Abstract
Pulmonary fibrosis (PF) is an intractable disorder with a poor prognosis. Although lung fibroblasts play a central role in PF, the key regulatory molecules involved in this process remain unknown. To address this issue, we performed a time-course transcriptome analysis on lung fibroblasts of bleomycin- and silica-treated murine lungs. We found gene modules whose expression kinetics were associated with the progression of PF and human idiopathic PF (IPF). Upstream analysis of a transcriptome network helped in identifying 55 hub transcription factors that were highly connected with PF-associated gene modules. Of these hubs, the expression of Srebf1 decreased in line with progression of PF and human IPF, suggesting its suppressive role in fibroblast activation. Consistently, adoptive transfer and genetic modification studies revealed that the hub transcription factor SREBP-1c suppressed PF-associated gene expression changes in lung fibroblasts and PF pathology in vivo. Moreover, therapeutic pharmacological activation of LXR, an SREBP-1c activator, suppressed the Srebf1-dependent activation of fibroblasts and progression of PF. Thus, SREBP-1c acts as a protective hub of lung fibroblast activation in PF. Collectively, the findings of the current study may prove to be valuable in the development of effective therapeutic strategies for PF.
Keywords: Inflammation, Pulmonology
Keywords: Bioinformatics, Fibrosis, Mouse models
Time-course transcriptome network analysis of lung fibroblasts reveals the LXR-SREBP-1c axis as a protective hub of fibroblast activation in murine pulmonary fibrosis.
Introduction
Pulmonary fibrosis (PF) is a progressive, devastating lung pathology with limited therapeutic options (1, 2). PF is characterized by chronic deposition of the extracellular matrix (ECM) in lung tissue, which decreases lung compliance and leads to respiratory failure (3).
Activated lung fibroblasts (myofibroblasts) are principal effector cells in both wound healing and PF that act as a primary source of various ECM components, including type I collagen (4). ECM components provide scaffold for reepithelialization and mediate wound closure in physiological healing (5). However, in pathological fibrosis, dense ECM and activated fibroblasts accumulate in fibrotic lesions, eventually destroying lung architecture (6). Thus, intervention against aberrant activation of lung fibroblasts could be a therapeutic target against PF development (5, 6).
Several signaling pathways and transcription factors (TFs) are known to be involved in the activation of lung fibroblasts and PF pathology. The TGF-β1-Smad3 pathway activates lung fibroblasts and causes fibrogenic responses in the lung (6). Early growth response-1 (Egr-1), another downstream target of TGF-β1, enhances fibroblast activation and PF, and broadly regulates TGF-β and ECM-related genes (7). Activation of PPAR-γ, a lipogenic TF, inhibits lung fibroblast activation and bleomycin-induced PF and restores lipogenic properties of lung fibroblasts (8–10). However, global transcriptional landscape and its regulatory networks of lung fibroblast activation during initiation, resolution, and progression of PF remain insufficiently understood.
Transcriptional profiling is an effective approach to investigate the landscape of physiological/pathological states of certain cell subsets and organs in various contexts. In particular, weighted coexpression network analysis (WGCNA) (11) is widely used to identify gene modules and centrally connected “hub” genes involved in a particular biological context, such as T cell exhaustion/memory (12), macrophage activation (13), neuronal differentiation (14), and pancreatic cancer (15). Time-course transcriptional profiling is another effective way to investigate biological event-associated gene clusters (16). Indeed, Bauer Y. et al. compared time-course microarray data of bleomycin-treated rat lungs and that of idiopathic PF (IPF) lungs and revealed that IPF-related gene signatures are mostly changed at 7 days after induction (dpi) of bleomycin injury (17). Based on the above knowledge, we assumed that time-course transcriptional profiling and the WGCNA approach are suitable for identifying PF pathology–associated gene modules and their hub TFs in activated lung fibroblasts.
By using 3′ serial analysis of gene expression sequencing (SAGE-seq) (18), we performed time-course transcriptome analysis of the fibroblasts purified from the lungs of bleomycin- and silica-treated Col1a2-GFP reporter mice (Col-GFP mice) and used WGCNA to examine these data. Then, we applied upstream analysis to WGCNA coexpression network and identified 55 hub TFs, including lipid-related TFs (Srebf1, Nr1h3 [Lxra], and Rxra), of PF-associated network in activated lung fibroblasts. Finally, we investigated the role of lipid-related TFs in fibroblast activation and PF pathology by genetic and pharmacological interventions.
Results
Transcriptomic landscape of lung fibroblasts in bleomycin- and silica-induced pulmonary fibrosis.
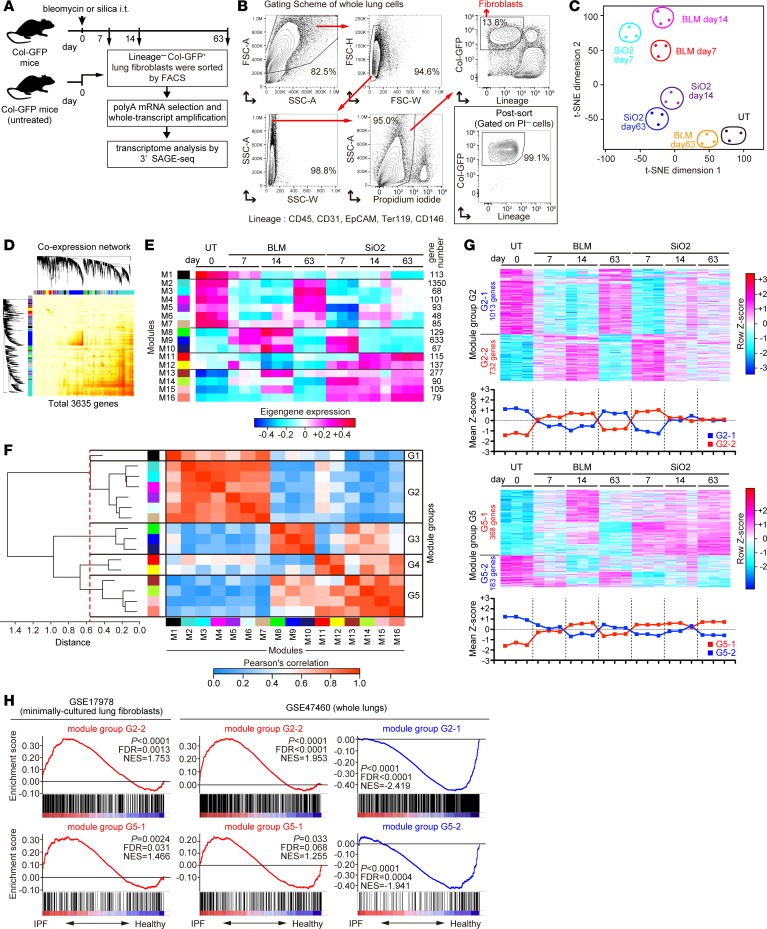
To clarify transcriptomic landscape of lung fibroblasts throughout PF initiation, progression, and resolution, we induced reversible or progressive PF in Col-GFP mice by bleomycin or silica aspiration, respectively (19–21). In the bleomycin model, PF was initiated at 7–14 dpi, and it spontaneously receded at 63 dpi (22). In the silica model, PF was initiated at 7–14 dpi, and it continuously progressed until at least 63 dpi (21). Based on the kinetics of pathological changes in these models, we purified lineage– (CD45, CD31, EpCAM, CD146, and Ter119) Col-GFP+ lung fibroblasts by flow cytometry at 0, 7, 14, and 63 dpi in both models and performed transcriptome analysis by 3′ SAGE-seq (Figure 1, A and B). t-Distributed stochastic neighbor embedding (t-SNE) analysis (23) of the transcriptome data revealed that the dissimilarity between steady-state and activated fibroblasts declined at 63 dpi in the bleomycin model but was maintained from 14–63 dpi in the silica model, consistent with pathological courses of these PF models (Figure 1C). To identify PF-associated transcriptomic signatures of activated fibroblasts, we applied WGCNA to 3,635 differentially expressed genes with adjusted P < 0.05 and a fold change of ≥2 between at least 2 samples (Figure 1D). WGCNA identified 16 coexpressed gene modules (Figure 1E), and hierarchical clustering of the module eigengenes classified these modules into 5 distinct groups (Figure 1F). In these groups, gene expression changes of module groups G2 and G5 positively or negatively correlated with PF initiation, progression, and resolution (Figure 1, E and G). In contrast, changes in gene expression of module groups G3, G4, and G1 were observed only in the initiation phase of both models, observed in the progression phase of the silica model, or not clearly associated with PF pathology, respectively (Figure 1E and Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.122163DS1).
Figure 1. Transcriptomic landscape of lung fibroblasts in bleomycin- and silica-induced pulmonary fibrosis.
(A) Experimental scheme. Lungs were collected from untreated, bleomycin-treated, and silica-treated Col-GFP mice at multiple time points. Lineage–Col-GFP+ lung fibroblasts were purified by cell sorting. Whole transcripts of purified fibroblasts were amplified, and transcriptome analysis was performed by the 3′ SAGE-seq method (n = 3 for each time point). (B) Gating scheme for lung fibroblasts and purity of fibroblasts after cell sorting. Representative plots of n = 7 from 2 independent experiments of the silica model data at 14 dpi are shown. (C) t-Distributed stochastic neighbor embedding (t-SNE) plot of 3′ SAGE-seq data of activated lung fibroblasts. (D) Coexpression network plot of differentially expressed 3,635 genes in activated lung fibroblasts. (E) Heatmap representation of eigengene kinetics of gene modules identified by the weighted coexpression network analysis. Each column represents group and time point, whereas each row represents an individual eigengene. Module size is shown on the right side of the heatmap. (F) Hierarchical clustering of eigengenes. Grouping threshold is shown as red dashed line. Module group is shown on the right side of the correlation heatmap. (G) Heatmap representation of the genes in module groups G2 and G5. Mean Z score of module groups is shown on the bottom of the heatmap. (H) Gene set enrichment analysis of fibrosis-associated module groups for the samples from IPF-derived lung fibroblasts (GSE17978) and idiopathic pulmonary fibrosis whole lungs (E-GEOD-47460). Enrichment plots for module groups G2-1, G2-2, G5-1, and G5-2 are shown. Black bars represent gene positions in the ranked gene list. NES, normalized enrichment score. P values and FDRs were determined using a permutation test–based method implemented in the GSEA software. Col-GFP mice, Col1a2-GFP reporter mice; SAGE, serial analysis of gene expression; FSC, forward scatter; SSC, side scatter; PI, propidium iodide; BLM, bleomycin model; SiO2, silica model; UT, untreated;IPF, idiopathic pulmonary fibrosis; NES, normalized enrichment score.
To assess the clinical relevance of identified module groups, we performed gene set enrichment analysis (GSEA) (24) of public transcriptome data from minimally cultured lung fibroblasts derived from human idiopathic pulmonary fibrosis (IPF) and healthy lungs, as well as a separate dataset from whole human IPF and healthy lungs (17, 25). CLICK clustering (26) subgrouped the genes in module groups G1–G5 into 2 distinct groups by their positive correlation. The genes in module groups G2-2 and G5-1, which were upregulated in line with PF progression, were significantly enriched both in IPF lung fibroblasts and whole IPF lungs (Figure 1H). In contrast, the genes in module groups G2-1 and G5-2, which were downregulated in line with PF progression, were significantly enriched in healthy whole lungs (Figure 1H). Other gene module groups did not show a consistent enrichment pattern in either murine or human PF (Supplemental Figure 2). These data indicate that genes in module groups G2 and G5 might be transcriptomic signatures of activated lung fibroblasts conserved both in murine and human PF.
