Summary
Bone marrow fibrosis (BMF) develops in various hematological and non-hematological conditions and is a central pathological feature of myelofibrosis. Effective cell-targeted therapeutics are needed, but the cellular origin of BMF remains elusive. Here, we show using genetic fate tracing in two murine models of BMF that Gli1+ mesenchymal stromal cells (MSCs) are recruited from the endosteal and perivascular niche to become fibrosis-driving myofibroblasts in the bone marrow. Genetic ablation of Gli1+ cells abolished BMF and rescued bone marrow failure. Pharmacological targeting of Gli proteins with GANT61 inhibited Gli1+ cell expansion and myofibroblast differentiation, and attenuated fibrosis severity. The same pathway is also active in human BMF and Gli1 expression in BMF significantly correlates with the severity of the disease. In addition, GANT61 treatment reduced the myofibroblastic phenotype of human MSCs isolated from patients with BMF, suggesting that targeting of Gli proteins could be a relevant therapeutic strategy.
Introduction
Bone marrow fibrosis is characterized by the increased deposition of reticulin fibers or collagen fibers. A number of hematologic and non-hematologic disorders are associated with increased bone marrow fibrosis (Kuter et al., 2007) which is a central pathological feature and WHO major diagnostic criterion of myelofibrosis (MF). Myelofibrosis (MF) refers to BCR-ABL1-negative myeloproliferative neoplasms (MPN)(Tefferi et al., 2007). The majority of patients with MF carry mutations that activate JAK–STAT signaling; 60% of patients with MF harbor the JAK2V617F mutation, approximately 30% carry a calreticulin mutation (CALR), and 8% carry a myelo-proliferative leukemia virus oncogene (MPL) mutation (Klampfl et al., 2013; Levine, 2012; Levine and Gilliland, 2008; Nangalia et al., 2013; Tefferi et al., 2014). PMF is the least common of the three classic MPNs; however, it is the most aggressive and is associated with a significantly shortened survival (Mehta et al., 2014; Tefferi, 2011). PMF is characterized by malignant clonal hematopoiesis, bone marrow fibrosis, extramedullary hematopoiesis, splenomegaly and abnormal cytokine expression leading to significant systemic symptoms, risk of transformation to acute leukemia, and reduced survival.
Although the somatic mutations that drive the development of MPN have been largely defined, the cellular targets of bone marrow fibrosis still remain obscure. In MPN, mesenchymal stromal cells (MSCs), key components of the HSC niche, have recently been shown to acquire a secretory, extracellular matrix remodelling phenotype and lose their hematopoiesis-supporting capacity (Schneider et al., 2014). A recent study using a knockin Jak2V617F MPN mouse model demonstrated that MPN progression in the bone marrow creates neuropathic changes in the BM niche, which affect the activity of perivascular MSCs and alter the function of the HSC niche (Arranz et al., 2014). Identifying the cells that drive the development of a fibrotic bone marrow niche with its detrimental consequences for the maintenance of HSCs is a prerequisite for the development of novel targeted therapeutics.
Multiple genetic fate tracing studies have been performed to elucidate the cellular origin of fibrosis driving myofibroblasts in solid organs (Kramann et al., 2013). The recent identification of perivascular Gli1+ MSC-like cells as a major cellular origin of organ fibrosis and as a relevant therapeutic target to prevent solid organ dysfunction after injury provides significant potential to identify the origin of fibrosis-driving cells in bone marrow fibrosis (Kramann et al., 2015b; Schepers et al., 2015). Given that the Hedgehog (Hh) signaling pathway regulates mesenchyme cell fate during development and in view of growing evidence implicating a critical role for Hh in solid organ fibrosis and cancer (Aberger and Ruiz, 2014; Kramann et al., 2013), these findings provide a rationale for potential targeting of the Hedgehog (Hh) pathway in bone marrow fibrosis.
Currently, the clonal myeloid neoplasm is the primary therapeutic target in MPN and the only potentially curative therapy for patients with PMF is allogeneic hematopoietic stem cell transplantation, a high risk procedure with significant associated morbidity and mortality. Establishing new modalities to directly block the cellular changes occuring in the malignant BM niche, including the inhibition of aberrant MSC differentiation into fibrosis-driving cells could have a substantial therapeutic impact in the treatment of bone marrow fibrosis.
Results
Perivascular and endosteal localization of Gli1+ cells in the bone marrow niche
Having identified Gli1 as a faithful marker for fibrosis-driving MSCs in solid organs (Kramann et al., 2015b), we sought to characterize Gli1+ cells in the bone marrow niche more thoroughly. Gli1CreERt2 driver mice were crossed to a tdTomato reporter for inducible genetic labeling. Gli1+ cells in the bone marrow either align against bone (Figure 1A) or are associated with the vasculature (Figure 1B-C). Quantification of Gli1+ cell distribution in bigenic Gli1CreER;tdTomato mice indicated that the majority of Gli1+ cells reside in the endosteal niche whereas a smaller fraction is associated with bone marrow sinusoids and arterioles (Fig 1D). Clearing of a sternal bone with deep imaging of the bone marrow further illustrated the endosteal and perivascular localization of Gli1+ cells (Figure S1A-B). We next assessed the expression of markers that have been reported for stromal and perivascular cells by immunostaining and confocal microscopy. We observed high CD105 and moderate NG2 expression in perisinusoidal and periarteriolar Gli1+ cells while expression of both markers was almost absent in endosteal Gli1+ cells (Figure 1E-G). Nestin expression was detected in many endosteal and perivascular Gli1+ cells; however, we also observed Gli1+ cells in both niches that did not co-express Nestin (Figure 1H). Of note, we did not detect leptin receptor expression in Gli1+ cells (Figure S1C). While we detected sympathic innvervation of periarteriolar and to a lesser extent perisinusoidal Gli1+ cells, endosteal Gli1+ cells did not show adjacent sympathic nerves (Figure 1I). Similarly, many perivascular but not endosteal Gli1+ cells were adjacent to Schwann cells (Figure 1J). We further studied the surface profile of Gli1+ cells isolated from bone chips of bigenic Gli1CreER;tdTomato mice by flow cytometry. We observed high expression of pericyte markers such as CD73, mesenchymal markers such as platelet derived growth factor receptor alpha PDGFRα (CD140a), the mesenchymal stem cell markers CD105, CD44, CD29, Sca1 and CD51 while we did not observe expression of the endothelial cell lineage marker CD31 (Figures 1K, S1D). To study Gli1 expression in human bone marrow, we validated Gli1 staining in human glioblastoma multiforme tissue that had been EDTA treated (Figure S1E). Immunostaining of human bone marrow indicates Gli1 expression in spindle shaped stromal cells (1) aligning the bone (endosteal niche) or (2) associated with the vasculature (Figure 1L). Image quantification of human bone marrow demonstrated that 68.1±2.7% of Gli1+ cells are in an endosteal and 31.9±2.7% in perivascular localization, comparable to the findings in murine bone marrows. Co-staining for Nestin and Gli1 indicated co-expression in some but not all Gli1 expressing cells (Figure S1F), whereas leptin receptor and NG2 were not expressed (Figure S1F). When isolated from human bone marrow, Gli1+ cells express CD90, CD105 and Nestin, consistent with their pericyte/MSC origin (Figure 1M, S1G).
Figure 1. Gli1+ cells reside in the perivascular and endosteal bone marrow niche in human and mice.
(A) Representative images of bone marrows (BM) from bigenic Gli1CreERt2;tdTomato mice 10 days after tamoxifen application (3x10mg p.o.). Brightfield (BF) and Fetuin A stained images. Scale bars 50μm (B-C) Representative images and Z-Stack analyses of the perivascular BM niche. Scale bars 50μm. (D) Distribution of Gli1+ cells in the endosteal (endost), perisinusoidal (perisin) and periarteriolar (periart) niche; mean±SEM. (E-H) CD105, NG2 and Nestin expression by Gli1+ cells in the endosteal, perisinusoidal and periarteriolar niche; mean±SEM; arrows indicate co-expression in Gli1+ cells; arrowheads indicate no-expression in Gli1+ cells; scale bars 50μm in E-F, 25μm in G-H; mean±SEM. (I) Representative Z-Stack images and quantification of Gli1+ cell innervation by tyrosine hydroxylase (TyrHydr)-positive sympathic nerve fibres. Scale bars 25μm; mean±SEM. (J) Representative Z-Stack image and quantification of Gli1+ cells adjacent to glial fibrillary acidic protein (GFAP)-expressing glia cells. Scale bars 25μm. (K) Surface expression pattern of tdTomato+ cells from bigenic Gli1CreERt2; tdTomato mice as assessed by flow cytometry; mean±SEM. (L) Representative images of human bone marrows stained for Gli1. Arrows indicate Gli1 expressing cells. Scale bars: 50μm. (M) Surface expression profile of human Gli1+ MSCs; mean±SEM. See also Figure S1.
Gli1+ cells are myofibroblast precusors in myelofibrosis
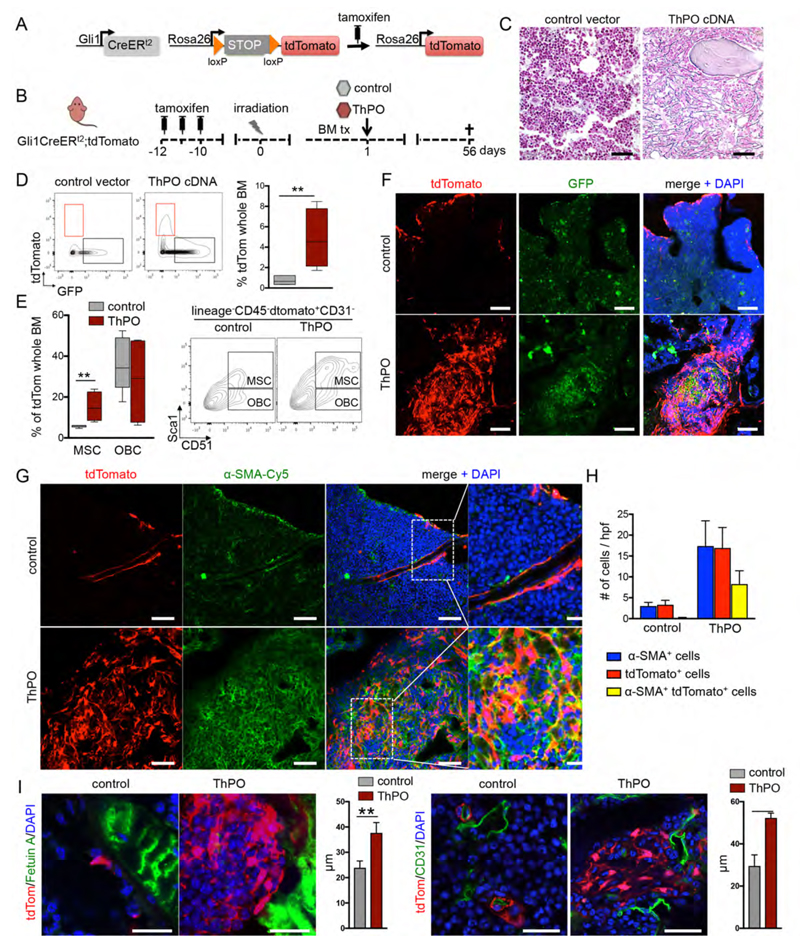
To determine if Gli1+ cells have a fibrosis-inducing role in bone marrow fibrosis, we performed genetic fate tracing experiments in a murine model of thrombopoeitin (ThPO)-induced myelofibrosis (Villeval et al., 1997; Yan et al., 1996). To trace the fate of Gli1+ cells, bigenic Gli1CreERt2+/-;tdTomato+/- recipient mice received tamoxifen to induce cell-specific expression of the tdTomato fluorochrome and were subjected to lethal irradiation followed by transplantation of ThPO overexpressing or control bone marrow (Figure 2A, B). We transduced hematopoietic stem and progenitor cells from Gli1CreERt2-/-;tdTomato-/- (wildtype-WT littermates) with ThPO overexpressing cDNA (pRRL.PPT.SFFV.Thpo.iGFP) or control (pRRL.PPT.SFFV.MCS.iGFP). The irradiation was performed at least 10 days after the last tamoxifen dose to eliminate any possibility of Cre recombination after injury.
Figure 2. Gli1+ cells expand and become myofibroblasts in thrombopoietin (ThPO) induced bone marrow fibrosis.
(A-B) Bigenic Gli1CreER;tdTomato mice were injected with tamoxifen (3x10mg p.o.) at 8 weeks of age, lethally irradiated at 10 days after the last tamoxifen dose and received c-kit enriched hematopoietic stem cells from wildtype littermates expressing either Thrombopoietin-cDNA (ThPO, n=5, 3 males) or control cDNA (control, n=5, 3 males; both lentiviral SFFV-iGFP vector backbone). Mice were sacrificed at 56 days after transplantation. (C) Representative images of reticulin stained bone marrows (BM) from control and ThPO group 56 days after transplantation. Scale bars 200μm. (D) Representative flow cytometric plots and quantification of tdTomato+ cells in the BM of control and ThPO group 56 days after transplantation. **p<0.01 by t-test. (box plot and whiskers, min to max). (E) Representative flow cytometric plots and quantification of the contribution of tdTomato+ cells to MSCs, and OBCs contained in the endosteal (Lin-/CD45-) BM stromal fraction in the control and ThPO group 56 days after transplantation. MSCs are tdTom+lin-CD45-CD31-Sca1+CD51+; OBCs are tdTom+lin-CD45- CD31-Sca1-CD51+; **p<0.01 by t-test. (box plot and whiskers, min to max). (F) Representative confocal images of BM from control and ThPO group. Scale bars 50μm. (G-H) Representative images and quantification of BM from the control and ThPO group stained for alpha smooth muscle actin (α-SMA). *p<0.05, ***p<0.001 versus the control group by t-test; scale bars 50μm, inserts 20μm. (I) Representative images of BM from the control and ThPO group stained for Fetuin A (left panel) and CD31 (right panel). The distance of Gli1+ cells to bone (Fetuin A+) and vasculature (CD31+) was quantified. **p<0.01 by t-test; mean±SEM; scale bars 25μm. See also Figure S2.
