Abstract
Background:
As is known, exosomes play an important role in promoting progression of cancers by increasing its invasive potential. The aim of this study was to evaluate the levels of tetraspanine-associated (ADAM-10) and tetraspanine-nonassociated proteases (20S proteasomes) in exosomes from culture medium, plasma exosomes of patients with breast tumors and plasma and ascites of ovarian tumor patients.
Methods:
MCF-7 and SVO-3 culture mediums and blood samples from healthy females (n = 30, HFs), patients with diffuse dyshormonal dysplasia of the breast (n=28, BBTPs), breast cancer patients (n=32, BCPs), borderline ovarian tumor patients (n=20, BOTPs) and blood and ascites samples ovarian cancer patients (n=35, OCPs) were included in the study. Exosomes from plasma, ascites and culture mediums were isolated and characterized in according to Extracellular Vesicles Society. The expression levels of 20S proteasome and ADAM-10 in exosomes were determined using flow cytometry and western blot analysis, correspondingly.
Results:
The subpopulation composition of the exosomes from MCF-7 culture medium and from blood plasma of HFs and breast diseases patients is similar, however CD9/CD24 subpopulation significantly increased at cell supernatant. The similar results was obtained for exosomes from SVO-3 medium and blood plasma and ascites of ovary tumor patients, but CD9/CD24 subpopulation significantly decreased at cells and illness samples, however CD63/CD24 exosomes increased significantly from cell supernatant. 20S proteasome level is significantly increased in exosomes from MCF-7 and SVO-3 culture medium, breast tumor patients and OCPs plasma in comparison to HUVEC culture medium and HFs plasma samples. At CD9-positive exosomes from BCPs plasma and MCF-7 was reveal a high expression of ADAM-10 and low expression is from BBDPs plasma and ovarian tumor patients plasma/ascites samples. Exosomes from ascites OCP had high expression of ADAM-10 in the CD24-positive subpopulation.
Conclusion:
Breast and ovarian cancer development is connected with functioning of immune proteasome forms in plasma and ascites exosomes, while increased ADAM10 expression at CD9-positive exosome was associated with breast cancer and at CD24-positive subpopulation – with ovarian cancer. Obtained data confirm role of exosomal proteases in tumor progression.
Keywords: Exosomes, ADAM-10, 20S proteasome, plasma, ascites, breast cancer, ovarian cancer
Introduction
Breast cancer and ovarian cancer are the most common types of tumor worldwide among women. Despite the active distribution of mammography, the proportion of primary-identified breast cancer patients (BCPs) at an advanced stage of the disease remains high (Witten and Parker, 2018). Most ovarian cancer patients (OCPs) are diagnosed at an advanced stage also and 70% of patients present with lymph node metastasis and ascites fluids (Dong et al., 2014). Cancer patients with metastasis have a higher rate of treatment failure and mortality (Ferlay et al., 2018). Therefore, it is crucial to explore the mechanisms of tumor metastasis and discover valuable factors for early diagnosis of female cancers and novel therapeutic strategies for metastasis.
As is known, exosomes play an important role in promoting progression of cancers by increasing its invasive potential (Azmi et al., 2013). Exosomes are small (30–100 nm) membrane vesicles that are released into the extracellular environment after fusion of multivesicular bodies with the plasma membrane (Yanez-Mo et al., 2015). Secreted cancer cells exosomes are carried through the blood and lymph circulation and can be detected in the body far away from the localization of the parent cells (Whiteside, 2018). These vesicular structures are stable for a long time and are involved in key physiological processes through the horizontal transfer of RNA and proteins (Bryzgunova et al., 2016; Li et al., 2017), thus affecting both the development of the primary tumor and the distant metastasis.
As is proposed by Mathivanan et al., most frequently occurring exosomal proteins are enzymes – 32 % (Mathivanan et al., 2010), among which the key role in the functional activity of vesicles is played by proteases. Exosomal proteases can be divided into two groups: tetraspanine-associated (structurally and functionally linked) and tetraspanine-nonassociated proteases (Yunusova et al., 2018).
Tetraspanin-associated proteases include ADAMs (a disintegrin and metalloproteases), MMPs (matrix metalloproteases) and EMMPRIN, of which ADAM10 is the most prominent member of surface expressed metalloproteinase (ADAM)-family (Yunusova et al., 2018). ADAM10 is involved in the ectodomain shedding of various substrates, including receptors of growth factors, adhesion receptors and cadherins, which as a result induces cell mobility and increases the metastatic potential of tumor cells (Lee et al., 2010).
Tetraspanine-nonassociated proteases include PAPP-A (Pregnancy-associated plasma protein-A) and 20S proteasome (Yunusova et al., 2018). Proteasomes, possessing trypsin-like, chymotrypsin-like and caspase-like activities, are of great importance for intracellular protein degradation, which causes their important role in origin and development malignant neoplasms (Mofers et al., 2017). It is shown that proteasomes are involved in the regulation of proliferation, apoptosis, cell migration, gene transcription and immune response, destroying or activating proteins - components of signaling pathways, factors transcription and other functionally significant molecules (Shashova et al., 2014; Kondakova et al., 2014).
