Abstract
Multicellular organisms receive extracellular signals at the surface of a cell by using receptors. The extracellular domains (ECDs) of cell surface receptors serve as interaction platforms, and as regulatory modules of receptor activation1,2. Understanding how interactions between ECDs produce signal-competent receptor complexes is challenging because of their low biochemical tractability3,4. In plants, discovery of ECD interactions is complicated by the massive expansion of receptor families, which creates tremendous potential for changeover in receptor interactions5. The largest of these families in Arabidopsis thaliana consists of 225 evolutionarily-related leucine-rich repeat receptor kinases (LRR-RKs)5, that function in microbe sensing, cell expansion, stomata development and stem cell maintenance6–9. While the principles governing LRR-RK signalling activation are emerging1,10, the systems-level organization of this family of proteins is totally unexplored. To address this, we interrogated 40,000 potential ECD interactions via a sensitized high-throughput interaction assay3, and produced an LRR-based Cell Surface Interaction network (CSILRR) comprising 567 interactions. To demonstrate the power of CSILRR for detecting biologically relevant interactions, we predicted and validated the function of uncharacterized LRR-RKs in plant growth and immunity. In addition, we show that CSILRR operates as a unified regulatory network in which the LRR-RKs most critical for its overall structure are required to prevent aberrant signalling of receptors that are several network-steps away. Thus, plants have evolved LRR-RK networks to process extracellular signals into carefully balanced responses.
LRR-RKs are modular proteins that feature an ECD with numerous LRR repeats, a transmembrane domain, and an intracellular kinase domain1. LRR-RKs sense a wide array of endogenous and exogenous ligands, including peptides and small molecule hormones, to regulate development and immunity in plants7,10. Stereotypical LRR-RKs include the steroid receptor BRASSINOSTEROID INSENSITIVE1 (BRI1) as well as the immune receptors FLAGELLIN SENSING 2 (FLS2) and PEP RECEPTORS 1/2 (PEPR1/2)1,11. Ligand-induced activation of BRI1, FLS2 or PEPR1/2 signaling requires physical interaction with the LRR-RK co-receptor BRI1-ASSOCIATED KINASE 1 (BAK1)12–15. In heterotypic LRR-RKs complexes, interactions between ECDs can activate or repress signalling pathways2. Yet, the full range of these interactions remains unmapped.
We cloned the ECDs of 200 LRR-RKs from Arabidopsis into bait and prey expression vectors for recombinant protein production in Drosophila Schneider S2 cells (Extended Data Fig.1, Supplementary Table 1). We then implemented the extracellular interaction assay established by Özkan et al. 3 and performed an all-by-all screen of the 200 ECDs (Extended Data Fig. 2). Since the Arabidopsis genome encodes 225 LRR-RKs5, we interrogated the extracellular LRRs interaction space to a completeness of 79%. This screen resulted in a CSILRR map containing 2,145 bidirectional interactions of which only 26.4% (567 high-confidence interactions (HCi)) passed our extremely stringent statistical cut-offs for network construction (Fig. 1a, Supplementary Text 1, Supplementary Table 2). To verify our screen results, the ECDs from the 567 HCi and from a random set of 248 low-confidence interactions (LCiCSI) were independently re-expressed and retested. To benchmark the retest screen, we assembled a positive reference set (PRS) of 20 literature-curated LRR-RK interaction pairs that complied with the criteria defined by Braun et al.4,16 (Supplementary Table 3). In the retest, the PRS, the HCi and the LCiCSI scored positive at a rate of 100%, 92% and 12.5%, respectively (Extended Data Fig. 3, Supplementary Text 2, Supplementary Table 4). As expected for a high-quality set., the confirmation rates of the HCi and the PRS are statistically indistinguishable (two-tailed Fisher’s Exact Test, P =0.3894).
Figure 1. CSILRR interaction map and functional validation.
a, Interaction heat map organized by phylogenetic subgroups of LRR-RKs (roman numeral, XIV and XV are merged)5. The colour scale bar on top shows interaction score values. b, Hypocotyl length ratios of seedlings grown in the presence (T) or absence (NT) of 500 nM brassinolide (BL). n= the numbers of biologically independent hypocotyls for all genotypes are in the Supplementary Methods. rlk *P =3.17 x 10-3, all others *P =3.2 x 10-15 and not significant (ns). c, flg22–induced SGI. n= numbers of biologically independent seedlings are indicated in the chart. mik1 *P =3.14 x 10-12, fls2 *P =2.8 x 10-15, bak1 *P =2.8 x 10-15, fir *P =2.88 x 10-10, pskr1 *P =2.88 x 10-10 and not significant (ns). b-c, Wild-type (WT; black), mutant lines targeting the HCi (top interactions; red) and LCi (bottom interactions; yellow) partners for BRI1 and FLS2 are indicated on the bottom and ordered by decreasing interaction score from left to right; Dots represent individual observations from six independent experiments; Box plots display the 1st and 3rd quartiles, split by the median; whiskers extend to include the max/min values. Statistical significance was determined using linear mixed effect modelling. The symbols on top of the boxes indicate the results of a post hoc unpaired two-sided t-test corrected with the Holm method for multiple testing. See Method section for information on genotypes. d, Western blot analyses of FLS2–BAK1 co-immunoprecipitations (Co-IP/IP) in seedlings treated with either water (-) or flg22 (+) for 10 min. anti-BAK1 or anti-FLS2 antibodies were used to analyse lysates from the genotypes indicated on the top. This experiment was repeated three times with similar results. e, flg22-induced oxidative bursts represented as total photon counts over 40 mins. Genetic backgrounds are indicated on the bottom. Dots represent individual observations from three independent experiments. Box plots and statistical significance as in b-c. n= numbers of biologically independent leaf discs and P values are indicated in the chart.
Paradigms for LRR-RK signalling posit that ECD interactions help bring together the intracellular domains (ICDs) for subsequent interaction and signal transduction2. We therefore tested whether ICDs from 372 HCi were more likely to interact than another set of 50 random LCiY2H via yeast two-hybrid assays (Y2H)17. The HCi and LCiY2H scored positive at a rate of 54,3% and 10%, respectively (Supplementary Table 5). Notably, of the ICD interactions assayed by Y2H, ten were present in our PRS, and all tested positive (Supplementary Table 3, Supplementary Text 3). We assign an extremely high level of confidence to interactions that occur at both the ECD and ICD level.
Next, we investigated the biological relevance of CSILRR interactions by studying the ligand-dependent activation of BRI1- and FLS2-mediated signaling1. We compiled a collection of 27 transfer DNA (T-DNA) insertion mutants18, targeting the HCi and LCi partners for both BRI1 and FLS2 (HCiBRI1/FLS2/LCiBRI1/FLS2) (Extended Data Figs. 4, 5, Supplementary Table 6-7). For these T-DNA lines, we used brassinosteroid (BR)-induced hypocotyl elongation or bacterial flagellin peptide (flg22)-induced seedling growth inhibition (SGI), peroxidase activity (POX), and luminol-based reactive oxygen species (ROS) assays to measure BRI1 and FLS2 activation, respectively19,20. While mutants corresponding to HCiBRI1/FLS2 partners showed altered signaling outputs (8/8 for BRI1; 3/5 for FLS2), mutants for the LCiBRI1/FLS2 partners were mostly indistinguishable from wild-type plants (6/7 for BRI1; 7/8 for FLS2) ((Fig. 1b, c, Extended Data Figs. 4b, 5b-e). Thus, we successfully used CSILRR to identify functionally relevant interactions for BRI1 and FLS2, and have as such expanded the repertoire of LRR-RKs that contribute to plant steroid signalling and flagellin-based immunity.
FLS2-INTERACTING RECEPTOR (FIR, AT2G27060), a previously uncharacterized LRR-RK, also interacted with the FLS2 co-receptor BAK1 in CSILRR, suggesting that FIR may influence the FLS2-BAK1 signalling complex in vivo. FLS2-BAK1 complex formation was reduced upon flg22 treatment in the fir mutant (Fig. 1d), and this correlated with a reduction in flg22-induced ROS burst and FLG22-INDUCED RECEPTOR KINASE1 (FRK1) gene expression (Fig. 1e, Extended Data Fig. 6a). We also measured flg22-induced root growth inhibition as well as resistance against the bacterium Pseudomonas syringae pv. tomato DC3000 (Pto DC3000), and found that both were significantly reduced in fir mutants (Extended Data Fig. 6b-d). Thus, FIR regulates FLS2 signalling and facilitates flg22-induced BAK1-FLS2 complex formation.
Next, we defined the key principles that govern interactions in CSILRR. LRR-RKs have large (>12 LRR repeats) or small ECDs (<12 LRR repeats), and the sizes are typically associated with roles in ligand perception or regulation, respectively1,21. We compared the experimental pattern of interactions between these groups to the expected distribution of interactions assuming random binding (Fig. 2a). The distributions between the subgroups significantly differed from each other (P <0.0001, chi-square test), indicating that binding events between ECDs in CSILRR are not random. We observed a 4- and 10-fold overabundance of homotypic interactions between large and small ECDs, respectively (Fig. 2a), and detected an increase as well in heterotypic interactions between small and large ECDs (Fig. 2a). We propose that plants have evolved small LRR-RKs to connect their otherwise unconnected larger counterparts.
Figure 2. CSILRR is defined by four distinct subnetworks and two critical nodes.
a, Expected and observed percentages of interactions organized by interaction types and ECD sizes. The expected % were calculated assuming random interaction between observed proteins. b, WalkTrap subnetworks are shown in orange. The diameter of the nodes (red circles) is proportional to their PageRank score. Numbers in each node are detailed in Extended Data Fig. 7. Edges (black lines) show interactions between nodes. BAK1 and APEX are marked in black and cyan, respectively. APs are surrounded by a white halo. c, Small ECDs have higher PageRank scores than large ECDs n= numbers of independent nodes (dots) and statistical significance determined by an unpaired two-sided t-test are indicated in the chart. The box plots contain the 1st and 3rd quartiles, split by the median; whiskers extend to include the max/min values. d, Representative rosettes of n=20 biologically independent 3-week-old Arabidopsis plants. Genetic backgrounds are indicated on the top.
Next, we used the Walktrap algorithm and identified four LRR-RKs subnetworks22 (Fig. 2b, Extended Data Fig. 7, Supplementary Table 8), of which at least one is biologically relevant (Supplementary Text 4). The PageRank algorithm was then used to compare the contributions of small and large ECDs to CSILRR connectivity23 (Supplementary Table 9). Nodes corresponding to small ECDs have significantly higher PageRank values and are thus more essential to the overall connectivity of the network (Fig. 2c). Notably, BAK1 (a small LRR-RK) was measured by PageRank as the most interconnected and important node in CSILRR.
