Abstract
The host immune system must constantly monitor the gastrointestinal tract for the presence of pathogens, while tolerating trillions of commensal microbiota. The mechanisms that maintain intestinal immune homeostasis are not fully understood, but it is clear that intestinal microbiota actively modulate the immune system to maintain a mutually beneficial relationship. Here we review recent findings on the ways in which intestinal microbiota shape immune development and responses. These have revealed that a range of microbiota derivatives can influence host immune functions, by targeting various cell types in the gut, including intestinal epithelial cells, mononuclear phagocytes, innate lymphoid cells and B and T lymphocytes. Although challenges remain, harnessing this knowledge could lead to new therapies for intestinal and systemic immune disorders.
Keywords: microbiota, commensals, mucosal immunity, immune regulation
The intestinal microbiota contributes to health and disease in the gut - and beyond
Advances in 16S ribosomal RNA sequencing have revealed the intestinal microbiota to be an incredibly complex community, comprising thousands of bacterial species in humans, which vary markedly in distinct regions of the intestinal tract [1, 2]. Although every human harbours a unique microbiota, there is a common pattern at the phyla level, with Bacteroidetes and Firmicutes (dominated by Clostridia) being the most abundant [2]. Novel metagenomic approaches have further clarified the composition of the microbiota, which will be useful in identifying potential roles that distinct bacteria play in intestinal homeostasis [3, 4].
Fundamental roles of the microbiota in mammalian physiology have been derived from studies of germ-free (GF) or antibiotic-treated animals, demonstrating that the microbiota aids in food digestion, nutrient supply and resistance to pathogenic infection [2]. Furthermore, GF animals exhibit impaired immune development, characterised by immature gut-associated lymphoid tissues (GALT), decreased numbers of intestinal lymphocytes and diminished levels of antimicrobial peptides and IgA, all of which are reversed upon colonization with commensal bacteria [5]. Additionally, maturation of the immune system appears dependent on host-specific commensals, as it did not occur in GF mice colonized with human microbiota [6], although this was recently challenged by the finding that selected species of human microbiota induced a population of regulatory T cells (Tregs) in the intestine of GF mice [7, 8]. Despite promoting GALT development, not all members of the intestinal microbiota are beneficial; some may act as opportunistic pathogens, overabundance of certain commensals may predispose to pathogenic infection [9], and antibiotic-mediated alterations in commensal microbiota can predispose to nosocomial infections [10]. Interactions with intestinal microbiota may even facilitate infection by other enteric pathogens, as has been demonstrated for certain viruses and parasites (Box 1).
Box 1. Exploitation of intestinal microbiota by enteric viruses and helminths.
Intestinal microbiota affect the ability of the host to deal with other classes of pathogens, as GF mice show enhanced susceptibility to many infections [130]. More recent studies have provided insight into some of the mechanisms involved. Antibiotic treatment to deplete intestinal microbiota revealed that tonic stimulation of innate immune circuits by the microbiota enhanced protective adaptive immune responses to influenza virus infection in the respiratory tract and to systemic infection with lymphocytic choriomeningitis virus [131, 132]. Conversely, other studies show that some enteric viruses exploit the commensal microbiota to promote infection. The retrovirus, mouse mammary tumor virus (MMTV), is transmitted to neonatal mice in the milk of infected mothers, however, ablation of the intestinal microbiota prevented MMTV transmission to the offspring [133]. Furthermore, MMTV bound microbiota-derived LPS forming a complex that triggered TLR4-dependent IL-10 secretion by myeloid cells, engendering tolerance toward the virus to facilitate transmission [133]. Similarly, antibiotic-mediated depletion of microbiota induced resistance to enteric infection with reovirus and poliovirus [134]. Mechanistically, binding of bacterial LPS enhances poliovirus infectivity by increasing virion stability and by enhancing binding to the poliovirus receptor [135]. Furthermore, the reactivation of pathogenic endogenous retroviruses observed in antibody-deficient mice was dependent on the presence of the microbiota [136]. These studies reveal new aspects of the microbiota’s influence on viral infections and suggest that the use of antibiotics and probiotics during viral infections should be carefully considered.
Recently, Iliev et al. performed high-throughput sequencing to define the mouse intestinal fungal ‘mycobiome’, containing over 200 fungal species [137]. Furthermore, they found that mice deficient in Dectin-1, a key PRR that senses fungal β-glucans, exhibited greater susceptibility to DSS-induced colitis, that was attenuated by treatment with an anti-fungal drug [137]. Finally, they identified a single nucleotide polymorphism (SNP) in Dectin-1 (CLEC7A) that was associated with more severe ulcerative colitis in humans [137]. Thus, impaired immunity to commensal fungi may exacerbate pathological inflammatory responses in the gut, but the mechanisms involved remain to be identified.
A key component of the intestinal fauna with which mammals have co-evolved are multi-cellular parasites, especially helminth worms that frequently colonize the gastrointestinal tract [138]. Helminth infections, or treatment with immune suppressive factors they produce, can inhibit various immunopathological disorders, including models of IBD [138]. Intestinal helminths may indirectly regulate immune responses against intestinal microbiota through ‘bystander’ immune suppression. In addition, mice infected with the parasitic nematode Heligmosomoides polygyrus had altered ileal microbiota composition [139]. Conversely, commensal bacteria may in some cases play a critical role in facilitating infection by metazoan parasites. Contact with intestinal microbiota was shown to promote hatching of eggs of the mouse intestinal nematode Trichuris muris and antibiotic-mediated depletion of the microbiota resulted in reduced worm burdens [140]. Thus, interactions with intestinal microbiota appear to trigger the parasite to hatch in the appropriate niche and type I fimbriae were shown to be capable of mediating this interaction [140]. Therefore, within the complex environment of the gut, both host-microbe and microbe-microbe interactions can profoundly influence local and systemic immune homeostasis.
Furthermore, accumulating evidence suggests a correlation between inflammatory bowel disease (IBD) - encompassing ulcerative colitis and Crohn’s disease - and altered microbiota, a state termed “dysbiosis”, although whether dysbiosis is a primary cause of IBD or arises as a consequence of chronic intestinal inflammation remains unclear [11]. Nevertheless, in individuals with predisposing genetic or environmental abnormalities, intestinal microbiota are the focus of the aberrant host immune responses that drive the chronic inflammation characteristic of IBD [11]. An important caveat of many experimental studies linking dysbiosis to disease susceptibility in particular genotypes is the demonstration that familial transmission of microbiota from mother to neonate can play a dominant role in conferring distinct microbiotas (Box 2).
Box 2. A cautionary note on microbiota associations with genotype and disease susceptibility.
Although a plethora of studies have implicated microbiota dysbiosis with increased disease phenotypes in gene knockout mice [141], such as those deficient in PRR signalling, a key study highlighted the risk of potential false-positive associations in these types of investigations. Ubeda et al. showed that Myd88-/- and Tlr-/- mice exhibited distinct microbiotas from each other, but that these were primarily shaped by maternal transmission rather than host genotype [142]. Thus, by crossing heterozygous Myd88-/+ mice or Tlr-/+ mice to generate WT and KO mice from the same litter, they demonstrated that there was little difference in the intestinal microbiota composition of WT and TLR-deficient littermates [142]. Indeed, the microbiota of WT littermates more closely resembled that found in their Myd88-/- littermates than that present in WT mice from other colonies [142]. Thus, some of the reported incidences of ‘dysbiosis’ between WT and KO mouse strains may in fact reflect divergence of microbiota that occurs during long-term breeding of isolated colonies, which is perpetuated by maternal transmission. This elegant study shows that littermate controls are crucial in differentiating between dysbiosis that may emerge stochastically and dysbiosis that is causally related to the genotype of the host, and therefore should be employed in future studies in this area.