Recently, ADAM-10 and 20S proteasomes were found in the composition of exosomes from mesenchymal stem cells (Lai et al., 2012), endothelial cells (Dieudé et al., 2015) and ovarian carcinoma cells (Stoeck et al., 2006). However, the level of these enzymes in the exosome of biological fluids of cancer patients remains unclear.
In the present article we conducted a comparative study the levels of ADAM-10 and 20S proteasomes in exosomes derived from culture medium, plasma exosomes of patients with breast tumors and plasma and ascites of ovarian tumor patients.
Materials and Methods
Cell cultures
HUVEC were obtained by treating the umbilical cord veins of women with a normal pregnancy and a healthy fetus with 0.1% collagenase (type IV, Invitrogen, Carlsbad, CA, USA) as described (Jaffe et al., 1973). All the experiments were performed with cells maintained in culture until passage 4. The human breast cancer cell line MCF-7 and ovarian cancer cell line SVO-3 were cultured in IM medium (Life Technologies, Alemeda, CA, USA) supplemented with 10% Fetal Bovine Serum (FBS) (Thermo Fisher Scientific) and Penicillin-Streptomycin (100 U/ml) (Thermo Fisher Scientific) in CO2-incubator (5% CO2) at 37°C up to 70-80% confluence. FBS taken for cell culturing was previously exosome depleted by ultracentrifugation for 3 h at 100,000 g.
Donors and Patients
Blood samples of HFs (n = 30, 44,4±3,7 years) and benign breast disease patients (BBDPs, diffuse dyshormonal dysplasia of the breast) (n=28, 41,5±2,5 years) were obtained from the Novosibirsk Central Clinical Hospital. HFs did not have any female disorders (dysplasia, endometriosis, etc.) or any malignant diseases. Blood samples of previously untreated BCPs (n=32, 60,3±7,1 years) were obtained from the Novosibirsk Regional Clinical Oncological Hospital (Table 1). Blood and ascites samples of borderline ovarian tumor patients (BOTPs) (n=20, 39,5±2,5 years) and previously untreated ovarian cancer patients (OCPs) (n=35, 56,5±2,5 years) (Table 2) were obtained from the Department of Gynecology of the Cancer Research Institute of Tomsk National Research Medical Center. The volume of ascites was measured clinically and by ultrasound examination using the LOGIQ S6 machine.
Table 1.
General Clinical Characteristics of the BCPs
| No (%) | |
|---|---|
| Histology | |
| Infiltrative ductal carcinoma | 22 (69) |
| Invasive carcinoma of no special type | 10 (31) |
| TNM staging | |
| T1 | 15 (47) |
| T2 | 17 (53) |
| N0 | 24 (75) |
| N1 | 6 (19) |
| N2 | 1 (3) |
| N3 | 1 (3) |
| M0 | 32 (100) |
| Tumor grade | |
| II | 28 (88) |
| III | 4 (12) |
| Hormone receptor status | |
| ER-positive | 22 (69) |
| ER-negative | 10 (31) |
| PR-positive | 20 (63) |
| PR-negative | 12 (37) |
| Growth factor status | |
| HER2-positive | 23 (72) |
| HER2-negative | 9 (28) |
| Triple-negative (ER, PR, HER2) | 6 (19) |
| Ki-67 | |
| ≤10% | 9 (28) |
| ≥ 10% | 23 (72) |
| Total no. of patients | 32 (100%) |
Table 2.
General Clinical Characteristics of the BOTPs and the OCPs
| BOTPs, No (%) | OCPs, No (%) | |
|---|---|---|
| Histology | ||
| Serous | 35 (100%) | |
| Other | 20 (100%) | |
| FIGO (2013) staging | ||
| I-II | 16 (80%) | |
| IIIB | 4 (20) | 5 (14%) |
| IIIC | 30 (86%) | |
| Tumor Grade | ||
| High-grade | 35(100%) | |
| Not specify | 20 (100%) | |
| BRCA status | ||
| Negative | 35 (100%) | |
| Not specify | 20 (100%) | |
| Total no. of patients | 20 (100%) | 35 (100%) |
Written informed consent was obtained from every participant and the studies were approved by the Ethical Committees of the Dispensary, Hospital and Institutes. Human samples used in this study were obtained according to the principles expressed in the Declaration of Helsinki.
Exosome isolation
Exosomes were isolated from medium collected from growing cell culture using differential centrifugation and ultrafiltration. Firstly, HUVEC-, MCF-7- or SVO-3-conditioned medium was centrifuged at 290 g for 20 min; the supernatant was further centrifuged at 17,000 g for 20 min. The resulting supernatant was filtered through a 100-nm pore-size filter (Minisart high flow, 16553-K, Sartorius) and the filtrate was centrifuged for 90 min at 100,000 g at 4°C. The pellet was suspended in 12 ml of phosphate buffered saline (pH 7.4) (PBS) and again centrifuged under the same condition listed above, ultracentrifugation and re-suspending steps were repeated three times. Supernatant was removed and the pellet containing exosomes was re-suspended in 150 μl of PBS.
Venous blood (9 ml) from all subjects was collected in K3EDTA spray-coated vacutainers (Vacuette, cat. no. 455036), immediately mixed using rotary mixer for vacutainers, placed at 4°C and fractionated into plasma and blood cells within 4 h after blood sampling. Ascites fluid (18–20 ml) was collected in K3EDTA spray-coated vacutainers during surgery of OCPs. Blood and ascites fluid samples from illness patients were obtained before any treatment.