ThPO overexpression induced significant myeloproliferative features including thrombocytosis and leukocytosis (Figure S2A). Hemoglobin (Hb) decreased over time in ThPO overexpressing mice and the percentage of ThPO-GFP+ overexpressing cells in these mice (gene marking) also significantly decreased suggesting the replacement of hematopoietic cells in the bone marrow by fibrosis (Figure S2A-B). The decrease of marrow hematopoiesis was accompanied by extramedullary hematopoeisis with significant increase in spleen weight (Figure S2C) and presence of lineagelowSca1+ckit+ hematopoietic stem and progenitor cells in the spleen (Figure S2D). Trichrome staining indicated marked remodeling of the spleen, however without development of collagen fibrosis (Figure S2E).
The bone marrow of ThPO overexpressing mice showed abundant reticulin and trichrome+ fibrosis (Figures 2C, S2F-G). Flow cytometric analysis of the genetically marked tdTomato+ population and GFP+ hematopoeisis demonstrated a significant increase in the frequence of Gli1+ stromal cells in the bone marrow cavity in ThPO-induced myelofibrosis (Figure 2D). Interestingly, while Gli1+ cells express markers of both MSCs and OBCs, primarily MSC marker expressing Gli1+ cells expanded in fibrosis (Figure 2E).
Confocal imaging further demonstrated that Gli1+ cells are activated by the malignant hematopoietic clone, undergo tremendous expansion, and acquire expression of alpha smooth muscle actin (α-SMA), indicating myofibroblast differentiation (Figure 2F-H, S2H). Of note, quantification indicated that approximatively half of all myofibroblasts were derived from the Gli1 lineage (Figure 2H). Quantification of Gli1+ cell distance to bone and endothelial cells by image analysis showed that Gli1+ cells are mobilized from their endosteal and perivascular niche (Figure 2I).
Genetic ablation of Gli1+ cells abolishes myelofibrosis and rescues bone marrow failure
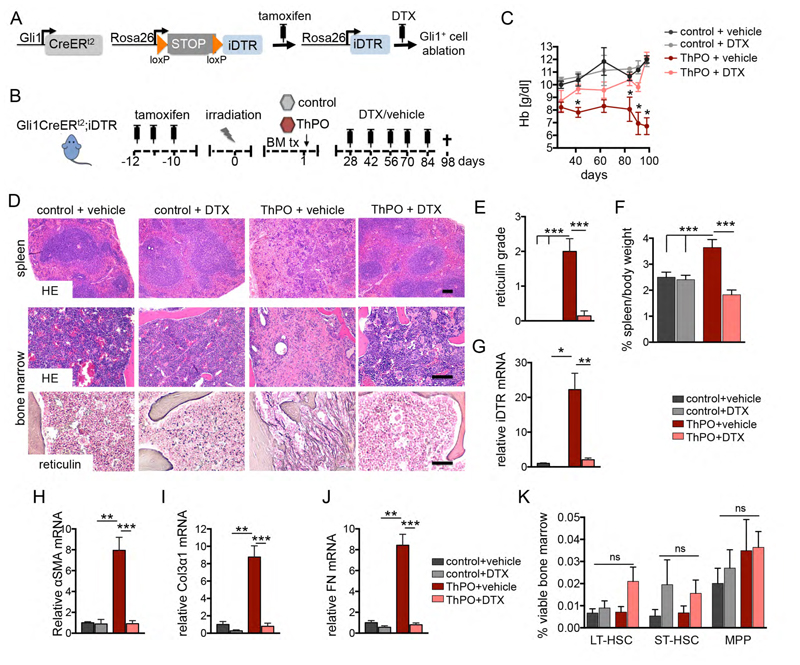
We next asked if Gli1+ cells are required for the development of bone marrow fibrosis and whether genetic ablation of Gli1+ cells affects fibrosis severity and bone marrow function. We generated a mouse model with inducible Gli1+ cell-specific expression of the human symian diphtheriatoxin receptor (iDTR) - Gli1CreERt2+/-;iDTR+/- (Figure 3A). In this model, Gli1+ cells express the iDTR after tamoxifen induced recombination and subsequent diphtheria toxin (DTX) injection will specifically ablate the Gli1+ cell population but no other cells as normal mouse cells lack expression of a diphtheria toxin receptor. Bigenic Gli1CreERt2+/-;iDTR+/- mice received tamoxifen and underwent bone marrow transplantation receiving ckit+ HSPCs expressing either the empty vector or ThPO overexpressing cDNA from wildtype littermates. Mice were randomized on their Hb level at 4 weeks after bone marrow transplantation and received DTX (50ng/kgBW) in order to ablate the Gli1+ cells (Figure 3B).
Figure 3. Genetic ablation of Gli1+ cells abolishes bone marrow fibrosis and rescues thrombopoietin induced bone marrow failure.
(A-B) Bigenic Gli1CreERt2;iDTR mice received tamoxifen (3x10mg p.o.), were lethally irradiated at 10 days after the last tamoxifen dose and transplanted with 2x105 c-kit purified bone marrow (BM) cells from wild type littermates transduced with either control or ThPO cDNA. Mice were injected with diphtheriatoxin in order to ablate Gli1+ cells (DTX, 50ng/kg body weight i.p.) or vehicle (PBS) as indicated (control + vehicle n=3, 2 males; control+DTX n=4, 2 males; ThPO + vehicle n=9, 5 males; ThPO + DTX n=6, 4 males). (C) Time course of hemoglobin counts. *p<0.05 by t-test; mean±SEM. (D) Representative hematoxylin & eosin (HE) and reticulin stained images from spleen and BM. Scale bars 300μm. (E) Scoring of reticulin fibrosis grade. ***p<0.001 by one way ANOVA with posthoc Tukey; mean±SEM. (F) Spleen weight relative to body weight among the groups. ***p<0.001 by one way ANOVA with posthoc Tukey; mean±SEM. (G) mRNA expression of the symian diphtheriatoxin receptor (iDTR). *p<0.05 **p<0.01 by one way ANOVA with posthoc Tukey; mean±SEM. (H-J) Relative mRNA expression for the fibrotic readouts alpha smooth muscle actin (α-SMA), collagen 3α1 (Col3α1) and fibronectin. **p<0.01, ***p<0.001 by one way ANOVA with posthoc Tukey; mean±SEM. (K) Analysis of the HSC compartment, defined as long-term (LT; linlowSca1+ckit+CD48-CD150+), short-term (ST; linlowSca1+ckit+CD48-CD150-) HSCs and multipotent progenitor cells (MPP, linlowSca1+ckit+CD48+CD150-) in the bone marrow within the four different groups. mean±SEM; ns=non-significant by one way ANOVA with posthoc Tukey. See also Figure S3.
Mice that received ThPO overexpressing marrow in the presence of Gli1+ cells (ThPO+vehicle) developed a severe bone marrow fibrosis, as indicated by significant decrease in the Hb counts over time and replacement of hematopoietic cells in bone marrow and spleen by reticulin fibrosis as well as significant splenomegaly, while the white blood cell and platelet counts were not affected (Figure 3C-F; Figure S3A). Ablation of Gli1+ cells completely abolished this fibrotic phenotype (ThPO+DTX, Figure 3C-F) as determined by stabilization of the Hb counts, complete absence of reticulin fibrosis in bone marrow and spleen, normalized spleen weights and reduced osteosclerosis (Figure S3B-C). mRNA expression of iDTR further demonstrated significant expansion of Gli1+ cells in ThPO-induced bone marrow fibrosis with sufficient ablation following DTX injection (Figure 3G). Genetic ablation of the Gli1+ cell population also reduced the fibrotic readouts α-SMA, collagen 3α1 (Col3α1) and fibronectin (Figure 3 H-J), which were all significantly increased following ThPO overexpression in the bone marrow.
These experiments provide the first in vivo functional evidence that Gli1+ cells are fibrosis-driving cells in bone marrow fibrosis and represent a novel cellular target. Importantly, the ablation of Gli1+ cells in normal hematopoiesis (control+DTX) did not have detrimental effects on differentiated lineages (Figure S3D-H) nor on HSPCs (Figure 3K). Furthermore, we did not observe a significant effect of Gli1+ cell ablation on marrow vascularization (CD31 surface area), sympathic nerve fibres (tyrosine hydroxylase surface area) or glia cells (GFAP surface area) (Figure S3I-K).
Pharmacological targeting of Gli1+ cells with GANT61 amelioreates myelofibrosis
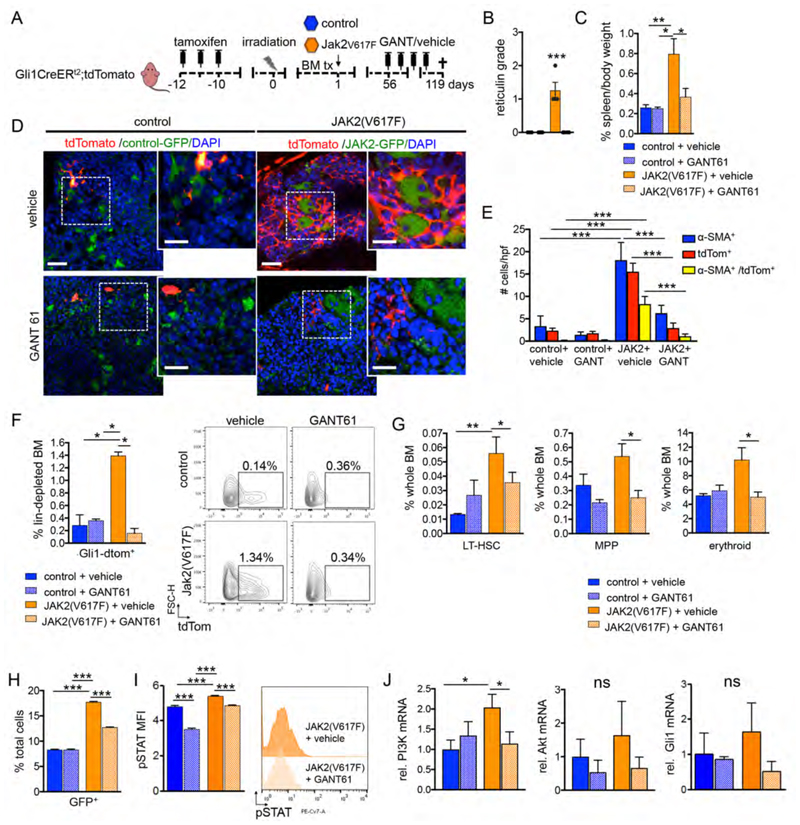
We next asked whether Gli1+ cells can be targeted pharmacologically in a more physiological myelofibrosis model. As 60% of patients with myelofibrosis harbor the JAK2(V617F) mutation, we introduced the mutation in a retroviral vector (MSCV-IRES-GFP, MIG) in ckit+ HSPCs (Wernig et al., 2008). Gli antagonist 61 (GANT61) was identified in a screen to be a direct small molecule compound inhibitor of Gli proteins (Lauth et al., 2007) and is a promising treatment of fibrosis in lung and kidney (Kramann et al., 2015a; Moshai et al., 2014). To elucidate whether pharmacologic inhibition of Gli proteins is a therapeutic strategy in myelofibrosis, we performed a genetic fate tracing experiment of Gli1+ cells in a Jak2(V617F) myelofibrosis model combined with GANT61 treatment. Bigenic Gli1CreERt2+/-;tdTomato+/- mice received tamoxifen and were transplanted with bone marrow of WT littermates retrovirally transduced with either WT Jak2 control vector or Jak2(V617F). Treatment with either GANT61 or vehicle was initiated at 8 weeks after onset of the disease in the Jak2(V617F) group (Figure 4A, S4A). Mice were randomized based on the Hb level to receive either GANT61 or vehicle every other day.
Figure 4. Gli1+ cells are myofibroblast precursors in JAK2(V617F) induced myelofibrosis and can be targeted pharmacologically by GANT61.
