Abstract
The lipid-enveloped influenza virus enters host cells during infection by binding cell surface receptors and, following receptor-mediated endocytosis, fusing with the membrane of the endosome, delivering the viral genome and transcription machinery into the host cell. These events are mediated by the haemagglutinin (HA) surface glycoprotein. At the low pH of the endosome, an irreversible conformational change in the HA, including the exposure of the hydrophobic fusion peptide, activates membrane fusion. Here we use electron cryomicroscopy and cryotomography to image influenza virus fusion with target membranes at low pH. We visualize structural intermediates of HA and their interactions with membranes during the course of membrane fusion as well as ultra-structural changes in the virus that accompany membrane fusion. Our observations are relevant to a wide range of protein-mediated membrane fusion processes and demonstrate how dynamic membrane events may be studied by cryomicroscopy.
Introduction
The influenza virus envelope contains two spike glycoproteins, the trimeric hemagglutinin (HA) which mediates receptor binding to sialic acid-containing receptors and membrane fusion with host membranes, and the tetrameric receptor-destorying neuraminidase (NA). The virus membrane also contains a proton channel, M2. HA is synthesized as a precursor, HA0, which is proteolytically cleaved to generate the disulphide-linked HA1 and HA2 subunits, which potentiates the membrane fusion activity1. HA2 subunits contain the C-terminal transmembrane anchors (TM) located in the viral membrane, and the N-terminal hydrophobic fusion peptides (FP) (Figure 1a). The X-ray structure of the soluble HA ectodomain at neutral pH (135 Å long) reveals globular head domains containing the receptor binding site on a helical stalk domain formed largely by HA2 with a long central coiled-coil and with the fusion peptides located 35 Å from the viral membrane2.
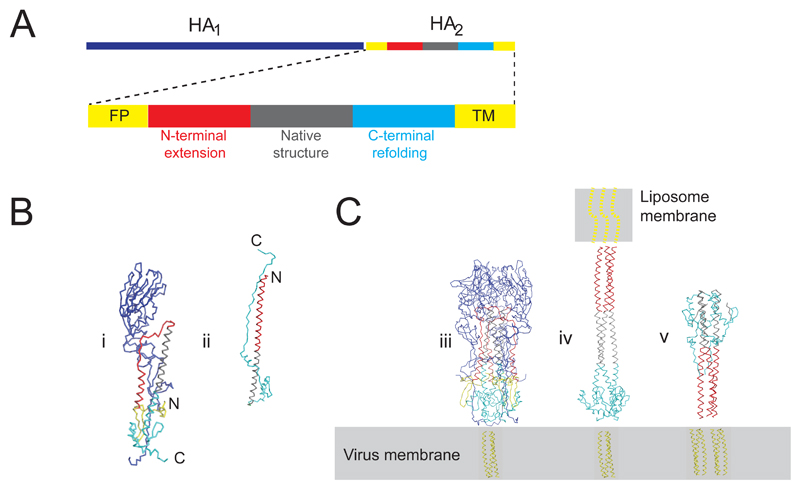
Figure 1. Atomic models for influenza hemagglutinin at neutral and low pH.
(a) Sequence diagram showing HA1 (328aa), HA2 (211aa), and polypeptide segments of HA2. (FP=fusion peptide, TM=transmembrane region). (b) X-ray models following same color scheme as in panel a (i) monomer model from the X-ray structure of soluble trimeric HA at neutral pH lacking TM (PDB 1HGE) with HA1 (blue) and HA2 colored according to sequence position: N-terminal fusion peptide (yellow) located toward bottom of the molecule near the C-terminal membrane attachment, N-terminal helix and loop (38-75, red), long helix (76-106, gray), C-terminal polypeptide (106-175, cyan). (ii) post-fusion “foldback” structure for HA2 (PDB 1Q1B “EHA2”) shows co-location of the N- and C- termini. Part of long helix that retains native neutral pH structure is gray. Numberings from X-31 strain HA.
(c) X-ray models following same color scheme as in panel a for the (iii) HA trimer (iv) putative extended HA intermediate containing HA2 residues 76-175 (grey, cyan) from the neutral pH HA2 structure and residues 30-75 (red) from the low pH structure as an N-terminal extension to the long-coiled-coil. (v) trimeric protein model for low pH “foldback” form in an orientation in which FPs and TMs are inserted into the virus membrane as in self-inserted HA2. Location of HA trans-membrane region of unknown structure indicated by cartoon in virus membrane (yellow). Fusion peptide shown as dotted yellow lines.
At the low pH of the endosome, an irreversible conformational change in the HA, including the exposure of the fusion peptide, activates membrane fusion3. The low pH transition is characterized by sensitivity to trypsin, which removes HA1 residues 28-328 that become mobile during the transition3,4. The HA conformational change has been studied by comparing the structures of the soluble ectodomain of HA at neutral pH to a trimeric fragment generated at low pH from viral HA (TBHA2) lacking TMs, FPs, and most of HA15,6 or to highly similar structures obtained by expression of HA2 38-175 (EBHA2) 7 and HA2- 23-185 (EHA2) 8.
Comparison (Fig. 1b) of the neutral pH HA structure 2 to structures of soluble forms of the HA2 subunit in the low pH conformation 5,6,8 reveals extensive rearrangements in HA2: (1) The loops and small helices N-terminal to the central α-helices refold to extend the triple stranded coiled coil. This extension would move the FPs over 100 Å to the membrane distal tip of the molecule where they could interact with a target membrane. (2) The C-terminal half of the central helices “fold back” anti-parallel to the newly formed helices and the more C-terminal region refolds to an extended structure that packs along grooves in the coiled-coils, thus co-locating the N- and C-termini of the HA2 subunit. FPs have been shown to interact with the target membrane during fusion 9, but in the absence of a target membrane, the FPs insert into their own viral membrane. Negative stain 4,10 and cryomicroscopy 11 studies suggest that the structure of the ectodomain of the resulting self-inserted HA2 is the same as that of the low pH crystal structures.
