Abstract
Smoking has historically been recognized as a negative prognostic factor in head and neck squamous cell carcinoma (HNSCC). This study aimed to assess the mutational differences between heavy smokers (>20 pack years) and never smokers among the HNSCC patients within The Cancer Genome Atlas (TCGA). Single nucleotide variation and copy number aberration differences between heavy smokers and never smokers were compared within human papillomavirus–positive (HPV-positive) (n = 67) and HPV-negative (n = 431) TCGA cohorts with HNSCC, and the impact of these mutations on survival were assessed. No genes were differentially mutated between smoking and never-smoking patients with HPV-positive tumors. By contrast, in HPV-negative tumors, NSD1 and COL1A11 were found to be more frequently mutated in heavy smokers, while CASP8 was more frequently altered in never smokers. HPV-negative patients with NSD1 mutations experienced significantly improved overall survival compared with NSD1 WT patients. This improved prognosis was validated in an independent cohort of 77 oral cavity cancer patients and a meta-analysis that included 2 additional data sets (688 total patients, hazard ratio for death 0.44, 95% CI, 0.30–0.65). NSD1 mutations are more common in HPV-negative heavy smokers, define a cohort with favorable prognosis, and may represent a clinically useful biomarker to guide treatment deintensification for HPV-negative patients.
Keywords: Genetics, Oncology
Keywords: Head & neck cancer
NSD1 is commonly mutated in heavy smokers with HPV-negative head and neck squamous cell carcinoma, and those with NSD1 mutations have a favorable prognosis.
Introduction
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common cancer worldwide, with over 550,000 new cases diagnosed each year (1). Human papillomavirus (HPV) infection causes a subset of HNSCCs that present with distinct clinical, pathological, and molecular features compared with the HPV-negative disease, which is typically associated with tobacco and alcohol use (2, 3).
Smoking history at the time of diagnosis is considered an important negative prognostic factor for both HPV-positive and -negative HNSCCs (4–6). Given the observed differences in outcomes based on smoking status, there is interest in understanding the molecular differences between never smokers and smokers in both HPV-positive and -negative diseases. A previous study by Yavorski and colleagues examined differences in mutation frequencies by smoking status in several smoking related cancers including lung, head and neck, pancreatic, and bladder cancer, using data from The Cancer Genome Atlas (TCGA) (7). They identified a difference in the frequency of single nucleotide variations (SNVs) associated with the cytoskeletal protein–related coding regions (CPRCRs) in both lung and head and neck cancers, as well as differential SNV rates in cancer driver genes in lung cancer. However, the investigators a priori limited their analysis to CPRCRs, tumor suppressors, and oncogenes, limiting the discovery potential of the study (7). Moreover, HNSCC data was not stratified by HPV status, which is another significant shortcoming.
Recently, Bratman and colleagues reanalyzed TCGA’s HNSCC RNA sequencing database for the presence of HPV transcripts to definitively identify the HPV status of each patient (8). Building on this new information, we have comprehensively explored the differences between the clinical characteristics, prognosis, and mutational profile of HPV-positive and -negative tumors within TCGA HNSCC data set, stratified by patients’ smoking history.
Results
NSD1, CASP8, and COL1A11 mutations were associated with smoking in HPV-negative HNSCC.
TCGA HNSCC cohort comprised 431 HPV-negative and 67 HPV-positive primary HNSCC samples with DNA mutation and survival information. Within the HPV-negative group, there were 205 heavy smokers and 90 never smokers. In the HPV-positive group, 21 of the patients were heavy smokers and 23 were never smokers. Patient demographic differences between heavy smokers and never smokers are provided in Supplemental Table 1 (supplemental material available online with this article; https://doi.org/10.1172/jci.insight.123443DS1).
