others have used connectivity-based clustering approaches to infer whether the human cortex follows similar organizational principles to that of the macaque (Tomassini et al. 2007, Beckmann et al. 2009). Finally, a growing number of studies has used diffusion MRI in multiple species to investigate the extent or projections of major white matter fibers across species (Rilling et al. 2008, Hecht et al. 2015).
Encouraging though these results are, the variety of approaches has meant that the results, or even the goals, of the different studies are often difficult to reconcile. Connectivity studies often yield very high-dimensional data that can be difficult to summarize. The purpose of the current paper is to provide a simple framework for comparing brain connectivity between species. The approach we present is based on matching connectivity fingerprints. Connectivity fingerprints were proposed by Passingham and colleagues (2002) as a way to summarize the important connections of a single cortical area with a selected set of other areas (Figure 1). They observed that the set of connections of each area is a unique identifier of a brain region. Importantly, the connections of an area give vital clues about its function by demonstrating the type of information an area has access to and the other brain regions it can influence. In their original paper, Passingham and colleagues used the example of macaque areas F3 and F5 which, although both premotor areas, not only differ substantially in their connections to the rest of the frontal cortex, but also show very distinct neuronal responses in various motor tasks. The goal of the fingerprint is thus to provide a diagnostic measure of an area that summarizes its most important anatomical features and has direct implications for the area’s functional relevance.
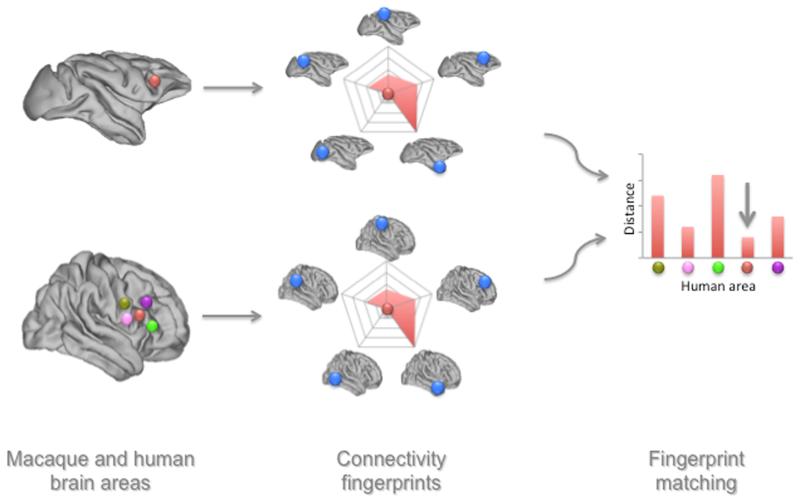
Figure 1. Schematic example of the connectivity fingerprint matching approach.
A macaque brain region (in red) is compared to multiple human brain regions (various colors in the left panel). For each of these regions, the connectivity with a predefined set of brain regions (in blue), for which the homology between the macaque and human has already been established. A difference measure between the macaque connectivity fingerprint and each of the human fingerprints is then determined. In this case, the human red area is the least dissimilar to the macaque brain region and thus the most likely candidate for homology.
Here, we will provide three examples of how connectivity fingerprints can be used as a tool to compare various aspects of brain organization across and within species. Using a unified framework of permutation testing, we will demonstrate (1) how the anatomical homologs or most similar areas can be selected from a set of candidate areas, (2) how putative homologs can be identified across the whole brain in an unbiased manner, and (3) how the specific connections that drive the differences between species can be uncovered.
Materials and methods
A comparison of connectivity profiles lies at the core of the proposed framework. Accordingly, the creation of a connectivity fingerprint is a crucial step aimed at summarizing the high-dimensional, often whole-brain, connectivity profile by a few ‘arms’ of the fingerprint (Figure 1). We refer to the area whose connectivity profile is displayed as the ‘seed’ area and to the areas on the arms of the fingerprint as the ‘target’ areas. The number of target areas should be sufficient to clearly demonstrate the diversity of connections of the seed area, but care should be taken to not include too many arms or the contribution of each arm will be too small and there will be a risk of overfitting the fingerprint. Importantly, the goal of the fingerprint is not to show only those areas with which the seed region is connected. Rather, the fingerprint should show a range of connection strengths, including the absence of a connection. As with any statistical test, it is important to have some variance to explain. Importantly, the fingerprint should be diagnostic for the seed areas under investigation (cf. Preuss (1995)).
Following the definition of the fingerprint, one needs to decide on the measure of comparison. Such a ‘distance measure’ will determine whether different fingerprints are either ‘close’ or ‘far’ from each other. Some measures emphasize the largest differences even further, while others actually downweight outliers. Importantly, if the target areas of a connectivity profile are carefully selected even simple distance measures often suffice, promoting interpretability, interchangability, and reproducability of the results. We have mostly used the city block or Manhattan distance
where p and q are two vectors representing the connectivity fingerprints to be compared and i indexes the n elements of the vectors, i.e., the n target areas of the fingerprint. Alternatively, sometimes it is more intuitive to use a measure of similarity, such as the cosine similarity:
In the following examples we created connectivity profiles using either resting state fMRI data or diffusion MRI data, but the approach is not limited to such data. The approach could in theory be used to compare fingerprints created using different types of data, for instance when comparing, on the one hand, a template derived from a database of connectivity studies using tracers to, on other hand, human diffusion MRI data. In such cases, the fingerprints need to be normalized in an appropriate fashion to ensure that any differences are not due to a difference in methods. For instance, in comparative studies using diffusion MRI, the voxel resolution will often differ both in absolute number and relative to the size of the brain. This can be addressed by normalizing the data to the maximum connection strength in the brain, such that any statement on connection strength becomes a statement of relative connection strength (Mars et al. submitted). In comparing connectivity fingerprints we follow the same logic. Rather than comparing absolute connections strengths, we normalize the fingerprint to a range between 0 (weakest connection with any of the target areas) and 1 (strongest connection with any of the target areas). We are thus comparing a pattern of connections with the target areas, rather than absolute numbers.
The data were analyzed using tools from FMRIB’s Software Library (FSL) (Smith et al. 2004) and custom tools in written in Matlab (the Mathworks, Natick, MA, USA) that form part of the in-house MR Comparative Analysis Toolbox (Mr Cat, www.neuroecologylab.org).