An HA2 structural re-arrangement in which the N-terminal extension of the coiled-coil positions FPs to interact with the target membrane while the TMs remains anchored in the virus membrane is therefore an attractive model for an HA intermediate in membrane fusion 12,13 (Fig. 1c) and has also been proposed as an intermediate in fusion by other enveloped viruses 14. The C-terminal refolding that brings the apposition of FP and TM may occur subsequently.
Cryomicroscopy and cryotomography 11,15–19 have shown previously that influenza viruses are capsular or filamentous particles of polar organization with the HA covering most of the virus surface and the receptor destroying NA most often in patches at one end of the virus11,17,20. A highly organized layer of the M1 matrix protein beneath the membrane maintains the virus shape, and an assembly of 8 RNPs packaging genetic segments is attached at one end of the particle 21–23.
Here we study the interaction of influenza particles with liposome targets to which they fuse at low pH to understand how changes in protein architecture and virus ultrastructure are coupled to membrane transformations. We interpret glycoprotein structures in images and tomograms by comparison with X-ray models for the HA shown in Fig. 1.
Results
Time course of influenza virus membrane fusion
Cryomicroscopy and cryotomography of the H3 N2 influenza virus A/Udorn/72 at pH7 showed the particles to be arranged as previously described11 with capsular or filamentous shape. The liposomes were content-less, sometimes multi-lamellar and of heterogeneous size. We mixed virus particle with liposomes at pH7 and observed no interaction, consistent with the absence of sialic acid receptors in the liposomes. (Fig 2a, Supplementary Fig1a).
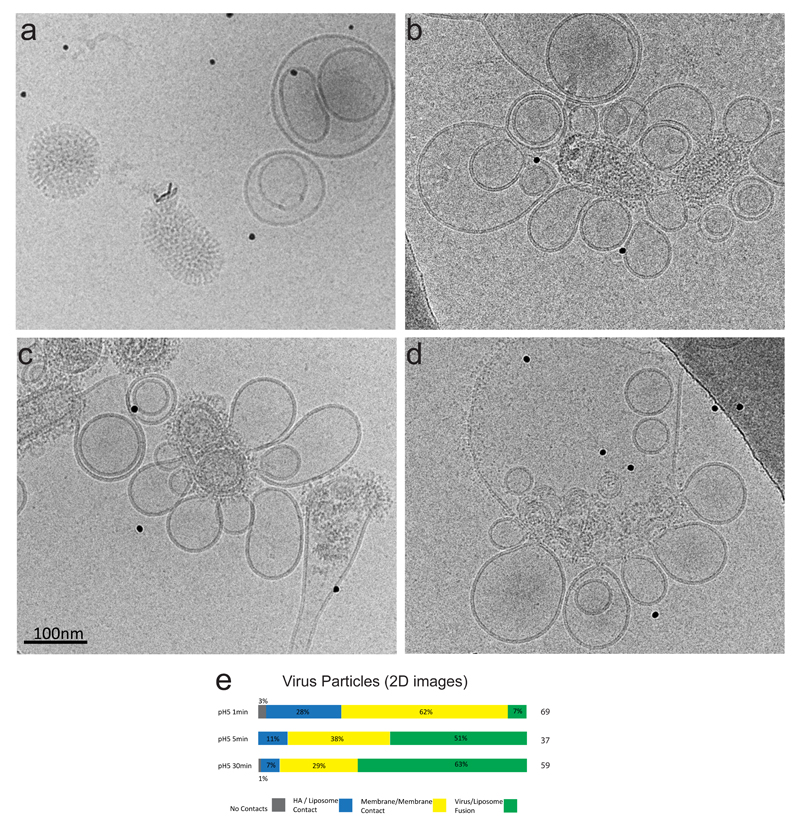
Figure 2. Timecourse of fusion studied by cryomicroscopy.
(a) pH7 images showing no interaction between virus, and liposomes. The virus is capsule-shaped with ordered glycoproteins, and the liposomes may be multi-lamellar and of heterogeneous size.
(b) pH5, 1min. images show viral particles interacting with many liposomes, but retaining their capsular shape and well-ordered M1 matrix layer. Their surface glycoproteins are disordered.
(c) pH5, 5mins. Viral particles have less internal order than after 1min and are contacting many liposomes. Some particles have fused with liposomes and viral contents can be observed in some liposomes.
(d) pH5, 30 mins. Large liposomes contain internal content of viruses, similar to those seen in low pH virions. Glycoproteins are widely distributed around the membrane and interactions with other liposomes are seen.
(e) 2D cryoEM image classification of all viral particle interactions with liposomes into 4 categories at the time intervals 1, 5, and 30 mins, and percentages given. Category 1: no liposome contact. Category 2: liposome contact via the HA alone with no evidence for membrane/membrane contact. Category 3: viral membrane/liposome membrane in contact. The liposome may be extruded into a dimple shape, but it is not possible to discern whether a fusion pore has formed. Category 4: full fusion and viral disassembly such that viral contents have become part of the liposomal structure.
We followed the fusion timecourse by incubating virus and liposomes (5 liposomes/virus) at pH 4.9, neutralizing pH after 1 minute, 5 minutes, and 30 minutes, and then imaging by cryomicroscopy.
After 1min at low pH, over 80% of virus particles interacted with liposomes, either via contact with the viral glycoprotein layer or where liposome and virus membranes became apposed, including through a previously described conical perturbation (pointed dimple) of liposomes by influenza virus 18. This is also similar to a structure observed by freeze-fracture EM 24. Viral particles looked intact and very few particles had fully fused with liposomes (Fig. 2b, Suppelmentary Fig.1).
After 5 minutes, more particles had fused with liposomes and viral contents were observed in the interior of some liposomes, but many particles retained a capsular shape, interacting with liposomes via glycoproteins (Fig. 2c, Suppelmentary Fig.1).
