Abstract
The cyclin dependent kinase (CDK) –retinoblastoma (RB) -E2F pathway plays a critical role in the control of cell cycle in estrogen receptor positive (ER+) breast cancer. Small molecule inhibitors of CDK4/6 have shown promise in this tumour type in combination with hormonal therapies, reflecting the particular dependence of this subtype of cancer on cyclin D1 and E2F transcription factors. mTOR inhibitors have also shown potential in clinical trials in this disease setting. Recent data has suggested cooperation between the phosphatidylinositol 3-kinase (PI3K)/mTOR pathway and CDK4/6 inhibition in preventing early adaptation and eliciting growth arrest, but the mechanisms of the interplay between these pathways have not been fully elucidated. Here we show that profound and durable inhibition of ER+ breast cancer growth is likely to require multiple hits on E2F mediated transcription. We demonstrate that inhibition of mTORC1/2 does not affect ER function directly, but does cause a decrease in cyclin D1 protein, RB phosphorylation and E2F mediated transcription. Combination of an mTORC1/2 inhibitor with a CDK4/6 inhibitor results in more profound effects on E2F dependent transcription, which translates into more durable growth arrest and a delay to the onset of resistance. Combined inhibition of mTORC1/2, CDK4/6 and ER delivers even more profound and durable regressions in breast cancer cell lines and xenografts. Furthermore, we show that CDK4/6 inhibitor resistant cell lines re-activate the CDK-RB-E2F pathway, but remain sensitive to mTORC1/2 inhibition, suggesting that mTORC1/2 inhibitors may represent an option for patients that have relapsed on CDK4/6 therapy.
Introduction
Hormone receptor positive (HR+) breast cancer is the most frequently occurring breast cancer subtype. Patients with HR+ advanced breast cancer typically respond well to endocrine therapy (1), but drug resistance remains a clinical challenge in this disease. Recent advances in elucidating the molecular mechanisms of pathway ‘cross-talk’ between the estrogen receptor (ER), cell cycle regulation and intracellular signalling pathways, such as the mTOR or the CDK-RB-E2F pathway, have provided the rationale for combining endocrine therapies with targeted agents (2–6).
The mammalian target of rapamycin (mTOR) pathway is frequently hyper-activated in estrogen receptor positive (ER+) breast cancer and a number of clinical studies have shown benefit from combining inhibition of mTOR with estrogen receptor targeting therapies (3, 4, 7, 8). The serine/threonine kinase mTOR integrates a wide variety of cellular signals, including mitogen and nutrient signals to control cell proliferation, cell cycle and cell size. mTOR kinase forms two distinct multiprotein complexes called mTORC1 and mTORC2. The distinct cellular functions of the two mTOR complexes are regulated by the presence of a number of different subunits, which define the assembly, sub-cellular localization, substrate binding and unique functions of mTORC1 and mTORC2 (9, 10). One of the inputs on the modulation of mTOR is the PI3K/AKT pathway which has been shown to activate the mTORC1 complex. In response to nutrient and growth factor availability, mTOR can activate catabolic processes, suppress autophagy and control protein translation. Moreover, mTOR orchestrates cell growth by stimulating anabolic pathways such as nucleotide and lipid synthesis (11, 12). Inhibition of both mTORC1 and 2 is hypothesised to be effective at inhibiting a broad range of mTOR functions, via inhibition of downstream substrates such as ribosomal protein S6, 4EBP1 and AKT (9, 10).
In ER+ breast cancer, the functional relationship between estrogen receptor signalling and mTOR has not been elucidated. A reciprocal feedback mechanism between PI3K and estrogen receptor has been suggested, whereby inhibition of PI3K results in an increase in estrogen receptor levels in the endocrine resistance setting (13). However, these reciprocal feedback mechanisms have not been demonstrated between mTORC1/2 and estrogen receptor to date. Furthermore, recent analysis of patients that have responded to the mTORC1 inhibitor everolimus, combined with the aromatase inhibitor exemestane has shown that progression free survival benefit with everolimus was maintained regardless of alteration status of any components of the PI3K pathway (7).
In addition to the mTOR pathway, endocrine resistance has often been associated with activation of CDK-RB-E2F signalling. The importance of this pathway in ER+ breast cancer is underscored by the frequent genomic aberrations in a number of components of this network. Cyclin dependent kinases (CDKs) are serine threonine kinases that modulate cell cycle progression. CDK4 and CDK6 together with D-type cyclins and cyclin E/CDK2 complexes control the commitment to cell cycle entry from quiescence and the G1 phase. These kinase complexes can phosphorylate RB, releasing the transcription factors E2F and modulating the expression of E2F target genes that are required for S phase entry (14–17). E2Fs are an evolutionarily conserved family of transcription factors that includes ten different proteins encoded by eight distinct genes. Their regulation and function is complex and highly context dependent. Mechanistically, phosphorylation of RB proteins by CDKs disables their function as transcriptional repressors and allows the activation of the E2F transcriptional program. These processes are negatively regulated by p15INK4 and p16INK4 proteins, which block the formation and activation of the cyclin D/CDK4/6 complexes (14–18). A number of CDK4/6 inhibitors, including palbociclib are now being investigated in clinical trials in ER+ breast cancer and have been approved by the FDA in this setting (5, 19, 20).
In pre-clinical models, inhibition of mTORC1/2 or CDK4/6 has been shown to result in a cytostatic phenotype and it remains unclear as to whether the effects elicited by single agent treatments targeting these pathways can result in effective long term control of tumour growth. Inhibition of either mTORC1/2 or CDK4/6 in combination with the ER down-regulator fulvestrant have also been shown to enhance efficacy in breast cancer models (21, 22). Moreover, combinations of therapies targeting PI3-kinases have been shown to synergise with CDK4/6 inhibitors through blockade of early adaptation (23–25) and these combinations have been proposed as a potential therapeutic modality to achieve more durable responses.
Here we show that inhibition of mTOR with the dual mTORC1/2 inhibitor vistusertib (AZD2014) (26), does not modulate ER binding to DNA, suggesting a different mechanism of interaction with the ER pathway to that described previously for PI3K inhibitors (13). Instead, we find that mTORC1/2 inhibition causes modulation of E2F mediated transcription and cooperates with the CDK4/6 inhibitor palbociclib in the inhibition of E2F function in estrogen receptor positive (ER+) breast cancer cells. The combination of the two agents does not result in an exacerbation of the senescence-like phenotype caused by palbociclib alone, but instead results in a prolonged and durable quiescent-like state. Finally, we demonstrate that the combination of inhibitors targeting the three independent pathways prevents the emergence of resistance and causes durable regressions in breast cancer xenografts. The data suggests cross talk between mTOR, CDK-RB-E2F and estrogen receptor pathways to control cell cycle progression.
Material and Methods
Cell lines and cell culture
All cell lines were maintained at 37°C and 5% CO2 in a humidified atmosphere. MCF-7 cells were grown in RPMI-1640 (phenol red-free, SIGMA) supplemented with 10% FCS and 2mmol/L glutamine. HCC-1428 cells were grown in MEM (Richter’s modification; phenol red-free, Life Technologies) supplemented with 10% FCS and 2mmol/L glutamine. MCF-7 LTED cells were grown in RPMI-1640 (phenol red-free, SIGMA) supplemented with 5% charcoal-stripped FCS and 2mmol/L glutamine. HCC-1428 LTED cells were grown in MEM (Richter’s modification; phenol red-free, Life Technologies) supplemented with 5% charcoal-stripped FCS and 2mmol/L glutamine. HCC-1428/LTED were obtained from C. Arteaga and grown as described in (26). All other cell line details and associated cell line identification procedures are summarized in Guichard et al (26). All cell lines were authenticated at AstraZeneca cell banking using DNA fingerprinting short-tandem repeat (STR) assays as previously described (26). MCF-7 cell line authentication date (STR fingerprinting): September 2015. MCF-7 LTED cell line authentication date (STR fingerprinting): September 2015. HCC-1428 cell line authentication date (STR fingerprinting): June 2015. HCC-1428 LTED cell line authentication date (STR fingerprinting): June 2015.
Compounds
AZD2014, palbociclib, and fulvestrant were obtained from AstraZeneca compound collection. RAD001 was purchased from Selleckchem. Staurosporine was purchased from Sigma. AZD2014, palbociclib and fulvestrant were dissolved in DMSO to a concentration of 10 mmol/L and stored under nitrogen.
Antibodies
Antibodies used for S6, p-S6 S235/236, P70S6K, p-P70S6K T389, 4EBP1, p-4EBP1S65, AKT, p-AKTS473, RB, p-RBS780, CDC6, E2F-1, TK1 and p21 were purchased from Cell Signalling Technology. RAD51 antibody was purchased from Santa Cruz. Cyclin D1 antibody was purchased from Abcam and vinculin antibody from SIGMA.
