Abstract
Recent molecular characterization of prostate cancer (PCa) identified novel genetic aberrations and disease subtypes. The frequencies of molecular aberrations show racial disparity. Clinical strategies and targeted therapies embracing these racial differences are required. Here we discuss ethnic differences in genetic alterations and their impact on the susceptibility, progression, and treatment of prostate cancer.
Introduction
PCa often harbors pathological and molecular intratumoral and interpatient heterogeneity (see Glossary) [1]. Racial differences in PCa prevalence have recently become the focus of many epidemiologic studies, but the underlying molecular mechanisms of ethnic disparity remain poorly understood. Studies by several groups demonstrated that African-American men with PCa tend to have higher serum estradiol and prostate-specific antigen (PSA) levels, advanced-stage cancer, and overall poorer survival than Caucasian-American men. By contrast, Asian men have lower disease prevalence compared with Asian-American or American PCa cohorts [2]. Despite lower PCa incidence, the Asian populations have a higher prevalence of advanced disease, probably due to the paucity of molecular and cancer screening methods, sensitive diagnostic tools, and strategies required for early detection. Heterogeneity in prostate cancer can be studied from biopsy specimens containing multiple focal regions with differing Gleason scores. The samples can be subcategorized based on known molecular biomarkers such as TMPRSS2–ERG fusion, SPINK1 overexpression, PTEN deletion, and c-MYC amplification status [3,4]. While much is known about interpatient molecular subtypes in Caucasian populations, detailed understanding of the molecular heterogeneity and disease progression in other ethnic groups is required.
Glossary.
Clinical sequencing
comprehensive examination of a patient's germline and somatic alterations within the genome using high-throughput sequencing techniques. Clinical sequencing has been instrumental in designing tailored medicine based on the individual's genotype, personal mutations or aberrations, and correlation with disease-susceptibility genes.
Genetic translocation
genetic rearrangement in which a segment breaks off from one chromosomal arm and fuses to the arm of another chromosome; may result in gene fusions.
Gleason score
a unique grading system used to evaluate PCa prognosis using biopsy specimens that is solely based on the architectural pattern of the tumor. Scoring is based on the differentiation pattern of the tumor biopsy, where a score of <6 means a well-differentiated tumor and >6 means a least-differentiated tumor.
Intergenic deletion
spontaneous deletion of a genomic region between genes resulting in genetic alterations.
Prostate-specific antigen (PSA)
kallikrein-like serine protease synthesized by prostate epithelial cells. It is a major protein found in semen and is involved in liquefaction of the seminal coagulum. Healthy men have serum PSA levels <4 ng/ml; this concentration is usually used as a cut off for estimating PCa risk.
Single-nucleotide polymorphism (SNP)
a single-nucleotide genetic variation at a specific position in the genome that may predispose an individual to a disease state.
Tumor heterogeneity
heterogeneity can exist within tumors of different individuals (intertumor heterogeneity) or be defined as subclonal diversity within the tumor of a single patient (intratumor heterogeneity).
PCa: Risk-Associated Susceptibility Genes
High-throughput studies identified genetic aberrations in PCa and their association with cancer susceptibility and familial risk. Single nucleotide polymorphisms (SNPs) in many risk-associated loci, including the 8q24 locus, have been positively associated with increased risk of cancer and higher PSA levels, Gleason score, and mortality. Furthermore, somatic amplifications at the 8q24 locus, a frequent genetic event, indicate that risk alleles in this locus may trigger the amplification of the entire region. Polymorphisms in the 8q24 locus confer increased susceptibility for PCa in Americans with African ancestry [5]. Fine mapping of the 3.8-Mb region at 8q24 demonstrated discrepancies across multiethnic populations with European, African-American, and Japanese ancestries [6]. It remains to be established whether the presence of risk alleles at 8q24 influences the regulation of neighboring genes such as c-MYC and FAM84B. Another possibility is the potential regulatory link between PCAT1, an oncogenic long non-coding RNA encoded by 8q24, and deregulation of c-MYC [7] and proximate genes.