After 30 minutes, more than 60% of virus particles had fused. The remaining particles had lost their capsular shape and matrix layer regularity as previously described for incubation of Udorn particles at low pH11 (Figure 2d, Supplementary Fig.1).
A count of the number of virus particles in 2D images at each time point showed a progression from glycoprotein-only contact with liposomes, to closer membrane/membrane apposition, through to fusion and redistribution of viral components, indicating that these categories are temporally-ordered events on the pathway to fusion (Fig. 2e).
We next applied electron cryotomography to characterize fusion events in three-dimensions for all the time points. Supplementary Fig. 2 shows sections of two tomograms of a 1 minute low pH incubation. The viral particles retain their capsular shape, with an intact M1 layer adjacent to the viral membrane, RNP segments are clearly visible in the interior, and NA clusters are observed at one end of the particles. The native HA pH 7 rod-like structure is less apparent and the particles are in association with liposomes.
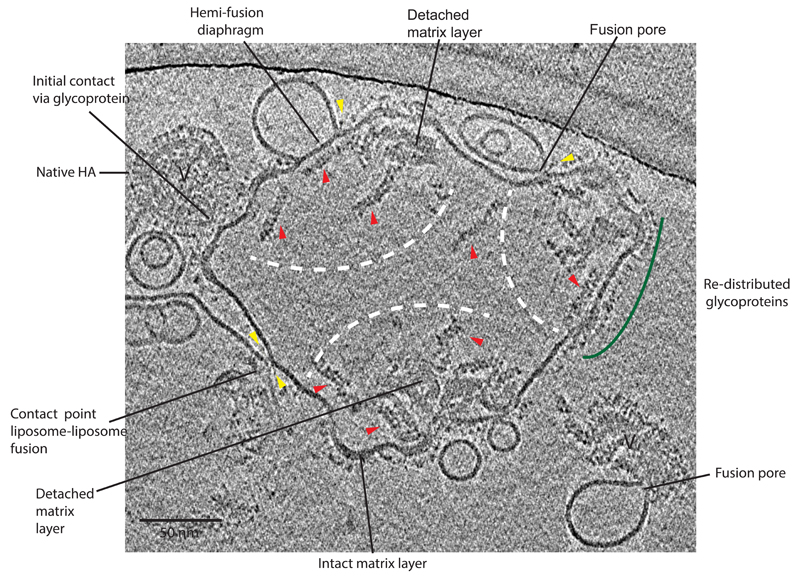
Fig. 3 shows the product of several fusion reactions, interpretable as 3 virions having fused completely with a single liposome target (30 minute time point). The glycoproteins are redistributed within the liposome membrane, though they may also remain densely packed in regions of membrane that retain an adjacent matrix layer that also shapes the surrounding membrane. Within the lumen of the fused structure, aggregations of M1 matrix layer 11,25 that have separated from the membrane can be identified. RNP assemblies have dispersed to free RNPs, some of which may remain associated, usually at one end, with the dense M1 aggregates. We also observe liposome-liposome contact and further fusion mediated by the HA redistributed in the post-fusion liposome. In addition, Fig. 3 shows viruses at the first steps of entry into the liposomes.
Figure 3. Tomogram section of 30 min. pH5 incubation time point.
3D tomogram section shows a large amorphous shaped liposome with an interior containing 3 separate areas, indicated by white dashed arcs, each containing material that resembles the content of a single virion. Glycoproteins are redistributed within the liposomal membrane (examples indicated by green arc). M1 matrix layer is detached from the membrane and lies close to portions of matrix layer still attached to membrane (labelled). Individual free RNP segments are visible (examples indicated by red arrows). Virus particles at earlier steps that have not fused are indicated (V). Yellow arrows indicate dense bar structures. Hemi-fusion diaphragm also has dense bars at ends of contact area.
Contact Zones
We refer to the HA-mediated interaction between membranes as a “contact zone”13. Prior to full fusion, a single virion typically interacted with several liposomes at separate contact zones on its surface. The example in Fig. 4a-d shows a single virus with 3 contact zones, each an extended area of glycoprotein interaction with the target. The segmentation in Figure 4d shows the perturbation of membranes.
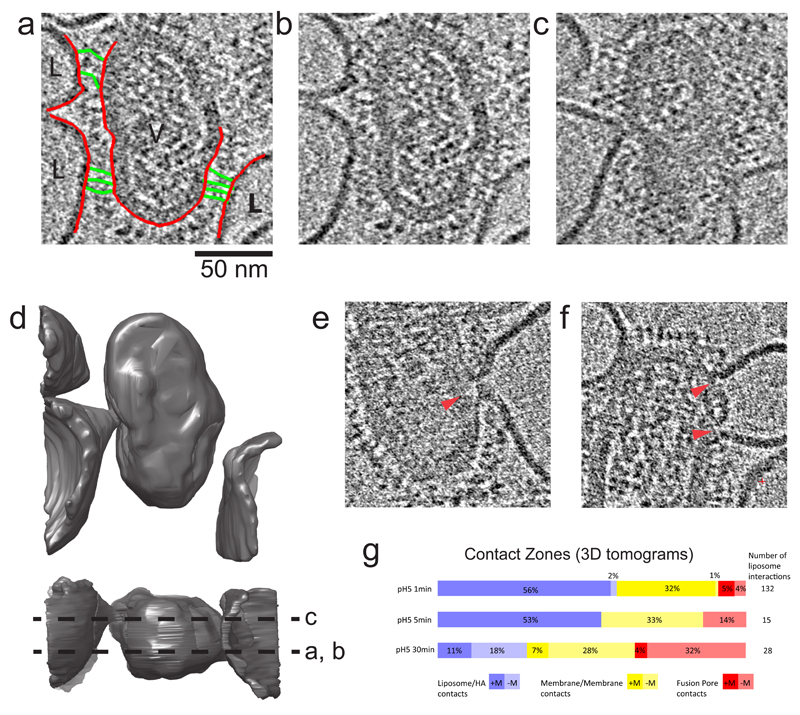
Figure 4. 3D Contact Zones.
