Abstract
In the past few years there has been a growth in the use of nano-particles for stabilizing lipid membranes with embedded proteins. These bionanoparticles provide a solution to the challenging problem of membrane protein isolation by maintaining a lipid bilayer essential to protein integrity and activity. We have described the use of an amphipathic polymer (Poly(styrene-co-maleic acid); SMA) to produce discoidal nanoparticles that contain a lipid bilayer with embedded protein. However the structure of the nanoparticle itself has not yet been determined. This leaves a major gap in understanding how the SMA stabilizes the encapsulated bilayer and how the bilayer relates physically and structurally to an unecapsulated lipid bilayer. In this paper we address this issue by describing the structure of the SMA Lipid Particle (SMALP) using data from small angle neutron scattering (SANS), electron microscopy (EM), attenuated total reflection Fourier transform infrared spectroscopy (ATR-FTIR), differential scanning calorimetry (DSC) and nuclear magnetic resonance spectroscopy (NMR). We show that the particle is disc shaped containing a polymer “bracelet” encircling the lipid bilayer. The structure and orientation of the individual components within the bilayer and polymer are determined showing that styrene moieties within SMA intercalate between the lipid acyl chains. The dimensions of the encapsulated bilayer are also determined and match those measured for a natural membrane. Taken together, the description of structure of the SMALP forms the foundation of future development and applications of SMALPs in membrane protein production and analysis.
Keywords: nanoparticles, lipid, polymer, membrane proteins, structure, detergent
Introduction
Membrane proteins make up approximately 40% of the proteome of most organisms, carrying out a range of diverse functions from catalysis to metabolite transport. This class of proteins also provides more than half of the therapeutic targets for modern drug discovery. However, up until 2013 only 1186 high resolution structures of membrane proteins had been deposited in the protein structure database compared to a total of 81209 for all types of protein (data from http://blanco.biomol.uci.edu/mpstruc/listAll/list#Latest). This slow progress regarding the structure and functions of membrane proteins results from several technical challenges that occur during production and subsequent handling of membrane protein containing samples. As membrane proteins are embedded in a lipid membrane continuum, the first step of purification is the solubilisation of the individual proteins. This process generally requires the use of surfactants (i.e. detergents) that replace the lipids surrounding the membrane protein (for review see [2]). However the surfactant:protein micelles that are formed do not adequately mimic the native membrane structure and typically lead to poor protein stability or even unfolding.
One strategy for overcoming the problem of membrane protein extraction has been to re-insert the membrane protein in a model lipid membrane, such as a proteo-liposome or disc-shaped lipid particles, such as bicelles and nanodiscs. These systems offer a potential solution but each suffers from significant issues. For example, bicelle formation requires very specific phospholipid compositions and tight control of the experimental conditions. These conditions are often not compatible with experimental processes and/or the native lipid conditions required for protein activity. A second, more successful method, relies on a particle containing a lipid core stabilized by an annulus of apolipoproteins or derived peptides (also called membrane scaffold proteins or MSPs) that protect the exposed hydrophobic edges of lipid membranes [3, 4]. Sligar and colleagues have led work on these systems and have established the molecular layout of the MSP discs using a range of methods including X-ray scattering, NMR, molecular dynamics simulations and atomic force microscopy (see [5] for a comprehensive review). This method has been shown to provide samples of a range of membrane proteins including cytochromes [6], 7TM proteins (including G-protein coupled receptors) [7] and blood clotting co-factors [8] with native-like activity, proving that the overall approach is workable. The observation that the MSP system produces active membrane proteins is a good indication that the lipid environment in the MSP has native-like biophysical characteristics. Calorimetric measurements established that the phase transitions of the lipid bilayer in the MSP show only a minimal perturbation compared to that of the bulk lipid [9].
The pioneering studies of the MSP system have demonstrated that the nanoencapsulation approach to membrane protein extraction may be a useful. However, the use of lipid-stabilizing peptides in the MSP limit the use of a number of biophysical techniques like circular dichroism (CD), Fourier transform infrared (FTIR) spectroscopy and nuclear magnetic resonance spectroscopy (NMR) as the signals from the stabilizing peptides can interfere with signals from the membrane protein. In addition preparation of the disc:peptide:membrane protein complex relies on the use of surfactants with the resulting removal of the native lipid environment which cannot easily be replaced.
In 2001 Tighe and Tonge showed that SMA could be used to produce nanoparticles that contained a synthetic lipid bilayer [10]. In 2009 we demonstrated that membrane proteins could be isolated as a nano-particle which also contains a lipid bilayer using a styrene maleic acid polymer which contained a 2:1 molar ratio of styrene to maleic acid [11]. We showed that this method did not require any surfactant providing significant advantages over other available methods [11]. The addition of this co-polymer of styrene and maleic acid to biological membrane and in the absence of any kind of surfactants, initiates the spontaneous formation of uniform 10.2 nm diameter discs (styrene maleic acid lipid particles, SMALPs), many of which contain membrane proteins. Although we have shown that the method can be used to purify and study a number of different membrane proteins, a detailed molecular characterisation of the structure of these particles is however still missing. Recent work by Watts and colleagues [12–14] have shown a preliminary analysis of similar particles produced using a 9.5 KDa SMA polymer with a styrene to maleic acid ratio of 3:1 in contrast to the 2:1 ratio used in our previous experiments. Their work suggests that these particles are structurally monodispersed and stabilized by ionic interactions between the head groups of the lipids and the SMA. They show that these discs can be used to solubilize a range of proteins including Complex IV from the inner membrane of mitochondria [14] and bacteriorhodopsin [13]. Unfortunately they also show that the phase transitions of the lipids in the nanoparticle are perturbed with the phase transition temperature being reduced by 7 °C compared to that for an equivalent bulk bilayer [12] indicating that the SMA can significantly alter the physical properties of the membrane.
Here we provide an in depth study of the structural and physical properties of SMALPs made using SMA with a 2:1 styrene to maleic acid ratio and the lipid 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC). The overall structure of the SMALP particle described in this study is assessed using small angle neutron scattering (SANS) combined with isotopic phase contrast and electron microscopy (EM). These data provide information on the dimensions of the particle and the position of the SMA polymer and lipids confirming that the membrane dimensions match those in the natural bilayer. Polarized attenuated total reflection FTIR spectroscopy is used to examine the orientation of the chemical groups from the polymer in the particle and to provide some information about the lipid organization in the disc. The interaction between the styrene groups and the lipid acyl chains is defined using NMR. Finally the effect of the SMALP formation on the lipid phase transitions is analysed using differential scanning calorimetry (DSC) showing that the formation of this particle perturbs the phase behavior to a lesser extent than the particles made using MSPs or with SMA containing 3:1 styrene:maleic acid ratio. Together this provides a comprehensive understanding of how these nanoparticles preserve a natural membrane microenvironment as a minimal functional unit.
1. Experimental
1.1. Materials
Poly(styrene-co-maleic anhydride) with a ratio of 2:1 styrene to maleic anhydride was used as described in Knowles et al. [11]. DMPC, 2H-DMPC and 2H-DPPC were purchased from Avanti Polar Lipids (Alabaster, AL) as a synthetic lipid in powder form. All other materials were purchased from Sigma Aldrich Ltd. (UK).
1.2. Preparation of styrene maleic copolymer
A 10% solution styrene maleic acid copolymer was prepared by dissolving 25g of styrene maleic anhydride copolymer in 250 ml of NaOH [1 M] in a round bottom flask on a magnetic stirrer overnight. The resulting solution was then refluxed for 2 hours and allowed to cool to room temperature. The material was then aliquoted into 50ml samples and placed in a freezer, and thawed and dialyzed against the buffer solution before use.
1.3. Preparation of styrene maleic acid lipid particles
For all experiments SMALPS were prepared by first preparing two solutions; the first solution was of 10% (wt/vol) of the required phospholipid prepared by suspending lipid powder in a known volume of phosphate buffer solution, then sonicating for ~ 5 minutes to disperse the lipid in the solution. A second solution consisting of 10% (wt/vol) copolymer in buffer was prepared as outlined above. The two solutions were then combined to give the desired concentrations, routinely this was 1% lipid and 2.5% SMA. This mixed solution was shaken for approximately 1 min until clear.
Where required Gel filtration purification of the initial disc suspensions was performed to remove any high molecular weight aggregates that might have affected data collection. SMALPs were purified at 6 °C using a Superdex 200 10/300 GL column attached to an AKTA ™ purifier FPLC purification system (GE Healthcare). A 50 mM phosphate buffer with 200 mM NaCl was used with a flow rate of 1 ml/min. Absorbance measurements were recorded at 254 nm.