Articulation points (APs) are nodes whose removal from a network results in the formation of at least two disconnected subnetworks24. Removal of AT5G63710 (hereafter named APEX), another small LRR-RK, resulted in the loss of the most nodes from the core structure of CSILRR, and was thus defined as the most important AP for network integrity (Supplementary Table 10). We predicted that genetic elimination of APEX and BAK1 would have obvious developmental consequences. To test this, we constructed apex bak1-5 double mutant plants25. While apex and bak1-5 single mutant plants were morphologically wild-type, apex bak1-5 double mutants were developmentally impaired (Fig. 2d). Thus, network properties defined in silico are relevant in living plants.
In our screen, APEX interacted with PEPR1/2. To test whether APEX associates with PEPR1/2 in the context of the full-length receptors, we performed co-immunoprecipitation assays. PEPR1/2 both associated with APEX in plant cells in the presence or absence of Pep2, a peptide ligand for PEPR1/226 (Fig. 3a, b, Supplementary Fig. 1). We next investigated whether the gene dosage of APEX would alter PEPR1/2 signalling (Extended Data Fig. 8a, b). apex knockout plants and two independent overexpression lines (35S::APEX) all displayed reduced Pep2-induced ROS burst (Fig. 3c). The further reduction in Pep2-triggered ROS burst in apex bak1-5 plants indicates that BAK1 and APEX are both required for wild-type PEPR1/2 signalling (Fig. 3c). Thus, APEX interacts with PEPR1/2 in a ligand-independent manner and a wild-type APEX dosage is required for appropriate Pep2-induced responses.
Figure 3. APEX interacts with PEPR1 and PEPR2 to regulate danger peptide signalling.
a-b, Nicotiana benthamiana leaves expressing FLAG-tagged variants of PEPR1/2 either alone or together with a YFP-tagged APEX were treated with water (-) or Pep2 (+). Western blot analyses of PEPR1/2-APEX (co-)immunoprecipitations (Co-IP/IP). anti-FLAG and anti-YFP antibodies were used to analyse lysates. These experiments were repeated three times with similar results. Full scans of the blots in Supplementary Fig.1. c, Pep2-induced oxidative bursts represented as total photon counts over 40 mins. Genetic backgrounds are indicated on the bottom. Dots represent individual observations from three independent experiments. n= numbers of biologically independent leaf discs are indicated in the chart. Box plots display the 1st and 3rd quartiles, split by the median (red line); whiskers extend to include the max/min values. Statistical significance was determined by linear mixed effect modelling. The letters on top of the boxes indicate the results of a post hoc Tukey test. Genotypes with the same letter are indistinguishable at >95% confidence.
Next, we predicted that changes in APEX dosage would impact the function of CSILRR as a coherent structural unit in vivo, thereby affecting the function of receptors without a direct physical interaction. To test this concept, we analysed whether the functions of BRI1 and FLS2, two receptors that reside several network-steps away from APEX, were affected in our set of APEX lines. The overexpression of APEX had either inconsistent or no effects on BR-induced hypocotyl elongation and flg22-induced ROS bursts (Fig. 4a, b, Extended Data Fig. 8c). In contrast, BRI1 and FLS2 functions were both altered in apex mutants, as indicated either by the low levels of hypocotyl elongation in response to BRs or by the enhanced flg22-induced ROS burst (Fig. 4a, b, Extended Data Fig. 8d-e). Notably, these aberrant ligand-induced signalling responses were both BAK1-dependent (Fig. 4a, b). Finally, apex mutants showed a striking increase in flg22-induced FLS2-BAK1 complex formation, mitogen-activated protein kinase (MAPK) activation and FRK1 expression (Fig. 4c-e, Supplementary Fig. 1). Thus, elimination of APEX has destabilizing effects in otherwise well-balanced LRR-RK signalling pathways.
Figure 4. CSILRR functions as a unified regulatory network.
a, Hypocotyl length ratios of seedlings grown in the presence (T) or absence (NT) of 500 nM brassinolide (BL). Genotypes are indicated on the top (Wild-type (WT; black)). Dots represent individual observations from three independent experiments. n = numbers of biologically independent hypocotyls are indicated in the chart n=T/n=NT. b, flg22-induced oxidative burst in leaf discs of the genetic backgrounds indicated on the top. Dots represent individual total photon counts over a 40-min time course; observations are from five independent experiments. n= numbers of biologically independent leaf discs are indicated in the chart. a-b, Box plots display the 1st and 3rd quartiles, split by the median (red line); whiskers extend to include the max/min values. Statistical significance was determined by linear mixed effect modelling. The letters on top of the boxes indicate the results of a post hoc Tukey test. Genotypes with the same letter are indistinguishable at >95% confidence. c, Western blot analyses of FLS2–BAK1 co-immunoprecipitations (Co-IP/IP) in seedlings treated with either water (-) or flg22 (+). anti-BAK1 or anti-FLS2 antibodies were used to analyse lysates from the genotypes indicated on the top. This experiment was repeated three times with similar results. d, flg22-induced activation of MAPKs in the genotypes indicated on top. The phosphorylated MPK3/6 proteins were detected with an anti-pERK antibody. This experiment was repeated four times with similar results. Colloidal brilliant blue (CBB) shows equal loading of the samples. c-d, Full scans of the blots are presented in Supplementary Fig. 1 e- Seedlings of the genotypes indicated on the top were treated with either water (NT) or flg22 (T) and changes in FRK1 transcripts quantified by qPCR. Dots represent individual observations from three independent experiments. n = numbers of biologically independent mRNA samples are n=9 (NT) and n=9 (T) for all genotypes. Box plots as in a-b. Statistical significance determined by an unpaired two-sided t-test, followed by multiple testing correction using the Holm method is indicated in the chart.
To support our contention that AP removal results in network disruption in vivo, we established that mutations in AT5G51560 (another predicted AP in CSILRR) altered BRI1 function (Extended Data Fig. 9). Our analysis has defined 16 additional LRR-RKs as APs of CSILRR (Supplementary Table 10). Although the removal of any one of these leads to the fragmentation of CSILRR into no more than 3 subnetworks, these APs therefore make tempting targets to study the LRR-RK family of receptors at the system level.
The minable resources introduced here have provided insights into the wiring diagram that underlays LRR-RK signalling. We propose that LRR-RKs operate in a unified regulatory network governed by the following key guiding tenets: (i) ligand-induced signalling is modulated locally by the presence and/or activities of other LRR-RKs, (ii) small LRR-RKs, in addition to their function as co-receptors, act as regulatory scaffolds and organize their larger counterparts into a signalling network, and (iii) coupling of LRR-RK signalling to the overall stability of the network ensures appropriate response modulation by network-feedback mechanisms, an overlooked determinant of response specificity.
Methods
Molecular Cloning of LRR-RK extracellular domains
For each ECD cloned in this work we determined the boundaries of signal peptides and transmembrane domains using a suite of bioinformatics tools27. A key step in defining the boundaries of each ECD was the identification of the N- and C-terminal cysteine-capping consensus motifs (CxxxxC and variations thereof) that borders most of the Arabidopsis ECDs. This was achieved by visual inspection of the primary amino acid sequences. These cysteine-caps are thought to play a role by capping the exposed edges of the hydrophobic core formed by the repetition of the LRRs and produce disulphide bonds that preserve the tertiary protein structure. In our hands, they were important for enhancing ECD solubility and preventing aggregation and proteolysis in vitro. For expression in Drosophila melanogaster Schneider 2 (S2) cells, each ECD was inserted into the pECIA-2 and the pECIA-14 expression vectors (a gift from Christopher K. Garcia)3. pECIA2/14 are derivatives of the pMT/BiP/V5 (Invitrogen, V4130-20), which uses a copper-inducible Drosophila metallothionein promoter and have the signal sequence of the Drosophila BiP protein. The ECDs were cloned by Sequence and Ligation Independent Cloning (SLIC) between the existing BiP signal sequence and the C-terminal epitope tags specific to each vector. Sanger sequencing confirmed the presence of each insert. Primers were designed to have a sequence partially homologous to the desired boundaries of the ECDs followed by extensions for RecA-mediated SLIC strategy attached (Supplementary Table 1). Amplification was done using Phusion Flash Mastermix (Thermo Scientific) according to the manufacturer’s instructions for 2-step Polymerase Chain Reaction. 176 ECDs out of 200 were cloned from plasmid templates available from the Arabidopsis Biological Resource Center (ABRC)28. Twenty-four ECDs were cloned from Arabidopsis seedlings and mature leaves using RT-PCR, followed by amplification as described above and by RecA-mediated SLIC cloning.
Secreted expression of LRR-RK extracellular domains
The ECDs cloned into the pECIA2 (for expression as bait) and pECIA14 (for expression as a prey) vectors were expressed using transient transfection of Drosophila Schneider 2 (S2) cells cultured at 27°C. Upon transfection using Effectene (Qiagen) the culturing temperature was changed to 21°C. Twenty-four hours after transfection protein expression was induced with 1 mM CuSO4 and supernatant was collected three days after induction. Protease inhibitors (Sigma) and 0.02% NaN3 were added to the medium (ESF 921, Expression Systems) containing the recombinant ECDs and then stored at 4°C prior to use. The cell supernatant was assessed for recombinant protein expression by western blotting using anti-V5 antibodies (Invitrogen) for the baits or by alkaline phosphatase activity quantification for the preys.
CSILRR primary screen
Pairwise Interaction assays were performed as detailed by Özkan et al.,3 for the extracellular interactome assay (ECIA) with the slight modifications indicated below. The Schneider’s medium containing recombinant ECDs was subjected to a 4-fold dilution in a phosphate-buffered saline (PBS) buffer containing 1 mM CaCl2, 1 mM MgCl2 (equilibration buffer) containing 0.1% Bovine Serum Albumin (BSA)-Sigma). Bait proteins fused to the Fc domain were captured directly on 96-well protein-A coated plates (Thermo Fisher Scientific) by overnight incubation at 4°C. Protein-A coated plates were washed in a PBS solution containing 0.1% Tween-20 prior to use. The bait-coated plates were blocked with the equilibration buffer containing 1% BSA for 3 hours at 4°C and subsequently washed. The prey proteins fused to the alkaline phosphatase (AP) were then added to the wells and incubated for 2 hours at 4°C and then washed away prior to adding the AP substrate (KPL 50-88-02). Upon addition of the substrate, plates were incubated for 2 hours at room temperature and AP activity monitored by measuring the absorbance at 650 nm using a Synergy H4 Multi-Mode plate reader (BioTek). Images of the 96-well plates were acquired for visual inspection. The complete set of raw absorbance values was combined into a binary dataset using an in-house designed script (Platero v0.1.4), and then subjected to post experimental statistical analysis to remove both false positive and false negative interactions.