(a,b) Identical sections of a tomogram showing glycoprotein attachments from a single virus particle to 3 liposomes. In (a), membranes are colored red, and HA extended intermediates are colored green. (c) Section of the same tomogram but 160 Å away from panel a,b showing membrane deformation of liposomes interacting with the virus particle. Glycoprotein-lipid contacts are also observed around these deformations indicating a wide zone of contact. (d) Surface rendering of virus and liposome membranes in tomogram giving rise to sections in panels b and c. The lower orthogonal view indicates the locations of sections shown in b and c by dotted lines. (e) Tomogram section showing contact zone with single fusion pore (full context in Supplementary Fig. 2) and (f) double fusion pore, local disruption of the matrix layer indicated by red arrows.
(g) Classification of 3D contact zones at the time intervals 1, 5, and 30 mins. Each of the 3 categories shown here are subdivided to show the presence (+) or absence (-) of the matrix protein (M1) layer at the localised point of virus/ liposome contact.
As in our 2D analysis, we classified the type of contact zone. Inspection in 3D showed that the membrane/membrane contact category shown in Fig. 2e could be further subdivided based on the presence or absence of a pore between the membranes (Figure 4e). The fusion pores resemble the dimple contact, but with a continuous connection between virus and liposome interior (e.g. locations indicated in Fig. 3, Fig. 4 e, f), and include pores found at liposome/liposome contacts mediated by HA. The pore connections had an average outer diameter of 14 nm +/- 5nm (n=52). We sometimes observed two pores in close proximity (Figure 4f). A classification of all the HA-mediated 3D contact zones for each of the incubation time points observed by cryotomography (Fig. 4g) revealed a progression from (1) glycoprotein-only contact with liposomes (e.g. Fig. 4a, b) to (2) membrane contact at a point via a dimple (e.g. Fig. 4 c, d) to (3) membrane-membrane contact via a pore (e.g. Fig. 4 e, f).
Typically, in observations of early virus:liposome interactions, the liposome membrane was deformed to a conical dimple which contacted the viral membrane at a point, but the virus membrane remained unperturbed with an adjacent, intact matrix layer (Fig. 4a-c). However, in the case of fusion pores we observed local disruption of the matrix layer (red arrows in Fig. 4 e, f), and the viral membrane curved towards the liposomal dimple, forming a symmetric neck. Thus local disruption or removal of the matrix layer may be important for formation of a symmetric pore.
We scored whether the matrix layer remained closely associated with viral membrane areas involved in all fusion contact zones and found it to be disrupted or absent in nearly half the fusion pores observed after 1 minute, and from most fusion pores and contacts after 5 and 30 minutes (Fig. 4g). These observations are consistent with studies reporting changes in the matrix layer15,16 and dissociation of the matrix layer and loss of filamentous shape when virions are incubated at pH 4.9 11.
HA architecture
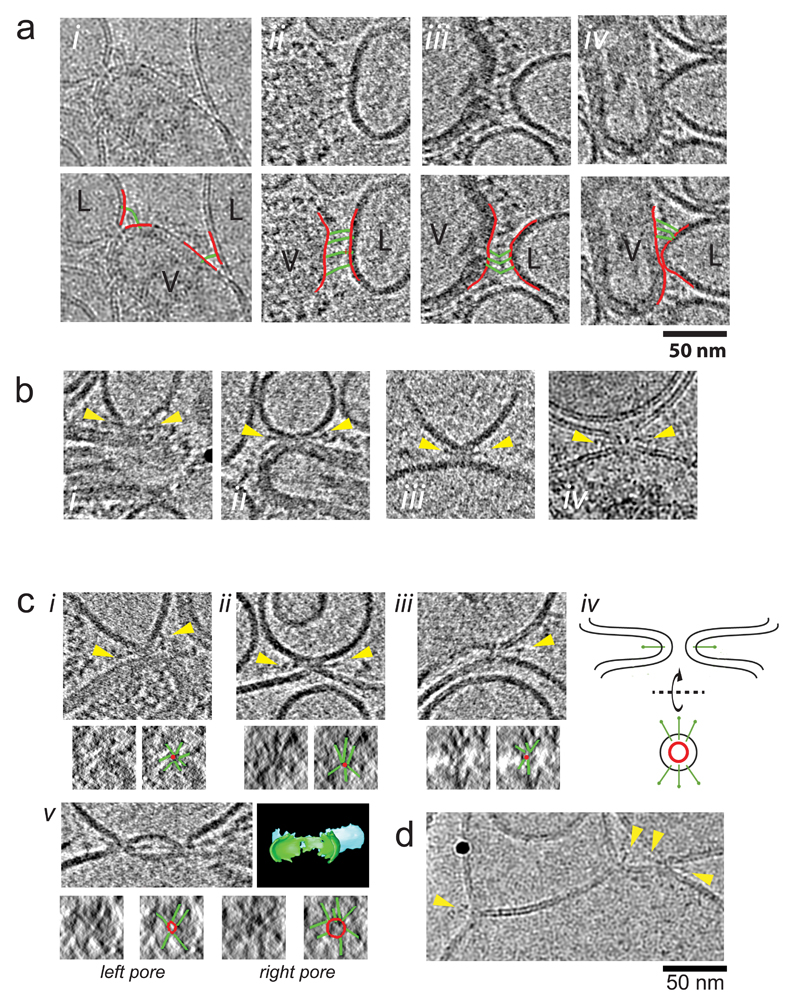
We examined the detailed protein architecture associated with the steps observed during the timecourse. In the tomogram section of the contact zone in Fig. 4 a-c and Fig. 5a, many thin densities are observed as a continuous line from the virus membrane to the liposome membrane, perpendicular to both membranes. See also Supplementary Fig. 3, 4. A histogram of similar densities shows a mean length 18.8 nm +/-3.2 nm (n=132). These are longer and thinner than measurements of glycoproteins assigned as pH7 HA which measure 13.2 nm +/-1.0 nm, similar to the X-ray structure (13.5 nm). See Supplementary Fig. 5. The distance between virus and liposome membranes at the point shown in Fig. 4a is too great to be spanned by neutral pH conformation HA anchored in the virus membrane. Rather, the length and thinness of the density, and its association with both membranes is consistent with an HA extended intermediate, such as the extended coiled-coil model iv shown in Fig. 1c, assuming the HA2 TMs are in the virus membrane and the FPs in the liposome membrane. Thus, as a first step in the progression toward fusion, glycoproteins extend beyond the neutral pH virus glycoprotein layer to interact with a target liposome, forming an initial contact zone between the virus and the liposome.