Blood was centrifuged at 290g for 20 min at 4°C, blood plasma was transferred into a new tube, and centrifuged a second time at 1,200 g for 20 min at 4°C. For isolation of exosomes, plasma samples were centrifuged at 17,000 g for 20 min at 4 °C to pellet cell debris. Supernatants were diluted as 1:5 by PBS and were passed through 100-nm pore-size filter, then the filtrates were ultracentrifuged under the same condition listed above, ultracentrifugation and re-suspending steps were repeated three times. Supernatant was removed and the pellet containing plasma exosomes was re-suspended in 150 μl of PBS.
Ascites fluids was centrifuged at 900g for 20 min at 4°C, supernatant was transferred into a new tube, and centrifuged to pellet cell debris, then supernatants were diluted as 1:3 by PBS and were filtered and ultracentrifuged under the same condition listed above. Supernatant was removed and the pellet containing ascites fluid exosomes was re-suspended in 150 μl of PBS.
Exosome samples from plasma, ascites fluid and from cell culture medium were aliquoted and stored either at −80°C or in liquid nitrogen. The aliquots were thawed once before use.
Electron microscopy of exosomes
For negative staining, a drop of exosomes was incubated for 1 min on copper grid, covered with formvar film, which was stabilized by carbon. Then the grids were exposed for 5-10 sec on a drop of 0.5 % uranyl acetate or 2% phosphotungstic acid.
The grids, holding adsorbed exosomes, were examined by transmission electron microscope (JEM 1400 (Jeol, Japan) supplied with digital camera Veleta (Olympus Corporation, Japan)). The measurements were made directly on camera screen using iTEM (Olympus Corporation, Japan) software.
Flow cytometry analysis
For the immunoprecipitation of exosomes and their subsequent analysis by fluorescence-activated cell sorting (FACS), 4 μm-diameter aldehyde/sulphate latex beads (Interfacial Dynamics) were incubated with purified anti-CD9 or anti-CD24 (BD Biosciences) or anti-ADAM-10 (Abcam, UK) antibodies at 22°C overnight at gentle agitation, as previously described. For FACS analysis, 30 μg of exosomes were incubated with 3 × 105 anti-CD9, anti-CD24 or anti-ADAM-10 beads in 150 μl of PBS at 4°C overnight at gentle agitation. The reaction was stopped by incubation of exosome-bead complexes in 0.2 M glycine for 30 min. The exosome-bead complexes were washed twice with FACS buffer (3% exosome depleted FBS in PBS). The beads-bound exosomes were then incubated with human IgG (BD Biosciences) at 4°C for 30 min with subsequent washing with FACS buffer and incubation with fluorescein-conjugated anti-CD9, anti-CD24, anti-CD63, anti-CD81 or isotype antibodies (all from BD Biosciences) for 40 min at room temperature at gentle agitation. The complexes were washed twice, suspended in 300 μl of FACS buffer and analyzed by flow cytometry using FACS Canto II (BD Biosciences) and FACS Diva 6.1 Software. The median fluorescence intensity (MFI) of the stained exosomes was analyzed as compared to isotype control.
Protein quantification
For estimation of protein concentration, 7.5 μl of exosomes were mixed with lysis buffer (0.25 M Tris-HCl, 8 % SDS, 0.2 M DTT, pH 6.8), incubated on ice (10 min) and then boiled (95°C, 10 min) and then cooled. After centrifugation (12,000g, 10 min), protein concentration was measured using a fluorometric protein assay (NanoOrange® Protein Quantitation Kit, Molecular Probes, USA) according to the manufacturer’s instructions.
Polyacrylamide Gel Electrophoresis and Western Blot
Immunoblotting was performed using specific polyclonal rabbit antibodies against human Proteasome 20S alpha and beta subunits (ab22673, Abcam, UK) as primary antibody. Thirty μl of exosomes sample was mixed with 7 μl of lysis buffer (125 mM Tris-HCl, pH 7.8, 750 mM NaCl, 0.5% SDS, 5% Triton X-100) and 3 μl of protease inhibitors cocktail (1.3 mM Aprotinin (Sigma, USA), 0.33 mM Pepstatin A (ICN, USA), 1 μg/ml Leupeptin (ICN, USA)), incubated for 90 min on ice, for 7 min at 95°C with sample buffer. After centrifugation (13,000 g, 5 min), the proteins resolved by 13% SDS-PAGE gels were blotted onto PVDF-membrane (0.45 μm, Immobylon, Millipore, USA). The membrane was blocked with 5% fat-free milk and treated with the primary antibody for 1 h (1/2,000 dilution). To detect the signal, the membranes were treated with goat anti-rabbit HRP-conjugated secondary antibody (Santa Cruz Biotechnology, UK) for 1 h (1/5,000 dilution). After three times washing, bound antibody was detected using the Amersham ECL-Plus Western Blotting Detection System (Amersham, USA) and ChemiDoc Touch imaging system (Bio-Rad, USA) according to the manufacturer’s protocol. Image densitometry was performed using “ImageJ”. Relative abundance of the 20S Proteasome was normalized to exosomal CD63 level. The results were expressed as a percentage of the content of 20S proteasome in exosomes from HUVEC culture medium and healthy donors’ plasma.
