Abstract
Kinetochores are specialised multi-protein complexes that play a crucial role in maintaining genome stability 1. They bridge attachments between chromosomes and microtubules during mitosis and they activate the spindle assembly checkpoint (SAC) to arrest division until all chromosomes are attached 2. Kinetochores are able to efficiently integrate these two processes because they can rapidly respond to changes in microtubule occupancy by switching localised SAC signalling ON or OFF 2–4. We show that this responsiveness arises because the SAC primes kinetochore phosphatases to induce negative feedback and silence its own signal. Active SAC signalling recruits PP2A-B56 to kinetochores where it antagonises Aurora B to promote PP1 recruitment. PP1 in turn silences the SAC and delocalises PP2A-B56. Preventing or bypassing key regulatory steps demonstrates that this spatiotemporal control of phosphatase feedback underlies rapid signal switching at the kinetochore by; 1) allowing the SAC to quickly transition to the ON state in the absence of antagonising phosphatase activity, and 2) ensuring phosphatases are then primed to rapidly switch the SAC signal OFF when kinetochore kinase activities are diminished by force-producing microtubule attachments.
The spindle assembly checkpoint (SAC) is globally activated at mitotic entry and only extinguished when all kinetochores have established force-producing microtubule attachments 2, 3. At each individual kinetochore however, the SAC responses are much more dynamic. Here, localised SAC signalling switches rapidly between the ON and OFF states depending on microtubule occupancy 2–6. Exactly how kinetochores manage to achieve this rapid signal switching remains unknown. To address this we initially focussed on characterising the kinetochore phosphatases responsible for SAC silencing in mammalian cells. We performed a targeted screen with siRNAs to 222 individual phosphatase subunits to identify those that regulate mitotic exit in mammalian cells. 48 hours after siRNA transfection, cells were synchronised in mitosis using the microtubule poison nocodazole, after which mitotic exit was forced by the small molecule MPS1 inhibitor reversine 7 for 1 hour. The fraction of cells remaining in mitosis was quantified and eight of the top 14 siRNAs that delayed mitotic exit targeted subunits of PP1 and PP2A-B56 (Fig.1a and Supplementary Table.1). PP1 is known to silence the SAC in S. pombe, S. cerevisiae and C. elegans 8–12 and therefore we initially focussed on PP2A-B56, a centromere- and kinetochore-localised phosphatase that maintains sister chromatid cohesion, regulates kinetochore-microtubule attachments and controls chromosome movements 13–15. To ensure that microtubule-associated functions of PP2A-B56 could not interfere with our analysis of SAC silencing, all subsequent experiments were performed in the presence of nocodazole (unless stated otherwise). A non-overlapping pool of siRNAs that collectively target all PP2A-B56 subunits 14 (hereafter referred to as B56) delayed mitotic exit following MPS1 inhibition in nocodazole (Fig.1b). Live monitoring of endogenous Cyclin B1 levels 16 showed that B56 depletion prevented efficient APC/C activation following MPS1 inhibition (Fig.1c). This indicated that PP2A-B56 depletion did not simply delay mitotic exit, but in fact prevented SAC silencing. PP2A-B56 has recently been shown to localise to the outer kinetochore via interaction with a short phosphorylated motif in BUBR1 (termed KARD) 17–19. We found that all B56 isoforms that we tested (B56α, β, γ1, γ3, δ, ε) localised to the centromere/kinetochores regions of mitotic chromosomes, with some more clearly enriched on kinetochores than others (B56γ1, γ3, δ; Supplementary Fig.1a). We next deleted the B56 binding motif from BUBR1 (BUBR1∆KARD; Supplementary Fig.1b-d), which specifically abolished B56 kinetochore localisation (Fig.1d, e and Supplementary Fig.1e), and delayed mitotic exit following MPS1 inhibition with either reversine (Fig.1f) or the distinct inhibitor AZ-3146 20 (Supplementary Fig.1f). These delays were accentuated by concomitant B56 depletion, which even allowed cells to mount a prolonged arrest with a high dose of reversine or AZ-3146 (Supplementary Figs.1g-j). This was unrelated to effects on centromeric PP2A-B56 because SGO1 depletion caused mitotic arrest due to reduced centromeric PP2A and loss of sister chromatid cohesion, as expected 13, but did not affect SAC silencing following MPS1 inhibition with reversine (Supplementary Fig.1k-m). Collectively, these data demonstrate that outer-kinetochore-localized PP2A-B56 is essential for SAC silencing in human cells.
Figure 1. Kinetochore PP2A-B56 is required for SAC silencing.
(a) Mitotic index screen of U2OS cells that were transfected with a panel of siRNAs to phosphatase subunits, arrested in prometaphase with nocodazole and subsequently treated with reversine (1 µM) for 1h. Graph shows each targeted gene ranked according to the mean fraction of mitotic cells persisting following MPS1 inhibition. (b) Time-lapse analysis of duration of mitotic arrest in nocodazole-treated Flp-in HeLa cells transfected with mock or a pool of PP2A-B56 siRNAs. Cells entered mitosis in the presence of reversine (500 nM). (c) Time-lapse imaging of endogenous Cyclin B-EYFP fluorescence in nocodazole-treated U2OS cells, transfected with mock siRNA or a pool of PP2A-B56 siRNAs, and treated with reversine (1 µM) at the indicated time. (d) Representative images and (e) quantification of kinetochore-localised HA-B56γ1 in nocodazole-arrested LAP-BUBR1WT or LAP-BUBR1∆KARD cells. (f) Time-lapse analysis of duration of mitotic arrest in nocodazole-treated LAP-BUBR1WT or LAP-BUBR1∆KARD-expressing cells that entered mitosis in the presence of reversine (500nM). Graphs in b and f show cumulative data from 50 cells from one experiment, which is representative of 3 independent experiments. Insets show magnifications of the boxed regions. Bar graph displays mean fold-change in kinetochore intensities (±SD) relative to mock treated LAP-BUBR1WT cells from 3 independent experiments with at least 10 cells quantified for each condition per experiment (see Supplementary Table 2). **** p<0.0001 (student t-test, unpaired).