In vitro cell proliferation measurements
To measure cell confluency, 2000 cells/well were seeded in 96-well plates (Costar) and dosed with compounds 24 hours later. Cell confluency was monitored at 4 hourly intervals for the duration of the experiment using an Incucyte Zoom platform with 10x objective (Essen Bioscience).
For live cell number analysis, one thousand MCF7 cells were seeded in 100 µl RPMI 1640, in each well of a 96 well clear bottom black plate (Costar), one plate per time point. Plates were incubated at 37°C, 5% CO2 for 24 hours prior to being treated with vehicle, AZD2014 or Palbociclib either alone or in combination. For long term experiments, medium was changed once a week and fresh compound was added. Number of live cells were determined at day 0, 1, 2, 3 and then twice weekly for 24 days using a sytox green assay. Briefly, 5mM sytox green nucleic acid dye (Invitrogen) was diluted 1:2500 in 500 mM TBS/EDTA solution and 7µl was added per well. Plates were then incubated in the dark at room temperature for 1 hour and the number of green cells in each well (dead cells) was measured using an Acumen Explorer high-throughput cell imager (TTP Labtech, Melbourn, UK). Next, 14µl of 0.25% w/v Saponin in TBS/EDTA solution was added overnight to permeabilise the cells therefore allowing a total cell count. Number of live cells was calculated by subtracting dead cell count from the total cell count.
Immunoblotting
Expression levels of total and phosphorylated protein were assessed using standard Western blotting techniques (NuPAGE Novex 4%-12% Bis-Tris gels). Cells were lysed in Pierce RIPA buffer Thermo Scientific), supplemented with Halt™ protease and phosphatase inhibitors (Thermo Scientific). Antibodies were diluted in 4% milk-PBS-Tween and signal detected using SuperSignal West Dura HRP substrate followed by visualization on a Syngene ChemiGenius Imager.
Selection of E2F dependent genes
Candidate E2F responsive genes were selected for transcriptional profiling on the basis of their reported association in the literature with E2F family transcription factors (28–31). A subset comprising 43 E2F dependent genes common to these reports was subsequently prioritised for further investigation (Supplementary Table S1).
Gene expression analysis
RNA was isolated from cell lines using the RNeasy MiniKit (Qiagen-RLT Buffer), with an additional DNAse treatment step, following the manufacturer’s protocol. Reverse transcription was performed using 50 ng of RNA with the High Capacity cDNA Reverse transcription kit (Applied Biosystems), following manufacturer’s instructions. A number of E2F, ER or FOXO modulated genes were selected from publications (27–30) and internal data (Supplementary Table S1). Targeted gene profiling was performed using the Fluidigm platform and cDNA was pre-amplified (14 cycles) using a pool of TaqMan primers (Life Technologies), following manufacturer’s instructions. Sample and assay preparation of the 96.96 Fluidigm Dynamic arrays was carried out according to the manufacturer’s instructions. Data was collected and analyzed using the Fluidigm Real-Time PCR Analysis 2.1.1 software.
Gene expression values and statistical analysis were calculated in Jmp-13 software and data represented in TIBCO™ Spotfire® 6.5.2. Data (Ct) were normalised to the average of the house keeping genes (ACTB, GAPDH, HPRT1, IPO8, PPIA, UBC, YWHAZ) to generate dCt; negative ddCt was calculated by subtracting treated dCt to DMSO dCt per matching treatment time point (negative ddCt-log2 fold change), and fold change after log2 transformation (2^ neg ddCt). Two technical replicates for each of the three biological replicate experiments were run and the mean and standard error of each group was calculated. A two-sided pairwise t-test was performed and data filtered comparing each treated group to DMSO control, and each combination group to the single agents with matching time points.
Beta-galactosidase staining
Beta-galactosidase staining was carried out as previously described (31). Briefly, cells were plated in 96-well plates and 8 days after compound treatment, cells were exposed to 100 nmol/L Bafilomycin A1 for 1h, followed by 2mmol/L C12FDG for a further 2h. Cells were then fixed with 4% formaldehyde, counterstained with Hoechst 33342 and analysed using the Cell Insight (Thermo Fisher Scientific). DMSO-treated cells and cells receiving doses of compound that were not cytostatic were re-plated 24-48 hours before Bafilomycin A1 treatment to achieve equal confluency in all samples. For image analysis, the cut off for low beta-gal activity was set to include over 90% of the DMSO population.
ChIP-seq
MCF7 cells, cultured in DMEM supplemented with 10% FBS, 1% pen/strep and 2mM L-glutamine, were treated for 2 hours with 500 nM RAD001, vistusertib (AZD2014), or vehicle (DMSO) at approximately 70-80% confluency. Cells were crosslinked by 1% formaldehyde for 10 min at RT, and the formaldehyde was subsequently quenched by 0.1M Glycine. Chromatin was sonicated using the Diagenode Bioruptor so the bulk of the DNA was approximately 100-500 bp. Immunoprecipitation was done overnight using a polyclonal ER antibody (sc-543, Santa Cruz). Beads were subsequently washed six times in RIPA buffer (50mM HEPES pH=7.6, 1mM EDTA, 0.7% Sodium deoxycholate, 1% NP-40, 0.5M LiCl), once in TE buffer and then eluted/de-crosslinked in elution buffer (50mM Tris-HCl, pH=8, 10mM EDTA, 1% SDS) ON at 65 °C. Samples were treated with RNase (Ambion) and protease K (Invitrogen), and DNA was purified by phenol/chloroform extraction. ChIP’ed DNA was subjected to Illumina sequencing on the HiSeq 2500 platform according to the manufacturer’s instructions. Sequence reads (50 bp) were mapped to the human genome (hg38) using Bowtie2 (32), peaks were identified by MACS2 (33) using reads from all replicates, and differential binding sites were identified by DiffBind (Stark R, Brown G. DiffBind : differential binding analysis of ChIP-Seq peak data. 2011; http://bioconductor.org/packages/devel/bioc/vignettes/DiffBind/inst/doc/DiffBind.pdf). De novo motif analyses were performed using MEME (version 4.9.1). Four independent biological replicates were performed. The data is now available in GEO (GSE103023).
Cell cycle distribution
MCF-7 cells were seeded in 25cm2 flasks at 4x105 cells/flask and allowed to adhere overnight before being dosed with vistusertib, alone or in combination with palbociclib at the doses indicated. After 6 or 24 hours compound treatment, media and cells were collected, and after washing in PBS cells were fixed in ice cold 70% ethanol for at least 24 hours. Cells were treated with 100ug/ml RNase (Sigma) and stained with 50ug/ml propidium iodide (Life Technologies) for 20 mins before being analysed on the BD FACS Calibur (BD Bioscience) collecting a total of 10,000 events. FL2-A was plotted as a histogram and gates established to calculate the proportion of cells in each stage of the cell cycle (G1, S and G2/M).
Annexin V assay
MCF7 cells were seeded into 96 well black with clear bottom assay plates at a density of 3000 cells per well. Twenty-four hours after incubation at 37°C/ 5% CO2, cells were treated with palbociclib, AZD2014 and/or fulvestrant using a HP D300 digital dispenser. Staurosporine was included as a positive control. In order to determine the percentage of apoptotic cells at each time point, cells were stained with Annexin V (CF568 conjugate, Biotium) and Hoechst 33258 (Life Technologies) at final dilutions of 1 in 500 and 1 in 1000 per well, respectively. Cells were subsequently analysed with a Cell Insight High Content Platform. Control wells were stained with Hoechst 33258 only.
In vivo xenograft experiments
Studies in MCF-7 xenograft models were performed at AZ and according to local regulations (Home Office UK), as previously described (26). Briefly, male SCID mice (Charles River UK) were implanted s.c. with a 0.5mg 21 day estrogen pellet then 24 hours later 5x106 MCF-7 cells (ICRF, London) in 50% Matrigel (BD Bioscience) in a 0.1ml injection volume were given s.c. in the left flank.
For efficacy studies, mice were randomized into groups of 8 to 15 when average tumour volume reached 0.3 to 0.4 cm3. Group sizes were determined by a statistical power calculation using historical data on variability of tumour growth for the cell line, typically set using a 40% TGI level. Mice were dosed for 3 or more weeks at defined doses and schedules as indicated in the figures. AZD2014 was orally dosed alone either at 7.5 mg/kg once daily or 10mg/kg twice daily on days 1 and 2, BID of a weekly cycle in 1% polysorbate. Fulvestrant was dosed at 5 mg per mouse, on day 1, 3, 5 each week in peanut oil subcutaneously. Palbociclib was given orally at 50 mg/kg once daily, orally in 1% polysorbate. Plasma samples were taken at the end of the study to measure PK and ensure we reached the expected exposure during the dosing period. Tumours were measured two to three times weekly by caliper and volume calculated using a formula assuming an elliptical tumour shape (pi/6 × width × width × length). Tumour growth inhibition (%TGI) from the start of treatment was assessed by comparison of the geometric mean change in tumour volume for the control and treated groups. Tumour regression was calculated as the percentage reduction in tumour volume from baseline (pre-treatment) value: % Regression = (1 − RTV) ×100 % where RTV = Geometric Mean Relative Tumour Volume. Statistical significance was evaluated using a two-tailed t test.