Functional characterization of transcriptomic signatures of activated lung fibroblasts in bleomycin- and silica-induced pulmonary fibrosis.
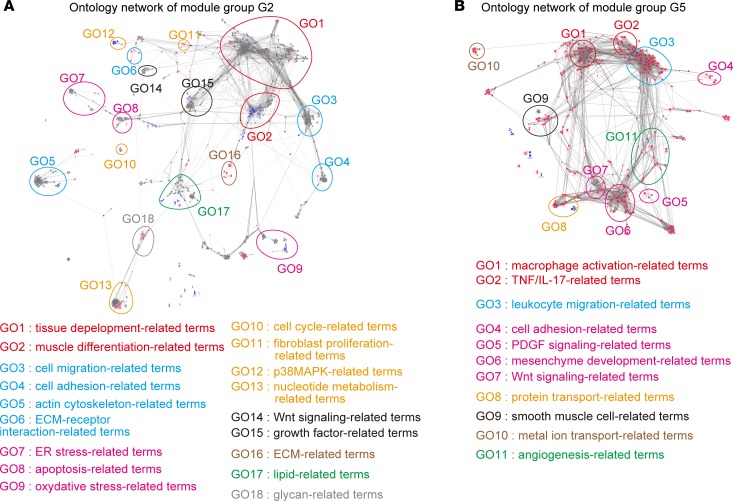
To clarify the biological functions associated with genes from PF-associated module groups G2 and G5, we performed their gene ontology (GO) network analysis to summarize overenriched GO terms by their similarities (27, 28) (Figure 2, A and B, and Supplemental Table 1). We revealed that fibrosis- and fibroblast activation–associated terms (tissue development–, cell adhesion/migration–, cell stress–, fibroblast proliferation–, and ECM-related terms) were highly enriched in module group G2 (Figure 2A). In contrast, macrophage-related terms (macrophage activation– and leukocyte migration–related terms) were highly enriched in module group G5 (Figure 2B), consistent with previous reports that macrophages regulated bleomycin- and silica-induced PF pathology (21, 29, 30). In addition, TGF-β and PDGF signaling–related terms (Wnt signaling–, growth factor–, PDGF signaling–, and mesenchyme development–related terms) were highly enriched in both module group G2 and G5 (Figure 2, A and B), consistent with the importance of these 2 cascades in the regulation of fibroblast activation and PF pathology (6, 31). These results indicate that PF-associated module groups G2 and G5 are functionally relevant to inflammation, PF progression, and fibroblast activation. Of note, lipid- and glycan-related terms were highly enriched in module group G2 (Figure 2A).
Figure 2. Functional characterization of transcriptomic signatures of lung fibroblasts in bleomycin- and silica-induced pulmonary fibrosis.
(A and B) Network of significantly enriched functional terms of module groups G2 (A) and G5 (B). Biological events associated with module groups G2 and G5 were explored and term network was clustered using Cytoscape 3.3.0 with ClueGO and Allegrolayout plugins. Each node and its size represent a functional term and enrichment significance, respectively. Red nodes represent functional terms for which genes from G2-2 (A) or G5-1 (B) comprised over 60% of all genes. Blue nodes represent functional terms for which genes from G2-1 (A) or G5-2 (B) comprised over 60% of all genes. Statistical significance was calculated for each term by using the 2-sided hypergeometric test with the Benjamini-Hochberg correction. ECM, extracellular matrix.
Transcriptome network analysis identifies hub transcription factors connected with fibrosis-associated gene modules in activated lung fibroblasts.
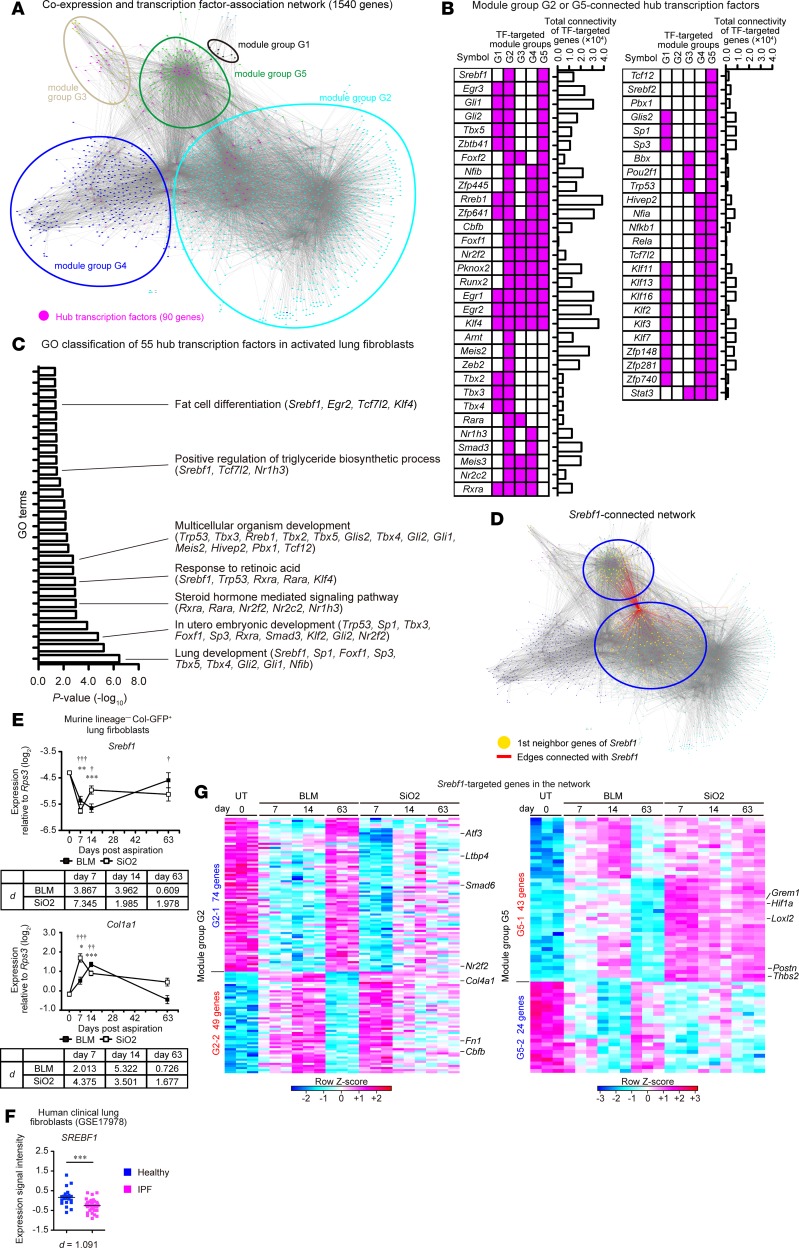
To elucidate regulatory pathways underlying PF-associated gene modules, we next sought to identify hub TFs that were located upstream of the PF-associated transcriptome network of activated lung fibroblasts. We performed TF binding motif enrichment analysis against the promoter regions of the genes (–1,000 to +100 bp from the transcriptional start sites) in each module group by using GeneXplain platform (32). GeneXplain platform identified 90 hub TFs, expressed in fibroblasts, which were highly connected with at least 1 of the module groups (Supplemental Table 2). Then, we merged the coexpression network that was defined by the WGCNA (threshold ≥ 0.25) and the connection between 90 hub TFs and their potential target genes identified by GeneXplain platform (Figure 3A). The transcriptome network contained 1,540 genes and 54,405 interactions, and network clustering revealed that genes in the same module groups closely positioned with each other (Figure 3A). Among hub TFs, we found that 55 TFs highly connected with PF-associated module groups G2 or G5 (Figure 3B). To assess the connectivity of TF-targeted genes in the network, we quantified total connectivity of each TF-targeted gene. The 55 TFs included well-known fibroblast activation regulator Egr1 and Smad3 (3, 7) (Figure 3B). The summation of the WGCNA-defined connectivity, which reflects the strength of the coexpression between each gene, of each of the 55 TF-targeted genes also revealed high and intermediate connectivity of Egr1-targeted and Smad3-targeted genes in the transcriptome network (Figure 3B). DAVID functional analysis (33) revealed that the 55 hub TFs comprised TFs downstream of sonic hedgehog signaling (Gli1 and Gli2) (34) and those essential for lung development (Tbx4, Tbx5, Foxf1, and Nfib) (35–37) (Figure 3C and Supplemental Table 3). Surprisingly, lipid-related TFs (steroid hormone–, retinoic acid–, triglyceride biosynthesis–, and fat cell differentiation–related TFs) were also highly enriched in hub TFs (Figure 3C). Notably, Srebf1, a master regulator of lipid metabolism (38), was the only hub TF that specifically connected with module groups G2 and G5 (Figure 3, B and D). In addition, Nr1h3 (Lxra) and Rxra, upstream regulators of Srebf1 (39, 40), were also identified as hub TFs of module group G2 (Figure 3B). Srebf1-, Nr1h3-, and Rxra-targeted genes possessed intermediate connectivity in the transcriptome network (Figure 3B). Quantitative PCR (qPCR) analysis revealed that Srebf1 expression was downregulated at 63 dpi in the silica model, but it returned to the steady-state level at 63 dpi in the bleomycin model (Figure 3E). The opposite pattern was observed with Col1a1, a marker of activated fibroblasts (Figure 3E). Importantly, SREBF1 expression was also downregulated in IPF lung fibroblasts compared with its expression in healthy lung fibroblasts (Figure 3F). Moreover, antifibrotic gens (Atf3 and Ltbp4) (41, 42), profibrotic genes (Grem1, Hif1a, Loxl2, Postn, and Thbs2) (43–47) and myofibroblasts differentiation–associated genes (Fn1 and Cbfb) (48, 49) were connected with Srebf1 (Figure 3G). These results indicate that Srebf1 is a centrally connected hub TF in the PF-associated transcriptome network of activated lung fibroblasts.
Figure 3. Transcriptome network analysis identifies hub transcription factors connected with fibrosis-associated gene modules in activated lung fibroblasts.
(A) Reconstructed transcriptome network of activated lung fibroblasts in pulmonary fibrosis. Each node represents genes, colors of each node represent module groups, and gray lines represent individual interactions. Hub transcription factors (TFs) are highlighted in magenta. (B) Pattern of the connection (magenta) between hub TFs and gene modules. Hub TFs that were connected with module groups G2 or G5 are shown. (C) Functional classification of 55 hub TFs using DAVID 6.8. Significantly enriched Gene Ontology (GO) terms and hub TFs included in GO terms are shown on the right of the graph. (D) Srebf1-connected network. Red line represents hub TF-target interaction. Targeted genes are highlighted in yellow. (E) qPCR analysis of Srebf1 and Col1a1 expression in activated lung fibroblasts. Data are presented as the mean ± SEM of n = 5 (UT, day 0; BLM, days 7, 63; SiO2, day 63), n = 6 (SiO2, day 7), n = 7 (BLM, day 14; SiO2, day 14). A representative result of 2 independent experiments is shown. Statistical significance is indicated as follows: *P < 0.05, **P < 0.01, ***P < 0.001 (untreated vs. BLM group); †P < 0.05, ††P < 0.01, †††P < 0.001(untreated vs. SiO2 group) according to 2-way ANOVA followed by the post hoc Tukey-Kramer’s multiple comparison test. Effect size (d) (compared with untreated group) is shown on the bottom of the graph. (F) Expression of SREBF1 in human lung fibroblasts derived from healthy (n = 19) or idiopathic pulmonary fibrosis (IPF) lungs (n = 37) (GSE17978). ***P = 0.0003 (2-tailed unpaired Student’s t test, t statistic [t] = 3.866, degree of freedom [df] = 54). Effect size (d) is shown on the bottom of the graph. (G) Heatmap representation of Srebf1-connected genes. Each column represents group and time point, whereas each row represents an individual gene. TF, transcription factor; Col-GFP, Col1a2-GFP reporter; BLM, bleomycin model; SiO2, silica model; UT, untreated.