More recently, it has become clear that the microbiota’s influence extends beyond the intestinal tract and affects the systemic immune system. For instance, GF mice exhibited resistance to experimental autoimmune encephalomyelitis (EAE), a mouse model of multiple sclerosis (MS), but disease susceptibility was restored upon colonization with microbiota, or by mono-colonization with segmented filamentous bacteria (SFB) [12–14]. Another interesting finding linked intestinal microbiota to gender-dependent susceptibility to type-1 diabetes (T1D) in the non-obese diabetic (NOD) mouse model [15]. Female NOD mice are more susceptible to T1D than males, a phenotype that is equalized under GF settings. Moreover, transfer of caecal contents from male into female NOD mice corrected the susceptibility in a manner that was dependent on androgen receptor signalling and correlated with increases in testosterone levels [15]. Further implicating a role for the microbiota in metabolic disease, shifts in microbiota composition were recently associated with the progression of the liver disorder non-alcoholic steatohepatitis (NASH) [16]. Thus, mice deficient in the inflammasome-associated, cytosolic innate immune receptors NLRP3 or NLRP6, displayed exacerbated NASH that was due to a dysbiotic microbiota, as WT mice co-housed with the inflammasome-deficient mice also showed increased susceptibility to NASH [16]. Additional examples of the influence of intestinal microbiota on autoimmune disorders include studies in mouse models of inflammatory arthritis and autoimmune polyglandular syndrome [17].
Together, these studies highlight that the microbiota have a wide range of effects on the development and responsiveness of the local and systemic immune system, but the mechanisms responsible are incompletely understood. Here, we will review recent progress that has begun to elucidate some of the cellular and molecular factors involved. We outline how structural moieties and metabolites derived from the intestinal microbiota act on intestinal epithelial cells (IEC) and local innate leukocytes to maintain barrier defence and regulate immune homeostasis. We then describe how microbiota-derived factors activate a multitude of pathways that control adaptive immunity in the gut, by promoting IgA secretion and regulating the balance between effector and regulatory T cells. Finally, we note some of the key issues that remain to be addressed in order to translate this improved understanding into novel treatments for infections and inflammatory diseases.
Sensing of microbiota by intestinal epithelial cells maintains intestinal homeostasis
Intestinal epithelial cells (IEC) form a single cell barrier layer on the surface of the intestinal mucosa and, although not considered bona fide immune cells, their interactions with intestinal microbiota influence the immune response and play a crucial role in maintaining homeostasis [18].
Commensal bacteria influence the epithelial barrier in a multitude of ways. Sensing of bacterial metabolites and structural components by IEC fortifies barrier integrity and protects from pathogen invasion [18]. For example, IEC sensing of commensals through toll like receptors (TLRs) protects from epithelial injury following administration of dextran sulphate sodium (DSS), in part, by regulating the secretion of cytoprotective factors (IL-6, TNF-α, KC-1, heat shock proteins) [19]. Recent studies further underlined the crucial role of pattern recognition receptor (PRR) activation in the intestinal epithelium, with NLRP3 inflammasome-mediated IL-18 production exerting protective effects against DSS-induced colitis and colon cancer [20, 21]. Although these studies highlight beneficial tissue-protective effects of the microbiota, chronic activation of IEC by a dysbiotic microbiota can exacerbate colon carcinogenesis by driving IL-17C production from IEC that acts in an autocrine fashion to inhibit apoptosis [22].
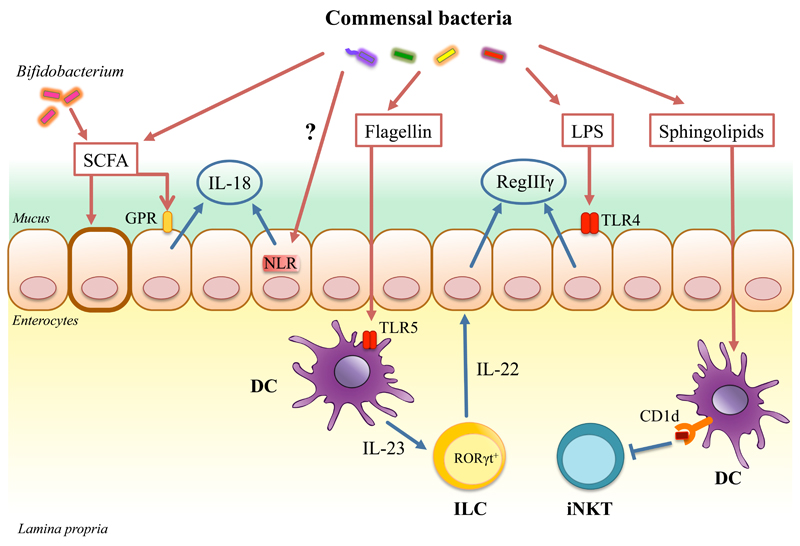
Apart from providing important energy sources for the intestinal epithelium, commensal metabolites also promote IEC homeostasis. Short chain fatty acids (SCFA) such as acetate, butyrate and propionate, which are generated by microbiota-mediated processing of dietary fibre and non-digestible carbohydrates, are important mediators in the communication between microbiota and IEC. For instance, microbiota-derived butyrate signals through the GPR109A receptor to induce IL-18 expression in IEC and this inhibited colitis-associated colon cancer (CAC) [23, 24]. The protective effects of butyrate-producing microbiota are emphasized by findings that these bacteria are reduced in IBD patients [25, 26]. Other work identified a role for Bifidobacterium-derived acetate in promoting anti-apoptotic responses in IEC, resulting in reduced mortality following challenge with enterohaemorrhagic E. coli infection [27]. Thus, microbiota-derived SCFA can have multiple protective effects on IEC following infection or insult (Figure 1).
Fig. 1. Examples of microbiota influence on the innate immune responses.
The microbiota regulates intestinal immune responses primarily through the production of PAMPs and metabolic by-products. Recognition of commensal-derived PAMPs e.g. LPS by the intestinal epithelial cells (IEC) induces secretion of antimicrobial peptide - RegIIIγ that mediates colonization resistance in the gut. RegIIIγ is also induced indirectly through flagellin recognition by CD103+ lamina propria dendritic cells (DC) that in turn activate innate lymphoid cells (ILC) to secrete IL-22, a strong AMP inducer. Microbiota derived signals induce IL-18 production from the IEC through activation of NLRs. The microbiota digests complex plant polysaccharides producing short chain fatty acids (SCFA) as by-products. These SCFA induce secretion of IL-18 form IEC via signaling through GPR109a receptor. Certain SCFAs such as acetate produced by Bifidobacterium promotes epithelial cell barrier function by inducing an anti-apoptotic response in the IEC. Microbiota derived sphingolipids presented on CD1d by DC inhibit colonic iNKT cells.
To bolster the epithelial barrier, specialized IEC known as goblet cells synthesize mucins that form a 150μm thick protective mucus gel, which covers the intestinal epithelium and is augmented with IEC-derived antimicrobial peptides (AMPs) [28]. In the colon, the mucus layer is divided into two zones: an inner dense layer that is firmly attached to the apical surface of IEC and is usually sterile, and a loose outer layer that provides a niche for certain commensals [28]. In animal models, defects in the mucus layer, such as genetic ablation of the major mucin MUC2, allow increased contact of commensal bacteria with IEC and lead to spontaneous colitis and colon cancer, emphasizing the importance of an intact mucus barrier in maintaining intestinal homeostasis [29–31]. Moreover, reduced mucus secretion is a common feature of human IBD, although it is again unclear whether this is a primary cause or a secondary consequence of the inflammatory setting [32]. Two recent studies have raised the intriguing possibility that mucus may also promote tolerogenic responses to food and commensal antigens [33]. Thus, MUC2, through interactions with galectin-3 and Dectin-1, conditioned dendritic cells to produce factors such as TGF-β, IL-10 and retinoic acid (RA) that promote Foxp3+ Treg cell induction [33]. In addition, goblet cells themselves have been proposed to promote intestinal tolerance by delivering luminal antigens to CD103+ dendritic cells, although whether this transport required interactions of such antigens with mucus was not determined [34]. However, further studies are required to ascertain the biological significance of antigen interactions with goblet cells and mucus and to determine whether these processes play an essential role in the induction of tolerance to intestinal antigens. Finally, given that degradation by some commensal species may influence the structure of the mucus layer [35, 36], it is possible that the composition of microbiota might also modulate functions of the host mucus.
While some AMPs, like α-defensins, are constitutively expressed by IEC, secretion of others is regulated by sensing of commensal-derived PAMPs [37]. Reg3γ, an inducible AMP, was recently shown to play a crucial role in establishing spatial segregation between commensal bacteria and the host epithelium [38]. Thus, mice lacking Reg3γ, or lacking MyD88 expression in IEC, lost the 50μm sterile inner mucus layer that segregates the microbiota from the epithelium, displayed higher faecal IgA levels and increased frequencies of intestinal IFN-γ+ CD4+ T cells [38]. The production of Reg3γ by IEC is directly induced by microbiota as oral administration of lipopolysaccharide (LPS) induced expression of Reg3γ only when TLR4 was expressed in nonhematopoietic cells (i.e. IEC) [39] (Figure 1). In addition to bactericidal activity, AMPs can exert modulatory functions on chemotaxis, TLR signaling and wound healing [40]. For example, expression of Reg3γ is increased in damaged mucosae and may regulate IEC proliferation [41].