(A) Bigenic Gli1CreERt2;tdTomato mice received tamoxifen (3x10mg p.o.) and were lethally irradiated 10 days after the last tamoxifen dose and transplanted with 2x105 c-kit purified bone marrow (BM) cells from wild type littermates that had been transduced with either control (n=10) or Jak2(V617F) (n=10) cDNA (both MCSV-IRES-GFP retroviral backbone vector). Mice were injected with GANT61 (50mg/kg body weight) or vehicle (corn oil / ethanol) every other day between 8 and 17 weeks after transplantation (control + vehicle n=3, 2 males; control+GANT61 n=5, 3 males; Jak2(V617F) + vehicle n=4, 3 males; Jak2(V617F) + GANT61 n=4, 3 males). (B) Grading of reticulin fibrosis. ***p<0.001 versus all other groups by one way ANOVA with posthoc Tukey; mean±SEM. (C) Spleen weight was determined as a percentage of the body weight. *p<0.05, **p<0.001 by one way ANOVA with posthoc Tukey; mean±SEM. (D) Representative images of sternal bones. Scale bars 50μm, inserts 25μm. (E) Quantification of all alpha smooth muscle actin expressing cells (α-SMA+), Gli1 cells (tdTomato+) and Gli1-derived myofibroblasts (α-SMA+/tdTomato+) in the BM of mice transplanted with Jak2(V617F) (Jak2) or control that have been treated with GANT61 or vehicle. ***p<0.001 by one way ANOVA with posthoc Tukey; mean±SEM. (F) Representative flow cytometric plots and quantification of Gli1+ cells and within the lineage depleted BM. *p<0.05 by one way ANOVA with posthoc Tukey; mean±SEM. (G) Analysis of long-term (LT; linlowSca1+ckit+CD48-CD150+) and multipotent progenitor cells (MPP, linlowSca1+ckit+CD48+CD150-) as well as erythroid cells (Gr1-CD11b-CD3-CD19-Ter119+) by flow cytometry. *p<0.05, **p<0.01 by one way ANOVA with posthoc Tukey; mean±SEM. (H) c-kit+ purified HSPCs from wild type mice were transduced with either control or Jak2(V617F) cDNA (both MCSV-IRES-GFP retroviral backbone vector). 48 hours after transduction, cells were treated with GANT61 or vehicle. The gene marking (GFP+) was quantified by flow cytometry 48 hours after treatment. ***p<0.001 versus all other groups by one way ANOVA with posthoc Tukey; mean±SEM. (I) Quantification of the mean fluorescence intensity (MFI) and representative flow cytometric analysis of levels of phospho-STAT5 (p-STAT5) in c-kit+ cells transduced with Jak2(V617F) or control (GFP+) 48 hours after treatment with GANT61 or vehicle. GFP+ cells are shown in the histogram. ***p<0.001 versus all other groups by one way ANOVA with posthoc Tukey; mean±SEM. (J) c-kit+ purified HSPCs were transduced with Jak2(V617F) or control cDNA, were sort-purified by GFP-expression 48 hours after transduction and treated with GANT61 or vehicle 24 hours later. 24 hours after treatment, cells were harvested for RNA isolation and qt-RT-PCRs. Relative mRNA expression for Phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K), Akt and Gli1 is shown. *p<0.05 ns=non-signifcant by one way ANOVA with posthoc Tukey; mean±SEM. See also Figure S4.
Mice transplanted with Jak2(V617F) had significantly elevated hemoglobin levels (Hb) compared to the control groups (Figure S4A), reflecting erythroid hyperplasia, while treatment with GANT61 rescued this phenotype with a significant decrease of Hb to levels comparable to the control groups. Analysis of the bone marrow at 17 weeks after transplantation showed a significant expansion of genetically tagged Gli1+ cells with development of reticulin fibrosis and splenomegaly (Figures 4B-F, S4B-C) in the Jak2(V617F) transplanted mice compared to the WT controls.
Co-staining with α-SMA demonstrated that Gli1+ cells differentiated into myofibroblasts and represented a significant part of the myofibroblast population (Figure 4E, F and S4C). Notably, treatment with GANT61 resulted in significantly reduced numbers of Gli1+-tdTomato+ and Gli1+-tdTomato+-αSMA+ cells and abolished development of reticulin fibrosis (Figure 4B-F, S4B-C). Distance measurements of Gli1+ cells to bone and endothelial cells revealed that the presence of a Jak2(V617F) hematopoietic clone resulted in Gli1+ cell mobilization from both the endosteal and the perivascular niche which was ameliorated by GANT61 treatment (Figure S4D-E).
Moreover, while Jak2(V617F) transplantation resulted in signifcantly increased spleen weight and numbers of long-term hematopoietic stem cells (LT-HSCs), multipotent progenitor cells (MPPs) and erythroid progenitors as typical for a polycythemia vera (PV), GANT61 reversed this phenotype (Figure 4G, S4F-G) suggesting an effect of GANT61 on both Gli1+ cells and the malignant hematopoietic clone. To assess the relative effect of GANT61 treatment on Jak2(V617F) expressing hematopoietic cells, we treated transduced (GFP+) and non-transduced (GFP-) c-kit+ HSPCs in vitro. GANT61 specifically reduced Jak2(V617F) expressing cells and induced significant apoptosis (Figure 4H and Figure S4H). We analyzed the down-stream effects of Jak2(V617) and observed a significant down-regulation of STAT5 phosphorylation by GANT61 treatment (Figure 4I). Jak2(V617F) induced expression of the phosphatidylinositol-3-kinase (PI3K), which was also inhibited by GANT61 treatment (Figure 4J). As the PI3K/Akt axis was shown to activate Gli in a non-canonical fashion, we analyzed Akt and Gli1 expression. Jak2(V617F) in HSPCs induced increased (but not significant) expression of both Akt and Gli1 which was in tendency decreased by GANT61 treatment (Figure 4J).
These data suggest that targeting Gli proteins by GANT61 is a novel therapeutic approach in myelofibrosis that affects both expansion of myelofibrosis driving Gli1+ myofibroblasts and the expansion of the malignant hematopoietic clone.
Gli1+ cells undergo distinct transcriptional changes in bone marrow fibrosis
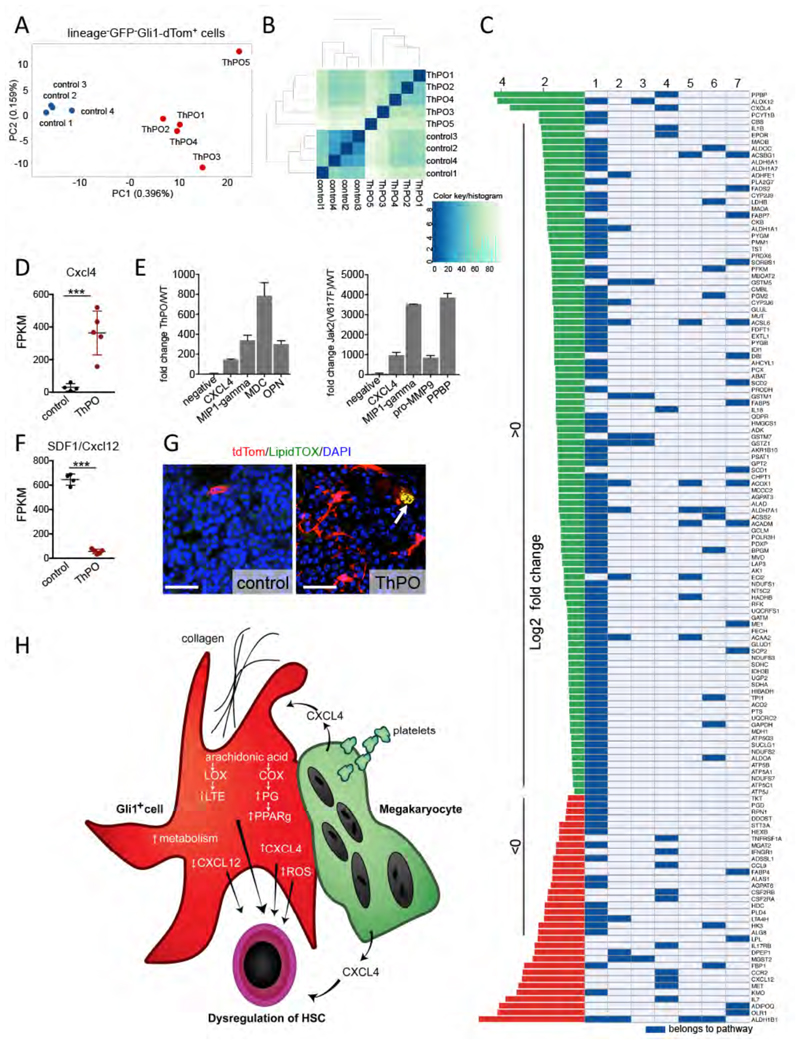
To determine whether and how Gli1+ stromal cells are altered in bone marrow fibrosis, we performed RNA sequencing on sort-purified lineage-GFP-Gli1-tdtom+ cells from controls and ThPO-overexpressing mice with bone marrow fibrosis (at 10 weeks after bone marrow transplantation). Principal component and hierarchical cluster analysis revealed that Gli1+ cells in bone marrow fibrosis are drastically distinct from Gli1+ cells in homeostasis (Figure 5A-B). Statistical analysis revealed that 1597 genes were differentially expressed between the Gli1+ cells in control conditions and Gli1+ cells in bone marrow fibrosis (0.5 log2 fold, Benjamini Hochberg-FDR corrected p-value 0.05), with 924 up-regulated and 673 down-regulated genes. To uncover differentially altered mechanisms in the Gli1+ stromal cells in bone marrow fibrosis and potential pathways of activation by the malignant hematopoietic clone, we performed gene set analysis using PIANO (Varemo et al., 2013) with a pathway collection adapted for mouse (Bares and Ge, 2015). We found up-regulated pathways that play a critical role in cytokine-cytokine receptor interaction but also in the inflammatory response, specifically the leukotriene (LOX) and prostaglandin (COX) pathways, both of which use the same primary precursor, arachidonic acid (Figures 5C, S5). There is evidence that bone marrow stromal cells actively metabolize arachidonic acid and that metabolites are important for the regulation of hematopoeisis (Abraham et al., 1991). Differential gene expression analysis demonstrated that megakaryocyte-associated genes were significantly up-regulated, in particular the chemokine Cxcl4 (cytokine-cytokine receptor interaction pathway, Figure 5C-D), which has been implicated to play a role in fibrosis (Burstein et al., 1984; van Bon et al., 2014; Zaldivar et al., 2010) and hematopoietic stem cell regulation (Bruns et al., 2014). In addition, we identified Cxcl4 among 4 up-regulated cytokines in a cytokine array from medium supernatant of both ThPO and Jak2(V617F) overexpressing HSPCs (Figure 5E).
Figure 5. Gli1+ cells in bone marrow fibrosis are transcriptionally distinct from Gli1+ cells in homeostasis.
Bigenic Gli1CreER;tdTomato mice were injected with tamoxifen (3x10mg p.o.) at 8 weeks of age, lethally irradiated at 10 days after the last tamoxifen dose and received c-kit enriched hematopoietic stem cells from wildtype littermates expressing either Thrombopoietin-cDNA (ThPO, n=5, 3 males) or control-cDNA (control, n=4, 3 males; both lentiviral SFFV-iGFP vector backbone). Mice were sacrificed at 70 days after transplantation. Gli1+ cells were sort-purified as lin-GFP-dtTomato+ and subjected to RNA sequencing. (A) Principal component analysis (PCA). (B) Heatmap representation with hierarchical clustering (C) Significantly directional enriched (median FDR >0.05) pathways. (D) Fragments per kilobase of exon per million fragments mapped (FPKM) values of CXCL4 in control and ThPO group; mean±SEM. (E) Cytokine analysis in serum and cytokine-free medium supernatant of ckit+ cells expressing ThPO or control (lentiviral SFFV-iGFP vector backbone) or Jak2(V617F) or control (both MCSV-IRES-GFP retroviral backbone). n=2. (F) FPKM value of stromal derived factor 1 (SDF 1/ CXCL12) in control and ThPO group; mean±SEM. (G) Representative images of bone marrow from Gli1CreERt2;tdTomato mice transplanted with bone marrow from wildtype littermates expressing either ThPO or control-cDNA stained with the neutral lipid staining LipidTOX. Scale bars 50μm. Arrow indicating LipidTOX positive Gli1+ cells. (H) Model for stroma-hematopoiesis interactions in bone marrow fibrosis. See also Figure S5.
In line with previous studies of niche cells in bone marrow fibrosis (Schepers et al., 2013), we found altered expression of many HSC-regulatory genes and cytokines, including a broad down-regulation of HSC retention factors, in particular Cxcl12 (Figure 5C,F).
We further observed increased metabolic activity in Gli1+ stromal cells in bone marrow fibrosis (Figure 5C), in particular of fatty acid metabolism. PPARy, the master switch of lipogenic differentiation in MSCs, was further significantly induced in bone marrow fibrosis and recently described to play a role in pulmonary fibrosis (El Agha et al., 2016). To identify a potential lipogenic phenotype of Gli1+ MSCs in bone marrow fibrosis, we performed co-staining with the neutral lipid stain (LipidTOX). We observed sparsely LipidTOX positive Gli1+ cells in bone marrow from ThPO-overexpressing mice (Figure 5G).
Based on previously published data and our findings, we propose a model of abnormal release of Cxcl4 from α-granules from atypical megakaryocytes and platelets in MPN (Vannucchi et al., 2005) (Figure 5H). In turn, Cxcl4 as a CXC chemokine contributes to the activation of Gli1+ cells and their transdifferentiation into α-SMA+ myofibroblasts and later to expression of Cxcl4 in Gli1+ cells. In fibrosis progression, thrombin and collagen contribute to the activation of arachidonic acid metabolism which transforms stromal cells into inflammatory, metabolically active cells which lose their hematopoiesis-supporting capacity, further contributing to abnormal hematopoeisis in MPN (Figure 5H).