Figure 5. Tomogram sections showing HA and membrane architecture during fusion.
(a) HA extended intermediate Panels show thin densities (HA extended intermediate) between virus (V) and liposome (L) membranes. (i) 2D image (ii-iv) tomograms sections (projections of 22 Å slab). Images in second row are identical to row above but indicate membranes (red lines) bridged by extended HA intermediate (green lines). More examples are provided in Supplementary Fig. 3, 4.
(b) Bars at Membrane Point Contact Panels show dense bar structures protruding from the point of membrane attachments (yellow arrows). (i-iii) tomograms sections (projections of 22 Å slab), (iv) 2D image.
(c) Bars radiating from point contacts and fusion pores. Panel i shows contact between virus and liposome. Panels ii and iii show pores between HA-containing liposomes devoid of matrix layer (products of full viral fusion) and liposomes. All these panels show dense bars protruding from the contact points (yellow arrows). When viewed orthogonally along the contact or pore axis (according to the schematic in iv), several dense bars are seen to radiate from the central lipid contact or pore in a ‘star’ arrangement. Images of these views are shown in the small image at the lower left of each panel. The small image at the lower right indicates the location of bars (green) and the point of contact or lipid pore (red). Panel v shows two fusion pores in close proximity between an HA-containing liposome devoid of matrix layer and a liposome along with a 3D segmentation model for membranes that shows continuity of pores connecting membranes. Orthogonal views for the left and right pores shown in small panels below. (d) 2D image of flat contact (hemi-fusion diaphragm) with bars indicated. Black sphere is gold fiducial marker.
We also observed lines of density connecting the virus membrane to liposomes similar in length and thinness to the extended intermediate, but these were bent at a point approximately midway along their length (e.g. Fig. 5a panel iii, Supplementary Fig. 3, 4, 6, 7). This occurred between membrane surfaces that were closer together than those where the extended intermediate exhibits its maximum length and between membranes that were not parallel to each other such as observed in the dimple. We observed that the curvature of the liposome membrane may vary which affects the number of HAs that can insert into the target membrane. We estimate 15-20 HAs interact with a liposome to form the contact zones in Figure 4a-c (Supplementary Fig. 6) and in Supplementary Fig. 7, similar to the number of HAs in an equivalent surface area on neutral pH virus23.
On examination of dimples and pores, we observed dense bars that were shorter and thicker than the extended intermediate and that projected radially from the membrane contact point or pore, roughly parallel to the virus membrane, and oblique to the liposome membrane (Fig. 5b, c and Supplementary Fig. 8).
We observed dense bar structures radiating from contacts between liposomes containing HA but devoid of an M1 protein layer (produced as a result of redistributed HA following fusion) and another liposome (Figure 5b,c, Supplementary Fig. 8). In membrane interactions between a liposome containing HA (but devoid of a matrix protein) and another liposome, both membrane contacts and fusion pores have symmetrical membrane profiles, consistent with a role for the matrix layer in maintaining the asymmetric dimple contact. Of 52 fusion pores, 26 had dense bars radiating from the neck region
The length of the bars surrounding both dimples and fusion pores, 11.5+/-1.1 nm (n=34), as well as their shape, was similar to the ectodomain of HA2 molecules (Fig. 1c v) based on our measurements of self-inserted HA in our tomograms (average 12.4 +/- 1.4 nm (n=116), Supplementary Fig. 5), comparison to earlier negative stain EM studies 10, and comparison to the low pH X-ray crystal structures. When viewed down the pore axis, the circumference of the pore had similar dense bars radiating from it (Figure 5c, small panels), resembling a star arrangement. The average length of these lines surrounding both dimple contacts and pores is 10.5 +/- 2.1 nm (n=35).
Occasionally, we observe extended, flat contacts of membranes comprised of a single leaflet from each liposome membrane, similar to hemi-fusion diaphragms reported during in vitro SNARE protein-mediated fusion 26,27. This occurs under conditions in which HAs are sparsely distributed in a membrane following fusion with liposome targets. A bar structure resembling an HA2 molecule is inserted at each end of the contact area (e.g. hemi-fusion diaphragm labeled in Figure 3, Fig. 5d).
