Abstract
The binding of the ruthenium-based anticancer drug candidates KP1019, NAMI-A, and RAPTA-T towards different double-stranded oligonucleotides was probed by electrospray ionisation-mass spectrometry (ESI-MS) and compared to the widely used platinum-based chemotherapeutics cisplatin, carboplatin and oxaliplatin. It was found that the extent of adduct formation decreased in the following order: cisplatin > oxaliplatin > NAMI-A > RAPTA-T > carboplatin > KP1019. In addition to the characterisation of the adducts formed with the DNA models, the binding sites of the metallodrugs on the oligonucleotides were elucidated employing top-down tandem mass spectrometry and found to be similar for all the metallodrugs studied, irrespective of the sequence of the oligonucleotide. A strong preference for guanine residues was established.
Keywords: Ruthenium, Platinum, Anticancer, Oligonucleotides, Mass Spectrometry
Introduction
Following the discovery that cisplatin inhibits cell division in bacteria by Rosenberg in the 1960s, and its subsequent identification as a potent anticancer agent, it quickly became, and still remains, the most widely used chemotherapeutic drug for the treatment of a wide range of malignancies. [1, 2] Although cisplatin is extremely effective against several cancers, e.g. testicular and ovarian cancers, its application is also accompanied by severe side effects such as nausea, vomiting and hearing loss, and many tumors show intrinsic or acquired resistance against the drug. These facts led to the development and clinical approval of platinum-based derivatives, i.e. carboplatin and oxaliplatin (Figure 1),[3] which mainly differ from cisplatin in their rate of aquation and therefore their overall reactivity. More recently, drugs based on metals other than platinum, such as ruthenium, gallium, titanium, and gold have been developed to treat cisplatin-resistant cancers,[4] and it is thought that these drugs exhibit a different mode of action compared to platinum-based drugs and could therefore widen the range of cancers that can be treated.
Figure 1.
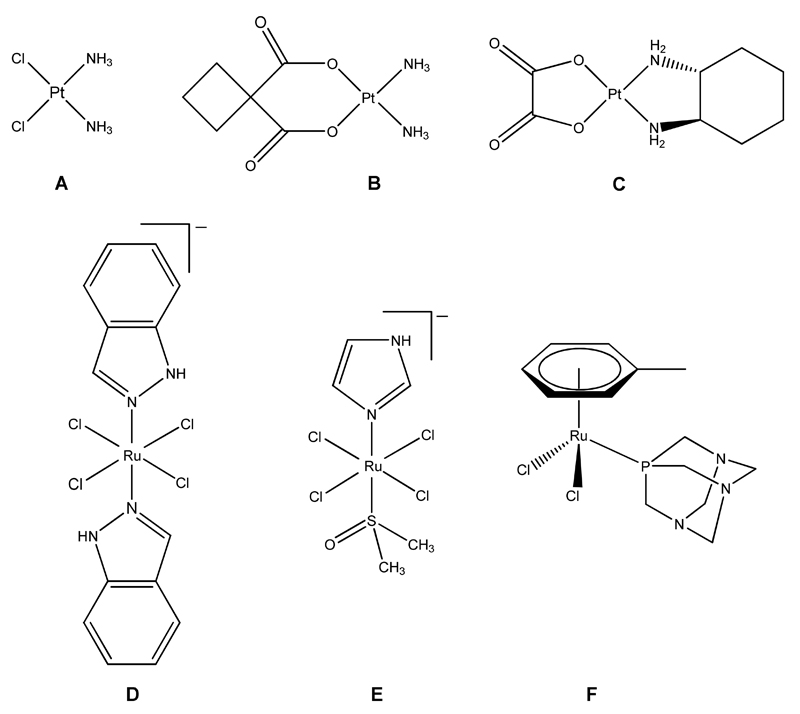
Platinum-based pharmaceuticals cisplatin (A), carboplatin (B) and oxaliplatin (C) and the ruthenium-based anticancer drug candidates KP1019 (D), NAMI-A (E) and RAPTA-T (F). Counter-ions have been omitted for clarity.
Indazolium trans-[tetrachloridobis(1H-indazole)ruthenate(III)] (KP1019), imidazolium trans-[tetrachlorido(1H-imidazole)(S-dimethylsulfoxide)ruthenate(III)] (NAMI-A), and [dichlorido(η6-toluene)(PTA)ruthenium(II)], where PTA = 1,3,5-triaza-7-phosphaadamantane (RAPTA-T), all of which incorporate ruthenium as the central atom, are among the most promising candidates (Figure 1).[5] KP1019 and NAMI-A have successfully completed phase 1 clinical trials; NAMI-A entered phase 2 clinical trials in 2008 (in combination with a cytotoxic drug), and KP1019 has followed in a modified formulation in 2009. NAMI-A and RAPTA-T seem to be highly effective against metastases, whereas KP1019 shows good activity towards primary tumours. For all of the compounds low general toxicity combined with excellent clearance rates was observed.
The antiproliferative activity of platinum-based anticancer agents relies on interactions with DNA, binding mainly to adjacent guanine moieties, which leads to kinks in the molecular structure and ultimately to inducing apoptosis.[6] Interactions with proteins, especially in the case of ruthenium, have also been suggested to play an important role, although direct interactions with DNA cannot be excluded.[7, 8] Over the years, virtually all the available bioanalytical/biophysical techniques including spectroscopy, chromatography, electrophoresis, X-ray diffraction analysis, isotopic labeling and mass spectrometry, as well as hyphenated techniques, have been applied to the study of metallodrug-(model) DNA interactions.[9] Following the development of ESI- and MALDI-MS techniques in the late 1980s, they quickly became invaluable for the analysis of nucleic acids and oligonucleotides,[10] and therefore also for the analysis of metal complex–DNA interactions.[11,12] Although ESI-MS analysis of duplex DNA–drug interactions have been reported on several occasions,[13–15] even the most recent studies focused primarily on the adducts formed with single strands and the mechanisms underlying dissociation in the gas phase.[16, 17] Cisplatin forms mainly DNA intrastrand-crosslinks with nucleobases,[18] but the formation of interstrand-crosslinks is also possible, and they might contribute significantly to the drug’s activity with enhanced repair being one of the causes for clinical acquired resistance.[19, 20] While ruthenium drugs have also been shown to bind to nucleotides and DNA,[21] adduct formation proceeds more slowly compared to cisplatin, therefore substantiating the hypothesis that DNA might not be the (only) target for this compound class.
In this paper, we describe a mass spectrometry-based technique for the analysis of adducts formated between duplex DNA and anticancer metallodrugs. Using a multistage mass spectrometric top-down approach the binding sites of the metal-based drugs have been elucidated.
Material and Methods
ESI-MS for characterisation of duplex DNA-metallodrug interactions was carried out on an Ultima II q-TOF mass spectrometer (Waters, Manchester, UK) operated in positive and negative ion mode. The instrument was calibrated daily using a 0.01% phosphoric acid solution in 50% acetonitrile. Determination of the extent of adduct formation was carried out by monitoring the 5+ and 6+ charge states of the duplex and the corresponding adducts and integrating their peak areas. Samples containing an effective complex/duplex ratio of 3:1 were freshly prepared in 20 mM ammonium acetate buffer at pH 7.4, and incubated in Eppendorf tubes for up to 120 h at 37 °C (duplex concentration 25 µM). Aliquots of 20 µL were taken after 1 h, 3 h, 6 h, 24 h and 120 h and stored at -20 °C until analysis. The samples were mixed with 100 mM ammonium acetate in MeOH at a ratio of 1:1 immediately prior to analysis to enhance spray formation and stability of the duplex DNA in the gas phase. Data acquisition and analysis were carried out using the MassLynx software bundle (Waters).
