Abstract
Background:
Breast cancer is most serious reasons of women death around worldwide result in increasing its morbidity and mortality. MicroRNAs are considered as significant regulators of cancer biological processes. The main aim of this study is restoration of miR-126 could lead to modulate breast cell line and impairs their proliferation by targeting vascular endothelial growth factor gene (VEGF-A).
Methods:
Breast cancer cell line (MCF7) was transfected by miR-126 lipofectamine and negative miR control for 24 hr. Cytotoxic effects of miR-126 lipofectamine were determined by cell viability assay. Cell proliferation and cell cycle were quantitatively measured using PicoGreen assay and DAPI stain-flow cytometer analysis. For further investigation, Taq-Man real time PCR assay was performed to detect relative VEGF-A and miRNA-126 level.
Results:
MiR-126 was overexpressed in treated breast cancer cell (MCF7) compared with control cells. miR-126 expression has been associated –with a decrease in cell proliferation and arrested MCF7 cells at G1 phase. The study found that vascular endothelial growth factor is regulated by miR-126. Hence, VEGF-A is considered as functional vital and direct target to miR-126 in breast cancer cell line (MCF7).
Conclusions:
This study provided that manipulated miR-126 level may suggest a novel therapeutic approach in breast cancer treatment. However, an animal models study is needed to address and prove predictive ability of miR-126 on breast cancer controlling.
Keywords: MCF7 cell, miR-126 lipofectamine, VEGF-A expression, cell cycle, Taqman real time PCR assay
Introduction
Breast cancer is still serious problems that experienced women in worldwide causing high mortality. It is well- known that vascular endothelial growth factor (VEGF) increases the permeability of blood vessels during the tumor growth which lead to create new blood vessels (Connolly et al. 1989). VEGF has a key role in promoting endothelial proliferation and tumor growth during tumor expansion production by growth of angiogenesis (Connolly et al., 1989).
In addition, vascular VEGF-A refer to representative factors of angiogenesis and that was similar to other representative factors such as VEGF-C/D (Adams and Alitalo, 2007). Consequently, previous study reported that the VEGF family induce angiogenesis in general in oral cancer (Sasahira et al., 2007; Sasahira et al., 2007; Sasahira et al., 2008; Sasahira et al., 2010; Sasahira et al., 2012). Conventional strategies of cancer treatment are focusing on chemotherapy exposure; meanwhile they have severed deleterious effects. Therefore, so many studies attempted to apply prevention approach in control cancer progression.
Several studies have proposed that miRNAs plays a crucial role in many of biological progressions in breast cancer throughout its action as a tumor suppressor gene in many malignant tumor cells, inhibiting the progression of some cancers via negative control of proliferation, migration, invasion and cell survival (Kayani et al., 2011; Mulrane et al., 2013; Serpico et al., 2014). It has been revealed that down regulation of VEGF is triggered by miR-126 exposure in oral squamous cell carcinoma (Sasahira et al., 2012) and lung cancer (Alevizakos et al., 2013).
In same context, it has been reported that the underlying mechanism of high expression miR-126 was associated with low expression of chemokine receptor CXCR4 and inhibits the MAPK, (proteinkinase) signalling pathway (Li et al., 2013). Although, former studies on miRNA administration have been reported in controlling cancer, however it needs to elucidate potential mechanism. Hence, our study was undertaken clarifying angiogenesis proceeding in breast cancer via miRNA supplementation.
Materials and Methods
Cell Culture
The cryostored breast cancer line (MCF7) was obtained from stocks in the School of Applied Sciences, RMIT, University (Australia). Cell line samples were in 100 frequents as biological samples. They were routinely cultured in DMEM medium adding 10% fetal bovine serum (Gibco, Australia) and incubated at 37°C in a humidified atmosphere containing 5 % CO2.
miRNA Preparation
MiR-126 was used as a model drug which was at sequences 5’UCGUACCGUGAGUAAUAAUGCG ’3 and 5’CAUUAUUACUUUUGGUACGCG’3 (Life technologists, Australia). A 1 µg anionic charge miR-126 was complexed with cationic charge lipofectamine® RNAMAX transfection reagent according to manufacturer instructions (Life technologists, Australia). The lipofecatmine vehicle was used in this experiment to protect miRNA degradation
MCF7 cell line transfection
Breast cancer cells were collected at 80% confluence, and assigned into three groups, (1) blank group (no treatment), (2) miR-126 mimics lipofectamine, (3) miR-126 negative control. All cell groups were transfected according to the manufacturer’s instructions (Life technologists, Australia). Briefly, MCF7 cells were seeded into 96 well plates until 60 % confluent transiently transfected with miR-126 mimics lipofectamine, and negative control prior for further analysis.