IEC contribute to mucosal immune tissue development in response to the microbiota. Peptidoglycan derived from the cell wall of commensal bacteria activates the cytoplasmic PRR NOD1 in IEC and induces β-defensin 3 and CCL20 secretion, which drive formation of isolated lymphoid follicles (ILFs) [42]. IEC also influence recruitment, activation and differentiation of immune cells by secretion of a variety of modulatory factors in response to commensal microbiota, including; thymic stromal lymphopoetin (TSLP), TGF-β, prostaglandin E2, RA and IL-25 [2]. For example, production of TSLP by IEC inhibits the secretion of IL-12/23p40 by CD11c+ CD11b+ dendritic cells, thus limiting pro-inflammatory Th1 and Th17 responses and promoting Th2 responses [43]. In addition, some commensal Clostridium species can promote the differentiation and function of colonic Treg cells, by increasing production of the active form of TGF-β [44] (Figure 2). Indeed Clostridium species were shown to enhance expression of molecules that convert latent TGF-β into its active form by IEC, such as matrix metalloproteinases (MMPs) [44]. However, more recent studies have linked the induction of TGF-β by Clostridium species to their production of SCFA, such as butyrate [7], which, as we will discuss later, may also act directly on T cells to promote FoxP3+ Treg cell differentiation [45].
Fig. 2. Examples of microbiota influence on the adaptive immune responses.
Signals from commensal bacteria induce production of BAFF, APRIL and TGF-β in the intestinal epithelial cells (IEC) and dendritic cells (DC), which in turn promotes the differentiation of B cells into IgA+ plasma cells. After activation by commensal bacteria follicular dendritic cells (FDC) also promote the differentiation of B cells into IgA+ plasma cells, as they are a main producers of TGF-β in the Payer’s patches. Commensals can regulate function of the innate lymphoid cells (ILC) which in turn promote T cell independent IgA induction through the interaction of membrane bound lymphotoxin (LTα1β2) with DC, whereas soluble form of ILC-derived lymphotoxin (sLTα3) supports T cell dependent IgA induction by promoting T cell homing to the lamina propria, presumably influencing Tfh population. Segmented filamentous bacteria (SFB) are found in close contact with IEC where they may induce SAA that stimulates DC and promotes the differentiation of Th17 cells. Presentation of SFB antigens by DC on MHC II is also needed for Th17 induction. Microbiota derived signals induce IL-1β production by mononuclear phagocytes that promotes Th17 differentiation. ATP produced by certain commensals activates DCs and leads to induction of Th17 cells. Th17 cells can differentiate into Tfh cell and therefore contribute to IgA production. Polysaccharide A (PSA) produced by Bacteroides fragilis directly promotes Treg cell differentiation via TLR2 or indirectly by conditioning DCs. Microbiota derived short chain fatty acids (SCFA), a by-product of metabolism, may promote Treg cell generation either directly through signalling via GPCR43 or indirectly via IEC. Clostridium species belonging to clusters IV, XIVa and XVIII induce TGF-β production in IEC, which promotes Treg differentiation in the colon.
Microbiota conditioning of intestinal mononuclear phagocytes
Intestinal mononuclear phagocytes (iMP) comprise dendritic cells (DC) and macrophages. They are key players in intestinal homeostasis, as they link innate and adaptive immunity, and participate in compartmentalisation of the systemic and mucosal immune system. iMP are heterogeneous and their origins and classification have been extensively reviewed [46, 47]. Here, we refer to CD103+ CD11c+ cells derived from classic DC precursors as CD103+ DC; and to CD11c+CD11b+CD103-CX3CR1+ cells derived from Ly6Chi blood monocytes as CX3CR1+ MP [46].
Distinct subsets of iMP exert different functions in the intestine. CD103+ DC express several PRRs and upon stimulation secrete cytokines and chemokines and migrate to mesenteric lymph nodes (MLN) to promote adaptive immune responses, that in homeostatic conditions are limited to the mucosa [46, 47]. CD103+ DC are crucial in promoting oral tolerance against food antigens [48]. Acquisition of the Foxp3+ Treg cell program can occur during development in the thymus (nTregs), or may be induced in peripheral locations (iTregs) [49], including the intestine. CD103+ DC promote iTreg cell differentiation in the MLN through secretion of RA and TGF-β [50–52] and by integrin αvβ8-mediated conversion of latent TGF-β into its active form [53]. Furthermore, secretion of RA by CD103+ DC also primes T cells and B cells to express the gut-homing receptors α4β7 and CCR9 [54]. Conversely, CX3CR1+ MP are thought to be non-migratory, and can sample intestinal luminal contents by extending dendrites through the epithelium and may be involved in the secondary expansion of Foxp3+ Treg cells in the lamina propria (LP), that were primed initially in gut-draining lymph nodes [55, 56]. This may be mediated via IL-10 production, since IL-10 secretion from iMP is required to maintain Foxp3 expression in Treg cells [57]. Interestingly, CX3CR1+ MP may also promote oral tolerance through the transfer of soluble food antigens to CD103+ DC via gap junctions, to facilitate iTreg induction by CD103+ DC [58].
Intestinal bacteria can directly modulate local iMP functions to regulate effector T cell responses in the gut, particularly Th17 CD4+ T cells, which are crucial for defence against extracellular pathogens. For example, ATP produced by commensal bacteria activates CX3CR1+ MP and leads to the induction of Th17 cells in a MyD88-independent manner, suggesting that TLR signaling may be dispensable [59] (Figure 2). Nevertheless, in other settings, ATP also acts on P2X7 purinergic receptors to activate the NLRP3 inflammasome that leads to IL-1β secretion [60], suggesting that NLR sensing could potentiate Th17 differentiation. Indeed, recent studies by Shaw et al. revealed that commensal-derived signals induced IL-1β production from iMP that was essential for the induction of small intestinal Th17 cells under homeostatic conditions [61]. However, conversely, the same group showed that iMP in the colonic LP produced IL-1β only in response to NLRC4-triggering pathogens but not to commensals [62]. This discrepancy may reflect the different environmental factors and distinct microbiota found in the small and large intestine. Furthermore, additional studies are required to decipher the role of direct microbial sensing by iMP, versus potential conditioning effects of IEC-derived factors, in IL-1β secretion by iMP in different intestinal locations. In models of bacterially-triggered IBD, IL-1β exacerbates pathology by promoting accumulation of IL-17-producing innate and adaptive leukocytes [63]. These findings parallel previous reports on IL-23, which is also produced by iMP in response to intestinal bacteria to boost local Th17 responses [64]. Indeed, IL-23 and IL-1β act synergistically to potentiate innate and adaptive Th17 responses in the gut [63]. Thus, the kinetics and context of iMP-derived IL-23 and IL-1β secretion may determine whether Th17 cells meditate protective or pathogenic effects.
GF mice harbour reduced frequencies of intestinal Th17 cells, which were restored upon mono-colonization with SFB, although this effect was mostly limited to small intestinal Th17 cells. [65, 66]. It was proposed that SFB colonization increased levels of the acute-phase protein serum amyloid A (SAA), which conditioned CD11c+ MP to prime Th17 cells [66]. However, recent studies reported that presentation of SFB antigens on MHC II molecules by LP CD11c+ DC led to the induction of SFB-specific intestinal Th17 cells [67–69]. It is possible that SFB may also modulate immune responses using other mechanisms, as recent genome sequencing of SFB revealed that they might produce SCFA [70–72], although whether this is of functional relevance remains to be established. However, others contend that SFB-induced Th17 cells may not constitute bona fide mutualistic Th17 cells, but rather represent a pathogenic subset [73], and whether SFB (or an equivalent) are present in humans remains unknown. Together, these studies suggest that microbial effects on distinct iMP populations may underlie the differential induction of Th17 and iTreg cells in the gut (Figure 2).