Several independent studies showed that PCa patients harboring the breast cancer 2, early onset (BRCA1/2) germline mutation are at greater risk for early disease onset and have a higher likelihood of developing locally advanced and metastatic disease. Multi-institutional integrative clinical sequencing of biopsies of metastatic castration-resistant PCa tumors revealed somatic and pathogenic germline alterations in BRCA2 (~12.7% of cases), with 90% of cases harboring biallelic loss of BRCA2 [8]. This finding suggests that treatment with poly(ADP-ribose) polymerase 1 (PARP1) inhibitors might be effective in patients with this molecular subtype. In light of the available treatment options, investigating BRCA1/2 mutation frequency among patients in other ethnic groups is necessary. Intriguingly, a recent study shows mutation in Homeobox B13 (HOXB13) (G84E, rs138213197) at a higher frequency among men of European descent with either early onset of disease or a positive family history. This association was not found in patients of African or Asian ancestry.
Incidence of TMPRSS2–ERG Fusion and SPINK1 Overexpression across Diverse Ethnic Groups
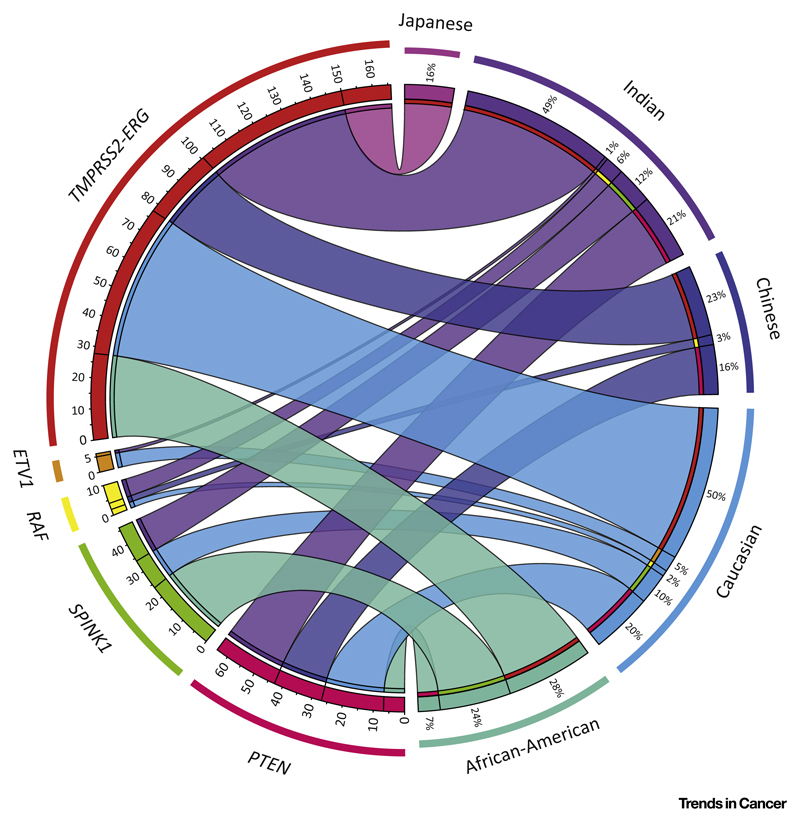
The genetic rearrangements involving erythroblast transformation-specific (ETS) transcription factors (ERG, ETV1, ETV4, ETV5, and FLI1) represent the primary oncogenic aberration in a majority of prostate cancers [3]. About 50% of PCa patients harbor the TMPRSS2–ERG genetic rearrangement involving the androgen-driven TMPRSS2 promoter, resulting in aberrant overexpression of the ERG oncoprotein [3]. Studies from several groups, including ours, have shown racial differences in the frequency of this genetic event. ERG aberrations are highly prevalent among Caucasian-American (~50%) and Indian (49%) patients. By comparison, African-American and Asian patients have significantly lower frequencies of around 30% and 10–20%, respectively (Figure 1) [4,9,10]. Conversely, African-American patients with ERG-negative status show higher-grade tumor indices and a less favorable clinical outcome [9]. A recent study that compared younger patients and older men with PCa suggested an association between age and higher incidences of TMPRSS2–ERG genetic rearrangement.