Cell viability
Breast cancer cells (MCF7) were seeded into 96 well plates with the density of 2 X 104 cell/ 100 µL and treated with miR-126 lipofectamine, miRNA-negative control and blank control. Then, the cells were incubated for 24 hr at 37°C and 5 % CO2. After that, cell viability measuring has been done using an AlamarBlue® assay according to (Alhasan et al., 2016). cell culture medium was added into each well and incubated at 37°C in humidified 5% CO2 for 4 h. Fluorescence was visualised under wavelength of 570 nm and an emission wavelength of 600 nm (SpectraMax® Paradigm, Molecular Devices LLC, Sunnyvale, CA, USA).
Relative miR-126 level
RNA extraction of all samples of breast cancer cells has been conducted using an RNeasy Mini Kit (Qiagen, Australia) according to the manufacturer’s protocol. The RNA concentration was determined by NanoDrop 2000 (Thermo Fisher Scientific Inc., Waltham, Massachusetts, USA), total RNA was converted into cDNA according to instructions of a TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, 433596). Relative miR-126 expression level was normalised to RNU44 (Applied Biosystems, assay ID 001094) as internal control in the tested samples.
TaqMan-qPCR assay
qPCR induction (Rotor-Gene, Qiagen Pty Ltd, Australia) with default settings was done using an activation start cycle for 10 min at 95°C, followed by 40 cycles of 15 seconds at 95°C and 60 seconds at 60°C and 30 seconds at 72°C. Relative mi-R 126 expression level was normalised to RNU44 (Applied Biosystems, assay ID 001094) and VEGF-A (Applied Biosystems, assay ID Hs00900055_m1) was normalised to GAPDH (Applied Biosystems, assay ID Hs02758991_g1). Experiment was conducted in triplicate for (6 samples for each sample of cancer cell line and in three technical samples). Folds values were determined by the 2-ΔΔCT method (Mw 2006).
Proliferation assay
The proliferation of the treated and untreated MCF7 cells were measured by quantifying their deoxyribonucleic acid (DNA) content using a Picogreen® assay according to manufacturer’s instructions. The cells were trypsinized after treatment after 24 h. The cells were then rinsed two times with cold DPBS, followed by centrifugation for 5 min at 1,200 rpm. Cell pellets were subsequently collected for lysis using a NP40 cell lysis buffer (Life Technologies, Australia) for 30 min on ice, and at 10-min intervals. The cell lysates were subjected to a freeze-thaw cycle prior to centrifugation at 13,000 rpm for 10 min at 4°C. A 100 µL portion of clear lysate was aliquoted to a clean 96-well plate and incubated for 5 min with 100 µl of PicoGreen® reagent at room temperature in the dark. Fluorescence was measured using the multi-mode microplate reader at an excitation wavelength of 480 nm and an emission wavelength of 520 nm. Cell numbers in each sample were determined by reference to a DNA standard curve. The DNA standard curve was obtained using cell lysates containing a number of control cells.
Flow cytometry analysis
All samples of Breast cancer cells (MCF7) were harvested by TrypLE Express (Life technologists), and washed 2X PBS to discard serum . Centrifugation has been done at 1,200 rpm, for 5min. The pellet was resuspend in 1 mL PBS (Ca and Mg free). A 95% Ethanol was added 3 mL and vortexing. Then, cells were fixed in 70 % Ethanol solution for 30 min. Cell suspension/dilute ETOH were spined at 2,000-2,200 rpm for 10 min, and washed
again in 15 mL PBS; and re-centrifuging at 2,000-2,200 rpm for 10 min. The cells were counted and resuspend in 0.5 µL/mL DAPI stain solution, and incubated for 30 min prior to flow cytometric analysis.