CTC174 (ER/PR positive, HER2 negative model) studies were conducted following implantation of a tumour fragment in female NSG mice (Jackson Laboratory). Tumour fragments were obtained by collecting a tumour from a donor mouse and cutting it into 50 mm3 pieces. Fragments were implanted orthotopically adjacent to mammary fat pad. The 0.18 mg 90 day 17β-estradiol pellets (Innovative Research of America) and were implanted at the same time into dorsal scapular region. Anaesthesia was maintained with isoflurane during the surgical procedures. Staples (9 mm) were used to close all incision sites and were removed 1 week after surgery. Mice were randomized into control and treatment groups once tumour sizes reached approximately 150mm3. All procedures were carried out in accordance with UK Home Office regulations and approved by an Institutional Animal Care and Use Committee (IACUC).
Results
mTOR modulates the CDK-RB-E2F pathway and E2F dependent gene transcription
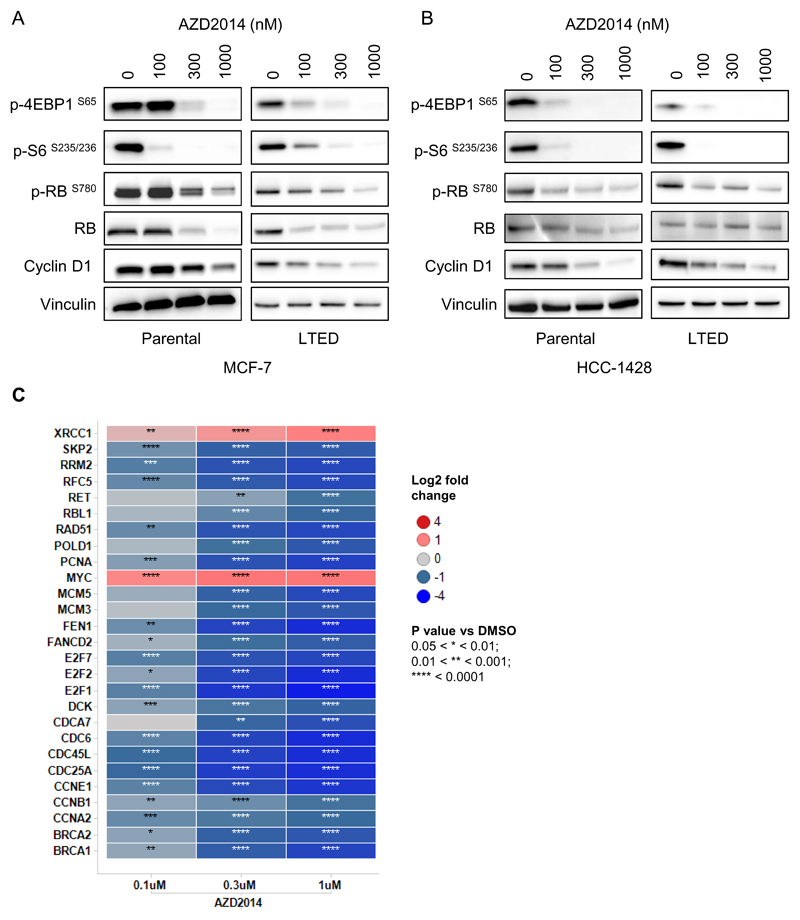
We have previously described that treatment of breast cancer cell lines with vistusertib (AZD2014) inhibits the phosphorylation of both mTORC1 and mTORC2 substrates (26). In this study, we also found that vistusertib (AZD2014) caused a decrease in the phosphorylation of RB at Serine 780 in a dose dependent manner in MCF-7 cells (Figure 1A). This was further confirmed in an additional ER positive cell line, HCC-1428 (Figure 1B) and in their long term estrogen deprived (LTED) variants, which have been described as a model of resistance to aromatase inhibitors (Figure 1A and B). The effects on RB phosphorylation were also measured 6 hrs following compound addition, suggesting that the effects on RB phosphorylation do not occur as a result of cell cycle arrest (Supplementary Figure S1). The effects on RB phosphorylation and cyclin D1 levels appeared to be consistent with the effects observed on the phosphorylation of 4EBP1, whilst effects on phosphorylation of the ribosomal S6 protein were evident at lower concentrations of vistusertib (AZD2014).
Figure 1. Vistusertib (AZD2014) inhibits E2F mediated gene transcription.
Effects of vistusertib (AZD2014) treatment (24 hours) on RB and mTORC1/2 substrates measured by Western blotting in the ER+ cell lines MCF-7 (A) and HCC-1428 (B). (C) Targeted gene expression profiling of 96 genes in MCF-7 cells, including 43 E2F dependent genes. Heatmap shows the 27 E2F dependent genes with a significant fold change modulation vs DMSO (-2 < fold change <2) and a p-value <0.05.
Since the CDK-RB-E2F pathway controls E2F mediated gene transcription, we assessed the effect of vistusertib (AZD2014) on the expression of a panel of E2F dependent genes by qPCR. Treatment of MCF-7 cells with vistusertib (AZD2014) caused changes in the mRNA levels of 27 out of 43 (63%) of a selected set of E2F target genes analysed (Figure 1C), again suggesting an effect of vistusertib (AZD2014) on the CDK-RB-E2F signalling axis.
The combination of the mTORC1/2 inhibitor vistusertib (AZD2014) and the CDK4/6 inhibitor palbociclib results in a decrease in growth and a decrease in E2F dependent gene transcription
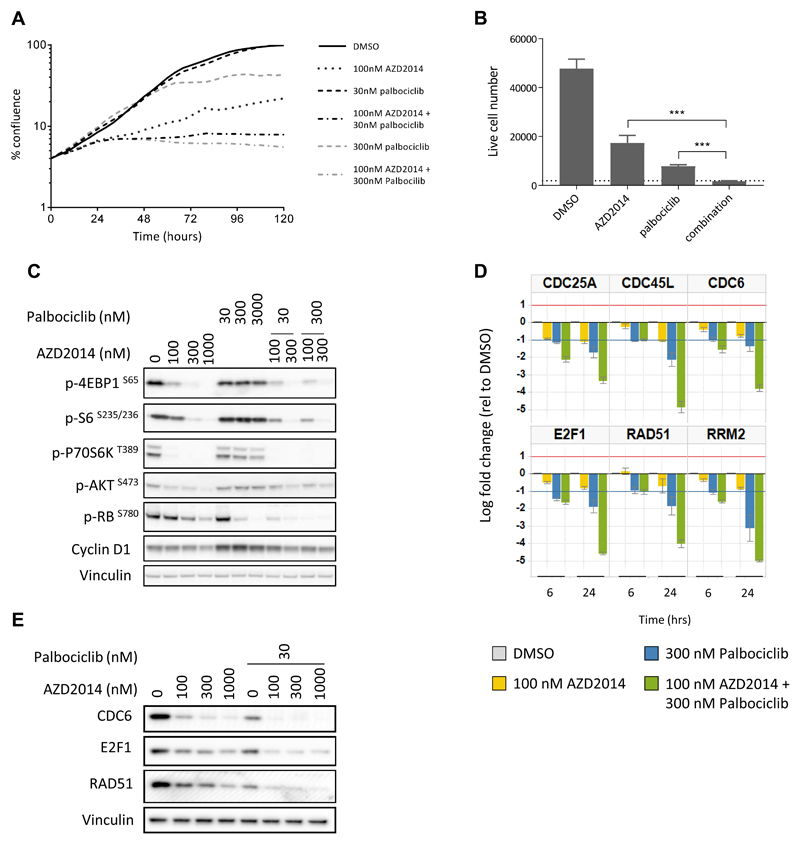
Following the discovery that vistusertib (AZD2014) treatment caused inhibition of the CDK-RB-E2F pathway, we investigated whether the CDK4/6 inhibitor palbociclib, would combine with vistusertib (AZD2014) to inhibit proliferation. Neither vistusertib (AZD2014) nor palbociclib, at either 30 or 300nM, caused complete growth inhibition as a monotherapy. However, both concentrations of palbociclib enhanced the growth inhibitory effect of vistusertib (AZD2014), resulting in complete loss of proliferation over the five day time course (Figure 2A). We confirmed these findings using a sytox green assay, which allows a more direct assessment of live cell number (Figure 2B), where the combination of 300nM palbociclib and 100nM vistusertib (AZD2014) caused a significant decrease in live cell number compared to either monotherapy alone. The allosteric mTORC1 inhibitor everolimus (RAD001) had a similar effect on cell number when used alone or in combination with palbociclib (Supplementary Figure S2).