1.4. Small angle neutron scattering
After gel filtration the SMALPs were dialysed sequentially into appropriate buffer solution (50 mM phosphate buffer with 200 mM NaCl, pH 8.0) in H2O, D2O or mixtures of H2O/D2O for SANS measurements. The SANS experiments were conducted on the D11 instrument located at the ILL laboratories, Grenoble (France). Samples were measured in single stopper rectangular Hellma quartz cuvettes with a 1 mm path length. This instrument uses a monochromatic neutron beam with a wavelength of 6 Å and the detector was placed at 1.2 m, 7 m and 13 m to collect a Q range between 0.004-0.5 Å-1. Water was used as the calibration standard. Measurements were taken at 25 °C. The experiment was performed using four different phosphate buffer contrasts, 100% H2O, 100% D2O, 60% D2O and 32% D2O.
1.5. Electron Microscopy
The SMALP sample was diluted in buffer solution (50 mM Sodium phosphate pH 7.0, 100 mM NaCl) to 1 X 10-5 mg/ml. The sample was then drop cast onto a glow discharged carbon coated electron microscopy grid and negatively stained with Uranyl acetate. Imaging was performed with an FEI Tecnai 12, 120 kV transmission electron microscope (TEM) with an FEI Eagle 4k x 4K CCD camera. All subsequent image processing was carried out using Fiji/ImageJ
1.6. Attenuated total reflection infrared spectroscopy (ATR-FTIR)
ATR-FTIR spectra were recorded at room temperature with a Bruker EQUINOX 55 FTIR spectrophotometer equipped with a liquid nitrogen-cooled mercury cadmium telluride (MCT) detector at a nominal resolution of 2 cm-1. The internal reflection element (IRE) was a diamond, with an incidence angle of 45° leading to one internal reflection. The spectrophotometer was continuously purged with air dried on a Hydrovane HV05.
1.7. Collection of FTIR spectra
A 1μl sample was deposited on the diamond element and dried under a nitrogen stream. Spectra were recorded either in the absence of polarization of the light, or with the incident light polarized parallel and perpendicular to the diamond surface. Dichroic spectra were computed by subtracting the parallel polarized spectrum from the perpendicularly polarized spectrum. Correction for differences in evanescent field intensity for each polarization was done by normalizing the spectrum with respect to the area of the lipid ester band at 1740 cm-1, estimated to be equal in both directions [15]. An upward deviation in the dichroic spectrum indicates a dipole having preferentially a near-normal orientation with respect to the diamond surface, while a downwards deviation on the dichroic spectra indicates a dipole oriented in the plane of the diamond surface. Spectra were corrected, if necessary, for water vapor, using water vapor spectra recorded with the same polarization of the light.
1.8. NMR Spectroscopy
Acyl-deuterated and non-deuterated DMPC SMALPs were prepared by adding 2.5% SMA in 50 mM sodium phosphate pH 8.0, 300 mM NaCl to a 2% (w/v) liposome suspension of either 1,2-dimyristoyl(d54)-sn-glycero-3-phosphocholine or 1,2-dimyristoyl-sn-glycero-3-phosphocholine in 50mM sodium phosphate pH 8.0, 300 mM NaCl and incubating at room temperature for 10 minutes. The resultant SMALPs were purified by size exclusion chromatography using a Superdex 200 10/300 GL column (GE Healthcare) with a buffer consisting of 50 mM sodium phosphate, 200 mM NaCl pH 7.5.
A 2D homonuclear NOESY experiment [16, 17] was obtained with a mixing time of 100ms on a Varian Inova 800 MHz spectrometer equipped with 5mm z-PFG cryogenic probe. Data were processed using NMRpipe [18] and spectra analysed using Sparky [19].
1.9. Differential Scanning Calorimetry
DMPC vesicles were prepared by solubilizing DMPC in chloroform and then drying under rotary evaporation to produce a thin layer of lipid. The lipid was then resuspended in 50 mM TrisHCl pH 8.0, 200 mM NaCl to a final concentration of 2 mM. 0.1 μm unimolecular vesicles were produced by subjecting the hydrated lipid suspension to 10 freeze/thaw cycles followed by extruding through 0.4, 0.2 and 0.1 μm membranes above the phase transition temperature of the lipid (for DMPC > 25 °C). DMPC SMALPs were prepared as described for NMR spectroscopy but concentrated down to a final lipid concentration of 6 mM.
A VP-DSC microcalorimeter (MicroCal, Northampton, MA) was used to carry out Differential Scanning Calorimetry. Before making the scans, the samples were degassed for 20 minutes at 4 °C. All samples were scanned from 5 °C to 95 °C, at a scan rate of 70 °/hour, with the prescan thermostat set for 15 minutes and the postscan thermostat 0 minutes. Thirty five scans were made for each sample in the high feedback mode with the post cycle thermostat set to 5 °C.
3. Results
It has already been shown by ourselves and others [13] that the SMALPs can provide a novel method for membrane protein extraction. However if this work is going to progress it is essential that an in-depth description of the structure of the particle itself is achieved. Such an understanding will enable future applications of the SMALP particle in studies of membrane proteins including pharmaceutical screening studies, structure determination and biochemical analyses. In addition a deeper understanding of the SMALP will provide baseline information for the continued development of new improved SMA based systems.
The study begins with analysis of SANS data from SMALPs containing either hydrogenated or deuterated lipids which provides information on the distribution of the polymer and the lipid components within the particle and on the overall dimensions of the SMALP structure. EM analysis adds to these observations and provides more detail on the size distributions of the discs. A Polarized FTIR spectroscopy study uncovers the orientation of the chemical moieties that make up the particle while NMR resolves interactions between the lipids and the polymer. In the last stage of the study calorimetric measurements show that the phase transition temperature of the lipid bilayer remains very close to that of the free membrane. These data together provide a detailed picture of the structure of the SMALP that lays the foundation for further studies using the SMALP system to investigate protein structure.
3.1. Small angle neutron scattering
The first goal was to ascertain the shape and size of the SMALP and the positions of the lipid and SMA constituents within the nanoparticle. This architecture is important for supporting a native-like membrane structure for solubilization of biologically intact states. SANS offers the ability to examine the structure of materials like SMALPs free in solution, providing dimensions of the overall particle, while solutions containing varying amounts of D2O as well as deuterated lipids could be used to determine the relative positions of the copolymer and lipids in the structure.
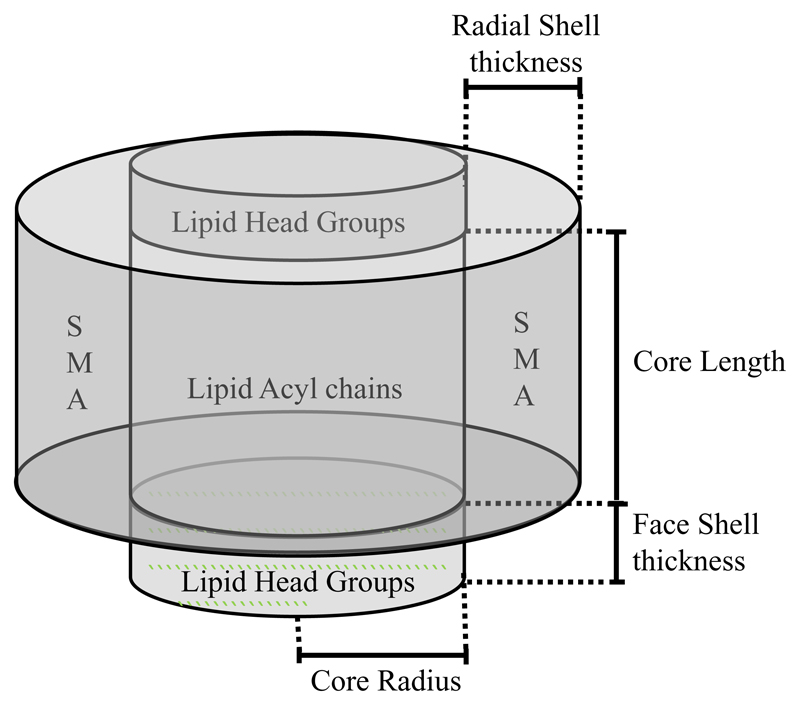
The data for four solution contrasts (100% H2O, 100% D2O, 60% D2O and 32% D2O) were fitted simultaneously using the NIST SANS Analysis Package [20] in the IGOR Pro fitting program to a model of core-shell cylinders with a face layer on top and bottom to account for the phospholipid headgroups (a schematic representation is given in Figure 1). The core radius was convoluted by a Schultz distribution to add polydispersity in the radius of the core of the particle. A Hayter-Penfold structure factor [21] for charged colloids in solution was used to account for electrostatic repulsion between the discs. The scattering length density (SLD) of the phospholipid headgroups was set to be 1.84×10-6 Å-2 based on the value cited by Smith et al. [22] who used molecular volumes obtained from molecular dynamics simulations to calculate this number. The water content of the headgroup layer was likewise set to a mole fraction 0.57 based on their results in fitting a deuterated DMPC bilayer. The SLD of the polymer was calculated to be 2.37 Å-2 and the SLD of the rim fitted by fitting the mole fraction of solvent in the rim based on this value and that of the phosphate buffer solution for each sample.