Example 1: Matching a set of connectivity fingerprints to a template
In the first example we were interested in finding out which of four potential medial frontal regions in the human brain has the most similiar connectivity fingerprint to area F6 in the macaque brain. The connectivity of macaque area F6 was determined using resting state fMRI from five anaesthetised animals belonging to a previously published dataset (Mars et al. 2011) (50 mins of BOLD fMRI at 3T, voxel size=2×2×2mm, TR=2s, TE=19ms, animals scanned under light anesthesia using isoflurane, carried out under authority of personal and project licenses in accordance with the UK Animals (Scientific Procedures) Act (1986) issued by the Home Office and approved by the University of Oxford Animal Care and Ethical Review Committee). The connectivity of human regions was determined using resting state fMRI data from 12 healthy volunteers from a dataset published previously (Neubert et al. 2014) (5 mins of BOLD fMRI at 3T, voxel size=3×3×3mm, TR=2.410s, carried out after written informed consent following procedures approved by the local ethics committee). Details of preprocessing are described in Neubert et al. (2014). In brief, the macaque resting state data was filtered to remove repiratory artifacts and submitted to denoising using independent component analysis as implemented in FSL’s MELODIC tool to remove non-physiological variance in the data. Human data were corrected for distortions due to magnetic field inhomogeneities using FSL’s FUGUE tool and the time courses of white matter, cortico-spinal fluid, and head movement were extracted for use as confound regressors.
A seed consisting of 3×3×3 voxels was drawn in macaque MNI standard space(Frey et al. 2011) at coordinates [1.25 11.25 16.25], placing it at the anterior border of F6 according to the atlas of Saleem and Logothetis (2012). As the human candidate regions to match to this macaque seed we defined in MNI 2mm standard space a 3×3×3 voxel region of interest (ROI) within the supplementary motor area (SMA) [10 4 58], the pre-supplementary motor area (pre-SMA) [14 22 52], and areas 9 [10 50 28], and 10 [16 58 4] as defined by Sallet et al. (2013). For the macaque area F6 and each of the four human test regions, the standard space ROI was transformed to an individual’s functional space and the correlation of that region’s dominant time course with that of each voxel in the rest of the brain was determined using FSL’s sbca tool (O’Reilly et al. 2010). The resulting correlation maps were back-transformed to standard space and converted to z-maps using the Fisher z transform. The macaque z-maps were averaged to create a ‘template’ F6 connectivity map, since the goal of this example is to compare multiple subject’s data (i.e., the human data) to a single template. To reiterate, this template could in theory have been based on any type of connectivity data.
Connectivity fingerprints were created for the macaque template and for each of the 12 human subjects’ four regions of interest by determining the average z-value of the functional connectivity map in five target regions placed in the anterior cingulate cortex (ACC), ventromedial prefrontal cortex (vmPFC), inferior parietal lobule (IPL), ventrolateral prefrontal cortex (vlPFC), and posterior cingulate cortex (PCC). Although this report is aimed at laying out a methodological framework, and not specifically to elucidate the human homolog of macaque F6, we refer to Sallet et al. (2013) for a more elaborate discussion of the connectivity fingerprints of the areas used in this example. Macaque target areas were drawn as 3×3×3 voxel regions of interest in macaque MNI space at the following coordinates: ACC [1.25 19.25 8.25], vmPFC [0.75 17.75 −2.75], IPL [16.25 −19.25 16.75], vlPFC [16.75 12.25 7.25], and PCC [1.75 −17.25 6.75]. Human target areas were drawn as 3×3×3 voxel regions of interest in MNI 2 mm space at the following coordinates: ACC [10 46 8], vmPFC [10 48 −20], IPL [48 −46 48], vlPFC [44 28 14], and PCC [10 −52 24]. We could then calculate the Manhattan distance between the normalized macaque template connectivity fingerprint and the normalized average human connectivity fingerprint of each region of interest.
Permutation testing (Nichols and Holmes 2007) was used to test the significance of the match between each of the human regions of interest and the macaque template F6. For each of the four human areas we tested the hypothesis that the difference between its connectivity fingerprint and that of the macaque template is on average smaller than expected by chance. The null hypothesis to reject is that the difference is on average the same independent of how we permute the target areas of the fingerprint. As such, for each human region of interest, the Manhattan distance between the template and the region of interest was calculated for 5000 different permutations of the fingerprint target areas.
Example 2: Matching a connectivity fingerprint across the cortex
An alternative approach for matching a connectivity fingerprint to a set of candidate fingerprints drawn from a range of areas is to search for a best fit to a template on a voxel-by-voxel basis across a large volume of cortex. In the second example we therefore calculated for each subject the whole-brain functional connectivity with the ACC, vmPFC, IPL, vlPFC, and PCC target areas used in Example 1. We calculated the whole-brain functional connectivity of each of these four areas using FSL’s sbca tool (O’Reilly et al. 2010). The resulting correlation maps were back-transformed to standard space and converted to z-maps using the Fisher z transform. We then used permutation testing as implemented in FSL’s randomise tool (Winkler et al. 2014), permuting over target areas, to test for each voxel how well the signal can be described as a combination of the five target areas’ functional connectivity according to the template. The results are thresholded at a voxel-wise FWE-corrected level of p<0.05. The resulting map shows which voxels have a connectivity fingerprint most similar to that of the template.
Example 3: Comparing frontal pole connectivity fingerprints
In the final example, we compare two groups of connectivity fingerprints with one another. We will use the example of the primate frontal pole as an illustration of this approach. Cytoarchitectonic work recently demonstrated the existence of a medial and a lateral subdivision of the human frontal pole (Bludau et al. 2014), which we will term FPm and FPl, respectively. Neubert and colleagues recently suggested that only the medial subdivision is homologous to the macaque frontal pole (Neubert et al. 2014). In this example we compared connectivity fingerprints to test both these assertions.