Cross regulation of microbiota and innate lymphoid cells (ILC)
ILC are a recently discovered lineage of innate leukocytes that exhibit lymphoid morphology but lack rearranged antigen receptors and are phenotypically distinct from myeloid lineages [74]. ILC are enriched in mucosal tissues and, based on phenotype and function, have been classified into three subgroups named ILC1, ILC2 and ILC3 [74].
ILC3 cells play important roles in the containment of commensals and in protective immune responses against enteric pathogens. ILC3 are the major innate source of IL-22 in the lamina propria, which limits systemic dissemination of commensals [75]. IL-22 signals through IL-22R on IEC to drive STAT3 activation and induction of AMPs, including Reg3γ and Reg3β, that protect against enteric pathogens such as Citrobacter rodentium [76–78]. A recent study suggested that ILC might also play a role in the maintenance of CD4+ T cell tolerance towards the commensal microbiota, as some were shown to express MHCII and were capable of antigen presentation [79]. ILC3s do not express co-stimulatory molecules and appeared to limit T cell responses, as deletion of MHCII specifically on RORγt+ ILC resulted in spontaneous T cell-mediated intestinal inflammation [79]. In contrast to these protective activities, excessive ILC activation can drive chronic intestinal inflammation and colitis associated cancer [80, 81]. In these models, ILC respond to IL-23 to produce high levels of IFN-γ, IL-17 and IL-22 that are pathogenic [80, 82].
Although numerous studies have shown the microbiota to be dispensable for the development of most ILC [82], NCR+ RORγt+ ILC are reduced in GF or antibiotic treated mice [83–85]. Commensals may regulate ILC either through direct recognition, or indirectly through the induction of cytokine secretion by other cells. For example, human RORγt+ ILC directly respond to TLR2 agonists by secreting IL-2, which acts in an autocrine manner to induce IL-22 expression [86]. In contrast, systemic administration of flagellin triggers TLR5 on lamina propria iMP, resulting in secretion of IL-23 that enhances IL-22 production by RORγt+ ILC, leading to Reg3γ release from IEC [87–89] (Figure 1). Conversely, another study reported that RORγt+ ILC constitutively produce IL-22, but that this is repressed by microbiota-driven IL-25 production from IEC [90]. A recent study revealed that ILC also interact cooperatively with iMP to regulate intestinal homeostasis [91]. Thus, steady state sensing of microbiota by intestinal macrophages drives IL-1β release, which in turn induces ILC3 to produce GM-CSF. ILC3-derived GM-CSF triggers DC and macrophages to produce IL-10 and RA that promote Treg expansion in the intestine, and ablation of GM-CSF production by ILC3 impaired tolerance induction to dietary antigen [91]. However, whether ILC3-mediated GM-CSF production plays any role in Treg cell-mediated tolerance towards the intestinal microbiota remains to be determined.
Dietary compounds, such as phytochemicals derived from the catabolism of cruciferous vegetables may also influence ILC function through activation of the aryl hydrocarbon receptor (Ahr) [92]. Indeed, Ahr-/- mice harbour reduced frequencies of NKp46+ ILC3, and have impaired IL-22 production [93, 94]. In addition, Ahr expression on RORγt+ ILC is essential for isolated lymphoid follicle (ILF) maturation [95]. Mechanistically, signaling through Ahr promoted ILC survival by reducing apoptosis and enhanced IL-22 production by binding to the IL-22 locus in synergy with RORγt [94]. Recent studies have uncovered how the microbiota regulates Ahr ligand availability and associated protective immunity. Mice fed tryptophan-enriched diets, displayed an outgrowth of particular Lactobacilli species that were able to utilise tryptophan as an energy source [96]. Tryptophan degradation by these Lactobacilli generates the metabolite indole-3-aldehyde, an Ahr ligand that promotes IL-22 transcription by ILC3, which confers colonization resistance against Candida albicans [96]. Another recent study showed that mice lacking Ahr in RORγt+ ILC3 exhibited a pronounced susceptibility to spontaneous intestinal inflammation, that was associated with an outgrowth of SFB and accumulation of IFNγ+IL-17+ CD4+ T cells [97]. Thus, Ahr-mediated regulation of IL-22 secretion by ILC3 may be important in preventing dysbiosis-driven intestinal inflammation.
Overall, these studies illustrate that ILCs can regulate the microbiota to maintain intestinal homeostasis, but they also play a key role by responding to signals from other cells that have sensed environmental perturbations induced by microbiota.
Microbiota stimulate multiple pathways to drive secretory IgA production
IgA constitutes about seventy-five percent of the total antibody production in mammals and the most abundant immunoglobulin in mucosal secretions is dimeric secretory IgA [98]. Studies in GF mice established that commensal bacteria are strong inducers of secretory IgA production, which is an important regulator of microbiota composition and of host exposure to intestinal bacteria [98]. For example, intestinal IgA production induced in response to orally-adminstered flagellin abrogated systemic activation of flagellin-specific CD4+ T cells [99].
Production of IgA in response to microbiota requires a high dosage of bacteria to be induced, but at the same time is very persistent [100]. In addition, it is flexible, as IgA specificity can rapidly change in response to shifts in microbiota composition [100]. Furthermore, a proportion of gut residing IgA+ plasma cells can acquire “myeloid-like” phenotypic and functional properties in response to microbial stimulation, such as expression of TNFα or inducible nitric oxide synthase (iNOS) [101]. These ‘multi-functional’ IgA+ plasma cells contributed to host protection against Citrobacter rodentium, and mice lacking TNFα and iNOS expression in B cells exhibited reduced IgA production and an altered microbiota [101]. Thus, IgA+ B cells may influence the microbiota through production of IgA or through elaboration of anti-microbial factors.
Development of gut resident B cells can also be influenced by the microbiota. Although the early stages of B cell development primarily occur in the bone marrow, a recent study found that, around the time of weaning, the intestinal LP contains a population of immature B cells that express Rag recombinase and undergo V(D)J recombination and B cell receptor editing [102]. Importantly, early B cell development in the intestine was promoted by commensals, as GF weanling mice had significantly decreased numbers of immature B cells in the LP [102]. These surprising findings might have implications not only for immunoglobulin repertoire diversification at mucosal sites, but also for establishing tolerance against commensal antigens during initial colonization.
Class switch recombination to IgA occurs mainly in mucosal associated lymphoid tissues, including Peyer’s patches (PPs), isolated lymphoid follicles and MLNs, through T cell-dependent or T cell-independent mechanisms [98]. In the T cell-independent pathway commensal bacteria induce IEC and iMP to secrete cytokines such as BAFF, APRIL and TGF-β, that promote IgA switching [98]. Some species of microbiota are more efficient in inducing an IgA response, presumably due to their closer contact with the intestinal epithelium, such as SFB [98]. Presentation of commensal bacteria antigens by CD103+ DC in the MLNs induces an IgA response [103]. In addition, follicular dendritic cells (FDC), stromal cells that are crucial organizers of B cell follicles and germinal center (GC) formation, can also support class switching to IgA in PPs, by secreting BAFF and TGF-β following stimulation by TLR and retinoic acid receptor agonists present in the intestinal environment [104]. Recent studies also identified a role for ILC in the induction of intestinal IgA, by providing distinct forms of the cytokine lymphotoxin [105]. Thus, ILC-derived soluble LT (LTα3) promotes T cell homing to the intestine to facilitate T cell-dependent IgA responses, whereas membrane-bound LT (LTα1β2) on ILC is critical for T cell-independent IgA responses, probably by regulating local DC activation [105]. Taken together, these studies indicate that a variety of stromal cells and innate leukocytes can contribute to microbiota-induced intestinal IgA production (Figure 2).
Foxp3+ Treg cells have been implicated in T cell-dependent IgA production, as transfer of Treg cells into T cell-deficient mice restored intestinal IgA secretion, through provision of TGF-β [99]. Consistent with these findings, Foxp3+ Treg cells can differentiate into follicular helper T (TFH) cells in the PPs [106]. However, these observations were recently challenged by fate mapping studies which demonstrated that a large proportion of TFH cells in PPs were derived from Th17 cells and not from Treg cells [107]. Furthermore, mice defective in IL-17R signaling have reduced expression of the intestinal polymeric Ig receptor (pIgR) and lower levels of intestinal IgA [108] and Th17-derived TFH cells were crucial for the induction of T cell-dependent intestinal IgA responses against cholera toxin [107]. The plasticity between Treg, Th17 and TFH cells in the gut clearly requires further investigation, but given that different species of microbiota are potent inducers of Treg or Th17 cells, they might modulate T cell-dependent intestinal IgA production.