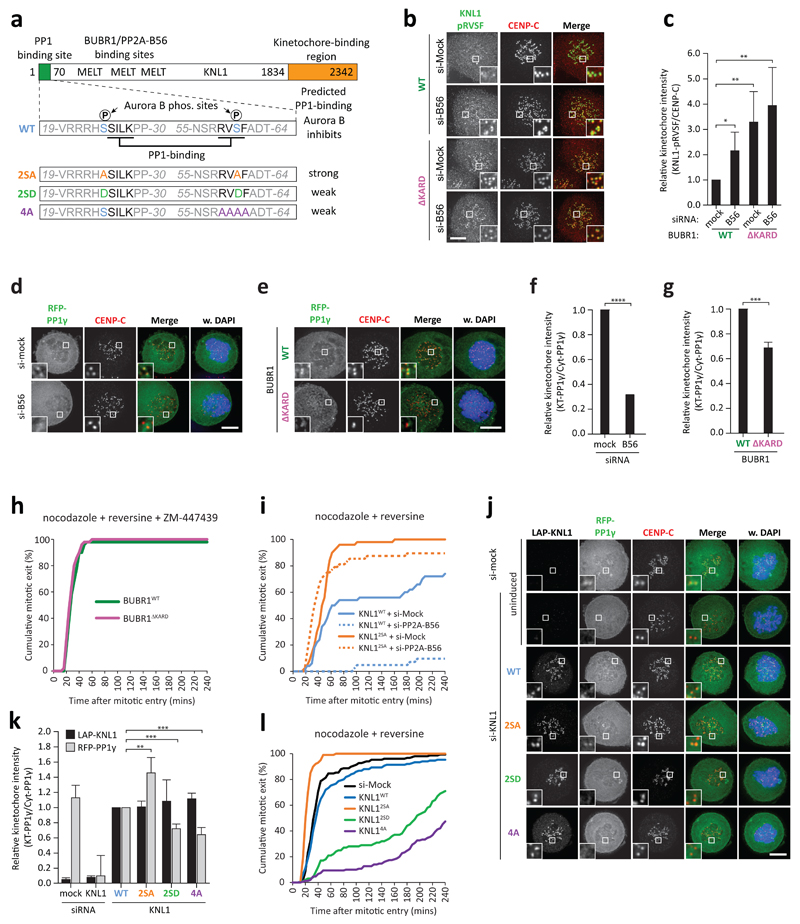
PP1 is required for SAC silencing in C. elegans, S. pombe and S. cerevisiae 8–12. PP1 and PP2A-B56 are known to bind to adjacent regions in the kinetochore scaffold KNL1: PP1 binds to conserved SSILK and RVSF motifs in the N-terminus of KNL1 21 and PP2A-B56 binds indirectly (via BUBR1) to MELT-like motifs scattered across the N-terminal half of KNL1 17–19, 22–24. BUBR1/PP2A-KNL1 interactions are promoted by MPS1-dependent phosphorylation of the MELT-like motifs 25–27 while PP1-KNL1 interaction is repressed by Aurora B-dependent phosphorylation of the SSILK/RVSF motifs 21 (Fig.2a). We hypothesised that PP2A-B56 may antagonise phosphorylation of the SSILK/RVSF motifs to induce PP1 kinetochore recruitment and SAC silencing. In agreement, PP2A-B56 depletion or BUBR1∆KARD expression elevated basal phosphoryation of the RVSF (p-Ser60) and SSILK (p-Ser24) motifs in nocodazole (Fig.2b, c and Supplementary Fig.2a-d), and this correlated with decreased kinetochore-PP1 (Fig.2d-g). Moreover, Aurora B inhibition with ZM-447439 or mutation of the SSILK/RVSF phosphorylation sites in KNL1 (KNL12SA) 21 (Supplementary Fig.2e, f) allowed efficient mitotic exit in BUBR1∆KARD-expressing or B56-depleted cells (Fig.2h, i and Supplementary Fig.3a-f). Conversely, mimicking Aurora B-dependent SSILK/RVSF phosphorylation sites (KNL12SD) or mutating the RVSF motif (KNL14A) 21 reduced kinetochore PP1 (Fig.2a, j, k and Supplementary Fig.2e, f), delayed mitotic exit following MPS1 inhibition in nocodazole (Fig.2l), and preserved cellular levels of the Mitotic Checkpoint Complex (MCC; the SAC effector) (Supplementary Fig.3g). The prolonged mitotic arrest in these cells also depended on MAD2 and BUBR1, which confirms that it was caused by persistent SAC activity (Supplementary Fig.3h). BUBR1∆KARD-expression did not significantly affect activity of relevant kinetochore kinases (MPS1, CDK1, Aurora B) (Supplementary Fig.4a). These data demonstrate that kinetochore-PP1 drives SAC silencing and that the balance between Aurora B and PP2A-B56 controls PP1 recruitment. It is of interest to note that other established kinetochore functions of PP2A-B56 14 may at least partially operate via PP1, because expression of KNL12SA also partially restored chromosome alignment defects upon B56 depletion and rescued the corresponding rise in Aurora B activity (Supplemental Fig.4b, c).
Figure 2. PP2A-B56 regulates the kinetochore-recruitment of PP1 to control SAC silencing.
(a) Schematic representation of the various KNL1 mutants used in this study. (b) Representative images and (c) quantification of immunolocalization of pRVSF(Ser60)-KNL1 and kinetochores (CENP-C) in nocodazole-treated LAP-BUBR1WT or LAP-BUBR1∆KARD-expressing cells treated with mock or PP2A-B56 siRNA, as indicated. (d, e) Representative images and (f, g) quantification of RFP-PP1γ localisation in nocodazole-arrested Flp-in HeLa cells treated with mock or B56 siRNA, or expressing LAP-BUBR1WT or LAP-BUBR1∆KARD, as indicated. (h) Time-lapse analysis of duration of mitotic arrest in nocodazole-treated LAP-BUBR1WT or LAP-BUBR1∆KARD-expressing cells that entered mitosis in the presence of ZM-447439 (2 µM) and a low dose of reversine (125 nM). (i) Time-lapse analysis of duration of mitotic arrest in nocodazole-treated Flp-in HeLa cells expressing KNL1WT or indicated KNL1 mutants and transfected B56 or mock siRNA. Cells entered mitosis in the presence of an intermediate dose of reversine (250 nM). (j) Representative images and (k) quantification of RFP-PP1γ localisation in nocodazole-treated Flp-in HeLa cells expressing KNL1WT or indicated KNL1 mutants. (l) Time-lapse analysis of duration of mitotic arrest in in Flp-in HeLa cells expressing KNL1WT or indicated KNL1 mutants. Cells entered mitosis in the presence of reversine (500 nM). Graphs in h, i and l show cumulative data from 50 cells from one experiment, which is representative of 3 independent experiments. Insets show magnifications of the boxed regions. Bar graphs display mean fold-change in kinetochore intensities (±SD) relative to mock treated LAP-BUBR1WT cells (c, f, g) or LAP-KNL1WT cell (k), from 3 (f, g) or 4 (c, k) independent experiments with at least 10 cells quantified for each condition per experiment (see Supplementary Table 2). Asterisks indicate significance (student t-test, unpaired). *: p<0.05, **: p<0.01, ***:p<0.001, ****: p<0.0001. DNA (DAPI) is shown in blue. Bars, 5 μm.
