Abstract
Circadian disruption is a common environmental and occupational exposure with public health consequences, but not much is known about whether circadian disruption affects in utero development. We investigated whether maternal circadian disruption, using night shift work as a proxy, is associated with variations in DNA methylation patterns of placental tissue in an epigenome-wide association study (EWAS) of night shift work. Here, we compared cytosine-guanosine dinucleotide (CpG) specific methylation genome-wide of placental tissue (measured with the Illumina 450K array) from participants (n = 237) in the Rhode Island Child Health Study (RICHS) who did (n = 53) and did not (n = 184) report working the night shift, using robust linear modeling and adjusting for maternal age, pre-pregnancy smoking, infant sex, maternal adversity, and putative cell mixture. Statistical analyses were adjusted for multiple comparisons and results presented with Bonferroni or Benjamini and Hochberg (BH) adjustment for false discovery rate. Night shift work was associated with differential methylation in placental tissue, including CpG sites in the genes NAV1, SMPD1, TAPBP, CLEC16A, DIP2C, FAM172A, and PLEKHG6 (Bonferroni-adjusted p<0.05). CpG sites within NAV1, MXRA8, GABRG1, PRDM16, WNT5A, and FOXG1 exhibited the most hypomethylation, while CpG sites within TDO2, ADAMTSL3, DLX2, and SERPINA1 exhibited the most hypermethylation (BH q<0.10). Functional analysis indicated GO-terms associated with cell-cell adhesion and enriched GWAS results for psoriasis. Night shift work was associated with differential methylation of the placenta, which may have implications for fetal health and development. This is the first study to examine the epigenetic impacts of night shift exposure, as a proxy for circadian disruption, on placental methylation in humans, and, while results should be interpreted with caution, suggests circadian disruption may have epigenetic impacts.
Introduction
Disruption of circadian rhythms is associated with negative health outcomes such as cancer, metabolic disorders, and neurological disorders in epidemiologic[1] and animal studies[2]; however, the impacts of circadian disruption during pregnancy on fetal development and child health have been largely overlooked. The core circadian clock consists of feedback loops of transcription factors (TF) that generate oscillating cycles of gene transcription and translation. These endogenously generated rhythms rely on cues, such as light, to synchronize patterns of physiological activity with the external environment. Light signals the suprachiasmatic nucleus (SCN) of the hypothalamus, the “master clock”, to set the body’s peripheral clocks[3].
There have been numerous studies to evaluate health outcomes associated with night shift work, an occupational proxy for circadian disruption, but it is unknown whether working the night shift before or during pregnancy poses health risks to the mother or child. This understudied exposure may have large public health consequences, as approximately 15% of American employees work outside of the traditional 9AM-5PM work schedule[4]. While some aspects of the circadian system may return to normal after a regular schedule of night shift work, studies suggest the majority of regular night shift workers (~97%) are not able to fully adapt their endogenous circadian rhythms to their work schedules[5].
Although there appear to be only small risks of negative reproductive health outcomes associated with shift work[6], not much is known about the impact of light or circadian rhythms in human pregnancy or on long-term fetal programming. The placenta, an organ responsible for mediating the maternal and fetal environment to regulate growth and development, may be affected by circadian disruption; yet, little attention has been paid to the impact of circadian disruption on placental function. Because the placenta is composed of fetal DNA, methylation of placental tissue may reflect fetal exposures and future health effects. Therefore, differences in placental methylation patterns between night shift workers and non-night shift workers may indicate altered fetal development and infant health in response to circadian disruption. In this study, we conducted an epigenome-wide association study (EWAS) to investigate whether night shift work is associated with differences in DNA methylation in the placental epigenome, which can impact long-term health outcomes in the offspring.
Methods
Study population—The Rhode Island Child Health Study (RICHS)
The Rhode Island Child Health Study (RICHS) is a hospital-based cohort study of mothers and infants in Rhode Island, described in detail elsewhere[7]. Briefly, from 2009 to 2014, women between the ages of 18–40 and their infants were enrolled at the Women and Infants Hospital of Rhode Island, oversampling for large and small for gestational age infants and matching each to an appropriate for gestational age control by maternal age (± 2 years), sex, and gestational age (± 3 days). RICHS enrolled only full-term (≥37 weeks), singleton deliveries without congenital or chromosomal abnormalities. All participants provided written informed consent under protocols approved by the Institutional Review Boards of Women and Infants Hospital and Emory University.
Demographic information was collected from a questionnaire administered by a trained interviewer and clinical outcome information was obtained from medical records. Information on night shift work was obtained from questionnaire by first asking, “Have you ever worked outside the home? (Yes/No)” and if “Yes”, participants were asked “If yes, please list all of the jobs you have had starting with your current job first. Please indicate whether you worked a swing shift or a night shift on any of these jobs”. To indicate shift jobs, the questionnaire included check boxes for “Yes” and “No” under a category for “Night Shift”. For this analysis, only the most recently reported job history was used and only those who reported “Yes” for night shift work were considered night shift workers; people who reported working a swing shift but not a night shift were not included as night shift workers.
To adjust for socioeconomic factors while avoiding multicollinearity, we used an adversity score index to adjust for household income, maternal education, marital status and partner support. The cumulative risk score ranged from 0 to 4, with 0 representing the lowest level of adversity and 4 representing the highest level of adversity. A higher risk score was given to women whose median household income (adjusting for the number of people in the household) fell below the federal poverty line for the year the infant was born (+1), to women whose household was larger than 6 (+1), to women who were single and did not receive support from a partner (+1) and to women whose highest level of education was high school or less (+1)[8].
Placental sample collection and measurement of DNA methylation
Genome-wide DNA methylation (measured with the Illumina 450k arrays) was obtained on 334 placentae parenchyma samples in RICHS as previously described[9], and of these, data from 237 samples were included in this analysis. The QA/QC process has been described elsewhere[10], including functional normalization, BMIQ, and ‘ComBAT’ to adjust for technical variations and batch effects in R[9, 11]. Briefly, we used the ‘minfi’ package in R to convert the raw methylation files to β values, a ratio of methylation ranging from 0 to 1, for analysis. Probes associated with the X or Y chromosomes, single nucleotide polymorphism (SNP)-associated (within 10bp of the target cytosine-guanosine dinucleotide (CpG) site and with minor allele frequency >1%), identified as cross-reactive or polymorphic by Chen et al[12], or with poor detection p-values were excluded, yielding 334,692 probes for analysis in this study[9]. DNA methylation array data for RICHS can be found in the NCBI Gene Expression Omnibus (GEO) database with the number GSE75248. Women with missing information on pre-pregnancy smoking status (“No”/”Yes”), defined as smoking 3 months prior to pregnancy, or adversity score were not included in the analysis. Women who did not provide an answer for the nightshift variable (n = 16) were recoded to “No”. This study included the 237 mother-infant pairs within RICHS for which DNA methylation data and the necessary demographic information were available.