Sterol-regulatory element binding protein 1, one of the hub transcription factors, suppresses lung fibroblast proliferation and PF pathology.
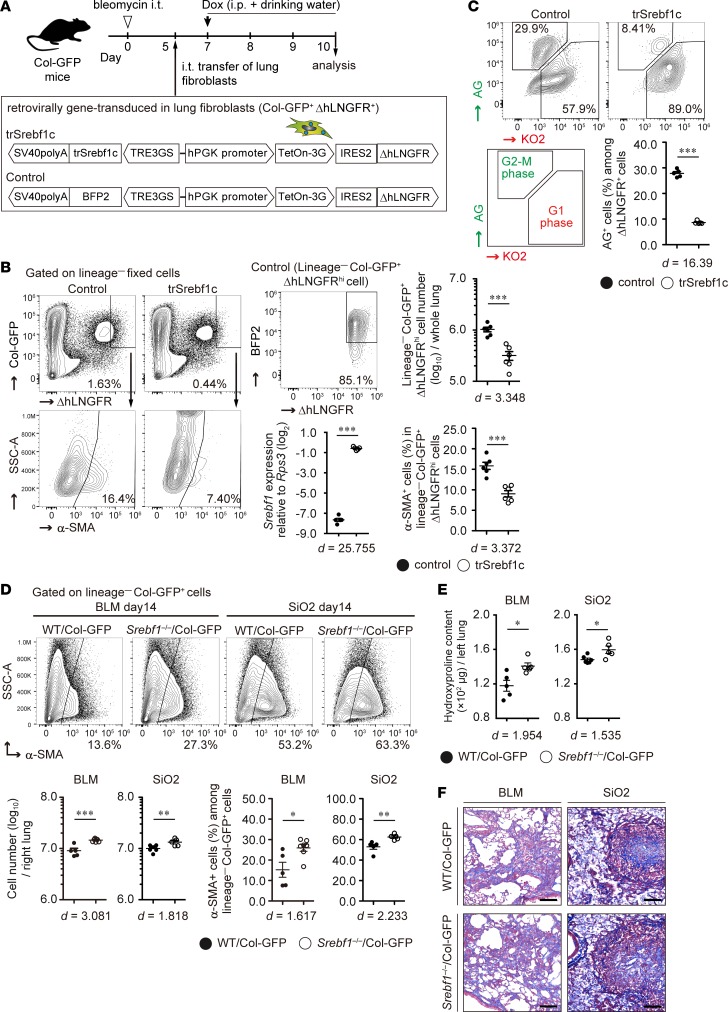
To clarify whether Srebf1 contributes to lung fibroblast activation, we retrovirally transduced Tet-One expression system, which induced the active form of Srebf1c (trSrebf1c) upon doxycycline administration, into lung fibroblasts. Transduced cells continuously expressed the truncated form of human low-affinity nerve growth factor receptor (ΔhLNGFR). Then, we intratracheally transferred gene-transduced fibroblasts into bleomycin-injured lungs (20) (Figure 4A). We previously reported that intratracheally transferred lung fibroblasts are engrafted into fibrotic lesions, accompanied by an upregulation in profibrotic genes and an increase in hydroxyproline levels in bleomycin-injured lungs (20). Thus, the intratracheal transfer method may help investigate the role of trSrebf1c specifically in lung fibroblast activation in vivo. We induced trSrebf1c expression on day 2 after transfer by doxycycline administration (i.e., at the time point when transferred fibroblasts become integrated into fibrotic lesions; ref. 20). As expected, Srebf1 expression levels were higher in trSrebf1c-expressing ΔhLNGFRhi donor fibroblasts than in control donor fibroblasts (Figure 4B). We found that ectopic expression of trSrebf1c decreased the number of ΔhLNGFRhi donor fibroblasts (Figure 4B). α-Smooth muscle actin (α-SMA) expression was also lower in trSrebf1c-expressing donor fibroblasts than in control donor fibroblasts (Figure 4B). In addition, by using lung fibroblasts isolated from fluorescence ubiquitin cell cycle indicator–transgenic (Fucci-transgenic) mice (50), we found that trSrebf1c expression arrested G1-to-S transition in lung fibroblasts in vitro (Figure 4C). These results indicate that trSrebf1c suppresses lung fibroblast proliferation and activation. To further assess the role of Srebf1 in fibroblast activation and PF pathology, we administered bleomycin or silica into Srebf1–/–Col-GFP mice (51). Consistent with the results of the adoptive transfer experiment, we found that the numbers of lung fibroblasts and α-SMA+ myofibroblasts increased in Srebf1–/–Col-GFP mice compared with those in WT Col-GFP mice at 14 dpi of both bleomycin and silica models (Figure 4D). Hydroxyproline content was also higher in the lungs of Srebf1–/–Col-GFP mice than in the lungs of WT Col-GFP mice at 14 dpi in both bleomycin and silica models (Figure 4E). Masson’s trichrome staining of fibrotic lung sections revealed that the area of collagen deposition was larger in the lungs of Srebf1–/–Col-GFP mice rather than WT Col-GFP mice at 14 dpi in the bleomycin model but not in the silica model (Figure 4F and Supplemental Figure 3). Furthermore, diffuse PF was not induced upon silica administration in Srebf1–/– mice, in contrast to that in Ccr2–/– and Myd88–/– mice (21, 52) (Figure 4F). These data suggest that Srebf1 represses fibroblast/myofibroblast proliferation and collagen deposition in both bleomycin- and silica-induced PF.
Figure 4. Effect of sterol regulatory element–binding protein 1 on lung fibroblast proliferation and pulmonary fibrosis pathology.
(A) Experimental scheme of the intratracheal transfer of genetically modified lung fibroblasts. (B) Role of active form of Srebf1c (trSrebf1c) in activated fibroblasts in bleomycin-injured lung. Donor fibroblasts were identified by flow cytometry. Graphs show the mean ± SEM (n = 6). A representative result of 2 independent experiments is shown. ***P < 0.001 (2-tailed unpaired Student’s t test). ΔhLNGFRhi gate was defined by induction rates of BFP2 ≥ 80% in all of the control donor fibroblasts. (C) Effect of trSrebf1c on lung fibroblast proliferation in vitro. Cell cycle of genetically modified lung fibroblasts was analyzed by the fluorescence ubiquitination cell cycle indicator. Graphs show the mean ± SEM (n = 5). A representative result of 2 independent experiments is shown. ***P < 0.001 (2-tailed unpaired Student’s t test). (D–F) Lung fibroblast activation and pulmonary fibrosis pathology in Srebf1–/–Col-GFP mice. (D) Number of lung fibroblasts and myofibroblasts in bleomycin- or silica-treated right lungs on day 14. Graphs show the mean ± SEM of n = 5 (BLM WT Col-GFP, SiO2 Srebf1–/–Col-GFP), n = 6 (SiO2 WT Col-GFP), n = 7 (BLM Srebf1–/–Col-GFP). A representative result of 3 independent experiments is shown. *P < 0.05, **P < 0.01, ***P < 0.001 (2-tailed unpaired Student’s t test). (E) Quantification of hydroxyproline content in the whole left lung of bleomycin- or silica-treated Srebf1–/–Col-GFP and WT Col-GFP mice. Graphs show the mean ± SEM of n = 5 (BLM WT Col-GFP, BLM Srebf1–/–Col-GFP, SiO2 Srebf1–/–Col-GFP), n = 6 (SiO2 WT Col-GFP). A representative result of 3 independent experiments is shown. *P < 0.05 (2-tailed unpaired Student’s t test). (F) Masson’s trichrome staining of bleomycin- or silica-treated left lung sections from Srebf1–/–Col-GFP and WT Col-GFP mice at day 14. Scale bars: 100 μm. Representative images of n = 8 (BLM WT Col-GFP, SiO2 WT Col-GFP, SiO2 Srebf1–/–Col-GFP), n = 9 (BLM Srebf1–/–Col-GFP) from 2 independent experiments are shown. (B, C, and D) Effect size (d) is shown on the bottom of the graph. Col-GFP, Col1a2-GFP reporter; ΔhLNGFR, truncated form of human low-affinity nerve growth factor receptor; hPGK, human phosphoglycerate kinase 1; IRES2, internal ribosomal entry site 2; BFP2, mTagBFP2; AG, azami green, KO2, kusabira orange 2; BLM, bleomycin model; SiO2, silica model.
SREBP-1c broadly regulates fibrosis-associated gene expression in activated lung fibroblasts.
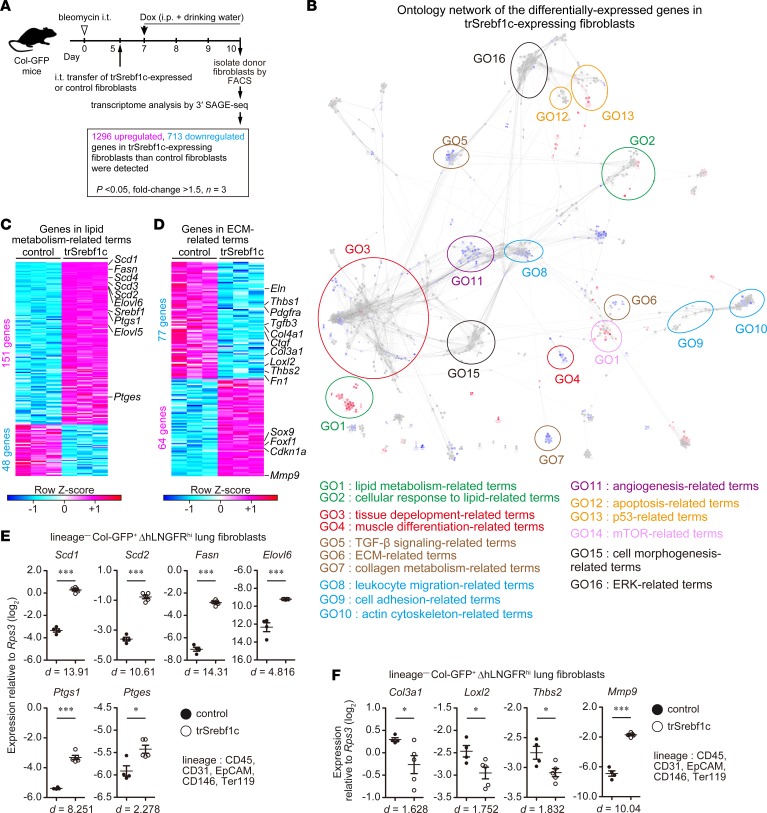
To further clarify the contribution of Srebf1 to fibroblast activation in PF, we isolated intratracheally transferred trSrebf1c-expressing or control fibroblasts from bleomycin-injured lungs and performed transcriptome analysis (Figure 5A). We identified 2,009 differentially expressed genes (1,296 upregulated and 713 downregulated) with a fold change of ≥ 1.5 and adjusted P < 0.05 in trSrebf1c-expressing fibroblasts compared with control fibroblasts; Figure 5A and Supplemental Table 4). GO network analysis revealed that lipid-related terms (lipid metabolism– and cellular response to lipid–related terms) were highly enriched in 1,296 upregulated genes (Figure 5B). Lipid-related genes included Scd1–Scd4, Fasn, Elovl5, Elovl6, Ptgs1, and Ptges (Figure 5C). Terms related to p53 were also highly enriched in the upregulated genes, consistent with our observation that trSrebf1c suppressed G1-to-S transition (Figure 5B and Figure 4C). Strikingly, fibrosis- and fibroblast activation–associated terms (tissue development–, TGF-β signaling–, ECM-, collagen metabolism–, cell adhesion–, and actin cytoskeleton–related terms) were highly enriched in the 713 downregulated genes (Figure 5B). ECM-related downregulated genes included Col3a1, Col4a1, Ctgf, Eln, Fn1, Loxl2, Tgfb3, Thbs1, and Thbs2, whereas ECM-related upregulated genes included Cdkn1a, Foxf1, and Mmp9 that inhibit fibroblast proliferation or profibrotic phenotype (53, 54) (Figure 5D). qPCR analysis also showed upregulation of the expression of lipid-related genes (Scd1, Scd2, Fasn, Elovl6, Ptgs1, and Ptges), downregulation of the expression of profibrotic genes (Col3a1, Loxl2, and Thbs2), and upregulation of antifibrotic gene Mmp9 by trSrebf1c expression (Figure 5, E and F). Consistent with the enrichment of actin cytoskeleton–related components in the genes downregulated by trSrebf1c, Srebf1–/–Col-GFP lung fibroblasts derived from the lungs of mice after 7 days of bleomycin administration displayed greater migration than corresponding WT Col-GFP lung fibroblasts in vitro (Supplemental Figure 4). In addition, after 72 hours of TGF-β1 stimulation, Srebf1–/–Col-GFP lung fibroblasts displayed a greater increase in their cell size and Col1a2 promoter activity than WT Col-GFP lung fibroblasts in vitro (Supplemental Figure 5, A and B). Consistent with the broad connection between Srebf1 and the genes in the fibrosis-associated module groups G2 and G5 in our reconstructed transcriptome network (Figure 3D), these results indicate that trSrebf1c broadly suppressed fibrosis-associated gene expression changes in activated lung fibroblasts in bleomycin-induced PF.