First, we tested whether two neighboring regions in the frontal cortex, namely the lateral and the medial frontal pole (FPl and FPm, respectively, see figure 4) are differentially connected to any of the major longitudinal fiber bundles in the brain. We performed tractography in 24 subjects from a previously published dataset (Neubert et al. 2014). Diffusion MRI data (60 isotropically distributed directions, voxel size 2×2×2 mm, b-value 1000 s×mm−2, eight b=0 volumes collected for reference) were collected and preprocessed as described previously (Neubert et al. 2014), following which a two fiber model was fitted to the data using FSL’s BedpostX (Behrens et al. 2007). Standard space masks of FPm and FPl (thresholded at 25% of the population) taken from the atlas of Neubert et al. (2014) (figure 3, top left) were warped to each individual’s diffusion space and probabilistic tractography was performed from 2500 sample streamlines seeded from each voxel within each individual’s seed mask using the following parameters: maximum of 2000 steps; step size of 0.5 mm; curvature threshold of 0.2. Each streamline followed local orientations sampled from the posterior distribution given by BedpostX, as described previously (Behrens et al. 2007). A visitation (streamline) map or tractogram was constructed for each individual. In order to allow comparison of these maps between participants the tractograms were normalized by log transforming the data and then dividing each voxel’s value by that of the maximum value in the map yielding voxel values between 0 and 1 (cf. Mars et al. (submitted)).
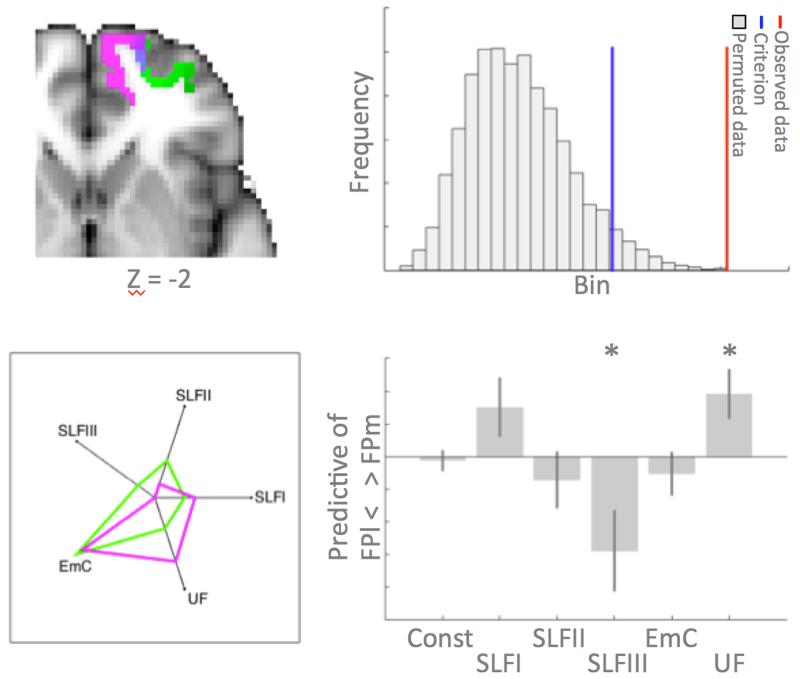
Figure 4. Connectivity fingerprint comparison between groups.
(top left) Location in MNI space of the medial (green) and lateral (pink) frontal pole. (bottom left) Connectivity fingerprint of the two areas with target areas in six longitudinal tracts. (top right) Permutation distribution of difference statistic between groups, i.e. FPm and FPl, showing the criterion value in blue and the actual statistic from the data in red. This indicates a significant difference in the connectivity fingerprint of the two areas. (bottom right) Beta weights of a logistic regression testing which tracts predict area membership (positive FPm, negative FPl), showing that SLFIII and UF explain differential variance between the two areas. * indicates significant predictive ability.
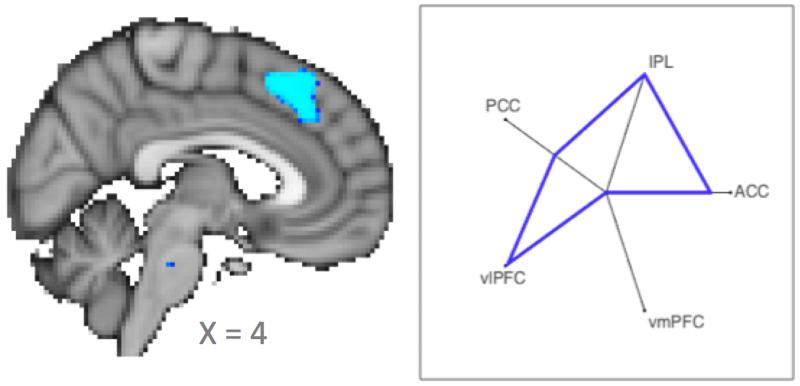
Figure 3. Matching macaque F6 connectivity fingerprint to voxels across a cortical territory.
(left panel) Statistical map, thresholded at p<0.05 correct, indicating a significant match with the macaque template in the human dorsal medial frontal cortex. (right panel) Connectivity fingerprint of the human significant cluster.
Targets were drawn as 3×3×3 voxel regions of interest in MNI 1mm standard space in the location of five major longitudinal fibers according to the atlas by Neubert et al. (in preparation): the first [14 15 52], second [30 15 36], and third [34 15 22] branches of the superior longitudinal fascicle (SLFI, SLFII, and SLFIII, respectively), the extreme capsule fiber complex (EmC) [30 15 −6], and the unicate fascicle (UF) [23 15 −17]. The average value of the FPl and FPm tractograms in each fiber’s target region of interest was taken as the value of the connectivity fingerprint. Permutation testing was then used to test for a difference between tracts. For illustration purposes only, we treated the two tracts as two different groups, permuting over group membership 5000 times and calculating the Manhattan distance beween the average connectivity fingerprints of the two groups every time. As before, for each test the connectivity fingerprints were normalized. In order not to bias the test, the normalization was performed over the combined arms of both fingerprints.
Having established whether the tracts differ in their connectivity fingerprint, we then aimed to quantify which tracts drive observed differences. Logistic regression was used to test which tracts test predict group membership (i.e., between FPm and FPl). Logistic regression does not make any assumptions on the distribution of the independent variables and is therefore suitable for this test.