Statistical Analysis
All experiments were represented as the average value ± standard deviation. Data were compared using One-Way Analysis of Variance (ANOVA). Two groups of data were compared using the Student’s t-test. Differences were considered statistically significant when p < 0.0001.
Results
Cell Viability
There was no significant increasing in cell viability numbers between treated and control and negative –miRNA cell line (p= 0.9874, 0.0562) however; the ratio of cell viability in treated sample appeared higher than control (Figure 1).
Figure 1.
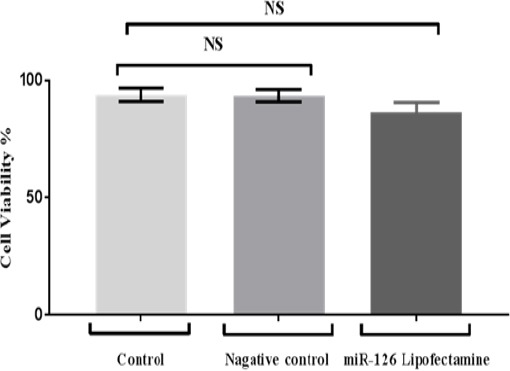
Shows Cell Viability of Treated MCF7 Cells by miR-126 Lipofectamine and Negative miRNA Control. Cells were examined 24 h using Alamar blue assay. Data were analysed by One Way Analysis of Variance (ANOVA) test.
Relative miR 126 expression levels in MCF7 cell line
Our findings found that miR-126 expression level in treated sample given a highest fold which attained 19 fold in comparing with two control samples which were 0.69 and 1.24 fold respectively. As well as statistical analysis showing that highly significant differences at p<0.0001 as shown in Figure 2.
Figure 2.
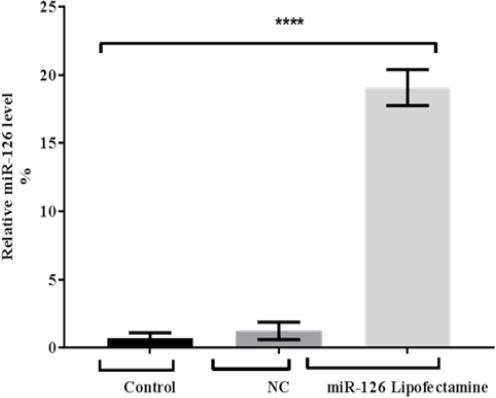
miR-126 Expression after Transfection with miR-126 Lipofectamine of MCF7 Cells. MiR-126 (normalised to RNU44) expression levels were detected by real time PCR. Data were mean and standard deviation (n=3) and were analysed by unpaired student’s t-test (one tailed). Significantly up regulated miR-126 levels in treated MCF cells are shown relative to miR-126 levels in adjacent control and negative miRNA control) (P < 0.0001).
Figure 3.
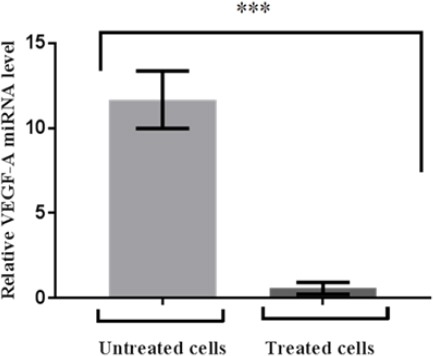
The VEGF-A Expression at 24 hr of MCF7 Cells. Cells were transfected with miR-126 lipofectamine. Relative miRNA VEGF-A (normalised to GAPDH) was detected by real time RT PCR. Data are mean ± standard error of mean (n=3) and were analysed by student’s t-test (one tailed). Significantly down regulated VEGF-A levels in treated breast cancer are shown relative to VEGF-A levels in adjacent untreated (control) cells
Relative VEGF-A expression level
Current study showing that evident down regulation of VEGF-A expression in miR-126-treated sample. Also high significant differences were in comparing folds values between treated and untreated samples which attained 11 and 2 respectively as shown in Figure 4.