Discussion
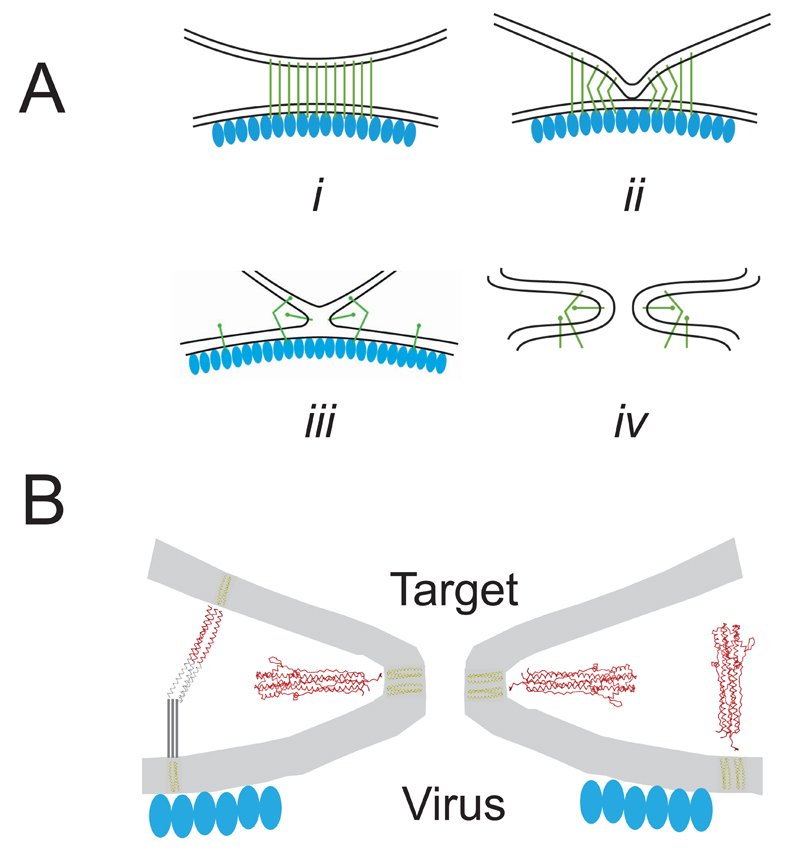
In our identification of the major structural steps in membrane fusion (Fig. 6a) observed here by cryotomography, interaction of the HA extended intermediate with the target liposome is an early event in the progression toward fusion and forms an extended contact zone (Figure 6a(i)). The dimpling of the liposome membrane is an effect of multiple FP insertions and is consistent with models for membrane curvature arising from hydrophobic protein insertion 28,29. While the extended intermediate is held at each end respectively by virus and target membrane, HA2 cannot refold into the post-fusion structure, which requires apposition of the N- and C- termini.
Figure 6. Summary of structural steps in membrane fusion.
(a) Schematics for sequential steps (i) initial contact zone between virus and target liposomal membrane, mediated through extended HA intermediate (green lines). Virus matrix layer is indicated by blue ovals. (ii) Dimpled liposome membrane with bent HA extended intermediate, and extended intermediate toward periphery of contact zone. (iii) membrane-membrane contact without a pore, with HA foldback intermediate radiating from contact point. (iv) fusion pore with symmetric membrane profile, HA foldback structure and virus matrix layer removed. (b) Protein architecture associated with membrane deformation and fusion including cartoon for bent HA extended intermediate and foldback structure inserted radially around fusion pore or self-inserted in the virus membrane. Virus matrix layer is disrupted at the location of the fusion pore.
The observed flexibility of the extended intermediate allows it to remain engaged with the dimpled membrane, and stabilize its close approach to the viral membrane (Figure 6a(ii)). The bending of the extended intermediate brings the FP and TM closer to each other, eventually coming together near the point of the dimple (Figure 6a(iii)). The dimpled profile of the target membrane serves as a force of constraint, guiding the metastable extended intermediate to a conformation and position in which the C-terminal polypeptide can pack against the N-terminal coiled-coil seen in the post-fusion conformation. The bars of density observed perpendicular to the axis of the dimples and fusion pores (Figure 6a(iv), 6b) suggest that the re-folding of the HA2 ectodomain into the low pH “foldback” structure is complete. The foldback structure also appears at membrane contact before a fusion pore is discernable in our experiments in which case the FP and TM must still be located in different membranes. This geometry, in which the ectodomain is oblique to either the virus or liposome membranes, may be facilitated by non-helical polypeptide between the coiled-coil of the foldback structure and the membrane-inserted segments as observed in the crystal structure of the low pH form 8 (Fig. 1b (ii)).
We estimate from the observed “star assemblies” (Fig. 5c) that 6-8 such bars can fit radial to the point at which the fusion pore forms. Membrane fusion may occur when enough foldback structures are arranged about the axis of the dimple at its point of contact. The extended hemi-fusions diaphragms that we observe under conditions in which there are very few HA’s in close proximity may result from an insufficient number of HA foldback structures available for full fusion. Membrane fusion may be driven by additional free energy available when the FP and TM become located in the same membrane and/or through the formation of conserved structures such as the N-cap at the end of the coiled-coil located near the membrane 8.
The observed membrane transformations and the proposed protein structural re-arrangements during membrane fusion are consistent with other virus fusion proteins where the same flexibility and refolding may be invoked30,31, and with SNARE-mediated membrane fusion which proceeds through a similar point contact between membranes26,32 and is driven by the flexible association of SNARE proteins on different membranes to form a coiled-coil assembly 27,33.
In addition, ultra-structural changes in virus architecture are important to membrane fusion. The matrix layer maintains the asymmetry of the dimple contact of the target with the virus membrane. Acidification of the virus interior via the M2 proton channel34, which has previously been shown to increase the rate of fusion35, is required for local disruption of the matrix layer enabling formation of a symmetrical fusion pore and continuity of the virus and target lumens.
Online Methods
Growth and Purification of Virus
A/Udorn/72 virus was grown in the allantoic cavity of 10-day old hens’ eggs under UK Home Office project license number PPL 80/2487. The allantoic fluid was harvested after 3 days, from which virus was purified over sucrose gradients as previously described 36.
Preparation of liposomes
A lipid mixture of phosphatidyl choline PC, phosphatidyl ethanolamine PE, phosphatidyl serine PS, phosphatidyl inositol PI, sphingomyelin S, cholesterol C in molar ratio 10:3:1:0.5:3:4 in chloroform was used to make small liposomes mimicking the composition of the plasma membrane10. The chloroform was evaporated under nitrogen gas and lipids re-suspended in PBS, freeze thawed and pressure extruded through a 0.1μm filter. The resulting liposomes are sometimes multi-lamellar and have a diameter in the range 200-500 nm.