Finally, we directly tested differences between human FPm and FPl connectivity and that of macaque FP. For this analysis we used resting state functional connectivity, as in Examples 1 and 2, since these data were available to us from both species and to illustrate that our methods are independent of the data modality employed. We used data from ten humans and ten macaques using the same scanning parameters as in the previous example. The human ROIs used in the previous diffusion MRI were used and a single macaque 3×3×3 voxel FP mask was draw at coordinates [1.75 25.75 4.75] in macaque MNI space. As above, the standard space ROIs were transformed to an individual’s functional space using non-linear warping and the correlation of that region’s dominant time course with that of each voxel in the rest of the brain was determined using FSL’s sbca tool (O’Reilly et al. 2010). The resulting correlation maps were back-transformed to standard space and converted to z-maps using the Fisher z transform.
Connectivity fingerprints were created with five targets areas: the IPL (macaque [16.25 −19.25 16.75], human [48 −46 48]), PCC (macaque [0.75 −17.25 6.75], human [10 −52 24]), vmPFC (macaque [1.75 15.5 −1.75], human [8 44 −14]), temporal pole (TP, macaque [18.75 7 −7.25], human [34 12 −36]), and dorsolateral prefrontal cortex area 9/46v (human [46 20 40], macaque [12.25 14.75 7.75]). As our dependent measure we used the Manatthan distance between the average human and average macaque connectivity fingerprint. We used permutation testing, permuting over group membership, to create a test distribution.
Results
Example 1: Matching a set of connectivity fingerprints to a template
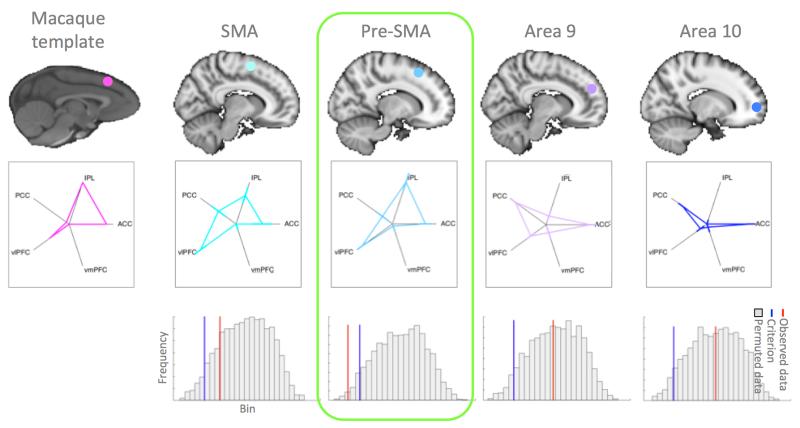
In the first example, we aimed to compare data from four candidate brain areas acquired in 12 subjects to a single macaque template. The functional connectivity fingerprint of macaque area F6 and of the four human candidate areas are displayed in figure 2. Permutation tests were used to determine whether the Manhattan distance between the template and the average fingerprint of each human region of interest were lower than expected by chance. The permutation distribution and criterion are indicated in the bottom panels of figure 2. The p-values of the actual distance for each of the four regions of interest were: SMA p=0.1718, pre-SMA p=0.0104, Area 9 p=0.4501, and Area 10 p=0.4887. This means only human pre-SMA presented a significant match with macaque area F6, even when correcting for the fact that we tested four areas. This result is as would be predicted from the literature (Picard and Strick 2001). The result was the same when, rather than using the Manhattan distance as our statistic, we looked at the cosine similarity between the macaque template and the human fingerprints and tested which are more similar than expected by chance. The p-values then became: SMA p=0.1152, pre-SMA p=0.0106, Area 9 p=0.5993, Area 10 p=0.5993. Again, only pre-SMA showed a significant match.
Figure 2. Matching macaque F6 connectivity fingerprint to four human areas.
(left column) Approximate anatomical location and connectivity fingerprint of the macaque template area at the anterior border of F6. (four right-hand columns) Connectivity fingerprints of four human seed areas (error bars represent ±SEM) and the results of the permutation tests showing a histogram of the distribution of the test statistic, the criterion value (blue line), and the value of the actual statistic given by the data (red line). Only in the case of pre-SMA is the actual statistic lower than the criterion, indicating a smaller difference between the area and the template than expected by chance.
Example 2: Matching a connectivity fingerprint across the cortex
Having shown that we can use connectivity fingerprints to match areas between species, we next took a more ambitious approach where we test whether there are any voxels in the human brain that show a similar connectivity profile to the template macaque area. In other words, rather than selecting four candidate medial wall areas, we here search across all voxels to see which show a similar connectivity profile, in essence providing a blind search across a cortical territory for the presence of a possible homolog. Although the test was performed at the whole-brain level, we here concentrate only on the results along the medial wall. There, we find a single contiguous cluster located dorsally to the cingulate cortex with a center of gravity of [4 28 45] (figure 3, middle panel). In the anterior-posterior dimension this cluster is closer to pre-SMA as defined in the atlas of Sallet al.(Sallet et al. 2013) than it is to the more posterior SMA or the more anterior Area 9.
To confirm the result, we next calculated the connectivity fingerprint of the region found in the analysis. The medial wall cluster of voxels with a p-value lower than 0.05 was warped to each individual participant’s function space and the whole-brain functional connectivity was calcalated in a manner anologous to that in Example 1. Whole brain z-maps in standard space were used to calculate the fingerprint with ACC, IPL, PCC, vlPFC, and vmPFC. The resulting connectivity fingerprint is shown in the right panel of figure 3, showing a good similarity with the macaque F6 template fingerprint.
Example 3: Comparing fingerprints between groups
Having matched connectivity fingerprints between datasets, in this final example we aim to test whether two different groups of connectivity fingerprints differ from on another and, if so, wether we can determine which connections are driving any difference. As an example case we look at the connectivity of the frontal pole. It has previously been suggested that human frontal pole contains separate subareas in the medial (FPm) and lateral (FPl) part (Bludau et al. 2014) and that only one of those has a homolog in the macaque (Neubert et al. 2014).
We first set out to determine whether FPm and FPl indeed have differential connectivity in the human brain. We determined the connectivity fingerprints of frontal areas FPm and FPl (figure 4, top left panel) with five longitudinal fiber bundles in the human brain using diffusion MRI tractography data. Permutation testing confirmed our suspicion, showing that the Manhattan distance between the groups was larger than expected by chance: p<0.001 (figure 4, top right panel). As can be seen in the connectivity fingerprints in figure 4 (lower left panel), both area tended to be connected most strongly with the extreme capsule fiber complex. However, there we noticable differences in the connections with other tracts, especially the unicate fascicle and the third branch of the superior longitudinal fascicle (SLFIII). To investigate which connections explained most of the variance between the FPm and FPl groups, we performed a logistic regression in which each arm, i.e., each tract, of the connectivity fingerprint was used to predict group membership. Two tracts significantly predicted group membership (figure 4, bottom row). A stronger SLFIII connection predicted FPl membership (p=0.017). In contrast, stronger uncinate fascicle connectivity predicted FPm membership (p=0.008).