Statistical analysis
Statistical analysis was performed using Statistica 6.0 software. All data were expressed as medians with interquartile ranges or as means with standard errors. To evaluate the difference, MannWhitney U-test was used. P-values < 0.05 were considered statistically significant.
Results
Characterization of exosomes from culture medium, blood plasma and ascites
To prepare the exosomes, we used a combination of ultrafiltration and repeated ultracentrifugation, which has been successfully applied for isolation and concentration of exosomes from various biological fluids (Grigor’eva et al., 2017). The resulting preparations of exosomes from SVO-3 and MCF-7 mediums were free of macromolecular aggregates and particles larger than 100 nm in size; more than 90% of the purified extracellular vesicles (EV) were represented by spherical and cup-shaped vesicles (40-100 nm) (Figure 1D, H). All the preparations of exosomes from plasma and ascites were characterized by the presence of ~20% contaminating structures: low electron density particles without limiting membrane and therefore could not be attributed to exosomes (“non-vesicles”). These “non-vesicles” corresponded to that of intermediate very low density lipoproteins, which were identical to exosomes by size (Figure 1A-C, E-G). The same contamination of exosome preparations obtained by the method of sequential centrifugation with lipoproteins has been demonstrated by Sodar (2016). The authors found the relationship between low-density lipoproteins and exosomes and showed that lipoproteins could not be removed from the exosome preparation by any of the known methods applicable for exosome isolation from biological fluids (Sodar et al., 2016), including centrifugation in a density gradient used for separation of lipoprotein fractions.
Figure 1.
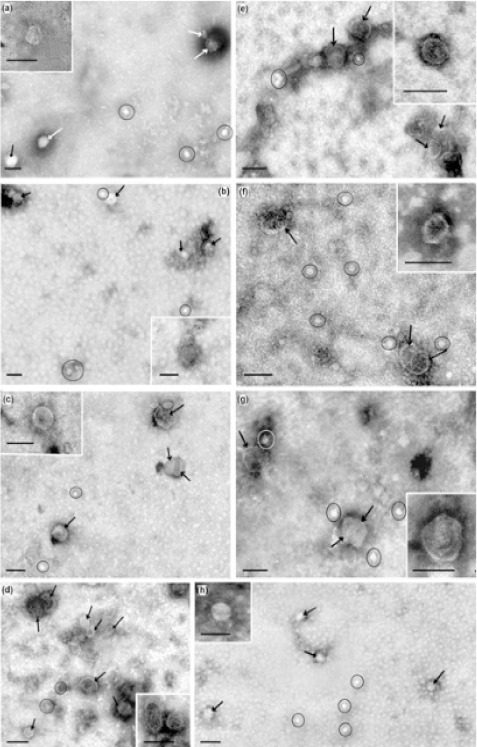
Total View of Exosome Preparation Obtained from: A, blood plasma of HFs; B, blood plasma of BBDPs; C, blood plasma of BCPs; D, culture medium of MCF-7 cells; E, ascetic fluid of OCPs; F, blood plasma of BOTPs; G, blood plasma of OCPs; H, culture medium of SVO-3 cells. Inserts shows exosomes. Arrows indicates exosomes, ellipses – «non-vesicles». Scale bars correspond to 100 nm. Electron microscopy, negative staining by phosphotungstate acid.
To demonstrate that purified EV were exosomes, we immobilized them onto latex beads covered with CD9 and CD24 antibodies and analyzed the presence of exosomal markers CD9, CD63 and CD81 on the EV surface. Detection of these markers clearly indicated that the collected EVs from culture medium, blood plasma and ascites were indeed exosomes (Table 3).
Table 3.
Expression of CD 24, CD 63 and CD 81 on the Surface of Exosomes from Culture Medium and Biological Fluids of HFs and Female Tumor Patients
| Source of exosomes | CD 9-positive exosomes | CD 24-positive exosomes | |||
|---|---|---|---|---|---|
| CD 63 expression | CD 81 expression | CD 9 expression | CD 63 expression | ||
| HFs | Plasma | 513±76 | 645±97 | 1048±120 | 523±75 |
| BBDPs | Plasma | 513±84 | 628±83 | 703±84* | 548±81 |
| BCPs | Plasma | 490±48 | 659±67 | 969±116 | 510±56 |
| BOTPs | Plasma | 690±110 | 760±120 | 642±95* | - |
| OCPs | Plasma | 580±78 | 740±104 | 704±96* | 590±81 |
| Ascites | 580±69 | 750±106 | 691±70* | 580±94 | |
| MCF-7 | Culture medium | 541±38 | 691±66 | 2151±205* | 560±52 |
| SVO-3 | Culture medium | 549±44 | 549±38 | 789±63* | 744±67* |
, differences were significant compared to HFs (P value<0.05); Data represent median fluorescence intensity (MFI) ± SEM.
Quantitative and Subpopulations analysis of exosomes from culture medium, blood plasma and ascites
The concentration of the exosomal protein may indirectly represent the quantity of exosomes in the biological sample. For quantitative analysis of exosomes from culture medium and biological fluids was used NanoOrange Protein Quantitation kitTM. The exosomal protein concentration from blood plasma did not differ between HFs and illness patients (Figure 2). The median of exosomal protein concentration in blood plasma preparations of HFs, BBDPs, BCPs, BOTPs and OCPs were 14.5, 12.2, 14.2, 11.2, 14.2 mkg/ml, respectively. However, the level of the exosomal protein was significantly higher in the ascites samples compared to that observed in the paired plasma samples (median 24.1 and 14.2 mkg/ml, respectively; Figure 2).