Placental RNA sequencing
Gene expression was measured using the Illumina HiSeq 2500 system in 199 placental samples from RICHS; methods have been previously described [13]. After standard QA/QC procedures, final data were normalized to log2 counts per million (logCPM) values. Raw data is available in the NCBI sequence read archive (SRP095910).
Statistical analyses
Because a reference panel for placental cell types does not yet exist, we used a validated reference-free method, the ‘RefFreeEWAS’ package in R, to adjust for heterogeneity in cell-type composition[14, 15]. We implemented the RefFree estimation via the same process described in detail in our lab’s prior work[16], and identified 8 components to represent the putative cell mixture in our placental samples. We also examined the outlier screening plots of the cell mixture array for extreme outliers. We then conducted an EWAS using robust linear modeling by regressing CpG methylation β-values on night shift work (“No”/”Yes”), adjusting for putative cell mixture, maternal age (years), pre-pregnancy smoking status (“No”/”Yes”) adversity score (0–4)[17], and sex of the infant (“Female”/”Male”). To adjust for multiple comparisons, we used the Bonferroni method and the Benjamini and Hochberg (BH) false discovery rate (FDR) methods. To evaluate the extent of in utero night shift exposure, we compared job and delivery date data. A sensitivity analysis using data from women who provided night shift job information (n = 221) without recoding missing to “No” was performed. We also conducted a sensitivity analysis to evaluate gestational diabetes mellitus (GDM) and CpG methylation outcomes, as numerous studies have found an association between night shift work and the development of obesity and metabolic diseases[18, 19], as well as GDM and altered methylation[20]. While DNA methylation is not expected to change on a day-to-night basis[21], a few recent studies of brain tissue have found diurnal differences in DNA methylation[22, 23]. To assess possible confounding by time of placenta sample collection, we categorized time of sample collection into 3-hour bins (7AM-9AM, 10AM-12PM, 1PM-3PM, and 4PM-5PM) and performed a Fisher’s exact test (one of the cells had <5 participants) to compare night shift and non-night shift workers. We also modelled sample collection time as a continuous outcome and night shift work as a categorical exposure (No/Yes).
Additionally, we investigated differentially methylated regions (DMRs) using the ‘Bumphunter’ package in R[24]. We modeled the β-values between non-night shift workers and night shift workers, controlling for the same variables as the individual CpG by CpG site genome-wide analysis. CpG sites within 500 base pairs were clustered together and β-values were modeled against a null distribution generated via bootstrapping; sites with differential methylation of 2% or more were considered to be possible DMRs.
To examine the functional implications of night shift work-associated DNA methylation (BH q<0.05), we also conducted an expression quantitative trait (eQTM) analysis using ‘MEAL’[25] in R to investigate whether methylation was associated with gene expression in the RICHS samples on which both DNA methylation and expression data were available (n = 199). Using robust linear modeling, we regressed the expression levels of genes within a 100kb window of the CpG site on methylation β-values (p<0.05).
Bioinformatic analyses
To better understand the biological significance of the EWAS results, we performed an enrichment analysis of the top 298 CpG sites (BH q<0.10) with GO-terms and KEGG pathways in R using the ‘missMethyl’ package[26]. We also evaluated whether the genes (n = 45) from overlapping CpG sites with BH q<0.05 were listed as rhythmic within the available CircaDB mouse databases[27]. We searched within CIRCA mouse experimental datasets using the JTK filter with a q-value probability cut-off of 0.05 and a JTK phase range of 0–40[28]. To investigate whether the CpGs from our EWAS results were within genomic regions that have been linked to traits from previous GWAS findings, windows of the top 298 CpG sites (BH q<0.10) and flanking 5kb regions of DNA were compared for overlap with SNP results (p<1x10-8) in the GWAS catalog of the National Human Genome Research Institute and the European Bioinformatics Institute (NHGRI-EBI)[29] using the TraseR package[30] in R. As background, we only included SNPs that were within 5kb of CpGs that were included in the EWAS study. If more than one top CpG site fell within the same 10kb window, they were clustered together and considered the same region. Only GWAS trait-associated SNP windows that overlapped with 2 or more of the top CpG windows and were statistically significant after Fisher’s exact test (BH q <0.05) were considered enriched.
Results
Demographic and medical information
Demographic information for the women (n = 237) and covariates included in the final model for the epigenome-wide analysis are provided in Table 1. A sensitivity analysis comparing results from women who provided night shift job information (n = 221) without recoding missing to “No” did not indicate any large differences in demographic features. Overall, women who reported working the night shift were more likely to be younger, smokers pre-pregnancy, cases of GDM, single and never married, lower household income, and higher adversity (p<0.05). While not statistically significant, women who worked the night shift trended towards a higher BMI and an evening chronotype. Of those included in the analysis, one participant reported taking melatonin and she was not a night shift worker. Additionally, 37 out of the 53 (70%) night shift workers reported working the night shift during pregnancy; time between working the night shift and the birth of the infant ranged from within a week to approximately 4.5 years, with a median value of 10 weeks.
Table 1. Demographic characteristics of participants included in the analysis (n = 237) by night shift work status.
| N | Non-night shift (n = 184) | Night shift (n = 53) | Statistical significance | |
|---|---|---|---|---|
| Maternal age*, mean ± SD | 237 | 30.7+/- 5.4 | 28.8+/- 5.1 | p<0.05 |
| Pre-pregnancy smoking*, % (n) | 237 | 12% (22) | 25% (13) | p<0.05 |
| Sex of the infant (male/female), % (n) | 237 | 48% / 52% (88/96) | 53% / 47% (28/25) | p = 0.6 |
| Gestational diabetes*, % (n) | 234 | 9% (16) | 21% (11) | p<0.05 |
| Marital status*, % (n) | 237 | p<0.05 | ||
| Single, never married | 24% (44) | 43% (23) | ||
| Separated or divorced | 3% (6) | 4% (2) | ||
| Married | 73% (134) | 53% (28) | ||
| Household income*, % (n) | 229 | p<0.01 | ||
| <$9–14,999 | 14% (25) | 24% (12) | ||
| $15–29,999 | 9% (17) | 20% (10) | ||
| $30–49,999 | 10% (18) | 18% (9) | ||
| $50–99,999 | 39% (69) | 30% (15) | ||
| .>$100,000 | 28% (50) | 8% (4) | ||
| Adversity score*, % (n) | 237 | p<0.05 | ||
| 0 | 78% (143) | 58% (31) | ||
| 1 | 12% (22) | 30% (16) | ||
| 2 | 9% (16) | 9% (5) | ||
| 3 | 1% (2) | 2% (1) | ||
| 4 | 1% (1) | 0% (0) | ||
| Maternal education*, % (n) | 237 | p<0.01 | ||
| <11th grade | 5% (10) | 4% (2) | ||
| High school | 15% (28) | 26% (14) | ||
| Junior college or equivalent | 22% (40) | 40% (21) | ||
| College | 36% (67) | 26% (14) | ||
| Any post-graduate | 21% (39) | 4% (2) |
*Signifies p-value <0.05 (using either χ2 test, Fisher’s exact test or 2-sided t-test) between non-night shift and night shift workers.