The PP1 binding site in KNL1 lies in close proximity to the MELT-like motifs that are phosphorylated by MPS1 21, 25–27, contributing to kinetochore recruitment of essential SAC effectors such as MAD1, MAD2, BUB1 and BUBR1 22, 24, 28. Expression of KNL12SD or KNL14A to reduce kinetochore PP1, elevated basal MELT motif phosphorylation and limited MELT dephosphorylation following MPS1 inhibition in nocodazole (Fig.3a, b and Supplementary Fig.4d). This correlated with an increase in kinetochore BUB1 and a corresponding increase in MAD1 (Supplementary Fig.4e-j). KNL12SD or KNL14A similarly prevented MELT dephosphorylation and BUB1 loss at metaphase as well (Supplementary Fig.4k, l). Conversely, expression of KNL12SA to enhance kinetochore PP1 (Fig.2j, k), decreased basal MELT phosphorylation and inhibited kinetochore association of BUB1 (Fig.3a, b and Supplementary Fig.4g, h). Thus, in agreement with other studies 24, 26, KNL1-bound PP1 antagonises MPS1 signalling at kinetochores. It is important to note that other pools of PP1 clearly exist at kinetochores (Fig.2j, k), as observed previously by others 29, but these cannot potently regulate SAC silencing given the strong SAC silencing defect in KNL12SD and KNL14A cells (Fig.2l).
Figure 3. PP1 controls silencing of the SAC signal and removal of kinetochore PP2A-B56.
(a) Representative images and (b) quantification of relative kinetochore intensities of indicated antigens in nocodazole-arrested Flp-in HeLa cells expressing KNL1WT or indicated KNL1 mutants. Cells were treated, as indicated, with nocodazole, MG132, DMSO and/or reversine (500 nM). (c) Representative images and (d) quantification of relative kinetochore intensities of indicated antigens in nocodazole-arrested Flp-in HeLa cells expressing KNL1WT or indicated KNL1 mutants. (e) Representative images and (f) quantification of relative kinetochore intensities of indicated antigens in nocodazole-arrested Flp-in HeLa cells that entered mitosis in the presence of DMSO or the Aurora B inhibitors ZM-447439 (2 μM) or hesperadin (100 nM). (g-j) Representative images (g, i) and quantification (h, j) of relative kinetochore intensities of indicated antigens in nocodazole-arrested Flp-in HeLa cells expressing KNL1WT or indicated KNL1 mutants. Cells in i, j were treated with mock or PP2A-B56 siRNA, as indicated. Insets show magnifications of the boxed regions. Bar graphs display mean fold-change in kinetochore intensities (±SD) relative to mock treated KNL1WT (b, d, h, j) or DMSO-treated Flp-in HeLa (f) cells, from 3 (b, d, h) or 4 (f, j) independent experiments with at least 10 cells quantified for each condition per experiment (see Supplementary Table 2). Asterisks indicate significance (student t-test, unpaired). *: p<0.05, **: p<0.01, ****: p<0.0001. DNA (DAPI) is shown in blue. Bars, 5 μm.
Since MELT phosphorylation is crucial for BUBR1 kinetochore binding 22, 24 our data suggested that PP1 may remove BUBR1-associated PP2A-B56 from kinetochores. In support of this, expression of KNL12SD or KNL14A elevated kinetochore PP2A-B56, while KNL12SA decreased it (Fig.3c, d). Interestingly, Aurora B inhibition or KNL12SA expression inhibited phosphorylation of Ser670 within the KARD of BUBR1, which is required for efficient binding of PP2A-B56 to BUBR1 17, 18, while KNL12SD and KNL4A expression elevated KARD phosphorylation (Fig.3e-h). The various KNL1 mutants did not significantly affect kinetochore activity of the relevant kinases (Supplementary Fig.4m), thus we conclude that kinetochore-PP1 promotes removal of kinetochore-PP2A-B56 by dephosphorylating the MELT and KARD motifs. Importantly, KNL12SA expression was also able to inhibit the rise in MELT and KARD phosphorylation seen following PP2A-B56 depletion (Fig.3i, j), confirming that PP2A-B56 regulates SAC silencing and its own recruitment principally via PP1. Although incomplete KNL1 knockdown/replacement (Supplementary Fig.2c) is likely to contribute to the modest rise in MELT/KARD phosphorylation in KNL12SA cells (Fig.3j), we cannot formally exclude a small additional effect of PP2A-B56 on the MELT/KARD motifs directly, as suggested recently by others 30. We consider this unlikely, however, since high kinetochore B56 in cells expressing KNL14A or KNL12SD (Fig.3c, d) could not prevent an increase in MELT/KARD phosphorylation in those cells (Fig.3a, b, g, h) and could not remove BUB1 from kinetochores following 30 min reversine treatment (Supplementary Fig.4f). These data do pose an interesting biological conundrum however; namely how do PP1 and PP2A-B56 achieve any level of specificity when they display little (if any) substrate preference in vitro 31, 32, yet they localise to an almost identical molecular space in vivo and thus their respective substrates are in very close proximity? We hypothesise that dephosphorylation of their own recruitment motifs is restricted because 1) PP1 only binds KNL1 when the SSILK/RVSF motifs are already dephosphorylated, and 2) the kinetochore-localisation of PP2A-B56 requires interaction between p-MELT/BUB3 23 and p-KARD/PP2A 17, 18, thus potentially masking these motifs from dephosphorylation when PP2A-B56 is co-localised. A requirement for PP1 docking to the KNL1 N-terminus to allow dephosphorylation of the adjacent MELT/KARD motifs may also help to explain how other pools of kinetochore PP1 can exist that do not regulate SAC silencing (see KNL12SD and KNL14A cells in Fig.2j-l), but do control other process such as kinetochore-microtubule attachment 29.
Collectively, these data demonstrate spatial negative feedback between two kinetochore phosphatases; PP2A-B56 promotes the recruitment of PP1 to kinetochores, which subsequently antagonises the localisation of PP2A-B56. MPS1-dependent MELT phosphorylation thus both initiates the SAC signal and at the same time primes the silencing of that signal by recruiting PP2A-B56. We hypothesised that such a system could impart responsiveness to the SAC (i.e. the ability to switch rapidly between the ON and OFF states): When the SAC is OFF Aurora B is predicted to phosphorylate the SSILK/RVSF motifs unopposed, thereby repressing PP1 kinetochore binding and allowing efficient MPS1-dependent MELT phosphorylation. Conversely, when the SAC is ON PP2A-B56 is predicted to compete with Aurora B to enhance PP1 kinetochore binding, thus ensuring that the SAC is primed to silence rapidly when kinetochore Aurora B and MPS1 activities diminish upon microtubule attachment/tension 33, 34 (see Fig.4a for model).
Figure 4. Lack of phosphatase feedback during prophase allows rapid initiation of the SAC signal.