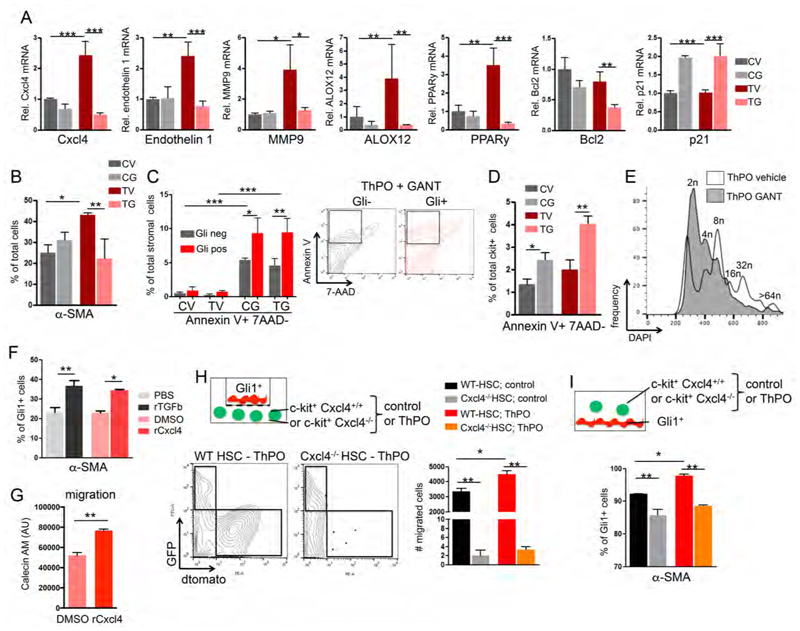
To validate the findings of the RNA sequencing and to dissect the direct interaction of stromal cells and hematopoietic cells in bone marrow fibrosis in vitro, we co-cultured ThPO overexpressing c-kit+ hematopoietic stem and progenitor cells (HSPCs) and Gli1+ isolated stromal cells. As GANT61 ameliorates bone marrow fibrosis, we further tested the effect of GANT61 treatment on differentially expressed genes (Figure 6A). The co-culture with ThPO overexpressing HSPCs significantly induced expression of Cxcl4 in stromal cells, which was completely normalized by GANT61 treatment. Endothelin 1 and MMP9 as pro-fibrotic readouts associated with increased Cxcl4 expression in organ fibrosis (van Bon et al., 2014), were significantly up-regulated in stromal cells exposed to ThPO overexpressing HSPCs and were normalized after GANT61 treatments (Figure 6A). ALOX12 and PPARy as targets of activated arachidonic acid were up-regulated in stromal cells in the presence of ThPO overexpressing HSPCs and normalized by GANT61 treatment. As previous studies suggested that GANT61 leads to apoptosis and cell cycle arrest, we analyzed the anti-apoptotic gene Bcl2 and the cyclin-dependent kinase inhibitor p21. Bcl2 was significantly decreased after treatment with GANT61 and p21 significantly induced, suggesting that GANT61 induces apoptosis and might affect cell-cycle (Figure 6A).
Figure 6. Cxcl4 induces migration and myofibroblastic differentiation of Gli1+ cells.
(A-E) tdTomato-Gli1+ stromal cells co-cultured with c-kit+ hematopoietic stem and progenitor cells (HSPCs) expressing either Thrombopoietin-cDNA (ThPO) or control cDNA (control vector=CV) were treated for 24 hours with vehicle or GANT61. CV= control vector, vehicle; CG= control vector and GANT61; TV= ThPO overexpression and vehicle; TG= ThPO overexpression and GANT61. (A) Relative mRNA expression in tdTomato-Gli1+ stromal cells is shown for platelet-factor 4 (Cxcl4), endothelin 1, matrix metalloproteinase 9 (MMP9), Arachidonate 12-lipoxygenase (ALOX12), Peroxisome proliferator-activated receptor gamma (PPARy), B-cell lymphoma gene 2 (Bcl2) and p21. **p<0.01, ***p<0.001 by one way ANOVA with posthoc Tukey; mean±SEM. n=3. (B) Quantification of Gli1+α-SMA+ myofibroblasts in stromal cells co-cultured with HSPCs (expressing control or ThPO cDNA) and treated with GANT61. *p<0.05, **p<0.01 by one way ANOVA with posthoc Tukey; mean±SEM. n=3. (C) Quantification and representative flow blots of early apoptosis (AnnexinV+7AAD-) in td-Tomato+ and td-Tomato- stromal cells co-cultured with HSPCs and treated with GANT61. *p<0.05, **p<0.01, ***p<0.001 by one way ANOVA with posthoc Tukey; mean±SEM. n=3. (D) Quantification of early apoptosis (AnnexinV+7AAD-) in c-kit+ HSPCs (expressing control or ThPO cDNA) co-cultured with stromal cells. *p<0.05, **p<0.01 by one way ANOVA with posthoc Tukey; mean±SEM. n=3. (E) Representative histogram showing ploidy in CD41+ ThPO-overexpressing megakaryocytes treated with GANT61 or vehicle. Representative data from one of three experiments. (F) Flow cytometric quantification of α-SMA+ in stromal cells treated with recombinant platelet factor 4 (rCxcl4) or TGFβ as a known stimulus for myofibroblastic differentiation (or respective controls). *p<0.05, **p<0.01 by t-test; mean±SEM. n=3. (G) Quantification of migration of tdTomato-Gli1+ stromal cells towards an rCxcl4 or control (DMSO) gradient. AU= arbitrary unit; mean±SEM; **p<0.01 by t-test. n=2 (H) Schematic, representative flow blots and quantification of tdTomato+ Gli1+ migration towards c-kit+ HSPCs isolated from Cxcl4-/- mice or littermate controls (Cxcl4+/+) expressing either ThPO or control cDNA (GFP+). **p<0.01 by one way ANOVA with posthoc Tukey; mean±SEM, n=3. (I) Schematic and quantification of α-SMA+ myofibroblast differentiation (flow cytometry) of tdTomato+ Gli1+ stromal cells co-cultured with c-kit+ HSPCs isolated from Cxcl4-/- mice or littermate controls expressing either ThPO or control cDNA (GFP+). *p<0.05, **p<0.01 by one way ANOVA with posthoc Tukey; mean±SEM. n=3. See also Figure S6
We validated in co-culture experiments that the exposure of Gli1+ cells with ThPO overexpressing HSPCs induces differentiation into α-SMA+ myofibroblasts and that GANT61 inhibits the differentiation or induces apoptosis in differentiated cells (Figure 6B). We analyzed apoptosis in Gli1+ and Gli1- stromal cells and confirmed that GANT61 induces apoptosis specifically in Gli1+ stromal cells (Figure 6C). GANT61 further induced apoptosis in hematopoietic cells, suggesting that part of the positive effect of GANT61 in bone marrow fibrosis can be explained by reduction of the malignant hematopoietic clone (Figure 6D). Interestingly, we observed a direct effect of GANT61 on megakaryocyte ploidy, suggesting inhibition of megakaryocyte maturation (Figures 6E, S6). This data further validates that GANT61 affects both Gli1+ stromal cells and the malignant hematopoietic clone.
Cxcl4 is necessary and sufficient for the migration of Gli1+ stromal cells and induces their myofibroblastic differentiation
To confirm that Cxcl4 induces migration and myofibroblastic differentiation of Gli1+ cells, we exposed Gli1+ cells to recombinant Cxcl4 and analyzed myofibroblast differentiation and cell migration towards a Cxcl4 gradient. Recombinant Cxcl4 (rCxcl4) significantly induced myofibroblast differentiation of Gli1+ cells comparable to induction with TGFβ, a known stimulus for differentiation of MSCs into myofibroblast (Figure 6F). In a chemotactic migration assay, Cxcl4 significantly increased the migration of Gli1+ cells towards the rCxcl4 gradient (Figure 6G) Having established that Cxcl4 induces migration and myofibroblast differentiation of Gli1+ cells, we sought to confirm the hypothesis that Cxcl4 in hematopoietic cells in bone marrow fibrosis induces the migration and differentiation process of Gli1+ stromal cells. We tested if genetic knockdown of Cxcl4 in hematopoeitic cells can ameliorate the migration and myofibroblastic differentiation of Gli1+ cells. We performed a migration assay with Gli1+ cells on a transwell membrane and hematopoietic cells isolated from Cxcl4-/- mice and WT controls, expressing control or ThPO cDNA, in the lower compartment as a chemotactic gradient (Figure 6H). The migration of Gli1+ cells was significantly increased in the presence of ThPO overexpressing cells and almost absent in conditions with Cxcl4-/- hematopoietic cells as the chemoattractant, demonstrating that Cxcl4 is necessary for the migration of Gli1+ stromal cells induced by ThPO overexpressing HSPCs (Figure 6H). We next analyzed whether the genetic knockout of Cxcl4 in hematopoietic cells had an effect on Gli1+ myofibroblasts in a direct co-culture model. The absence of Cxcl4 in hematopoeitic cells ameliorated myofibroblastic differentiation, but did not completely inhibit the differentiation process (Figure 6I), indicating that myofibroblastic differentiation results from multiple cytokines secreted by the hematopoietic clone. In summary, these data show that Cxcl4 in the hematopoietic clone is a strong chemotactic factor and contributes to fibrotic transformation.
Gli1+ cells expand in human MPN and can be targeted therapeutically
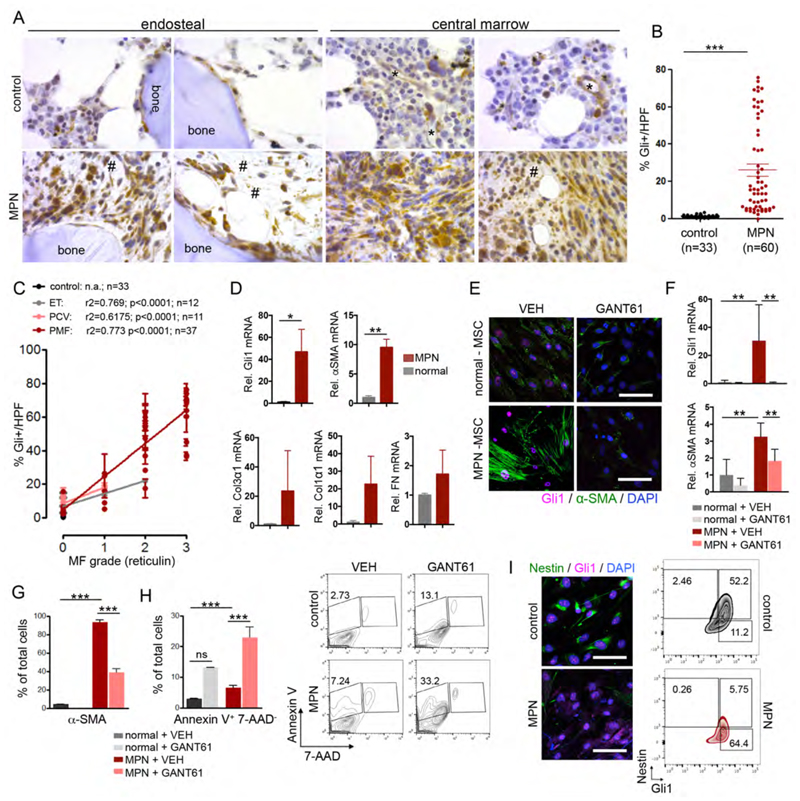
Given that Gli1+ cells are important myofibroblast progenitors and show efficacy as a therapeutic target in two distinct mouse models of myelofibrosis, we next asked whether the same pathway is active in human myelofibrosis. Bone marrow punch biopsies were collected from MPN patients (n=60) and age-matched, normal bone marrow biopsies without a primary hematological disease (healthy controls; n=33), stained for reticulin and scored blindly by pathologists for the severity of myelofibrosis and Gli1 expression (Figure 7A and S7A; Table S1). Gli1+ cells aligned the bone and sinusoids in healthy controls as demonstrated in Figure 7A. Comparable to the findings in murine MPN models, the frequency of spindle-shaped Gli1+ cells significantly increased in MPN (Figure 7A, B). Gli1+ cells were diffusely present in the hematopoietic marrow, and not restricted to their localization in the endosteal and vascular niche. Gli1+ cells were found in cellular areas or positioned within amorphous matrix deposits (Figure 7A). The frequency of Gli1+ cells positively correlated with an increase in reticulin grading of myelofibrosis, independent of the MPN classification (Figure 7C). The frequency of Gli1+ cells did not correlate with JAK2(V617F) or calreticulin gene (CALR) mutation status (Fig. S7B), suggesting a common pathobiological pathway that regulates the interaction between the malignant hematopoietic clone, atypical megakaryocytes and their niche.
Figure 7. Increased Gli1 frequency correlates with fibrosis grade in human bone marrows and can be targeted pharmacologically.