Figure 1.
The core shell model used to fit the SANS data showing the dimensions that can be calculated from the model.
The scattering length densities of the 100% H2O (-0.56×10-6 Å-2) and 100% D2O (6.3×10-6 Å-2) buffer solutions were also calculated and held during fitting. Other model parameters which were predetermined and held constant during fitting were the temperature (298 K), the dielectric constant of the solution (78), the monovalent salt concentration (250 mM based on the buffer preparation). The majority of other parameters were linked during fitting except for the SLDs of the two mixed H2O/D2O buffer solutions (to account for any incomplete exchange during dialysis), and the backgrounds for each sample. Fitting results are shown in Table 1 and the fitted data in Figure 2.
Table 1.
Fit parameters for fitting SANS data to a model of a charged core-shell cylinder with polydisperse core and headgroup regions at top and bottom of the cylinder. * calculated or set from literature values and held during fitting. Figures in brackets derived from 8 for a DMPC bilayer at 30 °C.
| Model Coefficients | 100%D2O | 60%D2O | 32%D2O | 0%D2O |
|---|---|---|---|---|
| Volume fraction | 0.019 ±0.01 | |||
| Mean core radius (Ǻ) | 38 ±2 | |||
| Radial polydispersity (sigma) | 0.35 ±0.05 | |||
| Core length (Ǻ) | 26 ±2 (26.2)† | |||
| Radial shell thickness (Ǻ) | 9 ±2 | |||
| Face shell thickness (Ǻ) | 10 ±2 (5.25) | |||
| Calculated Total Bilayer Thickness (Ǻ), =Core length + 2× face shell thickness | 46 (44) | |||
| Core SLD (Ǻ-2) | 6.5 ×10-6 ±0.05 ×10-6 | |||
| Mol% solvent in face | 0.57* | |||
| Mol% solvent in rim | 0.42 ±0.05 | |||
| Solvent SLD (Ǻ-2) | 6.29e×10-6 * | 3.86×10-6 ±0.05×10-6 | 1.87e×10-6 ±0.05×10-6 | -0.57×10-6 * |
| Charge on the disc | 0.31±0.05 | |||
| Incoherent background (cm-1) | 0.015±0.05 | 0.011±0.05 | 0.022±0.05 | 0.006±0.05 |
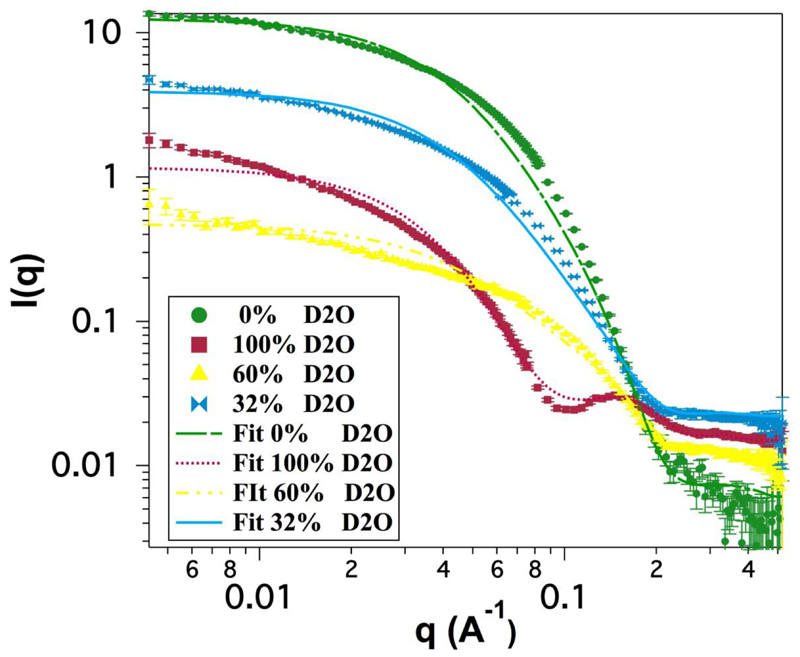
Figure 2.
Scattering patterns from SMALPs fitted to the model described in the text. Dotted lines represent the experimental results while plain lines represent the fitting.
These results show that the SMA polymer encircles a disc of lipid membrane with a radius of 38 ± 2 Å thus indicating an overall bilayer size of between 4071 and 5026 Å2. These data also show that the bilayer in the SMALP is supported on the edges by a 7 ± 2 Å thick annulus of styrene maleic acid co-polymer indicating that the annulus is likely to be made up of a single thickness of the polymer.
The thickness of the core of the disc (which represents the lipid bilayer) of 26 ± 2 Å agrees well with the expected thickness of the hydrophobic region of a DMPC bilayer of 26.2 Å at 30 °C [23]. The thickness of the face layer, being measured as 8 ± 2 Å, which would be expected to represent the headgroups of the lipids also agrees well with previous measurements of 9 Å [23].
3.2. Electron microscopy
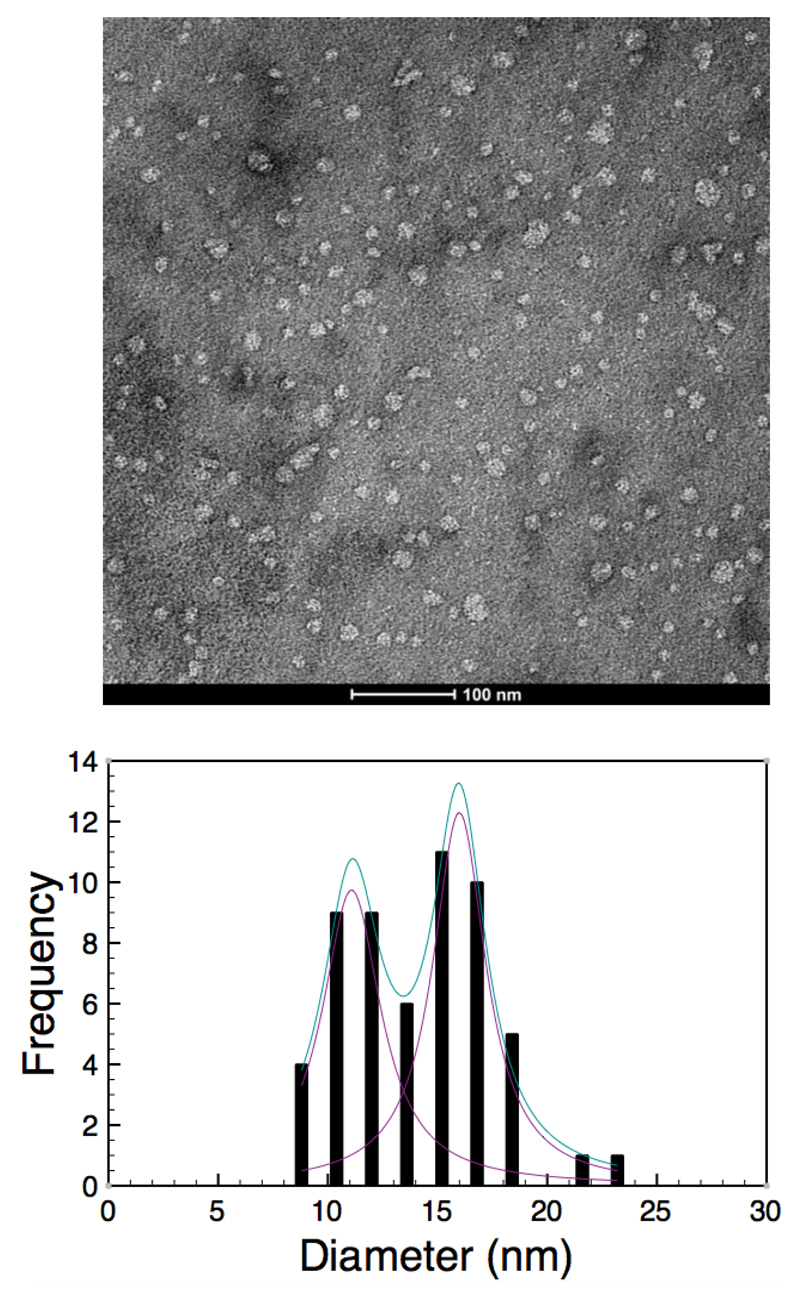
Electron microscopy studies of the structure of the SMALP provide information on the gross morphology and dimensions of the particle. Images from EM confirm the overall disc shaped nature of the SMALP (Figure 3). The dimensions of these particles clearly show some variability. A measurement of the diameters of a population of SMALPs using EM shows a bimodal distribution of diameters. Analysis of this distribution using a Gaussian distribution model shows the presence of two maxima at 11.1 ± 3.3 nm and 16.0 ± 3.0 nm. These dimensions are larger than those observed in the SANS experiments, which suggest a maximum diameter of 9.8 nm. It is likely that this disparity reflects the different preparation methods for the two samples, the SANS date being averaged in buffered solution while the EM images are taken of individal particles on a dried sample stained with uranyl acetate.