Fragmentation experiments for binding site elucidation were carried out on a linear ion trap MS ( LTQ XL, ThermoFisher Scientific, Bremen, Germany) specifically in negative ion mode due to its capability to perform multiple stage MS/MS experiments. For identification of unmodified fragments the web-based Mongo Oligo Mass calculator v2.06 (http://library.med.utah.edu/masspec/mongo.htm) was used.[22] Samples containing an effective complex:duplex ratio of 1:1 were prepared in ammonium acetate buffer at pH 7.4 and incubated at 37 °C for up to 72 h. For analysis, the samples were diluted 10-fold with a solution containing MeOH:water:n-propanol in a ratio of 65:20:5 and placed into a 96-well plate in an Advion TriVersa nano-ESI robot (AdvionBiosciences, Ithaca, NY, USA) equipped with a 5.5 μm-nozzle chip. The ESI robot was controlled with ChipSoft v7.2.0 software employing a gas pressure of 0.45 psi and a spray voltage of 1.7 kV. The mass spectrometer was operated in enhanced resolution mode and for fragmentation experiments an isolation width of 3.0 m/z and a relative collision energy of 35% were used. The Xcalibur software bundle was utilised for data acquisition and analysis (ThermoFisher Scientific).
MALDI-MS experiments were carried out in linear mode on an Axima CFR Plus (Kratos/Shimadzu Biotech, Kyoto, Japan) MALDI-TOF instrument. MALDI matrices (nicotinic acid, anthranilic acid, trihydroxyacetophenone, 3-hydroxypicolinic acid, picolinic acid) were obtained from Sigma-Aldrich (Buchs, Switzerland).
HPLC purified synthetic oligonucleotides were purchased from Microsynth (Balgach, Switzerland) and DNA Technology A/S (Risskov, Denmark). Duplex DNA was prepared by mixing 200 μM of the corresponding single strands in 100 mM aqueous ammonium acetate, heating at 70°C for 15 min and then annealing by slowly cooling to 4°C. Annealing was confirmed by gel electrophoresis. Ammonium acetate (>99.99%) and cisplatin were purchased from Sigma-Aldrich (Switzerland), and HPLC-grade solvents (water, acetonitrile, n-propanol, methanol) from Acros (Beel, Belgium). Oxaliplatin was purchased from Sequoia Research Products (Pangbourne, UK), KP1019, carboplatin, RAPTA-T and NAMI-A were synthesized as described elsewhere.[23–25]
Results and Discussion
Optimization of Conditions
In order to establish the optimum technique and conditions for the analysis of the metallodrug-modified duplex DNA, both ESI- and MALDI-MS were evaluated for their suitability. The first successful analysis of intact double stranded DNA by MALDI-MS was reported in 1995 [26] – but due to the considerable dissociation of the duplex into the single strands for short oligonucleotides (< 20mer), the approach has mainly been used to study longer DNA fragments.[27] Furthermore, MALDI-MS has also been used for analysis of interactions between single-stranded oligonucleotides with ruthenium drugs.[28]
For all three double strands (see Table 1 for sequences and molecular weights), MALDI-MS experiments with commonly used matrices containing either nicotinic acid combined with anthranilic acid,[29] 6-aza-thiothymine (ATT),[27, 30] trihydroxyacetophenone (THAP) [28] or a combination of 3-hydroxypicolinic (HPA) with picolinic acid (PA)[31] were carried out. Independent of the matrix composition, the ionised species consisted mainly of the single strands in both positive and negative ionisation modes (see supporting information). Also note that analysis could only be carried out in linear mode as additional ion flight time in the reflectron mode induced fragmentation of the oligonucleotides. Consequently, no further experiments probing the adducts formed between the duplexes and metallodrugs were carried out as dissociation of the double strands had to be expected as well for these systems.
Table 1.
Sequences and masses of the complementary oligonucleotides.
| Sequence | Monoisotopic Mass (Da) | |||
|---|---|---|---|---|
| DS1 | SS1a | 5’-d(GTATTGGCACGTA)-3’ | 3987.7 | 7904.4 |
| SS1b | 5’-d(TACGTGCCAATAC)-3’ | 3916.6 | ||
| DS2 | SS2a | 5’-d(TACGTGCCAATAC)-3’ | 4003.7 | 7905.4 |
| SS2b | 5’-d(TACACACCGGTAC)-3’ | 3901.7 | ||
| DS3 | SS3a | 5’-d(ATATTATGCTTAATTA)-3’ | 4867.8 | 9753.7 |
| SS3b | 5’-d(TAATTAAGCATAATAT)-3’ | 4885.9 | ||
Although it seems logical to carry out the analysis of oligonucleotides in negative ion mode due to their negatively charged phosphate backbone, good results for intact duplex DNA–drug interaction can also be obtained in the positive mode by ESI-MS.[14–15] In this case, dissociation of the double strand is probably hindered by the presence of ammonium ions, which may stabilize the duplex during ion transfer from solution to the gas phase in the process of electrospray. Still, the majority of publications dealing with ESI-MS of duplex DNA utilizes negative ion mode with high salt concentrations in the spraying solution [13], which has to be considered the more widely applicable instrument setting.
In order to establish the optimal conditions for our application, ESI-MS spectra of the oligonucleotides and metallodrugs were recorded in positive and negative ion mode with varying ammonium acetate concentrations between 0 and 100 mM and with methanol contents of up to 50%. Surprisingly, most satisfactory results were obtained in positive ion mode with a spraying solution containing 50 mM ammonium acetate in 50% MeOH, especially when analyzing metallodrug-olignucleotide adducts (note that positive charges are introduced by the metallodrug-fragments). Although the signal-to-noise ratio is poorer in positive ion mode, due to adduct formation between the oligonucleotides and ammonium ions as well as with sodium and potassium ions leading to additional peaks in the spectra, increased stabilisation of the duplexes was observed compared to the negative ion mode (see supporting information), especially when samples containing the oligonucleotides and metallodrugs are analysed. Therefore, this solvent system was chosen for all further measurements which focused on the analysis of intact double strands. Negative ion mode was selected to identify the preferred strand for binding of the metallodrugs incubated with duplex DNA in accordance with a study reported earlier.[16]
Interactions with dsDNA
Platinum Complexes
For platinum-based compounds, the main focus of attention is on their interactions with DNA since it is considered to be the ultimate target inside the cancer cell. A multitude of studies on this topic has been carried out over the years, although enzymes and proteins have also been shown to play a role in the inhibition of transcription and consequently in the induction of apoptosis.[32] The three platinum compounds were incubated with the selected oligonucleotides for up to 120 h under simulated physiological conditions. Following incubation, the samples were analyzed by ESI-MS and the obtained spectra compared to the pure duplexes.