CSILRR Data Analysis
The complete set of absorbance values for each pairwise interaction was combined into a data matrix. To make measurements comparable across plates and eliminate any bias in the data arising from the differential background binding capacities of the baits and preys we used a 2-way median polish29,30. The residuals were then used to calculate the median and median absolute deviation (MAD). The MAD is the median of the absolute values of the residuals (deviations) from the data’s median. The MAD was used for the calculation of modified Z-scores for each individual interaction measured. The modified Z-score used here is (i) nonparametric and makes minimal distributional assumptions, (ii) minimizes measurement bias due to positional effects and (iii) is resistant to statistical outliers. The modified Z-score usually excludes control measurements altogether under the assumption that most interactions in a screen such as CSILRR would be unproductive and thus serve as controls. However, during the primary screen each 96-well plate contained two mock prey negative control wells and one well with the positive control interaction pair BAK1-BIR431. To identify high-stringency bidirectional interactions we calculated the geometric mean modified Z-score of the interaction as measured in the bait-prey and prey-bait orientations. Any value for which the geometric mean product of the Z-scores was greater than 2.5 was considered significant for the purposes of network construction.
CSILRR retest screens
All of the HCi in CSILRR and a randomly selected subset of LCiCSI were independently retested. Each ECD was newly expressed and all retested interactions were assayed in both bait-prey orientations. For each interaction tested, three prey-only negative control wells were included, to control for non-specific binding. Thus, a total of six negative controls were tested for each bidirectional interaction. One well containing the positive control interaction pair BAK1-BIR4 was included on each plate. The 2-way median polish and modified Z-scoring system used in the initial screen depends upon large numbers of non-interactions to perform reliably. The low sample number, enriched with high or low performing protein pairs, led to an asymmetrical data distribution in the retest, making it inappropriate to implement our original hit calling method. Instead, we implemented a multi-stage hit calling process to ensure reliable data confirmation. The absorbance values were paired with the corresponding value from the CSILRR and subjected to an interquartile range (IQR) normalization step to ensure the two data-sets could be accurately compared (Extended Data Fig. 3). The geometric mean of the normalized absorbance values for each bidirectional interaction was then calculated. The threshold for inclusion in the positive interaction set was set to the lowest geometric mean absorbance value found in the 567 interactions present in the CSILRR (Abs=0.090989). Therefore, any interaction with a geometric mean Abs value >0.090989 was considered positive, while all others were considered negative.
CSILRR network construction and analysis. CSILRR network construction and analysis
The network was constructed using the igraph package (http://igraph.org/r/) in the R programming environment (https://www.r-project.org/). To identify clusters of interacting proteins in the network, we used the WalkTrap algorithm as implemented in igraph which is based on a concept that, if one performs random walks on a network, then the walks are more likely to stay within the densely connected parts of the network, thus corresponding to clusters with higher levels of interconnectedness22. The WalkTrap was implemented with edge weights corresponding to the interaction score and a length of random walk of 8. To measure the importance of each node within the network, we applied the PageRank algorithm as implemented in igraph that operates by counting the number and quality of links to a node hence establishing its importance and assigning a ‘weight’ value to it23. In simpler terms, PageRank measure node connectivity via the number of connections to other nodes. The PageRank algorithm is an example of link-analysis algorithms which are iterative and interactive data-analysis techniques and operate with the underlying assumption that nodes with higher scores are likely to be more connected to other nodes when compared to nodes with lower scores23. The PageRank implementation using the PRPACK library within the igraph package was used with edge weights corresponding to the interaction score, and a damping factor of 0.85, which is also the default. Finally, we identified the articulation points (or cut vertices) in the network. An articulation point is any node in a unidirectional network the removal of which disconnects the network.
Yeast two-hybrid assays with LRR-RK intracellular domains
The yeast two-hybrid experiment was conducted according to Mukhtar et al 17 with some modifications. Briefly, we used a collection of LRR-RK ICDs cloned in both bait and prey plasmids17. The ICDs of the LRR-RKs were fused to the GAL4 activation domain using a pDEST-AD-CHY2 vector with a tryptophan selection marker to form the prey constructs and to the GAL4 DNA binding domain using a pDEST-DB vector with a leucine selection marker to form the bait constructs. Target prey and bait constructs were transformed into S. cerevisiae strains Y8800 (MATa) and Y8930 (MATα), respectively. Transformations were confirmed by selecting the haploid yeast strains on their corresponding selective media (SD-T and SD-L). Haploid bait and prey strains were mated in liquid YEPD (yeast extract 10 g/L, peptone 20 g/L, dextrose 20 g/L, adenine 100 mg/L) media overnight at 30°C. The resulting diploid yeasts were selected in liquid SD-LT media for 48 hours at 30°C. Reconstitution of the GAL4 transcription factor through the interaction of the bait and prey led to the activation of a HIS3 reporter gene and subsequently biosynthesis of histidine. Since the pDEST-AD vector contains the CHY2 (a cycloheximide sensitive gene), any growth on the yeast media containing cycloheximide constitutes a false positive interaction. Equal amounts of diploid yeasts were transferred to solid SD-LTH (positive selection plates) and SD-LH+ cycloheximide (20 mg/L) media (de novo auto activation plates). Interactions were scored positive if there was growth on positive selection plates, but no growth on de novo auto activation plates. The retest on the random LCiY2H pairs was performed in similar conditions.
T-DNA insertions of top and bottom BRI1- and FLS2-interaction partners
Noting that our statistical cut-off for considering an interaction for network construction was set to a CSI score (Geometric mean modified Z-score) > 2.5 we compiled a list of ‘top-interactions’ (HCiBRI1/FLS2; CSI score >1.75) and ‘bottom-interactions’ (LCiBRI1/FLS2; CSI score=0) (Supplementary Table 6). We amassed a collection of T-DNA insertion lines from the Arabidopsis Biological Resource Centre (ABRC) for the HCiBRI1/FLS2 and LCiBRI1/FLS2 genes, focusing when possible on exon insertions closest to the 5’ end of each gene. Following tests for the presence of non-segregating (homozygous) T-DNA insertions in each target gene and PCR confirmation of altered target gene expression (Extended Data Figs. 4-5, Supplemental Table 7). For BRI1 we tested mutant lines targeting the following interaction partners: HCiBRI1 top genes: 1st Rank: STRUBBELIG-RECEPTOR FAMILY 9 (SRF9)32, 2nd Rank: ERECTA-LIKE 2 (ERL2)33, 3rd Rank: FIR/AT2G27060 (this study), 4th Rank: BAK1 (bak1-4 allele)25,34,35, 6th Rank: BARELY ANY MERISTEM 3 (BAM3)36, 7th Rank: SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE 4 (SERK4)35,37, 8th Rank: RECEPTOR-LIKE PROTEIN KINASE 1 (RPK1)38, 9th Rank: HAESA-LIKE 2 (HSL2)39–41. We were not able to test the following genes: 5th Rank: RECEPTOR-LIKE PROTEIN KINASE 2 (RPK2)42, and 10th Rank: BAK1 INTERACTING RECEPTOR 4 (BIR4)31. The mutant lines obtained from the SALK were annotated as homozygous for the T-DNA inserts but we genotyped both as wild-type plants. LCiBRI1 bottom genes: 191st Rank: RECEPTOR-LIKE KINASE (RLK)43, 193rd Rank: REDUCED IN LATERAL GROWTH1 (RUL1)44, 194th Rank: SENESCENCE-ASSOCIATED RECEPTOR-LIKE KINASE (SARK)45, 196th Rank: STERILITY-REGULATING KINASE MEMBER 1 (SKM1)46, 197th Rank: SUPPRESSOR OF BIR1-1 (SOBIR1)47, 198th Rank: STRUBBELIG-RECEPTOR FAMILY 4 (SRF4)32, 200th Rank: TRANSMEMBRANE KINASE LIKE 1 (TMKL1)48. The following genes were not tested: 192th Rank: RECEPTOR-LIKE KINASE 902 (RLK902)49 and 199th Rank: TRANSMEMBRANE KINASE 1 (TMK1)50. Although annotated as homozygous for the T-DNA insert in the SALK database, we genotyped both lines as wild-type plants.
For FLS2 we tested mutant lines targeting the following HCiFLS2 top genes: 1st Rank: MDIS1-INTERACTING RECEPTOR LIKE KINASE1 (MIK1)51, 2nd Rank: FLS2 as an internal control but as well as a self-interaction52, 3rd Rank: FIR/AT2G27060 (this study), 5th Rank: BAK1 (bak1-4 allele)25,34,35, 7th Rank: AT5G62710, 8th Rank: BARELY ANY MERISTEM 3 (BAM3)36, 13th Rank: RECEPTOR-LIKE PROTEIN KINASE 1 (RPK1)38, 14th Rank: STRUBBELIG-RECEPTOR FAMILY 9 (SRF9)32 and 15th Rank: AT2G27060. We did not test the following: 4th Rank: ERECTA and 6th Rank: ERECTA-LIKE 2 (ERL2) since the er mutant shows altered flg22-induced marker gene expression 33, 10th Rank: IMPAIRED-OOMYCETE SUSCEPTIBILITY 1 (IOS1) has been implicated in flg22-induced ROS burst, marker gene expression, and FLS2-BAK1 complex formation53. The following T-DNA lines were not tested: 5th Rank: RECEPTOR-LIKE PROTEIN KINASE 2 (RPK2)42, 11th Rank: RECEPTOR-LIKE KINASE 1 (RKL1)49 and 12th Rank: BAK1 INTERACTING RECEPTOR 4 (BIR4)31 because we genotyped as wild-type despite their annotation as homozygous for the presence of a T-DNA insert. LCiFLS2 bottom genes: 190th Rank: PHYTOSULFOKINE PEPTIDE RECEPTOR 1 (PSKR1)54–56, 191st Rank: PEPR257, 192nd Rank: AT3G46350, 194th Rank: AT3G14840, 195th Rank: AT2G01210, 196th Rank: PEPR157, 198th Rank: FEI231, 200th Rank: NSP-INTERACTING KINASE 3 (NIK3)58.
Brassinosteroid hypocotyl responses assays
POX flg22 responses assays
The peroxidase assay was carried out as in Mott et al.,20. Briefly, leaf discs were taken from 4-week-old A. thaliana plants. The discs were washed for 1 hour in 1 mL of 1X MS solution with agitation. After washing, discs were transferred to individual wells of a clear 96-well assay plate avoiding the use of the edge wells to minimize evaporation effects. Each well received 50 1µl of 1X MS buffer alone, or supplemented with 1µM of flg22 peptide. Plates were sealed with parafilm and incubated for 20 hours with agitation. The leaf discs were removed and each well received 50 µl of a 1 mg/mL solution of 5-aminosalicylic acid (A79809, Sigma-Aldrich) pH 6.0 with 0.01% hydrogen peroxide. The reaction proceeded for 1-3 min and was stopped by the addition of 20 µl 2N NaOH prior to reading the OD600 on a POLARstar OPTIMA microplate reader (BMG Labtech). The flg22 peptide was obtained from Genscript (NJ, USA).