(a) Schematic model for responsive SAC signalling at kinetochores. When the SAC is OFF (early prophase) signalling can be initiated rapidly due to unopposed kinase activity. When the SAC is ON (prometaphase) negative feedback is primed to silence, but then restricted by Aurora B activity. When kinase activities diminish (metaphase), phosphatases are primed to rapidly extinguish the SAC signal. (b) Representative images and (c) quantifications of relative kinetochore intensities of indicated antigens from nocodazole-treated Flp-in HeLa cells. Different mitotic phases where determined by nuclear morphology with early mitosis defined as dispersed lightly condensed chromatin and late mitosis as highly condensed chromatin balls typical of nocodazole-arrested cells. Insets display magnifications of the boxed regions. (d-f) Quantification of relative kinetochore intensities of indicated antigens from nocodazole-treated Flp-in HeLa cells following (d) endogenous BUBR1 knockdown and re-expression of LAP-BUBR1WT or LAP–BUBR1∆KARD (e) DMSO or ZM-447439 (2μM) treatment before mitotic entry, or (f) endogenous KNL1 knockdown and re-expression of LAP-KNL1WT or LAP–KNL12SA. All kinetochore intensities are relative to the maximum signal in each experiment, except in d, which are relative to the prophase signal in LAP-BUBR1WT cells. At least 10 cells were quantified for each condition per experiment and quantifications show the mean data (±SD) from 3-5 independent experiments (see Supplementary Table 2 for the specific n number for each treatment). Prophase (Pro), early mitosis (EM), late mitosis (LM). Asterisks indicate significance (student t-test, unpaired). NS: not significant, *: p<0.05, **: p<0.01, ***: p<0.001, ****: p<0.0001. Bars, 5 μm.
To test this hypothesis we first monitored key phosphorylation sites on KNL1 during mitotic entry in nocodazole, when the SAC signal is OFF and needs to quickly establish. RVSF and MELT motif phosphorylation were maximal during prophase of nocodazole-treated cells, and declined in early mitosis (Fig.4b, c). This decline, which still occurred in a dose of nocodazole known to prevent residual microtubules (3.3 μM 35; Supplementary Fig.5a), was associated with a corresponding decrease in SAC components at the kinetochore and coincided with elevated phosphorylation of the BUBR1 KARD and kinetochore recruitment of PP2A-B56 and PP1 (Fig.4b, c). Expression of BUBR1∆KARD halted the decline in RVSF and MELT phosphorylation (Fig.4d and Supplementary Fig.5b) confirming that kinetochore-PP2A-B56 antagonises Aurora B to induce negative feedback specifically following progression into prometaphase. If Aurora B effects are antagonised earlier, by direct Aurora B inhibition or KNL12SA expression, then MELT phosphorylation and SAC protein accumulation are both delayed (Fig.4e, f). Thus, the shielding of prophase kinetochores from PP2A-B56-mediated feedback, which is likely due to exclusion of BUBR1 from the nucleus, allows Aurora B to phosphorylate the SSILK/RVSF motifs unopposed and permit rapid initiation of SAC signalling. Furthermore, phosphorylation of the BUBR1 KARD 17, 18) was also markedly reduced by Aurora B inhibition or KNL12SA expression (Fig.4e, f). Thus, negative feedback from PP2A-B56 is also restricted during early mitosis until the SSILK/RVSF motifs are phosphorylated and PP1 is removed. Collectively, these data demonstrate that the absence of negative feedback from kinetochore phosphatases allows rapid establishment of SAC signalling during early mitosis.
The decline in RVSF and MELT phosphorylation in early and late mitosis was associated with PP2A-B56/PP1 recruitment and a decrease in SAC components at kinetochores (Fig.4b, c). We therefore examined whether elevated phosphatase levels at kinetochores during mitosis may prime the SAC for rapid silencing when Aurora B and MPS1 activities drop (for instance by tension-producing microtubule attachments). To this end, BUBR1WT and BUBR1∆KARD cells were arrested in nocodazole and a metaphase-like state was mimicked by combined addition of Aurora B and MPS1 inhibitors. We opted for this approach to circumvent indirect effects on SAC silencing by PP2A- and PP1-dependent regulation of kinetochore-microtubule attachments 14, 17, 21. While Aurora B and MPS1 inhibition caused rapid silencing of the SAC signal in control cells, dephosphorylation of the RVSF and MELT motifs (Fig.5a), and loss of kinetochore BUBR1 (Fig.5b), were both delayed by BUBR1∆KARD expression. This effect was even more pronounced by additional depletion of B56 (Fig.5c-e), which is likely to reflect a synergistic effect on kinetochore-B56 levels because targeting the centromeric pool of B56 directly (by SGO1 depletion) did not delay mitotic exit (Supplementary Fig.5c). Thus physical coupling between the SAC signal and PP2A-B56 ensures that kinetochore SAC signalling can be silenced rapidly following microtubule attachment/tension.
Figure 5. The priming of phosphatase-mediated feedback in prometaphase allows rapid SAC silencing.
(a) Quantification of relative kinetochore intensities and representative images of pRVSF- and pMELT-KNL1 in LAP-BUBR1WT or LAP–BUBR1∆KARD-expressing cells, arrested in prometaphase with nocodazole and MG132, and treated with reversine (1 µM) and ZM-447439 (2 µM) for the indicated times. (b) Quantification of relative LAP-BUBR1 kinetochore intensities from time-lapse images of LAP-BUBR1WT or LAP–BUBR1∆KARD cells treated as in a. (c, d) As in a, b except in LAP–BUBR1∆KARD cells depleted of PP2A-B56 and LAP-BUBR1WT treated with mock siRNA. (e) Representative images LAP-BUBR1 localisation from quantifications shown in d. (f) Schematic model of regulated negative feedback. MELT phosphorylation installs negative feedback (via PP2A-B56 recruitment), but Aurora B restricts this feedback by inhibiting PP1 recruitment. In (a, c) At least 10 cells were quantified for each condition per experiment and quantifications show the mean kinetochore intensities (±SD), relative to the 0 min timepoint in LAP-BUBR1WT cells, from 3 independent experiments (see Supplementary Table 2). In (b, d) at least 15 cells were quantified for each cell line per experiment and quantifications show the mean data (±SD), relative to the 0 min timepoint, from 3 independent experiments. Asterisks indicate significance (student t-test, unpaired). NS: not significant, **: p<0.01, ***: p<0.001. Bars, 5 μm
In summary, we show here that SAC responsiveness is due to localized negative feedback between PP1 and PP2A-B56. This ensures that the SAC signal can be switched ON rapidly, after which the SAC signal primes its own silencing to ensure kinetochores can rapidly switch SAC signalling OFF when needed. Once the SAC signal is silenced, negative feedback is locally uncoupled due to removal of PP2A-B56, which is predicted to return kinetochores to a state that permits rapid SAC initiation if required. Therefore, SAC responsiveness may not only be important during prophase and metaphase (when the SAC must be globally switched ON and OFF, respectively) but also during prometaphase when error-correction is continuously detaching chromosomes and re-establishing the SAC signal at individual kinetochores. The regulated negative feedback that we show here may be a common mechanism used by signalling networks to elicit responsiveness: the key is that the activating stimulus (Aurora B and MPS1 in our example) primes negative feedback, but then restricts this feedback until the appropriate time (Fig.5f). This is analogous to a similar feedback network at mitotic entry, when Cyclin B/CDK1 primes its own degradation by activating the APC/C, but then initiates the SAC to restrict this degradation until chromosome alignment is complete 36–39. The result is an active APC/C that can rapidly degrade Cyclin B as soon as the brake on negative feedback is released at metaphase. It will be important to determine whether such network topology is repeated in other signalling processes that must be similarly responsive.