The total SNV mutational load was significantly lower in HPV-positive patients (median = 72, 95% CI, 57–95) compared with HPV-negative patients (median = 110.5, 95% CI, 106–117; P = 0.0001, Supplemental Figure 1A). The correlation between total mutation count and pack year history of smoking was significant in HPV-negative patients (Spearman’s Rho = 0.28, 95% CI, 0.18–0.38, P < 0.0001) but not in HPV-positive patients (Spearman’s Rho = 0.05, 95% CI, –0.21–0.31, P = 0.68; Supplemental Figure 1B).
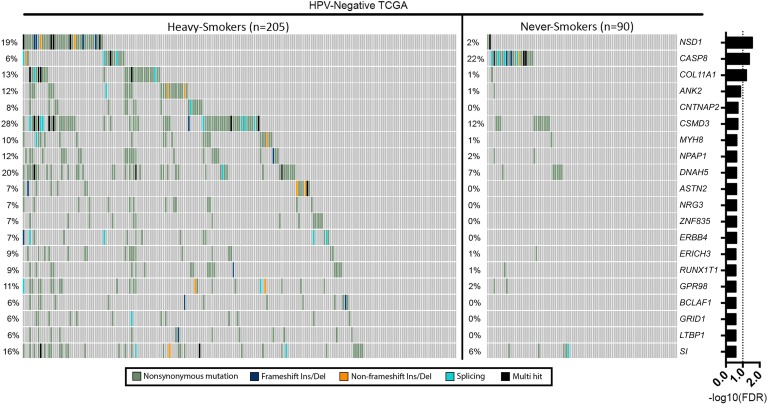
A comparison of SNVs in individual genes identified no significant differences between never smoker and heavy smoker samples in the HPV-positive group following correction for multiple hypothesis testing (Supplemental Figure 2). In contrast, the frequency of SNVs in NSD1, CASP8, and COL11A1 between the never smokers and heavy smokers in the HPV-negative group was significantly different (Figure 1).
Figure 1. Top 20 differentially mutated genes between never smokers and heavy smokers in the HPV-negative patient group.
NSD1, CASP8, and COL11A1 are mutated at significantly different frequencies after correction for the FDR. Fisher’s exact test was used for comparison, and FDR correction was done through Benjamini-Hochberg method.
The frequency of NSD1 mutations in HPV-negative light smokers (≤ 20 pack year) was greater than that of never smokers but less than that of heavy smokers (2.22% never smokers, 4.55% light, and 18.5% heavy smokers, respectively; Supplemental Table 2). The opposite pattern was seen for CASP8 and COL11A1 (Supplemental Table 2). The occurrence rates stratified by smoking status for all 3 genes were similar to those Stransky et al. reported (ref. 9 and Supplemental Table 2). Lists of the 100 most differentially mutated genes, based on smoking history, in HPV-positive and -negative patients are provided in Supplemental Tables 3 and 4, respectively.
Analysis of copy number aberrations (CNAs) did not reveal any statistically significant differences in large amplifications (e.g., amplifications larger than the arm-level amplifications observed for the sample; ref. 10) or deep deletions based on smoking status in either the HPV-positive or -negative subsets after FDR correction (Supplemental Table 5).
NSD1 mutations were associated with improved overall survival in HPV-negative patients.
There was no significant difference in overall survival between the never smokers and heavy smokers in either the HPV-positive or HPV-negative cohorts (Supplemental Figure 3 and Supplemental Table 6). However, in HPV-negative patients, those with NSD1 mutations experienced superior overall survival compared with NSD1 WT patients (Figure 2A and Supplemental Table 6). The impact of NSD1 mutation on survival within the entire TCGA HNSCC tumor cohort (n = 498, 55 NSD1 mutations) was comparable to that of HPV status (n = 67 HPV positive; hazard ratios [HRs] in Supplemental Figure 4). By contrast, mutations in CASP8 or COL11A1 were not associated with significant survival differences (Supplemental Figure 5). NSD1 functions as an H3 histone lysine 36 (H3K36) methyltransferase, and NSD1-mutant tumors exhibited an altered global DNA methylation pattern that was recapitulated in tumors with H3K36M mutations (10). Importantly, H3K36M mutation status was not correlated with overall survival (log-rank P = 0.99), ruling out a role for altered DNA methylation in the survival advantage associated with NSD1 mutation (Supplemental Figure 6).