Figure 3.
Top. Negative stain image of SMALPs. Bottom. Histogram showing the distribution of the diameters of SMALP particles.
3.3. Polarized infrared spectroscopy
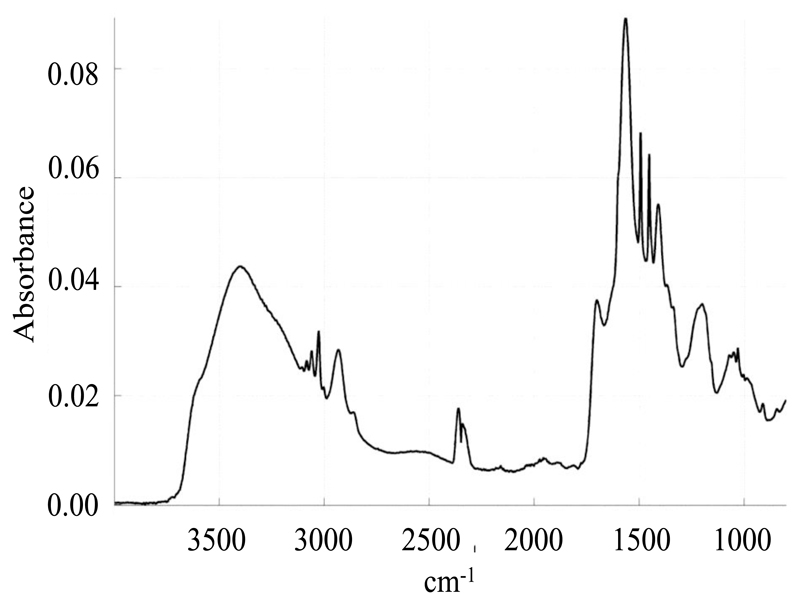
ATR-FTIR is a powerful tool to study biological samples, and in particular biological membranes. It allows a simultaneous study of the structure of the various biological and chemical components present in the system, and does not require the introduction of any probe, which could perturb the system’s integrity. Furthermore, when using polarized incident light, information about the orientation of the various chemical moieties of a molecule with respect to each other can be obtained [24]. Here we used infrared spectroscopy to study the orientation of the chemical moieties of the polymer with respect to the lipids. Assignment of the various bands of the SMA spectrum (Figure 4) is summarized in Table 2. Bands related to the styrene moiety were assigned on the basis of the polystyrene spectrum [1]. Bands related to the maleic acid moiety were assigned from the spectra of succinic acid and SMA both recorded at pH 8 and pH 1 (data not shown).
Figure 4.
IR spectrum of SMA
Table 2.
Assignment of the bands of the SMA spectrum. Reference spectra were the spectra of polystyrene [1] and of succinic acid.
| Position (cm-1) | Chemical Function | |
|---|---|---|
| SMA spectrum | reference spectrum | |
| 3083 | 3083 | C-H stretching in the styrene ring (in ortho and meta of the substituted carbon) |
| 3060 | 3061 | C-H stretching in the styrene ring (in para of the substituted carbon) |
| 3026 | 3029 | C-H stretching in the styrene ring (in ortho and meta of the substituted carbon) |
| 2932 | 2923 | antisymmetric C-H stretching of the polymer backbone CH2 |
| 2858 | 2851 | symmetric C-H stretching of the polymer backbone CH2 |
| 1705 | 1655 | COOH stretching in the maleic acid |
| 1602 | 1602 | ring vibration (C-C stretching) |
| 1565 | 1550 | COO- stretching in the maleic acid |
| 1493 | 1493 | ring vibration (C-C stretching) |
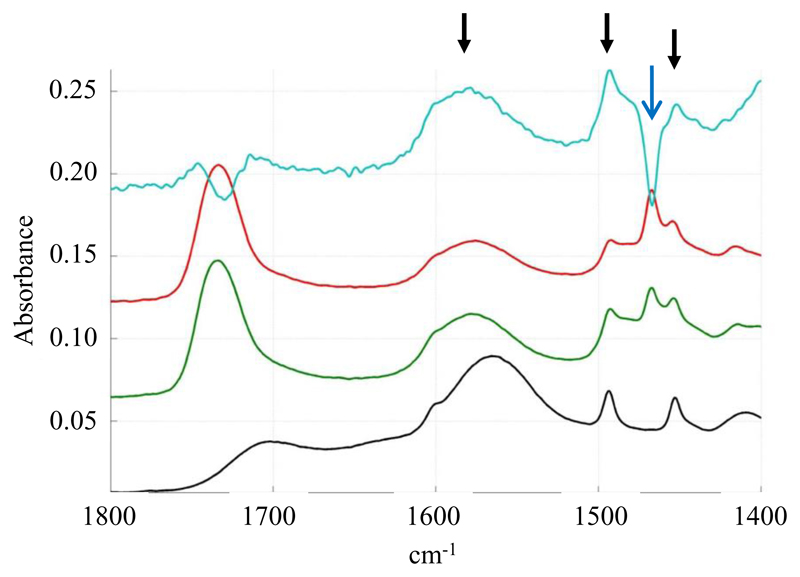
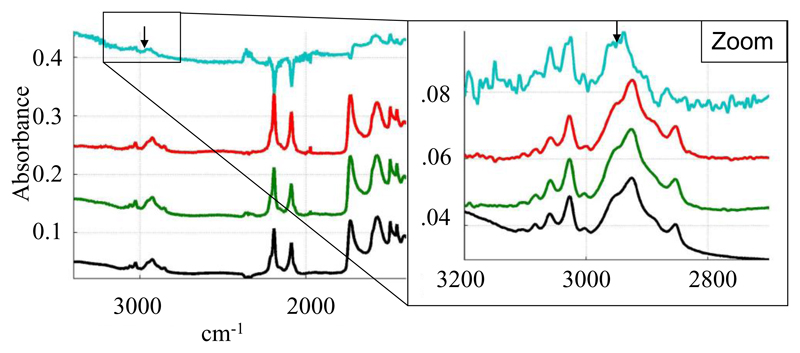
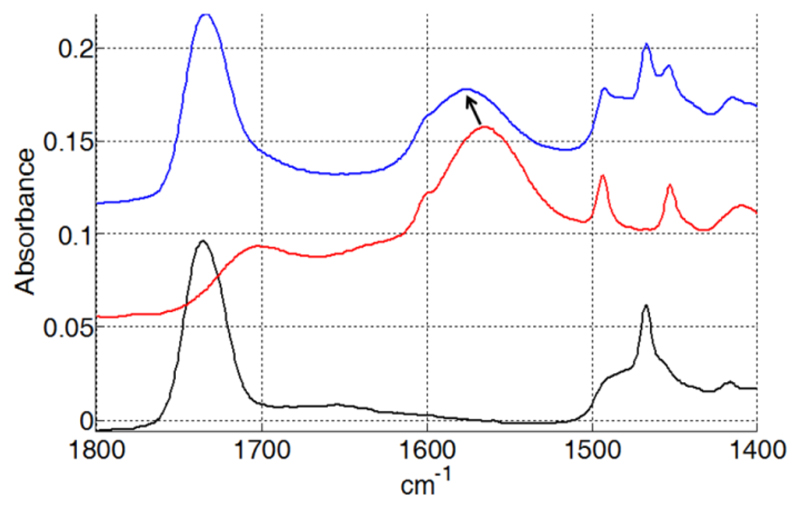
Polarized spectra of the SMALPs were recorded using parallel and perpendicular polarized light. A dichroic spectrum was computed by subtracting the parallel polarized spectrum from the perpendicularly polarized spectrum after normalization, and comparisons with free polymer controls were made. Several bands show a positive deviation on the SMALP dichroic spectrum (Figure 5). On the basis of the assignments made above, they correspond to the vibration of the styrene ring and to the COO- stretching. As the COO- transition dipole is parallel to the COO- bisector and as the dipole of the styrene ring is in the plane of the ring, it means that the two polymer functions orient perpendicularly to the membrane plane. This suggests that the ring of the styrene intercalate between the lipid chains in a manner analogous to cholesterol. As a control, we did not detect any orientation of the free polymer deposited on the IR plate. The band corresponding to the C-H stretching of the polymer carbon backbone is not visible in the spectrum due to the overlap with the lipid C-H stretching bands of the lipid acyl chains. In order to detect C-H stretching of the polymer backbone, SMALPs were prepared with deuterated DMPC. In these conditions, C-D bands from the lipid acyl chains shifted to around 2000 cm-1 and the C-H stretching bands of the polymer backbone at 2858 and 2932 cm-1 become visible (Figure 6). The polarized infrared spectrum shows a positive deviation of the band at 2932 cm-1. As the dipole of the C-H bond is perpendicular to the polymer backbone, it indicates an orientation perpendicular to the lipid acyl chain, compatible with the polymer forming a ring around the lipid in a manner that is analogous to the Apolipophorin III helices in HDL-like particles [25]. Interestingly, the COO- band of the polymer is strongly shifted in the presence of lipids (to 1575 cm-1 rather than 1565 cm-1) (Figure 7), suggesting a specific interaction between the carboxyl function of the polymer and the lipid.