Figure 1. Racial Differences in Prostate Cancer (PCa) Molecular Subtypes.
Integrated Circos plot shows the correlation between molecular subtypes of PCa and their prevalence in different ethnic groups. Individual ethnic groups (on the right) are represented in different colors and corresponding colored ribbons connect each ethnic group to various molecular aberrations (on the left). The width of the individual ribbon denotes the prevalence of a molecular subtype in a given ethnic group.
These proportions are nearly reversed for the subtype characterized by overexpression of SPINK1, which represents the second largest and more aggressive subset of PCa. Caucasian and Indian patients show ~10–15% frequency of SPINK1 overexpression in a largely mutually exclusive manner with ERG aberration [4,11]. By contrast, African-American men showed nearly twice the frequency (~24%) [11]. Mutations in Speckle-type POZ (SPOP) in ERG-negative tumors define another molecular subtype [12]. Recently, SPOP mutations were found in ~10% of patients with localized tumors, and there was an overlap between SPINK1-positive status, mutations in SPOP, and deletion of CHD1, a chromatin remodeling factor. This distinct molecular subtype also shows mutual exclusivity with ETS rearrangements [1]. SPOP mutations were less frequently observed among African-Americans than in Caucasians [11]. It will be compelling to investigate the SPOP and CHD1 mutation frequency in other ethnic groups. Taken together, these data suggest that molecular subclassification of PCa is highly warranted across different ethnicities and could help our understanding of disease heterogeneity and assist in the design of tailored therapies. Future studies will provide more detailed information on the pathobiology associated with these molecular subtypes.
Differential Genesis of TMPRSS2–ERG Fusion in Prostate Cancer
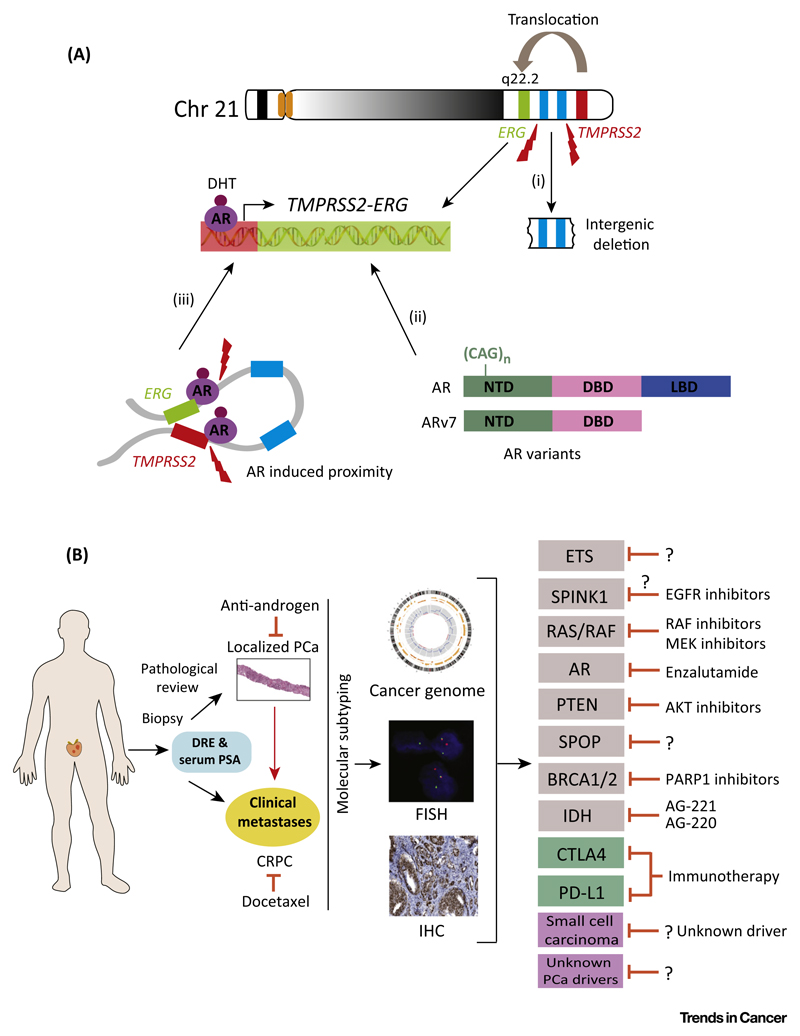
The genesis of TMPRSS2–ERG fusion through intergenic deletion is more prevalent in African-Americans than in Caucasian and Japanese patients [10]. TMPRSS2–ERG is associated with more aggressive tumors. Loss of ETS2, a potential tumor suppressor gene located between the TMPRSS2 and ERG genes on chromosome 21, is likely to contribute to this aggressive phenotype (Figure 2A).