Methods
Cell culture and reagents
U2OS cells, Cyclin B-EYFP U2OS cells 16 and HeLa Flp-in cells (a gift from S. Taylor), stably expressing a TetR, were cultured in DMEM supplemented with 9% tetracycline-approved FBS, 50 µg/ml penicillin/streptomycin and 2 mM L-glutamine. All cell lines were routinely screened (every 4-8 weeks) to ensure they were free from mycoplasma contamination. All HeLa Flp-in cells stably expressing a doxycycline-inducible construct were derived from the HeLa Flp-in cell line by transfection with pCDNA5/FRT/TO vector (Invitrogen) and pOG44 (Invitrogen) and cultured in the same medium but containing 200 µg /ml hygromycin and 4 µg /ml blasticidin. The HeLa Flp-in RFP-PP1γ D10 cell line, stably and constitutively expressing RFP-PP1γ, was created by transfection of HeLa Flp-in cells with pCDNA3-puro-2xRFP-PP1γ, puromycin selection and clonal isolation. Double positive LAP-KNL1/RFP-PP1γ cell lines were derived from this background by genomic integration of pCDNA5-LAP-KNL1 plasmids, as described above. HeLa Flp-in cells stably expressing doxycycline-inducible LAP-BUBR1WT or LAP-BUBR1∆KARD were infected twice with pSuperior retrovirus expressing a doxycycline-inducible short hairpin targeting BUBR1 (AGATCCTGGCTAACTGTTCtctcttgaa GAACAGTTAGCCAGGATCT). Stable polyclonal cell lines were generated following puromycin selection. To induce protein expression in the inducible cell lines, 1 µg/ml doxycycline was added for ≥36 h. Thymidine (2 mM), nocodazole (830 nM), MG132 (10 µM), reversine and puromycin were all obtained from Sigma-Aldrich. Hygromycin was purchased from Roche, ZM-447439 from Tocris Bioscience, Hesperadin from Selleck Chemicals, Blasticidin from PAA Laboratories. AZ-3146 was from Axon. Purified MPS1 was purchased from Life Technologies (PV3792).
Plasmids and cloning
pOG44 (Invitrogen) encodes a FLP recombinase expression vector. HA-PP2A-B56 plasmids were described previously (addgene plasmids 14532-14537; deposited by D. Virshup). pCDNA5-LAP-BUBR1WT, encoding and N-terminally LAP-tagged and siRNA-resistant wild type BUBR1, and pCDNA5-LAP-BUBR1ΔKARD, lacking amino acids 647-697, were subcloned from the respective pLAP-BUBR1 constructs 17. GST-KNL1-M3 and GST-KNL1-A3, which contain a KNL1818-1051 fragment encompassing 3 MELT motifs, were described previously 22. pCDNA5-LAP-KNL1WT encodes full length, N-terminally LAP-tagged and siRNA-resistant wild type KNL1 (modified codons 258 and 259) and was created by digestion of pEYFP-LAP-KNL1WT (a gift from I. Cheeseman) with XhoI and HpaI to isolate the full length KNLWT cassette, which was ligated into the XhoI and PmeI sites of pCDNA5/FRT/TO (Invitrogen). An N-terminal LAP-tag was introduced by subcloning the LAP-tag cassette from pCDNA3-LAP-MPS1Δ200 (Ref 34) into the KpnI and XhoI sites of the resulting plasmid. pCDNA5-LAP-KNL12SA and pCDNA5-LAP-KNL12SD were created by subcloning the KNL1 N-terminus from pEYFP-LAP-KNL12SA and pEYFP-LAP-KNL12SD (also gifts from I. Cheeseman) into pCDNA5-LAP-KNL1WT via the XhoI and EcoRV restriction sites. pCDNA5-LAP-KNL14A was created by site-directed mutagenesis of pCDNA5-LAP-KNL1WT. pCDNA3-puro-2xRFP-PP1γ was created by PCR of a tandem of tagRFP inserts from pPA-TAGRFP-H2B (Evrogen) and ligation into the BamHI and NotI restriction sites of the N-terminal LAP-tag of pLAP-PP1γ (a gift from I. Cheeseman). The resulting RFP-RFP-PP1γ cassette was subcloned into the BamHI and EcoRI restriction sites of pCDNA3-puro. The pSuperior BUBR1 inducible-short hairpin vector was generated by annealing synthesised primers and subsequent ligation into the pSuperior vector, as per manufacturer’s instructions (Oligoengine).
Phosphatase screen
U2OS cells were seeded in 96 well plates at 20% confluency, and transfected with 20 nM siRNA using Hiperfect (20nM final of four pooled siRNAs from a dharmacon ON-TARGETplus siRNA library targeting human phosphatases; GU103700). After 48 hours, cells were treated with nocodazole and allowed to accumulate in prometaphase for 16 hours. Finally, cells were driven out of mitosis by the addition of 1 µM of reversine for 1 hour. Cells were fixed with 4% PFA for 45 min at room temperature, washed with PBS and blocked with 2% BSA, supplemented with 0,2% TX100, for at least 1 hour. Cells were stained for pSer10-Histone-H3 to identify mitotic cells and DAPI. Image acquisition was performed using a Cellomics ArrayScan VTI (Thermo Scientific) using a 10× 0.50NA objective. Image analysis was performed using Cellomics ArrayScan HCS Reader (Thermo Scientific). In short, cells were identified on the basis of DAPI staining and they were scored as `mitotic' if the pSer10-Histone-H3 staining reached a pre-set threshold.
Knockdown and reconstitution experiments with LAP-KNL1 and LAP-BUBR1WT/∆KARD
For all KNL1 and BUBR1 mutant experiments, the endogenous mRNA was knocked down and replaced with an siRNA-resistant mutant using HeLa Flp-in cells, as stated below. For knockdown and reconstitution of KNL1 in HeLa Flp-in cells, cells were transfected with 20 nM KNL1 or mock siRNA and, in some experiments, 20 nM additional mock, MAD2, BUBR1 or B56 siRNA for 16 h after which the cells were arrested in early S phase for 24 h by addition of thymidine. Subsequently, cells were release from thymidine for 8-10 h and arrested by the addition of nocodazole and (for immunolocalisation experiments) subsequently treated with MG132 to prevent mitotic exit and (in some experiments) Aurora B or MPS1 inhibitors or DMSO for 20-30 minutes. LAP-KNL1 expression was induced by the addition of doxycycline during and following the thymidine block. For knockdown and reconstitution of BUBR1WT/∆KARD, stable cells were induced with doxycycline overnight (to simultaneously knockdown endogenous BUBR1 and induce the ectopic forms), then arrested in thymidine and doxycycline for a further 24 hours before release from thymidine for 8-10 h into nocodazole and doxycycline (for immunolocalisation or live imaging experiments).