In terms of how intestinal IgA regulates microbiota composition, AIDG23S transgenic mice, which harbour a point mutation in the enzyme activation-induced cytidine deaminase (AID), resulting in a defect in somatic hyper-mutation (SHM) but not in class switch recombination, enabled selective evaluation of the role of SHM in mucosal homeostasis [109]. Although mice unable to undergo SHM had normal levels of intestinal IgA and IgA+ B cells, they exhibited pronounced GC B cell hyperplasia in PPs, that was accompanied by outgrowth of microbiota in the small intestine, and had increased susceptibility to oral challenge with Yersinia enterocolitica and cholera toxin [109]. Furthermore, mice lacking the inhibitory co-receptor PD-1, a key regulatory molecule expressed by TFH cells, accumulated dysfunctional TFH cells in the GC of the PPs, which led to dysregulated selection of IgA+ B cells and an altered repertoire of intestinal IgA with reduced bacteria-binding capacity [110]. This in turn caused marked dysbiosis in PD-1-deficient mice [110]. Together with the reversible mono-colonization study by Hapfelmeier et al. [100], these reports underline the role of constant stimulation by microbiota for the induction of SHM in IgA+ B cells in the PP GCs that maintains diversified, high affinity IgA secretion and a balanced humoral response against commensals.
Sensing microbiota through innate signaling pathways is important in several aspects of systemic B cell responses. The lack of systemic antibody responses against the intestinal microbiota is considered a sign of well-maintained sequestration of commensals and mucosal homeostasis [98], but it proves to be beneficial in the absence of innate immune protection [111]. Furthermore, Kirkland et al. observed that B-cell intrinsic MyD88 signaling was required to provide protection from microbial dissemination following DSS-induced intestinal damage, through influencing IgM, but not IgA production [112]. Similarly, in Myd88-deficient mice, protection from bacteraemia caused by commensal bacteria was provided by systemic T cell-dependent IgG production directed against commensal antigens [113]. Recently Rauch et al. identified a new subset of innate response activator (IRA) B cells, derived from B1a cells, that produce GM-CSF and contribute to protection from septic shock [114]. Although IRA B cells depend on MyD88 signaling for their differentiation and expanded in response to LPS, they secreted IgM but not IgA [114], therefore further studies are required to assess their potential role in intestinal homeostasis.
Microbiota shape the differentiation and functions of intestinal T cells
The intestine is populated by a large number of T lymphocytes that perform various regulatory and effector functions [2]. Much recent interest has focused on how the microbiota modulate the balance among CD4+ T cell subsets and several examples of how commensal bacteria may indirectly influence Th17 and Treg cell induction, through conditioning of iMP and IEC, have been discussed above [33, 44, 59, 61, 66–69, 73]. In addition, T cells may also express PRR that allow them to directly sense and respond to microbial constituents. For example, cell-intrinsic TLR2 signalling potentiated Th17 differentiation and pathogenic effector responses [115]. Similarly, γδ T cells, which are enriched in the intraepithelial lymphocyte population (IEL) and are an important source of innate IL-17 production, can also directly respond to TLR2 agonists [116]. Furthermore, commensal bacteria modulate the abundance and activation status of γδ T cells, as GF mice or mice treated with antibiotics have fewer IL-17-producing γδ T cells in the intestine [117]. Another study reported that γδ+ IEL can produce AMPs in response to intestinal bacteria that penetrate the epithelial barrier, however sensing of bacteria in this case required activation of MyD88 signaling in IEC [118]. In addition, the abundance of IEL was also shown to be regulated by activation of Ahr, as Ahr-/- mice did not maintain IEL and had increased commensal loads in the small intestine [92]. These results, together with a study showing that Ahr signalling is required for ILC-driven isolated lymphoid follicle formation [95], highlight crucial links between diet, mucosal immune maturation and microbiota, as in both cases vegetable-derived ligands of Ahr mediated the reported immunomodulatory effects [92, 95]. The influence of commensals has also been extended to encompass invariant natural killer T cells (iNKT), by work showing that exposure to commensal microbiota during the neonatal period limits the accumulation of iNKT cells at mucosal sites [119]. Lack of commensal exposure during early life resulted in adult mice exhibiting increased mortality to oxazolone induced colitis (an iNKT-dependent disease) and increased susceptibility to allergic asthma [119]. Mechanistically, the inhibition of colonic iNKT cell development by commensal microbiota may at least in part be mediated by the interaction of iNKT cells with commensal-derived inhibitory sphingolipids [120] (Figure 1). Collectively, these findings illustrate that the microbiota have extensive and long-lasting effects on the development and activation of both innate and adaptive T cell populations in the gut.
How commensals influence Foxp3+ Treg cell induction has remained poorly understood, but recent findings have shed new light on this process (Figure 2). The capsular polysaccharide A (PSA) of the gram-negative anaerobic commensal Bacteroides fragilis can promote IL-10 producing Foxp3+ Treg cells [121] through direct interaction with TLR2 on T lymphocytes [122], or indirectly through TLR2–dependent conditioning of dendritic cells [123, 124]. Moreover, recent reports presented compelling evidence that SCFA metabolites derived from commensal microbiota, particularly butyrate, can directly induce the differentiation of intestinal Foxp3+ iTreg [45, 125, 126]. Mechanistically, butyrate appeared to promote iTreg induction by inhibiting histone deacetylases (HDAC), because butyrate-treated naive CD4+ T cells exhibited increased acetylation of the Foxp3 locus, including the key CNS1 enhancer region that is essential for iTreg differentiation [45, 125, 126]. A key question in immunology is how the immune system discriminates between commensals and pathogens, as both express similar PAMPs. These new data suggest that SCFA metabolites may serve as a ‘surrogate for symbiosis’ – a signal that enforces homeostasis and tolerance towards beneficial commensal microbiota.
The iTreg promoting activities of commensal metabolites fit well with a previous study which reported that Foxp3+ iTreg cells in the LP expressed a unique subset of T-cell receptors (TCRs) that recognized epitopes derived from commensal microbiota [127]. Furthermore, the TCR repertoire in Foxp3+ iTreg cells was distinct from those expressed by naïve and effector CD4+ T cells in the LP, suggesting that exposure to some commensal antigens may preferentially direct naïve CD4+ T cells to differentiate into iTreg cells [127]. However, a recent report challenged the concept that locally-induced iTreg play a dominant role in maintaining tolerance towards the intestinal microbiota [128]. Thus, single cell, high-throughput sequencing revealed that vast majority of TCRs expressed by colonic Foxp3+ Treg cells were shared with the thymic Foxp3+ Treg population, including many TCR that were able to recognise microbiota-derived antigens [128]. In addition, analogous to the flexible IgA responses described above, manipulation of the microbiota composition with antibiotics resulted in alterations in the TCR repertoire of colonic Foxp3+ Treg [128]. Although differences in methodological approach might have contributed to the conflicting results regarding the ontogeny of microbiota-specific intestinal Treg cells, the current evidence suggests that the microbiota may shape the pool of intestinal Treg cells in at least two ways; by providing factors that promote the de novo differentiation of iTreg and by providing antigenic peptides that stimulate particular clones of thymically-derived nTreg cells.
Conclusions and perspectives
The studies discussed here illustrate the diverse mechanisms through which commensal microbiota contribute to the development, maturation and regulation of the host immune system. They highlight the complex and dynamic nature of host-microbiota interactions and illustrate that immune responses are regulated at several levels by the microbiota. The first of these involves sensing of microbiota constituents and metabolites by IEC and iMP, which, under steady state conditions, helps maintain intestinal barrier function and conditions iMP to favour regulatory responses. On the next level, resident ILC remain poised to respond to perturbations in intestinal immune homeostasis, by rapidly detecting cytokines released by IEC and iMP during stress or infection. However, microbiota can also directly influence ILC function by producing metabolites that serve as Ahr ligands. Similarly, at the level of adaptive immunity, the unique range of T and B cell responses observed in the gut are not simply due to conditioning factors produced by IEC and local iMP, but also reflect direct effects of microbiota antigens and metabolites. Thus, a common theme to emerge is that microbiota can regulate the immune and inflammatory responses of diverse cellular compartments in the gut both directly and indirectly, and extensive interactions between these cells facilitate co-ordinated, flexible responses to different conditions or infections.