Figure 4.
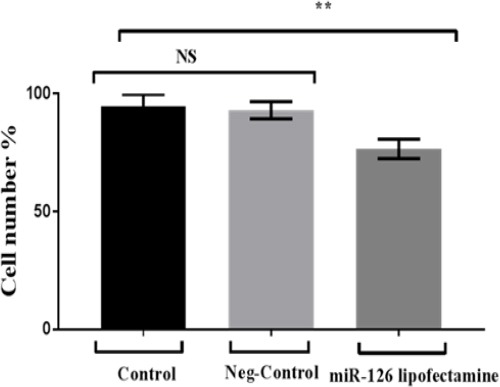
Proliferation Activity of Breast Cancer Cells (MCF7): Cells were treated with miR-126 lipofectamine compared to negative control and control. The data is represented in terms of a mean value (n = 3) ± standard deviation and analysed by One Way of Variance (ANOVA).
Proliferation and apoptosis incidence
Present data administrated that cell number in miR-126 treated cells were significant decrease in comparing to two controls. In which, cell number in miR-126 treated cells attained 76.7 %. Whereas the other two control samples attained 94.7 % and 93% moreover there was no significant differences between control samples. Over expression of miR-126 was significantly inhibiting the proliferative ability as shown in Figure 4.
As regarding to cell cycle phases, study showing that S phase cells proportion reduced and G1 phase cell proportion increased in MCF7 cells transfected with miR-126 mimic lipofectamine comparing to control after 24 h treatment (p<0.0001) (Figure 5).
Figure 5.
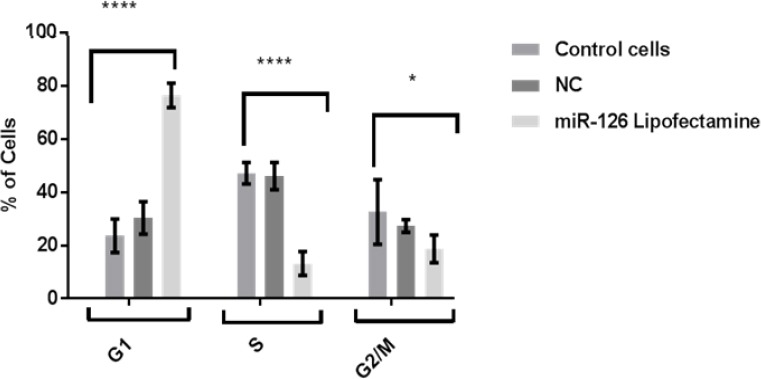
Effect of Mature miR-126 Lipofectamine on Breast Cancer Cell Cycle. Data were shown as mean and standard error of mean and analysed by One Way of Variance (ANOVA).
Discussion
Current study using miR-126 as controlling cancer progression, this is may consider promising strategy treatment. Furthermore, it has been found out that proliferation of cancer cells were inhibited via angiogenesis arresting. That was corresponding with prior studies have mentioned that miRNA aberration has a critical role in cancer metastasis (Huang et al., 2011; Raveche et al., 2007).
In spite of, this study referred that there was no significant differences between treated and control breast cancer cells (MCF7) in cell viability. Nevertheless, treated breast cancer cells were lower than control breast cancer in cell viability proportion. Probably, this was due to miR-126-treated cancer cells and effected on cell growth. Essentially, it has been mentioned by Musiyenko et al that microRNA-126 might reduce cancer cell proliferation by controlling PI3K signal pathway (Musiyenko et al., 2008).
It is known that the up-regulation of VEGF-A angiogenesis is a potential target of miR-126 and due to a study demonstrate that a strong involvement of VEGF-A in tumor metastasis via promoting the endothelial proliferation and tumor growth (Connolly et al., 1989). As predicted, the level expression of miR-126 was evidently up regulated in treated cancer cells whereas down regulation in control cancer this was positively responded to study supplementation cancer cells with miR-126. Consequently, our results have revealed that significant down regulation VEGF-A in treated cancer cell was due to high expression of miR-126. That was consistent with other studies have been explored that there was negative correlation between up regulation VEGFA expression level and down regulation miR-126 expression that was revealed in previous in vitro-experiments in lung cancer (Liu et al., 2009), oral cancer (Sasahiraet al., 2012) and esophagus cancer (Kong et al., 2016). So that VEGF-A may be a target gene of miR-126 which promotes growth and metastasis (Liu et al., 2009; Yang et al., 2012; Zhu et al., 2012).