Low pH incubation and trypsin-treatment of virus liposome mixtures
Samples were incubated at 4°C and the pH lowered to 4.9 with 0.1 M sodium citrate. After the specified time interval aliquots were removed and neutralized with 1M Tris pH 7.4. This was performed on 2 separate virus preps for each time point. In the case of further trypsin-treatment, samples were then incubated with trypsin (Sigma) at an enzyme:protein ratio of 1:50 for 15 mins at 20°C, before stopping the reaction with trypsin inhibitor (Sigma). This was also performed on 2 separate virus preps for each time point. Trypsin treatment removes the HA1 subunits of HA molecules that have undergone the low pH conformational change and facilitates imaging of the remaining HA2 subunits4.
Electron Cryomicroscopy
Samples were mixed with 10 nm Protein A gold particles (British-Biocell) and 4 μl applied to amylamine glow-discharged 200 mesh copper Quantifoil™ (R2/4) grids in the environment chamber (4 °C, 90% R.H.) of a Vitrobot Mark III (FEI), blotted on both sides with a double layer of paper for 4 seconds before plunging into liquid ethane. Grids were transferred to a Gatan 626 tomography holder or a Polara stage. Imaging was performed in an FEI Spirit TWIN microscope at 120 keV using a tungsten filament source and equipped with a cryobox around the sample, or in an FEI Polara FEG microscope operated at 200 keV. Images were recorded un-binned on an Eagle 2K camera (FEI, Spirit) at 30K (7.2 Å/pixel) or 52K (4.3Å/pixel) magnification, or un-binned on a 224HD detector (TVIPS, Polara) at 31K (4.3 Å/pixel) under a range of defocus conditions (-3 to -4.5 μm). Magnification was calibrated by recording images of catalase crystals under identical conditions. At least two grids were imaged for each preparation and each timepoint.
2D images were collected from the same grids as those used for tomograms. It was not possible to identify definitive virus/liposome complexes when scanning the grid at low magnification so images were recorded at each hole in the quantifoil carbon that appeared to have some content.
Tilt series for tomography were recorded automatically using Serial EM 37 up to ±60° in 3° steps, typically with a total dose less than 80 e-/Å2.
2D Image Analysis
All 2D images acquired were used when counting the number of particles involved in liposome complexes thus avoiding operator bias for different sized complexes when acquiring images of different experimental conditions. We manually inspected each virus particle and its interaction with liposomes to assess which of the following 4 categories it belonged to: 1) no virus – liposome contact, 2) virus – liposome contact via viral glycoproteins, 3) virus – liposome contact via membranes, 4) viral fusion with liposome.
2D images were filtered to the first zero of the contrast transfer function (~30 Å) except for Figure 2 and Supplementary Figure 1.
Image and Tilt Series Analysis
Tomographic tilt series were aligned using IMOD software 38. Alignment initially used cross correlation and then used gold particles as fiducial markers. Reconstructed 3D volumes were generated by back-projection or by the SIRT iterative alignment and reconstruction procedure. Length measurements were performed in 3D using IMOD. We analysed 38 tomograms to obtain the results presented here.
We manually inspected each virus – liposome contact observed in all tomograms to classify these contacts according to the following 3 categories: 1) contact via viral glycoproteins only, 2) contact of liposomal membrane abutting the viral membrane, 3) membrane – membrane contact via a pore between the membranes.
Supplementary Material
Acknowledgements
We are grateful to S. Wharton for advice on experiments, M. Maiorca for help in building segmentation models of the membranes, and J. Skehel and J. Molloy for discussions. Research was supported by the Francis Crick Institute, which receives its core funding from Cancer Research UK, the UK Medical Research Council (programme code U117581334 to P.B.R.), and the Wellcome Trust.
Footnotes
Accession Codes
Tomograms have been deposited in the Electron Microscopy Data Bank under accession code EMD-4034.
Author Contributions
L.C. and P.B.R designed experiments and wrote the manuscript. L.C. performed experiments and analyses.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–69. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 2.Wilson IA, Skehel JJ, Wiley DC. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature. 1981;289:366–73. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 3.Skehel JJ, et al. Changes in the conformation of influenza virus hemagglutinin at the pH optimum of virus-mediated membrane fusion. Proc Natl Acad Sci U S A. 1982;79:968–72. doi: 10.1073/pnas.79.4.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ruigrok RW, et al. Electron microscopy of the low pH structure of influenza virus haemagglutinin. EMBO J. 1986;5:41–9. doi: 10.1002/j.1460-2075.1986.tb04175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bullough PA, Hughson FM, Skehel JJ, Wiley DC. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 6.Ruigrok RW, et al. Studies on the structure of the influenza virus haemagglutinin at the pH of membrane fusion. J Gen Virol. 1988;69(Pt 11):2785–95. doi: 10.1099/0022-1317-69-11-2785. [DOI] [PubMed] [Google Scholar]
- 7.Chen J, et al. A soluble domain of the membrane-anchoring chain of influenza virus hemagglutinin (HA2) folds in Escherichia coli into the low-pH-induced conformation. Proc Natl Acad Sci U S A. 1995;92:12205–9. doi: 10.1073/pnas.92.26.12205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen J, Skehel JJ, Wiley DC. N- and C-terminal residues combine in the fusion-pH influenza hemagglutinin HA(2) subunit to form an N cap that terminates the triple-stranded coiled coil. Proc Natl Acad Sci U S A. 1999;96:8967–72. doi: 10.1073/pnas.96.16.8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsurudome M, et al. Lipid interactions of the hemagglutinin HA2 NH2-terminal segment during influenza virus-induced membrane fusion. J Biol Chem. 1992;267:20225–32. [PubMed] [Google Scholar]