Epigenome-wide methylation associations
DNA methylation at 298 CpG sites was found to be significantly different in night shift workers after FDR correction at the BH q<0.10, 57 CpG sites significant at the BH q<0.05 (Table 2), and 10 CpG sites at the Bonferroni-corrected p<0.05 (Table 2). CpG sites for the NAV1, SMPD1, TAPBP, CLEC16A, DIP2C, FAM172A, and PLEKHG6 genes had genome-wide significance after Bonferroni correction (p<0.05). The ADAMTS10, CLEC16A, CTBP1, EGFL8, GNAS, HDAC4, HEATR2, KCNA4, KDELC2, MFHAS1, MXRA8, NAV1, PLXND1, UBR5, WNT5A, and ZBTB22 genes had multiple CpG sites represented in the results.
Table 2. List of differentially methylated CpG sites in night shift workers compared to non-night shift workers after epigenome-wide analysis (BH q<0.05).
| UCSC Gene Name | Chromosome | Position | Probe ID | UCSC Gene Group | Enhancer | β1 | SE | P-value | BH q-value | Bonferroni |
|---|---|---|---|---|---|---|---|---|---|---|
| NAV1 | chr1 | 201708718 | cg14168733 | TSS1500 | NA | -0.04 | 0.007 | 2.53E-08 | 0.003 | 0.008 |
| NAV1 | chr1 | 201709135 | cg14377596 | 1stExon | TRUE | -0.04 | 0.007 | 2.98E-08 | 0.003 | 0.01 |
| SMPD1 | chr11 | 6412852 | cg14814323 | Body | NA | -0.016 | 0.003 | 2.97E-08 | 0.003 | 0.01 |
| NAV1 | chr1 | 201709390 | cg01411786 | Body | TRUE | -0.032 | 0.006 | 9.91E-08 | 0.004 | 0.033 |
| TAPBP | chr6 | 33273011 | cg03190911 | Body | NA | -0.014 | 0.003 | 9.94E-08 | 0.004 | 0.033 |
| chr6 | 27390647 | cg06667732 | NA | -0.023 | 0.004 | 9.35E-08 | 0.004 | 0.031 | ||
| CLEC16A | chr16 | 11073063 | cg08082763 | Body | TRUE | -0.023 | 0.004 | 7.21E-08 | 0.004 | 0.024 |
| DIP2C | chr10 | 560669 | cg21373996 | Body | NA | -0.019 | 0.004 | 1.06E-07 | 0.004 | 0.035 |
| FAM172A | chr5 | 93076910 | cg25342875 | Body | NA | -0.024 | 0.004 | 9.46E-08 | 0.004 | 0.032 |
| PLEKHG6 | chr12 | 6436676 | cg14858786 | Body | NA | -0.026 | 0.005 | 1.42E-07 | 0.005 | 0.047 |
| KRT15 | chr17 | 39675154 | cg11983245 | 5'UTR | NA | -0.024 | 0.005 | 1.84E-07 | 0.005 | 0.062 |
| NAV1 | chr1 | 201709675 | cg18539461 | Body | TRUE | -0.036 | 0.007 | 1.71E-07 | 0.005 | 0.057 |
| RHOT2 | chr16 | 717556 | cg04365973 | TSS1500 | NA | -0.019 | 0.004 | 2.58E-07 | 0.007 | 0.086 |
| NAV1 | chr1 | 201708888 | cg13877974 | TSS200 | NA | -0.043 | 0.009 | 4.11E-07 | 0.01 | 0.137 |
| ERI3 | chr1 | 44716226 | cg24373865 | Body | NA | -0.024 | 0.005 | 5.66E-07 | 0.013 | 0.189 |
| PTPN6 | chr12 | 7060187 | cg23147227 | TSS1500 | NA | -0.02 | 0.004 | 8.98E-07 | 0.019 | 0.301 |
| EGFL8 | chr6 | 32135718 | cg08759957 | Body | NA | -0.021 | 0.004 | 1.22E-06 | 0.023 | 0.407 |
| ZBTB22 | chr6 | 33284168 | cg14771240 | Body | NA | -0.02 | 0.004 | 1.18E-06 | 0.023 | 0.396 |
| chr10 | 22725309 | cg01422243 | NA | -0.019 | 0.004 | 1.51E-06 | 0.027 | 0.504 | ||
| UBR5 | chr8 | 103344822 | cg02530407 | Body | TRUE | -0.019 | 0.004 | 1.68E-06 | 0.027 | 0.561 |
| HDAC4 | chr2 | 240213173 | cg23601374 | Body | TRUE | -0.017 | 0.004 | 1.64E-06 | 0.027 | 0.549 |
| chr7 | 25702848 | cg03700230 | NA | 0.048 | 0.01 | 1.93E-06 | 0.029 | 0.645 | ||
| CYB5R2 | chr11 | 7694163 | cg05919312 | 5'UTR | NA | -0.018 | 0.004 | 2.03E-06 | 0.03 | 0.679 |
| RPS6KA4 | chr11 | 64139406 | cg07425109 | 3'UTR | NA | -0.016 | 0.003 | 2.15E-06 | 0.03 | 0.719 |
| MSI2 | chr17 | 55742491 | cg07618409 | Body | TRUE | -0.02 | 0.004 | 2.26E-06 | 0.03 | 0.755 |
| CDYL2 | chr16 | 80716710 | cg16713168 | Body | TRUE | -0.021 | 0.004 | 2.33E-06 | 0.03 | 0.78 |
| FAM118A | chr22 | 45705265 | cg06575572 | 5'UTR | NA | -0.02 | 0.004 | 2.50E-06 | 0.03 | 0.835 |
| LRRC2 | chr3 | 46618325 | cg07225641 | 5'UTR | NA | -0.027 | 0.006 | 2.61E-06 | 0.03 | 0.875 |