Figure 2. Overall survival comparison between NSD1-mutant and WT patients.
Survival rate in (A) the HPV-negative TCGA HNSCC cohort and (B) an independent cohort of 77 locally advanced HPV-negative oral cavity squamous cell carcinomas (OSCCs) from London Health Sciences Centre (LHSC).
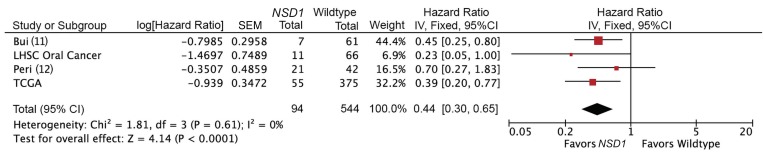
Targeted sequencing of NSD1 in an independent cohort of 77 oral cavity cancers also demonstrated improved overall survival for patients with NSD1 mutations in a multivariable survival model (P < 0.05, Figure 2B and Supplemental Table 7). We carried out a meta-analysis of 4 available data sets, including TCGA data set, Bui et al. (11), Peri et al. (12), and our validation cohort, comparing survival by NSD1 mutation status. The pooled estimate for these studies demonstrated strong evidence for improved overall survival for patients with NSD1-mutant tumors compared with patients with WT tumors (Figure 3, HR 0.44; 95% CI, 0.30–0.65; P < 0.0001). There was no evidence of heterogeneity (P = 0.61, I2 = 0%). A sensitivity analysis was performed, excluding incomplete data; this did not significantly change the pooled HR estimate (HR 0.40; 95% CI, 0.26–0.62; P < 0.0001).
Figure 3. Meta-analysis comparing overall survival stratified by NSD1 mutation status in HPV-negative tumors.
IV, Inverse Variance; df, degrees of freedom.
NSD1 mutations were associated with mutations in ECM genes.
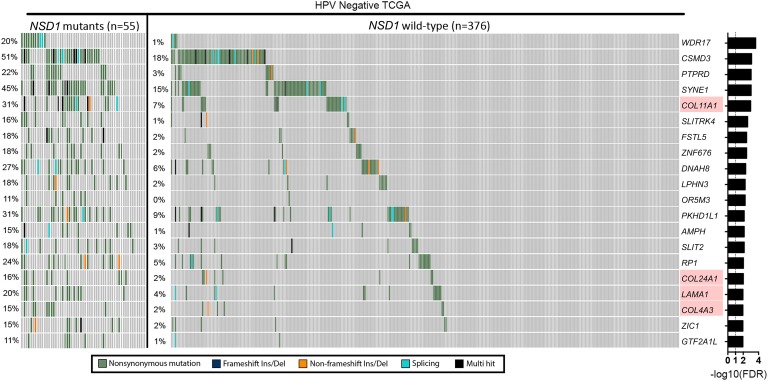
In TCGA HPV-negative cohort, NSD1-mutant tumors were associated with a heavier smoking history and were more prevalent in the laryngeal subsite compared with NSD1 WT tumors (Supplemental Table 8). In TCGA HPV-negative patients, total mutational load was higher in NSD1-mutant (median = 209, 95% CI, 163–336) tumors relative to NSD1 WT tumors (median = 105.5, 95% CI, 100–110), and this difference was statistically significant (Mann-Whitney U test, P < 0.0001). Comparison of gene-wise mutation status between NSD1-mutant and WT tumors revealed 180 genes to be mutated at different frequencies (FDR < 0.01; Figure 4). A list of the top 100 differentially mutated genes between NSD1-mutant and WT HPV-negative HNSCC samples is provided in Supplemental Table 9. Pathway analysis with these differentially mutated genes revealed multiple overrepresented pathways (top 20 overrepresented pathways in Supplemental Table 10), suggesting a potentially altered ECM environment in the NSD1-mutant compared with NSD1 WT HPV-negative HNSCC tumors. Analysis with the top mutated genes in HPV-negative HNSCC showed that NSD1 mutations had a pattern of co-occurrence with other highly mutated genes, and no mutually exclusive mutations were identified (Supplemental Table 11).