Figure 2.
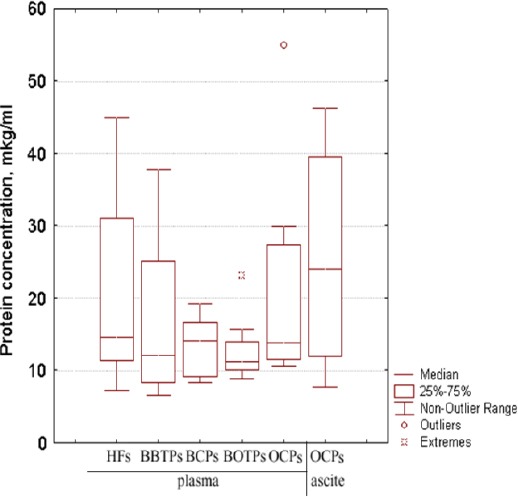
Tukey Box Plots of Protein Concentration in Preparations of Exosomes Isolated from the Biological Fluids of HFs and Tumor Patients.
Various types of the exosomes circulated in the blood of healthy donors and patients with different types of cancer, and the surface proteins, mainly represented by CD9, CD24, CD63 and CD81 were considered as the universal markers of blood exosomes. Using combination of unconjugated and conjugated antibodies were characterized subpopulations of exosomes from culture medium and biological fluids. It was found that the subpopulation composition of the exosomes from breast carcinoma culture medium and from blood plasma of healthy females and breast diseases patients is similar: CD24/CD9 > CD9/CD81 > CD24/CD63 > CD9/CD63 (Table 3). Thus, in the MCF-7 and plasma of HFs, BBDPs and BCPs the exosomes expressing CD9 and CD24 receptors are detected most frequently. During ovarian tumors development redistribution of various subpopulations of exosomes occurred: in plasma of patients with benign ovarian tumors - CD9/CD81 > CD9/CD63 > CD24/CD9, in plasma and ascites of OCPs - CD9/CD81 > CD24/CD9 > CD24/CD63 > CD9/CD63, in SVO-3 medium - CD24/CD9 > CD24/CD63 > CD9/CD81 = CD9/CD63 (Table 3). Thus, in biological fluids of ovarian tumors the exosomes expressing CD9 and CD81 receptors are detected most frequently. Thus, CD9, CD24 and CD 81-exosomes are apparently secreted by epiteliocytes, endotheliocytes and blood cells. The CD63 receptor was detectable on the surface of the smallest population of exosomes, and these exosomes simultaneously exposed other receptors (CD9, CD24).
The greatest changes in the subpopulations of exosomes were observed in the cell supernatants and in biological fluids of BBDPs and ovarian tumor patients. Compared with healthy women, in samples from illness patients and SVO-3 cells, the CD24/9 subpopulation of exosomes is decreased (Table 3). A decrease in the subpopulation of CD24/9 exosomes appears to be due to dedifferentiation of epithelial cells associated with the loss of surface markers of CD24 by cells. However this subpopulation was predominantly expressed in the exosomes obtained from the culture medium of MCF-7; on the contrary, CD24/CD9 and CD24/CD63 subpopulations in the culture medium of SVO-3 exosomes are presented equally (Table 3).
20S proteasome and ADAM-10 expression level
We analyzed expression levels of 20S proteasome and ADAM-10 in exosomes from culture medium, blood plasma and ascites. It was found that expression of the surface form of ADAM-10 significantly increased in the subpopulation of CD9-positive exosomes isolated from the BCP plasma and from MCF-7 culture medium compared with the HF samples (at 1.8 and 1.9 times, respectively). In contrast, in this subpopulation, the expression of the surface form of ADAM-10 in BOTPs plasma, as well as OCP plasma and ascites, is significantly reduced (Table 4). Analysis of the ADAM-10 expression on CD24- and CD81-positive exosomes from culture medium and biological fluids revealed a significant increase this metalloproteinase level at 1.6 times only in CD24-positive subpopulation from OCP ascites.
Table 4.
Expression of CD 9, CD 24 and CD 81 on the Surface of ADAM-10 Positive Exosomes from Culture Medium and Biological Fluids of HFs and Female Tumor Patients
| Source of exosomes | CD 9 expression | CD 24 expression | CD 81 expression | |
|---|---|---|---|---|
| HFs | Plasma | 844±99 | 884±124 | 621±84 |
| BBDPs | Plasma | - | 1,031±155 | - |
| BCPs | Plasma | 1531±162* | 926±94 | 696±68 |
| BOTPs | Plasma | 640±71* | 961±134 | - |
| OCPs | Plasma | 692±85* | 980±147 | 639±69 |
| OCPs | Ascites | 625±75* | 1,423±157*, ** | 620±68 |
| MCF-7 | Culture medium | 1,607±128* | 887±67 | 597±48 |
| SVO-3 | Culture medium | 743±59 | 871±74 | - |
, differences were significant compared to HFs;
, differences were significant compared to OCP plasma (P.value<0.05). Data represent median fluorescence intensity (MFI) ± SEM.