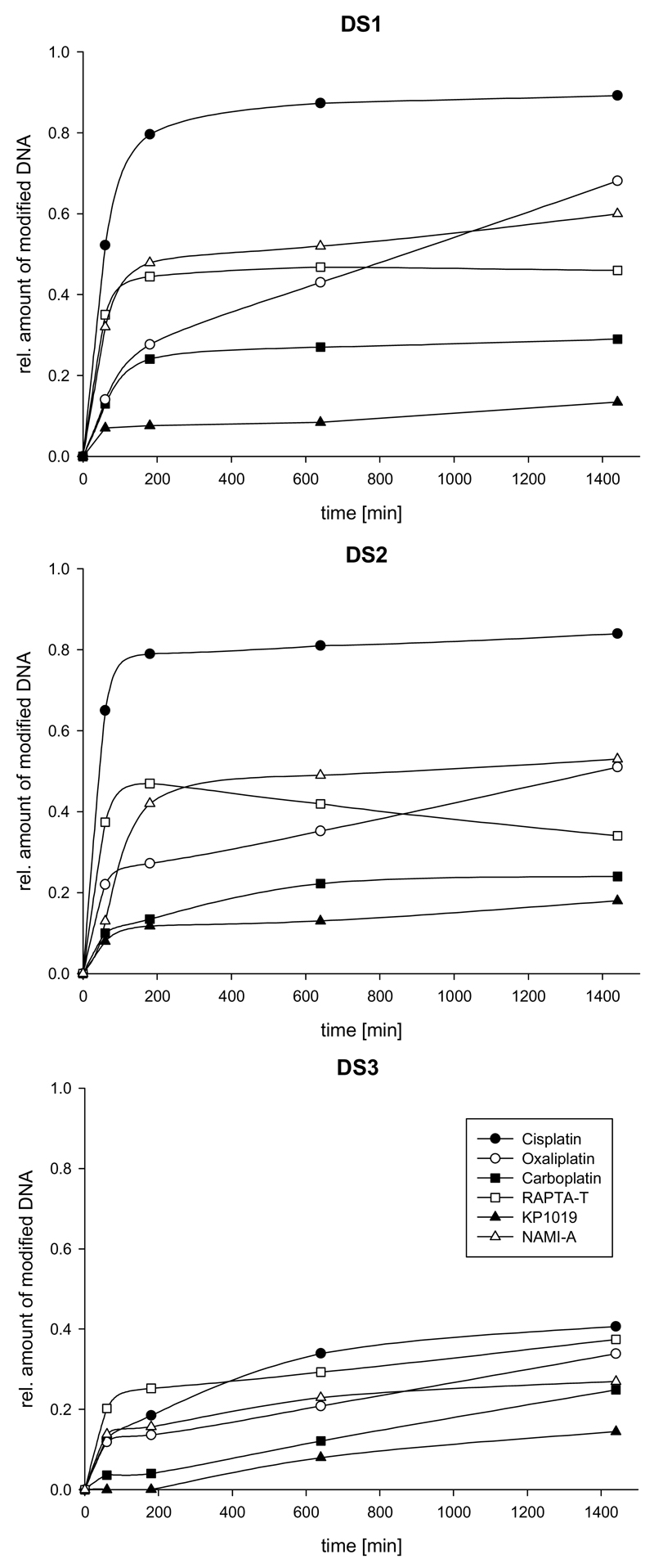
For samples containing the duplexes with multiple guanines (G; it is known that binding occurs mainly through interaction with the nucleophilic N7 of this nucleotide), i.e. DS1 and DS2, a fast decrease of the peaks corresponding to the unmodified oligonucleotides was observed when incubated with cisplatin. This was accompanied by an increase in the relative intensity of the peaks corresponding to [DS+Pt(NH3)2] and [DS+2Pt(NH3)2] species. If the number of guanines is reduced to one in each strand (DS3), consequently also rendering intrastrand G-G crosslinking impossible, only about 40% of the oligonucleotide is modified after 24 h of incubation, whereas this number is more than doubled for DS1 and DS2 with more than one G residue (Table 2). Note that the extent of modification as shown in Figure 4 and Table 2 represents only an estimate as adduct formation with the metal complexes, which introduce additional positive charges, might lead to different ionization characteristics compared to the unmodified oligonucleotides. However, an approximation of the relative reactivity of the different compounds can be derived from the data.
Table 2.
Assignment of adducts formed between oligonucleotides and the studied metallodrugs as well as the estimated percentage of oligonucleotide modified after an incubation period of 24 h.
| Drug | Adducts | Average Mass (Da) | Relative amount of modified duplexes after 24 h (%) | ||
|---|---|---|---|---|---|
| DS1 | DS2 | DS3 | |||
| Cisplatin | Pt(NH3)2 | 229.1 | 89 | 84 | 40 |
| Pt(NH3)2Cl | 264.6 | ||||
| Carboplatin | Pt(NH3)2 | 229.1 | 29 | 24 | 24 |
| Oxaliplatin | Pt(DACH) | 309.3 | 68 | 51 | 34 |
| RAPTA-T | Ru(PTA) | 258.2 | 46 | 37 | 36 |
| Ru(PTA)(T) | 350.4 | ||||
| Ru(PTA)(T)Cl | 385.8 | ||||
| KP1019 | Ru | 101.9 | 13 | 18 | 15 |
| Ru(Ind) | 219.2 | ||||
| Ru(Ind)(H2O) | 237.2 | ||||
| Ru(Ind)(H2O)2 | 255.2 | ||||
| Ru(Ind)2 | 337.3 | ||||
| NAMI-A | Ru | 101.9 | 60 | 53 | 27 |
| Ru(Im) | 169.1 | ||||
| Ru(Im)(H2O) | 187.1 | ||||
| Ru(Im)(H2O)2 | 205.1 | ||||
Figure 4.
Estimated extent of binding of the studied metallodrugs towards duplex DNA. Top: DS1. Middle: DS2. Bottom: DS3.
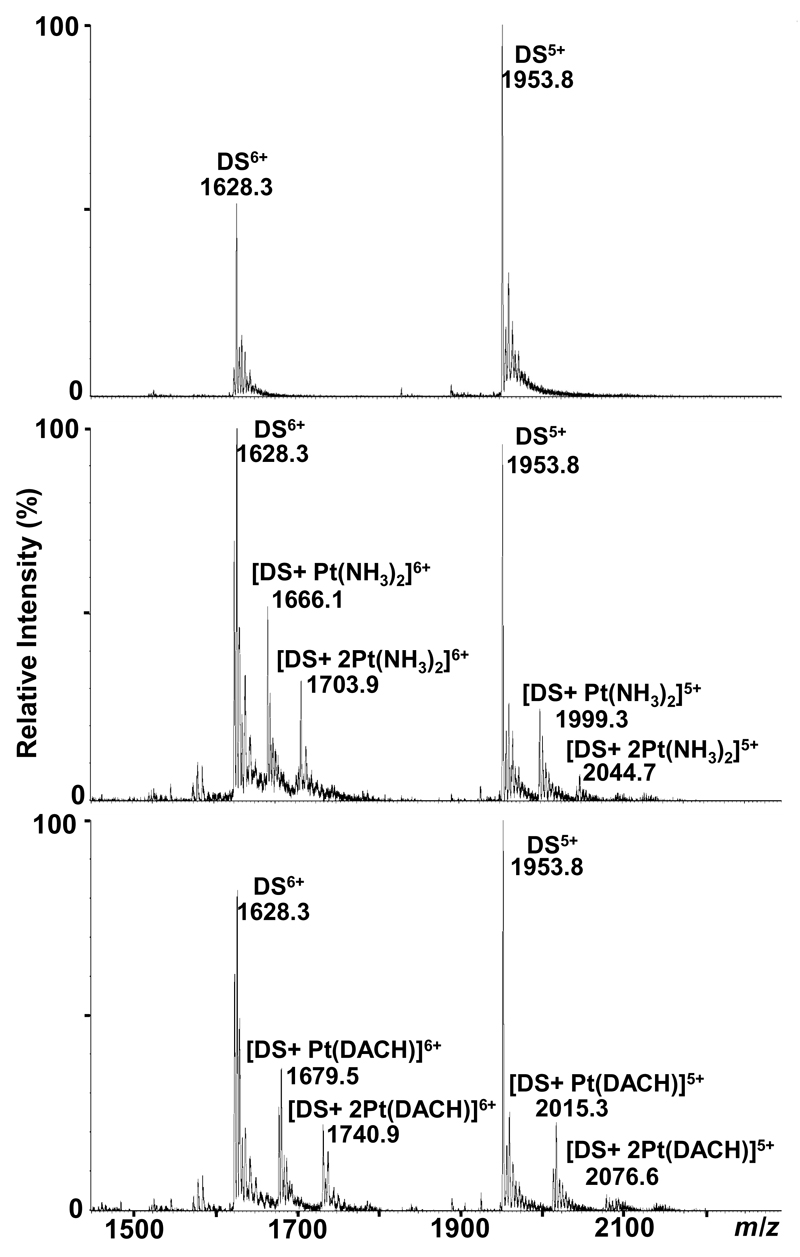
Interestingly, even in the case of DS3 (most prominent peaks correspond to the 6+ and 5+ species at 1628.3 and 1953.8 m/z, respectively; Figure 2), the major adduct is a bifunctional oligo-[Pt(NH3)2] species involving coordination of the metal center to the duplex as, for example, evidenced by the peak at 1999.3 m/z corresponding to [DS3+Pt(NH3)2]5+ (Figure 2). This infers that either adenosine serves as a second binding partner to form G-A or G-X-A (X=C, T) crosslinks, or that interstrand crosslinking between the two guanine residues on each strand takes place. Indeed, both of processes have been shown to be relevant in the biological environment.[33] When recording spectra in negative mode, the majority of signals is attributable to single stranded oligonucleotides, indicating that the relative amount of interstrand crosslinks lies below 5% as covalent bridging via the platinum moiety should lead to a stable double stranded species. Also bisadducts detected in positive ion mode ([DS3+2Pt(NH3)2]5+ at 2044.7 m/z), indicate binding of one metallodrug moiety to both of the strands.
Figure 2.