Figure 4. Analysis of somatic SNVs between NSD1-mutant and WT HPV-negative HNSCC tumors.
Genes associated with the ECM are highlighted in red. Fisher’s exact test was used for comparison, and FDR correction was done through Benjamini-Hochberg method.
Discussion
We have identified genes with SNVs that occur at significantly different frequencies in HPV-negative heavy smokers and never smokers: NSD1 and COL1A11 (more frequent in smokers) and CASP8 (more frequently in never smokers; Figure 1). Importantly, NSD1 mutations were associated with improved survival, independent of smoking status. Indeed, our meta-analysis of 4 independent cohorts comprising 638 patients confirmed the strong survival advantage for patients with NSD1-mutant tumors (Figure 3). This survival difference was profound, with an HR comparable to the prognostic significance of HPV (Supplemental Figure 4). Given these findings are reproducible across multiple independent data sets, it is likely that NSD1 represents a clinically useful biomarker that can be used to direct treatment intensity.
The NSD1 methyltransferase functions to monomethylate and dimethylate H3K36 (13). Mutations in NSD family proteins and dysregulation of methylation at H3K36 have both been linked to multiple human cancers (14). Recently, Bui and colleagues suggested that these epigenetic changes are responsible for the improved outcomes, citing increased sensitivity to cisplatin chemotherapy as a possible mechanism (11). However, using unsupervised hierarchical clustering of TCGA HNSCC DNA methylation data, we previously identified a unique cluster comprising 13% of HPV-negative patients that contained either recurrent mutations in NSD1 or mutations in K36M in multiple H3 genes (10). Despite similar methylation profiles, NSD1- and H3K36M-mutant tumors appeared markedly different from a prognostic perspective, with only patients with NSD1 mutation demonstrating favorable outcomes. H3K36M-mutant tumors behave similarly to the remainder of the tumors of the HPV-negative cohort, making methylation changes unlikely to be the sole source of treatment sensitivity.
In an effort to provide an alternative explanation for the improved survival observed in NSD1-mutant tumors, we compared the spectrum of mutations in NSD1-mutant and NSD1 WT tumors. This revealed multiple genes encoding ECM components, including collagen (COL4A3, COL11A1, COL19A1, COL22A1, COL24A1) and laminin (LAMA1 and LAMA2), that were more frequently altered in NSD1-mutated tumors (Figure 4 and Supplemental Table 9). Further functional studies are necessary to come to definitive conclusions regarding the role of the ECM and NSD1 mutations in tumor aggressiveness.
Recently, Peri et al. reported that NSD1 mutation status predicted improved outcomes in laryngeal cancer using TCGA data; however, they did not identify a difference in prognosis in HPV-negative tumors in other head and neck subsites (12). That study did not integrate definitive viral transcript identification to classify tumor HPV status and instead relied on p16 and in situ hybridization testing, which was carried out on only a subset of cases. This misclassifies a significant number of tumors and partially obscures the analysis of an HPV-negative cohort with previously unidentified HPV-positive samples (8). Tumor classification after HPV transcript analysis shows that only 2 of the HPV-positive tumors are from the larynx, suggesting that anatomical location specification in Peri and colleagues’ analysis may have served to isolate HPV-negative tumor samples. Our multivariate analysis with definitive HPV status assignment confirmed NSD1 mutation status as an independent prognostic factor in HPV-negative tumors regardless of tumor site (Supplemental Table 6). Our validation cohort of advanced oral cavity cancers as well as an additional cohort of HPV-negative patients from Bui et al. (11) confirm generalizability of the prognostic importance of NSD1, greatly broadening the impact of this finding.