Figure 5.
Dichroic spectrum of SMALPs: Black: non polarized spectrum of SMALPs (DMPC), Green/red: polarized (0° and 90°) spectra of SMALPs, Blue: Dichroic spectrum of SMALPs. Black arrows show the positive deviation of SMA bands while the blue arrow shows the negative deviation of the lipid CH2 bending band.
Figure 6.
Dichroic spectrum of deuterated DPPC SMALPs: Black: non polarized spectrum of SMALPs (DMPC). Green/red: polarized (0° and 90°) spectra of SMALPs, Blue: Dichroic spectrum of SMALPs. Arrows show the positive deviation of SMA bands
Figure 7.
IR spectrum of SMALPs: Black: IR spectrum of DMPC, Red: IR spectrum of SMA, Blue: IR spectrum of DMPC SMALPs. Arrow shows the shift from 1565 cm-1 to 1575 cm-1 of the maximum of the SMA band corresponding to the COO- stretching.
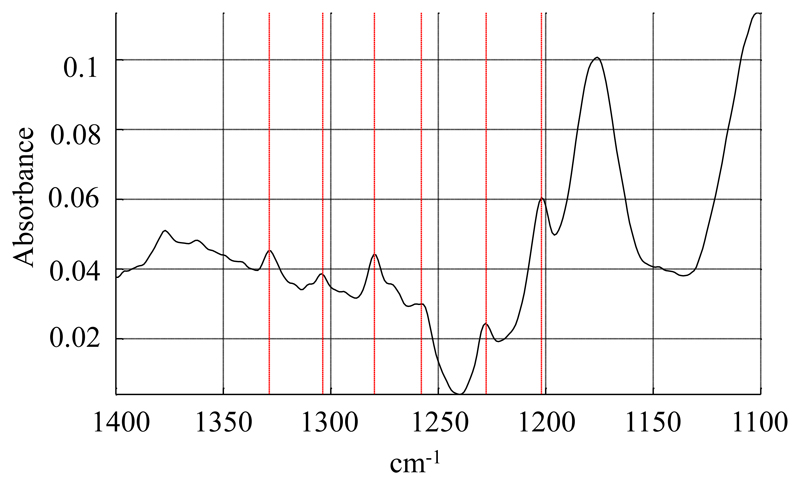
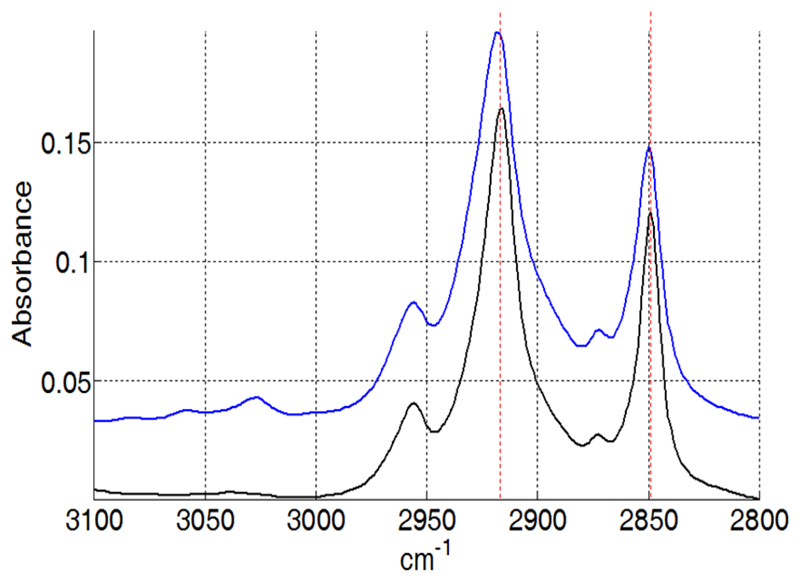
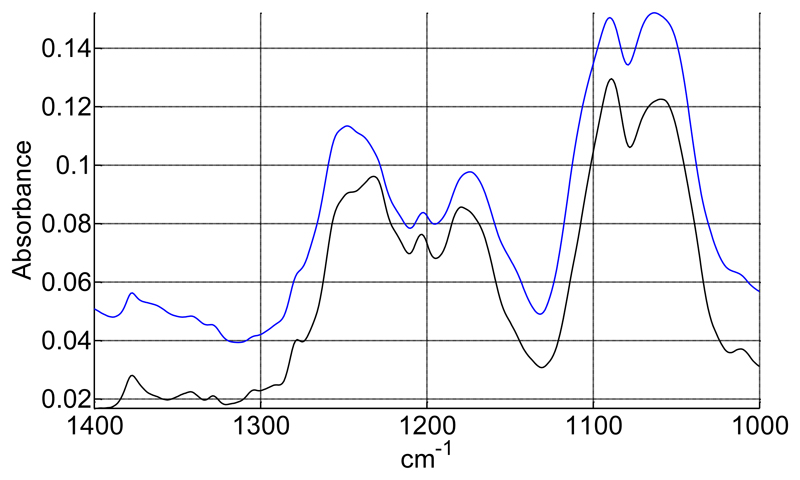
Lipid moieties present the typical signature of a bilayer organization, with a negative deviation in the dichroic spectrum for the symmetric and anti-symmetric stretching of the fatty acid CH2 at 2848 cm-1 and 2915 cm-1 respectively (data not shown), as well as a negative orientation of the CH2 bending band around 1467 cm-1 (Figure 5) and a positive orientation of the CH2 wagging bands located between 1200 and 1330 cm-1 (Figure 8) [25, 26]. However, small variations can be observed in comparison to the spectrum of pure DMPC. Upon interaction with SMALPs, the anti-symmetric and symmetric CH2 stretching bands are shifted to higher wavenumbers, going respectively from 2916.5 cm-1 to 2918 cm-1, and from 2849 cm-1 to 2850 cm-1 (Figure 9). Such shifts have also been observed for lipids upon phase transition, and are representative of the C-C bonds shifting from trans conformation to gauche conformation. This suggests that the presence of the polymer may force some of the lipid aliphatic chains to kink, inducing the formation of a C-C bond in gauche conformation. However a complete transition should induce a shift of about 3 cm-1 [27], indicating that only some of the aliphatic chains undergo this change of conformation. Similarly, the bands corresponding to the wagging of the C-H bonds of the aliphatic chain, that only appear when the aliphatic chain is in all-trans conformation, broaden and disappear when the temperature is increased due to the presence of bonds in the gauche conformation. In our spectra, bands located at 1180 (hidden by the absorption of the ester bond itself), 1203 and 1231 cm-1 (Figure 10) also decrease in intensity after interaction with the SMA polymer suggesting the appearance of bonds in gauche conformation [26]. The dichroic spectrum allows observation of an almost complete wagging series of 6 bands (normally n/2 wagging bands, where n= 14, the number of carbon in the aliphatic chain of DMPC), with positive deviation at 1202, 1228, 1258, 1280, 1304 and 1328 cm-1 (Figure 8). The dichroic ratio (i.e. the absorbance ratio of this band for spectra obtained with parallel and perpendicular polarized incident light) of the band at 1202 cm-1 can be calculated. A dichroic ratio of 5.7 is obtained for the DMPC bilayer, and increases to 11.7 for the SMALP disc, which suggests a straightening or a decreased mobility of the lipid acyl chains in the bulk phase of the nanoparticle (as this only concerns the acyl chains that are in all-trans conformation), potentially due to the constraint of the polymer scaffold. Similar behavior had previously been observed for lipids in HDL-like particles [28]. A decreased mobility of the lipids in SMALPs was also observed in a previous EPR study [12]. Although the authors attributed it to a decrease in lipid-chain gauche-trans isomerization, a straightening of the acyl chains could also explain their results.