Figure 2. Molecular Mechanism for TMPRSS2–ERG Genesis and Emerging Treatment Modalities for Prostate Cancer (PCa).
(A) Schematic representation of the genomic organization of TMPRSS2 and ERG on chromosome 21 and possible mechanisms supporting the genesis of TMPRSS2–ERG genetic rearrangement: (i) intergenic deletion or translocation between ERG and TMPRSS2; (ii) androgen-induced proximity between TMPRSS2 and ERG resulting in TMPRSS2–ERG fusion on gamma-irradiation-induced double-strand DNA breaks; (iii) CAG repeats in the N-terminal transactivation domain of the androgen receptor (AR) and a constitutively active AR isoform encoded by splice variant 7 (ARv7) lacking a ligand-binding domain (LBD). (B) Schematic representation of clinical strategies for the treatment and management of localized PCa and castration-resistant prostate cancer (CRPC). Sequential needle core biopsy prompted by an increase in serum prostate-specific antigen (PSA) and digital rectal examination (DRE) is routinely used for initial diagnosis. Molecular subtyping is done by fluorescence in situ hybridization (FISH), biomarker screening [immunohistochemistry (IHC)], and integrative genomic sequencing approaches. In localized PCa, treatment is largely determined by tumor stage and includes first-generation antiandrogens, castration, or watchful waiting. In advanced PCa, first-line treatment options such as antiandrogens or docetaxel may be combined with targeted therapies to improve patient outcome. Some of the evolving targeted therapies matched with respective genomic aberrations for PCa treatment are shown on the right.
Chinnaiyan and colleagues demonstrated that androgen signaling induced spatial proximity of the TMPRSS2 and ERG genomic loci (both located on 21q22) and that this is an important factor in gamma-irradiation-induced DNA double-strand breaks that lead to TMPRSS2–ERG fusion [13]. Several observations suggest that aberrant androgen receptor (AR) signaling may play a pivotal role in the genesis of this rearrangement (Figure 2A). Multiple genetic variants of AR [different numbers of the CAG repeat sequence in exon 1 or the splice variant 7 (ARv7)] can dictate the transcriptional activity of AR and resistance to antiandrogens. The number of CAG repeats and presence of TMPRSS2–ERG fusion associates with risk for prostate cancer. Tumors with shorter CAG repeats have higher tendency to harbor the TMPRSS2– ERG fusion [14]. Therefore, it could be speculated that the lower prevalence of PCa among Japanese and Asian patients may be due to longer CAG repeats. Moreover, cases of PCa in African-Americans, who tend to have a higher cancer risk and lower incidence of ERG rearrangement, could be dictated by AR signaling and its variants. Taking these findings together, the disparity observed in the TMPRSS2– ERG fusion incidence is likely to be influenced by racial background. Nevertheless, disparity in cancer could be affected by many factors and not limited to risk-associated SNPs, AR variants, CAG repeats, or the kind of genetic exchange that occurs between the TMPRSS2 and ERG loci. Therefore, further epidemiological studies addressing the exact mechanism of PCa disparity across different ethnic groups are highly warranted.