It is well established that miRNA have been shown to down-regulate or up-regulate as an oncogenes or tumor suppressors in various cancers. MiR-126 expression reduces or loss in many cancers such as breast, lung, colon and gastric cancer (Guo et al., 2008; Tavazoie et al., 2008) Hence, miR-126 is considered as a tumor suppressive genes.
Down-regulation of miR-126 has led to cell apoptosis, and enhance cell proliferation and tumor angiogenesis (Gong et al., 2017). Present study shown that miR-126 could reduction of cell proliferation, induce apoptosis, and inhibit tumor angiogenesis in MCF7 by down-regulating the VEGF-A signalling pathway. The high correlation between the number of cells and DNA content was referring to evident cellular proliferation rate. So that the treated cells proliferated slightly slower than untreated and negative miR- cells which were observed similarity in their proliferation ability. That was dealing with our results related to cell cycle which are representing in S phase cells proportion was decreased, and MCF7 cells treated with miR-126 were increased due to cells being arrested and delayed at G1 phase compared to the control and miR-NC groups. So that it has been suggested that evaluation of apoptotic cell and cell number is related to angiogenesis arresting. Results study concluded that miR-126 could act as tumor suppressor in MCF7 cells that obstacle growth and metastasis of cancer cells throughout angiogenesis inhibition.it may be regarded as promising therapy to increase the breast cancer survival. Nevertheless, further research on animal models trials is required to more investigation about the miR-126 role to confirm breast cancer controlling.
Conflict of interest
The author has no conflict of interest.
Acknowledgments
Layla is grateful to Higher Education and Scientific Research/ Iraqi Government for the provision of a scholarship.
References
- 1.Adams RH, Alitalo K. Molecular regulation of angiogenesis and lymphangiogenesis. Nat Rev Mol Cell Biol. 2007;8:464–78. doi: 10.1038/nrm2183. [DOI] [PubMed] [Google Scholar]
- 2.Alevizakos M, Kaltsas S, Syrigos KN. The VEGF pathway in lung cancer. Cancer Chemother Pharmacol. 2013;72:1169–81. doi: 10.1007/s00280-013-2298-3. [DOI] [PubMed] [Google Scholar]
- 3.Alhasan L, Qi A, Rezk AR, et al. Assessment of the potential of a high frequency acoustomicrofluidic nebulisation platform for inhaled stem cell therapy. Integr Biol (Camb) 2016;8:12–20. doi: 10.1039/c5ib00206k. [DOI] [PubMed] [Google Scholar]
- 4.Connolly DT, Olander JV, Heuvelman D. Human vascular permeability factor. Isolation from U937 cells. JBC. 1989;264:20017–24. [PubMed] [Google Scholar]
- 5.Gong C, Fang J, Li G, et al. Effects of microRNA-126 on cell proliferation, apoptosis and tumor angiogenesis via the down-regulating ERK signaling pathway by targeting EGFL7 in hepatocellular carcinoma. Oncotarget. 2017;8:52527–42. doi: 10.18632/oncotarget.17283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo C, Sah JF, Beard L, et al. The noncoding RNA, miR-126, suppresses the growth of neoplastic cells by targeting phosphatidylinositol 3-kinase signaling and is frequently lost in colon cancers. Genes Chromosom Cancer. 2008;47:939–46. doi: 10.1002/gcc.20596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang Z, Huang S, Wang Q, et al. MicroRNA-95 promotes cell proliferation and targets sorting Nexin 1 in human colorectal carcinoma. Cancer Res. 2011;7:1–9. doi: 10.1158/0008-5472.CAN-10-3032. [DOI] [PubMed] [Google Scholar]
- 8.Kayani M, Kayani MA, Malik FA, et al. Role of miRNAs in breast cancer. Asian Pac J Cancer Prev. 2011;12:3175–80. [PubMed] [Google Scholar]