Survival analysis of TCGA’s HPV-positive and -negative HNSCC patient cohorts did not show a significant difference between never smokers and heavy smokers. This is contrary to the reported prognostic significance of smoking history as an independent variable in both HPV-positive and -negative disease in clinical trials (4, 15). We speculate that this observation may result from the heterogeneity of treatment protocols within this cohort, which may obscure smaller survival differences.
After correction for multiple testing, significant differences in gene mutation frequency between never smokers and heavy smokers were identified but only in the HPV-negative cohort. A larger number of HPV-positive tumors may be necessary to better elucidate the mutational differences between never smokers and heavy smokers within this group.
This study shows that the link between smoking and disease outcomes in HPV-negative HNSCCs may be more complex than previously believed. Mutations in NSD1 strongly predict improved overall survival, and genes involved with the ECM may be a possible mediator for this effect. Further studies are needed to understand the mechanisms of the improved survival experienced by this subset of patients.
Methods
Data collection.
Patient data from TCGA, including level 3 DNA mutation packager calls data, CNAs, and merged clinical data sets, were retrieved using The Broad Institute’s Firehose databases (16). Firehose GISTIC2 copy number databases were used for CNA comparisons (17). The HPV status of samples was assigned based on the detection of viral transcripts by Bratman et al. (8). The resulting data sets included 431 HPV-negative and 67 HPV-positive primary HNSCC samples with DNA mutation data and survival information for all but 1 of the HPV-negative HNSCC cases. Patients were stratified as never smokers or heavy smokers (>20 pack year history) based on similar research in the literature (18–20). Patients with between 1 and 20 pack years (n = 59), patients with no smoking history information (n = 10), and patients who were identified as current or former smokers but lacked pack year history information (n = 90) were excluded. An additional data set from Stransky et al. (9) was used to assess the distribution of gene SNVs by smoking history.
Validation cohort.
Tumor samples from patients with advanced T stage (T3–4) oral cavity cancers were obtained from 77 patients undergoing definitive primary surgery at LHSC. Matched blood was available for 74 patients (96%). Patient demographics and survival information were prospectively collected. Tumor cellularity of greater than 70% was confirmed by pathologist review of frozen sections. DNA was extracted from blood and tumor samples using Qiagen kits. The coding region of the NSD1 gene was targeted for sequencing using an Illumina primer pool. Samples were processed at London Regional Genomics Centre (LRGC) on the Illumina MiSeq platform, with a targeted depth of 800×. FASTQ files were downloaded from LRGC and aligned to GRCh38 (GenBank accession: GCA_000001405.15, with decoy hs38d1) using BWA-MEM (v0.7.15). Duplicates were not marked due to the targeted nature of the data. Indel realignment and recalibration were performed using Genome Analysis Toolkit (GATK) (v3.7.0). For tumors with a matched normal, germline SNPs were called using GATK’s HaplotypeCaller (v3.7.0) and filtered for quality and read depth. Somatic SNVs were identified using MuTect (v1.1.7). For tumors without a matched normal, a panel of normals (generated using 438 normal samples from TCGA HNSCC data set — BWA v0.7.12 with hs37d5, GATK v3.4.0, and MuTect v1.1.6, converted to GRCh38 coordinates using Picard v2.7.1) was used to remove probable germline variants (along with the following filters: SNV passed quality control, had a minimum read depth of 50×, and was present in fewer than 4 samples in the panel of normals).
Statistics.