| CLDN9 | chr16 | 3063894 | cg10492999 | 1stExon | NA | -0.026 | 0.006 | 2.70E-06 | 0.03 | 0.905 |
| LOC645323 | chr5 | 87955859 | cg13982098 | Body | NA | -0.028 | 0.006 | 2.65E-06 | 0.03 | 0.886 |
| C2orf54 | chr2 | 241827789 | cg21333033 | Body | NA | -0.019 | 0.004 | 2.93E-06 | 0.032 | 0.981 |
| SLC41A1 | chr1 | 205780033 | cg00762738 | 5'UTR | NA | -0.017 | 0.004 | 3.08E-06 | 0.032 | 1 |
| MXRA8 | chr1 | 1290712 | cg00040588 | Body | NA | -0.051 | 0.011 | 3.49E-06 | 0.032 | 1 |
| EGFL8 | chr6 | 32135715 | cg12305588 | Body | NA | -0.019 | 0.004 | 3.35E-06 | 0.032 | 1 |
| BAIAP2 | chr17 | 79022879 | cg12472449 | Body | NA | -0.016 | 0.004 | 3.43E-06 | 0.032 | 1 |
| FBXW7 | chr4 | 153437193 | cg13536107 | 5'UTR | TRUE | -0.022 | 0.005 | 3.35E-06 | 0.032 | 1 |
| BAT2 | chr6 | 31599646 | cg25371129 | Body | NA | -0.005 | 0.001 | 3.61E-06 | 0.033 | 1 |
| MIRLET7A3 | chr22 | 46508563 | cg04063235 | TSS200 | NA | -0.019 | 0.004 | 3.71E-06 | 0.033 | 1 |
| HDLBP | chr2 | 242174625 | cg11221200 | Body | NA | -0.014 | 0.003 | 3.90E-06 | 0.033 | 1 |
| chr11 | 22454301 | cg23181580 | TRUE | -0.031 | 0.007 | 4.22E-06 | 0.035 | 1 | ||
| BATF3 | chr1 | 212874153 | cg00168835 | TSS1500 | NA | 0.005 | 0.001 | 4.42E-06 | 0.036 | 1 |
| chr22 | 50221949 | cg08174792 | NA | -0.034 | 0.007 | 4.91E-06 | 0.039 | 1 | ||
| MFHAS1 | chr8 | 8749074 | cg01022370 | 1stExon | TRUE | -0.023 | 0.005 | 5.20E-06 | 0.04 | 1 |
| ZNF284 | chr19 | 44575547 | cg05333740 | TSS1500 | NA | -0.023 | 0.005 | 5.83E-06 | 0.042 | 1 |
| DPEP2 | chr16 | 68027297 | cg06866814 | 5'UTR | NA | 0.002 | 0 | 5.50E-06 | 0.042 | 1 |
| GALNTL4 | chr11 | 11438208 | cg16337763 | Body | TRUE | -0.022 | 0.005 | 5.67E-06 | 0.042 | 1 |
| AZI1 | chr17 | 79184968 | cg20296990 | Body | NA | -0.02 | 0.004 | 5.84E-06 | 0.042 | 1 |
| GALNTL1 | chr14 | 69725831 | cg00080706 | TSS1500 | NA | -0.019 | 0.004 | 5.99E-06 | 0.042 | 1 |
| MFHAS1 | chr8 | 8749278 | cg01784220 | 1stExon | TRUE | -0.022 | 0.005 | 6.21E-06 | 0.042 | 1 |
| C11orf2 | chr11 | 64863151 | cg13626866 | TSS1500 | NA | -0.026 | 0.006 | 6.37E-06 | 0.043 | 1 |
| BANF1 | chr11 | 65770987 | cg17985854 | Body | NA | -0.023 | 0.005 | 6.49E-06 | 0.043 | 1 |
| IQGAP2 | chr5 | 75784957 | cg23289545 | Body | TRUE | -0.019 | 0.004 | 6.62E-06 | 0.043 | 1 |
| chr17 | 43222106 | cg00625783 | TRUE | -0.025 | 0.006 | 7.26E-06 | 0.045 | 1 | ||
| TUBGCP2 | chr10 | 135120640 | cg04070692 | 5'UTR | NA | -0.019 | 0.004 | 7.21E-06 | 0.045 | 1 |
| BAT1 | chr6 | 31502388 | cg10895184 | Body | NA | -0.018 | 0.004 | 7.53E-06 | 0.045 | 1 |
| HAPLN1 | chr5 | 83016779 | cg18024167 | 1stExon | NA | -0.023 | 0.005 | 7.44E-06 | 0.045 | 1 |
| PKHD1L1 | chr8 | 110374866 | cg19906741 | 1stExon | TRUE | 0.018 | 0.004 | 7.77E-06 | 0.046 | 1 |
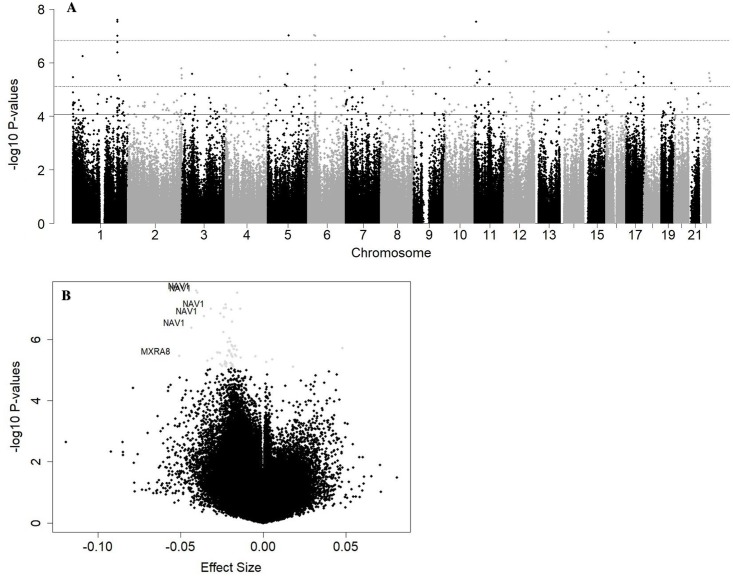
The Manhattan plot of the results indicated a number of differentially methylated sites that distributed across the genome, with some occurring in the same regions (Fig 1A). There was also an overall trend towards hypomethylation (Fig 1B). CpG sites for NAV1, MXRA8, GABRG1, PRDM16, WNT5A, and FOXG1 were among the 10 sites with the most hypomethylation, while CpG sites for TDO2, ADAMTSL3, DLX2, and SERPINA1 were among the 10 sites with the most hypermethylation To more rigorously examine the co-located CpG sites associated with night shift work, we employed a ‘Bumphunter’ analysis and identified 6584 ‘bumps’, with areas of the NAV1, PURA, C6orf47, and GNAS genes as DMRs (BH q<0.10)(Table 3). Of these, CpGs for the NAV1 and GNAS genes were also differentially methylated in the CpG by CpG analysis (S1 Table).