ESI TOF mass spectra showing the interactions between DS3 and platinum-based chemotherapeutics. Top: unmodified DS3. Middle: DS3+cisplatin (incubation with carboplatin yields the same adducts albeit in lower relative intensity). Bottom: DS3+oxaliplatin.
Results comparable to cisplatin were obtained for carboplatin and oxaliplatin, although adduct formation proceeds more slowly compared to cisplatin (Figure 4, Table 2). This correlates to the slower aquation kinetics of carboplatin and oxaliplatin relative to that of cisplatin (the aquated species are believed to be essential intermediates during the reaction).[33] The formation of adducts occurs after exchange of the dicarboxylato and oxalato ligands, respectively, resulting in mainly bifunctional crosslinks, as evidenced e.g. by the peak at 2015.3 m/z corresponding to [DS3+Pt(DACH)]5+ (DACH=1,2-diaminocyclohexane) for oxaliplatin in Figure 2; also the formation of a bisadduct similar to cisplatin can be observed ([DS3+2Pt(DACH)]5+ at 2076.6 m/z); for carboplatin, the spectrum resembles that of cisplatin (although with lower intensities for the metallodrug adducts relative to the free unmodified oligonucleotides), with a major peak at 1999.3 m/z corresponding to [DS3+Pt(NH3)2]5+ (not shown). For reactions with DS3, no significant formation of interstrand crosslinks was observed, as evidenced by spectra recorded in negative ion mode. A list of metallodrug-species bound to the oligonucleotides is given in Table 2.
Even though dissociation of the double strands in the negative ion mode seems disadvantageous, it provides information on the distribution of the metal complexes between the single strands. For DS1, a slight preference for binding towards SS1a was observed, for DS2 a preference for SS2a, and for DS3, adduct formation to both strands proceeds at approximately the same rate. These preferences can be explained by the higher number of G residues in the respective strands, and additionally, for example, by the presence of the advantageous sequence TGGC in SS1a compared to GTG in SS1b.[34]
Ruthenium Complexes
The reactivity of ruthenium compounds towards nucleotides has already been demonstrated,[35] although a different mode of action as compared to the platinum drugs including protein interactions has been proposed by several authors.[36] Such interactions might explain their activity towards cisplatin-resistant cancer cells as well as the antimetastatic properties of NAMI-A and RAPTA-T.[37, 38]
Analogous to the experiments with the platinum compounds, all measurements were carried out in both positive and negative ion mode. When comparing the spectra of the unmodified to the ruthenated oligonucleotides, it can be deduced from the differences in mass that the compounds have to undergo ligand exchange before or on binding to the oligonucleotides. For RAPTA-T, the major adducts were assigned to [Ru(PTA)]- and [Ru(PTA)(T)]-containing species. For NAMI-A, the most prominent adduct peaks correspond to the oligonucleotides with the [Ru(Im)]-moiety attached, whereas [Ru(Ind)]- and [Ru(Ind)2]-adducts are found for samples incubated with KP1019. As a series of aquated species are detected for NAMI-A and KP1019, which results in a higher number of possible adducts, data analysis is complicated compared to the samples with platinum compounds. The apparently multidentate nature of the binding to DNA severely changes its secondary structure, leading to dissociation of the duplex into its single strands upon binding of the ruthenium-based drugs. Consequently, almost no adduct formation between intact duplex DNA and the drugs could be detected and negative ion mode provided similar information to positive ion mode, although at higher signal-to-noise ratios (see supporting information). Additionally, the nature of the adducts is confirmed by measurements in negative ion mode as loss of the chlorido ligands could otherwise be interpreted as an artifact during ionisation in positive ion mode. Furthermore, data on the preferred strand is gained due to the dissociation of the double strand, showing a higher extent of adduct formation for single strands SS1a and SS2a similar to the platinum compounds. Again, no difference was observed for SS3a and SS3b. It was expected that RAPTA-T would form the greatest number of adducts since the ruthenium centre in this compound is in the more reactive +II oxidation state compared to +III in KP1019 and NAMI-A.[7] Surprisingly, the double strands are modified to the greatest extent by NAMI-A, whereas KP1019 only shows very little interaction with the oligonucleotides (Figure 4, Table 2). These observations might be explained by the hydrolysis kinetics of the compounds; according to previous studies,[39, 40] NAMI-A is aquated more rapidly than RAPTA compounds.
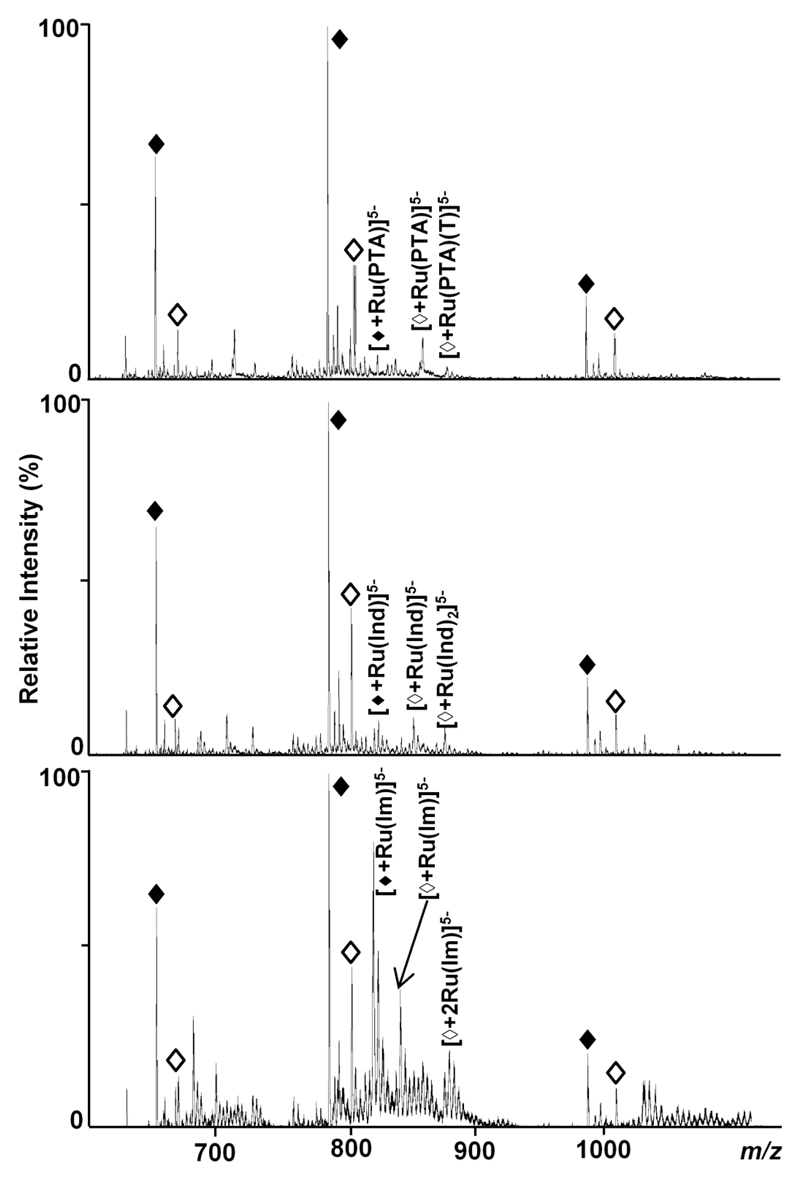
As depicted in Figure 3 for DS2, the most intense ions are found for the -5 charge state, with the monoisotopic peaks at 799.8 and 779.4 m/z assigned to unmodified SS2a and SS2b, respectively, when operating the instrument in negative ion mode. Therefore, the following assignments correspond to the -5 charge state. Minor peaks stemming from adduct formation with sodium, potassium and ammonium ions are also present in the spectra but have not been labelled for clarity. For RAPTA-T, the two chlorido ligands are lost on binding, but during prolonged incubation also the aromatic toluene ring is displaced, as noted previously for other RAPTA compounds,[41, 42] resulting in peaks at m/z values of 832.4 and 850.9 for [SS2b+Ru(PTA)]5- and [SS2b+Ru(PTA)(T)]5- (Figure 3, top). Similarly, peaks with slightly higher relative intensities at 852.9 and 871.3 m/z were assigned to [SS2a+Ru(PTA)]5- and [SS2a+Ru(PTA)(T)]5-, respectively. Adducts containing the [Ru(PTA)(T)Cl]--moiety are only present at very low intensities for incubation times longer than 6h.