Recent progress has facilitated a conceptual shift in understanding immune-microbe interactions in the gut, expanding the focus beyond PRR sensing of PAMPs, to incorporate the effects of microbial metabolites. The identification of regulatory metabolites such as SCFA, the key commensal bacteria that produce them, and insights into the molecular pathways involved, offer new possibilities for targeting intestinal diseases like IBD. However, several challenges remain before the translational potential of this new knowledge can be realized. The pleiotropic effects of microbiota metabolites such as SCFA make it desirable that strategies be developed to allow efficient, targeted delivery to the relevant cell type, eg. IEC versus lamina propria T cells. In addition, the synergistic and/or antagonistic effects of these metabolites with one another, and with other immune modulator factors, such as cytokines, remain to be fully characterized. Furthermore, whether these metabolites exhibit similar immune regulatory potential for the control of systemic inflammatory diseases requires further investigation.
Although beyond the scope of this review, characterization of host microbiota composition in healthy and diseased individuals continues apace, and should further inform the development of novel treatment regimes. For example, heterologous fecal microbiota transplant (FMT) has already demonstrated efficacy for the treatment of recurrent Clostridium difficile infections, although the precise components of the microbiota responsible remain to be determined [129]. Additional advances in this area, when merged with the improved molecular understanding derived from experimental studies described above, should allow the identification of a new generation of beneficial commensal species in humans. However, therapeutic administration of defined microbiota may be dependent on overcoming several technical limitations, including establishing new culture techniques to enable growth and selection of beneficial species, as well as devising protocols to ensure efficient delivery and, when desired, long-term persistence in the gut. Continued progress in this field is essential in order to advance potential treatments for inflammatory disorders and for developing improved vaccines against mucosal pathogens.
Acknowledgement
We thank Dr O. Harrison for insight and comments on the manuscript and A. Szafranski for help with generating figures. A.M.K is funded by a Wellcome Trust IITM Programme and K.J.M is funded by a Wellcome Trust New Investigator award.
References
- 1.Eckburg PB, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hill DA, Artis D. Intestinal bacteria and the regulation of immune cell homeostasis. Annu Rev Immunol. 2010;28:623–667. doi: 10.1146/annurev-immunol-030409-101330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arumugam M, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chinen T, Rudensky AY. The effects of commensal microbiota on immune cell subsets and inflammatory responses. Immunol Rev. 2012;245:45–55. doi: 10.1111/j.1600-065X.2011.01083.x. [DOI] [PubMed] [Google Scholar]
- 6.Chung H, et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149:1578–1593. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Atarashi K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 8.Faith JJ, et al. Identifying gut microbe-host phenotype relationships using combinatorial communities in gnotobiotic mice. Sci Transl Med. 2014 doi: 10.1126/scitranslmed.3008051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stecher B, et al. Like will to like: abundances of closely related species can predict susceptibility to intestinal colonization by pathogenic and commensal bacteria. PLoS Pathog. 2010 doi: 10.1371/journal.ppat.1000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ubeda C, et al. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J Clin Invest. 2010;120:4332–4341. doi: 10.1172/JCI43918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kostic AD, et al. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology. 2014;146:1489–1499. doi: 10.1053/j.gastro.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu HJ, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee YK, et al. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci USA. 2011;108:4615–4622. doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berer K, et al. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature. 2011;479:538–541. doi: 10.1038/nature10554. [DOI] [PubMed] [Google Scholar]
- 15.Markle JG, et al. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science. 2013;339:1084–1088. doi: 10.1126/science.1233521. [DOI] [PubMed] [Google Scholar]
- 16.Henao-Mejia J, et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482:179–185. doi: 10.1038/nature10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mathis D, Benoist C. Microbiota and autoimmune disease: the hosted self. Cell Host Microbe. 2011;10:297–301. doi: 10.1016/j.chom.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 18.Goto Y, Kiyono H. Epithelial barrier: an interface for the cross-communication between gut flora and immune system. Immunol Rev. 2012;245:147–163. doi: 10.1111/j.1600-065X.2011.01078.x. [DOI] [PubMed] [Google Scholar]
- 19.Rakoff-Nahoum S, et al. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 20.Dupaul-Chicoine J, et al. Control of intestinal homeostasis, colitis, and colitis-associated colorectal cancer by the inflammatory caspases. Immunity. 2010;32:367–378. doi: 10.1016/j.immuni.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 21.Zaki MH, et al. The NLRP3 inflammasome protects against loss of epithelial integrity and mortality during experimental colitis. Immunity. 2010;32:379–391. doi: 10.1016/j.immuni.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song X, et al. Alterations in the microbiota drive interleukin-17C production from intestinal epithelial cells to promote tumorigenesis. Immunity. 2014;40:140–152. doi: 10.1016/j.immuni.2013.11.018. [DOI] [PubMed] [Google Scholar]
- 23.Kalina U, et al. Enhanced production of IL-18 in butyrate-treated intestinal epithelium by stimulation of the proximal promoter region. Eur J Immunol. 2002;32:2635–2643. doi: 10.1002/1521-4141(200209)32:9<2635::AID-IMMU2635>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 24.Singh N, et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity. 2014;40:128–139. doi: 10.1016/j.immuni.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frank DN, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Willing BP, et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology. 2010;139:1844–1854. doi: 10.1053/j.gastro.2010.08.049. [DOI] [PubMed] [Google Scholar]
- 27.Fukuda S, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–547. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 28.Johansson ME, et al. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4659–4665. doi: 10.1073/pnas.1006451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Velcich A, et al. Colorectal cancer in mice genetically deficient in the mucin Muc2. Science. 2002;295:1726–1729. doi: 10.1126/science.1069094. [DOI] [PubMed] [Google Scholar]
- 30.Van der Sluis M, et al. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology. 2006;131:117–129. doi: 10.1053/j.gastro.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 31.Fu J, et al. Loss of intestinal core 1-derived O-glycans causes spontaneous colitis in mice. J Clin Invest. 2011;121:1657–1666. doi: 10.1172/JCI45538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johansson ME, et al. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut. 2014;63:281–291. doi: 10.1136/gutjnl-2012-303207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shan M, et al. Mucus enhances gut homeostasis and oral tolerance by delivering immunoregulatory signals. Science. 2013;342:447–453. doi: 10.1126/science.1237910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McDole JR, et al. Goblet cells deliver luminal antigen to CD103+ dendritic cells in the small intestine. Nature. 2012;483:345–349. doi: 10.1038/nature10863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sonnenburg JL, et al. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 36.Koropatkin NM, et al. How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol. 2012;10:323–335. doi: 10.1038/nrmicro2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol. 2010;10:159–169. doi: 10.1038/nri2710. [DOI] [PubMed] [Google Scholar]
- 38.Vaishnava S, et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brandl K, et al. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature. 2008;455:804–807. doi: 10.1038/nature07250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gallo RL, Hooper LV. Epithelial antimicrobial defence of the skin and intestine. Nat Rev Immunol. 2012;12:503–516. doi: 10.1038/nri3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miyaoka Y, et al. Transgenic overexpression of Reg protein caused gastric cell proliferation and differentiation along parietal cell and chief cell lineages. Oncogene. 2004;23:3572–3579. doi: 10.1038/sj.onc.1207333. [DOI] [PubMed] [Google Scholar]
- 42.Bouskra D, et al. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature. 2008;456:507–510. doi: 10.1038/nature07450. [DOI] [PubMed] [Google Scholar]
- 43.Taylor BC, et al. TSLP regulates intestinal immunity and inflammation in mouse models of helminth infection and colitis. J Exp Med. 2009;206:655–667. doi: 10.1084/jem.20081499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Atarashi K, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Furusawa Y, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 46.Varol C, et al. Securing the immune tightrope: mononuclear phagocytes in the intestinal lamina propria. Nat Rev Immunol. 2010;10:415–426. doi: 10.1038/nri2778. [DOI] [PubMed] [Google Scholar]