Western-blot analysis showed that 20S proteasome level from cancer culture medium exosome samples were significantly higher than in the corresponding exosomes from HUVEC culture medium (Figure 3A). Moreover, similar data was obtained from exosomes of BBTPs, BCPs and OCPs plasma in comparison with normal plasma, while this level from plasma BOTPs was significantly below (Figure 3B, D). Comparative analysis was reveal elevated 20S proteasome level in samples of exosomes from plasma malignant and non-malignant breast tumors than in exosomes from MCF-7 culture medium (2,28±0,46, 1,81±0,34 and 1,25±0,08 related units, correspondingly). In contrast, the highest 20S proteasome level was reveal in samples of exosomes from SVO-3 culture medium than in exosomes from plasma OCPs and BOTPs as well as ascites of OCPs (2,41±0,17, 1,4±0,25, 0,22±0,04 and 0,93±0,16 related units, correspondingly). In paired samples from OCPs (plasma and ascites) was detected the same 20S proteasome level (Figure 3C).
Figure 3.
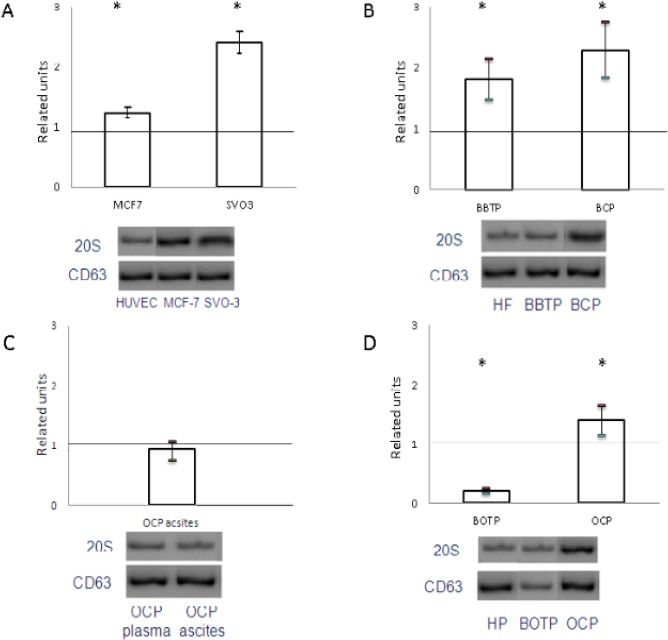
20S Proteasomes Level in Exosomes from Cancer Culture Medium, Plasma and Ascites of Tumor Patients Compared to Control Samples. Proteasomes level in exosomes from MCF-7 and SVO-3 culture medium compared to HUVEC culture medium (A). Proteasomes level in exosomes from plasma of BBTPs and BCPs (B) and BOTPs and OCPs (D) compared to HFs. 20S proteasomes level in paired samples exosomes from plasma and ascites of OCPs (C). The results were standardized taking into account the level of CD63 in exosomes and were expressed as a related units of the level of the target protein in the exosomes in the control samples.
Discussion
Recent reviews, which devoted to function of exosomes, are focused on cell-to-cell communication, cellular adhesion, migration, invasion, angiogenesis and growth of tumor cells (Azmi et al., 2013; Tai et al., 2018). Therefore, they can no longer be considered simply as garbage bags for throwing out unwanted protein cargo from the cell as originally suggested, necessitating a recalibration of our understanding and of their potential functions in physiological processes. In present study were done to shed light on the change of protease cargo in exosomes, which are secreted by cancer cells to culture medium or plasma and ascites.
There are contradictory data on the amount exosomes in patients’ plasma with different types of cancer and no publications on the number of exosomes in the blood of BCPs or OCPs. A few data about the subpopulation composition of exosomes from blood of BCPs and OCPs have been found. In particular the increased expression of CD151 has been correlated with higher tumor grade and node metastasis in patients with invasive ductal carcinoma of the breast (Hassuna et al., 2009; Sadej et al., 2014). CD82 has been shown to strongly associate with a poor prognosis in patients with ovarian and breast cancer (Malla et al., 2018). Another group indicates that ovarian cancer–derived exosomes transfer CD44 to HPMCs, facilitating cancer invasion. (Nakamura et al., 2017). The combination of plasma exosomal markers CA-125, EpCAM, and CD24 provided desirable diagnostic accuracy for non-invasive, early detection of ovarian cancer (Zhao et al., 2016). Our data do not confirm the over-expression of CD24 in blood exosomes of breast cancer patients, which probably is due to the differences in stages and types of the malignant disease. Particularly, in the current work we studied the expression of the CD receptors in blood of primary breast cancer patients at first stages of the disease (stages I–II, without regional or remote metastases).