Transient Expression in Nicotiana benthamiana
Agrobacterium tumefaciens GV3101 strains were grown in LB medium supplemented with appropriate antibiotics overnight. Cultures were spun down and resuspended in 10 mM MgCl2 to OD600 = 0.1. Agrobacterium strains carrying the pB35GWF binary plant expression vector for the expression of the full-length coding regions of PEPR1 (S1G73080BFF)28 and PEPR2 (S1G17750BFF)28 fused to a C-terminal Flag epitope tag were constructed and used for immunoprecipitation and western blot assays. pDONR-Zeo vector (Life Technologies) containing the cDNA of APEX (N5G63710ZEF) was used for gateway recombination in the binary plant expression vector pEarleyGate101 vector to generate the C-terminal YFP-HA tag fusion vector expressing APEX-YFP-HA under the control of the CaMV35S promoter. For each of the protein interaction pairs tested, the respective sets of agrobacterium strains were mixed 1:1 and syringe infiltrated into 3-week-old N. benthamiana leaves of plants grown in short-day conditions (12h light/12 h dark). Samples for protein extraction were harvested 3 days after infiltration before flash-freezing in liquid nitrogen.
Protein Extraction and Immunoprecipitation in N. benthamiana
Leaves were ground in liquid nitrogen and extraction buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10% glycerol, 10 mM DTT, 10 mM EDTA, 1 mM NaF, 1 mM Na2MoO4•2H2O, 1x [v/v] cOmplete Tablets, EDTA-free [Roche], and 1% [v/v] IGEPAL CA-630 [Sigma-Aldrich]) was added at 2 mL/g tissue powder. Samples were homogenized by alternate rounds of Polytron and incubated in extraction buffer for 1 hour at 4°C. Samples were the clarified by a 20-min centrifugation step at 4°C and 16.000 g. Supernatants (3 mL) were adjusted to 2 mg/mL protein and incubated for 3h at 4°C with 30 μL GFP Trap-A beads (Chromotek) with slow but constant rotation. Following incubation, beads were washed four times with washing buffer containing 50 mM Tris-HCl pH7.5, 150 mM NaCl, 1% PMSF, and 0.1% IPEGAL. One hundred microliters of 5x SDS Laemmli buffer was added to the beads, and the beads were heated at 95°C for 10 min and subjected for further SDS PAGE and immunoblotting analysis.
Plant Cultivation, transgenic plants and mutants
The wild-type (WT) used in all experiments was A. thaliana accession Columbia (Col-0). If not specified otherwise the apex-1 allele was used in this work (Extended Data Fig. 8a). Plants were grown on soil or vertically on Petri dishes containing 0.5X Murashige and Skoog medium in long-day light conditions (16 h light/8 h dark). For Pto DC3000 pathogen assay and callose deposition upon flg22 treatment, plants were grown in short-day conditions (12h light/12 h dark). The mutant plant genotypes used in this work are listed in Supplemental Table 7. For overexpression studies the 35S::APEX-YFP-HA construct was transformed separately into WT plants and more than 20 independent T1 lines were isolated and between three and eight representative mono-insertion lines were selected in the T2 generation. DNA genotyping, epifluorescence microscopy and protein extraction were performed on segregating T2 to obtain homozygous T3 generation lines with maximal expression levels (Extended Data Fig. 8b). The double mutant apex-1 bak1-5 was generated by crosses and genotyped for homozygosity using allele-specific primers for apex-1 and dCAPS marker for bak1-5 as described in 25. Genotyping was repeated for two consecutive generations and confirmed by Sanger sequencing. Primers are listed in Supplementary Table 7.
Protein Extraction and immunoprecipitation in Arabidopsis
Fifteen to twenty seedlings were grown in each well of a 6-well plate for 2 weeks. Subsequently, seedlings were transferred to water and incubated overnight. The next day, flg22 was added at a final concentration of 100 nM and incubated for 10 min. Seedlings were than frozen in liquid nitrogen and subjected to protein isolation. To analyse FLS2-BAK1 receptor complex formation, proteins were isolated in 50 mM Tris-HCl pH 7.5, 150 mM NaCl, 10% glycerol, 5 mM dithiothreitol, 1% protease inhibitor cocktail (Sigma Aldrich), 2 mM Na2MoO4, 2.5 mM NaF, 1.5 mM activated Na3VO4, 1 mM phenylmethanesulfonyl fluoride, and 1 % IGEPAL. For immunoprecipitations, anti-rabbit Trueblot agarose beads (eBioscience) coupled with anti-FLS2 antibodies and incubated with the crude extract for 2-3 h at 4°C. Subsequently, beads were washed 3 times with wash buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM phenylmethanesulfonyl fluoride, and 0.5 % IGEPAL) before adding Laemmli sample buffer and incubating for 10 min at 95°C. Analysis was carried out by SDS-PAGE and western blots using anti-FLS2 and anti-BAK1 antibodies25.
Protein analysis
In all our protein manipulations, equal loading was ensured by Bradford protein quantification prior to loading and by CBB or Red Ponceau staining of the membrane post-protein transfer. anti-GFP-HRP (MACS) and anti-FLAG- HRP (Sigma-Aldrich) were used according to manufacturer’s instructions. Polyclonal anti-FLS2 and anti-BAK1 antibodies were used as in25. Signal detection was achieved through chemiluminescence (SuperSignal™ West Pico Chemiluminescent Substrate, ThermoFisher) and detected using autoradiography films (CL-XPosure Film, ThermoFisher).
RNA isolation, cDNA synthesis and real-time PCR analysis
Total RNA was isolated from 1-week-old seedlings grown on 1/2 MS plates using either the GeneMATRIX Universal RNA Purification Kit (EURx) or TRI Reagent (Sigma-Aldrich), followed by DNaseI treatment (Thermo Scientific). Reverse transcription reactions were performed using up to 2 μg of total RNA and a reverse transcription kit (Applied Biosystems or Life Technologies). The cDNAs were used as a template for quantitative real-time PCR. Real-time PCR analyses were performed using a Roche LightCycler96 instrument (Roche Applied Science, Mannheim, Germany) and data analysed by the LightCycler 96 version 1.1 software or BioRad C1000 thermal cycler (BioRad). FastStart Essential DNA Green Master (Roche) or Maxima SYBR Green/ROX qPCR Master Mix (Thermo Scientific) were used according to manufacturer’s instructions. Material from wild type plant served as the calibrator, and ACTIN or UBQ10 was used as a reference. Relative gene expression levels were calculated using the 2-ΔΔCT method. The amplification protocol comprised: 95°C for 1 min, (95°C for 10 sec, 55-62°C for 10 sec, 72°C for 20 sec) x 44 cycles. The relative mRNA levels were determined by normalizing the PCR threshold cycle number with Actin or UBQ10. All experiments were repeated three times independently, and the mean was calculated. The specificity of the amplification products was verified by melting curve analysis.
MAMP and DAMP responses assays
flg22 (QRLSTGSRINSAKDDAAGLQIA) and pep2 (DNKAKSKKRDKEKPSSGRPGQTNSVPNAAIQVYKED) peptides were synthesized at >95% purity by the in-house protein chemistry facility and dissolved to a 10mM stock in pure water. For ROS burst assays, Leaf disks (Ø 6mm) were cut out from 4- to 5-week-old plants. Single disks were placed adaxial side up into 96-well microtiter plates in which every well contained 200 µL of sterile MonoQ water. Floated disks were then vacuum infiltrated for 10 min. The plates were incubated on a rocking table at 45 rpm in continuous light, at 21°C for 5 hours. The mix for elicitation was freshly prepared in falcon tubes wrapped with aluminum foil on ice. For 10 wells, 99,1 µL of sterile MonoQ water was mixed together with 4 µL of 500x HRP, 4 µL of 500x L-012 and appropriate peptide at a final concentration of 1 µM. For each well the water was carefully removed and replaced immediately with 100 µL of elicitation solution using a multichannel pipette. Relative luminescence measurements were started immediately after adding the elicitation mix using a BiTec Synergy 4 microplate reader. Horseradish Peroxidase (HRP) was purchased from Sigma-Aldrich and prepared at a 10 mg/mL (500X) concentration in sterile MonoQ water. L-012 was purchased from Wako Chemicals GmbH. Preparation of a 500x L-012 stock solution containing 17mg/mL L-012 in sterile MonoQ water and was subsequently protected from light. Solutions were stored at -20°C. For the analysis of ROS burst data, the models were constructed using the total RLU measured for the first 39 time points to ensure comparability across experiments. Root inhibition ratios were calculated on 7-day-old seedlings grown on plates left untreated or treated with 1μM flg22.
Seedling Growth Inhibition Assay
Seedlings of the noted A. thaliana lines were grown for 5 days on MS-Agar plates with 1% sucrose prior to transfer of up to 10 seedlings to each well of a 6-well plate containing 1 ml of 0.5x MS medium with 1% sucrose. The seedlings were treated with water (NT) or 100 nM flg22 (T) peptide and grown further for 7 days. The seedlings were removed, briefly dried, and weighed (fresh weight). The percentage of seedling growth inhibition was calculated by dividing the weight of individual treated seedlings by the mean weight of the NT seedlings of the same genotype. The percent seedling growth inhibition was calculated by dividing the weight of individual treated seedlings by the mean weight of 10 NT seedlings of the same genotype. A maximum of 10 seedlings of each genotype were treated and the experiment was performed six times.
Pathogens assays
Assays with Pseudomonas syringae pv. tomato DC3000 (Pto DC3000) have been previously described19. Bacterial growth in plant leaves was assessed by inoculating 4-week-old plants with a bacterial inoculum of 105 cfu/ml. Growth inhibition of Pto DC3000 by 1µM flg22 was conducted as described19. Leaves were either infiltrated with water or with an elicitor solution containing 1 μM flg22. For each sample, four leaf discs were pooled and three samples were taken per data point (12 leaf discs in total). Leaf discs were bored from the infiltrated area and ground to homogeneity in 10 mM MgCl2. The bacterial titer was determined by plating and serial dilution.