- 47.Cerovic V, et al. Intestinal macrophages and dendritic cells: what’s the difference? Trends Immunol. 2014;35:270–277. doi: 10.1016/j.it.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 48.Pabst O, Mowat AM. Oral tolerance to food protein. Mucosal Immunol. 2012;5:232–239. doi: 10.1038/mi.2012.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Josefowicz SZ, et al. Regulatory T cells: mechanisms of differentiation and function. Annu Rev Immunol. 2012;30:531–564. doi: 10.1146/annurev.immunol.25.022106.141623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Coombes JL, et al. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. J Exp Med. 2007;204:1757–1764. doi: 10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun CM, et al. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J Exp Med. 2007;204:1775–1785. doi: 10.1084/jem.20070602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Benson MJ, et al. All-trans retinoic acid mediates enhanced T reg cell growth, differentiation, and gut homing in the face of high levels of co-stimulation. J Exp Med. 2007;204:1765–1774. doi: 10.1084/jem.20070719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Worthington JJ, et al. Intestinal dendritic cells specialize to activate transforming growth factor-beta and induce Foxp3+ regulatory T cells via integrin alphavbeta8. Gastroenterology. 2011;141:1802–1812. doi: 10.1053/j.gastro.2011.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Coombes JL, Powrie F. Dendritic cells in intestinal immune regulation. Nat Rev Immunol. 2008;8:435–446. doi: 10.1038/nri2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Niess JH, et al. CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science. 2005;307:254–258. doi: 10.1126/science.1102901. [DOI] [PubMed] [Google Scholar]
- 56.Hadis U, et al. Intestinal tolerance requires gut homing and expansion of FoxP3+ regulatory T cells in the lamina propria. Immunity. 2011;34:237–246. doi: 10.1016/j.immuni.2011.01.016. [DOI] [PubMed] [Google Scholar]
- 57.Murai M, et al. Interleukin 10 acts on regulatory T cells to maintain expression of the transcription factor Foxp3 and suppressive function in mice with colitis. Nat Immunol. 2009;10:1178–1184. doi: 10.1038/ni.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mazzini E, et al. Oral Tolerance Can Be Established via Gap Junction Transfer of Fed Antigens from CX3CR1(+) Macrophages to CD103(+) Dendritic Cells. Immunity. 2014;40:248–261. doi: 10.1016/j.immuni.2013.12.012. [DOI] [PubMed] [Google Scholar]
- 59.Atarashi K, et al. ATP drives lamina propria T(H)17 cell differentiation. Nature. 2008;455:808–812. doi: 10.1038/nature07240. [DOI] [PubMed] [Google Scholar]
- 60.Pelegrin P, Surprenant A. Pannexin-1 mediates large pore formation and interleukin-1b release by the ATP-gated P2X7 receptor. EMBO J. 2006;25:5071–5082. doi: 10.1038/sj.emboj.7601378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shaw MH, et al. Microbiota-induced IL-1beta, but not IL-6, is critical for the development of steady-state TH17 cells in the intestine. J Exp Med. 2012;209:251–258. doi: 10.1084/jem.20111703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Franchi L, et al. NLRC4-driven production of IL-1beta discriminates between pathogenic and commensal bacteria and promotes host intestinal defense. Nat Immunol. 2012;13:449–456. doi: 10.1038/ni.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Coccia M, et al. IL-1beta mediates chronic intestinal inflammation by promoting the accumulation of IL-17A secreting innate lymphoid cells and CD4+ Th17 cells. J Exp Med. 2012;209:1595–1609. doi: 10.1084/jem.20111453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maloy KJ, Powrie F. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature. 2011;474:298–306. doi: 10.1038/nature10208. [DOI] [PubMed] [Google Scholar]
- 65.Gaboriau-Routhiau V, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 66.Ivanov II, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goto Y, et al. Segmented filamentous bacteria antigens presented by intestinal dendritic cells drive mucosal Th17 cell differentiation. Immunity. 2014;40:594–607. doi: 10.1016/j.immuni.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lecuyer E, et al. Segmented filamentous bacterium uses secondary and tertiary lymphoid tissues to induce gut IgA and specific T helper 17 cell responses. Immunity. 2014;40:608–620. doi: 10.1016/j.immuni.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 69.Yang Y, et al. Focused specificity of intestinal T17 cells towards commensal bacterial antigens. Nature. 2014;510:152–156. doi: 10.1038/nature13279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kuwahara T, et al. The lifestyle of the segmented filamentous bacterium: a non-culturable gut-associated immunostimulating microbe inferred by whole-genome sequencing. DNA Res. 2011;18:291–303. doi: 10.1093/dnares/dsr022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sczesnak A, et al. The genome of th17 cell-inducing segmented filamentous bacteria reveals extensive auxotrophy and adaptations to the intestinal environment. Cell Host Microbe. 2011;10:260–272. doi: 10.1016/j.chom.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Prakash T, et al. Complete genome sequences of rat and mouse segmented filamentous bacteria, a potent inducer of Th17 cell differentiation. Cell Host Microbe. 2011;10:273–284. doi: 10.1016/j.chom.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 73.Geuking MB, et al. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 2011;34:794–806. doi: 10.1016/j.immuni.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 74.Spits H, et al. Innate lymphoid cells--a proposal for uniform nomenclature. Nat Rev Immunol. 2013;13:145–149. doi: 10.1038/nri3365. [DOI] [PubMed] [Google Scholar]
- 75.Sonnenberg GF, et al. Innate lymphoid cells promote anatomical containment of lymphoid-resident commensal bacteria. Science. 2012;336:1321–1325. doi: 10.1126/science.1222551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zheng Y, et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14:282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 77.Pickert G, et al. STAT3 links IL-22 signaling in intestinal epithelial cells to mucosal wound healing. J Exp Med. 2009;206:1465–1472. doi: 10.1084/jem.20082683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sonnenberg GF, et al. CD4(+) lymphoid tissue-inducer cells promote innate immunity in the gut. Immunity. 2011;34:122–134. doi: 10.1016/j.immuni.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hepworth MR, et al. Innate lymphoid cells regulate CD4+ T-cell responses to intestinal commensal bacteria. Nature. 2013;498:113–117. doi: 10.1038/nature12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Buonocore S, et al. Innate lymphoid cells drive interleukin-23-dependent innate intestinal pathology. Nature. 2010;464:1371–1375. doi: 10.1038/nature08949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kirchberger S, et al. Innate lymphoid cells sustain colon cancer through production of interleukin-22 in a mouse model. J Exp Med. 2013;210:917–931. doi: 10.1084/jem.20122308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sonnenberg GF, Artis D. Innate lymphoid cell interactions with microbiota: implications for intestinal health and disease. Immunity. 2012;37:601–610. doi: 10.1016/j.immuni.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Satoh-Takayama N, et al. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity. 2008;29:958–970. doi: 10.1016/j.immuni.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 84.Sanos SL, et al. RORgammat and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nat Immunol. 2009;10:83–91. doi: 10.1038/ni.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vonarbourg C, et al. Regulated expression of nuclear receptor RORgammat confers distinct functional fates to NK cell receptor-expressing RORgammat(+) innate lymphocytes. Immunity. 2010;33:736–751. doi: 10.1016/j.immuni.2010.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Crellin NK, et al. Regulation of cytokine secretion in human CD127(+) LTi-like innate lymphoid cells by Toll-like receptor 2. Immunity. 2010;33:752–764. doi: 10.1016/j.immuni.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 87.Kinnebrew MA, et al. Bacterial flagellin stimulates Toll-like receptor 5-dependent defense against vancomycin-resistant Enterococcus infection. J Infect Dis. 2010;201:534–543. doi: 10.1086/650203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Van Maele L, et al. TLR5 signaling stimulates the innate production of IL-17 and IL-22 by CD3(neg)CD127+ immune cells in spleen and mucosa. J Immunol. 2010;185:1177–1185. doi: 10.4049/jimmunol.1000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kinnebrew MA, et al. Interleukin 23 Production by Intestinal CD103(+)CD11b(+) Dendritic Cells in Response to Bacterial Flagellin Enhances Mucosal Innate Immune Defense. Immunity. 2012;36:276–287. doi: 10.1016/j.immuni.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sawa S, et al. RORgammat+ innate lymphoid cells regulate intestinal homeostasis by integrating negative signals from the symbiotic microbiota. Nat Immunol. 2011;12:320–326. doi: 10.1038/ni.2002. [DOI] [PubMed] [Google Scholar]
- 91.Mortha A, et al. Microbiota-dependent crosstalk between macrophages and ILC3 promotes intestinal homeostasis. Science. 2014 doi: 10.1126/science.1249288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li Y, et al. Exogenous stimuli maintain intraepithelial lymphocytes via aryl hydrocarbon receptor activation. Cell. 2011;147:629–640. doi: 10.1016/j.cell.2011.09.025. [DOI] [PubMed] [Google Scholar]