Recent findings suggest that upregulated ADAM-10 expression participates in the progression of multiple types of cancer, including breast cancer (Lendeckel et al., 2005). Later it was shown that the inclusion of mature ADAM10 in exosomes and its proteolytic activity is regulated by tetraspanins CD9, CD81, CD82 (Arduise et al., 2008). In our study we detected increased level of ADAM-10 in exosomes from BCP’s plasma and from MCF-7 supernatant at CD9-positive subpopulation and from OCP’s ascites at CD24-positive subpopulation. The findings confirm the hypothesis of the importance of the circulating exosomes protease cargo in the tumor progression. Moreover, exosome proteomics revealed distinct integrin expression patterns, in which the exosomal integrins α6β4 and α6β1 were associated with lung metastasis, while exosomal integrin αvβ5 was linked to liver metastasis. Targeting the integrins α6β4 and αvβ5 decreased exosome uptake, as well as lung and liver metastasis, respectively (Hoshino et al., 2015). Since ADAM-proteases of exosomes interact with integrins, it can be assumed that the association of ADAM-integrins supports the organotropic nature of metastases.
Data on the expression of 20S in exosomes from plasma of cancer patients are absent. A comprehensive proteomic analysis reveals a secretory path-and status-dependent signature of the exosomes released from tumor-associated macrophages associated with 20S proteasome subunits and ribosomal proteins (Zhu et al., 2015). Obtained our data revealed the high level of expression of 20S proteasomes in exosome samples from culture mediums, breast tumors patients and OCPs plasma is evidently due to increased expression of 20S proteasomes in tumor tissue. In particular, it was found that the level of total chymotrypsin-like proteasome activity in breast malignant tissue was higher in comparison with adjacent normal tissue in 76 of 106 patients and correlated with the status of steroid prescriptions in the tumor (Shashova et al., 2014). A revealed high level of 20S proteasome in exosomes in patients with breast cancer and OC appears to be necessary for recipient cells to realize a multitude of proteasome functions in tumor cells and a tumor microenvironment (Sharova and Zakharova, 2008).
In conclusion, subpopulations composition of exosomes and their protease cargo are different in culture medium and biological fluids of healthy woman and female tumor patients. The revealed redistribution of tetraspanin-associated and tetraspanine-nonassociated proteases in subpopulation of exosomes at different cancer types could be point to various mechanisms for sorting biologically active molecules into vesicles, most of which are not fully understood. What is clear is that the exosome composition will determine the outcome of the communication. Exosome-mediated communication is very important for tumor cells, which secrete exosomes constitutively. Further understanding of the role of different subpopulation of exosomes in tumor microenvironment, cancer metastasis and metastatic niche is expected to lead to development of tumor specific therapeutics.
Conflicst of interest statement
The authors whose names are listed immediately below certify that they have NO affiliations with or involvement in any organization or entity with any financial interest (such as honoraria; educational grants; participation in speakers’ bureaus; membership, employment, consultancies, stock ownership, or other equity interest; and expert testimony or patent-licensing arrangements), or non-financial interest (such as personal or professional relationships, affiliations, knowledge or beliefs) in the subject matter or materials discussed in this manuscript.
Acknowledgments
This work was supported by the Russian Foundation for Basic Research and Government of Novosibirsk region of the Russian Federation, grant 18-415-540012.
References
- 1.Arduise C, Abache T, Li L, et al. Tetraspanins regulate ADAM10-mediated cleavage of TNF-alpha and epidermal growth factor. J Immunol. 2008;181:7002–13. doi: 10.4049/jimmunol.181.10.7002. [DOI] [PubMed] [Google Scholar]
- 2.Azmi AS, Bao B, Sarkar FH. Exosomes in cancer development, metastasis, and drug resistance:a comprehensive review. Cancer Metastasis Rev. 2013;32:623–42. doi: 10.1007/s10555-013-9441-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bryzgunova OE, Zaripov MM, Skvortsova TE, et al. Comparative study of extracellular vesicles from the urine of healthy individuals and prostate cancer patients. PLoS One. 2016;11:e0157566. doi: 10.1371/journal.pone.0157566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dieudé M, Bell C, Turgeon J, et al. The 20S proteasome core, active within apoptotic exosome-like vesicles, induces autoantibody production and accelerates rejection. Sci Transl Med. 2015;7:318ra200. doi: 10.1126/scitranslmed.aac9816. [DOI] [PubMed] [Google Scholar]
- 5.Dong X, Men X, Zhang W, et al. Advances in tumor markers of ovarian cancer for early diagnosis. Indian J Cancer. 2014;51:e72–6. doi: 10.4103/0019-509X.154049. [DOI] [PubMed] [Google Scholar]
- 6.Ferlay J, Colombet M, Soerjomataram I, et al. Cancer incidence and mortality patterns in Europe:Estimates for 40 countries and 25 major cancers in 2018. Eur J Cancer. 2018 doi: 10.1016/j.ejca.2018.07.005. S0959-8049(18)30955-9. [DOI] [PubMed] [Google Scholar]
- 7.Grigor'eva AE, Dyrkheeva NS, Bryzgunova OE, et al. Contamination of exosome preparations, isolated from biological fluids. Biochem Moscow Suppl Ser B. 2017;11:265. doi: 10.18097/PBMC2017630191. [DOI] [PubMed] [Google Scholar]