- 10.Wharton SA, et al. Electron microscopy of antibody complexes of influenza virus haemagglutinin in the fusion pH conformation. EMBO J. 1995;14:240–6. doi: 10.1002/j.1460-2075.1995.tb06997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calder LJ, Wasilewski S, Berriman JA, Rosenthal PB. Structural organization of a filamentous influenza A virus. Proc Natl Acad Sci U S A. 2010;107:10685–90. doi: 10.1073/pnas.1002123107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carr CM, Kim PS. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell. 1993;73:823–32. doi: 10.1016/0092-8674(93)90260-w. [DOI] [PubMed] [Google Scholar]
- 13.Ivanovic T, Choi JL, Whelan SP, van Oijen AM, Harrison SC. Influenza-virus membrane fusion by cooperative fold-back of stochastically induced hemagglutinin intermediates. Elife. 2013;2:e00333. doi: 10.7554/eLife.00333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim YH, et al. Capture and imaging of a prehairpin fusion intermediate of the paramyxovirus PIV5. Proc Natl Acad Sci U S A. 2011;108:20992–7. doi: 10.1073/pnas.1116034108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fontana J, Cardone G, Heymann JB, Winkler DC, Steven AC. Structural changes in Influenza virus at low pH characterized by cryo-electron tomography. J Virol. 2012;86:2919–29. doi: 10.1128/JVI.06698-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fontana J, Steven AC. At low pH, influenza virus matrix protein M1 undergoes a conformational change prior to dissociating from the membrane. J Virol. 2013;87:5621–8. doi: 10.1128/JVI.00276-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harris A, et al. Influenza virus pleiomorphy characterized by cryoelectron tomography. Proc Natl Acad Sci U S A. 2006;103:19123–7. doi: 10.1073/pnas.0607614103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee KK. Architecture of a nascent viral fusion pore. EMBO J. 2010;29:1299–311. doi: 10.1038/emboj.2010.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vijayakrishnan S, et al. Cryotomography of budding influenza A virus reveals filaments with diverse morphologies that mostly do not bear a genome at their distal end. PLoS Pathog. 2013;9:e1003413. doi: 10.1371/journal.ppat.1003413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chlanda P, et al. Structural Analysis of the Roles of Influenza A Virus Membrane-Associated Proteins in Assembly and Morphology. J Virol. 2015;89:8957–66. doi: 10.1128/JVI.00592-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fournier E, et al. A supramolecular assembly formed by influenza A virus genomic RNA segments. Nucleic Acids Res. 2012;40:2197–209. doi: 10.1093/nar/gkr985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noda T, et al. Architecture of ribonucleoprotein complexes in influenza A virus particles. Nature. 2006;439:490–2. doi: 10.1038/nature04378. [DOI] [PubMed] [Google Scholar]
- 23.Wasilewski S, Calder LJ, Grant T, Rosenthal PB. Distribution of surface glycoproteins on influenza A virus determined by electron cryotomography. Vaccine. 2012;30:7368–73. doi: 10.1016/j.vaccine.2012.09.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanaseki T, Kawasaki K, Murata M, Ikeuchi Y, Ohnishi S. Structural features of membrane fusion between influenza virus and liposome as revealed by quick-freezing electron microscopy. J Cell Biol. 1997;137:1041–56. doi: 10.1083/jcb.137.5.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruigrok RW, Calder LJ, Wharton SA. Electron microscopy of the influenza virus submembranal structure. Virology. 1989;173:311–6. doi: 10.1016/0042-6822(89)90248-1. [DOI] [PubMed] [Google Scholar]
- 26.Diao J, et al. Synaptic proteins promote calcium-triggered fast transition from point contact to full fusion. Elife. 2012;1:e00109. doi: 10.7554/eLife.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hernandez JM, et al. Membrane fusion intermediates via directional and full assembly of the SNARE complex. Science. 2012;336:1581–4. doi: 10.1126/science.1221976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chernomordik LV, Kozlov MM. Mechanics of membrane fusion. Nat Struct Mol Biol. 2008;15:675–83. doi: 10.1038/nsmb.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ford MG, et al. Curvature of clathrin-coated pits driven by epsin. Nature. 2002;419:361–6. doi: 10.1038/nature01020. [DOI] [PubMed] [Google Scholar]
- 30.Bose S, Jardetzky TS, Lamb RA. Timing is everything: Fine-tuned molecular machines orchestrate paramyxovirus entry. Virology. 2015;479-480:518–31. doi: 10.1016/j.virol.2015.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harrison SC. Viral membrane fusion. Virology. 2015;479-480:498–507. doi: 10.1016/j.virol.2015.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bharat TA, et al. SNARE and regulatory proteins induce local membrane protrusions to prime docked vesicles for fast calcium-triggered fusion. EMBO Rep. 2014;15:308–14. doi: 10.1002/embr.201337807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sollner TH. Intracellular and viral membrane fusion: a uniting mechanism. Curr Opin Cell Biol. 2004;16:429–35. doi: 10.1016/j.ceb.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 34.Jackson DC, et al. Electron microscopic evidence for the association of M2 protein with the influenza virion. Arch Virol. 1991;118:199–207. doi: 10.1007/BF01314030. [DOI] [PubMed] [Google Scholar]
- 35.Wharton SA, Belshe RB, Skehel JJ, Hay AJ. Role of virion M2 protein in influenza virus uncoating: specific reduction in the rate of membrane fusion between virus and liposomes by amantadine. J Gen Virol. 1994;75(Pt 4):945–8. doi: 10.1099/0022-1317-75-4-945. [DOI] [PubMed] [Google Scholar]
- 36.Skehel JJ, Schild GC. The polypeptide composition of influenza A viruses. Virology. 1971;44:396–408. doi: 10.1016/0042-6822(71)90270-4. [DOI] [PubMed] [Google Scholar]
- 37.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 38.Mastronarde DN. Dual-axis tomography: An approach with alignment methods that preserve resolution. J Struct Biol. 1997;120:343–352. doi: 10.1006/jsbi.1997.3919. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.