TCGA exome sequencing data were analyzed using the maftools package (version 1.2.3) (21) within the Bioconductor framework in R statistical environment (version 3.4.0). The total mutation loads of HPV-positive and HPV-negative patients were compared using Mann-Whitney U test. The correlation between total SNV count and number of pack years was calculated using Spearman’s rank correlation, and 2-tailed P values were reported. Synonymous mutation variants were excluded from downstream analyses. Fisher’s exact tests were used to identify the differentially mutated genes between never smokers and heavy smokers. Only genes that were mutated in at least 10% of patients in at least 1 arm of the comparison were evaluated. Derived P values were adjusted for FDR using the Benjamini-Hochberg method, and an FDR threshold of 0.1 was set for significance.
CNAs were compared for significant differences in deep deletions or large amplifications, as assigned by TCGA’s GISTIC2 analysis. Deep deletions were defined as deletions greater than the minimum arm-level deletion observed for the sample, and large amplifications as amplifications larger than the arm-level amplifications observed for the sample. Fisher’s exact tests were used for statistical testing of differences of CNAs by smoking status in HPV-positive and -negative cohorts separately, and FDR adjustment was done as previously described.
The total mutation load, SNVs, and CNAs were similarly compared using Fisher’s exact tests with FDR adjustment to identify differences between NSD1-mutant and WT patients in the HPV-negative TCGA HNSCC cohort. The top 100 genes were defined as those with the largest differences in mutation frequency between NSD1-mutant and WT HPV-negative HNSCCs and were analyzed for pathway overrepresentation using the Reactome Knowledgebase online software (22).
Association of NSD1 mutations with the top 25 mutated genes in HPV-negative HNSCC was done using Fisher’s exact test comparing the proportion of samples with NSD1 mutation within a cohort with another gene mutation to the proportion of NSD1-mutant samples in the entire HPV-negative TCGA cohort. FDR correction was done using the Benjamini-Hochberg method, and a threshold of 0.1 was set for significance.
χ2 Goodness of fit and Mann-Whitney-Wilcoxon tests were used to compare the distribution of clinical features between never smokers and heavy smokers in HPV-positive and -negative groups. The same analysis was applied to compare the distribution of clinical features in the HPV-negative cohort, stratified by tumor NSD1 mutation status.
Overall survival outcomes were compared between never smokers and heavy smokers in both HPV-positive and HPV-negative subsets of patients using the log-rank statistical approach. Analyses were performed using the survival package in R (v2.41-3) (23). For each of the identified genes (NSD1, COL1A11, CASP8), a survival analysis was done using the log-rank test to compare outcomes in HPV-negative patients with mutations to those without. A Cox regression model was used for multivariate analysis to assess the association of each clinical variable, smoking status, and NSD1 mutation status with overall survival. The P values derived from the Wald test on the survival coefficients of smoking history were reported, and the proportional hazards assumptions of Cox models were tested, based on the scaled Schoenfeld residuals, to search for a significant relationship between residuals and time. Survival analysis was also carried out, stratifying by H3K36 mutation status because Papillon-Cavanagh et al. (10) previously identified this in the HPV-negative TCGA cohort.
Somatic SNVs were filtered to remove nonfunctional (intronic/intergenic) variants before downstream analyses. Kaplan-Meier curves were constructed to compare overall survival for patients with NSD1-mutant versus NSD1 WT tumors. A Cox proportional hazards model was fit to carry out multivariate survival analysis for NSD1 mutation status with clinical covariates, including gender; age; smoking history (heavy, light, or never smoker); alcohol abuse (described history of alcoholism or abuse by the patient or over 21 drinks per week); N-staging category; and margins (positive or negative). Multivariate analysis was performed via backward and forward stepwise analysis to attain the best model of survival, and the final multivariate model of survival with HRs associated with each variable was reported.