Figure 5. SREBP-1c broadly suppresses fibrosis-caused gene expression changes in activated lung fibroblasts.
(A) Experimental scheme of transcriptome analysis of intratracheally transferred genetically modified lung fibroblasts. In total, 2,009 genes were identified as differentially expressed genes as a result of ectopic expression of trSrebf1c. (B) Network of significantly enriched functional terms of the 2,009 genes. Biological events associated with the 2,009 genes were explored and term network was clustered using Cytoscape 3.3.0 with ClueGO and Allegrolayout plugins. Each node and its size represent functional term and enrichment significance, respectively. Red nodes represent functional terms for which genes from upregulated 1,296 genes comprised over 60% of all genes. Blue nodes represent functional terms for which genes from downregulated 713 genes comprised over 60% of all genes. Statistical significance was calculated for each term by using the 2-sided hypergeometric test with the Benjamini-Hochberg correction. (C and D) Details of lipid-related genes (C) and extracellular matrix–related (ECM-related) (D) genes identified by gene ontology network analysis. Each column represents group, whereas each row represents an individual gene. (E and F) qPCR analysis of expression changes of lipid-related (E) and ECM–related (F) genes. Graphs show the mean ± SEM of n = 4 (control) and n = 5 (trSrebf1c). A representative result of 2 independent experiments is shown. *P < 0.05, ***P < 0.001 (2-tailed unpaired Student’s t test). Effect size (d) is shown on the bottom of the graph. Col-GFP, Col1a2-GFP reporter; trSrebf1c active form of Srebf1c; ΔhLNGFR, truncated form of human low-affinity nerve growth factor receptor; SAGE, serial analysis of gene expression.
Pharmacological activation of LXR, an upstream activator of SREBP-1c expression, suppresses fibroblast activation and collagen deposition in murine PF.
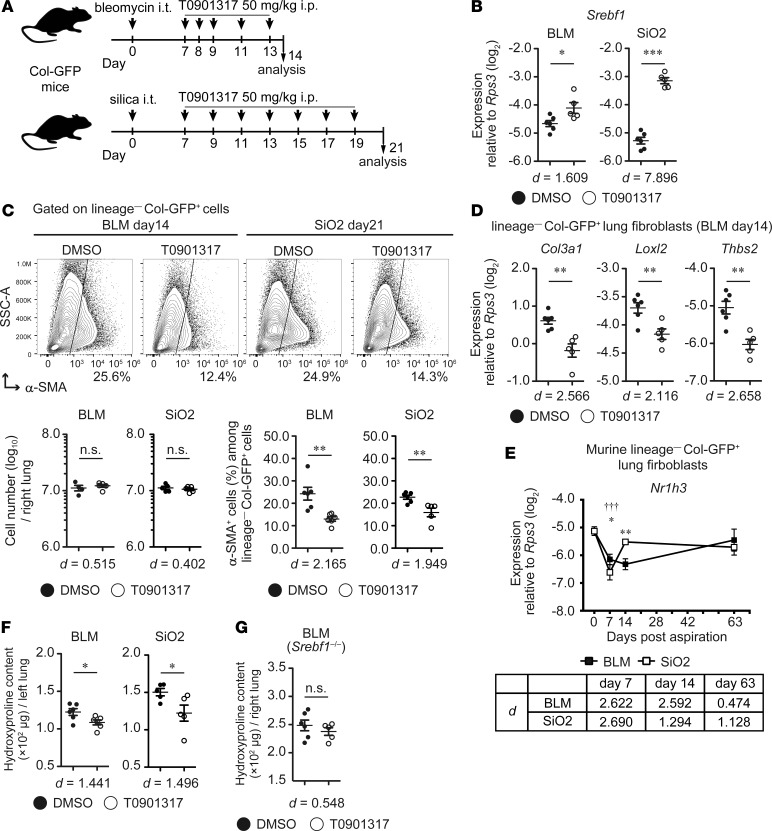
Next, we assessed whether an intervention against SREBP-1c expression could ameliorate fibroblast activation and PF pathology. It has been reported that liver X receptor/retinoid X receptor (LXR/RXR) heterodimer directly regulates SREBP-1c expression, and the LXR agonist T0901317 enhanced SREBP-1c expression in vivo (39, 40). However, the effect of T0901317 on fibroblast activation in PF development remains elusive. To solve this issue, we administered T0901317 in bleomycin- and silica-induced PF models from 7 dpi (Figure 6A), at whole-lung fibroblast activation, and changes in IPF-associated gene expression peaked (17) (Figure 1C). Dosage of T0901317 (50 mg/kg) was determined from a previous study reporting that the dosage effectively induces the expression of LXR/RXR-targeted gene Abca1 and downregulates α-SMA in the lungs of a mouse model of OVA-induced asthma (55). We found that administration of T0901317 (50 mg/kg) at 7–14 dpi in the bleomycin model increased Srebf1 expression (Figure 6B) and suppressed α-SMA expression in lung fibroblasts (Figure 6C). Similar observation was made in the silica model when 50 mg/kg T0901317 was administered at 7–21 dpi (Figure 6, B and C). T0901317 administration also decreased the expression of Col3a1, Loxl2, and Thbs2 in lung fibroblasts of bleomycin-treated lung (Figure 6D). Notably, expression levels of these genes were also suppressed by trSrebf1c in the bleomycin model (Figure 5D), and Nr1h3 (Lxra) was downregulated in lung fibroblasts, similarly to Srebf1 (Figure 6E and Figure 3E) and was maintained moderately but not significantly at 63 dpi in the silica model (effect size [d] = 1.128, Figure 6E). Hydroxyproline content was also decreased by the administration of T0901317 in both bleomycin- and silica-treated lungs (Figure 6F). Moreover, in the bleomycin model, the suppression of hydroxyproline content by T0901317 was not observed in Srebf1–/– mice (Figure 6G). These data demonstrate that pharmacological activation of LXR, an upstream inducer of SREBP-1c expression, suppresses fibroblast activation and fibrosis development in bleomycin- and silica-induced PF.
Figure 6. Pharmacological activation of LXR suppresses fibroblast activation and collagen deposition in murine pulmonary fibrosis.
(A) Experimental scheme of the treatment of bleomycin- and silica-induced pulmonary fibrosis by the LXR agonist T0901317. (B) qPCR analysis of Srebf1 expression in lung fibroblasts. (C) Changes in the number of lung fibroblasts and myofibroblasts in bleomycin- or silica-treated right lungs following T0901317 administration. (D) qPCR analysis of expression changes of trSrebf1c-targeted genes (detected by intratracheal transfer model of bleomycin-treated lungs; see Figure 5D) in the fibroblasts of bleomycin-treated lungs. (E) qPCR analysis of Nr1h3 expression in activated lung fibroblasts. (F) Changes in hydroxyproline content in the whole left lung of bleomycin- or silica-treated Col-GFP mice following T0901317 administration. (G) Changes in hydroxyproline content in the whole right lung of bleomycin-treated Srebf1–/–Col-GFP mice following T0901317 administration. (B and D) Graphs show the mean ± SEM (n = 6, DMSO; n = 5, T0901317). (C) Graphs show the mean ± SEM (n = 4, BLM day14 DMSO fibroblast number; n = 5, BLM day14 T0901317 and SiO2 day21 fibroblast number; n = 6, BLM day14 and SiO2 day21 DMSO, % of α-SMA; n = 5, SiO2 day21 T0901317, % of α-SMA). (E) Graphs show the mean ± SEM of n = 5 (UT, day 0; BLM, days 7, 63; SiO2, day 63), n = 6 (SiO2, day 7), n = 7 (BLM, day 14; SiO2, day 14). (F) Graphs show the mean ± SEM (n = 6, BLM, day14; n = 5, SiO2, day21). (G) Graphs show the mean ± SEM (n = 6, DMSO; n = 5, T0901317). Representative results of 3 (B–D and F) or 2 (E and G) independent experiments is shown. (B–D, F, and G) Statistical significance is indicated as follows: *P < 0.05; **P < 0.01, ***P < 0.001 (2-tailed unpaired Student’s t-test). Effect size (d) is shown on the bottom of the graph. (E) Statistical significance is indicated as follows: *P < 0.05, **P < 0.01 (untreated vs. bleomycin group), and †††P < 0.001(untreated vs. silica group) from 2-way ANOVA followed by the post hoc Tukey-Kramer’s multiple comparison test. Effect size (d) (compared with untreated group) is indicated at the bottom of the graph. Col-GFP, Col1a2-GFP reporter; BLM, bleomycin model, SiO2, silica model; SSC, side scatter.
Discussion
Clarifying PF-common molecular signatures and their regulators in activated lung fibroblasts is a major step for understanding PF pathogenesis. Here, we identified gene signatures in activated lung fibroblasts associated with PF. GO network and TF motif enrichment analysis identified biological events and hub TFs possibly involved in lung fibroblast activation. Particularly, lipid-related biological events/TFs centrally interconnected PF-associated gene module groups. Consistent with the network analysis results, we revealed protective roles of the LXR/SREBP-1c axis in lung fibroblast activation and PF development. Collectively, these results suggested LXR/SREBP-1c axis as one of the key molecular pathways in lung fibroblast activation and PF development.