(A) Representative images of human bone marrow biopsies from control patients (MF=0) and patients with myelofibrosis stained for Gli1. *= sinusoids; #= amorphous matrix. Scale bars: 200μm. (B) Quantification of Gli1+ cell frequency in human bone marrows from healthy control patients (healthy) and MPN patients. 3 high power fields (HPF) (400x) were counted for each patient and the mean value calculated; each dot represents one patients (n=33 control; n=60 MPN patients p<0.05, ****p<0.0001 by t-test; mean±SEM. (C) Correlation of Gli1+ cell frequency and myelofibrosis (MF) grade in human bone marrow; mean±SEM. The myelofibrosis (MF)/reticulin grade was graded by hematopathologists at different institutions. Each dot represents one patient. ET: Essential Thrombocythemia; PCV: Polycythemia Vera; PMF: Primary Myelofibrosis. (D) Relative mRNA expression of Gli1 and the fibrotic readouts alpha smooth muscle actin (α-SMA), collagen3α1 (Col3α1), collagen1α1 (Col1α1) and fibronectin (FN) in human mesenchymal stem cells (MSCs) isolated from bone marrows of patients with myeloproliferative neoplasia (MPN, n=4) or healthy controls (control, n=4). *p<0.05, **p<0.001 by t-test; mean±SEM, (E) Representative imagesof human mesenchymal stem cells (MSCs) from bone marrow biopsies of patients with myeloproliferative neoplasia (MPN-MSCs, n=4) or healthy controls (control MSCs, n=4) stained for alpha-smooth muscle actin (α-SMA) and Gli1 24 hours after treatment with GANT61 (10μM) or vehicle (DMSO, VEH). **p<0.01 by one way ANOVA with posthoc Tukey. Scale bars 20μm. (F) Relative Gli1 and alpha smooth muscle actin (α-SMA) expression in human MSCs from healthy donors (control) and MPN patients after treatment with GANT61 (n=4 each) or vehicle (DMSO, VEH, n=4 each). **p<0.01 by one way ANOVA with posthoc Tukey. (G) Flow cytometric quantification of αSMA after treatment with GANT61 (n=3 each) or vehicle (DMSO, VEH, n=3 each). ***p<0.001 by one way ANOVA with posthoc Tukey. (H) Flow cytometric quantification and representative flow blots of Annexin V staining after treatment with GANT61 (n=4 each) or vehicle (DMSO, VEH, n=4 each). ***p<0.001 by one way ANOVA with posthoc Tukey. (I) Representative images and flow cytometric plots of MSCs from healthy donors (control) and MPN patients co-stained for Gli1 and Nestin. Scale bars 20μM mean±SEM. See also Figure S7 and supplementary table S1/S2 for patient characteristics.
Next, we isolated MSCs from bone marrow biopsies of MPN patients (MPN-MSC; n=4) and healthy control donors (n=4). Interestingly, we observed both up-regulation of Gli1 and αSMA mRNA in MSCs from MPN patients when compared to MSCs of healthy donors, just as observed in the murine models (Figure 7D). MSCs from MPN patients also up-regulated mRNA expression of extracellular matrix proteins such as collagens and fibronectin (Figure 7D). Cells with myofibroblast morphology were only observed in MPN as shown by staining of α-SMA+ stress fibres (Figure 7E), α–SMA gene expression (Figure 7F) and an α-SMA population by flow cytometry (Figure 7G). Strikingly, treatment with GANT61 significantly reduced both α–SMA and Gli1-expression in MPN-MSCs on protein and gene expression level (Figure 7F-G). We observed a significant increase in apoptosis (AnnexinV+7AAD-) specifically in MPN-MSCs treated with GANT61 (Figure 7H). These results suggest that GANT61 induces apoptosis in Gli1-expressing myofibroblasts. Expression of the MSC marker nestin has been shown to be significantly reduced in MPN (Arranz et al., 2014). We asked how Gli1 and nestin expression are related in myelofibrotic transformation. Flow cytometry confirmed that Gli1 cells express nestin under steady state conditions and that nestin expression is lost in MPN wheras Gli1 expression is increased, indicating that Gli1 promotes the myelofibrotic transformation (Figure 7I). In summary, these data suggest that targeting Gli1+ MSCs by GANT61 might be a potential novel therapy in human bone marrow fibrosis.
Discussion
Our studies strongly suggest a critical role for Gli1+ stromal cells in the pathogenesis of bone marrow fibrosis. Genetic ablation and pharmacological targeting of Gli1+ cells in two mouse models of bone marrow fibrosis/myelofibrosis and in human samples, reversed the myofibroblast phenotype and matrix production, two central mechanisms in the progression of bone marrow fibrosis.
Across different organ systems most investigators agree that myofibroblasts cause fibrosis, however the functional contribution of myofibroblasts in bone marrow fibrosis remained unclear. Electron-microscopy studies from the last century already suggested an increase of myofibroblasts in human bone marrow fibrosis (Biagini et al., 1985; Thiele et al., 1991). Our inducible genetic fate tracing data and ablation studies show a functional contribution of Gli1+ stromal cells as progenitors of myofibroblasts in bone marrow fibrosis. The fact that genetic ablation of Gli1+ cells abolishes bone marrow fibrosis and restores hematopoiesis indicates that Gli1+ MSCs are a promising cellular therapeutic target.
Frenette et al. reported Nesperi cells: periarteriolar Nestin GFP+ (bright) cells along arterioles that co-express NG2 and are associated with sympathic nerves and GFAP+ glia (Kunisaki et al., 2013; Mendez-Ferrer et al., 2010). They further report Nesreti cells: Nestin-GFP+ (dim) cells aligned with sinusoids and co-expression of leptin receptor. While there might be an overlap of periarteriolar Gli1+ cells and Nesperi cells, we did not observe leptin receptor expression in Gli1+ cells (Figure S1C). The majority of Gli1+ cells in the endosteal niche was not associated with GFAP+ glia or sympathic nerve fibres and only partially expressed Nestin. Thus, these cells might be partially distinct from the reported Nestin+ populations.
Gli1+ cells have trilineage (multipotent) differentiation potential including osteogenic differentiation (Kramann et al., 2015b). A subpopulation of Gli1+ MSCs expresses CD51 (and Sca1low), the marker combination for osteoblastic lineage cells (OBC). Schepers et al. elegantly demonstrated that OBCs are derived from multipotent stromal cells and that the BCR-ABL+ clone in chronic myelogenous leukemia (CML) leads to expansion of OBCs resulting in matrix production, bone marrow fibrosis and trabecular thickening (Schepers et al., 2013). We hypothesize that Gli1+ cells are precursor cells for myofibroblasts and OBCs, providing a mechanistic basis for both myelofibrosis as well as also preceeding osteosclerosis.
It was recently shown that HSCs carrying a JAK2(V617F) mutation induce significant neuroglial damage, reduction of Nestin+ MSCs and Schwann cell death that contributes to the pathogenesis of the disease by causing a neuropathic changes (Arranz et al., 2014). Both Schepers et al. and Arranz et al. point to the potential for the microenvironment as a critical cooperator in the malignant process in MPN and emphasize a two-way perturbation process required for MPN. HSCs acquire a mutation (e.g., JAK2-V617F, BCR-ABL mutation) that leads to cell expansion and the mutant HSC perturbs the bone marrow niche, which further drives HSCs into neoplasia. It remained an open question which cells are the origin of bone marrow fibrosis. We demonstrate that in steady state conditions, a subfraction of Gli1+ cells co-expresses nestin+. Upon myelofibrotic transformation, Gli1+ cells expand while nestin+ cells decrease, suggesting that neuropathic changes lead to dysregulation of the niche accompanied by unregulated expansion and myofibroblast differentiation of Gli1+ MSCs.
Already 30 years ago studies indicated that abnormal megakaryocytes in myelofibrosis stimulate the proliferation of fibrosis-driving fibroblasts, partially mediated by Cxcl4 (Burstein et al., 1984). A central role of Cxcl4 in fibrosis was confirmed in solid organs and it was demonstrated that Cxcl4 is not only secreted by activated platelets but also plasmacytoid dendritic cells and fibroblasts (van Bon et al., 2014; Zaldivar et al., 2010). We demonstrated that Cxcl4 in MPN HSPCs induces migration of Gli1+ stromal cells and myofibroblastic differentiation. We further observed significant down-regulation of the chemokine stromal-cell derived factor (SDF)-1 (CXCL12), in line with previous studies characterizing niche cells in MPN (Arranz et al., 2014; Schepers et al., 2013). Deletion of Cxcl12 in stromal cells was shown to increase circulating platelets (Arranz et al., 2014; Tzeng et al., 2011). Based on our data and previous studies, we propose that Gli1+ stromal cells are activated from their niche by atypical platelets/megakaryocytes (mediated by Cxcl4), which leads to a cascade of myofibroblastic differentiation, metabolic reprogramming and down-regulation of CXCL12.
Currently, the standard therapies for MF can be divided into two catergories: clonal eradication or targeting bone marrow fibrosis. In PMF, the only therapy with a curative potential is allogenic hematopoietic stem cell transplantation (Rondelli et al., 2005). With the introduction of selective JAK inhibitors over the last 10 years, initial expectation of clonal suppression as measured by elimination of molecular and karyotypic abnormalities was dampened by the observations of persistent clonal hematopoiesis and unaltered MPN bone marrow pathological features. Although JAK inhibitors have been shown to improve patient constitutional symptoms and reduce splenomegaly, JAK inhibitor monotherapy does not significantly reduce mutant allele burden in the majority of MPN patients (Harrison et al., 2012; Harrison et al., 2016; Verstovsek et al., 2012).
The therapeutic window for JAK inhibitors is limited due to the essential role of the JAK-STAT signaling pathway in normal hematopoiesis, which has been observed in the clinic where these inhibitors have been associated with dose limiting toxicities. Therefore, there is a need to identify additional pathways that might be involved in the development and maintenance of MPN mutant clones, which could be targeted in combination with JAK2 for improved therapeutic benefit.
An increase in the expression of hedgehog target genes has been observed in granulocytes isolated from MPN patients (Bhagwat et al., 2013). So far, the exact role of the Hh pathway in MF and its contribution to bone marrow fibrosis is not fully understood. Here, we show that Gli1-expression is significantly increased in stromal cells from MPN patients and that Gli1+ MSCs are activated from their niche in the bone marrow upon expansion of the malignant clone, migrate into the hematopoietic marrow where they differentiate into matrix-producing myofibroblasts. Combinations of JAK2 inhibitors with inhibitors of the Hh pathway could provide an avenue of targeting stem cell–derived clonal myeloproliferation (which evades JAK2-targeted monotherapy). Preclinical and clinical data suggest that hedgehog pathway inhibitors have therapeutic activity in MF. To date, only smoothened (SMO)-inhibitors have been tested in MF with varying success (Sasaki et al., 2015). We hypothesize that Gli proteins in Gli1+ cells can be activated independent of canonical Hh signaling, explaining the mixed response in patients with MF, e.g. by PI3K/AKT signaling. Our data provide a rationale for targeted therapy of Gli1+ cells in bone marrow fibrosis, as monotherapy or combined therapy with other agents.
Experimental Model and Subject Details
Mice
All mouse experiments were approved by the responsible institutional and governmental ethics board i.e. Animal Care and Use Committee at Harvard University and Boston Childrens Hospital and the German government authorities (Landesamt für Natur, Umwelt und Verbraucherschutz Nordrhein Westfalen). Gli1CreERt2 (i.e. Gli1tm3(re/ERT2)Alj/J, JAX Stock #007913), Rosa26tdTomato (i.e. B6-Cg-Gt(ROSA)26Sorttm(CAG-tdTomato)Hze/J JAX Stock # 007909) and iDTR mice (i.e. C57BL/6-Gt(ROSA)26Sortm1(HBEGF)Awai/J, JAX Stock # 007900) were purchased from Jackson Laboratories (Bar Harbor, ME, USA). Offspring were genotyped by PCR according to the protocol from the Jackson laboratory. Males and females were used for the experiments as stated in the figure legends. Animals were maintained on the same 12hr light-dark cycle and received water and standard chow at libitum. For lineage tracing studies, 6-7 week old mice received 3x10 mg tamoxifen in corn oil / 3% ethanol (Sigma) via oral gavage 10 days before lethal irradiation (1050 Rads). Mice received 5x105 cells from 8 week old wild type littermates that had been harvested 48 hours prior to transplantation and were transduced with (1) Jak2(V617F) (JAK2) or control (control; WT Jak2) cDNA retrovirus or (2) ThPO or control (empty vector) lentivirus. Mice were randomly assigned to groups by choosing from a list of animal numbers with four digits. Mice were sacrificed 8 weeks after bone marrow transplantation in the ThPO fate tracing experiments and 17 weeks after bone marrow transplantation in the JAK2 experiments, when a significant decrease in the hemoglobin counts was observed. For RNA-Sequencing mice were sacrificed 10 weeks after bone marrow transplantation.
For the ablation experiments, bigenic Gli1CreERt2; iDTR mice received tamoxifen (3x 10mg per oral gavage) 10 days prior to irradiation and bone marrow transplantation and were injected with diphtheriatoxin dissolved in PBS (List Biological Laboratories) at a dose of 50ng/g body weight intraperitoneally (i.p.) as indicated (Figure 3B). Mice were sacrificed 14 weeks after bone marrow transplantation. Mice were equally assigned to DTX or vehicle groups based on their Hb to control for significant differences before start of the treatment (at 4 weeks after bone marrow transplantation). GANT61 Cayman Chemical) was dissolved in ethanol and stored at -80°C. The ethanol solution was further diluted in corn oil (1:4) immediately before injections. Mice were injected with 50mg/kg-body weight of GANT61 (s.c.) or vehicle (corn oil / ethanol) every other day starting 8 weeks after bone marrow transplantation until sacrifice at 119 days after transplantation.