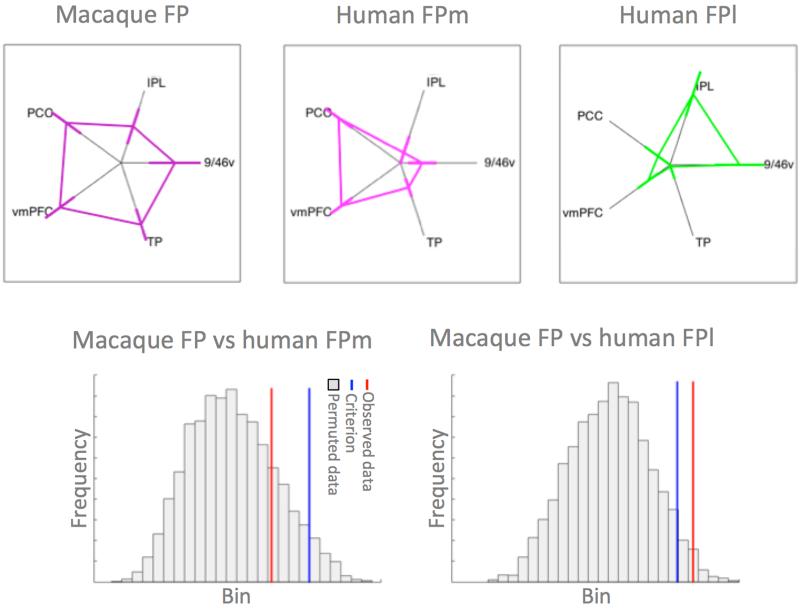
Having established the differential connectivity of human FPm and FPl, we set out to test whether the connectivity of human FPm was similar to that of macaque FP, whereas that of human FPl was different from macaque FP. For this analysis we created resting state functional connectivity fingerprints in ten humans and ten macaques with target areas in the vmPFC, PCC, temporal pole, and area 9/46v. Again, we used a permutation test where we permuted over group membership (human or macaque) and used the Manhattan distance between the average human and average macaque connectivity fingerprint as our dependent variable. Macaque FP and human FPl were significantly different (p=0.017), whereas macaque FP and human FPm did not differ significantly from one another (p=0.191) (figure 5).
Figure 5. Comparison of human and macaque connectivity fingerprints.
(top row) Connectivity fingerprints of macaque FP (left), human FPm (middle), and human FPl (right) (error bars represent ±SEM). (bottom row) Histogram of the distribution of the test statistic comparing macaque FP and human FPm (left) and macaque FP and human FPl (right).
Discussion
We set out to demonstrate the use of connectivity fingerprint matching across a variety of scenarios. As the examples above show, connectivity fingerprint matching using a permutation framework is a straight-forward approach to analyzing comparative connectivity data. In the first two examples, we compared the organization of the frontal cortex between humans and macaques in both a hypothesis-driven and a data-driven approach. In the first example, following Sallet et al. (2013), we tested which of four human medial frontal regions SMA, pre-SMA, Area 9, and Area 10 had the most similar connectivity profile to a macaque template region at the anterior border of macaque area F6. Pre-SMA provided the only significant match with the macaque template, even though in our simple connectivity fingerprint SMA and pre-SMA were not strickingly different. In the second example an unbiased search across the human medial frontal cortex revealed a large area to match with the macaque template. Interestingly, this area was located a bit anterior to the cortical territory normally identified with pre-SMA in the human (Picard and Strick 1996, Johansen-Berg et al. 2004), perhaps reflecting the slightly anterior placement of our F6 seed.
Demonstrating that this approach to connectivity fingerprint matching is a viable technique for comparative neuroscience, it was recently used in a number of studies, albeit sometimes in a more qualitative fashion. Sallet and colleagues (2013) matched ten areas of the dorsal frontal cortex to areas in the macaque. On the medial surface, they identified between-species homologs of SMA, pre-SMA, and Areas 9 and 10. The data in Example 1 show a quantitative version of their analysis. The approach used in Example 2, to fit a connectivity profile across a large part of cortex was developed in Mars et al. (2013). The authors searched for an anatomical homolog of the human temporoparietal junction area that is often identified in studies of human social cognition (Mars et al. 2012, Bzdok et al. 2013), identifying it in the middle part of the superior temporal sulcus. This result, although initially surprising, fits with earlier work on the neural basis of macaque social cognition (Perrett et al. 1992, Sallet et al. 2011) and suggest a possible relationship between human higher-order social cognition and social information processing of the non-human primate—although care should be taken that anatomical homology does not imply complete functional analogy (Krubitzer and Disbrow 2010). Employing a similar approach, Neubert and colleagues (2015) recently compared areas in the cingulate and orbitofrontal cortex between species. There has been controversy whether some of the areas often studied in human and macaque decision making neuroscience are in fact homologous (Rushworth et al. 2011, Wallis 2011). Neubert and colleagues investigated whether regions identified in macaque neurophysiological studies have anatomical homologs in the human and, vice versa, whether some of the characteristic areas identified in human decision neuroscience exist in the macaque brain, showing that between-species homologs exist for the majority of areas implicated even in high-level decision making.
In our final example, we compared the connectivity fingerprint of two human frontal polar areas that were identified as separate cortical territories in two recent, independent studies (Bludau et al. 2014, Neubert et al. 2014). We showed that these regions have significantly different connections to the major longitudinal white matter fiber bundles in the human brain and that these differences were reflected in stronger SLFIII connections for the lateral frontal pole, dove-tailing with earlier observations that human lateral frontal pole has stronger inferior parietal connections (Mars et al. 2011), and stronger UF connections for the medial frontal pole. We then applied the method to directly compare the macaque and human connectivity fingerprints. Corroborating the results obtained by Neubert et al. (2014) we did not observe a significant difference between macaque and human FPm, whereas macaque FP differed significantly from human FPl.