Figure 3.
Negative ion mode ESI TOF mass spectra showing the interactions between DS2 and ruthenium-based compounds after an incubation period of 6 h. Dissociation into the single strands SS2a (◊) and SS2b (♦) (charge states -6 to -4) takes place under the selected conditions and the different extent of adduct formation depending on the strand and the compound is demonstrated (top: RAPTA-T, middle: KP1019, bottom: NAMI-A). Only the most intense adduct peaks for each compound are labelled for the -5 charge state.
NAMI-A undergoes a more complicated series of hydrolysis reactions prior to the formation of stable adducts with biomolecules. Indeed, in a previous study characterising the interactions of NAMI-A with cytochrome c, it was shown that the chlorido ligands have to be substituted (partially by aqua ligands) in order to form a stable adduct.[43] Consequently, the peaks in Figure 3 (bottom spectrum) at 814.6, 818.2 and 821.8 m/z have been assigned to [SS2b+Ru(Im)]5-, [SS2b+Ru(Im)(H2O)]5- and [SS2b+Ru(Im)(H2O)2]5-, respectively, whereas peaks at 832.6, 836.2 and 839.8 m/z correspond to the similar adducts formed with SS2a. Furthermore, peaks at 868.8, 872.4, 876.0 and 879.6 m/z indicate binding of even two partially aquated Ru(Im) containing moieties to SS2a, which in general seemed to be the preferred binding partner due to the higher number of guanosine residues.
Although the rate of aquation of the chlorido ligands of KP1019 is comparable to NAMI-A, there is a second nitrogen-containing heterocycle coordinated to ruthenium which is more strongly coordinating than the DMSO ligand in NAMI-A. As these nitrogen donor ligands are in a trans position to each other, binding to the oligonucleotide could be sterically hindered, therefore explaining the lower extent of adduct formation. The major peaks in Figure 3 for the KP1019-DS2 interaction (middle spectrum) where assigned as follows: 824.5 m/z corresponds to [SS2b+Ru(Ind)]5-, 848.6 m/z to [SS2b+Ru(Ind)2]5-, 845.0 m/z to [SS2a+Ru(Ind)]5- and 868.5 m/z to [SS2a+Ru(Ind)2]5-. Also note that there is a series of minor peaks corresponding to the aquated species of the Ru(Ind) moiety.
For both KP1019 and NAMI-A, even adducts with only the ruthenium centre (loss of all original ligands) coordinated to the oligonucleotides were detected after incubation times of 5 days, whereas the PTA ligand stays coordinated to ruthenium in RAPTA-T. This result is rather surprising as loss of the nitrogen-containing ligands in KP1019 and NAMI-A has not been observed in earlier protein binding experiments; in our study, the ligand exchange could be induced sterically due to the flexibility of the oligonucleotide. A list of metallodrug-species found bound to the oligonucleotides is given in Table 2.
Binding Site Determination
In order to establish the binding site of the metallodrugs, i.e. the metalated nucleobases, tandem mass spectrometry experiments using collision induced dissociation (CID) were carried out. CID of double stranded DNA yields mainly the intact single strands, and their further fragmentation in positive ion mode results only in large oligonucleotide fragments (the phosphate groups on the nucleotides induce low ionisation efficiency for short sequences in positive ion mode) that give little structural information. Therefore, fragmentation experiments were carried out in negative ion mode on an ion trap instrument capable of MSn analysis. Due to the characteristic isotopic pattern, induced by coordination of the metals to the oligonucleotides, metal-free species could easily be distinguished from the modified oligonucleotides.
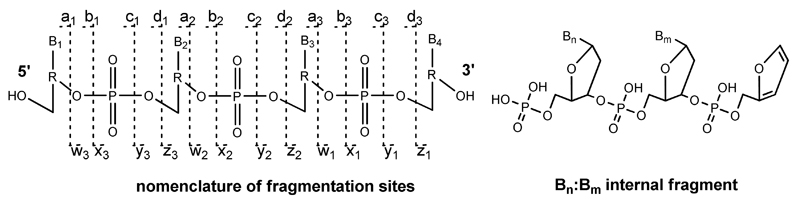
The commonly applied nomenclature for oligonucleotide fragments is summarised in Figure 5.[44] For n-type fragments, Bn (B is one of the nucleobases A,C,G, or T) is usually lost by an elimination reaction prior to strand breaking which is caused by a second elimination reaction, leading to a furan ring system.[44] Internal fragments, resulting from two strand breaks at the a/w-site, possess a phosphate group at their 5' terminus, whereas the 3' terminus carries a furan system.
Figure 5.
Nomenclature of oligonucleotide fragments observed by tandem MS; a-d fragments correspond to fragments with an intact 5' terminus and w–z-type fragments have an intact 3′ terminus. Internal fragments resulting from double fragmentation usually occur at the a/w site.
Platinum Complexes
The interaction of cisplatin with DS1 and DS2 was already characterized by ESI-MS/MS earlier, revealing GG and GTG as the major binding sites.[16] In this study, we wanted to verify that the preference towards guanine residues is maintained even if adenine and thymine are present in large excess as in DS3, and additionally, establish the preferred binding sites for the platinum-based pharmaceuticals oxaliplatin and carboplatin. Previous studies have shown that both oxaliplatin and carboplatin form adducts with DNA comparable to those of cisplatin, showing also a strong preference for guanosine as the major binding partner.[46, 47]
For this purpose, the ions of the Pt(NH3)2 (for cisplatin and carboplatin) and Pt(DACH) (for oxaliplatin) adducts of the single stranded oligonucleotides were isolated and subjected to CID. As expected, binding towards the oligonucleotides takes place mainly at guanine containing residues, even in the case of DS3. The formation of complementary modified and unmodified (an-Bn), wn and internal (Bn:Bm) fragments comparable to peptide fragmentation experiments in proteomics gives the exact metal binding site. As the coordination of the metal might result in fragmentation mechanisms additional to standard single fragmentation pathways, i.e. a, b, c, d- and w, x, y, z-fragments as well as internal a/w type fragments, not all the peaks could be assigned.
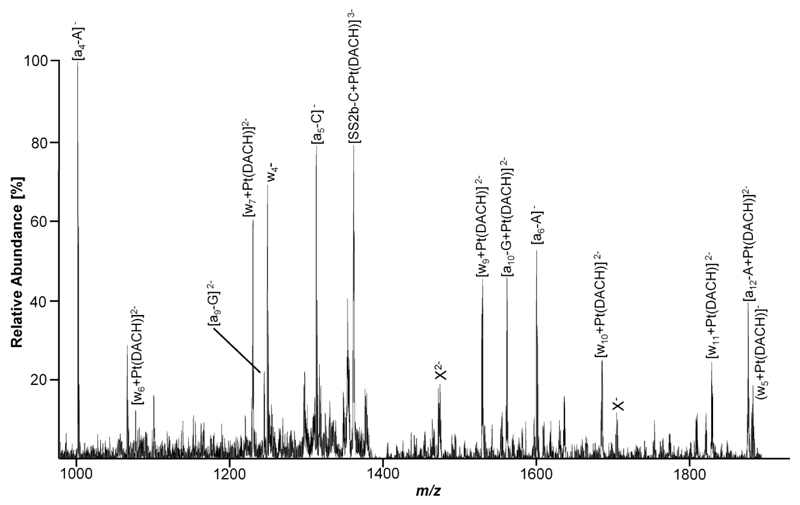
In Figure 6, an expanded segment of a representative CID spectrum for the binding site determination on DS2 incubated with a metallodrug (in this case oxaliplatin) is depicted. In this case, the peak corresponding to [SS2b+Pt(DACH)]3- at 1402.2 m/z was isolated in the ion trap of the instrument and subjected to fragmentation. The major peaks in the resulting CID spectrum correspond to [SS2b-C+Pt(DACH)]3- at 1365.0 m/z, as well as the non-platinum containing fragments [a4-A]- and [a5-C]- at 1003.1 and 1316.0 m/z, respectively. It can be seen that all (an-Bn)-type fragments from 5' terminus up to the guanine residue in position 9 remain unmodified (as evidenced, for example, by the non-platinum peak at 1247.6 m/z which was assigned to [a9-G]2-), whereas all (an-Bn) fragments from there on exhibit an isotopic pattern characteristic for platinum. This can be attributed to the attachment of the Pt(DACH) moiety, which leads to a mass increase of 307.1 Da (neutral species). Consequently, the peak at 1566.0 m/z can be identified as [a10-G+Pt(DACH)]2-. Complementary, all wn-type fragments from the 3' terminus up to w4 (1252.1 m/z) remain unmodified, whereas all further wn-fragments (which include the G9 residue) are modified by Pt(DACH). This is evidenced by, for example, the singly charged peak at 1888.1 m/z with an isotopic pattern characteristic for platinum which can be assigned to (w5+Pt(DACH)]-. By combining the information gained from the modified and unmodified fragments from both the 3' and 5' terminus, the G9 residue can be unambiguously identified as the nucleotide mainly involved in the Pt-binding. Note that not all detected fragment ions are contained in the expanded segments of the spectrum (Figure 6) for clarity. Calculated and experimental m/z values for the peaks in Figure 6 are listed in Table 3.