Program used for modelling and statistical analysis
Statistical analysis was performed using linear mixed effect modelling in the R programming environment (https://www.r-project.org/). Prior to modeling, data from independent experiments were combined and outliers were removed using the ROUT method, as implemented in GraphPad PRISM 7.0 (Q = 0.1%) (GraphPad Software, La Jolla California USA, www.graphpad.com). Each data set was checked for normality to ensure accurate modeling. qPCR data was analyzed as fold induction, while all other data were log10 transformed prior to modeling to improve fit. Linear mixed effect models were constructed using the lme4 package: https://cran.r-project.org/package=lme4, using the genotype as a fixed effect and the individual experiment as a random effect. The resulting models were inspected for fit and further outlier checks were accomplished by examining both the Cook’s distance and dfbeta distributions using the LMER Convenience Functions and influence. ME packages (http://cran.r project.org/package=LMER Convenience Functions; http://cran.rproject.org/package=influence.ME) and statistical significance was determined using the lmerTest package https://cran.r-project.org/package=lmerTest using the Satterthwaite approximation and the resulting p-values were corrected for multiple testing using the Holm method. In cases where pairwise comparisons were required, the adjusted p-values were calculated using Tukey’s HSD as implemented in the multcomp package (https://cran.r-project.org/package=multcomp).
To calculate the expected binding frequencies of a random network, we classified each node based on its ECD. Assuming equal frequency of a given node binding to any other node, the frequency for each class of binding event was calculated and divided into self-interactions between small ECDs (small-small homotypic), self-interactions between large ECDs (large-large homotypic), interactions between two different small ECDs (small heterotypic), interactions between two different large ECDs (large heterotypic), and interactions between one small and one large ECD (small-large heterotypic).
To estimate the reliability of the estimates provided by the retest screen (Extended Data Fig.3d), the observed rate of interactions found in the CSI and Retest sets were used for a Monte Carlo simulation. Sets of observations were selected at random from these populations, with the number of observations equal to the number present in the retest sets. This process was completed 100,000 times. These values were used to calculate the mean and standard deviations of the samplings.
Details about the linear mixed effect modeling can be found in the Supplementary Method file.
Data and software accessibility
The authors declare that the data supporting the findings of this study are available within the paper and its supplementary information files. Source data for Fig.1, 2, 3, 4, Extended Data Fig.2, 3, 4, 5, 6, 8 and 9 are provided with the paper. All the raw absorbance reads related to the ECD interaction screen are available in the Supplementary Table 11. The high-confidence LRR-RKs interaction dataset is publically available online at the Botany Array Resource under accession (MI:2189(BAR)/ PubMed placeholder 123456789): http://bar.utoronto.ca/interactions. The custom PLATERO script used for concatenating the interaction absorbance values is available upon request from the corresponding author (Y.B) or from https://github.com/AdamMott/platero-code. There are no restrictions on data availability
Extended Data
Extended Data Figure 1. Expression profiles of LRR-RK ECDs produced as recombinant baits with the Drosophila S2 cells protein expression system.
a-o, Western blot analyses of raw supernatants from S2 cells transfected with ECD expression vectors (WB: anti-V5). Blots were cropped and arranged to match the phylogenetic tree of the LRR-RK gene family. The family subclasses and AGIs are indicated on the top of the blots. For lanes showing no obvious anti-V5 signals, a mild concentration of the S2 cell media and/or purification on protein-A coated 96-well plates allowed for confirmation of expression and secretion of the ECDs. This experiment was conducted one time with the full set of 200 ECDs. The expression of 130 independently expressed ECDs was tested one additional time with similar results.
Extended Data Figure 2. Calibration of the CSILRR screen conditions on ligand-dependent (FLS2-BAK1) and ligand-independent (BAK1-BIR4) interaction pairs.
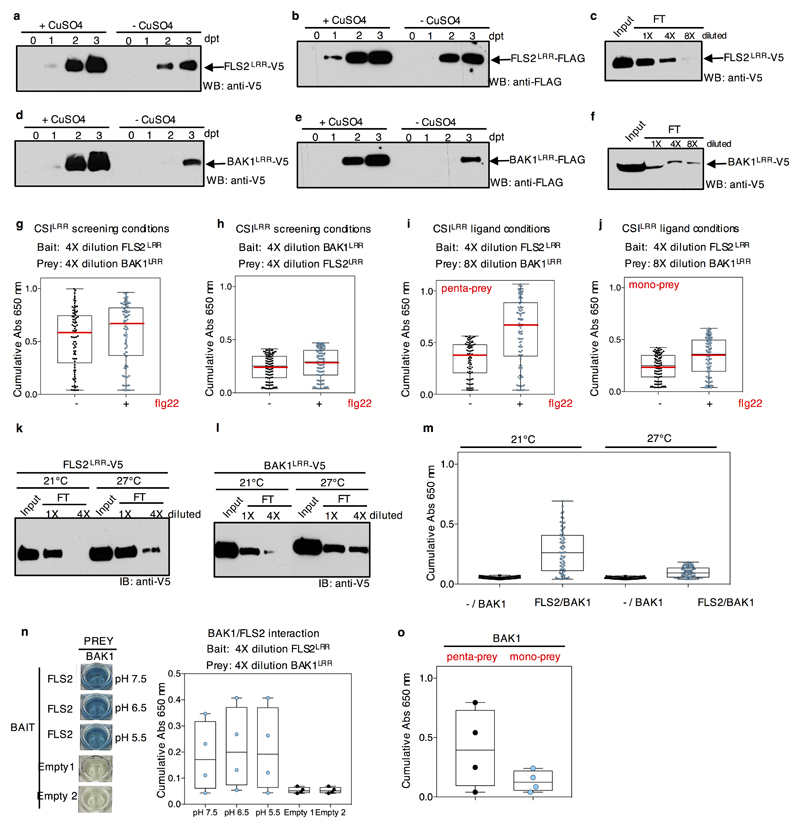
a-b, Western blot analyses of raw supernatants from S2 cells transfected with prey and bait expression vectors for the ECD of FLS2 (Bait, WB: anti-V5; Prey, WB: anti-FLAG). Drosophila S2 cells left untreated (-) or treated with CuSO4 (+). Days post induction are indicated on top (dpt). The experiment was repeated independently two times with similar results. c, Binding of the FLS2 ECD to the protein-A coated 96-well plates. A 4-fold dilution (4X) of the insect cell media containing the ECD of FLS2 saturates the binding sites of protein-A coated wells as indicated by immunoblots of the flow-through (FT). The experiment was repeated independently two times with similar results. d-f, same as a-c- but for BAK1. The experiment was repeated independently two times with similar results. g, Plate interaction assays between the ECDs of BAK1 (prey) and FLS2 (bait) represented as cumulative absorbance (Abs 650) over 18 hours. Dots represent individual observations at each hour from five technical replicates. Box plots display the 1st and 3rd quartiles, split by the median (red line); whiskers extend to include the max/min values. The presence of flg22 (+) in 4-fold diluted CSILRR screening conditions promotes weakly the interaction between the two ECDs. h, Technical replicates and box plots as in g, but with BAK1 (bait) and FLS2 (prey). i, Technical replicates and box plots as in g but with BAK1 (prey 8-fold diluted) and FLS2 (bait 4-fold diluted). In these conditions, the binding between the ECDs of BAK1 and FLS2 is largely enhanced by the presence of flg22 (+), indicating that the proteins produced in our expression system can interact in a ligand-dependent manner and are thus functional. j, Technical replicates and box plots as in g, but using a prey variant of BAK1 that can no longer pentamerize due to the deletion of the COMP domain (BAK1 mono-prey). Binding between the two ECDs is still observed, but at a reduced level, thus indicating the importance of the pentamerization motif for detecting transient and low affinity interactions in the absence of ligand. k-l, Binding of FLS2 and BAK1 ECDs to protein-A coated 96-well plates (as indicated by immunoblots of the flow-through (FT)) when proteins are produced from S2 cells growing either at 21°C or 27°C. Immunoblots show a slight increase in protein production at 27°C with similar binding capacities to the protein-A coated plate. The protein expression levels at the two temperatures were assessed more than three times with similar results. The plate saturation experiment for proteins produced at 27°C was conducted once. m, Plate interaction assays between BAK1 (prey) and FLS2 (bait) (in 4-fold diluted conditions) represented as cumulative absorbance (Abs 650) over a 150-mins time course. Dots represent individual observations made every 10 mins from four technical replicates. Box plots display the 1st and 3rd quartiles, split by the median; whiskers extend to include the max/min values. While slightly more abundant, proteins produced at 27°C do not interact as well as when produced at 21°C. Protein expression for the CSILRR screen was performed at 21°C. n, The FLS2-BAK1 interaction is insensitive to changes in pH conditions. Left panel, the interaction between FLS2 (bait) and BAK1 (prey) was observed in the pH range from 5.5 to 7.5. This experiment was conducted once. Right, plate interaction assays between BAK1 (prey) and FLS2 (bait) (in 4-fold diluted conditions) represented as cumulative absorbance (Abs 650) over a 3-hour time course. Dots represent individual observations at each hour from one technical replicate. The CSILRR screen was performed at the pH of the conditioned S2 cells supernatant (~pH 7.5). o, Plate interaction assays between BAK1 (as mono-prey (blue dots) or penta-prey (black dots)) and BIR4 represented as cumulative absorbance (Abs 650) over a 3-hour time course. Dots represent individual observations at each hour from one technical replicate. This experiment was conducted once. The data indicates that the pentamerization of the prey is a key requirement for enhancing the interaction detection sensitivity, without disrupting the functionality of the ECDs. BAK1 and BIR4 are ligand-independent interaction partners and the screening conditions used are also appropriate to detect this interaction.
Extended Data Figure 3. Comparison of the primary and retest screens parameters.
a, Geometric mean of the normalized absorbance values for the HCi (red dots) and LCi (yellow dots) obtained from the primary screen (CSI), the validation screen (Retest) and the negative prey controls (NC) associated with the two screens. n = numbers of bidirectional interactions: HCi CSI (n=567), HCi Retest (n=567), LCi CSI (n=248), LCi Retest (n=248), and NC (n=618). The box plots contain the 1st and 3rd quartiles, split by the median (yellow or red lines indicated by the arrow on the left of the boxes); whiskers extend to include the max/min values. Statistical significance was determined using unbalanced one-way ANOVA by Tukey’s HSD for all pairwise comparisons. Data sets with the same letter are indistinguishable at >95% confidence. b, Plots of a linear regression for the entire set of normalized absorbance values obtained from the retest screens (Absorbance Retest; y-axis) and the corresponding values from the from the primary screen (Absorbance CSI; x-axis). The thick, straight red line is the linear regression that best describes the entire set of data points (Spearman r: 0.7696; indicated on top). The fine red dashed lines represent the 95% confidence intervals of the regression. n = 815 bidirectional interactions. c, Comparison of the geometric mean of normalized absorbance values for selected interactions. Values from the primary screen (Absorbance CSI; y-axis) and the validation screen (Absorbance Retest; x-axis) are shown for the LCi set (yellow dots) and for 20 interactions selected at random from the HCi set (red dots). The number of interactions shown for each set was selected to approach the numbers present in the entire interaction search space. The red lines show the absorbance values corresponding to the FLS2-BAK1 interaction in both screens. d, Retest assay performance parameters interpreted within the performance window measured by PRS and LCi calibration. To estimate the reliability of the estimates provided by the retest, the observed rate of interactions found in the HCi and LCi sets were used for a Monte Carlo simulation. n =100,000 independent sets of observations selected at random from these populations, with the number of observations equal to the number present in the retest sets. These values were used to calculate the mean and standard deviations of the samplings, which are presented as error bars.