The methods presented here can be used to match connectivity fingerprints, and by proxy regions, between species, but also to quantify differences in connectivity fingerprints. Although these two aims might be considered opposing, this does not necessarily preclude the use of both methods on the same data. The difference between different areas (i.e., between FPm and FPl in the human) can be expected to be substantially larger than the differences in the connections of homologous areas between species (i.e., between human FPm and the macaque frontal pole). Moreover, although the matching of fingerprints requires a number of known homologous target areas between species, differences in connections to other parts of the cortex can then be investigated post hoc within the same framework. As an example of this approach, Neubert et al. (2014) matched areas of the ventrolateral prefrontal cortex between human and macaque by looking at the connectivity with parietal, frontal, and temporal pole areas and subsequently investigated differences in the strength of connections between homologous ventrolateral prefrontal areas and auditory association cortex. They reported human-macaque homologs for all areas in the cortical territory often referred to as ‘Broca’s complex’ (Xiang et al. 2010), but also showed that the auditory association cortex is preferentially interacting with these areas in the human only, whereas in the macaque its interactions with the frontal cortex are predominantly with cingulate areas.
In summary, the fingerprint matching results were quantifiable, reliable, and consistent with previous observations in the literature. All of the analyses performed here were done using either existing tools (Winkler et al. 2014) or following previously established methods (Nichols and Holmes 2007) implemented in a Matlab toolbox. The analyses presented here used resting state fMRI data and diffusion MRI data, but there is no reason in principle why this approach cannot be extended to other types of data. The connectivity matching approach can be used in a variety of scenarios, matching brain areas across species, searching for homologs, or describing differences between connections of homologous areas between species.
The main decision that an experimenter has to make to employ the connectivity fingerprint matching approach successfully is to determine which areas form the target areas, i.e., the ‘arms’ of the connectivity fingerprint. There is a potential trade-off between using more target regions to reduce selection bias and using fewer regions to minimize noise by excluding those with weak or unspecific connections, such that any included region has a substantial effect on the statistics. The selected target areas should not only be diagnostic for the seed area, but they should also be identifiable in all the species under investigation. In our previous studies comparing human and macaques we have taken a relativley conservative approach and mostly relied on a restricted number of areas in the premotor, prefrontal, and parietal cortex as target areas, rather than areas in the temporal cortex, for which homologies are less well understood (Mars et al. 2013). The choice of target areas and their homology between the species under study should be justified by the experimenter. More exploratory techniques for interrogating connectivity data (Beckmann and Smith 2004, O’Muircheartaigh et al. 2011) could help in identifying candidate target areas and future work will focus on using an iterative search to optimize target area selection across species. Over time, as more homologs are identified, new targets regions can be used to iteratively build on existing knowledge. However, it should be acknowledged that there might be parts of the cortex where this approach might be less successful if, for instance, there are few connections to known homologs or the reorganization has been too substantial.
Finally, it is informative to compare our connectivity fingerprint matching approach to other methods that have been employed to compare brain organization between species or between individuals and to discuss some potential future extensions. Graph theoretical approaches have been used to derive both summary measures of whole networks and to charaterise particular nodes within the networks (Bullmore and Sporns 2009). In a comparative context such measure have been applied to compare human, chimpanzee, and macaque brains (Li et al. 2013). These methods present a powerful and complementary approach to the one advocated here. Our approach is mostly hypothesis-driven, but—as discussed above—needs to be preceded by more explorative approaches. Graphic theoretic methods provide the possibility of more explorative comparisons as well as quantifiable network statistics. In turn, our approach could complement graph theoretical methods by allowing one to provide an anatomical explanation for observed differences and similarities in graph quantifications. Future approaches are likely to take into account not just the identity of brain areas and their possible homologs, i.e., the network nodes in graph theoretical terms, and a diagnostic set of connections, i.e., edges, but also the patterns of connections. The spatial distributions and topologies of connections can provide vital information regarding the function of parts of the cortex (Jbabdi et al. 2013) and comparing these between species provide a new avenue for comparative neuroscience. The current approach makes use of the patterns of connections of a single region, but has not yet been applied to study patterns of connections along a sheet of cortex. We note that we have here used the connectivity fingerprint matching approach to compare only two species’ brains, but the approach easily extended to larger numbers. Indeed, studying larger numbers of species is essential if one wants to analyse these data within a full phylogenetic framework (cf. Mars et al. (2014)).
The problem of dealing with high-dimensional datasets that need to summarized and quantitatively compared is of course not unique to comparative neuroscience. For instance, Buckholtz and Meyer-Lindenberg (2012) argued for the use of connectivity fingerprints as a diagnostic tool in psychopathology. Many psychopathologic syndromes are associated with alterations in functional interactions between brain areas and connectivity fingerprints present a convenient way to quantify such alterations. But the advantages of fingerprints to quickly visualise complex dataset has also been appreciated outside the study of brain connectivity, for instance, in the development of ‘cognitive profiles’ of neurological patients (Arnould et al. 2013, Njomboro et al. 2014). Some of the approaches presented in this communication will be useful for addressing these questions as well.
Acknowledgements
This work was supported by a VIDI grant of the Dutch Organization for Scientific Research NWO (452-13-015) to R.B.M., a British Academy Small Research Grant to R.B.M., a Marie Curie Intra-European Fellowship within the European Union’s 7th Framework Programme (MC-IEF-623513) to L.V., a Gustav Born Scholarship to F.X.N., a Christopher Welch Scholarship to F.X.N., and a Wellcome Trust Sir Henry Dale fellowship to J.S.