Figure 2. Combined vistusertib (AZD2014) and palbociclib treatment results in more profound growth inhibition and enhanced CDK-RB-E2F pathway modulation.
(A) The effect of vistusertib (AZD2014) and palbociclib, alone or in combination, on the growth of MCF-7 cells over 120 hours, measured using an Incucyte Zoom. (B) The effect of vistusertib (AZD2014, 100nM) and palbociclib (300nM), alone or in combination, on cell number (6 days). Dotted line represents cell number at day 0. ***<0.0001 by Student t-test. (C) Analysis of mTOR pathway and CDK-RB-E2F pathway biomarkers following 24 hours treatment with increasing concentrations of vistusertib (AZD2014) or palbociclib, alone and in combination. (D) qPCR analysis of six E2F dependent genes. Bar charts represent log fold change mRNA expression relative to DMSO control following 6 and 24 hours exposure to 100nM AZD2014, 300nM palbociclib or the combination. (E) E2F dependent protein levels measured by western blotting, following 24 hours treatment using stated concentrations of vistusertib (AZD2014) with or without palbociclib.
Investigation of the effects of the treatments on downstream signalling pathways, revealed that both vistusertib (AZD2014) and palbociclib caused a decrease in the levels of RB phosphorylation, and this was further enhanced in the combination (Figure 2C). Furthermore, the addition of vistusertib (AZD2014) was able to prevent the apparent increase in cyclin D1 levels caused by palbociclib treatment. Consistent with these findings, both vistusertib (AZD2014) and palbociclib caused downregulation in the mRNAs of a range of E2F target genes (including E2F1 itself), which were further reduced in the combination treatment (Figure 2D and Supplementary Table S2). These effects were observed at both 6 and 24 hours following the start of treatment, suggesting a direct effect on the E2F pathway rather than an indirect effect on gene expression through cell cycle inhibition. The changes in gene expression were confirmed at the protein level for a number of E2F dependent genes (Figure 2E). Interestingly, despite reports that CDK4/6 inhibition leads to increased phosphorylation of AKT through RB suppression of mTORC2 activation (24, 25), we found no evidence of increased AKT phosphorylation or altered mTOR signalling by palbociclib in our experiments (Figure 2C).
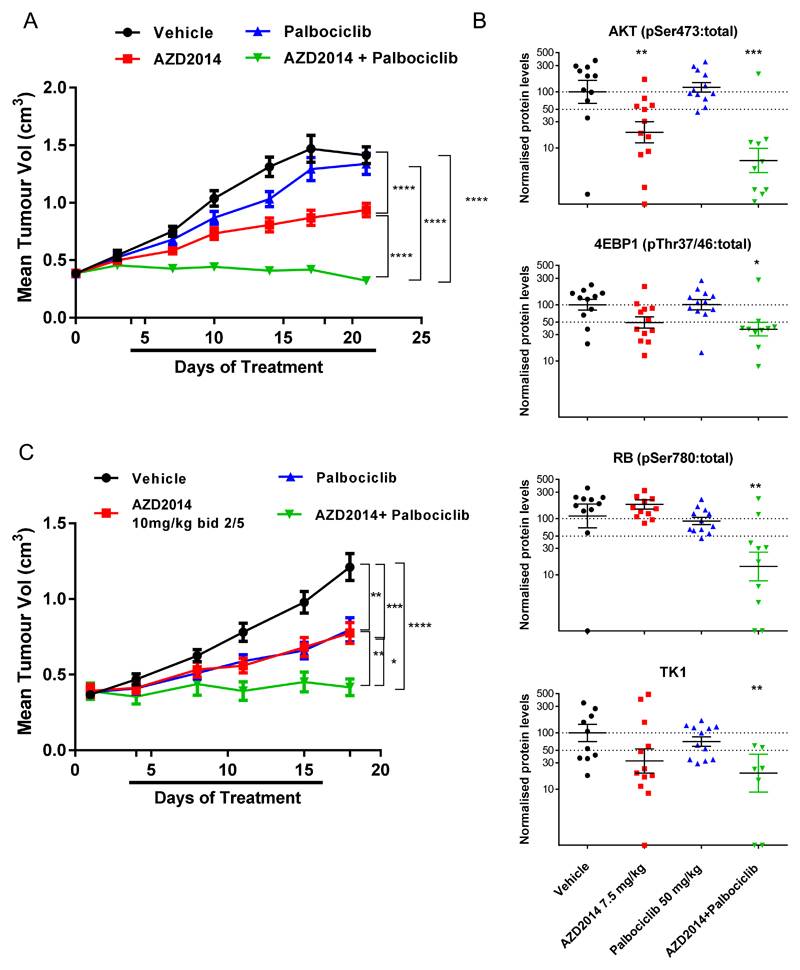
We next examined whether the combination of vistusertib (AZD2014) and palbociclib was efficacious in vivo, using mice implanted with an MCF-7 xenograft model. The combination of palbociclib and vistusertib (AZD2014), each dosed at sub-efficacious doses (50 mg/kg and 7.5 mg/kg orally once daily, respectively) resulted in profound tumour growth inhibition, greater than either agent alone (106.4% tumour growth inhibition cf. 46.5% AZD2014 alone or 7.0% palbociclib alone), measured on day 22 of dosing (Figure 3A). Our data suggests that combination efficacy could be achieved using sub-efficacious doses of either compound. We confirmed the effects of mTORC1/2 inhibition by measuring phosphorylation of downstream markers AKT(S473) and 4EBP-1(Thr37/46) and the effects of CDK4/6 inhibition by measuring phosphorylation of RB(Ser780). Inhibition of both CDK4/6 and mTORC1/2 caused a greater decrease in the phosphorylation of RB(Ser780) than either agent alone. Furthermore, the combination decreased the levels of thymidine kinase 1 (TK1), an E2F dependent gene (Figure 3B), confirming our in vitro findings and suggesting a convergence of the two pathways on E2F mediated transcription in vivo. To confirm the in vivo efficacy effects observed with the continuous dosing schedule, we also tested a well-tolerated, sub-efficacious intermittent dose of vistusertib (10mg/kg BID 2 days on/ 5 off) in combination with palbociclib (7.5mg/kg, as above). This combination confirmed the efficacy benefit over a similar dosing period (Figure 3C). Similar results were obtained when testing the combination in the CTC-174 patient derived model (Supplementary Figure S3).
Figure 3. Combined vistusertib (AZD2014) and palbociclib treatment results in significant tumour growth inhibition in ER+ breast cancer xenografts.
(A) Efficacy combination study of vistusertib (AZD2014, 7.5mg/kg once daily p.o.) and palbociclib (50 mg/kg once daily p.o.) compared to either agent alone in MCF-7 xenograft grown in male SCID mice. (B) Biomarker analysis of MCF-7 xenografts treated with vistusertib (AZD2014) and palbociclib. Tumours were excised at the end of the study and protein expression was analysed by Western blotting. (C) Efficacy combination study of vistusertib (AZD2014, 10mg/kg twice daily p.o., 2 days on/5 days off) and palbociclib (50 mg/kg once daily p.o.) compared to either agent alone in MCF-7 xenograft grown in male SCID mice. p values were calculated using two-sided Student’s t test. * p<0.05; ** p<0.01; *** p<0.001.