Figure 8.
Dichroic spectrum of DMPC SMALPs. CH2 wagging bands are indicated by red lines.
Figure 9.
Shift of the C-H stretching bands in SMALPs. Black: IR spectrum of DMPC, Blue: IR spectrum of DMPC SMALPs. Red dashed lines have been added to better visualize the shift.
Figure 10.
decrease of the CH2 wagging bands in SMALPs. Black: IR spectrum of DMPC. Blue: IR spectrum of DMPC SMALPs.
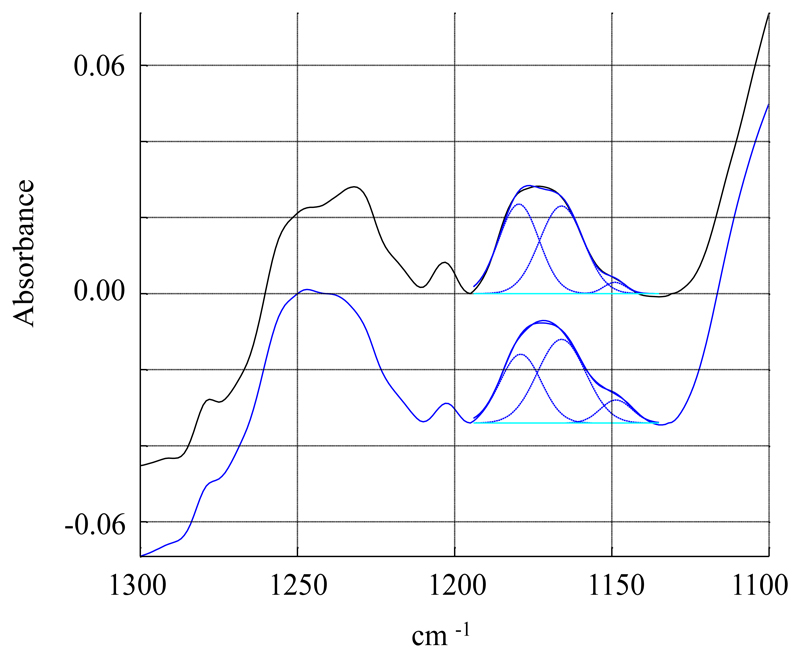
Further support for conformational constraints induced by the polymer can be deduced from the ester bond C-O stretching that absorbs either at 1180 cm-1 (planar conformation), or at 1165 cm-1 (gauche conformation). In a bilayer, the ester in sn-1 position is planar, and the ester in sn-2 position is gauche. Deconvolution of this peak into two components centered at 1180 and 1165 cm-1 respectively confirm this, as 48% is found in planar conformation and 52% is found in gauche conformation (Figure 11). On the contrary, in DMPC SMALPs, there is a decrease of the planar component to 41%, suggesting that some ester bonds may change slightly their conformation upon interaction with the polymer. Similar behavior has already been observed for lipids in HDL-like particles [28].
Figure 11.
Deconvolution of the ester C-O stretching band. Black: IR spectrum of DMPC. Blue: IR spectrum of DMPC SMALPs. Dashed blue lines represent the fitting of the two components at 1165 and 1180 cm-1.
3.4. Nuclear magnetic resonance spectroscopy
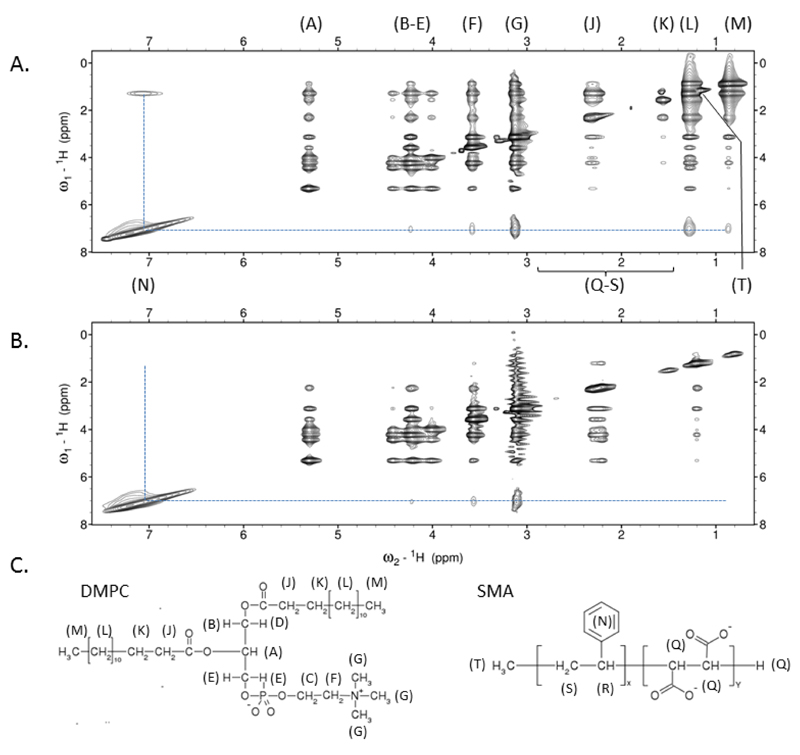
The results from the scattering and infrared experiments suggest that the polymer is closely associated with the acyl chains of the lipids protecting them from the polar solvent. If this was the case it would be predicted that the styrene moieties on the SMA could pack into the hydrophobic interior of the bilayer, although other arrangements could be envisaged. To detect the interactions a two dimensional NOESY experiment was performed (Figure 12). This experiment shows through-space interactions between nuclei within a distance of 5Å. For DMPC-SMALP strong proton NOEs were noted between the styrene of SMA and the acyl chain of DMPC (Positions L & M) confirming the styrene group is positioned closely to the DMPC acyl chains. Interestingly, further NOEs were observed to the choline head group of DMPC of similar intensity to that of the acyl terminal methyl (M), suggesting that the styrene group is likely intercalated approximately midway along the acyl chain, thus able to contact both ends of the DMPC molecule. Confirmation of acyl assignments was provided by utilizing acyl deuterated DMPC, in this situation all NOEs from the styrene to DMPC proton positions L and M were lost thus unambiguously showing that the styrenes of the polymer pack against the acyl chains of the lipid. Of note, very weak NOEs were observed from the methylene groups of DMPC to its headgroup in the D54-form of DMPC suggesting incomplete deuteration of the lipid acyl chains. This was later confirmed by 1D analysis of D-54 DMPC (Supplementary Figure 1).
Figure 12.
The SMA styrene groups in SMALPs show close proximity to the acyl chains of DMPC. (A) Homonuclear 2D dpfgse-NOESY of DMPC-SMALP in 50mM sodium phosphate, 200mM NaCl, pH 7 showing NOE interactions (dotted line) between the styrene group of SMA and DMPC. Signals for SMA groups Q-S are not observed due to the broadness of signal (B) As in (A) but using 1,2-dimyristoyl-d54-sn-glycero-3-phosphocholine. The absence of NOEs at positions L and M confirm the predominant NOEs observed in (A) are to the acyl chain CH2 groups and to a lesser extent the terminal CH3 of DMPC. (C) Schematic representation of SMA and DMPC with groups labelled according to (A).