Transfections and siRNA
Plasmids were transfected into Flp-in HeLa cells using Fugene HD (Promega) according to the manufacturer’s instructions. siRNAs used in this study were as follows: si-MPS1, 5’-GACAGAUGAUUCAGUUGUA-3’ (custom; Thermo Fisher Scientific); si-mock (Luciferase GL2 duplex; D-001100-01-20; Thermo Fisher Scientific); si-KNL1, 5’-GCAUGUAUCUCUUAAGGAA-3’ (CASC5#5; J-015673-05; Thermo Fisher Scientific); si-BUBR1, 5’-AGAUCCUGGCUAACUGUUC-3’ (custom; Thermo Fisher Scientific); si-MAD2, 5’-UACGGACUCACCUUGCUUG-3’ (custom; Thermo Fisher Scientific); si-SGO1, 5’-GAUGACAGCUCCAGAAAUU-3’ (custom; Thermo Fisher Scientific). The B56 family siRNA pool was composed of 5 individual siRNAs that together targeted all B56 isoforms as described previously 14. The siRNAs were mixed at an equimolar ratio and transfected at a total concentration of 20 nM. The siRNAs used (all from Thermo Fisher Scientific) were B56α (PPP2R5A), 5’-UGAAUGAACUGGUUGAGUA-3’; B56β (PPP2R5B), 5’-GAACAAUGAGUAUAUCCUA-3’; B56γ (PPP2R5C), 5’-GGAAGAUGAACCAACGUUA-3’; B56δ (PPP2R5D), 5’-UGACUGAGCCGGUAAUUGU-3’; B56ε (PPP2R5E), 5’-GCACAGCUGGCAUAUUGUA-3’. All siRNAs were transfected using HiPerFect (Qiagen) at 20 nM according to the manufacturer’s instructions.
Immunoprecipitation and immunoblotting
Flp-in HeLa cells were treated with thymidine for 24 h and subsequently released into nocodazole for 14 h. Cells were treated with reversine (500 nM, unless stated otherwise) or DMSO and MG132 to prevent mitotic exit for 1 hour. Mitotic cells were isolated by mitotic shake off and lysed in lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1% TX-100, 2mM MgCl2, 5 mM EDTA, 1 mM Na3VO4, 1 mM β-glycerophosphate, 1 mM NaF and complete protease inhibitor [Roche]) on ice. The cleared extract was incubated with 10% protein A-agarose beads (Roche)/antibody mix for 2 hours at 4°C on a rotating wheel. The beads were washed four times with lysis buffer. Supernatant and beads were processed for SDS-PAGE and the proteins were transferred to nitrocellulose membranes for immunoblotting. Immunoblotting was performed using standard protocols; the signal was visualized and analyzed on a scanner (ImageQuant LAS 4000; GE Healthcare) using enhanced chemiluminescence.
Antibodies
The pMELT-KNL1 antibody, directed against Thr943 and Thr1155 of human KNL1 (which have identical sequences), was raised in rabbits using the peptide MEIpTRSHTTALEC (Genscript). The antibody was used at 1:2000 dilution in the presence of nonphosphorylated peptide (1 ng/ml) in all experiments. The pSILK-KNL1 (pSer24-KNL1) and pRVSF-KNL1 (pSer60-KNL1) antibodies (custom rabbit polyclonals, characterised previously 40; gifts from Iain Cheeseman), were used at 1:2000 dilution in the presence of a nonphosphorylated peptide (1 ng/ml) in all experiments. The pKARD antibody recognizes pSer670-BUBR1 (custom rabbit polyclonal) and was used at 1:2000 dilution. The following primary antibodies were used for immunofluorescence and/or immunoblotting at indicated dilutions: α-tubulin (clone B-5-1-2, T5168, Sigma-Aldrich. 1:10000), Aurora B pT232 (Rockland; 600-401-677S. 1:4000), BUB1 (A300-373A; Bethyl. 1:2000), BUBR1 (A300-386A; Bethyl. 1:2000), BUBR1 (custom sheep polyclonal. 1:30000), BUBR1 (clone 8G1, 05-898; Upstate/Millipore. 1:2500), CDC20 (clone E-7, sc-13162; Santacruz Biotechnology), CENP-C (PD030; MBL. 1:5000), CENP-T (D286-3; MBL. 1:2000), CENP-T pS47 (custom rabbit polyclonal, a gift from I. Cheeseman 41. 1:2000), CREST (Cortex Biochem. 1:2000), GFP (clones 7.1 and 13.1, #11814460001; Roche. 1:1000), GFP (clone 4E12/8; a gift from Peter Parker. 1:1000), GFP (clone LGB-1, ab291, Abcam. 1:2000), GFP (custom rabbit polyclonal. 1:10000), GST (clone B1-14, sc-138; Santacruz Biotechnology. 1:1000), HA (clone Y-11, sc-805; Santacruz Biotechnology. 1:1000), pSer10-Histone 3 (06-570, Millipore. 1:2000), KNL1 (ab70537, Abcam. 1:1000), MAD1 (clone BB3-8, custom mouse monoclonal, a gift from A. Musacchio. 1:100), MAD2 (custom rabbit polyclonal), MPS1 pThr676 (custom rabbit polyclonal. 1:1000) 42, PP2A-B56α (clone 23, sc-136045; Santacruz Biotechnology. 1:1000), SGO1L1 (clone 3C11, H00151648-M01; Abnova. 1:2000), TFR (13-6890; Invitrogen. 1:2500). Secondary antibodies for immunofluorescence experiments were goat anti-rat Alexa Fluor 647, high-cross absorbed goat-anti-guinea pig and anti-human Alexa Fluor 647, donkey anti-sheep Alexa Fluor 568 and goat anti-rabbit and anti-mouse Alexa Fluor 488 and Alexa Fluor 568 (Molecular Probes); all used at 1:1000 dilution.
Live-cell imaging and immunofluorescence
For live-cell imaging for time-lapse analysis, cells were plated in 24-well glass-bottom plates (MatTek Corporation), transfected, and imaged in a heated chamber (37°C and 5% CO2) using a 20x/0.5 NA UPLFLN objective (Olympus) on a microscope (IX-81; Olympus) controlled by Cell-M software (Olympus). Images were acquired using an ORCA-ER camera (Hamamatsu Photonics) and processed using Cell-M software.
For live-cell imaging of LAP-BUBR1, cells were plated in 8 well chamber slides (Ibidi) in the presence of doxycycline (to replace endogenous BUBR1 with LAP-BUBR1). 16h later Thymidine was added for a further 24h before release into Leibovitz L-15 media (Invitrogen) supplemented with 10% FCS, 2 mM L-glutamine, 100 U/ml penicillin and streptomycin, doxycycline and nocodazole. Cells arrested in mitosis 12h later were treated as indicated and imaged on a DeltaVision core system equipped with a heated 37°C chamber, with a 100x/1.40 NA U Plan S Apochromat objective using softWoRx software. Images were acquired using a camera (CoolSNAP HQ2; Photometrics) and processed using softWorx software and ImageJ (National Institutes of Health).