Fig 1. Results of placental DNA methylation and night shift work EWAS.
A, Manhattan plot of CpG results, adjusted for maternal age, pre-pregnancy smoking, adversity score, sex of the infant, and estimated cell mixture. The dashed upper boundary line denotes p-value of 1.49x10-7 as the significance threshold after Bonferroni adjustment (p<0.05), the dashed middle boundary line denotes the p-value of 7.7x10-6 as the approximate significance threshold of BH q<0.05, and the solid boundary line at denotes the p-value of 8.8x10-5 as the approximate significance threshold of BH q<0.10. B, Volcano plot of results, adjusted for maternal age, pre-pregnancy smoking, adversity score, sex of the infant, and estimated cell mixture. Gray dots signify CpG sites with BH q<0.05 and CpG sites with both absolute beta coefficients of 0.03 or greater and BH q<0.05 are labelled with UCSC gene names.
Table 3. ‘Bumphunter’ results of significant DMRs (BH q<0.10).
| Gene | Chromosome | Start | End | β1 | Area | L | clusterL | P-value | FWER | P-value Area | FWER Area | BH q-value | Bonferroni |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NAV1 | chr1 | 201708500 | 201709675 | -0.038 | 0.452 | 12 | 12 | 3.29E-05 | 0.166 | 1.74E-04 | 0.606 | 0.054 | 0.217 |
| PURA | chr5 | 139493486 | 139494006 | -0.054 | 0.544 | 10 | 10 | 2.54E-05 | 0.131 | 6.80E-05 | 0.31 | 0.054 | 0.167 |
| C6orf47 | chr6 | 31627678 | 31627678 | -0.112 | 0.112 | 1 | 38 | 3.24E-05 | 0.163 | 1.17E-02 | 1 | 0.054 | 0.213 |
| GNAS | chr20 | 57463325 | 57463725 | -0.034 | 0.482 | 14 | 30 | 9.35E-06 | 0.05 | 1.31E-04 | 0.512 | 0.054 | 0.062 |
A sensitivity analysis with GDM included as an additional covariate shared many of the top CpG sites with the primary results, suggesting GDM is not a confounder of these associations. An additional analysis evaluating GDM as the primary exposure shared no top genes with the EWAS results. In comparing time of placenta sample collection, there was no significant difference between the two groups when compared categorically (p = 0.547) or continuously (p = 0.945), with a mean collection time around 11AM. In another sensitivity analysis comparing the beta coefficients from models that utilized in utero night shift work exposure as the independent variable (n = 37) to the beta coefficients from models that included all night shift workers (n = 53), the differences were small; only 1 CpG site, cg24373865, had an absolute difference in beta coefficients greater than 0.01, at 0.011. We also re-examined our results removing those with missing data on night shift work and the findings were substantially similar.
Functional analyses
Comparing the 298 significant CpG sites (BH q<0.10) to the remaining 334,394 CpG sites, there was a higher frequency of top CpG sites within enhancer regions (χ2 = 13.48, df = 1, p-value = 0.0002). Because transcription factors (TFs) can bind to enhancer regions to alter gene expression, we assessed whether CpG methylation was associated with expression levels in nearby genes. The eQTM analysis found the expression of 18 genes to be associated with 14 CpG sites (p<0.05). Of these, the expression levels of ACBD4 were associated with methylation in cg00625783 (β1 = 2.515, p-value = 1.94E-05) and the expression levels of KRT15 were associated with methylation in cg11983245 (β1 = 7.895, p-value = 8.04E-05)(S2 Table). For both of these genes, increasing methylation of the CpG sites was associated with increased gene expression. The cg00625783 CpG is not annotated to a gene, but is located within an enhancer region, and cg11983245 is annotated to the 5’ untranslated region (5’UTR) and 1st exon of the KRT15 gene. Methylation of cg11983245 was also associated (p<0.05) with increased KRT19 (β1 = 4.404, p-value = 3.87E-03) and LINC00974 (β1 = 6.011, p-value = 3.40E-02) expression levels.
We analyzed the top 298 CpG sites (BH q<0.10) for enrichment of KEGG pathways and GO-terms. The GO-terms “cell-cell adhesion”, “cell-cell adhesion via plasma-membrane adhesion molecules”, and “hemophilic cell adhesion via plasma membrane adhesion molecules” were found to be significant after FDR correction (BH<0.05)(S3 Table). The top KEGG pathway results were “valine, leucine and isoleucine biosynthesis”, “mucin type O-glycan biosynthesis” and “melanogenesis”, but they were not significant after correcting for FDR (S3 Table). Surprisingly, PER1 was the only core circadian gene represented among the 298 CpG sites. However, we evaluated whether the 45 genes of the top 57 CpG sites exhibited circadian rhythmicity within the CircaDB mouse expression database[27] and found 27 out of the 45 genes (60%) displayed rhythmic expression[28](S4 Table). Of these genes, BAIAP2, GALNTL1, HDLBP, NAV1, and TAPBP displayed rhythmicity in mouse SCN tissue. We then tested for trait-associated SNP enrichment within 10kb regions surrounding the top 298 CpG sites (BH q<0.05) among GWAS SNPs (p<1x10-8) in NHGRI-EBI GWAS catalog[29] using a Fisher’s exact test in TraseR[30]. These regions were significantly enriched (FDR < 5%) for the following traits: Psoriasis, Systemic Lupus Erythematosus, Type 1 Diabetes Mellitus, and Multiple Sclerosis (S5 Table).
Discussion
We identified a number of CpG sites exhibiting differential methylation associated with night shift work in newborn placental tissue. While the average absolute differences for the 298 CpG site corresponded to a roughly 1.7% change in methylation, even a small change in methylation may have physiologically-relevant effects, and these magnitudes of association are comparable to others reported for exposures including toxic trace elements and maternal smoking during pregnancy[31]. The overall trend of hypomethylation with night shift work may be due to increased TF binding to DNA, leading to chromatin changes establishing the hypomethylated state[32]. Because one of the core components of the circadian clock, CLOCK, acts as a histone acetyltransferase[33], it is also possible that circadian disruption impacts the epigenetic activity of CLOCK, affecting chromatin state and accessibility. However, there is still much to discover about circadian interactions with methylation and developmental processes.