Figure 6.
An expanded segment of a LTQ CID mass spectrum of SS2b modified by oxaliplatin in the range from 950 to 1900 m/z that contains the majority of platinum containing fragments. Unidentified fragments exhibiting an isotopic distribution characteristic for platinum are labelled X.
Table 3.
Calculated and experimental m/z values for fragment ions found in Figure 6 (monoisotopic peaks).
| Ion | Calculated (m/z) | Experimental (m/z) |
|---|---|---|
| [a4-A]- | 1003.2 | 1003.1 |
| [a5-C]- | 1316.2 | 1316.1 |
| [a6-A]- | 1605.3 | 1605.0 |
| [a9-G]2- | 1247.7 | 1247.6 |
| [a10+Pt(DACH)]2- | 1565.8 | 1566.0 |
| [a12+Pt(DACH)]2- | 1882.3 | 1882.1 |
| w4- | 1252.2 | 1252.1 |
| [w5+Pt(DACH)]- | 1888.4 | 1888.1 |
| [w6+Pt(DACH)]2- | 1088.2 | 1088.5 |
| [w7+Pt(DACH)]2- | 1232.7 | 1232.6 |
| [w9+Pt(DACH)]2- | 1533.8 | 1533.6 |
| [w10+Pt(DACH)]2- | 1690.3 | 1690.0 |
| [w11+Pt(DACH)]2- | 1834.8 | 1835.0 |
| [SS2b-C+Pt(DACH)]3- | 1365.3 | 1365.1 |
Fragmentation experiments with the other oligonucleotides and platinum complexes were carried out in a similar fashion and revealed guanine as the major binding partner for the other platinum complexes and all other single strands: for SS1a, the neighbouring G-G residues in position 6 and 7 were identified, for SS1b G6 appears to be preferred over G4. For SS2a, which contains five guanine residues, again adjacent G-G residues in position 6 and 7 show stronger interaction than G-T-G sequences. For SS3a and SS3b, despite the fact that there is only one guanine residue present, no binding to A and T residues was detected. In all cases, mainly (an-Bn), wn and internal (Bn:Bm) fragments were observed.
Ruthenium Complexes
NMR and gel electrophoresis studies suggest that NAMI-A, RAPTA and KP1019 exhibit similar reactivity towards adenine and guanine in plasmid DNA,[35, 48] and capillary electrophoresis inductively coupled plasma (CE-ICP) MS studies have shown that RAPTA compounds and KP1019 can react with the DNA model compound 5'-dGMP.[39] In NMR studies with different Ru-arene complexes with single nucleotides, thymine residues were also suggested as binding partners in addition to guanosine. However, under these conditions, steric effects resulting from the three dimensional structure of oligonucleotides, which makes the N3 of thymine virtually inaccessible, are not taken into account, which is the major disadvantage of single nucleotide DNA models.[49] Subsequent 2D NMR studies employing 14 mer oligonucleotides confirmed adduct formation with guanines and suggested intercalation and stacking interactions of the arene ligand with adjacent thymine residues.[21] Although DNA might not be the main target of ruthenium compounds in terms of their anticancer activity, reactions with single nucleotides, and also oligonucleotides and RNA molecules present in the cytoplasm cannot be ruled out.
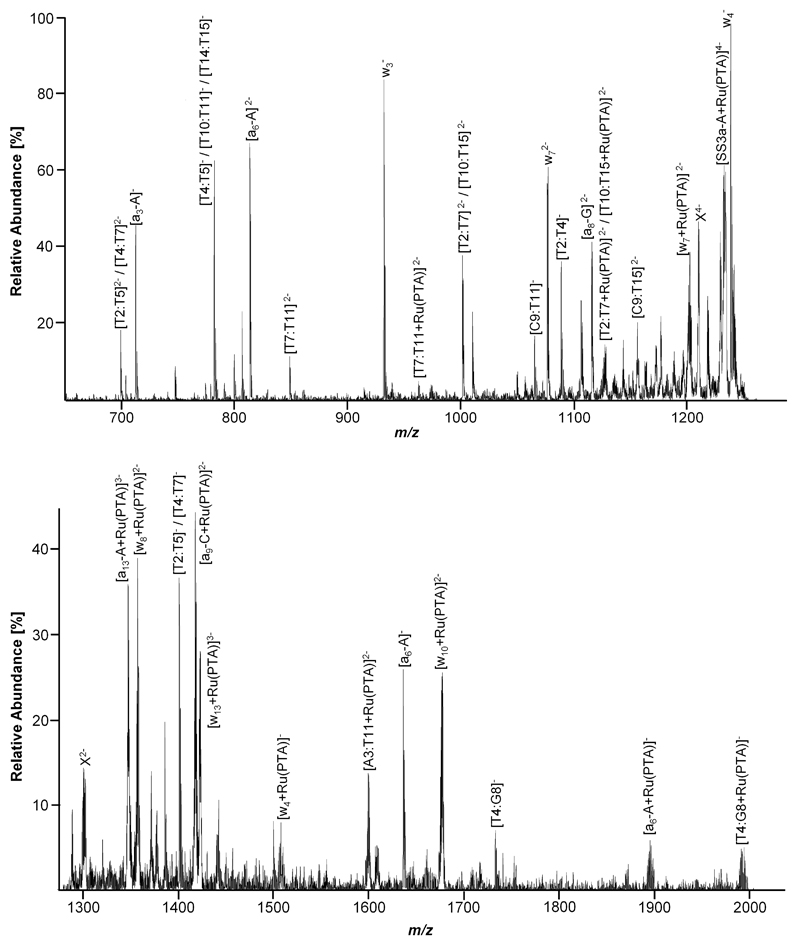
As an example, expanded segments of a CID spectrum corresponding to a [SS3a+Ru(PTA)]4- parent ion (containing only one guanine residue) fragmentation are depicted in Figure 7. Assignment of the fragment ions is conducted in a similar fashion as for the platinum compounds, and the majority of CID product ions could be assigned to wn and (an-Bn) type strand breaks as well as internal (Bn:Bm) fragments. Again, some product ions could not be assigned, most probably due to a different fragmentation mechanism induced by the bound metal
Figure 7.
Expanded segments of a LTQ CID mass spectrum of [SS3a+Ru(PTA)]4-. Top: m/z range 650 to 1275; bottom: m/z range 1275 to 2000. Unidentified fragments displaying an isotopic pattern characteristic for ruthenium are labelled X. Note that not all detected fragment ions are contained in the expanded segments for clarity.