Extended Data Figure 4. Characterization of BRI1 interaction partners.
a, Quantitative real-time PCR analyses showing altered gene expression in T-DNA lines targeting the interaction partners of BRI1 (Fig. 1b). Genotypes are indicated at the bottom of the chart. Relative expression levels were calculated and ACTIN was used as reference gene to control for cDNA amount in each reaction. The box plots contain the 1st and 3rd quartiles, split by the median; whiskers extend to include the max/min values. n = 4 biologically independent mRNA samples for all genotypes, except for bak1-4, skm1, and sobir1 where n=3. Statistical significance was estimated by an unpaired two-sided t-test and is indicated on top of the boxes: ns (not significant), erl2 *P =0.0012, fir *P =5.3508 x 10-6, bak1-4 *P =3.08212 x 10-7, bam3 *P =1.9378 x 10-5, serk4 *P =0.0108, hsl2 *P =2.06945 x 10-5, sark *P =0.0259, rlk *P =2.12971 x 10-10, rul1 *P =7.49918 x 10-5, srf4 *P =3.08212 x 10-7, skm1 *P =5.5911 x 10-6, sobir1 *P =0.0001. b, T-DNA insertions targeting the HCi (Top interactions) and LCi (bottom interaction) partners of BRI1. Morphology of representative seedlings grown for 7-days in the absence (NT) or presence of 500 nM brassinolide (BL), the most potent brassinosteroid. Genotypes are indicated on the top of the picture. The experiment was conducted six times with similar results.
Extended Data Figure 5. Characterization of FLS2 interaction partners.
a, Quantitative real-time PCR analyses showing altered gene expression in T-DNA lines targeting the interaction partners of FLS2 (Fig. 1c). Genotypes are indicated at the bottom of the chart. Relative expression levels were calculated and ACTIN was used as reference gene to control for cDNA amount in each reaction. The box plots contain the 1st and 3rd quartiles, split by the median; whiskers extend to include the max/min values. n = 9 biologically independent mRNA samples for all tested genotypes. Statistical significance was estimated by an unpaired two-sided t-test and is indicated on top of the boxes: ns (not significant), mik1 *P =8.17192 x 10-6, pskr1 *P =0.007, pepr2 *P =0.007, at3g14840 *P =0.005, at2g01210 *P =0.0032, pepr1 *P =1.16519 x 10-5, fei1 *P =0.005, nik3 *P =0.0015. b, Oxidative burst represented as total photon counts, triggered by 1μM flg22 in wildtype (WT; black) and in mutant lines targeting the HCi (top; red) and LCi (bottom, yellow) partners for FLS2. Genotypes are indicated on the bottom of each graph. Dots represent individual observations from four independent experiments; Box plots display the 1st and 3rd quartiles, split by the median; whiskers extend to include the max/min values. n = numbers of biologically independent leaf discs: WT (n=36), mik1 (n=36), fls2 (n=28), pskr1 (n=27), pepr2 (n=38), at3g46350 (n=39). Statistical significance was determined using linear mixed effect modelling. The symbols on top of the boxes indicate the results of a post hoc unpaired two-sided t-test corrected with the Holm method for multiple testing: ns (not significant), mik1 *P =4.32 x 10-2, fls2 *P =1 x 10-15. c, as in (b,) except: WT (n=32), fls2 (n=27), bak1 (n=39), at3g14840 (n=33), at2g01210 (n=38), pepr1 (n=40). ns (not significant), bak1 *P =1 x 10-15, fls2 *P =1 x 10-15. d, as in b, and c, except: WT (n=43), fls2 (n=29), bam3 (n=33), fir (n=39), srf9 (n=32), fei2 (n=45), nik3 (n=32). ns (not significant), fir *P =1.38 x 10-3, fls2 *P =1.2 x 10-15, nik3 *P =1.38 x 10-3. b-d, The ROS burst assays were performed on independent plates (set number) and every plate contained WT and fls2 controls, as well as randomly assigned mutant lines. e, flg22–induced peroxidase assay (POX) in wildtype (WT; black bar) and in mutant lines targeting the HCi (top interactions; red) and LCi (bottom interactions, yellow) partners for FLS2. Genotypes are indicated on the bottom of the graph. Leaf disks from 4-week-old plants were treated with water (NT) or 1 µM flg22 (T). The level of flg22-induced POX was normalized to the corresponding NT control. The level of POX present in WT was set to 100 for easier interpretation. Box plots contain the 1st and 3rd quartiles, split by the median; whiskers extend to include the max/min values. n = numbers of biologically independent leaf discs from two independent experiments: WT (n=44), mik1 (n=10), fls2 (n=17), bak1 (n=31), bam3 (n=42), srf9 (n=18), fir (n=55), pskr1 (n=24), pepr2 (n=12), at3g46350 (n=36), at3g14840 (n=12), at2g01210 (n=18), pepr1 (n=12), fei2 (n=11), nik3 (n=15). Statistical significance was estimated using a paired two-sided t-test for each genotype, corrected for multiple tests using the Holm-Bonferroni correction and is indicated in top of the boxes: ns (not significant), mik1 *P =5.71 x 10-4, fls2 *P =0.046, bak1 *P =0.0039, fir *P =0.0048, pskr1 *P =9.49 x 10-5.
Extended Data Figure 6. FIR regulates flg22-induced responses.
a, Seedlings of the genotypes indicated on the bottom were treated with either water (NT) or flg22 (T) and changes in FRK1 transcripts quantified by quantitative real-time PCR analyses. Dots represent individual observations from three independent experiments. n = numbers of biologically independent mRNA samples: WT (n=9 (NT), n=9 (T)), fir (n=9, n=9) and fls2 (n=6, n=6). Box plots display the 1st and 3rd quartiles, split by the median; whiskers extend to include the max/min values. Statistical significance was determined using linear mixed effect modelling followed by comparison of each genotype to the WT control using unpaired two-sided t-test followed by multiple testing correction using the Holm method and is indicated on top of the boxes: ns (not significant), fir *P =1.42 x 10-7, fls2 *P =4 x 10-16. b, Growth of Pto DC3000 on the genetic backgrounds indicated at the bottom of the chart. Four-week-old plants were infiltrated with 105 cfu/ml in the absence (black bars) or presence (grey bars) of 1 μM flg22. The number of bacteria per area of leaf (cfu/ml) was plotted on a log10 scale for day 0 (open bars) and day 3 (closed bars). Dots represent individual observations from two independent experiments. n = numbers of samples each including 4 biologically independent leaf discs : For Day 0- WT(n=6), fir (n=6), fls2 (n=6); Day 3- WT (n=6), fir (n=6), fls2 (n=6); for Day 3 + flg22- WT (n= 6), fir (n= 6), fls2 (n=6). Box plots display the 1st and 3rd quartiles, split by the median; whiskers extend to include the max/min values. Statistical significance for bacterial growth was estimated by two-way ANOVA and is indicated on top of the boxes: ns (not significant), relevant P values are indicated in the chart. A third experiment performed at an inoculum of 106 cfu/ml corroborated these results. c, Morphology of 7-day-old seedlings grown in the absence (-) or presence (+) of 1 μM flg22. Genotypes are indicated on top of the panel. The experiment was conducted two times with similar results. d, Primary root length (cm) from seedlings grown in the presence (T) or absence (NT) of 1μM flg22. Fold changes are T/NT ratios. Dots represent individual observations from two independent experiments. n = the following numbers of biologically independent roots: WT (n=32 (NT), n=36 (T)), fir (n=34, n=32), fls2 (n=27, n=26). Box plots display the 1st and 3rd quartiles, split by the median; whiskers extend to include the max/min values. Statistical significance for two biological replicates was determined using linear mixed effect modelling followed by comparison of each genotype to the WT control using unpaired two-sided t-test followed by multiple testing correction using the Holm method and is indicated on top of the bars: fir *P =2.02 x 10-6, fls2 *P =2.02 x 10-6.
Extended Data Figure 7. CSILRR network representation and table of nodes with their corresponding identification numbers or acronyms.
The network construction and other features are the same as shown in Fig. 2b. The nodes surrounded by white halos are articulation points. The numbers in each node corresponding to the ECD of specific LRR-RKs IDs are detailed in the bottom table.
Extended Data Figure 8. Characterization of independent apex mutant and 35S::APEX transgenic lines.