Concluding Remarks
The frequencies of molecular driver events of PCa such as TMPRSS2–ERG fusion, SPINK1 overexpression, and PTEN deletion show racial disparity. A better understanding of population-specific cancer subtypes can help in drug development, in devising better clinical translational strategies, and in improving population-centric health-care policies. Low-frequency occurrence of rare aberrations poses a challenge for racial studies, but it is essential to screen for RAS/RAF fusions, IDH mutations, and BRCA1/2 mutations due to the availability of targeted therapies (Figure 2B). Although not discussed here, geographical treatment disparities (orchiectomy versus chemical castration) also exist. Further research is required to understand aggressive small-cell carcinoma of the prostate and cancers that lack known driver alterations, a topic that is outside the scope of this article. Currently, CTLA-4- and PD-1/PD-L1-based immunotherapy holds promise in PCa. However, its utility in the metastatic setting needs to be carefully evaluated for novel treatment opportunities. Understanding how racial differences might influence immunotherapy will be crucial. In summary, emerging knowledge on racial disparity in disease molecular subtypes and new advances in therapeutic approaches will aid in strategizing the treatment options and management of PCa in the near future.
Acknowledgments
The authors apologize to those whose relevant research was not cited owing to limitations of space in this Forum article. They thank Saravana Mohan Dhanasekaran for critically reading the manuscript. B.A. is an Intermediate Fellow of the Wellcome Trust/DBT India Alliance and a Young Investigator of the DST-FAST Track scheme. This work is supported by a Wellcome Trust/DBT India Alliance grant [IA/I(S)/12/2/500635 to B.A.].
References
- 1.Cancer Genome Atlas Research Network. The molecular taxonomy of primary prostate cancer. Cell. 2015;163:1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ito K. Prostate cancer in Asian men. Nat Rev Urol. 2014;11:197–212. doi: 10.1038/nrurol.2014.42. [DOI] [PubMed] [Google Scholar]
- 3.Tomlins SA, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 4.Ateeq B, et al. Molecular profiling of ETS and non-ETS aberrations in prostate cancer patients from northern India. Prostate. 2015;75:1051–1062. doi: 10.1002/pros.22989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freedman ML, et al. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc Natl Acad Sci USA. 2006;103:14068–14073. doi: 10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amundadottir LT, et al. A common variant associated with prostate cancer in European and African populations. Nat Genet. 2006;38:652–658. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 7.Ahmadiyeh N, et al. 8q24 prostate, breast, and colon cancer risk loci show tissue-specific long-range interaction with MYC. Proc Natl Acad Sci USA. 2010;107:9742–9746. doi: 10.1073/pnas.0910668107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robinson D, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–1228. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosen P, et al. Differences in frequency of ERG oncoprotein expression between index tumors of Caucasian and African American patients with prostate cancer. Urology. 2012;80:749–753. doi: 10.1016/j.urology.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Magi-Galluzzi C, et al. TMPRSS2–ERG gene fusion prevalence and class are significantly different in prostate cancer of Caucasian, African-American and Japanese patients. Prostate. 2010;71:489–497. doi: 10.1002/pros.21265. [DOI] [PubMed] [Google Scholar]
- 11.Khani F, et al. Evidence for molecular differences in prostate cancer between African American and Caucasian men. Clin Cancer Res. 2014;20:4925–4934. doi: 10.1158/1078-0432.CCR-13-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barbieri CE, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mani RS, et al. Induced chromosomal proximity and gene fusions in prostate cancer. Science. 2009;326:1230. doi: 10.1126/science.1178124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bastus NC, et al. Androgen-induced TMPRSS2: ERG fusion in nonmalignant prostate epithelial cells. Cancer Res. 2010;70:9544–9548. doi: 10.1158/0008-5472.CAN-10-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]