For immunofluorescence, cells plated on 12-mm coverslips were pre-extracted with 0.1% Triton X-100 in PEM (100 mM Pipes, pH 6.8, 1 mM MgCl2 and 5 mM EGTA) for 45s before fixation with 4% paraformaldehyde in PBS for 10 min. Coverslips were washed with PBS and blocked with 3% BSA in PBS for 30 min, incubated with primary antibodies for 2-4 h at room temperature or 16 h at 4°C, washed with PBS and incubated with secondary antibodies for an additional hour at room temperature. Coverslips were then incubated with DAPI for 2 min, washed and mounted using antifade (ProLong; Molecular Probes). For imaging of RFP-PP1γ, cells were treated as above, but fixed with 4% in PBS for 10 min, washed with PBS and permeabilised with 0.5% Triton X-100 for 15 min. RFP-PP1γ was imaged by acquiring the direct fluorescence of the tagRFP moiety. For alignment assays, cells were treated as before, but were treated with MG132 for 30 min and fixed with 3.7% Shandon Zinc Formal-Fixx (Thermo Scientific) for 10 min, washed with PBS and permeabilised with 0.5% Triton X-100 for 15 min. All images were acquired on a deconvolution system (Deltavision RT or Deltavision Elite; Applied Precision) with a 100x/1.40 NA U Plan S Apochromat objective (Olympus) using softWoRx software (Applied precision). Images are maximum intensity projections of deconvolved stacks. All displayed immunofluorescence images were chosen to most closely represent the mean quantified data.
Image quantification
For quantification of immunostainings, all images of similarly stained experiments were acquired with identical illumination settings and analysed using ImageJ (for experiments in which ectopic proteins were expressed, cells with comparable levels of exogenous protein were selected for analysis). An ImageJ macro was used to threshold and select all kinetochores and all chromosome areas (excluding kinetochores) using the DAPI and anti- kinetochore antibodies channels as described previously 42. This was used to calculate the relative mean kinetochore intensity of various proteins ([kinetochores-chromosome arm intensity (test protein)]/[kinetochores-chromosome arm intensity (CENP-C/CENP-T/CREST)]. For the quantification of RFP-PP1γ, a maximal intensity projection was generated from a selected, kinetochore-dense 2 µm region within the deconvolved stack to isolate the weak kinetochore signal from the cytoplasmic signal above and below the chromatin. Kinetochores were selected and measured as above, but RFP-PP1γ kinetochore intensity was calculated as a ratio of the cytosolic signal to correct for fluctuations in expression. For quantification of live LAP-BUBR1 kinetochore levels, two regions were selected for each time-point: a region encompassing all kinetochores [KT] and a region immediately adjacent in the cytoplasm [BG]. Relative change in kinetochore intensity ([mean KT]-[mean BG]) was calculated over time. For quantification of HA-B56 localisation, a line was drawn through KT pairs lying in the same Z-section (using ImageJ). The plot profile function was used to measure intensities across the line, after placing the first KT peak at a fixed 0.2 μM distance from the start. The LAP-BUBR1 channel was used to choose 5 random kinetochore pairs per cell for intensity measurements.
Statistical tests
Two-tailed, unpaired t-tests were performed to compare experimental groups in immunofluorescence quantifications (using Prism 6 software). The comparisons most pertinent for the conclusions are shown in the figures and legends, and a more complete set of comparisons is given in the source data file.
Supplementary Material
Acknowledgments
We thank Mathijs Vleugel and Manja Omerzu for help with the pMELT-KNL1 antibody, Iain Cheeseman, Andrea Musacchio, Anna de Antoni, Peter Parker and Stephen Taylor for reagents. We thank the Kops, Lens, Saurin, Swedlow and Griffis labs for discussions. This work is supported by the European Research Council (ERC-StG KINSIGN to GJPLK), by the Netherlands Organisation for Scientific Research (NWO-Vici 865.12.004 to GJPLK), by the KWF Kankerbestrijding (UU-2012-5427 to GJPLK), by TiPharma (T3-503 to GJPLK) and by funding to ATS (from the Ninewells Cancer Campaign, Leng Charitable Trust and Tenovus Scotland Tayside).
Footnotes
Author contributions
WN, GJPLK and ATS conceived the study, designed experiments and interpreted data. WN, GV, AvdD and ATS performed experiments. GJPLK and ATS wrote the manuscript, with input from WN.
References
- 1.Santaguida S, Musacchio A. The life and miracles of kinetochores. The EMBO journal. 2009;28:2511–2531. doi: 10.1038/emboj.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Foley EA, Kapoor TM. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nature reviews Molecular cell biology. 2013;14:25–37. doi: 10.1038/nrm3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kops GJ, Shah JV. Connecting up and clearing out: how kinetochore attachment silences the spindle assembly checkpoint. Chromosoma. 2012;121:509–525. doi: 10.1007/s00412-012-0378-5. [DOI] [PubMed] [Google Scholar]
- 4.Funabiki H, Wynne DJ. Making an effective switch at the kinetochore by phosphorylation and dephosphorylation. Chromosoma. 2013;122:135–158. doi: 10.1007/s00412-013-0401-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bomont P, Maddox P, Shah JV, Desai AB, Cleveland DW. Unstable microtubule capture at kinetochores depleted of the centromere-associated protein CENP-F. The EMBO journal. 2005;24:3927–3939. doi: 10.1038/sj.emboj.7600848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dick AE, Gerlich DW. Kinetic framework of spindle assembly checkpoint signalling. Nature cell biology. 2013;15:1370–1377. doi: 10.1038/ncb2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Santaguida S, Tighe A, D'Alise AM, Taylor SS, Musacchio A. Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. J Cell Biol. 2010;190:73–87. doi: 10.1083/jcb.201001036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Espeut J, Cheerambathur DK, Krenning L, Oegema K, Desai A. Microtubule binding by KNL-1 contributes to spindle checkpoint silencing at the kinetochore. J Cell Biol. 2012;196:469–482. doi: 10.1083/jcb.201111107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vanoosthuyse V, Hardwick KG. A novel protein phosphatase 1-dependent spindle checkpoint silencing mechanism. Current biology : CB. 2009;19:1176–1181. doi: 10.1016/j.cub.2009.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meadows JC, et al. Spindle checkpoint silencing requires association of PP1 to both Spc7 and kinesin-8 motors. Dev Cell. 2011;20:739–750. doi: 10.1016/j.devcel.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosenberg JS, Cross FR, Funabiki H. KNL1/Spc105 recruits PP1 to silence the spindle assembly checkpoint. Current biology : CB. 2011;21:942–947. doi: 10.1016/j.cub.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pinsky BA, Nelson CR, Biggins S. Protein phosphatase 1 regulates exit from the spindle checkpoint in budding yeast. Current biology : CB. 2009;19:1182–1187. doi: 10.1016/j.cub.2009.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gutierrez-Caballero C, Cebollero LR, Pendas AM. Shugoshins: from protectors of cohesion to versatile adaptors at the centromere. Trends in genetics : TIG. 2012;28:351–360. doi: 10.1016/j.tig.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 14.Foley EA, Maldonado M, Kapoor TM. Formation of stable attachments between kinetochores and microtubules depends on the B56-PP2A phosphatase. Nature cell biology. 2011;13:1265–1271. doi: 10.1038/ncb2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu P, Virshup DM, Lee SH. B56-PP2A regulates motor dynamics for mitotic chromosome alignment. Journal of cell science. 2014 doi: 10.1242/jcs.154609. [DOI] [PubMed] [Google Scholar]