References
- Arnould A, Rochat L, Azouvi P, Van der Linden M. A multidimensional approach to apathy after traumatic brain injury. Neuropsychol Rev. 2013;23:210–233. doi: 10.1007/s11065-013-9236-3. [DOI] [PubMed] [Google Scholar]
- Bakker R, Wachtler T, Diesmann M. CoCoMac 2.0 and the future of tract-tracing databases. Front Neuroinform. 2012;6:30. doi: 10.3389/fninf.2012.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann CF, Smith SM. Probabilistic independent component analysis for functional magnetic resonance imaging. IEEE Trans Med Imaging. 2004;23:137–152. doi: 10.1109/TMI.2003.822821. [DOI] [PubMed] [Google Scholar]
- Beckmann M, Johansen-Berg H, Rushworth MF. Connectivity-based parcellation of human cingulate cortex and its relation to functional specialization. 2009;29:1175–1190. doi: 10.1523/JNEUROSCI.3328-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens TE, Johansen-Berg H, Jbabdi S, Rushworth MF, Woolrich MW. Probabilistic diffusion tractography with multiple fibre orientations: What can we gain? NeuroImage. 2007;34:144–155. doi: 10.1016/j.neuroimage.2006.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bludau S, Eickhoff SB, Mohlberg H, Caspers S, Laird AR, Fox PT, Schleicher A, Zilles K, Amunts K. Cytoarchitecture, probability maps and functions of the human frontal pole. NeuroImage. 2014;93:260–275. doi: 10.1016/j.neuroimage.2013.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckholtz JW, Meyer-Lindenberg A. Psychopathology and the human connectome: Toward a transdiagnostic model of risk for mental illness. Neuron. 2012;74:990–1004. doi: 10.1016/j.neuron.2012.06.002. [DOI] [PubMed] [Google Scholar]
- Bullmore E, Sporns O. Complex brain networks: Graph theoretical analysis of structural and functional systems. Nat Rev Neurosci. 2009;10:186–198. doi: 10.1038/nrn2575. [DOI] [PubMed] [Google Scholar]
- Bzdok D, Langner R, Schilbach L, Jakobs O, Roski C, Caspers S, Laird AR, Fox PT, Zilles K, Eickhoff SB. Characterization of the temporo-parietal junction by combining data-driven parcellation, complementary connectivity analyses, and functional decoding. NeuroImage. 2013;81:381–392. doi: 10.1016/j.neuroimage.2013.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey S, Pandya DN, Chakravarty MM, Bailey L, Petrides M, Collins DL. An MRI based average macaque monkey stereotaxic atlas and space (MNI monkey space) NeuroImage. 2011;55:1435–1442. doi: 10.1016/j.neuroimage.2011.01.040. [DOI] [PubMed] [Google Scholar]
- Hecht EE, Gutman DA, Bradley BA, Preuss TM, Stout D. Virtual dissection and comparative connectivity of the superior longitudinal fasciculus in chimpanzees and humans. NeuroImage. 2015;108:124–137. doi: 10.1016/j.neuroimage.2014.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jbabdi S, Sotiropoulos SN, Behrens TEJ. The topographic connectome. Curr Opin Neurobiol. 2013;23:207–215. doi: 10.1016/j.conb.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen-Berg H, Behrens TE, Robson MD, Drobnjak I, Rushworth MF, Brady JM, Smith SM, Higham DJ, Matthews PM. Changes in connectivity profiles define functionally distinct regions in human medial frontal cortex. Proc Natl Acad Sci USA. 2004;101:13335–13340. doi: 10.1073/pnas.0403743101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krubitzer L, Disbrow E. The evolution of parietal areas involved in hand use in primates. In: Dolins EL, Mitchell RW, editors. Spatial cognition, spatial perception. Cambridge University Press; Cambridge: 2010. pp. 357–374. [Google Scholar]
- Krubitzer L, Kaas J. The evolution of the neocortex in mammals: how is phenotypic diversity generated? Curr Opin Neurobiol. 2005;15:444–453. doi: 10.1016/j.conb.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Li L, Hu X, Preuss TM, Glasser MF, Damen FW, Qiu Y, Rilling J. Mapping putative hubs in human, chimpanzee and rhesus macaque connectomes via diffusion tractography. NeuroImage. 2013;80:462–474. doi: 10.1016/j.neuroimage.2013.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies DS, Petrides M. Distinct parietal and temporal connectivity profiles of ventrolateral frontal areas involved in language production. J Neurosci. 2013;33:16846–16852. doi: 10.1523/JNEUROSCI.2259-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mars RB, Foxley S, Jbabdi S, Sallet J, Noonan MP, Neubert FX, Andersson JL, Verhagen L, Croxson PL, Dunbar RIM, Khrapichev A, Sibson N, Miller KL, Rushworth MFS. The extreme capsule fiber complex in humans and macaques: A comparative diffusion MRI tractography study. Manuscript submitted for publication. doi: 10.1007/s00429-015-1146-0. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mars RB, Jbabdi S, Sallet J, O’Reilly JX, Croxson PL, Olivier E, Noonan MP, Bergmann C, Mitchell AS, Baxter MG, Behrens TE, Johansen-Berg H, Tomassini V, Miller KL, Rushworth MF. Diffusion-weighted imaging tractography-based parcellation of the human parietal cortex and comparison with human and macaque resting-state functional connectivity. J Neurosci. 2011;31:4087–4100. doi: 10.1523/JNEUROSCI.5102-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mars RB, Neubert FX, Verhagen L, Sallet J, Miller KL, Dunbar RIM, Barton RA. Primate comparative neuroscience using magnetic resonance imaging: Promises and challenges. Front Neurosci. 2014;8:298. doi: 10.3389/fnins.2014.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mars RB, Sallet J, Neubert FX, Rushworth MF. Connectivity profiles reveal the relationship between brain areas for social cognition in human and monkey temporoparietal cortex. Proc Natl Acad Sci USA. 2013;110:10806–10811. doi: 10.1073/pnas.1302956110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mars RB, Sallet J, Schüffelgen U, Jbabdi S, Toni I, Rushworth MF. Connectivity-based subdivisions of the human right “temporoparietal junction area”: evidence for different areas participating in different cortical networks. Cereb Cortex. 2012;22:1894–1903. doi: 10.1093/cercor/bhr268. [DOI] [PubMed] [Google Scholar]