After assigning the peaks to their corresponding fragmentation pathways and confirming characteristic isotopic patterns for modified residues, the major binding partner was identified. Again starting from the 5' terminus, it becomes clear that up to [a8-G]2- (1125.6 m/z) mainly unmodified residues are detected, whereas from there on, all found (an-Bn) fragments, which also incorporate the guanine residue, contain the Ru(PTA) moiety (mass increase of 256.0 Da for the neutral species), e.g., evidenced by [a9-C+Ru(PTA)]2- at 1418.5 m/z. Complementary information is obtained by assigning the fragments starting from the 3' terminus: up to w7, the peaks can be assigned to unmodified fragments, whereas w72- was detected in both the unmodified (1085.6 m/z) and [w7+Ru(PTA)]2- (1213.6 m/z) forms. All further wn-fragments with n>7 were found with the Ru(PTA) moiety attached. This suggests that the T10 residue might serve as an additional binding partner for the ruthenium drug to form a mulitdentate adduct. Additionally, modified internal fragments such as [T7:T11+Ru(PTA)]2- (969.5 m/z), [A3:T11+Ru(PTA)]2- (1598.5 m/z) and [T4:G8+Ru(PTA)]- (1987.0 m/z), all of which contain the G8 residue, were detected. Although small peaks attributable to, for example, [w4+Ru(PTA)]- (1508.0 m/z) and [a6-A+Ru(PTA)]- (1891.9 m/z) indicate that adduct formation is also possible with adenine or thymine residues (as they do not contain G8), the majority of the modified fragment ions points to G8 as the preferred binding partner. Interestingly, no loss of the PTA ligand was observed during CID. Some of the peaks correspond to more than one species, as, for example, internal [T2:T5] and [T4:T7] fragments exhibit the same mass (peaks at 700.6 and 1402.0 m/z for singly and doubly charged species, respectively). In Table 4, the calculated and observed m/z values for the modified fragment ions from Figure 7 are listed.
Table 4.
Calculated and experimental m/z values for fragment ions of SS3a modified by RAPTA-T obtained from the spectra shown in Figure 7 (monoisotopic peaks).
| Ion | Calculated (m/z) | Experimental (m/z) |
|---|---|---|
| [a6-A+Ru(PTA)]- | 1891.3 | 1891.0 |
| [a9-C+Ru(PTA)]2- | 1418.2 | 1418.0 |
| [a13-A+Ru(PTA)]3- | 1348.5 | 1348.6 |
| [w4+Ru(PTA)]- | 1507.2 | 1507.2 |
| [w7+Ru(PTA)]2- | 1213.6 | 1213.6 |
| [w8+Ru(PTA)]2- | 1358.2 | 1358.5 |
| [w10+Ru(PTA)]2- | 1674.7 | 1674.5 |
| [w13+Ru(PTA)]3- | 1423.2 | 1423.4 |
| [T2:T7+Ru(PTA)]2- | 1137.1 | 1137.0 |
| [T10:T15+Ru(PTA)]2- | ||
| [A3:T11+Ru(PTA)]3- | 1598.2 | 1598.5 |
| [T4:G8+Ru(PTA)]- | 1987.2 | 1986.9 |
| [T7:T11+Ru(PTA)]2- | 981.1 | 981.0 |
| [SS3a-A+Ru(PTA)]4- | 1246.2 | 1246.0 |
For RAPTA-T, adducts of Ru(PTA) with the other single strands were further analysed, and Ru(Im) and Ru(Ind) adducts were subjected to CID for NAMI-A and KP1019, respectively. Surprisingly, even for DS3 which contains mainly A and T residues, a behaviour comparable to the platinum-based chemotherapeutics (i.e., preference for guanine) was observed. Similar observations were made for KP1019 and NAMI-A.
Also for the other strands, the results did not differ strongly from those obtained with the platinum-based complexes: in SS1a, G6 and G7 are preferred binding partners, G6 in SS1b, G6 and G7 in SS2a and G9 in SS2b. In addition, low intensity peaks attributable to interactions with other residues were detected.
Discussion
In this paper, we present a comparative study of the adduct formation between duplex DNA and platinum- and ruthenium-based anticancer drugs by mass spectrometry. Note that this is the first example of a top-down tandem mass spectrometry approach for the elucidation of binding sites on DNA fragments using CID of selected adducts for ruthenium-based pharmaceuticals, as well as for oxaliplatin and carboplatin. Interestingly, mass spectrometry performed in the positive ion mode was more informative for the characterisation of the double strand–metallodrug adducts than negative ion mode data. Positively charged adducts appear to be more stable in the gas phase under the employed experimental conditions for this application. Time course experiments confirmed cisplatin with its monodentate chlorido ligands to be the most efficient compound in terms of adduct formation over the other platinum complexes with their bidentate biscarboxylates, whereas NAMI-A was more reactive towards the double stranded oligonucleotides than RAPTA-T and KP1019 for ruthenium-based compounds. However, in order to confirm the applicability of the proposed method for accurate quantitative kinetics, validation with a complementary analytical technique such as spectroscopy, chromatography or (capillary) electrophoresis would have to be carried out.
The top-down approach used to determine the binding partner on the oligonucleotides has several advantages in comparison to enzymatic digestion: primarily, exonucleases normally used for the removal of terminal nucleotides do not recognize nucleotides modified by metallodrugs, therefore inhibiting the digestion reaction and yielding an incomplete data set. The combination of enzymes simultaneously attacking from the 3'- and 5'-ends partially circumvents this problem, but in the case of multiple binding, again only incomplete data is obtained. Moreover, the top-down approach is much more rapid than the enzymatic digestion procedure which might also require change of pH and addition of catalytically active cations which could interfere with the interaction of the metallodrug with the oligonucleotide. Several factors have to be considered in the multistage mass spectrometric approach: the adducts formed must be sufficiently stable to not be cleaved during CID experiments, therefore requiring covalent bonding to the oligonucleotide. Furthermore, positive charges are introduced by the metals, making it difficult to characterise short modified oligonucleotides (consisting of less than three bases) in negative ion mode, usually employed for this kind of experiment. Nevertheless, as CID yields complementary fragments, i.e. ions from the 3’-and 5'-end of the biomolecule, complete sequence coverage can be achieved and internal fragments can substantiate assumptions on specific binding partners. The results described herein show that guanine is the preferred binding partner for both platinum- and ruthenium-based metallodrugs, even if other bases are present in large excess, although minor peaks indicate that adenine or thymine could also serve as a binding partner for the ruthenium compounds. This is in good agreement with data obtained by other techniques including 2D-NMR and biochemical assays which also showed that adduct formation between DNA and both platinum and ruthenium compounds occurs mainly via adduct formation with guanosine.[21]
Supporting Information
Supporting information containing MALDI-MS data for the analysis of the double strands as well as ESI-MS and CID mass spectra for determining metallodrug-oligonucleotide interactions are provided.
Acknowledgements
The authors thank Dr. Laure Menin for guidance in operating the q-TOF instrument. M.G. thanks the Austrian Science Foundation for financial support (Schrödinger Fellowship J2882-N19).