3.5. Differential Scanning Calorimetry
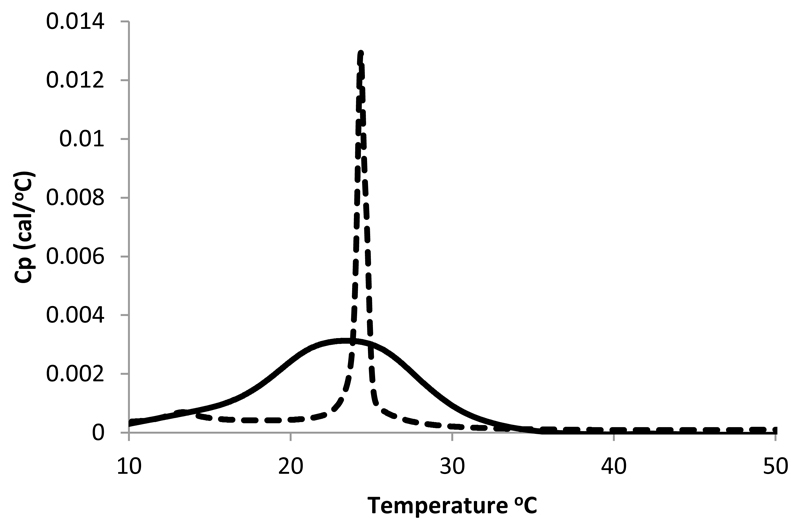
Results from SANS, NMR and polarized infrared spectroscopy suggest an intimate contact between the SMA and the lipid bilayer in SMALP particles. The polarized IR data indicates both that the styrene moiety is oriented in such a manner to suggest that it may intercalate between the acyl chains of the lipids. In addition, the signals for the acyl chains suggest that the presence of the SMA may induce some lipids to change their conformation. To assess whether the SMA has indeed disrupted the phase transition behavior of DMPC, differential scanning calorimetry (DSC) experiments were carried out on SMA (The SMA alone showed no transition, See supplementary figure 2) and the DMPC SMALP and the results compared to those for DMPC vesicles. DSC data from free DMPC bilayers (Figure 13 left) shows the presence of a clear transition at 24 °C which represents gel to liquid phase transition alongside a smaller transition around 13 °C. These observations agree well with those measured for DMPC in the past [29] [30]. Examination of the DSC data for the DMPC bilayer incorporated into the SMALP show clear perturbation of the gel to liquid transition behavior (Figure 13 right). In the SMALP the gel to liquid transition broadens significantly and the transitions temperature decreases marginally to 23 °C. A similar broadening of the gel-liquid transition has been observed for other lipid disc encapsulations systems. These have included studies of lipid discs stabilized by membrane stabilizing peptides (MSP) [9] and those stabilized by 3:1 SMA polymer [12]. In the MSP study a similar broadening of the liquid to gel phase transition is observed while the transition temperature of the MSP stabilized discs in this case increased to 28 °C. This change in behavior of the lipid phase transition is explained by Shaw et al. [9] as resulting from a loss of cooperativity within the phase transition perhaps resulting from the presence of a boundary of non-melting lipids on the interface with the polymer. For the 3:1 SMA polymer stabilized discs a lowering of the transition temperature of DMPC in SMALPs was also recorded. We interpret our data in the same way considering the identification of close interactions between the polymer and the lipid identified in previous sections. These interactions are likely to alter the dynamics of the lipids immediately adjacent to the polymer annulus thereby reducing the cooperative nature of the phase transition. Interestingly the studies of MSP noted that repeated scanning of the sample in the DSC (which involves repeated heating of the sample to 90°C) led to a re-appearance of the sharp transition observed for the non-encapsulated lipid [9]. This appearance of this transition could indicate the structural integrity of the disc has been disrupted by elevated temperature. To test whether a similar effect could occur with the SMALP system we carried out a similar experiment by repeatedly scanning the DSC instrument 35 times from 5-90 °C (data not shown). These data show no such return of the sharp transition. This indicates that, unlike the MSPs, the structural integrity of the SMALPs is resistant to elevated temperature. We note that that unlike our observations with the 2:1 SMA polymer, Orwick et al. [12] observed a lowering of the transition temperature of DMPC in SMALPs made using 3:1 SMA.
Figure 13.
DSC data for DMPC vesicles (dotted line) showing two transitions at 14 °C and 24 °C and of the SMALP (solid line) showing a broadening of the gel to liquid transition (23 °C).
4. Discussion
Our membrane protein extraction method based on styrene maleic acid co-polymer [11] resolved one of the major limitations encountered with other nano-encapsulation methods. The SMALP method does not require a detergent to extract the membrane protein from the biological membrane before its reinsertion into the membrane nanoparticle and can be used to understand the structure and function of membrane proteins. However before these studies can be undertaken the physical characteristics of the SMALP have to be determined in order to optimize and engineer their properties and capabilities.
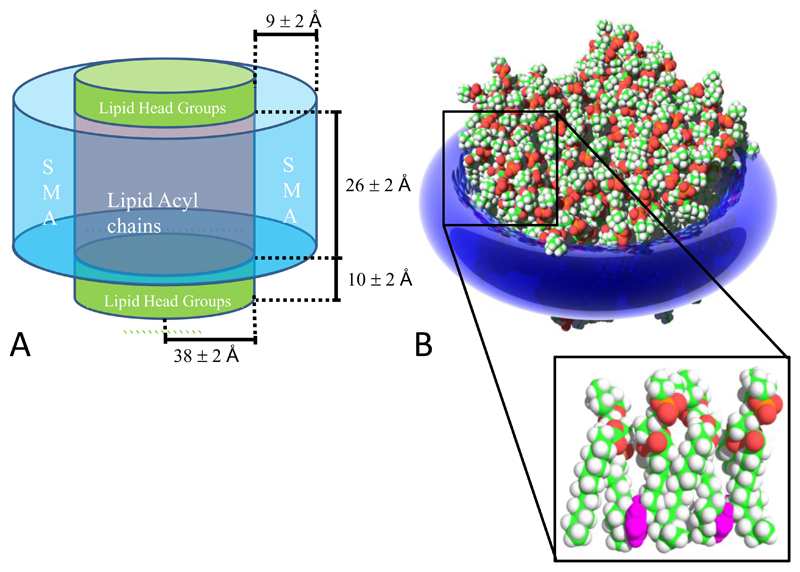
In this study we present a more detailed structure of the SMALP (Figure 14). These data show that the structure of SMALPs produced from DMPC bilayer is formed by SMA encircling the edge of the membrane disc. The study also shows that the geometric structure of the membrane within the disc is maintained. This is essential if the isolated membrane is going to provide an environment that mimics the native membrane lamella. Analysis of the size distribution of the disc showed a bimodal distribution of diameters centered on ~11 and ~16 nm. These results can be compared well with a similar electron microscopy study of SMALPs made using the 3:1 polymer which showed that diameter of the particles varied between 5-15 nm [12]. Our observation of bimodality may also indicate something about the how the polymer interacts with the bilayer during formation. For example it may mean that there are discrete numbers of lipids that can be stabilized by the SMA to form a viable disc.
Figure 14.
A) The dimensions of a SMALP as measured using SANS. B) A model showing the orientation and interactions of the benzene moieties (purple) within SMALPS
The dimensions of the SMALP defined by the SANS experiments provides a clear picture of the partition of space in the particle between the polymer and lipid. These data allow an estimation of the number of lipids that are encapsulated within the particle. If it is assumed that the area of each DMPC molecule within the bilayer is 59.6 Å2 [23] then each leaflet of the bilayer within the SMALP contains approximately 70 DMPC molecules or 140 in the bilayer. This value is similar to figures calculated for the MSP stabilized lipid discs developed by Sligar and colleagues [9]. These data also allow us to assess the size of a membrane protein that can be included in a SMALP. Assuming a maximum radius for the membrane in the SMALP of 40 Å and for an α-helix of 2.7 Å, a single SMALP would have the potential to encapsulate up to 40 transmembrane helices.
Both SANS and ATR-FTIR measurements suggest that the SMA polymer takes the form of a “bracelet” encircling the lipid membrane with the styrene moieties oriented parallel to the membrane normal. NMR confirms that the styrene moieties interact directly with the acyl chains of the lipids and it is likely that they intercalate approximately midway along the acyl chains. Our data also shows that the maleic acid groups are oriented in the same direction as the styrene moieties but are likely to interact with the lipid headgroups confirming the work of Orwick et al [12]. These data suggest that one of the major driving forces for the spontaneous formation of the SMALPs is likely to be the burial of the styrene groups into the hydrophobic core of the membrane.
The picture of interactions between the polymer and lipid poses the question as to whether the lipid membrane is still able to effectively mimic that of a lipid bilayer. To examine this we have carried out a study of the membrane phase transitions using DSC. These data indicate that like other disc systems the main phase transition is broadened indicating a loss of cooperativity. This is likely to be produced by the interactions between the lipids and the polymer. We also observe a perturbation of the transition temperature by 1 °C. This contrasts with calorimetry data reported by Orwick et al. [12] which indicated that the 3:1 SMA caused the transition temperature to reduce considerably (by approximately 10 °C) while a study of the MSP system reported an increase in transition temperature of 3 °C. These changes in transition temperature further indicate a perturbation of the lipid bilayer. The lowering of the transition temperature in the SMA 3:1 discs is significant and could be of some concern if membrane fluidity is important in the conformational flexibility of any proteins contained in the encapsulated membrane. This is particularly important for proteins where a conformational change is important for function (i.e. G protein coupled receptors). Our DSC study also indicated that the structure of the SMALP is resistant to repeated exposure to elevated temperatures when compared to the MSP system. This observation suggests that the SMA systems may provide a more resilient encapsulation method compared to that offered by the MSP. Interestingly, the differences observed between the two SMA polymers so far studied offers the intriguing possibility that other formulations of the SMA polymer may provide SMALPs with different physical characteristics. It is therefore important that further studies are carried out to assess other SMA polymers with different molecular weights and compositions that may offer an improved solution to this important problem.