Combined mTOR and CDK4/6 inhibition prevents early adaptation and the onset of resistance
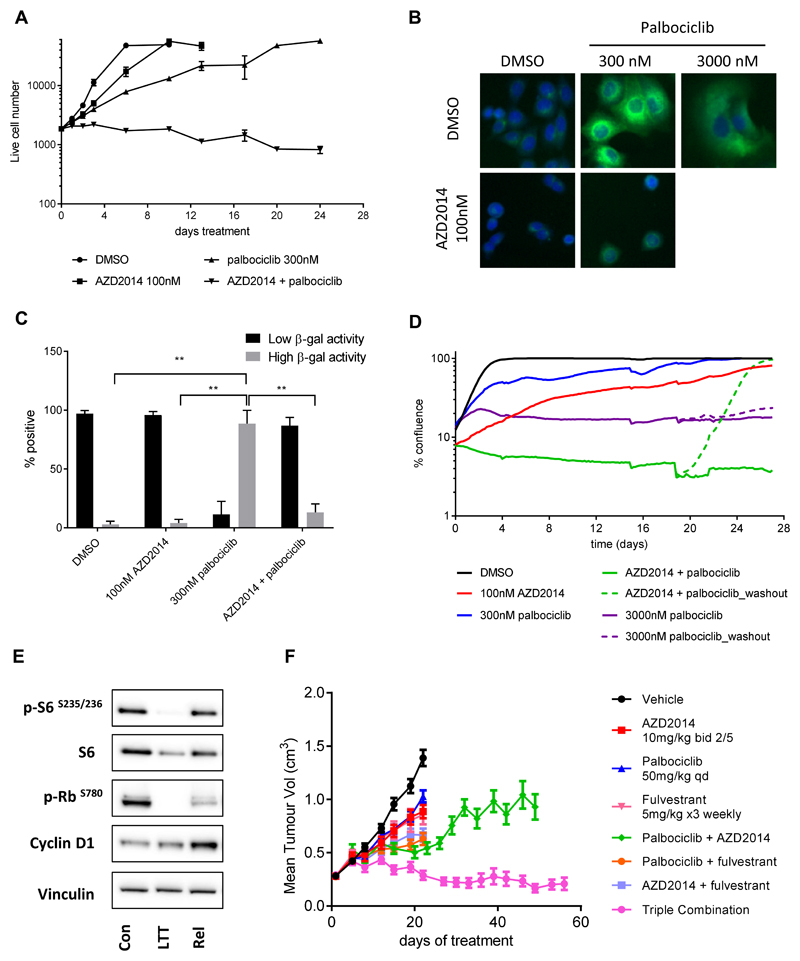
It has been reported that inhibition of CDK4/6 causes short term growth inhibition and early adaptation in ER+ breast cancer cells and that this adaptation can be prevented by combination with PI3K inhibitors. We therefore tested the effects of long term mTOR inhibition alone and in combination with CDK4/6 inhibition. In agreement with Herrera-Abreu et al. (21), we found that after an original phase of slow growth, the cultures of palbociclib treated cells (300nM) eventually reached confluence (Figure 4A). Similarly, treatment with vistusertib (AZD2014) alone (100nM) slowed the growth of the cells, compared to DMSO control. In contrast, the combination of vistusertib (AZD2014) with palbociclib resulted in complete growth arrest, which was maintained for at least 24 days in culture (Figure 4A).
Figure 4. Inhibition of mTORC1/2 and CDK4/6 signalling causes long term growth arrest via inhibition of E2F signalling.
(A) MCF-7 cells were treated with vistusertib (AZD2014) and/or palbociclib and cell number was assessed at the indicated times using a sytox green endpoint. (B) β-galactosidase activity (green) and the nucleus (blue) were stained following treatment of MCF-7 cells as indicated (8 days). (C) Bar charts representing the proportion of cells expressing low or high levels of β-galactosidase activity. Each bar represents the mean +/- s.e.m from three independent experiments. (D) MCF-7 cells were treated with vistusertib (AZD2014) and/or palbociclib as indicated and % confluency was measured using an Incucyte Zoom. For some treatments, parallel wells were set up and after 19 days compound was removed from half the wells (washout) and % confluency measured for a further 8 days. (E) MCF-7 cells were treated with 100nM vistusertib (AZD2014) plus 300nM palbociclib for 21 days. Cells were then left in media containing the drugs (long term treated; LTT) or washed a returned to drug free media (release; Rel) for a further 72 hours before being lysed and subject to immunoblot with the antibodies indicated; (F) Efficacy combination study of vistusertib (AZD2014, 10mg/kg twice daily p.o., 2 days on/5 days off), palbociclib (50 mg/kg once daily p.o.) and fulvestrant (5 mg/kg/ week, s.c.) compared to each agent alone in MCF-7 xenograft grown in male SCID mice. p values were calculated using two-sided Student’s t test. * p<0.05; ** p<0.01; *** p<0.001
In order to investigate the mechanism of the long term growth inhibition observed when combining inhibition of mTORC1/2 and CDK-RB-E2F pathway, we assessed whether vistusertib (AZD2014) could exacerbate the reported effects of palbociclib on senescence induction. As a control for our experiments, we used a high concentration (3000nM) of palbociclib, reported to induce a senescence like state (29). Although both agents induced cell cycle arrest (Supplementary Figure S4A), palbociclib treatment induced a concentration dependent ‘large flat cell’ morphology (Supplementary Figure S4B) and increased β-galactosidase activity (Figure 4B and C), indicative of the induction of a senescence-like phenotype (31, 34, 35). In contrast, vistusertib (AZD2014) did not induce a large flattened morphology but rather a decrease in cell size (Supplementary Figure S4B). Furthermore, cells treated with the combination of vistusertib (AZD2014) plus palbociclib had a significantly lower percentage of high β-galactosidase staining compared to cells treated with palbociclib alone (13% cf. 89%, Figure 4B, 4C), consistent with the reported role of mTOR in regulating cell size and in modulating the senescence phenotype (35, 36). To confirm our findings, we also measured the levels of induction of p21. Whilst palbociclib caused an increase in p21 levels (as expected), the combination of palbociclib and vistusertib (AZD2014) did not (Supplementary Figure S5).
To confirm that combined inhibition of CDK4/6 and mTORC1/2 did not result in an exacerbated senescence-like phenotype, inhibitors were removed from the culture approximately three weeks into the study and the growth of the cells was monitored for a further 8 days. Following drug withdrawal, individual cells could be observed to start dividing as early as 24 hours and the cell population reached confluence within 7-8 days (Figure 4D). This was in contrast to a high dose palbociclib treatment (3000nM) which was known to induce senescence, where cells did not regrow following compound removal and remained in a senescence-like state (Figure 4D and Supplementary Figure S4C).
To further characterise the long term treated cells which remain viable in cell culture, we analysed the levels of several key biomarkers by western blot analysis. As expected, cells treated for 21 days with a combination of 100nM vistusertib (AZD2014) and 300nM palbociclib maintained inhibition of the mTORC1/2 and CDK-RB-E2F signalling pathways as evidenced by low levels of phosphorylation of S6 and RB (Figure 4E). Cells ‘released’ from the drug treatment showed levels of phosphorylation of S6 similar to control cells, and appeared to reactivate the CDK-RB-E2F signalling pathway, measured by phosphorylation of RB and cyclin D1 levels (Figure 4E).
Estrogen receptor function has been shown to impact E2F gene transcription through multiple mechanisms, including direct effects on E2F1 promoter function (37, 38). We therefore hypothesised that a triple combination of vistusertib (AZD2014), palbociclib and the ER down regulator fulvestrant, would be required to achieve maximum growth inhibition. Herrera-Abreu et al (21) had previously reported that the triple combination of a PI3K inhibitor with fulvestrant and palbociclib resulted in a significant reduction of colony formation and xenograft tumour growth. Upon treatment of MCF-7 xenografts with vistusertib (AZD2014), palbociclib and fulvestrant, we observed prolonged tumour regressions (110.2% tumour growth inhibition at day 48), suggesting benefit of the triple treatment combination over the other combinations (Figure 4F). Over a period of 58 days, tumours failed to re-grow in the presence of the triple combination compared to vistusertib (AZD2014)/ palbociclib treatment, suggesting that the triple combination is necessary to maintain the growth inhibition in vivo.
Analysis of tumours treated with triple combination at the regression stage (day 30) suggested both mTOR and RB-E2F pathways were profoundly suppressed (Supplementary Figure S6A and B). To determine whether the enhanced effect of the triplet was due to the induction of apoptosis we assessed the levels of cleaved PARP in these tumours but saw no changes in cleaved PARP levels. We hypothesize that this might be due to in vivo clearance of apoptotic cells. Further and more comprehensive time course studies would be needed to address the kinetics of tumour cell induced apoptosis in vivo. An assessment of apoptosis induction in vitro revealed a very small induction of apoptosis in both double and triple treatment groups (Supplementary Figure S6C).
In summary, combination dosing of vistusertib (AZD2014), palbociclib and fulvestrant, delivered significant efficacy in MCF-7 xenografts. The combination of all three agents was required to induce tumour regression and to maintain long term inhibition of growth in vivo.