References
- 1.Lippert B. Cisplatin. Chemistry and biochemistry of a leading anticancer drug. VHCA; Zürich: 1999. [Google Scholar]
- 2.Dyson PJ, Sava G. Dalton Trans. 2006:1929–1933. doi: 10.1039/b601840h. [DOI] [PubMed] [Google Scholar]
- 3.Kelland L. Nat Rev Cancer. 2007;7:573–584. doi: 10.1038/nrc2167. [DOI] [PubMed] [Google Scholar]
- 4.Ott I, Gust R. Arch Pharm (Weinheim) 2007;340:117–126. doi: 10.1002/ardp.200600151. [DOI] [PubMed] [Google Scholar]
- 5.Ang WH, Dyson PJ. Eur J Inorg Chem. 2006:4003–4018. [Google Scholar]
- 6.Reedijk J. Eur J Inorg Chem. 2009:1303–1312. [Google Scholar]
- 7.Clarke MJ. Coord Chem Rev. 2003;236:209–233. [Google Scholar]
- 8.Wang D, Lippard SJ. Nat Rev Drug Discov. 2005;4:307–320. doi: 10.1038/nrd1691. [DOI] [PubMed] [Google Scholar]
- 9.Bosch ME, Sanchez AJ, Rojas FS, Ojeda CB. J Pharm Biomed Anal. 2008;47:451–459. doi: 10.1016/j.jpba.2008.01.047. [DOI] [PubMed] [Google Scholar]
- 10.Nordhoff E, Kirpekar F, Roepstorff P. Mass Spec Rev. 1996;15:67–138. doi: 10.1002/(SICI)1098-2787(1996)15:2<67::AID-MAS1>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 11.Hadjiliadis N, Sletten E, editors. Metal Complex-DNA Interactions. Blackwell Publishing Ltd; 2009. [Google Scholar]
- 12.Iannitti-Tito P, Weimann A, Wickham G, Sheil MM. Analyst. 2000;125:627–33. doi: 10.1039/a908920i. [DOI] [PubMed] [Google Scholar]
- 13.Beck JL, Colgrave ML, Ralph SF, Sheil MM. Mass Spectrom Rev. 2001;20:61–87. doi: 10.1002/mas.1003. [DOI] [PubMed] [Google Scholar]
- 14.Gupta R, Kapur A, Beck JL, Sheil MM. Rapid Commun Mass Spectrom. 2001;15:2472–2480. doi: 10.1002/rcm.524. [DOI] [PubMed] [Google Scholar]
- 15.Rosu F, Pirotte S, De Pauw E, Gabelica V. Int J Mass Spectrom. 2006;253:156–171. [Google Scholar]
- 16.Egger AE, Hartinger CG, Ben Hamidane H, Tsybin YO, Keppler BK, Dyson PJ. Inorg Chem. 2008;47:10626–10633. doi: 10.1021/ic801371r. [DOI] [PubMed] [Google Scholar]
- 17.Nyakas A, Eymann M, Schurch S. J Am Soc Mass Spectrom. 2009;20:792–804. doi: 10.1016/j.jasms.2008.12.018. [DOI] [PubMed] [Google Scholar]
- 18.Fichtinger-Schepman AM, van der Veer JL, den Hartog JH, Lohman PH, Reedijk J. Biochemistry. 1985;24:707–713. doi: 10.1021/bi00324a025. [DOI] [PubMed] [Google Scholar]
- 19.Heringova P, Woods J, Mackay FS, Kasparkova J, Sadler PJ, Brabec V. J Med Chem. 2006;49:7792–7798. doi: 10.1021/jm0606692. [DOI] [PubMed] [Google Scholar]
- 20.Wynne P, Newton C, Ledermann JA, Olaitan A, Mould TA, Hartley JA. Br J Cancer. 2007;97:927–933. doi: 10.1038/sj.bjc.6603973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pizarro AM, Sadler PJ. Biochimie. 2009 doi: 10.1016/j.biochi.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ni J, Pomerantz C, Rozenski J, Zhang Y, McCloskey JA. Anal Chem. 1996;68:1989–1999. doi: 10.1021/ac960270t. [DOI] [PubMed] [Google Scholar]
- 23.Mestroni G, Alessio E, Sava G. Patent No. WO 98/00431. Italy: 1998. [Google Scholar]
- 24.Scolaro C, Bergamo A, Brescacin L, Delfino R, Cocchietto M, Laurenczy G, Geldbach TJ, Sava G, Dyson PJ. J Med Chem. 2005;48:4161–4171. doi: 10.1021/jm050015d. [DOI] [PubMed] [Google Scholar]
- 25.Lipponer KG, Vogel E, Keppler BK. Met Based Drugs. 1996;3:243–260. doi: 10.1155/MBD.1996.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lecchi P, Pannell LK. J Am Soc Mass Spectrom. 1995;6:972–975. doi: 10.1016/1044-0305(95)00524-H. [DOI] [PubMed] [Google Scholar]
- 27.Kirpekar F, Berkenkamp S, Hillenkamp F. Anal Chem. 1999;71:2334–2339. doi: 10.1021/ac990018w. [DOI] [PubMed] [Google Scholar]
- 28.Ang WH, Daldini E, Scolaro C, Scopelliti R, Juillerat-Jeannerat L, Dyson PJ. Inorg Chem. 2006;45:9006–9013. doi: 10.1021/ic061008y. [DOI] [PubMed] [Google Scholar]
- 29.Zhang LK, Gross ML. J Am Soc Mass Spectrom. 2000;11:854–865. doi: 10.1016/S1044-0305(00)00161-6. [DOI] [PubMed] [Google Scholar]
- 30.Lecchi P, Le HMT, Pannell LK. Nucleic Acids Res. 1995;23:1276–1277. doi: 10.1093/nar/23.7.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taranenko NI, Chung CN, Zhu YF, Allman SL, Golovlev VV, Isola NR, Martin SA, Haff LA, Chen CH. Rapid Commun Mass Spectrom. 1997;11:386–392. doi: 10.1002/(SICI)1097-0231(19970228)11:4<386::AID-RCM867>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 32.Todd RC, Lippard SJ. Metallomics. 2009;1:280–291. doi: 10.1039/b907567d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reed E. In: Cancer Chemotherapy and Biotherapy. Chabner BC, Longo DL, editors. Williams & Wilkins; Philadelphia: 2001. pp. 332–343. [Google Scholar]
- 34.Bregadze VG. In: Metal Ions in Biological Systems. Sigel A, Sigel H, editors. Marcel Dekker; New York: 1996. pp. 419–451. [PubMed] [Google Scholar]
- 35.Dorcier A, Hartinger CG, Scopelliti R, Fish RH, Keppler BK, Dyson PJ. J Inorg Biochem. 2008;102:1066–1076. doi: 10.1016/j.jinorgbio.2007.10.016. [DOI] [PubMed] [Google Scholar]
- 36.Scolaro C, Chaplin AB, Hartinger CG, Bergamo A, Cocchietto M, Keppler BK, Sava G, Dyson PJ. Dalton Trans. 2007:5065–5072. doi: 10.1039/b705449a. [DOI] [PubMed] [Google Scholar]
- 37.Morbidelli L, Donnini S, Filippi S, Messori L, Piccioli F, Orioli P, Sava G, Ziche M. Br J Cancer. 2003;88:1484–1491. doi: 10.1038/sj.bjc.6600906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bergamo A, Masi A, Dyson PJ, Sava G. Int J Oncol. 2008;33:1281–1289. [PubMed] [Google Scholar]
- 39.Groessl M, Hartinger CG, Dyson PJ, Keppler BK. J Inorg Biochem. 2008;102:1060–1065. doi: 10.1016/j.jinorgbio.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 40.Groessl M, Reisner E, Hartinger CG, Eichinger R, Semenova O, Timerbaev AR, Jakupec MA, Arion VB, Keppler BK. J Med Chem. 2007;50:2185–2193. doi: 10.1021/jm061081y. [DOI] [PubMed] [Google Scholar]
- 41.Dorcier A, Ang WH, Bolano S, Gonsalvi L, Juillerat-Jeannerat L, Laurenczy G, Peruzzini M, Phillips AD, Zanobini F, Dyson PJ. Organometallics. 2006;25:4090–4096. [Google Scholar]
- 42.Dorcier A, Dyson PJ, Gossens C, Rothlisberger U, Scopelliti R, Tavernelli I. Organometallics. 2005;24:2114–2123. [Google Scholar]
- 43.Casini A, Mastrobuoni G, Terenghi M, Gabbiani C, Monzani E, Moneti G, Casella L, Messori L. J Biol Inorg Chem. 2007;12:1107–1117. doi: 10.1007/s00775-007-0280-4. [DOI] [PubMed] [Google Scholar]
- 44.McLuckey SA, Vanberkel GJ, Glish GL. J Am Soc Mass Spectrom. 1992;3:60–70. doi: 10.1016/1044-0305(92)85019-G. [DOI] [PubMed] [Google Scholar]
- 45.Hoffmann Ed. Mass spectrometry: principles and applications. John Wiley & Sons Ltd; Chichester: 2007. [Google Scholar]
- 46.Blommaert FA, Van Dijk-Knijnenburg HCM, Dijt FJ, Denengelse L, Baan RA, Berends F, Fichtinger-Schepman AMJ. Biochemistry. 1995;34:8474–8480. doi: 10.1021/bi00026a031. [DOI] [PubMed] [Google Scholar]
- 47.Kasparkova J, Vojtiskova M, Natile G, Brabec V. Chem-Eur J. 2008;14:1330–1341. doi: 10.1002/chem.200701352. [DOI] [PubMed] [Google Scholar]
- 48.Malina J, Novakova O, Keppler BK, Alessio E, Brabec V. J Biol Inorg Chem. 2001;6:435–445. doi: 10.1007/s007750100223. [DOI] [PubMed] [Google Scholar]
- 49.Zorbas-Seifried S, Hartinger CG, Meelich K, Galanski M, Keppler BK, Zorbas H. Biochemistry. 2006;45:14817–14825. doi: 10.1021/bi061063i. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.