Taken together our study provides useful information on the structure of the SMALP for those planning on using nanoparticle systems to study proteins and membranes, and highlights the distinguishing properties that offer advantages for native state solubilization and characterization.
Supplementary Material
Acknowledgements
TD, MO, MJ, TK, NS, YPL, RF and KJE would like to acknowledge the support of the BBSRC through grants BB/G010412/1, BB/I020349/1, BB/I019960/1, BB/I013865 and BB/J017310/1 and Follow on Funding. VG and CG were supported by the F.R.S. – FNRS. KJE and II acknowledge the ILL for provision of beamtime on D11 (experiment 9-13-344) and the assistance of Dr Ralf Schweins in collecting and reducing the SANS data. II acknowledges the STFC BioMemembrane Network (BMN10-01) and the University of Bath for funding. We thank Erik Goormaghtigh for discussions about interpretation of the IR data. We thank Cameron Neylon for discussions about interpretation of the SANS data
References
- 1.L CY, K S. Infrared Spectra of High Polymers. VI. Polystyrene. Journal of Polymer Science. 1958;27:241–254. [Google Scholar]
- 2.Lichtenberg D, et al. Detergent solubilization of lipid bilayers: a balance of driving forces. Trends in biochemical sciences. 2013;38(2):85–93. doi: 10.1016/j.tibs.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 3.Bayburt TH, Carlson JW, Sligar SG. Reconstitution and imaging of a membrane protein in a nanometer-size phospholipid bilayer. Journal of structural biology. 1998;123(1):37–44. doi: 10.1006/jsbi.1998.4007. [DOI] [PubMed] [Google Scholar]
- 4.Borch J, et al. Nanodiscs for immobilization of lipid bilayers and membrane receptors: kinetic analysis of cholera toxin binding to a glycolipid receptor. Analytical chemistry. 2008;80(16):6245–52. doi: 10.1021/ac8000644. [DOI] [PubMed] [Google Scholar]
- 5.Bayburt TH, Sligar SG. Membrane protein assembly into Nanodiscs. FEBS letters. 2010;584(9):1721–7. doi: 10.1016/j.febslet.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denisov IG, et al. The ferrous-dioxygen intermediate in human cytochrome P450 3A4. Substrate dependence of formation and decay kinetics. The Journal of biological chemistry. 2006;281(33):23313–8. doi: 10.1074/jbc.M605511200. [DOI] [PubMed] [Google Scholar]
- 7.Leitz AJ, et al. Functional reconstitution of Beta2-adrenergic receptors utilizing self-assembling Nanodisc technology. BioTechniques. 2006;40(5):601–2. doi: 10.2144/000112169. 604, 606, passim. [DOI] [PubMed] [Google Scholar]
- 8.Shaw AW, et al. The local phospholipid environment modulates the activation of blood clotting. The Journal of biological chemistry. 2007;282(9):6556–63. doi: 10.1074/jbc.M607973200. [DOI] [PubMed] [Google Scholar]
- 9.Shaw AW, McLean MA, Sligar SG. Phospholipid phase transitions in homogeneous nanometer scale bilayer discs. FEBS letters. 2004;556(1–3):260–4. doi: 10.1016/s0014-5793(03)01400-5. [DOI] [PubMed] [Google Scholar]
- 10.Tonge SR, Tighe BJ. Responsive hydrophobically associating polymers: a review of structure and properties. Advanced drug delivery reviews. 2001;53(1):109–22. doi: 10.1016/s0169-409x(01)00223-x. [DOI] [PubMed] [Google Scholar]
- 11.Knowles TJ, et al. Membrane proteins solubilized intact in lipid containing nanoparticles bounded by styrene maleic acid copolymer. Journal of the American Chemical Society. 2009;131(22):7484–5. doi: 10.1021/ja810046q. [DOI] [PubMed] [Google Scholar]
- 12.Orwick MC, et al. Detergent-free formation and physicochemical characterization of nanosized lipid-polymer complexes: Lipodisq. Angewandte Chemie. 2012;51(19):4653–7. doi: 10.1002/anie.201201355. [DOI] [PubMed] [Google Scholar]
- 13.Orwick-Rydmark M, et al. Detergent-free incorporation of a seven-transmembrane receptor protein into nanosized bilayer Lipodisq particles for functional and biophysical studies. Nano letters. 2012;12(9):4687–92. doi: 10.1021/nl3020395. [DOI] [PubMed] [Google Scholar]
- 14.Long AR, et al. A detergent-free strategy for the reconstitution of active enzyme complexes from native biological membranes into nanoscale discs. BMC biotechnology. 2013;13:41. doi: 10.1186/1472-6750-13-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bechinger B, Ruysschaert JM, Goormaghtigh E. Membrane helix orientation from linear dichroism of infrared attenuated total reflection spectra. Biophysical journal. 1999;76(1 Pt 1):552–63. doi: 10.1016/S0006-3495(99)77223-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dalvit C, Ramage P, Hommel U. Heteronuclear X-filter 1H PFG double-quantum experiments for the proton resonance assignment of a ligand bound to a protein. Journal of magnetic resonance. 1998;131(1):148–53. doi: 10.1006/jmre.1997.1342. [DOI] [PubMed] [Google Scholar]
- 17.Hwang TL, Shaka AJ. Water Suppression That Works. Excitation Sculpting Using Arbitrary Wave-Forms and Pulsed-Field Gradients. J Magn Resn. 1995;A112:275–279. [Google Scholar]
- 18.Delaglio F, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. Journal of biomolecular NMR. 1995;6(3):277–93. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 19.Goddard TD, Kneller DG. SPARKY 3. 2004.
- 20.Kline SR. Reduction and analysis of SANS and USANS data using IGOR Pro. Journal of Applied Crystallography. 2006;39:895–900. [Google Scholar]
- 21.Hayter JB, Penfold J. An Analytic Structure Factor for Macroion Solutions. Mol Phys. 1981;42(1):109–118. [Google Scholar]
- 22.Smith MB, et al. Neutron reflectometry of supported hybrid bilayers with inserted peptide. Soft Matter. 2010;6(5):862–865. doi: 10.1039/b915800f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagle JF, Tristram-Nagle S. Structure of lipid bilayers. Biochimica et biophysica acta - Reviews on Biomembranes. 2000;1469(3):159–195. doi: 10.1016/s0304-4157(00)00016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goormaghtigh E, Raussens V, Ruysschaert JM. Attenuated total reflection infrared spectroscopy of proteins and lipids in biological membranes. Biochimica et biophysica acta. 1999;1422(2):105–85. doi: 10.1016/s0304-4157(99)00004-0. [DOI] [PubMed] [Google Scholar]
- 25.Raussens V, et al. Alignment of the apolipophorin-III alpha-helices in complex with dimyristoylphosphatidylcholine. A unique spatial orientation. The Journal of biological chemistry. 1995;270(21):12542–7. doi: 10.1074/jbc.270.21.12542. [DOI] [PubMed] [Google Scholar]
- 26.Fringeli UP, Gunthard HH. Infrared membrane spectroscopy. Molecular biology, biochemistry, and biophysics. 1981;31:270–332. doi: 10.1007/978-3-642-81537-9_6. [DOI] [PubMed] [Google Scholar]
- 27.Lewis RN, Pohle W, McElhaney RN. The interfacial structure of phospholipid bilayers: differential scanning calorimetry and Fourier transform infrared spectroscopic studies of 1,2-dipalmitoyl-sn-glycero-3-phosphorylcholine and its dialkyl and acyl-alkyl analogs. Biophysical journal. 1996;70(6):2736–46. doi: 10.1016/S0006-3495(96)79843-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wald JH, et al. Investigation of the lipid domains and apolipoprotein orientation in reconstituted high density lipoproteins by fluorescence and IR methods. The Journal of biological chemistry. 1990;265(32):20044–50. [PubMed] [Google Scholar]
- 29.Heimburg T. A model for the lipid pretransition: coupling of ripple formation with the chain-melting transition. Biophysical journal. 2000;78(3):1154–65. doi: 10.1016/S0006-3495(00)76673-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blume A. Apparent molar heat capacities of phospholipids in aqueous dispersion. Effects of chain length and head group structure. Biochemistry. 1983;22:5436–5442. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.