Short term inhibition of mTOR signalling does not affect binding of the estrogen receptor to chromatin
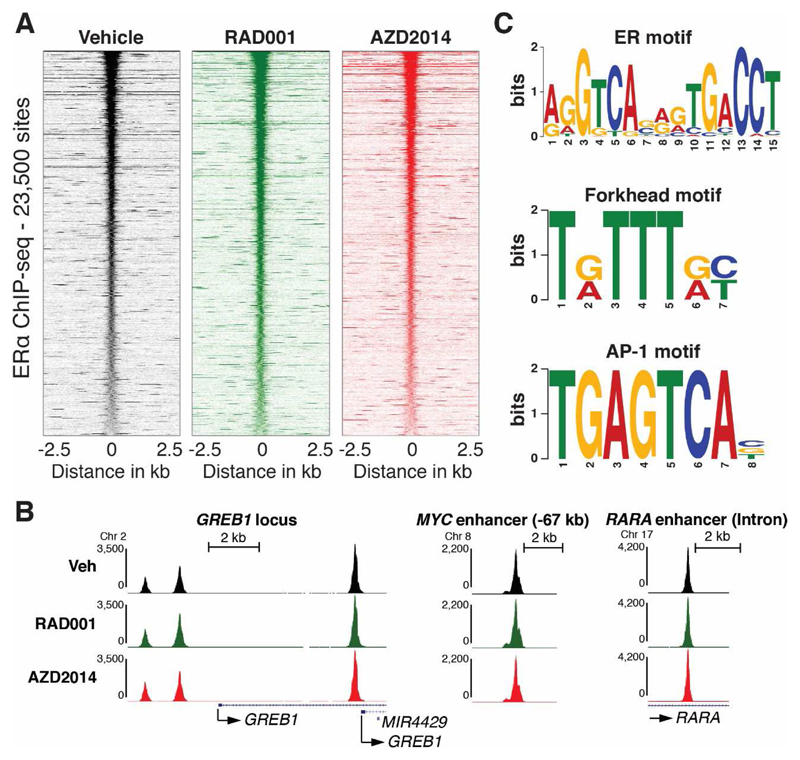
Cross talk between the PI3K pathway and ER signalling has been previously described (13, 39, 40). These findings led us to investigate if binding of ER to chromatin is directly affected by inhibition of mTORC1/2. We performed chromatin immunoprecipitation combined with deep sequencing (ChIP-seq) for ER upon 2 hours of treatment of MCF-7 cells with the mTORC1 inhibitor everolimus (RAD001) or the mTORC1/2 inhibitor vistusertib (AZD2014) in full media, where ER is fully activated by estrogens. Under these conditions, vistusertib (AZD2014) potently inhibits both mTORC1 and mTORC2, whereas everolimus only inhibits mTORC1 function (Supplementary Figure S7A), consistent with previous findings (26). Surprisingly, differential binding analysis revealed as few as 37 out of 23,537 binding sites, where ER binding is affected by everolimus and no binding sites affected by vistusertib (AZD2014) treatment (Figure 5A and Supplementary Figure S7B). The absence of any effect of everolimus or vistusertib (AZD2014) on ER binding is also clear at individual well known ER binding sites near GREB1, MYC, and RARA (Figure 5B). Importantly, the identified ER binding sites are highly enriched for the ER response element (ERE, Figure 5C), validating the ER ChIP-seq. Furthermore, we identify motifs for known ER-cooperating transcription factors (Figure 5C and 43), including the forkhead motif that is occupied by FOXA1 in breast cancer cells, where it plays an essential role as a pioneer factor for ER (41, 42, 43). Taken together, these genome-wide analyses demonstrate that inhibition of mTOR signalling, does not affect ER binding to chromatin and impinges on ER mediated growth via mechanisms that are downstream of ER.
Figure 5. Inhibition of mTOR signalling does not affect binding of ER to chromatin.
(A) ChIP-seq signal in a 5 Kb window around 23,500 ER binding sites that are common between MCF-7 cells treated with vehicle, RAD001, or vistusertib (AZD2014, 500 nM) for two hours (see Supplementary Figure S2B for Venn diagram of the differential binding analysis of the three conditions). (B) Examples of ER binding at well-known ER binding sites from the UCSC genome browser (http://genome.ucsc.edu/). (C) Selected enriched motifs in the 17,561 ER binding sites identified by MACS2 (49) in vehicle treated MCF-7 cells. Similar motifs were identified in ER binding sites identified in MCF-7 cells treated with RAD001 or vistusertib (AZD2014).
Palbociclib resistant cells reactivate the CDK-RB-E2F pathway and are sensitive to mTORC1/2 inhibition
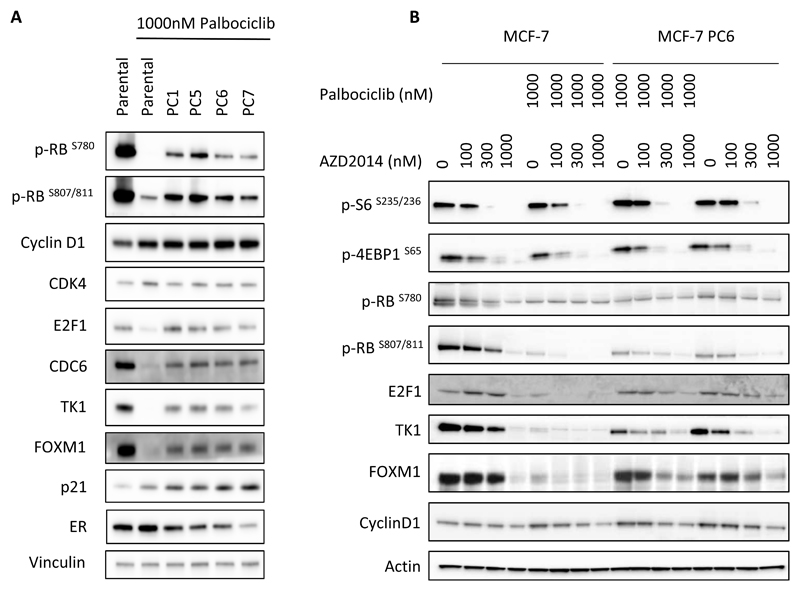
It has been previously reported that cells can readily adapt to prolonged treatment with palbociclib and develop resistance through a number of mechanisms, including CCNE1 amplification or RB1 loss (21–23). We therefore developed a panel of MCF-7 cell lines with acquired resistance to palbociclib. Ten discrete populations of MCF-7 cells were cultured in increasing concentrations of palbociclib over the course of 4-9 months until they were able to grow in 1000nM palbociclib (MCF-7_PC1 to 10). We selected four of these populations (PC1, PC5, PC6 and PC7) for further evaluation and measured the levels of phosphorylation of RB compared to control cells. The palbociclib resistant cell populations retained varying levels of RB phosphorylation as well as E2F transcriptional activity, even in the presence of 1000nM palbociclib (Figure 6A). No mutations in the CDK4 or CDK6 were identified in the resistant cell populations. Some of the cell populations were found to express lower levels of estrogen receptor, in agreement with previous reports (22).
Figure 6. Palbociclib resistant cells reactivate CDK-RB-E2F signalling and retain sensitivity to vistusertib (AZD2014).
(A) MCF-7 cells, (parental or palbociclib resistant PC1,5,6,7) were treated with 1000nM palbociclib for 24 hours before lysis and Western blotting. (B) Parental MCF-7 cells and palbociclib resistant MCF-7 cells (PC6) were treated with increasing concentrations of vistusertib (AZD2014) in the presence or absence of 1000nM palbociclib before lysis and Western blotting.
The palbociclib resistant cell lines were also found to be resistant to other CDK4/6 inhibitors, (Supplementary Table S3). Significantly, vistusertib (AZD2014) treatment caused significant growth inhibition in these populations (Supplementary Table S3). Furthermore, treatment of palbociclib resistant cells with vistusertib (AZD2014) resulted in inhibition of RB phosphorylation and decreased expression of E2F dependent genes such as E2F1, TK1 and the downstream effector FOXM1 (Figure 6B), similarly to what was observed in parental cells. Therefore, vistusertib (AZD2014) was able to modulate the CDK-RB-E2F axis even under conditions where cells have become resistant to palbociclib, suggesting that treatment with an mTOR inhibitor could be beneficial in patients that have relapsed on palbociclib treatment.
Discussion
A major limitation of some of the targeted therapies which are currently being tested in clinical trials in ER+ breast cancer is the short term, cytostatic nature of the inhibition of tumour growth. Early adaptation has been described as a phenomenon that may limit the effectiveness of both mTORC1 and CDK4/6 inhibitors clinically (21–23). Here we describe the effects of combination therapies in estrogen receptor positive breast cancer models, where convergence on inhibition of E2F dependent transcription is required to deliver durable responses. Inhibition of either mTOR signalling or CDK-RB-E2F signalling causes a decrease in E2F dependent transcription and the expression of genes required for S phase entry. Optimal inhibition of E2F mediated transcription is only achieved upon blockade of both pathways and the convergent effects of multiple pathway inhibition in repressing E2F activity is likely to be required to achieve sustained and durable growth inhibitory effects. Hence, ‘triple’ combinations of mTOR inhibitors (dosed either as a continuous or intermittent schedule), CDK4/6 inhibitors and endocrine therapies may represent the most effective way to optimally inhibit E2F activity and the G1/S transition in this tumour type.