Survival data across cohorts was pooled and analyses carried out as above comparing patients with NSD1-mutant and NSD1 WT tumors. The pooled outcome was total mortality expressed as an HR. If the studies did not quote HRs, we calculated the log10 HR and SEM from the available summary statistics or Kaplan-Meier curves, according to the methods proposed by Parmar and Tierney (24). For 1 source (12), we were unable to calculate the HR using these methods because the study did not report the observed events or the median and range of follow-up. We therefore made the assumption that the 2-year mortality reported in the Kaplan-Meier curve was the observed death rate. We assessed the significance of any discrepancies in the estimates of the treatment effects from the different trials using Cochrane’s test for heterogeneity and the I2 statistic: I2 values over 50% indicated moderate to high heterogeneity, as described previously (25). We considered heterogeneity statistically significant if the P value was less than 0.10 for the χ2 test. We combined HRs for survival data, using a fixed-effect model. We used Review Manager 5.3 (The Cochrane Collaboration) to enter HR data into the meta-analysis using the inverse variance method.
Study approval.
For the local oral cancer cohort, ethics approval was obtained from the University of Western Ontario Health Sciences Research Ethics Board (REB 16579). Informed consent was received from patients.
Author contributions
FG, PCB, and ACN designed the study. FG, SDP, SFG, and PCB carried out bioinformatics. NM, JWB, NP, KMR, and MIK collected clinical data, extracted nucleic acid, performed HPV testing, and identified samples. DM, AS and DAP performed the meta-analyis. CH, WS, and PP performed the pathology review. FG, JY, KF, EW, JSM and ACN prepared the manuscript. All authors provided scientific advice and reviewed the paper.
Supplementary Material
Acknowledgments
This study was funded by a Canadian Institutes for Health Research (CIHR) grant MOP 340674 to ACN and PCB. PCB was supported by CIHR and Terry Fox Research New Investigator Awards and by the Ontario Institute for Cancer Research. ACN was supported by the Wolfe Surgical Research Professorship in the Biology of Head and Neck Cancers Fund.
Version 1. 01/10/2019
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
License: Copyright 2019, American Society for Clinical Investigation.
Reference information: JCI Insight. 2019;4(1):e123443. https://doi.org/10.1172/jci.insight.123443.
Contributor Information
Farhad Ghasemi, Email: fghasemi2019@meds.uwo.ca.
Stephenie D. Prokopec, Email: Stephenie.Prokopec@oicr.on.ca.
Neil Mundi, Email: Neil1.Mundi@lhsc.on.ca.
William Stecho, Email: wstecho@gmail.com.
Paul Plantinga, Email: Paul1.Plantinga@lhsc.on.ca.
Nicole Pinto, Email: npinto4@uwo.ca.
Kara M. Ruicci, Email: kruicci@uwo.ca.
John Yoo, Email: john.yoo@lhsc.on.ca.
Kevin Fung, Email: kevin.fung@lhsc.on.ca.
Axel Sahovaler, Email: axel.sahovaler1@gmail.com.
Eric Winquist, Email: eric.winquist@lhsc.on.ca.
Joe S. Mymryk, Email: jmymryk@uwo.ca.
John W. Barrett, Email: john.barrett@lhsc.on.ca.
Anthony C. Nichols, Email: anthony.nichols@lhsc.on.ca.