- 9.Kong R, Ma Y, Feng J, et al. The crucial role of miR-126 on suppressing progression of esophageal cancer by targeting VEGF-A. Cell Mol Biol Lett. 2016;21:1–10. doi: 10.1186/s11658-016-0004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Z, Li N, Wu M, et al. Expression of miR-126 suppresses migration and invasion of colon cancer cells by targeting CXCR4. Mol Cell Biochem. 2013;381:233–42. doi: 10.1007/s11010-013-1707-6. [DOI] [PubMed] [Google Scholar]
- 11.Liu B, Peng XC, Zheng XL, et al. MiR-126 restoration down-regulate VEGF and inhibit the growth of lung cancer cell lines in vitro and in vivo. Lung Cancer. 2009;66:169–75. doi: 10.1016/j.lungcan.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 12.Mulrane L, McGee SF, Gallagher WM, et al. miRNA dysregulation in breast cancer. Cancer Res. 2013;73:6554–62. doi: 10.1158/0008-5472.CAN-13-1841. [DOI] [PubMed] [Google Scholar]
- 13.Musiyenko A, Bitko V, Barik S. Ectopic expression of miR-126*, an intronic product of the vascular endothelial EGF-like 7 gene, regulates prostein translation and invasiveness of prostate cancer LNCaP cells. J Mol Med. 2008;86:313–22. doi: 10.1007/s00109-007-0296-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raveche ES, Salerno E, Scaglione BJ, et al. Abnormal microRNA-16 locus with synteny to human 13q14 linked to CLL in NZB mice. Blood. 2007;109:5079–86. doi: 10.1182/blood-2007-02-071225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sasahira T, Kirita T, Bhawal UK, et al. The expression of receptor for advanced glycation end products is associated with angiogenesis in human oral squamous cell carcinoma. Virchows Archiv. 2007;450:287–95. doi: 10.1007/s00428-006-0359-2. [DOI] [PubMed] [Google Scholar]
- 16.Sasahira T, Kirita T, Bhawal UK, et al. Receptor for advanced glycation end products (RAGE) is important in the prediction of recurrence in human oral squamous cell carcinoma. Histopathology. 2007;51:166–72. doi: 10.1111/j.1365-2559.2007.02739.x. [DOI] [PubMed] [Google Scholar]
- 17.Sasahira T, Kirita T, Kurihara M, et al. MIA-dependent angiogenesis and lymphangiogenesis are closely associated with progression, nodal metastasis and poor prognosis in tongue squamous cell carcinoma. EJC. 2010;46:2285–94. doi: 10.1016/j.ejca.2010.04.027. [DOI] [PubMed] [Google Scholar]
- 18.Sasahira T, Kirita T, Oue N, et al. High mobility group box-1-inducible melanoma inhibitory activity is associated with nodal metastasis and lymphangiogenesis in oral squamous cell carcinoma. Cancer Sci. 2008;99:1806–12. doi: 10.1111/j.1349-7006.2008.00894.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sasahira T, Kurihara M, Bhawal UK, et al. Downregulation of miR-126 induces angiogenesis and lymphangiogenesis by activation of VEGF-A in oral cancer. Br J Cancer. 2012;107:700–6. doi: 10.1038/bjc.2012.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Serpico D, Molino L, Di Cosimo S. microRNAs in breast cancer development and treatment. Cancer Treat Rev. 2014;40:595–4. doi: 10.1016/j.ctrv.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 21.Tavazoie SF, Alarcón C, Oskarsson T. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–52. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang J, Lan H, Huang X, et al. MicroRNA-126 inhibits tumor cell growth and its expression level correlates with poor survival in non-small cell lung cancer patients. PLoS One. 2012;7:e42978. doi: 10.1371/journal.pone.0042978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu X, Li H, Long L, et al. miR-126 enhances the sensitivity of non-small cell lung cancer cells to anticancer agents by targeting vascular endothelial growth factor A. Acta Biochim Biophys Sin (Shanghai) 2012;44:519–26. doi: 10.1093/abbs/gms026. [DOI] [PubMed] [Google Scholar]