- Neubert FX, Mars RB, Sallet J, Rushworth MFS. Connectivity reveals relationship of brain areas for reward-guided learning and decision making in human and monkey frontal cortex. Proc Natl Acad Sci USA. 2015;112:E2695–E2704. doi: 10.1073/pnas.1410767112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubert FX, Mars RB, Thomas AG, Sallet J, Rushworth MF. Comparison of human ventral frontal cortex areas for cognitive control and language with areas in monkey frontal cortex. Neuron. 2014;81:700–713. doi: 10.1016/j.neuron.2013.11.012. [DOI] [PubMed] [Google Scholar]
- Neubert FX, Verhagen L, Jbabdi S, Sallet J, Rushworth MFS, Mars RB. Comparing the projections of white matter association fibers between humans and macaques. Manuscript in preparation. in preparation. [Google Scholar]
- Nichols T, Holmes A. Non-parametric procedures. In: Friston K, Ashburner J, Kiebel S, Nichols T, Penny W, editors. Statistical parametric mapping: The analysis of functional brain images. Elsevier; Amsterdam: 2007. pp. 253–272. [Google Scholar]
- Njomboro P, Humphreys GW, Deb S. Exploring social cognition in patients with apathy following acquired brain damage. BMC Neurol. 2014;14:18. doi: 10.1186/1471-2377-14-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Muircheartaigh J, Vollmar C, Traynor C, Barker GJ, Kumari V, Symms MR, Thompson PM, Duncan JS, Koepp MJ, Richardson MP. Clustering probabilistic traograms using independent component analysis applied to the thalamus. NeuroImage. 2011:2020–2032. doi: 10.1016/j.neuroimage.2010.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly JX, Beckmann CF, Tomassini V, Ramnani N, Johansen-Berg H. Distinct and overlapping functional zones in the cerebellum defined by resting state functional connectivity. Cereb Cortex. 2010;20:953–965. doi: 10.1093/cercor/bhp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passingham RE, Stephan KE, Kotter R. The anatomical basis of functional localization in the cortex. Nat Rev Neurosci. 2002;3:606–616. doi: 10.1038/nrn893. [DOI] [PubMed] [Google Scholar]
- Perrett DI, Hietanen JK, Oram MW, Benson PJ, Rolls ET. Organization and funtions of cells responsive to faces in the temporal cortex. Phil Trans R Soc B. 1992;335:23–30. doi: 10.1098/rstb.1992.0003. [DOI] [PubMed] [Google Scholar]
- Picard N, Strick PL. Motor areas of the medial wall: A review of their location and functional activation. Cereb Cortex. 1996;6:342–353. doi: 10.1093/cercor/6.3.342. [DOI] [PubMed] [Google Scholar]
- Picard N, Strick PL. Imaging the premotor areas. Curr Opin Neurobiol. 2001;11:663–672. doi: 10.1016/s0959-4388(01)00266-5. [DOI] [PubMed] [Google Scholar]
- Preuss TM. Do rats have prefrontal cortex? The rose-woolsey-akert program reconsidered. J Cogn Neurosci. 1995;7:1–24. doi: 10.1162/jocn.1995.7.1.1. [DOI] [PubMed] [Google Scholar]
- Rilling JK, Glasser MF, Preuss TM, Ma X, Zhao T, Hu X, Behrens TEJ. The evolution of the arcuate fasciculus revealed with comparative DTI. Nat Neurosci. 2008;11:426–428. doi: 10.1038/nn2072. [DOI] [PubMed] [Google Scholar]
- Rushworth MF, Noonan MP, Boorman ED, Walton ME, Behrens TE. Frontal cortex and reward-guided learning and decision-making. Neuron. 2011;70:1054–1069. doi: 10.1016/j.neuron.2011.05.014. [DOI] [PubMed] [Google Scholar]
- Saleem KS, Logothetis . A combined MRI and histology atlas of the rhesus monkey brain in strerotaxic coordinates. Elsevier; Amsterdam: 2012. [Google Scholar]
- Sallet J, Mars RB, Noonan MP, Andersson JL, O’Reilly JX, Jbabdi S, Croxson PL, Jenkinson M, Miller KL, Rushworth MF. Social network size affects neural circuits in macaques. Science. 2011;334:697–700. doi: 10.1126/science.1210027. [DOI] [PubMed] [Google Scholar]
- Sallet J, Mars RB, Noonan MP, Neubert FX, Jbabdi S, O’Reilly JX, Filippini N, Thomas AG, Rushworth MF. The organization of dorsal frontal cortex in humans and macaques. J Neurosci. 2013;33:12255–12274. doi: 10.1523/JNEUROSCI.5108-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TE, Johansen-Berg H, Bannister PR, De Luca M, Drobnjak I, Flitney DE, Niazy RK, Saunders J, Vickers J, Zhang Y, De Stefano N, Brady JM, Matthews PM. Advances in functional and structural MR image analysis and implementation as FSL. NeuroImage. 2004;23(Suppl 1):S208–219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- Striedter GF, Belgard TG, Chen CC, Davis FP, Finlay BL, Gunturkun O, Hale ME, Harris JA, Hecht EE, Hof PR, Hofmann HA, Holland LZ, Iwaniuk AN, Jarvis ED, Karten HJ, Katz PS, Kristan WB, Macagno ER, Mitra PP, Moroz LL, Preuss TM, Ragsdale CW, Sherwood CC, Stevens CF, Stuttgen MC, Tsumoto T, Wilczynski W. NSF workshop report: discovering general principles of nervous system organization by comparing brain maps across species. Brain Behav Evol. 2014;83:1–8. doi: 10.1159/000360152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiebaut de Schotten M, Dell’Acqua F, Valabregue R, Catani M. Monkey to human comparative anatomy of the frontal lobe association tracts. Cortex. 2012;48:82–96. doi: 10.1016/j.cortex.2011.10.001. [DOI] [PubMed] [Google Scholar]
- Tomassini V, Jbabdi S, Klein JC, Behrens TE, Pozzilli C, Matthews PM, Rushworth MF, Johansen-Berg H. Diffusion-weighted imaging tractography-based parcellation of the human lateral premotor cortex identifies dorsal and ventral subregions with anatomical and functional specializations. J Neurosci. 2007;27:10259–10269. doi: 10.1523/JNEUROSCI.2144-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis JD. Cross-species studies of orbitofrontal cortex and value-based decision-making. Nat Neurosci. 2011;15:13–19. doi: 10.1038/nn.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler AM, Ridgway GR, Webster MA, Smith SM, Nichols TE. Permutation inference for the general linear model. NeuroImage. 2014;92:381–397. doi: 10.1016/j.neuroimage.2014.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang HD, Fonteijn HM, Norris DG, Hagoort P. Topographical functional connectivity pattern in the perisylvian language networks. Cereb Cortex. 2010;20:549–560. doi: 10.1093/cercor/bhp119. [DOI] [PubMed] [Google Scholar]