a, Top: Rosette morphology of 4-week-old wildtype (WT), apex-1 and apex-2 knockdown, and apex-3 knockout, lines grown under long-day photoperiod, at 22°C. Genetic backgrounds are indicated on the top. No obvious changes in rosette morphology are observed. The experiment was conducted three times with similar results. Bottom: Quantitative real-time PCR analyses showing fold reduction of APEX transcripts in the independent mutant lines. Relative expression levels were calculated and ACTIN was used as reference gene to control for cDNA amount in each reaction. Dots represent individual observations from three independent experiments. n =9 biologically independent mRNA samples for each genotype. Box plots display the 1st and 3rd quartiles, split by the median; whiskers extend to include the max/min values. Statistical significance was determined using linear mixed effect modelling followed by comparison of each genotype to the WT control using unpaired two-sided t-test followed by multiple testing correction using the Holm method and is indicated on top of the boxes: apex-1 *P =6 x 10-16, apex-2 *P =5.33 x 10-15, apex-3 *P =6 x 10-16. b, Top: Rosette morphology of 3-week-old WT, 35S::APEX line1 and line2 lines grown under long-day photoperiod, at 22°C. Genetic backgrounds are indicated on the top. Rosettes of 35S::APEX lines are slightly larger than WT under long-day photoperiod, at 22°C. The experiment was conducted three times with similar results. Middle: Quantitative real-time PCR analyses showing fold induction of the APEX transgene in the overexpression lines used in this study. Relative expression levels were calculated and ACTIN was used as reference gene to control for cDNA amount in each reaction. Dots represent individual observations from two independent experiments. n=6 biologically independent mRNA samples for each genotype. Box plots display the 1st and 3rd quartiles, split by the median; whiskers extend to include the max/min values. Statistical significance was determined using linear mixed effect modelling followed by comparison of each genotype to the WT control using an unpaired two-sided t-test followed by multiple testing correction using the Holm method and is indicated on top of the boxes: 35S::APEX line 1 *P =3.38 x 10-14, 35S::APEX line2 *P =7.77 x 10-14. Bottom: Detection of APEX-YFP in stable transgenic T3 lines by Western Blot using anti-GFP antibody. c, Modulation of BRI1 signalling by APEX gene dosage. Morphology of representative seedlings corresponding to Fig. 4a. Genotypes are indicated on the top of the panel. The experiment was conducted over three times with similar results. d, Hypocotyl length ratios of seedlings grown in the presence (T) or absence (NT) of 500 nM brassinolide (BL). Genotypes are indicated on the top. Dots represent individual observations from three independent experiments. n = numbers of biologically independent hypocotyls: WT (n=43 (NT), n=33 (T)), apex-1 (n=31, n=35), apex-2 (n=32, n=33), apex-3 (n=39, n=38), bri1 (n=28, n=32). Box plots display the 1st and 3rd quartiles, split by the median (red line); whiskers extend to include the max/min values. Statistical significance was determined using linear mixed effect modelling followed by comparison of each genotype to the WT control using unpaired two-sided t-test followed by multiple testing correction using the Holm method and is indicated on topof the boxes: apex-1 *P =2.53 x 10-14, apex-2 *P =1.10 x 10-5, apex-3 *P =1.55 x 10-12, bri1 *P =8 x 10-16. e, flg22-induced oxidative bursts represented as total photon counts over 40 mins. Genetic backgrounds are indicated on the top. Dots represent individual observations from three independent experiments. n= numbers of biologically independent leaf discs: WT (n=31), apex-1 (n=19), apex-2 (n=23), apex-3 (n=25), fls2 (n=15). Box plots display the 1st and 3rd quartiles, split by the median (red line); whiskers extend to include the max/min values. Statistical significance was determined using linear mixed effect modelling followed by comparison of each genotype to the WT control using an unpaired two-sided t-test followed by multiple testing correction using the Holm method and is indicated on top of the boxes apex-1 *P =2.99 x 10-3, apex-2 *P =2.84 x 10-2, apex-3 *P =2.84 x 10-2, fls2 *P =8 x 10-16.
Extended Data Figure 9. Modulation of BR signalling by AT5G51560.
a, Morphology of representative seedlings grown for 7 days in the absence (NT) or presence of 500 nM brassinolide (BL), the most potent brassinosteroid. Genotypes are indicated on the top of the picture. The experiment was conducted two times with similar results. b, Hypocotyl length fold changes corresponding to panel (a,). Genotypes are indicated on the top of the chart. Dots represent individual observations from two independent experiments. n= numbers of biologically independent hypocotyl: WT (n=39 (NT), n=29 (T)), at5g51560 line 1 (n=36, n=26), at5g51560 line 2 (n=39, n=34), bri1 (n=25, n=27). Box plots display the 1st and 3rd quartiles, split by the median; whiskers extend to include the max/min values. Statistical significance was determined using linear mixed effect modelling followed by comparison of each genotype to the WT control using an unpaired two-sided t-test followed by multiple testing correction using the Holm method and is indicated on top of the boxes: at5g51560 line 1 *P =3.75 x 10-6, at5g51560 line 2 *P =2.26 x 10-12, bri1 *P =6 x 10-16.
Supplementary Material
Acknowledgments
We thank: A. Pasha for uploading the CSILRR interaction dataset to the Bio-Analytic Resource for Plant Biology, Y. Dagdas for comments on the manuscript, CK. Garcia for the pECIA2/14 vectors, E. Özkan for his protocols, Y. Saijo, K. Tori and M. Butenko for the pepr1/2, erl2 and hsl2 mutant lines, respectively, A. Bindeus for help with programming software. This work was supported by grants from the Austrian Academy of Science through the Gregor Mendel Institute (Y.B and W.B); the Natural Sciences and Engineering Research Council of Canada Discovery Grants to D.S.G. and D.D; a Canada Research Chair in Plant-Microbe Systems Biology (D.D.) or Comparative Genomics (D.S.G.); and the Centre for the Analysis of Genome Evolution and Function (D.D. and D.S.G.). This research was also funded by the Gatsby Charitable Foundation (C.Z.) and the European Research Council (grant “PHOSPHinnATE”) (C.Z.). E.S.L is supported by a Hertha Firnberg Programme post-doctoral fellowship (T-947) from the FWF Austrian Science Fund. M.S. is supported by a post-doctoral fellowship (STE 2448/1) from the Deutsche Forschungsgemeinschaft (DFG). This work was supported by the National Science Foundation (IOS-1557796) to M.S.M. P.S.B. acknowledge funding from the Austrian Federal Ministry of Science, Research & Economy, and the City of Vienna through the Vienna Biocenter Core Facilities (VBCF). We would like to thank, P. Serrano Drozdowskyj, A. Aszodi and A. Gyenesei from the VBCF Biocomputing facility for developing the Platero software. We also thank the VBCF Plant Sciences facilities for the plant growth chambers.
Footnotes
Author Contributions. E.S.L., G.A.M., K.G., and Y.B conceived and designed the experiments for the CSI screen. J.N. and A.L. cloned and expressed all the ECDs with inputs from E.S.L., P.S.B. and Y.B.; E.S.L. performed all the ECD interaction assays. T.C.H. conceived, designed and performed the Y2H assays under M.S.M. supervision; G.A.M., E.S.L., K.P, characterized and tested all the T-DNA insertion lines under the supervision of D.D., D.S.G, and Y.B.; G.A.M., E.S.L., K.P., N.W., K.G., J.K., genotyped and bulked all the T-DNA insertion lines. G.A.M. analysed and implemented the computational and statistical analysis of all the data with inputs from D.S.G. and Y.B.; M.L. and G.A.M. conceived, designed and performed the network analysis with inputs from D.D., D.S.G, N.J.P and Y.B.; M.S. designed and performed the BAK1-FLS2 co-IP assays under C.Z. supervision; K.P. organized and performed the APEX-PEPRs co-IP experiment with guidance from E.S.L.; E.S.L and K.G. generated the apex bak1 double mutants and the 35S::APEX transgenic lines. S.B.S. and W.B. contributed and characterized the fir T-DNA mutant. E.S.L conceived, organized and performed the physiological assays with BRs, Pep2, flg22. D.M and M.M devised the synthesis of the flg22 and Pep2 peptides. G.A.M. and Y.B wrote the manuscript with major input from E.S.L., M.S.M., C.Z., D.D, and D.S.G.; all authors commented and agreed on the manuscript.
Author Information Reprints and permissions information is available at www.nature.com/reprints. Readers are welcome to comment on the online version of the paper. Correspondence and requests for materials should be addressed to (Y.B), (youssef.belkhadir@gmi.oeaw.ac.at)
The authors declare no competing financial interests.
References
- 1.Belkhadir Y, Yang L, Hetzel J, Dangl JL, Chory J. The growth-defense pivot: crisis management in plants mediated by LRR-RK surface receptors. Trends in biochemical sciences. 2014 doi: 10.1016/j.tibs.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jaillais Y, Belkhadir Y, Balsemao-Pires E, Dangl JL, Chory J. Extracellular leucine-rich repeats as a platform for receptor/coreceptor complex formation. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:8503–8507. doi: 10.1073/pnas.1103556108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ozkan E, et al. An extracellular interactome of immunoglobulin and LRR proteins reveals receptor-ligand networks. Cell. 2013;154:228–239. doi: 10.1016/j.cell.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braun P, et al. An experimentally derived confidence score for binary protein-protein interactions. Nature methods. 2009;6:91–97. doi: 10.1038/nmeth.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun J, Li L, Wang P, Zhang S, Wu J. Genome-wide characterization, evolution, and expression analysis of the leucine-rich repeat receptor-like protein kinase (LRR-RLK) gene family in Rosaceae genomes. BMC genomics. 2017;18:763. doi: 10.1186/s12864-017-4155-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shiu SH, et al. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. The Plant cell. 2004;16:1220–1234. doi: 10.1105/tpc.020834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hohmann U, Lau K, Hothorn M. The Structural Basis of Ligand Perception and Signal Activation by Receptor Kinases. Annual review of plant biology. 2017 doi: 10.1146/annurev-arplant-042916-040957. [DOI] [PubMed] [Google Scholar]
- 8.Zipfel C, Oldroyd GE. Plant signalling in symbiosis and immunity. Nature. 2017;543:328–336. doi: 10.1038/nature22009. [DOI] [PubMed] [Google Scholar]
- 9.Soyars CL, James SR, Nimchuk ZL. Ready, aim, shoot: stem cell regulation of the shoot apical meristem. Current opinion in plant biology. 2016;29:163–168. doi: 10.1016/j.pbi.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 10.Song W, Han Z, Wang J, Lin G, Chai J. Structural insights into ligand recognition and activation of plant receptor kinases. Current opinion in structural biology. 2017;43:18–27. doi: 10.1016/j.sbi.2016.09.012. [DOI] [PubMed] [Google Scholar]
- 11.Boutrot F, Zipfel C. Function, Discovery, and Exploitation of Plant Pattern Recognition Receptors for Broad-Spectrum Disease Resistance. Annual review of phytopathology. 2017;55:257–286. doi: 10.1146/annurev-phyto-080614-120106. [DOI] [PubMed] [Google Scholar]
- 12.Santiago J, Henzler C, Hothorn M. Molecular mechanism for plant steroid receptor activation by somatic embryogenesis co-receptor kinases. Science (New York, N.Y.) 2013;341:889–892. doi: 10.1126/science.1242468. [DOI] [PubMed] [Google Scholar]
- 13.Sun Y, et al. Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science (New York, N.Y.) 2013;342:624–628. doi: 10.1126/science.1243825. [DOI] [PubMed] [Google Scholar]