- 8.Hassuna N, Monk PN, Moseley GW, et al. Strategies for targeting tetraspanin proteins:potential therapeutic applications in microbial infections. BioDrugs. 2009;23:341–59. doi: 10.2165/11315650-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoshino A, Costa-Silva B, Shen TL, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527:329–35. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jaffe EA, Nachman RL, Becker CG, et al. Culture of human endothelial cells derived from umbilical veins:identification by morphologic and immunologic criteria. J Clin Invest. 1973;52:2745–56. doi: 10.1172/JCI107470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kondakova IV, Yunusova NV, Spirina LV, et al. Association between intracellular proteinase activities and the content of locomotor proteins in tissues of primary tumors and metastases of ovarian cancer. Russ J Bioorgan Chem. 2014;40:681–7. doi: 10.1134/s1068162014060089. [DOI] [PubMed] [Google Scholar]
- 12.Lai RC, Tan SS, Teh BJ, et al. Proteolytic potential of the MSC exosome proteome:implications for an exosome-mediated delivery of therapeutic proteasome. J Proteomics. 2012;12:971907. doi: 10.1155/2012/971907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee SB, Schramme A, Doberstein K, et al. ADAM10 is upregulated in melanoma metastasis compared with primary melanoma. J Invest Dermatol. 2010;130:763–73. doi: 10.1038/jid.2009.335. [DOI] [PubMed] [Google Scholar]
- 14.Lendeckel U, Kohl J, Arndt M, et al. Increased expression of ADAM family members in human breast cancer and breast cancer cell lines. J Cancer Res Clin Oncol. 2005;131:41–8. doi: 10.1007/s00432-004-0619-y. [DOI] [PubMed] [Google Scholar]
- 15.Li W, Li C, Zhou T, et al. Role of exosomal proteins in cancer diagnosis. Mol Cancer. 2017;16:145. doi: 10.1186/s12943-017-0706-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malla RR, Pandrangi S, Kumari S, et al. Exosomal tetraspanins as regulators of cancer progression and metastasis and novel diagnostic markers. Asia Pac J Clin Oncol. 2018 doi: 10.1111/ajco.12869. doi:10.1111/ajco.12869. [DOI] [PubMed] [Google Scholar]
- 17.Mathivanan S, Ji H, Simpson RJ. Exosomes:extracellular organelles important in intercellular communication. J Proteomics. 2010;73:1907–20. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 18.Mofers A, Pellegrini P, Linder S, D'Arcy P. Proteasome-associated deubiquitinases and cancer. Cancer Metastasis Rev. 2017;36:635–53. doi: 10.1007/s10555-017-9697-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakamura K, Sawada K, Kinose Y, et al. Exosomes promote ovarian cancer cell invasion through transfer of CD44 to peritoneal mesothelial cells. Mol Cancer Res. 2017;15:78–92. doi: 10.1158/1541-7786.MCR-16-0191. [DOI] [PubMed] [Google Scholar]
- 20.Sadej R, Grudowska A, Turczyk L, et al. CD151 in cancer progression and metastasis:a complex scenario. Lab Invest. 2014;94:41–51. doi: 10.1038/labinvest.2013.136. [DOI] [PubMed] [Google Scholar]
- 21.Sharova N, Zakharova L. Multiple forms of proteasomes and their role in tumor fate. Recent Pat Endocr Metab Immune Drug Discov. 2008;2:152–61. [Google Scholar]
- 22.Shashova EE, Lyupina YV, Glushchenko SA, et al. Proteasome functioning in breast cancer:Connection with clinical-pathological factors. PLoS One. 2014;9:e109933. doi: 10.1371/journal.pone.0109933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sodar BW, Kittel A, Paloczi K, et al. Low-density lipoprotein mimics blood plasma-derived exosomes and microvesicles during isolation and detection. Sci Rep. 2016;6:24316. doi: 10.1038/srep24316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stoeck A, Keller S, Riedle S, et al. A role for exosomes in the constitutive and stimulus-induced ectodomain cleavage of L1 and CD44. Biochem J. 2006;393:609–18. doi: 10.1042/BJ20051013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tai YL, Chen KC, Hsieh JT, et al. Exosomes in cancer development and clinical applications. Cancer Sci. 2018 doi: 10.1111/cas.13697. doi:10.1111/cas.13697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whiteside TL. The emerging role of plasma exosomes in diagnosis, prognosis and therapies of patients with cancer. Contemp Oncol (Pozn) 2018;22:38–40. doi: 10.5114/wo.2018.73882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Witten M, Parker CC. Screening mammography:recommendations and controversies. Surg Clin North Am. 2018;98:667–75. doi: 10.1016/j.suc.2018.03.003. [DOI] [PubMed] [Google Scholar]
- 28.Yanez-Mo M, Siljander PR, Andreu Z, et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. 2015;4:27066. doi: 10.3402/jev.v4.27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yunusova NV, Tugutova EA, Tamkovich SN, et al. The role of exosomal tetraspanins and proteases in tumor progression. Biochem (Moscow), Suppl Series B:Biomed Chem. 2018;12:191–202. doi: 10.18097/PBMC20186402123. [DOI] [PubMed] [Google Scholar]
- 30.Zhao Z, Yang Y, Zeng Y, et al. A microfluidic exosearch chip for multiplexed exosome detection towards blood-based ovarian cancer diagnosis. Lab Chip. 2016;16:489–96. doi: 10.1039/c5lc01117e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu Y, Chen X, Pan QJ, et al. A сomprehensive proteomics analysis reveals a secretory path- and status-dependent signature of exosomes released from tumor-associated macrophages. Proteome Res. 2015;14:4319–31. doi: 10.1021/acs.jproteome.5b00770. [DOI] [PubMed] [Google Scholar]