Light at night and night shift work exposure can cause altered hormonal signaling and endocrine disruption; because hormone receptors can act as TFs, it is possible that circadian disruption causes increased hormonal signaling and increased TF binding. Animal studies of in utero circadian disruption suggest that circadian disruption may negatively affect the health and development of offspring[34]. For example, chronic changes in the photoperiod of pregnant rats caused increased leptin levels, insulin secretion, fat deposition, and decreased glucose tolerance of offspring in adulthood[35]. Additionally, mice exposed to a 22-hour light-dark cycle, instead of the normal 24-hour cycle, had altered methylation patterns in the SCN and altered circadian behavior; differential methylation was also found for genes related to axonal migration, synaptogenesis, and neuroendocrine hormones[36].
We identified a DMR and multiple individual CpG sites within and nearby to NAV1 that were consistently represented among the top results. In general, the functions of NAV1, particularly in the placenta, are not well characterized. NAV1 is homologous to the unc-53 gene in C.elegans, which plays a role in axonal migration[37]. The mouse homolog also appears to play a role in neuronal migration; NAV1 is enriched in growth cones and associates with microtubule plus ends[38], and the deficit of Nav1 causes loss of direction in leading processes[39]. Research has also found increased embryonic lethality, decreased birthweight, and infertility in female offspring for Nav1-/- mice[40], suggesting an important role for Nav1 in fetal development and health. Our mouse tissue query of the CircaDB database revealed that Nav1 specifically displayed circadian rhythmicity in mouse SCN tissue (S4 Table). This suggests NAV1 may play a role in the mammalian SCN. A DMR was also identified in GNAS, which is imprinted in the paraventricular nucleus of the hypothalamus and encodes the Gsα G-protein, which regulates cAMP generation and metabolism. Gnas is implicated in REM and NREM sleep and the browning of white adipose tissue for thermogenesis[41]. Additionally, in a microarray analysis of retina samples from an rd/rd mouse model, Gnas was implicated in melanopsin signaling[42]. Therefore, GNAS may be important in integrating light and metabolic cues.
The top 298 CpG results (BH q <.05) were enriched for traits related to psoriasis, lupus, type 1 diabetes, and multiple sclerosis, all of which involve the immune system and/or inflammation. Interestingly, a large study of night shift workers in the Nurse’s Health Study found an increased risk of psoriasis among night shift workers[43]. Another study that analyzed two separate cohorts also found an association between engaging in shift work before 20 years old and multiple sclerosis[44]. The skin has circadian rhythms that may affect the development of psoriasis[45], during which abnormal activity of keratinocytes and T cells can cause lesions. Because adhesion molecules may play an important role in this process, this GWAS trait may explain the KEGG-pathway and GO-term enrichment analysis, among which “cell-cell adhesion” and “melanogenesis” were some of the top results.
A possible limitation of this analysis is the moderate sample size of night shift workers (n = 53). Because placenta samples were only collected during daytime hospital hours (7AM-5PM), we are also limited in our ability to fully evaluate diurnal differences in DNA methylation. Additionally, the adjustment for cell-type heterogeneity is an estimation, so there is a possibility of residual confounding by cell type. On the other hand, the results may be a conservative estimate of the true association, as this analysis occurred in full-term pregnancies and approximately 30% of the women included as night shift workers did not have in utero exposure. While a sensitivity analysis of in utero night shift work exposure did not find large differences in the magnitudes of association, exposure to circadian disruption at different windows of development could have different magnitudes of effect. Prior research has found that shift workers continue to have chronic health effects even after they switch to a day shift schedule. For example, researchers found that a history of shift work was associated with a decrease in cognitive ability that took 5 years or more after cessation of shift work to recover[46]; this suggests recovery from regular shift work may take an extended period of time and a history of shift work may have a prolonged influence on health.
This is the first study to examine the epigenetic impacts of night shift exposure on placental methylation in humans, and results should be interpreted with caution. Methylation of placental tissue, an indicator of the in utero epigenetic landscape, reflects functional activities of the placenta, which can impact various aspects of fetal development, including neurodevelopment. The findings that the methylation of NAV1 differed by night shift work exposure and that Nav1 is rhythmically expressed in mouse SCN suggests NAV1 may play a role in the human circadian system. Because the circadian system coordinates an array of physiological systems, alterations to circadian system development could affect immune response, sleep patterns, behavior, metabolism, and future health status. We have found night shift work to be associated with variation in methylation of placental tissue, which has implications for fetal development and future health. However, these findings may also be relevant for people who experience circadian disruption due to common exposures such as light at night[47].
In conclusion, night shift work is associated with differential methylation patterns in placental tissue. NAV1 may be an important component in the development of the human circadian system. Night shift work is a complex exposure encompassing altered hormonal signaling, eating and activity patterns, light exposure, and sleep patterns. Therefore, it is difficult to tease apart which aspects of night shift work contribute to which result. However, night shift work is a prevalent exposure in the workforce and, more generally, circadian disruption is a common facet of modern life. Circadian disruption may contribute to immune-mediated and inflammatory disease, but it is still unclear how this exposure may affect fetal development and infant health. These findings warrant further investigation to evaluate the effects of in utero circadian disruption and possible impacts on fetal and child health, as well as the role of the circadian system in the function of the placenta.
Supporting information
(XLSX)
The data used in this analysis came from RICHS samples that had both methylation and RNAseq data available (n = 199).
(XLSX)
N refers to the overall number of genes annotated to the term or pathway and DE refers to the number of genes from the top CpG sites within that term or pathway.
(XLSX)
Gene list was entered and analyzed using a JTK q-value filter with a probability cut-off of 0.05 and JTK phase range of 0–40 of all available CIRCA mouse databases.
(XLSX)
(XLSX)
Data Availability
We have submitted the data to the NCBI GEO repository, accession number GSE75248. The methylation data and associated covariates used in the analysis are available here: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE75248.
Funding Statement
This work was supported by funding from the National Institutes of Health (NIH-NIEHS R01ES022223 [to CJM, JC, KH], NIH-NIEHS R24ES025145 [to CJM], NIH-NIEHS P01 ES022832 [to CJM], NIH-NICHD F31 HD097918 [to DACT], and NIH-NIEHS T32 ES012870 [to DACT]) (https://www.nih.gov/) and by the United States Environmental Protection Agency (US EPA grant RD83544201 (https://www.epa.gov/) [to CJM]). The study sponsors did not have any role in the study design, collection, analysis, and interpretation of the data, the writing of the report, or the decision to submit the paper for publication.