The convergence of different interdependent pathways on the modulation of E2F transcription in ER+ breast cancer has been previously described (28, 38). Both cyclin D1 and E2F1 are estrogen regulated genes, therefore the combination of ER signalling blockade with CDK4/6 inhibition would be expected to result in lower cyclin D1 levels, lower E2F1 levels and cooperation on the inhibition of the CDK-RB-E2F pathway to block proliferation and entry into the cell cycle from G1 phase. The effectiveness of mTORC1 inhibition in combination with aromatase inhibitors in estrogen receptor positive breast cancer may predict the basis for functional interactions between mTOR signalling, ER signalling and CDK-RB-E2F pathway activity. In this study, we determine that the mTOR pathway converges onto the CDK-RB-E2F signalling module. We demonstrate that the effects of mTOR inhibition in breast cancer are not mediated through modulation of ER function, and that ER binding to chromatin is not affected by mTORC1/2 inhibitor treatment. This is unlike the findings reported by Bosch et al (13), whereby PI3K inhibitors were shown to directly affect ER function and increase estrogen receptor binding to DNA. Instead, we show that inhibition of mTORC1 and 2 results in a decrease in cyclin D1 levels (possibly through effects on protein translation) and a decrease in the phosphorylation of RB. These effects result in the modulation of E2F mediated transcription by mTOR, consistent with a role of mTOR in modulating the G1/S checkpoint and the commitment to cell cycle entry from quiescence. In this setting, growth signalling and entry into S phase can only be triggered upon production of new nucleotides and other building blocks to accommodate an increase in RNA and DNA synthesis needed for ribosome biogenesis and anabolic growth (11, 12). The findings are also consistent with the notion that luminal estrogen receptor positive breast cancer cells are highly dependent on the G1/S transition modulated by cyclin D1 and CDK4/6. Cyclin D1 is often activated either by amplification or by other mechanisms in this disease setting (14–17).
The mechanisms that can contribute to modulation of E2F mediated transcription are not fully characterised to date. The family of E2F transcription factors and their functional properties is complex, with some members acting as transcriptional activators and others as transcriptional repressors during quiescence or early G1 phase (37). Some of the E2F transcription factors are also known to act as transcriptional modulators at later stages of the cell cycle (37). Therefore, the mechanism of modulation of E2F mediated transcription is likely to be complex and involve multiple players.
CDK4/6 inhibitors have been shown to cause a senescence-like phenotype in tumour cells (29). As common pathways are involved in the modulation of cell cycle arrest in senescence and quiescence (44), we wanted to establish whether the combination of mTORC1/2 inhibition and CDK4/6 inhibition caused an exacerbation of the senescence-like phenotype. The role of cellular senescence in tumours is complex and modulated by both cell autonomous and non-cell autonomous effects (34). Therefore, understanding how this process is affected by the combinations of different therapeutics will be key to understand how different therapies affect both cancer cells and the surrounding tissue homeostasis. In this study, we confirm the role of CDK4/6 inhibition in the induction of senescence, but demonstrate that the combination of CDK4/6 inhibition with mTORC1/2 inhibition does not result in the exacerbation of a senescence-like phenotype, but rather causes cells to enter into a long term quiescent state.
The ability of cells to survive drug treatments by developing a long term quiescent state has been described in other tumour types (45) upon treatment with kinase inhibitors. These cells, sometimes referred to as ‘drug tolerant persisters (DTPs), are characterised by their ability to revert back to their original phenotype upon drug withdrawal. Transcriptional reprogramming and epigenetic changes have been strongly implicated in maintaining this subpopulation of cells in the presence of drug. In this study, we show that cells treated with CDK4/6 and mTOR inhibitors remain viable in culture for extended periods of time, in a similar fashion to that described for other kinase inhibitor therapeutics (45).
A network of signalling pathways appears to converge onto the G1/S transition and E2F dependent transcription in many ER+ breast cancers. Estrogen receptor signalling activates the CCND1 promoter and cyclin D1 is often expressed at high levels in this tumour type, in the presence or absence of CCND1 gene amplification. Cyclin D1 has also been reported to bind and facilitate estrogen receptor transcriptional activity (15–17). Our findings confirm clinical observations that these tumours remain dependent on E2F transcription to drive proliferation, even when they become resistant to CDK4/6 inhibitors, confirming the exquisite dependence of ER+ breast cancer cells on this checkpoint. In the CDK4/6 resistance setting, despite the reactivation of E2F mediated signalling, mTOR inhibition was able to effectively block proliferation. Furthermore, in palbociclib resistant cells, vistusertib (AZD2014) treatment was able to modulate the phosphorylation of RB and cause a decrease in E2F function.
Recent pivotal phase III trials investigating CDK4/6 inhibitors have demonstrated a substantial improvement in progression free survival in patients with advanced breast cancer (1). As mechanisms of resistance to these agents begin to emerge, combination therapies which target the pivotal regulators of the G1/S checkpoint will have to be utilised for the successful inhibition of tumour growth. In this study, we suggest that treatment with an mTOR inhibitor may represent an option for patients that have relapsed on CDK4/6 inhibitor therapy, especially on a background of an anti-hormonal therapy.
Supplementary Material
Acknowledgments
R.S. is funded by a fellowship from the Novo Nordisk Foundation (NNF 14136).
Footnotes
CM, CC, OD, LSC, ADS, UMP, ML, RM and SCC are employees of AstraZeneca.
The authors declare no potential conflicts of interest
References
- 1.Turner NC, Neven P, Loibl S, Andre F. Advances in the treatment of advanced oestrogen-receptor positive breast cancer. Lancet. 2017;389(10087):2403–2414. doi: 10.1016/S0140-6736(16)32419-9. [DOI] [PubMed] [Google Scholar]
- 2.Jerusalem G, Bachelot T, Barrios C, Neven P, Di Leo A, Janni W, et al. A new era of improving progression-free survival with dual blockade in postmenopausal HR(+), HER2(-) advanced breast cancer. Cancer Treat Rev. 2015;41:94–104. doi: 10.1016/j.ctrv.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 3.Chumsri S, Sabnis G, Tkaczuk K, Brodie A. mTOR inhibitors: changing landscape of endocrine-resistant breast cancer. Future Oncol. 2014;10:443–456. doi: 10.2217/fon.13.178. [DOI] [PubMed] [Google Scholar]
- 4.Baselga J, Campone M, Piccart M, Burris HA, 3rd, Rugo HS, Sahmoud T, et al. Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer. N Engl J Med. 2012;366:520–529. doi: 10.1056/NEJMoa1109653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palanisamy RP. Palbociclib: A new hope in the treatment of breast cancer. J Cancer Res Ther. 2016;12(4):1220–1223. doi: 10.4103/0973-1482.168988. [DOI] [PubMed] [Google Scholar]
- 6.Boér K. Impact of palbociclib combinations on treatment of advanced estrogen receptor-positive/human epidermal growth factor 2-negative breast cancer. Onco Targets Ther. 2016;9:6119–6125. doi: 10.2147/OTT.S77033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hortobagyi GN, Chen D, Piccart M, Rugo HS, Burris HA, 3rd, Pritchard KI, et al. Correlative Analysis of Genetic Alterations and Everolimus Benefit in Hormone Receptor-Positive, Human Epidermal Growth Factor Receptor 2-Negative Advanced Breast Cancer: Results From BOLERO-2. J Clin Oncol. 2016;34:419–26. doi: 10.1200/JCO.2014.60.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yardley DA, Noguchi S, Pritchard KI, Burris HA, 3rd, Baselga J, Gnant M, et al. Everolimus plus exemestane in postmenopausal patients with HR(+) breast cancer: BOLERO-2 final progression-free survival analysis. Adv Ther. 2013;30(10):870–84. doi: 10.1007/s12325-013-0060-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu K, Liu P, Wei W. mTOR signaling in tumorigenesis. Biochim et Biophys Acta. 2014;1846:638–654. doi: 10.1016/j.bbcan.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang K, Fingar DC. Growing knowledge of the mTOR signaling network. Semin Cell Dev Biol. 2014;36:79–90. doi: 10.1016/j.semcdb.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Howell JJ, Ricoult SJ, Ben-Sahra I, Manning BD. A growing role for mTOR in promoting anabolic metabolism. Biochem Soc Trans. 2013;41(4):906–12. doi: 10.1042/BST20130041. [DOI] [PubMed] [Google Scholar]