Human specimen
Patients samples originated form RWTH Aachen University Hospital and Hannover Medical School and the study was approved by the Ethical board of the RWTH Aachen Medical Faculty (ethical votes 173/06, 206/09, 300/13, 94/16) and the Hannover Medical School (ethical vote 3381-2016). Samples were deidentified at the time of inclusion. All patients provided informed consent and the study was performed in accordance with the Declaration of Helsinki. Bone marrow biopsies for histological examination (tissue microarrays and whole punch biopsies) were chosen from archived patient samples of paraffin-embedded tissue from the Institute of Pathology at RWTH Aachen University Hospital, Aachen, Germany and from the Biobank of Dr. G. Büsche at the Department of Pathology, Hannover Medical School, Hannover, Germany. Biopsies were primarily taken during earlier hospitalization. Patient characteristics of biopsy specimen are reported in Table S1. Isolated human MSCs were studied from 4 healthy donors (age 62±10, 2 males) and 4 MPN patients (51±10, 2 males) as outlined in Table S2. To establish the Gli1 antibody staining human glioblastoma multiforme tissue was accessed from the RWTH Aachen Biobank.
Method Details
Human cell isolation and GANT61 treatment
Human MSCs (MPN patients or controls from hip replacement surgeries) were isolated by tissue culture plastic adherence after the mononuclear cell fraction was separated by Ficoll. Cells were seeded at a density of 5x106 cells/flask. MSCs were maintained in medium (Mesenpan; PAN Biotech) containing 10% fetal bovine serum (Life Technologies), 1% penincilin-streptomycin-glutamine (Life Technologies). Medium was changed every third day and cells were split at near confluence. MSC surface phenotype was assessed at passage 1 using antibodies against the following antigens: CD45 (clone HI30), CD105 (SN6), CD73 (AD2). Antibodies were purchased from BD Biosciences (San Jose, USA) or eBiosciences (San Diego, USA). To study the effect of GANT61, cells were treated for 24-48h with 10 μM GANT61 (Sigma) in DMSO versus vehicle (DMSO).
Isolation of Gli1+ cells
Gli1+ cells were isolated from the compact bone of 8-10 week old bigenic, male, Gli1CreER;tdTomato mice after receiving 3x10mg Tamoxifen p.o. Compact bones including femur, tibia, pelvis and vertebrae were crushed by mortar and pestle until only small fragments were left. The fragments were digested at 37°C in 10ml collagenase 2 (1mg/ml, Invitrogen) DMEM (Gibco) solution with 10% FBS (Gibco) for 90min. Following digestion the fragments were washed three times with PBS and then transferred to a 15cm cell-culture dish. The bone chips were cultured in alpha MEM (GlutaMAX, Life Technologies) containing 20% MSC qualified FBS (Life Technologies), 2% Penicillin Streptomycin (Life Technologies),1ng/ml murine basic fibroblast growth factor (Thermo Fisher Scientific) and 5ng/ml murine epidermal growth factor (Peprotech). To study the effect of TGFβ or CXCL4, primary Gli1+ bone marrow cells were seeded at a density of 60.000 cells/well in 6-well plates and cytokine treatment was started 24h later. The cytokines were added in fresh medium every 24 hours, for a total of 72 hours, at the following concentrations: TGFβ1 (100-21-10UG, Peprotech) 10ng/ml medium, CXCL4 (795-P4-025, R&D Systems) 400ng/ml. PBS (solvent for TGFβ) and DMSO (solvent for CXCL4) were added as controls. After 72 hours cells were subjected to flow cytometry for α-SMA as described below.
Viral transduction
For retro- and lentiviral transductions, ckit+ cells from wild type littermates were isolated using the autoMACS pro Separator after crushing the compact bone (Miltenyi Biotec). Ckit+ bone marrow cells were pre-stimulated for 24 hours in StemSpan™ Serum-Free Expansion Medium (Stem Cell Technology, Vancouver, Canada) supplemented with murine thrombopoietin (m-tpo) (50 ng/mL; Peprotech), murine stem cell factor (m-scf) (50 ng/mL; Peprotech), IL-6 ng/ml and 10ng/ml IL-3 (both Peprotech). Oncoretroviral vectors were pseudotyped with ecotropic envelope and produced using standard protocols. Retroviral transduction was performed on retroNectin (Takara Bio) coated cell culture dishes loaded with unconcentrated virus. Cells were then resuspended in virus containing medium in the presence of 10μg/ml polybrene (final concentration). Lentiviral particles were produced by transient transfection with lentiviral plasmid together with pSPAX and VSVG packaging plasmids using TransIT-LT (Mirus). Lentiviral and retroviral particles were concentrated using ultracentrifugation. Lentiviral transduction was performed with concentrated lentiviral supernatant in the presence of 2 μg/ml Polybrene using spin-infection for 90 minutes at 2,200rpm at 37°C.
Co-culture of HSCs and MSCs
For co-culture of Gli1+ stromal cells with c-kit+ hematopoietic stem and progenitor cells expressing control cDNA or ThPO/Jak2(V617F), c-kit+ cells were isolated and pre-stimulated for 24 hours in StemSpan™ Serum-Free Expansion Medium (Stem Cell Technology, Vancouver, Canada) supplemented with murine thrombopoietin (m-tpo) (50ng/mL; Peprotech), and murine stem cell factor (m-scf) (50ng/mL; Peprotech). C-kit cells were then transduced with concentrated lentivirus and 4μg/ml polybrene for 24 hours. Next, the cells were washed and co-cultured with Gli1+ stromal cells which were seeded at a density of 50 000 cells/well in alpha MEM (GlutaMAX, Life Technologies) containing 10% MSC qualified FBS (Life Technologies). For the analysis of stromal cells, hematopoietic cells were washed away (or harvested) and stromal cells were recovered by trypsinization. For the GANT61 experiments cells were treated for the last 48h with 10μM GANT61 in DMSO versus vehicle (DMSO) alone.
Migration assay
Lentivirally transduced c-kit+ cells from wild type littermates of Gli1CreER;tdTomato mice were seeded at 50,000 cells/well in a 24 well plates 24 hours after transduction. Primary Gli1+ bone marrow cells were seeded at 50,000 cells/well in transwells of the InnoCyte Cell Migration Assay (Millipore, #CBA017) above the ckit+ cells (bottom well). Cell migration was measured by flow cytometry from the bottom well. Alpha MEM (GlutaMAX, Life Technologies) containing 20% MSC qualified FCS (Life Technologies), 2% Penicillin Streptomycin (Life Technologies) was used as medium in this system.
In another set of experiments, solely Gli1+ cells were cultured in the transwell (600,000 cells/well) and recombinant Cxcl4 (95-P4-025, R&D Systems) in DMSO was added at 400ng/ml to the medium in the bottom well versus DMSO alone. After 12 hours of incubation, cell migration was measured by either flow cytometry or according to the manufacturer’s instructions on a plate reader (BMG Labtech, Clariostar).
Cytokine array
C-kit+ HSC were isolated from 8 week old wild type C57Bl6/J mice (males) as described above and lentivirally or retrovirally transduced with ThPO or Jak2(V617F) versus controls (empty backbone or WT Jak2, respectively). Cells were cultured in StemSpan™ serum-free expansion medium (Stem Cell Technology, Vancouver, Canada) supplemented with murine thrombopoietin (m-tpo) (50ng/mL; Peprotech), and murine stem cell factor (m-scf) (50ng/mL; Peprotech). After 7 days the cells were washed 3 times in PBS and starved in RPMI (Gibco) with 0.5% MSC-FCS (Life Technologies). The supernatant was harvested and incubated with the membranes of the mouse cytokine array (C2000 AAM-CYT-2000-2, Ray Biotech) according to the manufacturer’s instructions. Imaging was achieved by chemiluminescence detection with the ChemiDoc imager (BioRad) and quantification by analyzing the pictures with the Protein Array Analyzer plugin for ImageJ.
RNA-Sequencing and bioinformatics
For RNA-Sequencing 6-7 week old bigenic Gli1CreER;tdTomato mice were injected with tamoxifen (3x10mg p.o.), subjected to lethal irradiation 10 days after the last tamoxifen dose and received bone marrow of wildtype littermates that had been transduced with ThPO or control (empty backbone) lentivirus as described above. 10 weeks after bone marrow transplantation mice were sacrificed, whole bone marrow samples were lineage-depleted using paramagnetic microbeads and an autoMACS pro magnetic separator (Miltenyi Biotec). Gli1+ cells were sorted as lineage-, GFP-, tdTomato+ on a FACS Aria II (BD Bioscience) into PBS (5% FBS) and frozen in liquid nitrogen as pellets. RNA was extracted using TRIZOL Reagent (Life Technologies) according to the manufacturer’s instructions. cDNA libraries were generated using the Smart-Seq V4 ultra low input RNA kit (Clontech Laboratories) according to manufacturer instructions. Subsequently, amplified cDNA libraries were further processed using the Truseq Nano DNA sample prep guide (Illumina) generating Illumina Hiseq compatible pooled libraries that were sequenced paired-end (2x75 cycles) on a Hiseq2500 platform (Illumina).
The unprocessed data resulting from Hiseq2500 (Ilumina) were demultiplexed using casava (Ilumina). Quality metrics of the resulting fastq files were measured using the programs fastqc (Babraham bioinformatics) and multiqc (http://multiac.info). The later program summarizes the result of the first. An insert size estimate was performed by aligning the reads to a reference transcriptome using BWA (http://bio-bwa.sourceforge.net) and the actual insert size estimate was done with the program collectInsertSizeMetrics from the picard tools suite. The splicing aware alignment was performed using Tophat2 (https://ccb.jhu.edu/software/tophat/index.shtml) using the previously computed estimate for the insert size. Prior to the alignment the SMARTer adapters were trimmed. Gene and isoform abundance estimation was performed using cufflinks (http://cole-trapnell-lab.github.io/cufflinks/). Gene counts that were used in the differential expression analysis, clustering and principle component plots were obtained using HTseq counts (http://www-huber.embl.de/HTSeq/doc/count.html) using the intersect strict settings. Finally principle component analysis and clustering was performed using R (https://www.r-project.org/).
Directional pathway enrichment analysis was performed using the PIANO R package (Bioconductors, https://bioconductor.org/packages/release/bioc/html/piano.html, (Varemo et al., 2013) and a collection of pathway adapted to mus musculus (GSKB R package, Bioconductors, https://bioconductor.org/packages/release/data/experiment/html/gskb.html). PIANO compute the significance of the directional enrichment of a pathway by aggragating (here through the median) the FDR corrected p-values yielded by a set of enrichment methods. The method used here are : mean, median, sum, maxmean, stoufer, fisher, tail strength, wilcoxon and PAGE. The detailed explanation of the different methods can be found in (Varemo et al., 2013). The collection of pathway used here as gene sets is derived from MSigDB canonical pathway complemented by a few other currated pathway databases and metabolic reaction networks (see GSKB R package). Reactome was discarded from the pathway sources as it was too much redundant with itself and other pathway ressources. The script of the pathway enrichment analysis is available at https://github.com/adugourd/PIANO_kramann_2017.
Flow cytometry
Bone marrow (BM) cells were isolated by flushing and crushing pelvis and hind leg bones with PBS (GIBCO) + 2% FBS + Penicillin/Streptomycin (GIBCO). Whole bone marrow was lysed on ice with red blood cell (RBC) lysis solution (Invitrogen/Life Technologies), and washed in PBS (GIBCO) + 2% FBS. Single-cell suspensions of spleen were prepared by pressing tissue through a cell strainer followed by red blood cell lysis. Cells were labeled with the following monoclonal, directly fluorochrome conjugated antibodies: anti-mouse: Ter119 (eBioscience efluor 450), Gr1 eBioscience (efluor 450), CD11b (eBioscience, efluor 450), B220 (efluor 450 eBioscience), CD3 (efluor 450 eBioscience), CD150 (PeCy7 BioLegend), CD48 (APCCy7 Biolegend), CD34 (Alexa Fluor 700 eBioscience), ckit (APC eBioscience), Sca1 (PE, eBioscience), Gr1 (Alexa Fluor 700, eBioscience), CD11b (APC, eBioscience), CD3 (PE, eBioscience), CD19 (PeCy7 eBioscience), Ter119 (FITC, eBioscience), CD31 (APC, Biolegend), CD34 (FITC, eBioscience), CD29 (APC, eBioscience), Sca1 (APC-Cy7, Biolegend), CD44 (PE-Cy7 eBioscience); CD105 (PE-Cy7, Biolegend), CD73 (APC, Biolegend), PDGFRα (APC, Biolegend), CD31 (APC, Biolegend), CD34 (FITC, eBioscience), CD29 (APC, eBioscience), Sca1 (APC-Cy7, Biolegend), CD44 (PE-Cy7 eBioscience); CD105 (PE-Cy7, Biolegend), CD73 (APC, Biolegend), PDGFRα (APC, Biolegend). Anti-human: CD90 (PeCy7, BD Pharmingen), CD105 (V450, BDPharmingen), Nestin (PE, RnD Systems). Further the following primary non-conjugated antibodies were used: Nestin (1:100, Abcam), CD51 (1:100, Biolegend), αSMA (1:100 Sigma), Gli1 (1:100, Novus biological). For flow cytometry, cells were stained in antibodies diluted as 1:100 in 2% FBS/PBS for 30 min on ice.