References
- 1.Bass J, Lazar MA. Circadian time signatures of fitness and disease. Science. 2016;354(6315):994–9. Epub 2016/11/26. 10.1126/science.aah4965 . [DOI] [PubMed] [Google Scholar]
- 2.Opperhuizen AL, van Kerkhof LW, Proper KI, Rodenburg W, Kalsbeek A. Rodent models to study the metabolic effects of shiftwork in humans. Frontiers in pharmacology. 2015;6:50 Epub 2015/04/09. 10.3389/fphar.2015.00050 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berson DM, Dunn FA, Takao M. Phototransduction by retinal ganglion cells that set the circadian clock. Science. 2002;295(5557):1070–3. 10.1126/science.1067262 [DOI] [PubMed] [Google Scholar]
- 4.Straif K, Baan R, Grosse Y, Secretan B, El Ghissassi F, Bouvard V, et al. Carcinogenicity of shift-work, painting, and fire-fighting. The Lancet Oncology. 2007;8(12):1065–6. Epub 2007/12/01. 10.1016/S1470-2045(07)70373-X . [DOI] [PubMed] [Google Scholar]
- 5.Folkard S. Do permanent night workers show circadian adjustment? A review based on the endogenous melatonin rhythm. Chronobiol Int. 2008;25(2–3):215–24. 10.1080/07420520802106835 [DOI] [PubMed] [Google Scholar]
- 6.Palmer KT, Bonzini M, Harris EC, Linaker C, Bonde JP. Work activities and risk of prematurity, low birthweight and pre-eclampsia: an updated review with meta-analysis. Occupational and environmental medicine. 2013;70(4):213–22. 10.1136/oemed-2012-101032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Appleton AA, Murphy MA, Koestler DC, Lesseur C, Paquette AG, Padbury JF, et al. Prenatal Programming of Infant Neurobehaviour in a Healthy Population. Paediatric and perinatal epidemiology. 2016;30(4):367–75. Epub 2016/03/24. 10.1111/ppe.12294 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Appleton AA, Armstrong DA, Lesseur C, Lee J, Padbury JF, Lester BM, et al. Patterning in Placental 11-B Hydroxysteroid Dehydrogenase Methylation According to Prenatal Socioeconomic Adversity. PLoS One. 2013;8(9):e74691 10.1371/journal.pone.0074691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paquette AG, Houseman EA, Green BB, Lesseur C, Armstrong DA, Lester B, et al. Regions of variable DNA methylation in human placenta associated with newborn neurobehavior. Epigenetics. 2016;11(8):603–13. 10.1080/15592294.2016.1195534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maccani JZ, Koestler DC, Lester B, Houseman EA, Armstrong DA, Kelsey KT, et al. Placental DNA Methylation Related to Both Infant Toenail Mercury and Adverse Neurobehavioral Outcomes. Environ Health Perspect. 2015;123(7):723–9. Epub 2015/03/10. 10.1289/ehp.1408561 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics (Oxford, England). 2007;8(1):118–27. Epub 2006/04/25. 10.1093/biostatistics/kxj037 . [DOI] [PubMed] [Google Scholar]
- 12.Chen YA, Lemire M, Choufani S, Butcher DT, Grafodatskaya D, Zanke BW, et al. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics. 2013;8(2):203–9. Epub 2013/01/15. 10.4161/epi.23470 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deyssenroth MA, Peng S, Hao K, Lambertini L, Marsit CJ, Chen J. Whole-transcriptome analysis delineates the human placenta gene network and its associations with fetal growth. BMC Genomics. 2017;18(1):520 10.1186/s12864-017-3878-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Houseman EA, Kile ML, Christiani DC, Ince TA, Kelsey KT, Marsit CJ. Reference-free deconvolution of DNA methylation data and mediation by cell composition effects. BMC Bioinformatics. 2016;17(1):259 10.1186/s12859-016-1140-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teschendorff AE, Zheng SC. Cell-type deconvolution in epigenome-wide association studies: a review and recommendations. Epigenomics. 2017;9(5):757–68. 10.2217/epi-2016-0153 [DOI] [PubMed] [Google Scholar]
- 16.Everson TM, Punshon T, Jackson BP, Hao K, Lambertini L, Chen J, et al. Cadmium-associated differential methylation throughout the placental genome: epigenome-wide association study of two US birth cohorts. bioRxiv. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lam LL, Emberly E, Fraser HB, Neumann SM, Chen E, Miller GE, et al. Factors underlying variable DNA methylation in a human community cohort. Proc Natl Acad Sci U S A. 2012;109:17253–60. 10.1073/pnas.1121249109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Froy O. Metabolism and Circadian Rhythms—Implications for Obesity. Endocr Rev. 2010;31(1):1–24. 10.1210/er.2009-0014 [DOI] [PubMed] [Google Scholar]
- 19.Scheer F, Hilton MF, Mantzoros CS, Shea SA. Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc Natl Acad Sci U S A. 2009;106(11):4453–8. 10.1073/pnas.0808180106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haertle L, El Hajj N, Dittrich M, Müller T, Nanda I, Lehnen H, et al. Epigenetic signatures of gestational diabetes mellitus on cord blood methylation. Clin Epigenetics. 2017;9(1):28 10.1186/s13148-017-0329-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425 10.1038/nature05918 [DOI] [PubMed] [Google Scholar]
- 22.Coulson RL, Yasui DH, Dunaway KW, Laufer BI, Vogel Ciernia A, Zhu Y, et al. Snord116-dependent diurnal rhythm of DNA methylation in mouse cortex. Nat Commun. 2018;9(1):1616 10.1038/s41467-018-03676-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim ASP, Klein H-U, Yu L, Chibnik LB, Ali S, Xu J, et al. Diurnal and seasonal molecular rhythms in human neocortex and their relation to Alzheimer’s disease. Nat Commun. 2017;8:14931 https://www.nature.com/articles/ncomms14931#supplementary-information. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jaffe AE, Murakami P, Lee H, Leek JT, Fallin MD, Feinberg AP, et al. Bump hunting to identify differentially methylated regions in epigenetic epidemiology studies. International journal of epidemiology. 2012;41(1):200–9. Epub 2012/03/17. 10.1093/ije/dyr238 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruiz-Arenas C GJ. MEAL: Perform methylation analysis. 2018.