Our time-course transcriptome analysis identified module groups of genes whose expression levels were associated with the initiation or progression of PF. PF progression–associated gene modules contained many macrophage activation–associated genes. Because lung macrophages regulate PF pathology and the activation state of lung tissue cells (21, 29, 56, 57), interactions between activated fibroblasts and lung macrophages might be important in the regulation of lung fibroblast activation. PF progression–associated gene modules also contained glycan-related genes and the gene encoding the hyaluronan receptor CD44. It has been reported that the glycosaminoglycan hyaluronan and CD44 exacerbate fibroblast activation and bleomycin-induced PF (58), whereas the proteoglycan decorin suppresses TGF-β1–mediated PF (59). Because glycosaminoglycan and proteoglycan are components of IPF ECM (60), the glycan composition of the ECM possibly affects lung fibroblast activation.
We identified 90 hub TFs in the transcriptome network of activated lung fibroblasts in PF. These hub TFs showed several distinct patterns of connection with WGCNA-identified gene module groups, possibly reflecting the coverage of the aspects of lung fibroblast activation by each hub TF. Among the 90 hub TFs, 55 hub TFs connected with both fibrosis-associated module groups G2 and G5. This set contained TFs that are essential for the development of murine lung but which have not been ascribed a role in PF (Tbx4, Tbx5, Foxf1, and Nfib). Tbx4 and Tbx5 are lung fibroblast–specific TFs (61) that play essential roles in lung development (35). Foxf1 enhances Tbx5 expression and is critical for the formation of pulmonary vasculogenesis and alveologenesis (36). FOXF1 also inhibits human lung fibroblast proliferation and COL1 expression in vitro (54). Nfib is essential for mouse lung maturation after embryonic day 15.5 (37). Considering the overenrichment of tissue development–related GO terms in module groups G2 and G5, our results possibly point out the similarity of lung fibroblast activation states during development and during PF progression.
Our transcriptome network/in vivo analysis suggests that the LXR/SREBP-1c axis acts as a protective hub of lung fibroblast activation and PF pathology. Since both Lxra and Srebf1 were downregulated simultaneously with lung fibroblast activation in bleomycin- and silica-induced PF, repression of the LXR/SREBP-1c axis might reflect PF-associated activation of lung fibroblasts. Similarly to hepatic stellate cells (HSCs, Ito cells) in the liver, major progenitors of activated fibroblasts in liver fibrosis (62, 63), some lung fibroblasts contain lipid droplets and are referred to as lipofibroblasts (64). Activation of the LXR/SREBP-1c axis reversed HSC activation (65), and PPAR-γ stimulation reversed the activation of fibroblasts in bleomycin-induced PF (8, 10). Our results and those reports suggest the importance of lipid-related TFs in the regulation of lung fibroblast activation and PF development. Indeed, administration of retinoic acid, a ligand of LXR/RXR heterodimer (66), ameliorated bleomycin-induced PF (67), whereas retinoid deficiency was associated with poor prognosis of cystic fibrosis (68). Moreover, pirfenidone, an approved drug for IPF treatment, ameliorated LXR downregulation in myocardial infarction–induced heart fibrosis (69, 70). Because several LXR agonists are now in clinical trials (71) and T0901317 also exerted a preventive effect on bleomycin-induced lung fibrosis (72), the development of a safe LXR/SREBP-1c axis–targeting drug might become a novel prevention and treatment strategy against PF development. Because LXR is expressed not only in lung fibroblasts, but also in other cell subsets, including lung macrophages, epithelial cells, B cells, and endothelial cells (73), the effects of LXR agonist on nonfibroblasts warrant further investigation. Administration of another LXR agonist GW3965 reportedly exacerbates bleomycin-induced PF (74). Kurowska-Stolarska M. et al. (74) reported that GW3965 facilitates interactions between LXR and coactivators/corepressors, whereas T0901317 mediates interactions exclusively between LXR and coactivators (75). Therefore, the latter LXR agonists are favorable candidates to regulate lung fibroblast activation and for PF treatment.
An intratracheally transferred model revealed that overexpression of trSrebf1c not only suppressed profibrotic gene expression changes, but also enhanced the expression of lipid metabolism–related genes, including Scd1–Scd4, Elovl5, Elovl6, Ptgs1, and Ptges. Scd1–Scd4, Elovl5, and Elovl6 promote the production of polyunsaturated fatty acids (76–78), and Ptgs1 and Ptges catalyze the synthesis of Prostaglandin E2 (PGE2) (79). Therefore, synthesis of polyunsaturated fatty acids and PGE2 might be increased by the overexpression of trSrebf1c in lung fibroblasts. It has been reported that PGE2, docosahexaenoic acid (a polyunsaturated fatty acid), and its metabolite resolvin D1 play protective roles in bleomycin-induced lung fibrosis (80–83). In addition, it has also been reported that bleomycin-induced PF is worsened in Elovl6-KO or Ptges-KO mice (84, 85). Considering these reports and our results, SREBP-1c exert protective role in PF, possibly by mediating the synthesis of above antifibrotic lipid mediators.
Together, our results highlight the existence of characteristic PF transcriptional network in activated lung fibroblasts and the importance of SREBP-1c in the regulation of fibroblast activity. Further investigation of the molecular mechanisms that maintain the LXR/SREBP-1c axis in the suppressed state in activated lung fibroblasts, as well as the elucidation of the role of lipid metabolism in lung fibroblast activation, may lead to the discovery of novel therapeutic strategies against PF.
Methods
Mice.
C57BL/6J (B6) female mice were purchased from Japan SLC. Col-GFP female mice (B6 background for > 10 generations) were generated in a previous study (19). Srebf1–/– mice (allele B, B6 background for > 10 generations) were generated in a previous study (51). Srebf1–/–Col-GFP double-transgenic female mice were generated by crossing Col-GFP mice and Srebf1–/– mice. FucciG1-#639/FucciS/G2/M-#474 double-transgenic mice were provided by A. Miyawaki and M. Tomura (Laboratory for Cell Function and Dynamics, Brain Science Institute, RIKEN, Saitama, Japan) (50). All mice were bred at specific pathogen-free facilities at the University of Tokyo and were 7–10 weeks old at the commencement of experiments.
Administration of silica or bleomycin into mouse lungs.
Mice were administered with a single dose of crystalline silica particles (400 mg/kg suspended in 50 μl of sterile saline solution; MIN-U-SIL5; mass-median-diameter [d50] Z: 1.4 μm; US Silica) or bleomycin sulfate (2 mg/kg dissolved in 50 μl of sterile saline solution; Toronto Research Chemical) by oropharyngeal aspiration under deep anesthesia, as described previously (20, 21). Silica particles were pretreated as described previously (21).
Lung cell preparation, flow cytometry, and cell sorting.
Lung cells were prepared as described previously (21). Single cell suspensions were blocked with Fc block (anti-CD16/32, clone 2.4G2, BioXcell) and stained with appropriate antibody mixtures diluted with PBS supplemented with 2% FBS (Moregate Biotech). For α-SMA staining, stained cells were fixed and permeabilized by using a Fixation/Permeabilization Solution Kit (BD Biosciences) and stained with an allophycocyanin (APC) anti–α-SMA antibody (clone 1A4, R&D Systems). Data were collected on a Gallios flow cytometer (Beckman Coulter) and analyzed using FlowJo software v10.1. For cell sorting of lung fibroblasts, lung single cell suspensions were blocked with Fc block, stained for lineage (CD45, CD31, EpCAM, CD146, and Ter119) markers in DMEM (MilliporeSigma) supplemented with 5% FBS and 10 mM HEPES (GE Healthcare), and magnetically labeled with anti-biotin Maxibeads (Miltenyi Biotec). For cell sorting of intratracheally transferred lung fibroblasts, lung single cell suspensions were blocked with Fc block, stained for lineage (CD45, CD31, EpCAM, CD146, and Ter119) markers and R-phycoerythrin–Cy7 (PE-Cy7) anti-ΔhLNGFR in DMEM supplemented with 5% FBS and 10 mM HEPES, and magnetically labeled with anti-biotin Maxibeads. Labeled cells were magnetically depleted using a DynaMag-2 separator (Thermo Fisher Scientific). Finally, lineage–propidium iodide–GFP+ live fibroblasts or lineage–propidium iodide–GFP+ΔhLNGFR+ donor fibroblasts were sorted with a FACSAria cytometer (BD Biosciences) equipped with a 70-μm tip. The fibroblasts were sorted in 500 μl of DMEM supplemented with 50% FBS and 20 mM HEPES for the verification of the purity of isolated fibroblasts and directly sorted in 500 μl of cell lysis buffer (1% LiDS [MilliporeSigma],100 mM Tris-HCl pH 7.5 [NipponGene], 500 mM LiCl [MilliporeSigma], 10 mM EDTA [NipponGene], and 5 mM DTT [MilliporeSigma]) and stored at −80°C for further qPCR and transcriptome analyses. Purity of isolated fibroblasts among live cells routinely comprised >98% for all experiments.
Antibodies.
Fluorochrome-, biotin-conjugated, or purified antibodies and fluorochrome-conjugated streptavidin were purchased from BD Biosciences, BioLegend, eBioscience, R&D Systems, and Tonbo Biosciences (see Supplemental Table 5 for a detailed list of antibodies and streptavidin used for flow cytometry).
Amplification of the whole fibroblast transcript.
The whole transcript of lung fibroblasts and intratracheally transferred fibroblasts was amplified according to the previous report with some modifications (86). Details of the modified procedure have been deposited in the library construction protocol section in NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo; GSE110533). Primer sequences are shown in Supplemental Table 6. All oligos were HPLC purified and purchased from Integrated DNA Technologies or Sigma-Genosys.
3′ SAGE-seq library generation and sequencing.
Generation of 3′ SAGE-seq library was performed as described in a previous report, with some modifications (18). Details of the modified procedure have been deposited in the library construction protocol section in NCBI GEO (accession no. GSE110533). Sequencing was performed by using an Ion Hi-Q Chef kit, an Ion PI v3 Chip kit, and an Ion Proton Sequencer (Thermo Fisher Scientific) according to the manufacturer’s instructions; however, the input library concentration was 100 pM. Raw data from these experiments have been deposited in the NCBI GEO (accession nos. GSE110533 and GSE110540). Sequences of the primers and adapters are shown in Supplemental Table 6.
Transcriptome data analysis.
Adapter trimming and quality filtering of sequencing data were performed by using Trimommatic-v0.36 (87) and PRINSEQ-0.20.4 (88). Filtered reads were mapped to Refseq mouse mRNA sequences by using Bowtie2-2.2.5 (89) with the following parameters: -t -p 11 -N 1 -D 200 -R 20 -L 20 -i S,1,0.50 --norc. Reads that were not mapped to NlaIII sites were removed, and tag numbers of each gene were quantified as the expression level of each gene. Between-sample normalization was performed against raw count data by using Microsoft R open 3.3.3. (https://mran.microsoft.com/open/) and TCC package (EEE-E method) (90, 91). Analysis of differential gene expression in experimental groups was performed by using Microsoft R open 3.3.3 and glmLRT formula with the Benjamini-Hochberg correction of edgeR package (92), which was integrated in TCC package. In time-course transcriptome data of activated lung fibroblasts, genes for which adjusted P < 0.05, fold change ≥ 2, and maximum expression ≥ 50 were identified as statistically significant differentially expressed genes. In the transcriptome data of intratracheally transferred lung fibroblasts, genes for which adjusted P < 0.05, fold change ≥ 1.5, and maximum expression ≥ 50 were identified as statistically significant differentially expressed genes.
Detection of coexpressed gene modules.