- 93.Lee JS, et al. AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nat Immunol. 2012;13:144–151. doi: 10.1038/ni.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Qiu J, et al. The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity. 2012;36:92–104. doi: 10.1016/j.immuni.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kiss EA, et al. Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles. Science. 2011;334:1561–1565. doi: 10.1126/science.1214914. [DOI] [PubMed] [Google Scholar]
- 96.Zelante T, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39:372–385. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 97.Qiu J, et al. Group 3 innate lymphoid cells inhibit T-cell-mediated intestinal inflammation through aryl hydrocarbon receptor signaling and regulation of microflora. Immunity. 2013;39:386–399. doi: 10.1016/j.immuni.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Macpherson AJ, et al. The habitat, double life, citizenship, and forgetfulness of IgA. Immunol Rev. 2011;245:132–146. doi: 10.1111/j.1600-065X.2011.01072.x. [DOI] [PubMed] [Google Scholar]
- 99.Cong Y, et al. A dominant, coordinated T regulatory cell-IgA response to the intestinal microbiota. Proc Natl Acad Sci USA. 2009;106:19256–19261. doi: 10.1073/pnas.0812681106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hapfelmeier S, et al. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science. 2010;328:1705–1709. doi: 10.1126/science.1188454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fritz JH, et al. Acquisition of a multifunctional IgA+ plasma cell phenotype in the gut. Nature. 2012;481:199–203. doi: 10.1038/nature10698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wesemann DR, et al. Microbial colonization influences early B-lineage development in the gut lamina propria. Nature. 2013;501:112–115. doi: 10.1038/nature12496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kinnebrew MA, Pamer EG. Innate immune signaling in defense against intestinal microbes. Immunol Rev. 2012;245:113–131. doi: 10.1111/j.1600-065X.2011.01081.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Suzuki K, et al. The sensing of environmental stimuli by follicular dendritic cells promotes immunoglobulin A generation in the gut. Immunity. 2010;33:71–83. doi: 10.1016/j.immuni.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 105.Kruglov AA, et al. Nonredundant function of soluble LTalpha3 produced by innate lymphoid cells in intestinal homeostasis. Science. 2013;342:1243–1246. doi: 10.1126/science.1243364. [DOI] [PubMed] [Google Scholar]
- 106.Tsuji M, et al. Preferential generation of follicular B helper T cells from Foxp3+ T cells in gut Peyer’s Patches. Science. 2009;323:1488–1492. doi: 10.1126/science.1169152. [DOI] [PubMed] [Google Scholar]
- 107.Hirota K, et al. Plasticity of Th17 cells in Peyer’s patches is responsible for the induction of T cell-dependent IgA responses. Nat Immunol. 2013;14:372–379. doi: 10.1038/ni.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cao AT, et al. Th17 cells upregulate polymeric Ig receptor and intestinal IgA and contribute to intestinal homeostasis. J Immunol. 2012;189:4666–4673. doi: 10.4049/jimmunol.1200955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wei M, et al. Mice carrying a knock-in mutation of Aicda resulting in a defect in somatic hypermutation have impaired gut homeostasis and compromised mucosal defense. Nat Immunol. 2011;12:264–270. doi: 10.1038/ni.1991. [DOI] [PubMed] [Google Scholar]
- 110.Kawamoto S, et al. The inhibitory receptor PD-1 regulates IgA selection and bacterial composition in the gut. Science. 2012;336:485–489. doi: 10.1126/science.1217718. [DOI] [PubMed] [Google Scholar]
- 111.Lochner M, et al. Microbiota-induced tertiary lymphoid tissues aggravate inflammatory disease in the absence of RORgamma t and LTi cells. J Exp Med. 2011;208:125–134. doi: 10.1084/jem.20100052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kirkland D, et al. B cell-intrinsic MyD88 signaling prevents the lethal dissemination of commensal bacteria during colonic damage. Immunity. 2012;36:228–238. doi: 10.1016/j.immuni.2011.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Slack E, et al. Innate and adaptive immunity cooperate flexibly to maintain host-microbiota mutualism. Science. 2009;325:617–620. doi: 10.1126/science.1172747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rauch PJ, et al. Innate response activator B cells protect against microbial sepsis. Science. 2012;335:597–601. doi: 10.1126/science.1215173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Reynolds JM, et al. Toll-like receptor 2 signaling in CD4(+) T lymphocytes promotes T helper 17 responses and regulates the pathogenesis of autoimmune disease. Immunity. 2010;32:692–702. doi: 10.1016/j.immuni.2010.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Martin B, et al. Interleukin-17-producing gammadelta T cells selectively expand in response to pathogen products and environmental signals. Immunity. 2009;31:321–330. doi: 10.1016/j.immuni.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 117.Duan J, et al. Microbial colonization drives expansion of IL-1 receptor 1-expressing and IL-17-producing gamma/delta T cells. Cell Host Microbe. 2010;7:140–150. doi: 10.1016/j.chom.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ismail AS, et al. Gammadelta intraepithelial lymphocytes are essential mediators of host-microbial homeostasis at the intestinal mucosal surface. Proc Natl Acad Sci USA. 2011;108:8743–8748. doi: 10.1073/pnas.1019574108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Olszak T, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.An D, et al. Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell. 2014;156:123–133. doi: 10.1016/j.cell.2013.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci USA. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Round JL, et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Shen Y, et al. Outer membrane vesicles of a human commensal mediate immune regulation and disease protection. Cell Host Microbe. 2012;12:509–520. doi: 10.1016/j.chom.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dasgupta S, et al. Plasmacytoid dendritic cells mediate anti-inflammatory responses to a gut commensal molecule via both innate and adaptive mechanisms. Cell Host Microbe. 2014;15:413–423. doi: 10.1016/j.chom.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Arpaia N, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Smith PM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lathrop SK, et al. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478:250–254. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cebula A, et al. Thymus-derived regulatory T cells contribute to tolerance to commensal microbiota. Nature. 2013;497:258–262. doi: 10.1038/nature12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Smith LP, et al. Therapeutic potential of fecal microbiota transplantation. Gastroenterology. 2013;145:946–953. doi: 10.1053/j.gastro.2013.08.058. [DOI] [PubMed] [Google Scholar]
- 130.Smith K, et al. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin Immunol. 2007;19:59–69. doi: 10.1016/j.smim.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 131.Ichinohe T, et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci USA. 2011;108:5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Abt MC, et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity. 2012;37:158–170. doi: 10.1016/j.immuni.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kane M, et al. Successful transmission of a retrovirus depends on the commensal microbiota. Science. 2011;334:245–249. doi: 10.1126/science.1210718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kuss SK, et al. Intestinal microbiota promote enteric virus replication and systemic pathogenesis. Science. 2011;334:249–252. doi: 10.1126/science.1211057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Robinson CM, et al. Bacterial lipopolysaccharide binding enhances virion stability and promotes environmental fitness of an enteric virus. Cell Host Microbe. 2014;15:36–46. doi: 10.1016/j.chom.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Young GR, et al. Resurrection of endogenous retroviruses in antibody-deficient mice. Nature. 2012;491:774–778. doi: 10.1038/nature11599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Iliev ID, et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science. 2012;336:1314–1317. doi: 10.1126/science.1221789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.McSorley HJ, Maizels RM. Helminth infections and host immune regulation. Clin Microbiol Rev. 2012;25:585–608. doi: 10.1128/CMR.05040-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Walk ST, et al. Alteration of the murine gut microbiota during infection with the parasitic helminth Heligmosomoides polygyrus. Inflamm Bowel Dis. 2010;16:1841–1849. doi: 10.1002/ibd.21299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hayes KS, et al. Exploitation of the intestinal microflora by the parasitic nematode Trichuris muris. Science. 2010;328:1391–1394. doi: 10.1126/science.1187703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Blumberg R, Powrie F. Microbiota, disease, and back to health: a metastable journey. Sci Transl Med. 2012 doi: 10.1126/scitranslmed.3004184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ubeda C, et al. Familial transmission rather than defective innate immunity shapes the distinct intestinal microbiota of TLR-deficient mice. J Exp Med. 2012;209:1445–1456. doi: 10.1084/jem.20120504. [DOI] [PMC free article] [PubMed] [Google Scholar]