- 26.Phipson B, Maksimovic J, Oshlack A. missMethyl: an R package for analyzing data from Illumina’s HumanMethylation450 platform. Bioinformatics (Oxford, England). 2016;32(2):286–8. 10.1093/bioinformatics/btv560 [DOI] [PubMed] [Google Scholar]
- 27.Pizarro A, Hayer K, Lahens NF, Hogenesch JB. CircaDB: a database of mammalian circadian gene expression profiles. Nucleic Acids Res. 2013;41(Database issue):D1009–13. Epub 2012/11/28. 10.1093/nar/gks1161 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hughes ME, Hogenesch JB, Kornacker K. JTK_CYCLE: an efficient non-parametric algorithm for detecting rhythmic components in genome-scale datasets. J Biol Rhythms. 2010;25(5):372–80. 10.1177/0748730410379711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacArthur J, Bowler E, Cerezo M, Gil L, Hall P, Hastings E, et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 2017;45(Database issue):D896–D901. 10.1093/nar/gkw1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen L, Qin ZS. traseR: an R package for performing trait-associated SNP enrichment analysis in genomic intervals. Bioinformatics (Oxford, England). 2016;32(8):1214–6. Epub 2015/12/20. 10.1093/bioinformatics/btv741 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Breton CV, Marsit CJ, Faustman E, Nadeau K, Goodrich JM, Dolinoy DC, et al. Small-Magnitude Effect Sizes in Epigenetic End Points are Important in Children’s Environmental Health Studies: The Children’s Environmental Health and Disease Prevention Research Center’s Epigenetics Working Group. Environ Health Perspect. 2017;125(4):511–26. 10.1289/EHP595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin EM, Fry RC. A cross-study analysis of prenatal exposures to environmental contaminants and the epigenome: support for stress-responsive transcription factor occupancy as a mediator of gene-specific CpG methylation patterning. Environmental epigenetics. 2016;2(1). Epub 2016/04/12. 10.1093/eep/dvv011 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doi M, Hirayama J, Sassone-Corsi P. Circadian Regulator CLOCK Is a Histone Acetyltransferase. Cell. 2006;125(3):497–508. 10.1016/j.cell.2006.03.033 [DOI] [PubMed] [Google Scholar]
- 34.Smarr BL, Grant AD, Perez L, Zucker I, Kriegsfeld LJ. Maternal and Early-Life Circadian Disruption Have Long-Lasting Negative Consequences on Offspring Development and Adult Behavior in Mice. Sci Rep. 2017;7:3326 10.1038/s41598-017-03406-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Varcoe TJ, Wight N, Voultsios A, Salkeld MD, Kennaway DJ. Chronic Phase Shifts of the Photoperiod throughout Pregnancy Programs Glucose Intolerance and Insulin Resistance in the Rat. PLoS One. 2011;6(4). 10.1371/journal.pone.0018504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Azzi A, Dallmann R, Casserly A, Rehrauer H, Patrignani A, Maier B, et al. Circadian behavior is light-reprogrammed by plastic DNA methylation. Nat Neurosci. 2014;17(3):377–82. 10.1038/nn.3651 [DOI] [PubMed] [Google Scholar]
- 37.Maes T, Barcelo A, Buesa C. Neuron navigator: a human gene family with homology to unc-53, a cell guidance gene from Caenorhabditis elegans. Genomics. 2002;80(1):21–30. Epub 2002/06/25. . [DOI] [PubMed] [Google Scholar]
- 38.van Haren J, Draegestein K, Keijzer N, Abrahams JP, Grosveld F, Peeters PJ, et al. Mammalian Navigators are microtubule plus-end tracking proteins that can reorganize the cytoskeleton to induce neurite-like extensions. Cell motility and the cytoskeleton. 2009;66(10):824–38. Epub 2009/04/28. 10.1002/cm.20370 . [DOI] [PubMed] [Google Scholar]
- 39.Martinez-Lopez MJ, Alcantara S, Mascaro C, Perez-Branguli F, Ruiz-Lozano P, Maes T, et al. Mouse neuron navigator 1, a novel microtubule-associated protein involved in neuronal migration. Molecular and cellular neurosciences. 2005;28(4):599–612. Epub 2005/03/31. 10.1016/j.mcn.2004.09.016 . [DOI] [PubMed] [Google Scholar]
- 40.Kunert S. The role of neuron navigator 1 in vascular development: Humboldt-Universität zu Berlin, Mathematisch-Naturwissenschaftliche Fakultät I; 2014.
- 41.Lassi G, Ball ST, Maggi S, Colonna G, Nieus T, Cero C, et al. Loss of Gnas Imprinting Differentially Affects REM/NREM Sleep and Cognition in Mice. PLoS Genet. 2012;8(5):e1002706 10.1371/journal.pgen.1002706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peirson SN, Oster H, Jones SL, Leitges M, Hankins MW, Foster RG. Microarray analysis and functional genomics identify novel components of melanopsin signaling. Current biology: CB. 2007;17(16):1363–72. Epub 2007/08/19. 10.1016/j.cub.2007.07.045 . [DOI] [PubMed] [Google Scholar]
- 43.Li W-Q, Qureshi AA, Schernhammer ES, Han J. Rotating night-shift work and risk of psoriasis in US women. The Journal of investigative dermatology. 2013;133(2):565–7. Epub 08/30. 10.1038/jid.2012.285 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hedström AK, Åkerstedt T, Hillert J, Olsson T, Alfredsson L. Shift work at young age is associated with increased risk for multiple sclerosis. Annals of Neurology. 2011;70(5):733–41. 10.1002/ana.22597 [DOI] [PubMed] [Google Scholar]
- 45.Ando N, Nakamura Y, Aoki R, Ishimaru K, Ogawa H, Okumura K, et al. Circadian Gene Clock Regulates Psoriasis-Like Skin Inflammation in Mice. J Invest Dermatol. 2015;135(12):3001–8. Epub 2015/08/21. 10.1038/jid.2015.316 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marquié J-C, Tucker P, Folkard S, Gentil C, Ansiau D. Chronic effects of shift work on cognition: findings from the VISAT longitudinal study. Occupational and Environmental Medicine. 2015;72(4):258–64. 10.1136/oemed-2013-101993 [DOI] [PubMed] [Google Scholar]
- 47.Chang AM, Aeschbach D, Duffy JF, Czeisler CA. Evening use of light-emitting eReaders negatively affects sleep, circadian timing, and next-morning alertness. Proc Natl Acad Sci U S A. 2015;112(4):1232–7. 10.1073/pnas.1418490112 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
The data used in this analysis came from RICHS samples that had both methylation and RNAseq data available (n = 199).
(XLSX)
N refers to the overall number of genes annotated to the term or pathway and DE refers to the number of genes from the top CpG sites within that term or pathway.
(XLSX)
Gene list was entered and analyzed using a JTK q-value filter with a probability cut-off of 0.05 and JTK phase range of 0–40 of all available CIRCA mouse databases.
(XLSX)
(XLSX)
Data Availability Statement
We have submitted the data to the NCBI GEO repository, accession number GSE75248. The methylation data and associated covariates used in the analysis are available here: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE75248.