- 16.Akopyan K, et al. Assessing kinetics from fixed cells reveals activation of the mitotic entry network at the S/G2 transition. Molecular cell. 2014;53:843–853. doi: 10.1016/j.molcel.2014.01.031. [DOI] [PubMed] [Google Scholar]
- 17.Suijkerbuijk SJ, Vleugel M, Teixeira A, Kops GJ. Integration of kinase and phosphatase activities by BUBR1 ensures formation of stable kinetochore-microtubule attachments. Dev Cell. 2012;23:745–755. doi: 10.1016/j.devcel.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 18.Kruse T, et al. Direct binding between BubR1 and B56-PP2A phosphatase complexes regulate mitotic progression. Journal of cell science. 2013;126:1086–1092. doi: 10.1242/jcs.122481. [DOI] [PubMed] [Google Scholar]
- 19.Xu P, Raetz EA, Kitagawa M, Virshup DM, Lee SH. BUBR1 recruits PP2A via the B56 family of targeting subunits to promote chromosome congression. Biology open. 2013;2:479–486. doi: 10.1242/bio.20134051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hewitt L, et al. Sustained Mps1 activity is required in mitosis to recruit O-Mad2 to the Mad1-C-Mad2 core complex. J Cell Biol. 2010;190:25–34. doi: 10.1083/jcb.201002133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu D, et al. Regulated targeting of protein phosphatase 1 to the outer kinetochore by KNL1 opposes Aurora B kinase. J Cell Biol. 2010;188:809–820. doi: 10.1083/jcb.201001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vleugel M, et al. Arrayed BUB recruitment modules in the kinetochore scaffold KNL1 promote accurate chromosome segregation. J Cell Biol. 2013;203:943–955. doi: 10.1083/jcb.201307016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Primorac I, et al. Bub3 reads phosphorylated MELT repeats to promote spindle assembly checkpoint signaling. eLife. 2013;2:e01030. doi: 10.7554/eLife.01030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang G, Lischetti T, Nilsson J. A minimal number of MELT repeats supports all the functions of KNL1 in chromosome segregation. Journal of cell science. 2014;127:871–884. doi: 10.1242/jcs.139725. [DOI] [PubMed] [Google Scholar]
- 25.Yamagishi Y, Yang CH, Tanno Y, Watanabe Y. MPS1/Mph1 phosphorylates the kinetochore protein KNL1/Spc7 to recruit SAC components. Nature cell biology. 2012 doi: 10.1038/ncb2515. [DOI] [PubMed] [Google Scholar]
- 26.London N, Ceto S, Ranish JA, Biggins S. Phosphoregulation of Spc105 by Mps1 and PP1 Regulates Bub1 Localization to Kinetochores. Current biology : CB. 2012 doi: 10.1016/j.cub.2012.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shepperd LA, et al. Phosphodependent Recruitment of Bub1 and Bub3 to Spc7/KNL1 by Mph1 Kinase Maintains the Spindle Checkpoint. Current biology : CB. 2012 doi: 10.1016/j.cub.2012.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krenn V, Overlack K, Primorac I, van Gerwen S, Musacchio A. KI motifs of human Knl1 enhance assembly of comprehensive spindle checkpoint complexes around MELT repeats. Current biology : CB. 2014;24:29–39. doi: 10.1016/j.cub.2013.11.046. [DOI] [PubMed] [Google Scholar]
- 29.Posch M, et al. Sds22 regulates aurora B activity and microtubule-kinetochore interactions at mitosis. J Cell Biol. 2010;191:61–74. doi: 10.1083/jcb.200912046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Espert A, et al. PP2A-B56 opposes Mps1 phosphorylation of Knl1 and thereby promotes spindle assembly checkpoint silencing. J Cell Biol. 2014;206:833–842. doi: 10.1083/jcb.201406109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bollen M, Peti W, Ragusa MJ, Beullens M. The extended PP1 toolkit: designed to create specificity. Trends in biochemical sciences. 2010;35:450–458. doi: 10.1016/j.tibs.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barr FA, Elliott PR, Gruneberg U. Protein phosphatases and the regulation of mitosis. Journal of cell science. 2011;124:2323–2334. doi: 10.1242/jcs.087106. [DOI] [PubMed] [Google Scholar]
- 33.Liu D, Vader G, Vromans MJ, Lampson MA, Lens SM. Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science. 2009;323:1350–1353. doi: 10.1126/science.1167000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nijenhuis W, et al. A TPR domain-containing N-terminal module of MPS1 is required for its kinetochore localization by Aurora B. J Cell Biol. 2013;201:217–231. doi: 10.1083/jcb.201210033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Santaguida S, Vernieri C, Villa F, Ciliberto A, Musacchio A. Evidence that Aurora B is implicated in spindle checkpoint signalling independently of error correction. The EMBO journal. 2011;30:1508–1519. doi: 10.1038/emboj.2011.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pines J. Cubism and the cell cycle: the many faces of the APC/C. Nature reviews Molecular cell biology. 2011;12:427–438. doi: 10.1038/nrm3132. [DOI] [PubMed] [Google Scholar]
- 37.Morin V, et al. CDK-Dependent Potentiation of MPS1 Kinase Activity Is Essential to the Mitotic Checkpoint. Current biology : CB. 2012 doi: 10.1016/j.cub.2011.12.048. [DOI] [PubMed] [Google Scholar]
- 38.D'Angiolella V, Mari C, Nocera D, Rametti L, Grieco D. The spindle checkpoint requires cyclin-dependent kinase activity. Genes & development. 2003;17:2520–2525. doi: 10.1101/gad.267603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vazquez-Novelle MD, et al. Cdk1 inactivation terminates mitotic checkpoint surveillance and stabilizes kinetochore attachments in anaphase. Current biology: CB. 2014;24:638–645. doi: 10.1016/j.cub.2014.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Welburn JP, et al. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Molecular cell. 2010;38:383–392. doi: 10.1016/j.molcel.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gascoigne KE, et al. Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell. 2011;145:410–422. doi: 10.1016/j.cell.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saurin AT, van der Waal MS, Medema RH, Lens SM, Kops GJ. Aurora B potentiates Mps1 activation to ensure rapid checkpoint establishment at the onset of mitosis. Nat Commun. 2011;2:316. doi: 10.1038/ncomms1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.