- 14.Sun Y, et al. Structure reveals that BAK1 as a co-receptor recognizes the BRI1-bound brassinolide. Cell research. 2013;23:1326–1329. doi: 10.1038/cr.2013.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang J, et al. Structural basis for recognition of an endogenous peptide by the plant receptor kinase PEPR1. Cell research. 2015;25:110–120. doi: 10.1038/cr.2014.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Braun P. Interactome mapping for analysis of complex phenotypes: insights from benchmarking binary interaction assays. Proteomics. 2012;12:1499–1518. doi: 10.1002/pmic.201100598. [DOI] [PubMed] [Google Scholar]
- 17.Mukhtar MS, et al. Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science. 2011;333:596–601. doi: 10.1126/science.1203659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alonso JM, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science (New York, N.Y.) 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 19.Belkhadir Y, et al. Brassinosteroids modulate the efficiency of plant immune responses to microbe-associated molecular patterns. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:297–302. doi: 10.1073/pnas.1112840108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mott GA, et al. Genomic screens identify a new phytobacterial microbe-associated molecular pattern and the cognate Arabidopsis receptor-like kinase that mediates its immune elicitation. Genome biology. 2016;17:98. doi: 10.1186/s13059-016-0955-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shinohara H, Mori A, Yasue N, Sumida K, Matsubayashi Y. Identification of three LRR-RKs involved in perception of root meristem growth factor in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:3897–3902. doi: 10.1073/pnas.1522639113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu W, Pellegrini M, Wang X. Detecting communities based on network topology. Scientific reports. 2014;4:5739. doi: 10.1038/srep05739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li XQ, Xing T, Du D. Identification of Top-ranked Proteins within a Directional Protein Interaction Network using the PageRank Algorithm: Applications in Humans and Plants. Curr Issues Mol Biol. 2016;20:13–28. [PubMed] [Google Scholar]
- 24.Tian L, Bashan A, Shi DN, Liu YY. Articulation points in complex networks. 2017;8:14223. doi: 10.1038/ncomms14223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwessinger B, et al. Phosphorylation-dependent differential regulation of plant growth, cell death, and innate immunity by the regulatory receptor-like kinase BAK1. PLoS genetics. 2011;7:e1002046. doi: 10.1371/journal.pgen.1002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Couto D, Zipfel C. Regulation of pattern recognition receptor signalling in plants. Nat Rev Immunol. 2016;16:537–552. doi: 10.1038/nri.2016. [DOI] [PubMed] [Google Scholar]
- 27.McWilliam H, et al. Analysis Tool Web Services from the EMBL-EBI. Nucleic acids research. 2013;41:W597–600. doi: 10.1093/nar/gkt376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gou X, et al. Genome-wide cloning and sequence analysis of leucine-rich repeat receptor-like protein kinase genes in Arabidopsis thaliana. BMC genomics. 2010;11:19. doi: 10.1186/1471-2164-11-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malo N, Hanley JA, Cerquozzi S, Pelletier J, Nadon R. Statistical practice in high-throughput screening data analysis. Nature biotechnology. 2006;24:167–175. doi: 10.1038/nbt1186. [DOI] [PubMed] [Google Scholar]
- 30.Brideau C, Gunter B, Pikounis B, Liaw A. Improved statistical methods for hit selection in high-throughput screening. Journal of biomolecular screening. 2003;8:634–647. doi: 10.1177/1087057103258285. [DOI] [PubMed] [Google Scholar]
- 31.Halter T, et al. The leucine-rich repeat receptor kinase BIR2 is a negative regulator of BAK1 in plant immunity. Current biology : CB. 2014;24:134–143. doi: 10.1016/j.cub.2013.11. [DOI] [PubMed] [Google Scholar]
- 32.Eyuboglu B, et al. Molecular characterisation of the STRUBBELIG-RECEPTOR FAMILY of genes encoding putative leucine-rich repeat receptor-like kinases in Arabidopsis thaliana. BMC plant biology. 2007;7:16. doi: 10.1186/1471-2229-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jorda L, et al. ERECTA and BAK1 Receptor Like Kinases Interact to Regulate Immune Responses in Arabidopsis. Frontiers in plant science. 2016;7:897. doi: 10.3389/fpls.2016.00897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Russinova E, et al. Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1) The Plant cell. 2004;16:3216–3229. doi: 10.1105/tpc.104.025387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roux M, et al. The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. The Plant cell. 2011;23:2440–2455. doi: 10.1105/tpc.111.084301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hazak O, et al. Perception of root-active CLE peptides requires CORYNE function in the phloem vasculature. 2017;18:1367–1381. doi: 10.15252/embr.201643535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meng X, et al. Differential Function of Arabidopsis SERK Family Receptor-like Kinases in Stomatal Patterning. Current biology : CB. 2015;25:2361–2372. doi: 10.1016/j.cub.2015.07.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nodine MD, Yadegari R, Tax FE. RPK1 and TOAD2 are two receptor-like kinases redundantly required for arabidopsis embryonic pattern formation. Developmental cell. 2007;12:943–956. doi: 10.1016/j.devcel.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 39.Wang X, et al. IDL6-HAE/HSL2 impacts pectin degradation and resistance to Pseudomonas syringae pv tomato DC3000 in Arabidopsis leaves. The Plant journal : for cell and molecular biology. 2017;89:250–263. doi: 10.1111/tpj.13380. [DOI] [PubMed] [Google Scholar]
- 40.Meng X, et al. Ligand-Induced Receptor-like Kinase Complex Regulates Floral Organ Abscission in Arabidopsis. Cell reports. 2016;14:1330–1338. doi: 10.1016/j.celrep.2016.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Santiago J, et al. Mechanistic insight into a peptide hormone signaling complex mediating floral organ abscission. 2016;5 doi: 10.7554/eLife.15075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nimchuk ZL. CLAVATA1 controls distinct signaling outputs that buffer shoot stem cell proliferation through a two-step transcriptional compensation loop. PLoS genetics. 2017;13:e1006681. doi: 10.1371/journal.pgen.1006681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cole SJ, Diener AC. Diversity in receptor-like kinase genes is a major determinant of quantitative resistance to Fusarium oxysporum f.sp. matthioli. The New phytologist. 2013;200:172–184. doi: 10.1111/nph.12368. [DOI] [PubMed] [Google Scholar]
- 44.Agusti J, Lichtenberger R, Schwarz M, Nehlin L, Greb T. Characterization of transcriptome remodeling during cambium formation identifies MOL1 and RUL1 as opposing regulators of secondary growth. PLoS genetics. 2011;7:e1001312. doi: 10.1371/journal.pgen.1001312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiao D, et al. SENESCENCE-SUPPRESSED PROTEIN PHOSPHATASE Directly Interacts with the Cytoplasmic Domain of SENESCENCE-ASSOCIATED RECEPTOR-LIKE KINASE and Negatively Regulates Leaf Senescence in Arabidopsis. 2015;169:1275–1291. doi: 10.1104/pp.15.01112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kang YH, Hardtke CS. Arabidopsis MAKR5 is a positive effector of BAM3-dependent CLE45 signaling. 2016;17:1145–1154. doi: 10.15252/embr.201642450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Albert I, et al. An RLP23-SOBIR1-BAK1 complex mediates NLP-triggered immunity. 2015;1:15140. doi: 10.1038/nplants.2015.140. [DOI] [PubMed] [Google Scholar]
- 48.Valon C, Smalle J, Goodman HM, Giraudat J. Characterization of an Arabidopsis thaliana gene (TMKL1) encoding a putative transmembrane protein with an unusual kinase-like domain. Plant molecular biology. 1993;23:415–421. doi: 10.1007/BF00029017. [DOI] [PubMed] [Google Scholar]
- 49.Tarutani Y, et al. Molecular characterization of two highly homologous receptor-like kinase genes, RLK902 and RKL1, in Arabidopsis thaliana. Bioscience, biotechnology, and biochemistry. 2004;68:1935–1941. doi: 10.1271/bbb.68.1935. [DOI] [PubMed] [Google Scholar]
- 50.Chang C, et al. The TMK1 gene from Arabidopsis codes for a protein with structural and biochemical characteristics of a receptor protein kinase. The Plant cell. 1992;4:1263–1271. doi: 10.1105/tpc.4.10.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang T, et al. A receptor heteromer mediates the male perception of female attractants in plants. Nature. 2016;531:241–244. doi: 10.1038/nature16975. [DOI] [PubMed] [Google Scholar]
- 52.Sun W, et al. Probing the Arabidopsis flagellin receptor: FLS2-FLS2 association and the contributions of specific domains to signaling function. The Plant cell. 2012;24:1096–1113. doi: 10.1105/tpc.112.095919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yeh YH, Panzeri D. The Arabidopsis Malectin-Like/LRR-RLK IOS1 Is Critical for BAK1-Dependent and BAK1-Independent Pattern-Triggered Immunity. 2016;28:1701–1721. doi: 10.1105/tpc.16.00313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Igarashi D, Tsuda K, Katagiri F. The peptide growth factor, phytosulfokine, attenuates pattern-triggered immunity. The Plant journal : for cell and molecular biology. 2012;71:194–204. doi: 10.1111/j.1365-313X.2012.04950.x. [DOI] [PubMed] [Google Scholar]
- 55.Mosher S, et al. The tyrosine-sulfated peptide receptors PSKR1 and PSY1R modify the immunity of Arabidopsis to biotrophic and necrotrophic pathogens in an antagonistic manner. The Plant journal : for cell and molecular biology. 2013;73:469–482. doi: 10.1111/tpj.12050. [DOI] [PubMed] [Google Scholar]
- 56.Wang J, et al. Allosteric receptor activation by the plant peptide hormone phytosulfokine. Nature. 2015;525:265–268. doi: 10.1038/nature14858. [DOI] [PubMed] [Google Scholar]
- 57.Postel S, et al. The multifunctional leucine-rich repeat receptor kinase BAK1 is implicated in Arabidopsis development and immunity. European journal of cell biology. 2010;89:169–174. doi: 10.1016/j.ejcb.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 58.Sakamoto T, et al. The tomato RLK superfamily: phylogeny and functional predictions about the role of the LRRII-RLK subfamily in antiviral defense. BMC plant biology. 2012;12:229. doi: 10.1186/1471-2229-12-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cusick ME, et al. Literature-curated protein interaction datasets. Nature methods. 2009;6:39–46. doi: 10.1038/nmeth.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gao M, et al. Regulation of cell death and innate immunity by two receptor-like kinases in Arabidopsis. Cell host & microbe. 2009;6:34–44. doi: 10.1016/j.chom.2009.05.019. [DOI] [PubMed] [Google Scholar]
- 61.Ma C, et al. Structural basis for BIR1-mediated negative regulation of plant immunity. Cell research. 2017 doi: 10.1038/cr.2017.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fabregas N, et al. The brassinosteroid insensitive1-like3 signalosome complex regulates Arabidopsis root development. The Plant cell. 2013;25:3377–3388. doi: 10.1105/tpc.113.114462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee JS, et al. Competitive binding of antagonistic peptides fine-tunes stomatal patterning. Nature. 2015;522:439–443. doi: 10.1038/nature14561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Karlova R, et al. The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 protein complex includes BRASSINOSTEROID-INSENSITIVE1. The Plant cell. 2006;18:626–638. doi: 10.1105/tpc.105.039412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang D, et al. BKI1 Regulates Plant Architecture through Coordinated Inhibition of the Brassinosteroid and ERECTA Signaling Pathways in Arabidopsis. Molecular plant. 2017;10:297–308. doi: 10.1016/j.molp.2016.11.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.