Coexpressed gene modules among differentially expressed genes of activated lung fibroblasts were detected by using WGCNA package (11) in Microsoft R open 3.3.3. Variance-stabilizing transformation of TCC-normalized count data was performed by using DESeq2 package (93) in Microsoft R open 3.3.3, and the transformed data were used as input of WGCNA package. For WGCNA, the power value was 13, the merge_thre value was 0.25, the threshold value for the output of coexpression interactions was 0.25, and other calculation settings were set to defaults. The genes in the gene module groups detected by WGCNA were further clustered into positively and negatively correlated gene groups by the CLICK method (26) and visualized by using Microsoft R open 3.3.3.
GSEA.
Public transcriptome data for human clinical lung samples were downloaded from the GEO or ArrayExpress (http://www.ebi.ac.uk/arrayexpress/experiments). Public microarray data used in this study were as follows: lung fibroblasts from IPF and healthy lungs (GEO accession no. GSE17978; ref. 25) and IPF and healthy whole lungs (array no. E-GEOD-47460; ref. 17). The values of duplicated genes were averaged. GSEA was performed by using each clustered gene list and human lung expression data as gene sets and expression datasets, respectively. For the comparison of IPF versus healthy lung fibroblasts or IPF versus healthy whole lungs, the permutation type was gene set, and other calculation settings were set to defaults.
Functional annotation of gene module groups and GO network construction.
Functional analysis of gene module groups and differential expression of genes caused by trSrebf1c expression was performed by using Cytoscape 3.3.0 with ClueGO plugin (v2.4.3) (28, 94). Significantly enriched GO terms (27) (GO-biological process, GO levels 3–8) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway terms (95) in gene module groups G2-1 and G2-2, G5-1 and G5-2, or the differentially expressed 2,009 genes were explored and grouped, and a term network was constructed based on the overlap of their elements (κ score = 0.3). Leading terms within each group were defined as the most significantly enriched term in each group. Terms not connected with any other term were excluded. Clustering of the GO network was performed by using Cytoscape 3.3.0 with AllegroLayout plugin with Allegro Edge-Repulsive strong clustering algorithm. We used versions of the GO term database and KEGG pathway term database that were current on September 21, 2017.
Identification of hub TFs in the transcriptome network.
Enrichment analysis of TF binding motifs in the promoters of the genes from the gene module groups G1–G5 was performed by using GeneXplain platform (GeneXplain GmbH). Promoter regions were defined as the regions of –1,000 to +100 bp from the transcriptional start sites of each gene. A profile vertebrate_mouse_p0.001 was used as TF motif profile for the enrichment analysis, and other calculation settings were set to defaults. Significantly enriched motifs were annotated by corresponding TF genes, and the detected TFs for which the maximum expression value in the time-course 3′ SAGE-seq data was under 50 were filtered out. TFs remaining after such filtration were defined as hub TFs of corresponding gene module groups. Functional classification of the identified hub TFs was performed by using DAVID 6.8 (GOTERM_BP_DIRECT) (33).
Construction of the transcriptome network of activated lung fibroblasts in murine PF.
Interactions between the identified hub TFs and their potential targets were extracted by GeneXplain platform and combined with the interactions with the WGCNA-defined coexpression network (threshold = 0.25). The final combined network, which contained 1,540 nodes and 54,405 interactions, was visualized by using Cytoscape 3.3.0 with the AllegroLayout plugin with the Allegro Edge-Repulsive strong clustering algorithm. Total connectivity of the targeted genes of each TF was calculated by the summation of Ktotal values defined by WGCNA.
qPCR analysis.
qPCR analysis was performed by using a Thunderbird SYBR qPCR Mix (Toyobo) on an ABI 7500 real-time PCR system (Thermo Fisher Scientific) or a QuantStudio6 real-time PCR system (Thermo Fisher Scientific). Sequences of the primers used for qPCR are shown in Supplemental Table 7. The expression levels of all target mRNAs were normalized against the expression level of Rps3 in each sample.
Construction of the Tet-One retroviral expression vector.
Tet-One inducible expression system (Takara Bio) was cloned into the pMYs retroviral vector (gifted by Toshio Kitamura, Division of Cellular Therapy/Division of Stem Cell Signaling, The Institute of Medical Sciences, The University of Tokyo). The internal ribosomal entry site 2–coding (IRES2-coding) sequence was PCR amplified from pMYs retroviral vector. IRES2 and ΔhLNGFR reporter gene were cloned into a sequence downstream of the Tet-On 3G sequence of the Tet-One construct. The mTagBFP2-coding sequence was PCR amplified from pBAD-mTagBFP2 plasmid (Addgene) (96). trSrebf1c (XM_006532716.2, 113–1462 nucleotides, just before the S2P cleavage recognition site) was PCR amplified from the reverse-transcribed mRNA of lung fibroblast of Col-GFP mice. Then, the TagBFP2 or trSrebf1c sequence was cloned into the multicloning site (EcoRI restriction site) of the Tet-One construct. Detailed information about the sequence and annotation is shown in Supplemental File 1 (control vector) and Supplemental File 2 (trSrebf1c vector). Cloning reaction was performed by using an In-Fusion Cloning Kit (Takara Bio). Plasmid nucleotide sequences were verified by Sanger sequencing by using BigDye 3.1 terminator and an ABI3730 sequencer (Applied Biosystems) according to the manufacturer’s instructions. The pCMV-VSV-G envelope vector was provided by Kenzo Hirose (Department of Neurobiology, Graduate School of Medicine, The University of Tokyo). The plasmids for the production of each retroviral vector were purified by using Nucleobond Xtra Midi EF columns (Takara Bio). Retrovirus vectors were produced with GP2-293T cells, as described previously (97). Viral supernatant was passed through 0.2-μm filters and concentrated using an Amicon Ultra-15 filter unit (100 kDa MWCO, MilliporeSigma) to 50× final concentration. The virus solution was flash-frozen with liquid nitrogen and stored at −80°C.
Retroviral gene transduction and intratracheal transfer of fibroblasts.
Intratracheal transfer was performed as described previously (20). Lung single cell suspensions were blocked with Fc block, stained for lineage (CD45, CD31, EpCAM, CD146, and Ter119) markers, and magnetically labeled with anti-APC microbeads (Miltenyi Biotec). Labeled cells were magnetically depleted using an AutoMACS separator (Miltenyi Biotec). Isolated cells (3 × 106) were cultured in a 10-cm culture dish in DMEM supplemented with 10% FBS and 10 mM HEPES for 48 hours. Nonadherent cells were removed from the culture, and retroviral transduction (final concentration of the viral supernatant was 5×) was performed for 24 hours. Fibroblasts were passaged to a 15-cm culture dish and cultured in DMEM supplemented with 10% FBS and 10 mM HEPES for 8 hours, and another retroviral transduction was performed for 24 hours. Infected fibroblasts were stained with an APC anti-ΔhLNGFR antibody, and gene-transduced fibroblasts were magnetically isolated by using anti-APC microbeads (Miltenyi Biotec) and LS Columns (Miltenyi Biotec). Resulting fibroblasts were cultured for another 7 days, with passaging 3 times within a 2-day interval. The cell suspensions (3 × 106 cells in 50 μl sterile saline solution) were intratracheally transferred to Col-GFP mice at 5 dpi of bleomycin injury. Doxycycline hyclate (2 mg/head; LKT Laboratories) was intraperitoneally injected at day 2 after intratracheal transfer, and doxycycline was continuously administered via drinking water (2 mg/ml in sterile water supplemented with 1% sucrose) to mice for the duration of the experiment. Doxycycline-containing water was replaced every 2 days and protected from light.
Measurement of hydroxyproline content.
Lung hydroxyproline content was measured as described previously (21).
Masson’s trichrome staining.
Masson’s trichrome staining of lung sections was performed as described previously (21). Images were acquired and collagen-deposited area was quantified by BZ-X710 or BZ-X810 all-in-one microscope (Keyence).
Fibroblast migration assay.
Lung fibroblasts were isolated from Srebf1–/–Col-GFP and WT Col-GFP mice 7 days after bleomycin administration. Their migratability was assessed using an Oris cell migration assay kit (fibronectin-coated; Platypus Technologies), as described previously (20). The wells were imaged using an SP-5 confocal microscope (Leica Microsystems) at 0, 24, and 72 hours after removing the stopper. The cell-free area at the center of each well was quantified using Photoshop CC (Adobe Systems Incorporated). The relative area size against that at 0 hours in each well was calculated as the extent of lung fibroblast migration.
TGF-β1 stimulation assay.
Purified lung fibroblasts were isolated from untreated Srebf1–/–Col-GFP and WT Col-GFP murine lungs. The fibroblasts were cultured for 72 hours with DMEM supplemented with 10% FBS, 10 mM HEPES, and 10 U/ml penicillin-streptomycin (MilliporeSigma); passaged; and cultured 72 hours with DMEM supplemented with 5% FBS, 10 mM HEPES, and 10 U/ml penicillin-streptomycin. The fibroblasts were passaged and cultured 24 hours with DMEM supplemented with 1% FBS, 10 mM HEPES, and 10 U/ml penicillin-streptomycin, and they were then stimulated with or without recombinant human TGF-β1 (10 ng/ml, Peprotech) for 72 hours. The fibroblasts were collected and analyzed by a Gallios flow cytometer. Cell size and Col1a2 promoter activity of the fibroblasts were assessed by their mean fluorescence intensity of forward scatter (FSC) and Col-GFP.
Administration of LXR agonist.
The LXR agonist T0901317 (Cayman Chemical) was dissolved in sterile DMSO (Nacalai Tesque) to 50 mg/ml and stored at −20°C, protected from light. To prevent toxicity resulting from long-term administration of T0901317 in the bleomycin model, mice were i.p. administered with T0901317 (50 mg/kg in 50 μl DMSO) or vehicle (50 μl DMSO) at 7, 8, 9, 11, or 13 dpi of bleomycin injury and analyzed at 14 dpi. To prevent toxicity resulting from long-term administration of the T0901317 in silica model, mice were i.p. administered T0901317 (50 mg/kg in 50 μl DMSO) or vehicle (50 μl DMSO) every 2 days from 7–19 dpi of silica injury and analyzed at 21 dpi.
Statistics.
The significance of differential expression of genes in TCC-normalized 3′ SAGE-seq data was calculated by using TCC package (glmLRT formula in edgeR package) (90, 92) in Microsoft R open 3.3.3. The significance parameters of overenrichment of gene module groups in IPF and healthy lung fibroblasts or in IPF or healthy lung were calculated using GSEA 3.0 software. The significance of GO term enrichment was calculated using Cytoscape 3.3.0 with ClueGO plugin (v2.3.4) or with DAVID 6.8. Correction for multiple comparisons was performed using the Benjamini-Hochberg method. Other statistical analyses were performed using 1-way ANOVA with the Tukey-Kramer’s multiple comparison post hoc test or (for more than 3 groups) or 2-tailed unpaired Student’s t test (for pairwise comparisons) in GraphPad Prism software v5.0a (Graphpad Software). Effect size (Cohen’s d) (98) was calculated by using mes function of compute.es package (http://cran.r-project.org/web/packages/compute.es) in Microsoft R open 3.3.3. Statistics are shown in Supplemental Table 8. All statistical analyses were conducted with a significance level of α = 0.05 (P < 0.05).
Study approval.
All animal experiments were reviewed and approved by the Animal Care and Use Committee of the University of Tokyo (approval no. Medical P12-35) and the Animal Experiment Committee of Tokyo University of Science (approval no. S17034 and S18029).