The staining of the intracellular antigens was performed after fixation and permeabilization of the cells (Cytofix/Cytoperm solution, BD Biosciences). Following incubation with primary antibodies, cells were washed in PBS and incubated with the appropriate AF647/AF488 secondary antibodies (Jackson Immuno) at 1:200 for 15min on ice. For detection of apoptosis, the APC Annexin V antibody (BD Bioscience, #550474) was used together with the Annexin V Binding Buffer (BD Bioscience, #556454) and co-staining for DAPI or 7AAD (BD Bioscience, #559925) was performed according to the manufacturer’s instructions. For staining of pSTAT5, cells were fixed in 4% paraformaldehyde (Sigma) for 10 minutes followed by permeabilization with 90% ice cold methanol (Sigma) for 30 minutes followed by staining with the pSTAT5 antibody (PE-Cy7, 1:100, BD Bioscience #560117) in PBS. To increase the flow efficiency, whole bone marrow samples were lineage-depleted (for HSC staining and stromal cells) or –enriched (lineages) using paramagnetic microbeads and an autoMACS pro magnetic separator (Miltenyi Biotec). All flow cytometric analyses were performed at the LSR II Flow Cytometer or Canto II (both BD Biosciences). For flow cytometric analyses DAPI (1mg/ml 1:1000) was added in order to exclude dead cells. Data were analyzed by using Flow Jo software (Version 9.6.2, Tree Star Inc).
Viral vector cloning
The murine Jak2 cDNA was cloned into the retroviral vector MSCV-IRES EGFP (a gift from Tannishtha Reya Addgene #20672). The V617F mutation in Jak2 was achieved using site-directed mutagenesis (Quikchange-XL; Agilent). The murine Thrombopoietin cDNA (MGC clone 3593885, GE Dharmacon) was PCR amplified using the primer sequences 5'-AGACCGGTGCCACCATGGAGCTGACTGATTTGCTCC-3' and 5'-TCACGCGTCTATGTTTCCTGAGACAAATTCC-3'. Subsequently, the PCR product was digested with AgeI and XhoI and cloned into the pRRL.PPT.SFFV.IRES2.EGFP.PRE (kindly provided by A. Schambach and C. Baum, Hannover Medical School, Hannover, Germany).
Histological an immunohistochemical analysis
For histological and immunohistochemical analyses, murine organs were fixed in 4% formaldehyde overnight, dehydrated and prepared for paraffin embedding. H&E staining and reticulin staining was done according to routine protocols. Human bone marrow biopsies were fixed for 24 hours using 12% buffered formaldehyde plus 64% methanol, decalcified (EDTA), dehydrated and embedded in paraffin. For immunohistochemistry, samples were deparaffinized and stained as follows.
Immunohistochemical analysis was performed using a primary antibody for Gli1 (Novus biological, NG-600-600, 1:100). Antigen demarcation was achieved by pressure cooker treament in citrate buffer (Vector Antigen unmasking solution). After blocking (Blocking Kit; Vector Laboratories) and 3%H2O2 treatment, sections were incubated with the primary antibody. Biotinylated monoclonal goat-anti-rabbit (Vector Laboratories) was used as a secondary antibody. Detection was performed using the Vectastin ABC Kit (Vector Laboratories). Slides were counterstained with hematoxylin. Cell numbers and co-staining was evaluated in 400x magnification at the Keyence BZ9000 by a blinded pathologist. Co-staining by immunohistochemistry in human bone marrow biopsies was achieved by using the regular staining as described above for the first antigen (Leptin Receptor, 1:100, Santa Cruz sc-1834; Nestin 1:100, Abcam Ab134017; NG2 1:100, Millipore AB5320) followed by staining of the second antigen (Gli1 1:100, Novus biological, NG-600-600) with the ImmPRESS-AP Anti Rabbit IgG (alkaline phosphatase) polymer detection kit (Vector MP-5401) and the Vector Red Alkalkine Phosphatase (Red AP) Substrate Kit (#SK5100).
For immunofluorescence studies, tissue was fixed in 4% paraformaldehyde on ice for 1 hour, then incubated in 30% sucrose in PBS at 4°C overnight. OCT-embedded (Sakura Finetek) tissues were cryosectioned into 6μm sections and mounted on Superfrost slides (Fisher Scientific). Sections were washed in 1X PBS, blocked in 10% normal goat serum (Vector Labs) and incubated with primary antibodies specific for alpha SMA (1:200, Sigma, # A2547), CD31 (1:100, ebioscience, #14-0311 and AF3628 1:100 R&D Systems), leptin receptor (1:100; sc-1834, Santa Cruz), CD105 (1:100, MAB 1320, R&D Systems), Nestin (1:100, abcam ab134017), NG2 (AB5320 Millipore tyrosine hydroxylase (1:100, #AB152 millipore), GFAP (1:100; DAKO Z0334). Secondary antibodies were FITC-, or Cy5-conjugated (Jackson ImmunoResearch). To stain lipid droplets OCT sections were incubated with LipidTox Deep Red neutral lipid stain (Invitrogen, H34477) at 1:200 in PBS for 1 hour. To stain bone, we used Fetuin A from fetal bovine serum (Sigma aldrich #F2379) which was purified over HiLoad 16/60 Superdex 200pg columns (GE Healthcare #28989335) in an ÄKTA Purifier (GE Healthcare #28406264) and then conjugated with Alexa Fluor 647 NHS Ester (Invitroagen #A37573), according to the manufacturer’s instructions. All sections were counterstained with DAPI (4´,6´-´diamidino-2-phenylindole) and mounted in Prolong Gold (Life Technologies). Images were obtained by confocal microscopy (Nikon C1 eclipse and Zeiss LSM 710).
Clearing and imaging of mouse bone
Solvent-based optical clearing was performed using a sequence of dehydration steps in ethanol (50% for 12 hours, 70% for 6 hours, and 100% for 6 hours), followed by direct immersion on ethyl cinnamate (Sigma, #112372) for 12 hours in order to render the bone transparent. Vasculature was labeled with a CD31-AF647 antibody (BioLegend, #102516). Imaging was performed using an upright scanning confocal microscope (Leica TCS SP8; Leica Microsystems, Wetzlar, Germany). Optical sections were acquired with an optimised step-size of 2μm for a total depth of 120μm. Image post-processing and 3D rendering was performed with Imaris (Bitplane, v8.1).
Real Time PCR Experiments
Tissue was harvested and immediately snap frozen in liquid nitrogen. RNA from was extracted according to the manufacturer instructions using the RNeasy Mini Kit (Qiagen) and 600ng of total RNA was reverse transcribed with iScript (BioRad). During the RNA extraction DNA was removed by a DNAse digestion step (Life Technologies). Quantitative polymerase chain reactions were carried out with iQ-SYBR Green supermix (BioRad) and the Applied Biosystems, 7300 Teal-Time PCR System. Cycling conditions were 95°C for 3 minutes then 40 cycles of 95°C for 15 seconds and 60°C for 1 minute, followed by one cycle of 95°C for 10 seconds. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a housekeeping gene. Data was analyzed using the 2-ΔΔct method. Primers are listed in tables S3-S4.
Immunocytochemistry
Human MSCs (n=4 healthy; n=4 MPN patients) were seeded at a density of 1x105 cells/well in chamber slides (Nunc, lab tek, Thermo Fischer Scientific, Darmstadt, Germany), the next day cells were treated for 24 hours with 10 μM Gant61 (Sigma, Munich, Germany). After washing and fixation steps, cells were stained with α-SMA antibody (Sigma, Munich, Germany) at 1:400 for 3h at RT, and co-stained with the fluorescent secondary-labeled antibody donkey anti-mouse Alexa Fluor 488 (Thermo Fischer Scientific, Darmstadt, Germany) at 1:200 for 1h at RT. After washing, cells were stained with the primary unconjugated antibody Gli1 (Novus Biologicals, Wiesbaden Nordenstadt, Germany) at 1:100 overnight at 4 degree and stained with the fluorescent secondary-labeled antibody goat anti-rabbit Alexa Fluor 633 (Thermo Fischer Scientific, Darmstadt, Germany) at 1:200 for 1h at RT. Nuclear staining was performed with DAPI at 1ug/mL for 15 minutes at RT and mounting with Vectashield (#H-1000, Biozol, Eching, Germany). Images were acquired with a confocal laser scanning microscope (LSM 710; Zeiss, Germany) running the Zen 2012 (Zeiss) software.
Image Quantification
The distance to either CD31+ endothelial cells or Fetuin A+ bone was performed by ImageJ. Random images were acquired by the LSM710 confocal microscope (Zeiss) with a magnification of 800x (at least 3-4 images of 3 mice per group). All images were processed by ImageJ. First, all images were split into RGB channels. Gaussian blur was applied to adjust background noise in all channels. Auto-threshold was used to convert intensity values of CD31 staining and tdTomato+ area into binary data. Then a distance map was created from the binary data. One maximum intensity point was selected as the representative point of each nucleus. These points were used to divide the binary tdTomato+ area into smaller areas representing single tdTomato+ cells. Selected nuclei and the divided tdTomato+ areas were overlayed on a CD31 or Fetuin A generated distance map. The distance value from each nucleus to the nearest CD31 or Fetuin A positive area was calculated.
Quantification of GFAP+ and tyrosine hydroxylase+ areas was performed by Image J using the Gaussian blur and auto-threshold function. The number of tdTomato+, αSMA+ and αSMA+/tdTomato+ cells were calculated by Image J using split single channels (RGB). Auto-threshold was again used to identify αSMA+ and tdTomato+ binary area and the overlying point of maximal intensity of each nucleus was used to quantify the number of each cell population. CD31 surface area from immunohistochemistry was calculated using the number of stained pixels per total pixels in Adobe Photoshop CS5 (Adobe Systems Inc.). Reticulin staining (myelofibrosis grade) was quantified by a blinded hematopathologist based on the WHO criteria. Gli1+ cells in human bone marrow biopsies were counted by a blinded hematopathologist in at least 3 random high power fields (400x) per patient. The association to bone and vasculature was noted during counting. Gli1-tdTomato+ cell distribution and co-localization with marker expression was counted in backbone and sternal bone sections (at least 4/mouse) using the LSM710 confocal microscope.
Quantification and Statistical Analysis
Data are presented as mean±SEM. Comparison of two groups was performed using unpaired t-test. Paired t-test was used for comparsion of repeated measured in the same group. For multiple group comparison analysis of variance with posthoc Tukey correction was applied. Statistical analyses were performed using GraphPad Prism 5.0c (GraphPad Software Inc., San Diego, CA). A p value of less than 0.05 was considered significant.
Data and Software Availability
RNA Sequencing data is available online through the European Nucleotide Archive, which is hosted by the European Bioinformatics Institute (EBI). Accession number: ERA829202
Supplementary Material
Acknowledgements
This work was supported by grants: to RKS German Research Foundation (SCHN 1188/5-1) Interdisciplinary Center for Clinical Research RWTH Aachen (O1-5), the European Hematology Association, Deutsche Krebshilfe (111750); to RK German Research Foundation (KR-4073/3-1, SFBTRR57, SCHN 1188/5-1), European Research Council (ERC-StG 677448), START RWTH Aachen (101/15), State of North Rhine-Westphalia (Return to NRW); to BDH, NIH/NIDDK (DK088923, DK103740 and DK103050), Established Investigator Award of the American Heart Association (EIA14650059); to DH Deutsche Krebshilfe (11743); to AD European Union´s Horizon 2020 research and innovation program (675585 Marie-Curie ITN “SymBioSys”). AD is is a Marie-Curie ESR at RWTH (Germany). The work was supported by the confocal microscopy facility IZKF Aachen. We thank Willi Jahnen-Dechent (RWTH Aachen) for kindly providing labeled Fetuin A. We thank Peggy Jirak, Katrin Härthe, Nina Graff, Susanne Ziegler and Flavia Ribezzo for excellent technical support.
Footnotes
Author contributions
R.K.S. designed and carried out experiments, analyzed results and wrote the manuscript, A.M., B.L.E. and B.D.H. contributed to experimental design, data interpretion and writing of the manuscript, J.S.R, R.H., A.D. and J.W. analyzed data and reviewed the manuscript, G.B. selected patient specimen, analyzed and interpreted histopathology and reviewed the manuscript, G.M.N. contributed to confocal imaging and reviewed the manuscript, P.M.H.V-S, E.M.B., F.P, M.V.F., D.F. and V.P. carried out experiments, analyzed the data and reviewed the manuscript, R.K. designed and carried out experiments, analyzed results and wrote the manuscript.
Contact for Reagent and Resource Sharing
As Lead Contact, Rebekka K Schneider (Erasmus Medical Center, Rotterdam) is responsible for all reagent and resource requests. Please contact Rebekka K Schneider at r.k.schneider@erasmusmc.nl with requests and inquiries.