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127(12):2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Mehanna H, et al. Prevalence of human papillomavirus in oropharyngeal and nonoropharyngeal head and neck cancer — systematic review and meta-analysis of trends by time and region. Head Neck. 2013;35(5):747–755. doi: 10.1002/hed.22015. [DOI] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Network Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517(7536):576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ang KK, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363(1):24–35. doi: 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hafkamp HC, et al. Marked differences in survival rate between smokers and nonsmokers with HPV 16-associated tonsillar carcinomas. Int J Cancer. 2008;122(12):2656–2664. doi: 10.1002/ijc.23458. [DOI] [PubMed] [Google Scholar]
- 6.Gillison ML, et al. Tobacco smoking and increased risk of death and progression for patients with p16-positive and p16-negative oropharyngeal cancer. J Clin Oncol. 2012;30(17):2102–2111. doi: 10.1200/JCO.2011.38.4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yavorski JM, Blanck G. Smoking correlates with increased cytoskeletal protein-related coding region mutations in the lung and head and neck datasets of the cancer genome atlas. Physiol Rep. 2016;4(24):e13045. doi: 10.14814/phy2.13045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bratman SV, et al. Human papillomavirus genotype association with survival in head and neck squamous cell carcinoma. JAMA Oncol. 2016;2(6):823–826. doi: 10.1001/jamaoncol.2015.6587. [DOI] [PubMed] [Google Scholar]
- 9.Stransky N, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333(6046):1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Papillon-Cavanagh S, et al. Impaired H3K36 methylation defines a subset of head and neck squamous cell carcinomas. Nat Genet. 2017;49(2):180–185. doi: 10.1038/ng.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bui N, et al. Disruption of NSD1 in head and neck cancer promotes favorable chemotherapeutic responses linked to hypomethylation. Mol Cancer Ther. 2018;17(7):1585–1594. doi: 10.1158/1535-7163.MCT-17-0937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peri S, et al. NSD1- and NSD2-damaging mutations define a subset of laryngeal tumors with favorable prognosis. Nat Commun. 2017;8(1):1772. doi: 10.1038/s41467-017-01877-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wagner EJ, Carpenter PB. Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol. 2012;13(2):115–126. doi: 10.1038/nrm3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morishita M, di Luccio E. Cancers and the NSD family of histone lysine methyltransferases. Biochim Biophys Acta. 2011;1816(2):158–163. doi: 10.1016/j.bbcan.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 15.Sharp L, McDevitt J, Carsin AE, Brown C, Comber H. Smoking at diagnosis is an independent prognostic factor for cancer-specific survival in head and neck cancer: findings from a large, population-based study. Cancer Epidemiol Biomarkers Prev. 2014;23(11):2579–2590. doi: 10.1158/1055-9965.EPI-14-0311. [DOI] [PubMed] [Google Scholar]
- 16. doi: 10.7908/C11G0KM9. Broad-Institute-TCGA-Genome-Data-Analysis-Center. Broad Institute of MIT and Harvard; 2016. [DOI]
- 17. doi: 10.1186/gb-2011-12-4-r41. Broad-Institute-TCGA-Genome-Data-Analysis-Center. Analysis-ready standardized TCGA data from Broad GDAC Firehose 2016_01_28 run. 2016. [DOI]
- 18.Gillison ML, et al. Survival outcomes by tumor human papillomavirus (HPV) status in stage III-IV oropharyngeal cancer (OPC) in RTOG 0129. J Clin Oncol. 2009;27(15S):6003 [Google Scholar]
- 19.Nichols AC, et al. The epidemic of human papillomavirus and oropharyngeal cancer in a Canadian population. Curr Oncol. 2013;20(4):212–219. doi: 10.3747/co.20.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nichols AC, et al. High frequency of activating PIK3CA mutations in human papillomavirus-positive oropharyngeal cancer. JAMA Otolaryngol Head Neck Surg. 2013;139(6):617–622. doi: 10.1001/jamaoto.2013.3210. [DOI] [PubMed] [Google Scholar]
- 21. doi: 10.1101/052662. Mayakonda A, Koeffler HP. Maftools: Efficient analysis, visualization and summarization of MAF files from large-scale cohort based cancer studies. bioRxiv . [DOI]
- 22.Fabregat A, et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2018;46(D1):D649–D655. doi: 10.1093/nar/gkx1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Therneau TM, Grambsch PM. Modeling Survival Data: Extending the Cox Model. New York, New York, USA: Springer-Verlag; 2000. [Google Scholar]
- 24.Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat Med. 1998;17(24):2815–2834. doi: 10.1002/(SICI)1097-0258(19981230)17:24<2815::AID-SIM110>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 25.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.