Author contributions
SS, SU, JA, and KM designed the study. SS performed all the experiments with the support from SH, SD, and MO for 3′ SAGE-seq experiment and transcriptome data analysis. YI provided Col-GFP mice. HS provided Srebf1–/– mice. SS, SU, JA, SH, TT, MKK, FHWS, YI, HS, and KM analyzed data and wrote the manuscript. KM supervised the study. SS and TN performed retroviral gene transduction analysis. SS, MO, and FHWS designed the logical flow of the manuscript.
Supplementary Material
Acknowledgments
We thank the IMSUT FACS Core Laboratory for the assistance with cell sorting, Graduate Program for Leaders in Life Innovation for the assistance with next-generation sequencing, and A. Yamashita, S. Fujita, and S. Aoki for their technical assistance. SS was supported by the Tadamitsu Kishimoto Fellowship Program. This work was supported by the Japan Agency for Medical Research and Development CREST program (KM) and the Japan Society for the Promotion of Science Grant-in-Aid for Scientific Research on Innovative Areas program (Inflammation Cellular Sociology, 17H06392, KM), Grants-in-Aid for Scientific Research (B) (16H05203, KM), and Grant-in-Aid for Challenging Research (Exploratory) (17K19546, KM).
Version 1. 01/10/2019
Electronic publication
Footnotes
Conflict of interest: The authors declare no conflicts of financial interest.
License: Copyright 2019, American Society for Clinical Investigation.
Reference information: JCI Insight. 2019;4(1):e122163. https://doi.org/10.1172/jci.insight.122163.
Contributor Information
Shigeyuki Shichino, Email: shichino@m.u-tokyo.ac.jp.
Satoshi Ueha, Email: ueha@m.u-tokyo.ac.jp.
Shinichi Hashimoto, Email: hashimot@m.u-tokyo.ac.jp.
Mikiya Otsuji, Email: otsuji16@gmail.com.
Jun Abe, Email: j.abe1140@gmail.com.
Tatsuya Tsukui, Email: tsukutatsu@gmail.com.
Shungo Deshimaru, Email: deshimaru@rs.tus.ac.jp.
Takuya Nakajima, Email: t.nakajima0404@gmail.com.
Mizuha Kosugi-Kanaya, Email: eaudefeuille@gmail.com.
Yutaka Inagaki, Email: yutakai@is.icc.u-tokai.ac.jp.
Hitoshi Shimano, Email: shimano-tky@umin.ac.jp.
Kouji Matsushima, Email: koujim@m.u-tokyo.ac.jp.
References
- 1.Raghu G, et al. An Official ATS/ERS/JRS/ALAT Clinical Practice Guideline: Treatment of Idiopathic Pulmonary Fibrosis. An Update of the 2011 Clinical Practice Guideline. Am J Respir Crit Care Med. 2015;192(2):e3–19. doi: 10.1164/rccm.201506-1063ST. [DOI] [PubMed] [Google Scholar]
- 2.King TE. Clinical advances in the diagnosis and therapy of the interstitial lung diseases. Am J Respir Crit Care Med. 2005;172(3):268–279. doi: 10.1164/rccm.200503-483OE. [DOI] [PubMed] [Google Scholar]
- 3.Wynn TA. Integrating mechanisms of pulmonary fibrosis. J Exp Med. 2011;208(7):1339–1350. doi: 10.1084/jem.20110551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang K, Rekhter MD, Gordon D, Phan SH. Myofibroblasts and their role in lung collagen gene expression during pulmonary fibrosis. A combined immunohistochemical and in situ hybridization study. Am J Pathol. 1994;145(1):114–125. [PMC free article] [PubMed] [Google Scholar]
- 5.Klingberg F, Hinz B, White ES. The myofibroblast matrix: implications for tissue repair and fibrosis. J Pathol. 2013;229(2):298–309. doi: 10.1002/path.4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Araya J, Nishimura SL. Fibrogenic reactions in lung disease. Annu Rev Pathol. 2010;5:77–98. doi: 10.1146/annurev.pathol.4.110807.092217. [DOI] [PubMed] [Google Scholar]
- 7.Bhattacharyya S, Fang F, Tourtellotte W, Varga J. Egr-1: new conductor for the tissue repair orchestra directs harmony (regeneration) or cacophony (fibrosis) J Pathol. 2013;229(2):286–297. doi: 10.1002/path.4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Milam JE, et al. PPAR-gamma agonists inhibit profibrotic phenotypes in human lung fibroblasts and bleomycin-induced pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol. 2008;294(5):L891–L901. doi: 10.1152/ajplung.00333.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burgess HA, et al. PPARgamma agonists inhibit TGF-beta induced pulmonary myofibroblast differentiation and collagen production: implications for therapy of lung fibrosis. Am J Physiol Lung Cell Mol Physiol. 2005;288(6):L1146–L1153. doi: 10.1152/ajplung.00383.2004. [DOI] [PubMed] [Google Scholar]
- 10.El Agha E, et al. Two-Way Conversion between Lipogenic and Myogenic Fibroblastic Phenotypes Marks the Progression and Resolution of Lung Fibrosis. Cell Stem Cell. 2017;20(2):261–273.e3. doi: 10.1016/j.stem.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doering TA, Crawford A, Angelosanto JM, Paley MA, Ziegler CG, Wherry EJ. Network analysis reveals centrally connected genes and pathways involved in CD8+ T cell exhaustion versus memory. Immunity. 2012;37(6):1130–1144. doi: 10.1016/j.immuni.2012.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xue J, et al. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity. 2014;40(2):274–288. doi: 10.1016/j.immuni.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mariani J, et al. FOXG1-Dependent Dysregulation of GABA/Glutamate Neuron Differentiation in Autism Spectrum Disorders. Cell. 2015;162(2):375–390. doi: 10.1016/j.cell.2015.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bailey P, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531(7592):47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 16.Bar-Joseph Z, Gitter A, Simon I. Studying and modelling dynamic biological processes using time-series gene expression data. Nat Rev Genet. 2012;13(8):552–564. doi: 10.1038/nrg3244. [DOI] [PubMed] [Google Scholar]
- 17.Bauer Y, et al. A novel genomic signature with translational significance for human idiopathic pulmonary fibrosis. Am J Respir Cell Mol Biol. 2015;52(2):217–231. doi: 10.1165/rcmb.2013-0310OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsumura H, Urasaki N, Yoshida K, Krüger DH, Kahl G, Terauchi R. SuperSAGE: powerful serial analysis of gene expression. Methods Mol Biol. 2012;883:1–17. doi: 10.1007/978-1-61779-839-9_1. [DOI] [PubMed] [Google Scholar]
- 19.Higashiyama R, et al. Negligible contribution of bone marrow-derived cells to collagen production during hepatic fibrogenesis in mice. Gastroenterology. 2009;137(4):1459–66.e1. doi: 10.1053/j.gastro.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 20.Tsukui T, Ueha S, Shichino S, Inagaki Y, Matsushima K. Intratracheal cell transfer demonstrates the profibrotic potential of resident fibroblasts in pulmonary fibrosis. Am J Pathol. 2015;185(11):2939–2948. doi: 10.1016/j.ajpath.2015.07.022. [DOI] [PubMed] [Google Scholar]
- 21.Shichino S, et al. Reduced supply of monocyte-derived macrophages leads to a transition from nodular to diffuse lesions and tissue cell activation in silica-induced pulmonary fibrosis in mice. Am J Pathol. 2015;185(11):2923–2938. doi: 10.1016/j.ajpath.2015.07.013. [DOI] [PubMed] [Google Scholar]
- 22.Redente EF, et al. Tumor necrosis factor-α accelerates the resolution of established pulmonary fibrosis in mice by targeting profibrotic lung macrophages. Am J Respir Cell Mol Biol. 2014;50(4):825–837. doi: 10.1165/rcmb.2013-0386OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van der Maaten LH, Hinton G. Visualizing Data using t-SNE. Journal of Machine Learning Research. 2008;9:2579–2605. [Google Scholar]
- 24.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Emblom-Callahan MC, et al. Genomic phenotype of non-cultured pulmonary fibroblasts in idiopathic pulmonary fibrosis. Genomics. 2010;96(3):134–145. doi: 10.1016/j.ygeno.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 26.Sharan R, Maron-Katz A, Shamir R. CLICK and EXPANDER: a system for clustering and visualizing gene expression data. Bioinformatics. 2003;19(14):1787–1799. doi: 10.1093/bioinformatics/btg232. [DOI] [PubMed] [Google Scholar]
- 27.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bindea G, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25(8):1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang J, et al. A macrophage subpopulation recruited by CC chemokine ligand-2 clears apoptotic cells in noninfectious lung injury. Am J Physiol Lung Cell Mol Physiol. 2012;302(9):L933–L940. doi: 10.1152/ajplung.00256.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gibbons MA, et al. Ly6Chi monocytes direct alternatively activated profibrotic macrophage regulation of lung fibrosis. Am J Respir Crit Care Med. 2011;184(5):569–581. doi: 10.1164/rccm.201010-1719OC. [DOI] [PubMed] [Google Scholar]
- 31.Chaudhary NI, et al. Inhibition of PDGF, VEGF and FGF signalling attenuates fibrosis. Eur Respir J. 2007;29(5):976–985. doi: 10.1183/09031936.00152106. [DOI] [PubMed] [Google Scholar]
- 32.Koschmann J, Bhar A, Stegmaier P, Kel AE, Wingender E. “Upstream Analysis”: An Integrated Promoter-Pathway Analysis Approach to Causal Interpretation of Microarray Data. Microarrays (Basel) 2015;4(2):270–286. doi: 10.3390/microarrays4020270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 34.Park HL, et al. Mouse Gli1 mutants are viable but have defects in SHH signaling in combination with a Gli2 mutation. Development. 2000;127(8):1593–1605. doi: 10.1242/dev.127.8.1593. [DOI] [PubMed] [Google Scholar]
- 35.Arora R, Metzger RJ, Papaioannou VE. Multiple roles and interactions of Tbx4 and Tbx5 in development of the respiratory system. PLoS Genet. 2012;8(8):e1002866. doi: 10.1371/journal.pgen.1002866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kalinichenko VV, et al. Defects in pulmonary vasculature and perinatal lung hemorrhage in mice heterozygous null for the Forkhead Box f1 transcription factor. Dev Biol. 2001;235(2):489–506. doi: 10.1006/dbio.2001.0322. [DOI] [PubMed] [Google Scholar]
- 37.Steele-Perkins G, et al. The transcription factor gene Nfib is essential for both lung maturation and brain development. Mol Cell Biol. 2005;25(2):685–698. doi: 10.1128/MCB.25.2.685-698.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shimano H, Sato R. SREBP-regulated lipid metabolism: convergent physiology - divergent pathophysiology. Nat Rev Endocrinol. 2017;13(12):710–730. doi: 10.1038/nrendo.2017.91. [DOI] [PubMed] [Google Scholar]
- 39.Repa JJ, et al. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRalpha and LXRbeta. Genes Dev. 2000;14(22):2819–2830. doi: 10.1101/gad.844900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshikawa T, et al. Identification of liver X receptor-retinoid X receptor as an activator of the sterol regulatory element-binding protein 1c gene promoter. Mol Cell Biol. 2001;21(9):2991–3000. doi: 10.1128/MCB.21.9.2991-3000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bultmann-Mellin I, et al. Role of LTBP4 in alveolarization, angiogenesis, and fibrosis in lungs. Am J Physiol Lung Cell Mol Physiol. 2017;313(4):L687–L698. doi: 10.1152/ajplung.00031.2017. [DOI] [PubMed] [Google Scholar]