References
- Aberger F, Ruiz IAA. Context-dependent signal integration by the GLI code: the oncogenic load, pathways, modifiers and implications for cancer therapy. Seminars in cell & developmental biology. 2014;33:93–104. doi: 10.1016/j.semcdb.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham NG, Feldman E, Falck JR, Lutton JD, Schwartzman ML. Modulation of erythropoiesis by novel human bone marrow cytochrome P450-dependent metabolites of arachidonic acid. Blood. 1991;78:1461–1466. [PubMed] [Google Scholar]
- Arranz L, Sanchez-Aguilera A, Martin-Perez D, Isern J, Langa X, Tzankov A, Lundberg P, Muntion S, Tzeng YS, Lai DM, et al. Neuropathy of haematopoietic stem cell niche is essential for myeloproliferative neoplasms. Nature. 2014;512:78–81. doi: 10.1038/nature13383. [DOI] [PubMed] [Google Scholar]
- Bares V, Ge X. gskb: Gene Set data for pathway analysis in mouse. 2015 R package version 161. [Google Scholar]
- Bhagwat N, Keller MD, Rampal RK, Shank K, de Stanchina E, Rose K, Amakye D, Levine RL. Improved Efficacy Of Comibnation Of JAK2 and Hedgehog Inhibitors in Myelofibrosis. Blood. 2013;122 [Google Scholar]
- Biagini G, Severi B, Govoni E, Preda P, Pileri S, Martinelli G, Visani G, Finelli C, Castaldini C. Stromal cells in primary myelofibrosis: ultrastructural observations. Virchows Arch B Cell Pathol Incl Mol Pathol. 1985;48:1–8. doi: 10.1007/BF02890110. [DOI] [PubMed] [Google Scholar]
- Bruns I, Lucas D, Pinho S, Ahmed J, Lambert MP, Kunisaki Y, Scheiermann C, Schiff L, Poncz M, Bergman A, et al. Megakaryocytes regulate hematopoietic stem cell quiescence through CXCL4 secretion. Nat Med. 2014;20:1315–1320. doi: 10.1038/nm.3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstein SA, Malpass TW, Yee E, Kadin M, Brigden M, Adamson JW, Harker LA. Platelet factor-4 excretion in myeloproliferative disease: implications for the aetiology of myelofibrosis. Br J Haematol. 1984;57:383–392. doi: 10.1111/j.1365-2141.1984.tb02912.x. [DOI] [PubMed] [Google Scholar]
- El Agha E, Moiseenko A, Kheirollahi V, De Langhe S, Crnkovic S, Kwapiszewska G, Kosanovic D, Schwind F, Schermuly RT, Henneke I, et al. Two-Way Conversion between Lipogenic and Myogenic Fibroblastic Phenotypes Marks the Progression and Resolution of Lung Fibrosis. Cell Stem Cell. 2016 doi: 10.1016/j.stem.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison C, Kiladjian JJ, Al-Ali HK, Gisslinger H, Waltzman R, Stalbovskaya V, McQuitty M, Hunter DS, Levy R, Knoops L, et al. JAK inhibition with ruxolitinib versus best available therapy for myelofibrosis. N Engl J Med. 2012;366:787–798. doi: 10.1056/NEJMoa1110556. [DOI] [PubMed] [Google Scholar]
- Harrison CN, Vannucchi AM, Kiladjian JJ, Al-Ali HK, Gisslinger H, Knoops L, Cervantes F, Jones MM, Sun K, McQuitty M, et al. Long-term findings from COMFORT-II, a phase 3 study of ruxolitinib vs best available therapy for myelofibrosis. Leukemia. 2016 doi: 10.1038/leu.2016.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klampfl T, Gisslinger H, Harutyunyan AS, Nivarthi H, Rumi E, Milosevic JD, Them NC, Berg T, Gisslinger B, Pietra D, et al. Somatic mutations of calreticulin in myeloproliferative neoplasms. N Engl J Med. 2013;369:2379–2390. doi: 10.1056/NEJMoa1311347. [DOI] [PubMed] [Google Scholar]
- Kramann R, DiRocco DP, Humphreys BD. Understanding the origin, activation and regulation of matrix-producing myofibroblasts for treatment of fibrotic disease. The Journal of pathology. 2013;231:273–289. doi: 10.1002/path.4253. [DOI] [PubMed] [Google Scholar]
- Kramann R, Fleig SV, Schneider RK, Fabian SL, DiRocco DP, Maarouf O, Wongboonsin J, Ikeda Y, Heckl D, Chang SL, et al. Pharmacological GLI2 inhibition prevents myofibroblast cell-cycle progression and reduces kidney fibrosis. J Clin Invest. 2015a;125:2935–2951. doi: 10.1172/JCI74929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramann R, Schneider RK, DiRocco DP, Machado F, Fleig S, Bondzie PA, Henderson JM, Ebert BL, Humphreys BD. Perivascular Gli1+ progenitors are key contributors to injury-induced organ fibrosis. Cell stem cell. 2015b;16:51–66. doi: 10.1016/j.stem.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunisaki Y, Bruns I, Scheiermann C, Ahmed J, Pinho S, Zhang D, Mizoguchi T, Wei Q, Lucas D, Ito K, et al. Arteriolar niches maintain haematopoietic stem cell quiescence. Nature. 2013;502:637–643. doi: 10.1038/nature12612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuter DJ, Bain B, Mufti G, Bagg A, Hasserjian RP. Bone marrow fibrosis: pathophysiology and clinical significance of increased bone marrow stromal fibres. Br J Haematol. 2007;139:351–362. doi: 10.1111/j.1365-2141.2007.06807.x. [DOI] [PubMed] [Google Scholar]
- Lauth M, Bergstrom A, Shimokawa T, Toftgard R. Inhibition of GLI-mediated transcription and tumor cell growth by small-molecule antagonists. Proc Natl Acad Sci U S A. 2007;104:8455–8460. doi: 10.1073/pnas.0609699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine RL. JAK-mutant myeloproliferative neoplasms. Curr Top Microbiol Immunol. 2012;355:119–133. doi: 10.1007/82_2011_170. [DOI] [PubMed] [Google Scholar]
- Levine RL, Gilliland DG. Myeloproliferative disorders. Blood. 2008;112:2190–2198. doi: 10.1182/blood-2008-03-077966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta J, Wang H, Iqbal SU, Mesa R. Epidemiology of myeloproliferative neoplasms in the United States. Leukemia & lymphoma. 2014;55:595–600. doi: 10.3109/10428194.2013.813500. [DOI] [PubMed] [Google Scholar]
- Mendez-Ferrer S, Michurina TV, Ferraro F, Mazloom AR, Macarthur BD, Lira SA, Scadden DT, Ma'ayan A, Enikolopov GN, Frenette PS. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 2010;466:829–834. doi: 10.1038/nature09262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshai EF, Wemeau-Stervinou L, Cigna N, Brayer S, Somme JM, Crestani B, Mailleux AA. Targeting the hedgehog-glioma-associated oncogene homolog pathway inhibits bleomycin-induced lung fibrosis in mice. American journal of respiratory cell and molecular biology. 2014;51:11–25. doi: 10.1165/rcmb.2013-0154OC. [DOI] [PubMed] [Google Scholar]
- Nangalia J, Massie CE, Baxter EJ, Nice FL, Gundem G, Wedge DC, Avezov E, Li J, Kollmann K, Kent DG, et al. Somatic CALR mutations in myeloproliferative neoplasms with nonmutated JAK2. N Engl J Med. 2013;369:2391–2405. doi: 10.1056/NEJMoa1312542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondelli D, Barosi G, Bacigalupo A, Prchal JT, Popat U, Alessandrino EP, Spivak JL, Smith BD, Klingemann HG, Fruchtman S, et al. Allogeneic hematopoietic stem-cell transplantation with reduced-intensity conditioning in intermediate- or high-risk patients with myelofibrosis with myeloid metaplasia. Blood. 2005;105:4115–4119. doi: 10.1182/blood-2004-11-4299. [DOI] [PubMed] [Google Scholar]
- Sasaki K, Gotlib JR, Mesa RA, Newberry KJ, Ravandi F, Cortes JE, Kelly P, Kutok JL, Kantarjian HM, Verstovsek S. Phase II evaluation of IPI-926, an oral Hedgehog inhibitor, in patients with myelofibrosis. Leuk Lymphoma. 2015;56:2092–2097. doi: 10.3109/10428194.2014.984703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepers K, Campbell TB, Passegue E. Normal and leukemic stem cell niches: insights and therapeutic opportunities. Cell stem cell. 2015;16:254–267. doi: 10.1016/j.stem.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepers K, Pietras EM, Reynaud D, Flach J, Binnewies M, Garg T, Wagers AJ, Hsiao EC, Passegue E. Myeloproliferative neoplasia remodels the endosteal bone marrow niche into a self-reinforcing leukemic niche. Cell stem cell. 2013;13:285–299. doi: 10.1016/j.stem.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider RK, Ziegler S, Leisten I, Ferreira MS, Schumacher A, Rath B, Fahrenkamp D, Muller-Newen G, Crysandt M, Wilop S, et al. Activated fibronectin-secretory phenotype of mesenchymal stromal cells in pre-fibrotic myeloproliferative neoplasms. Journal of hematology & oncology. 2014;7:92. doi: 10.1186/s13045-014-0092-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tefferi A. Primary myelofibrosis: 2012 update on diagnosis, risk stratification, and management. Am J Hematol. 2011;86:1017–1026. doi: 10.1002/ajh.22210. [DOI] [PubMed] [Google Scholar]
- Tefferi A, Lasho TL, Tischer A, Wassie EA, Finke CM, Belachew AA, Ketterling RP, Hanson CA, Pardanani AD. The prognostic advantage of calreticulin mutations in myelofibrosis might be confined to type 1 or type 1-like CALR variants. Blood. 2014;124:2465–2466. doi: 10.1182/blood-2014-07-588426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tefferi A, Thiele J, Orazi A, Kvasnicka HM, Barbui T, Hanson CA, Barosi G, Verstovsek S, Birgegard G, Mesa R, et al. Proposals and rationale for revision of the World Health Organization diagnostic criteria for polycythemia vera, essential thrombocythemia, and primary myelofibrosis: recommendations from an ad hoc international expert panel. Blood. 2007;110:1092–1097. doi: 10.1182/blood-2007-04-083501. [DOI] [PubMed] [Google Scholar]
- Thiele J, Kuemmel T, Sander C, Fischer R. Ultrastructure of bone marrow tissue in so-called primary (idiopathic) myelofibrosis-osteomyelosclerosis (agnogenic myeloid metaplasia). I. Abnormalities of megakaryopoiesis and thrombocytes. Journal of submicroscopic cytology and pathology. 1991;23:93–107. [PubMed] [Google Scholar]
- Tzeng YS, Li H, Kang YL, Chen WC, Cheng WC, Lai DM. Loss of Cxcl12/Sdf-1 in adult mice decreases the quiescent state of hematopoietic stem/progenitor cells and alters the pattern of hematopoietic regeneration after myelosuppression. Blood. 2011;117:429–439. doi: 10.1182/blood-2010-01-266833. [DOI] [PubMed] [Google Scholar]
- van Bon L, Affandi AJ, Broen J, Christmann RB, Marijnissen RJ, Stawski L, Farina GA, Stifano G, Mathes AL, Cossu M, et al. Proteome-wide analysis and CXCL4 as a biomarker in systemic sclerosis. N Engl J Med. 2014;370:433–443. doi: 10.1056/NEJMoa1114576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannucchi AM, Migliaccio AR, Paoletti F, Chagraoui H, Wendling F. Pathogenesis of myelofibrosis with myeloid metaplasia: lessons from mouse models of the disease. Semin Oncol. 2005;32:365–372. doi: 10.1053/j.seminoncol.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Varemo L, Nielsen J, Nookaew I. Enriching the gene set analysis of genome-wide data by incorporating directionality of gene expression and combining statistical hypotheses and methods. Nucleic Acids Res. 2013;41:4378–4391. doi: 10.1093/nar/gkt111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstovsek S, Mesa RA, Gotlib J, Levy RS, Gupta V, DiPersio JF, Catalano JV, Deininger M, Miller C, Silver RT, et al. A double-blind, placebo-controlled trial of ruxolitinib for myelofibrosis. N Engl J Med. 2012;366:799–807. doi: 10.1056/NEJMoa1110557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeval JL, Cohen-Solal K, Tulliez M, Giraudier S, Guichard J, Burstein SA, Cramer EM, Vainchenker W, Wendling F. High thrombopoietin production by hematopoietic cells induces a fatal myeloproliferative syndrome in mice. Blood. 1997;90:4369–4383. [PubMed] [Google Scholar]
- Wernig G, Kharas MG, Okabe R, Moore SA, Leeman DS, Cullen DE, Gozo M, McDowell EP, Levine RL, Doukas J, et al. Efficacy of TG101348, a selective JAK2 inhibitor, in treatment of a murine model of JAK2V617F-induced polycythemia vera. Cancer Cell. 2008;13:311–320. doi: 10.1016/j.ccr.2008.02.009. [DOI] [PubMed] [Google Scholar]
- Yan XQ, Lacey D, Hill D, Chen Y, Fletcher F, Hawley RG, McNiece IK. A model of myelofibrosis and osteosclerosis in mice induced by overexpressing thrombopoietin (mpl ligand): reversal of disease by bone marrow transplantation. Blood. 1996;88:402–409. [PubMed] [Google Scholar]
- Zaldivar MM, Pauels K, von Hundelshausen P, Berres ML, Schmitz P, Bornemann J, Kowalska MA, Gassler N, Streetz KL, Weiskirchen R, et al. CXC chemokine ligand 4 (Cxcl4) is a platelet-derived mediator of experimental liver fibrosis. Hepatology. 2010;51:1345–1353. doi: 10.1002/hep.23435. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA Sequencing data is available online through the European Nucleotide Archive, which is hosted by the European Bioinformatics Institute (EBI). Accession number: ERA829202