- 12.Ben-Sahra I, Howell JJ, Asara JM, Manning BD. Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science. 2013;339(6125):1323–8. doi: 10.1126/science.1228792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bosch A, Li Z, Bergamaschi A, Ellis H, Toska E, Prat A, et al. PI3K inhibition results in enhanced estrogen receptor function and dependence in hormone receptor-positive breast cancer. Sci Transl Med. 2015;7(283):283ra51. doi: 10.1126/scitranslmed.aaa4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Otto T, Sicinski P. Cell cycle proteins as promising targets in cancer therapy. Nat Rev Cancer. 2017;17:93–115. doi: 10.1038/nrc.2016.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sherr CJ, Beach D, Shapiro GI. Targeting CDK4 and CDK6: From Discovery to Therapy. Cancer Discov. 2016;6(4):353–67. doi: 10.1158/2159-8290.CD-15-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O'Leary B, Finn RS, Turner NC. Treating cancer with selective CDK4/6 inhibitors. Nat Rev Clin Oncol. 2016;13(7):417–30. doi: 10.1038/nrclinonc.2016.26. [DOI] [PubMed] [Google Scholar]
- 17.Johnson J, Thijssen B, McDermott U, Garnett M, Wessels LF, Bernards R. Targeting the RB-E2F pathway in breast cancer. Oncogene. 2016;35(37):4829–35. doi: 10.1038/onc.2016.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pare R, Shin JS, Lee CS. Increased expression of senescence markers p14 (ARF) and p16(INK4a) in breast cancer is associated with an increased risk of disease recurrence and poor survival outcome. Histopathology. 2016;69(3):479–91. doi: 10.1111/his.12948. [DOI] [PubMed] [Google Scholar]
- 19.Patnaik A, Rosen LS, Tolaney SM, Tolcher AW, Goldman JW, Gandhi L, et al. Efficacy and Safety of Abemaciclib, an Inhibitor of CDK4 and CDK6, for Patients with Breast Cancer, Non-Small Cell Lung Cancer, and Other Solid Tumors. Cancer Discov. 2016;6(7):740–53. doi: 10.1158/2159-8290.CD-16-0095. [DOI] [PubMed] [Google Scholar]
- 20.Hortobagyi GN, Stemmer SM, Burris HA, Yap YS, Sonke GS, Paluch-Shimon S, et al. Ribociclib as First-Line Therapy for HR-Positive, Advanced Breast Cancer. N Engl J Med. 2016;375(18):1738–1748. doi: 10.1056/NEJMoa1609709. [DOI] [PubMed] [Google Scholar]
- 21.Herrera-Abreu MT, Palafox M, Asghar U, Rivas MA, Cutts RJ, Garcia-Murillas I, et al. Early Adaptation and Acquired Resistance to CDK4/6 Inhibition in Estrogen Receptor-Positive Breast Cancer. Cancer Res. 2016;76(8):2301–13. doi: 10.1158/0008-5472.CAN-15-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang C, Li Z, Bhatt T, Dickler M, Giri D, Scaltriti M, et al. Acquired CDK6 amplification promotes breast cancer resistance to CDK4/6 inhibitors and loss of ER signaling and dependence. Oncogene. 2017;36(16):2255–2264. doi: 10.1038/onc.2016.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jansen VM, Bhola NE, Bauer JA, Formisano L, Lee KM, Hutchinson KE, et al. Kinome-Wide RNA Interference Screen Reveals a Role for PDK1 in Acquired Resistance to CDK4/6 Inhibition in ER-Positive Breast Cancer. Cancer Res. 2017;77(9):2488–2499. doi: 10.1158/0008-5472.CAN-16-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang J, Xu K, Liu P, Geng Y, Wang B, Gan W, et al. Inhibition of Rb Phosphorylation Leads to mTORC2-Mediated Activation of Akt. Mol Cell. 2016;62(6):929–42. doi: 10.1016/j.molcel.2016.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goel S, Wang Q, Watt AC, Tolaney SM, Dillon DA, Li W, et al. Overcoming Therapeutic Resistance in HER2-Positive Breast Cancers with CDK4/6 Inhibitors. Cancer Cell. 2016;29(3):255–69. doi: 10.1016/j.ccell.2016.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guichard SM, Curwen J, Bihani T, D'Cruz CM, Yates JW, Grondine M, et al. AZD2014, an Inhibitor of mTORC1 and mTORC2, Is Highly Effective in ER+ Breast Cancer When Administered Using Intermittent or Continuous Schedules. Mol Cancer Ther. 2015;14(11):2508–18. doi: 10.1158/1535-7163.MCT-15-0365. [DOI] [PubMed] [Google Scholar]
- 27.Vora SR, Juric D, Kim N, Mino-Kenudson M, Huynh T, Costa C, et al. CDK4/6 inhibitors sensitize PIK3CA mutant breast cancer to PI3K inhibitors. Cancer Cell. 2014;26:136–49. doi: 10.1016/j.ccr.2014.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller TW, Balko JM, Fox EM, Ghazoui Z, Dunbier A, Anderson H, et al. ERα-dependent E2F transcription can mediate resistance to estrogen deprivation in human breast cancer. Cancer Discov. 2011;1:338–51. doi: 10.1158/2159-8290.CD-11-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anders L, Ke N, Hydbring P, Choi YJ, Widlund HR, Chick JM, et al. A systematic screen for CDK4/6 substrates links FOXM1 phosphorylation to senescence suppression in cancer cells. Cancer Cell. 2011;20:620–34. doi: 10.1016/j.ccr.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ertel A, Dean JL, Rui H, Liu C, Witkiewicz AK, Knudsen KE, Knudsen ES. RB-pathway disruption in breast cancer: differential association with disease subtypes, disease-specific prognosis and therapeutic response. Cell Cycle. 2010;9:4153–63. doi: 10.4161/cc.9.20.13454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Debacq-Chainiaux F, Erusalimsky JD, Campisi J, Toussaint O. Protocols to detect senescence-associated beta-galactosidase (SA-beta gal) activity, a biomarker of senescent cells in culture and in vivo. Nat Protoc. 2009;4(12):1798–806. doi: 10.1038/nprot.2009.191. [DOI] [PubMed] [Google Scholar]
- 32.Langmead B, Trapnell C, Pop M, Salzberg S. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based Analysis of ChIP-Seq (MACS) Genome Biology. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pérez-Mancera PA, Young AR, Narita M. Inside and out: the activities of senescence in cancer. Nat Rev Cancer. 2014;14(8):547–58. doi: 10.1038/nrc3773. [DOI] [PubMed] [Google Scholar]
- 35.Tomimatsu K, Narita M. Translating the effects of mTOR on secretory senescence. Nat Cell Biol. 2015;17(10):1230–2. doi: 10.1038/ncb3244. [DOI] [PubMed] [Google Scholar]
- 36.Walters HE, Deneka-Hannemann S, Cox LS. Reversal of phenotypes of cellular senescence by pan-mTOR inhibition. Aging. 2016;8(2):231–44. doi: 10.18632/aging.100872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thurlings I, de Bruin A. E2F Transcription Factors Control the Roller Coaster Ride of Cell Cycle Gene Expression. Methods Mol Biol. 2016;1342:71–88. doi: 10.1007/978-1-4939-2957-3_4. [DOI] [PubMed] [Google Scholar]
- 38.Wang W, Dong L, Saville B, Safe S. Transcriptional activation of E2F1 gene expression by 17beta-estradiol in MCF-7 cells is regulated by NF-Y-Sp1/estrogen receptor interactions. Mol Endocrinol. 1999;13(8):1373–87. doi: 10.1210/mend.13.8.0323. [DOI] [PubMed] [Google Scholar]
- 39.Alayev A, Salamon RS, Berger SM, Schwartz NS, Cuesta R, Snyder RB, Holz MK. mTORC1 directly phosphorylates and activates ERα upon estrogen stimulation. Oncogene. 2015;35(27):3535–43. doi: 10.1038/onc.2015.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamnik RL, Digilova A, Davis DC, Brodt NZ, Murphy CJ, Holz MK. S6 Kinase 1 Regulates Estrogen Receptor in Control of Breast Cancer Cell Proliferation. JBC. 2009;284(10):6361–6369. doi: 10.1074/jbc.M807532200. [DOI] [PubMed] [Google Scholar]
- 41.Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, et al. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38(11):1289–97. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- 42.Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122(1):33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 43.Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet. 2011;43(1):27–33. doi: 10.1038/ng.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Terzi MY, Izmirli M, Gogebakan B. The cell fate: senescence or quiescence. Mol Biol Rep. 2016;43:1213–1220. doi: 10.1007/s11033-016-4065-0. [DOI] [PubMed] [Google Scholar]
- 45.Hata AN, Niederst MJ, Archibald HL, Gomez-Caraballo M, Siddiqui FM, Mulvey HE, et al. Tumor cells can follow distinct evolutionary paths to become resistant to epidermal growth factor receptor inhibition. Nat Med. 2016;22(3):262–9. doi: 10.1038/nm.4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.