- 42.Li Y, et al. Cardiac Fibroblast-Specific Activating Transcription Factor 3 Protects Against Heart Failure by Suppressing MAP2K3-p38 Signaling. Circulation. 2017;135(21):2041–2057. doi: 10.1161/CIRCULATIONAHA.116.024599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Myllärniemi M, et al. Gremlin-mediated decrease in bone morphogenetic protein signaling promotes pulmonary fibrosis. Am J Respir Crit Care Med. 2008;177(3):321–329. doi: 10.1164/rccm.200706-945OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Naik PK, et al. Periostin promotes fibrosis and predicts progression in patients with idiopathic pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol. 2012;303(12):L1046–L1056. doi: 10.1152/ajplung.00139.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chien JW, et al. Serum lysyl oxidase-like 2 levels and idiopathic pulmonary fibrosis disease progression. Eur Respir J. 2014;43(5):1430–1438. doi: 10.1183/09031936.00141013. [DOI] [PubMed] [Google Scholar]
- 46.Daniel C, Vogelbacher R, Stief A, Grigo C, Hugo C. Long-term gene therapy with thrombospondin 2 inhibits TGF-β activation, inflammation and angiogenesis in chronic allograft nephropathy. PLoS ONE. 2013;8(12):e83846. doi: 10.1371/journal.pone.0083846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koli K, et al. Gremlin-1 Overexpression in Mouse Lung Reduces Silica-Induced Lymphocyte Recruitment - A Link to Idiopathic Pulmonary Fibrosis through Negative Correlation with CXCL10 Chemokine. PLoS ONE. 2016;11(7):e0159010. doi: 10.1371/journal.pone.0159010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim W, et al. RUNX1 is essential for mesenchymal stem cell proliferation and myofibroblast differentiation. Proc Natl Acad Sci USA. 2014;111(46):16389–16394. doi: 10.1073/pnas.1407097111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Serini G, et al. The fibronectin domain ED-A is crucial for myofibroblastic phenotype induction by transforming growth factor-beta1. J Cell Biol. 1998;142(3):873–881. doi: 10.1083/jcb.142.3.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tomura M, et al. Contrasting quiescent G0 phase with mitotic cell cycling in the mouse immune system. PLoS ONE. 2013;8(9):e73801. doi: 10.1371/journal.pone.0073801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shimano H, et al. Elevated levels of SREBP-2 and cholesterol synthesis in livers of mice homozygous for a targeted disruption of the SREBP-1 gene. J Clin Invest. 1997;100(8):2115–2124. doi: 10.1172/JCI119746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Re SL, et al. Uncoupling between inflammatory and fibrotic responses to silica: evidence from MyD88 knockout mice. PLoS ONE. 2014;9(7):e99383. doi: 10.1371/journal.pone.0099383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cabrera S, et al. Overexpression of MMP9 in macrophages attenuates pulmonary fibrosis induced by bleomycin. Int J Biochem Cell Biol. 2007;39(12):2324–2338. doi: 10.1016/j.biocel.2007.06.022. [DOI] [PubMed] [Google Scholar]
- 54.Melboucy-Belkhir S, et al. Forkhead Box F1 represses cell growth and inhibits COL1 and ARPC2 expression in lung fibroblasts in vitro. Am J Physiol Lung Cell Mol Physiol. 2014;307(11):L838–L847. doi: 10.1152/ajplung.00012.2014. [DOI] [PubMed] [Google Scholar]
- 55.Shi Y, Xu X, Tan Y, Mao S, Fang S, Gu W. A liver-X-receptor ligand, T0901317, attenuates IgE production and airway remodeling in chronic asthma model of mice. PLoS ONE. 2014;9(3):e92668. doi: 10.1371/journal.pone.0092668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Okuma T, et al. C-C chemokine receptor 2 (CCR2) deficiency improves bleomycin-induced pulmonary fibrosis by attenuation of both macrophage infiltration and production of macrophage-derived matrix metalloproteinases. J Pathol. 2004;204(5):594–604. doi: 10.1002/path.1667. [DOI] [PubMed] [Google Scholar]
- 57.Moore BB, et al. Protection from pulmonary fibrosis in the absence of CCR2 signaling. J Immunol. 2001;167(8):4368–4377. doi: 10.4049/jimmunol.167.8.4368. [DOI] [PubMed] [Google Scholar]
- 58.Li Y, et al. Severe lung fibrosis requires an invasive fibroblast phenotype regulated by hyaluronan and CD44. J Exp Med. 2011;208(7):1459–1471. doi: 10.1084/jem.20102510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kolb M, Margetts PJ, Sime PJ, Gauldie J. Proteoglycans decorin and biglycan differentially modulate TGF-beta-mediated fibrotic responses in the lung. Am J Physiol Lung Cell Mol Physiol. 2001;280(6):L1327–L1334. doi: 10.1152/ajplung.2001.280.6.L1327. [DOI] [PubMed] [Google Scholar]
- 60.Bensadoun ES, Burke AK, Hogg JC, Roberts CR. Proteoglycan deposition in pulmonary fibrosis. Am J Respir Crit Care Med. 1996;154(6 Pt 1):1819–1828. doi: 10.1164/ajrccm.154.6.8970376. [DOI] [PubMed] [Google Scholar]
- 61.Horie M, et al. TBX4 is involved in the super-enhancer-driven transcriptional programs underlying features specific to lung fibroblasts. Am J Physiol Lung Cell Mol Physiol. 2018;314(1):L177–L191. doi: 10.1152/ajplung.00193.2017. [DOI] [PubMed] [Google Scholar]
- 62.Wynn TA. Cellular and molecular mechanisms of fibrosis. J Pathol. 2008;214(2):199–210. doi: 10.1002/path.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hautekeete ML, Geerts A. The hepatic stellate (Ito) cell: its role in human liver disease. Virchows Arch. 1997;430(3):195–207. doi: 10.1007/BF01324802. [DOI] [PubMed] [Google Scholar]
- 64.McGowan SE, Torday JS. The pulmonary lipofibroblast (lipid interstitial cell) and its contributions to alveolar development. Annu Rev Physiol. 1997;59:43–62. doi: 10.1146/annurev.physiol.59.1.43. [DOI] [PubMed] [Google Scholar]
- 65.She H, Xiong S, Hazra S, Tsukamoto H. Adipogenic transcriptional regulation of hepatic stellate cells. J Biol Chem. 2005;280(6):4959–4967. doi: 10.1074/jbc.M410078200. [DOI] [PubMed] [Google Scholar]
- 66.He Y, et al. The role of retinoic acid in hepatic lipid homeostasis defined by genomic binding and transcriptome profiling. BMC Genomics. 2013;14:575. doi: 10.1186/1471-2164-14-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Leem AY, et al. All-trans retinoic acid attenuates bleomycin-induced pulmonary fibrosis via downregulating EphA2-EphrinA1 signaling. Biochem Biophys Res Commun. 2017;491(3):721–726. doi: 10.1016/j.bbrc.2017.07.122. [DOI] [PubMed] [Google Scholar]
- 68.Hakim F, et al. Vitamins A and E and pulmonary exacerbations in patients with cystic fibrosis. J Pediatr Gastroenterol Nutr. 2007;45(3):347–353. doi: 10.1097/MPG.0b013e31804069e5. [DOI] [PubMed] [Google Scholar]
- 69.Noble PW, et al. Pirfenidone in patients with idiopathic pulmonary fibrosis (CAPACITY): two randomised trials. Lancet. 2011;377(9779):1760–1769. doi: 10.1016/S0140-6736(11)60405-4. [DOI] [PubMed] [Google Scholar]
- 70.Li C, et al. Pirfenidone controls the feedback loop of the AT1R/p38 MAPK/renin-angiotensin system axis by regulating liver X receptor-α in myocardial infarction-induced cardiac fibrosis. Sci Rep. 2017;7:40523. doi: 10.1038/srep40523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hong C, Tontonoz P. Liver X receptors in lipid metabolism: opportunities for drug discovery. Nat Rev Drug Discov. 2014;13(6):433–444. doi: 10.1038/nrd4280. [DOI] [PubMed] [Google Scholar]
- 72.Shi Y, Chen Q, Yan H, Gu W. The effect of a liver-X-receptor ligand on bleomycin induced pulmonary fibrosis in mice. Int Immunopharmacol. 2016;41:116–121. doi: 10.1016/j.intimp.2016.10.016. [DOI] [PubMed] [Google Scholar]
- 73.Du Y, et al. Lung Gene Expression Analysis (LGEA): an integrative web portal for comprehensive gene expression data analysis in lung development. Thorax. 2017;72(5):481–484. doi: 10.1136/thoraxjnl-2016-209598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kurowska-Stolarska M, et al. The role of microRNA-155/liver X receptor pathway in experimental and idiopathic pulmonary fibrosis. J Allergy Clin Immunol. 2017;139(6):1946–1956. doi: 10.1016/j.jaci.2016.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Albers M, et al. A novel principle for partial agonism of liver X receptor ligands. Competitive recruitment of activators and repressors. J Biol Chem. 2006;281(8):4920–4930. doi: 10.1074/jbc.M510101200. [DOI] [PubMed] [Google Scholar]
- 76.Green CD, Ozguden-Akkoc CG, Wang Y, Jump DB, Olson LK. Role of fatty acid elongases in determination of de novo synthesized monounsaturated fatty acid species. J Lipid Res. 2010;51(7):1871–1877. doi: 10.1194/jlr.M004747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Moon YA, Shah NA, Mohapatra S, Warrington JA, Horton JD. Identification of a mammalian long chain fatty acyl elongase regulated by sterol regulatory element-binding proteins. J Biol Chem. 2001;276(48):45358–45366. doi: 10.1074/jbc.M108413200. [DOI] [PubMed] [Google Scholar]
- 78.Matsuzaka T, et al. Crucial role of a long-chain fatty acid elongase, Elovl6, in obesity-induced insulin resistance. Nat Med. 2007;13(10):1193–1202. doi: 10.1038/nm1662. [DOI] [PubMed] [Google Scholar]
- 79.Murakami M, et al. Regulation of prostaglandin E2 biosynthesis by inducible membrane-associated prostaglandin E2 synthase that acts in concert with cyclooxygenase-2. J Biol Chem. 2000;275(42):32783–32792. doi: 10.1074/jbc.M003505200. [DOI] [PubMed] [Google Scholar]
- 80.Serhan CN, et al. Resolvins: a family of bioactive products of omega-3 fatty acid transformation circuits initiated by aspirin treatment that counter proinflammation signals. J Exp Med. 2002;196(8):1025–1037. doi: 10.1084/jem.20020760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yatomi M, et al. 17(R)-resolvin D1 ameliorates bleomycin-induced pulmonary fibrosis in mice. Physiol Rep. 2015;3(12):e12628. doi: 10.14814/phy2.12628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhao H, et al. Pulmonary delivery of docosahexaenoic acid mitigates bleomycin-induced pulmonary fibrosis. BMC Pulm Med. 2014;14:64. doi: 10.1186/1471-2466-14-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bozyk PD, Moore BB. Prostaglandin E2 and the pathogenesis of pulmonary fibrosis. Am J Respir Cell Mol Biol. 2011;45(3):445–452. doi: 10.1165/rcmb.2011-0025RT. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sunaga H, et al. Deranged fatty acid composition causes pulmonary fibrosis in Elovl6-deficient mice. Nat Commun. 2013;4:2563. doi: 10.1038/ncomms3563. [DOI] [PubMed] [Google Scholar]
- 85.Wei B, et al. Microsomal prostaglandin E synthase-1 deficiency exacerbates pulmonary fibrosis induced by bleomycin in mice. Molecules. 2014;19(4):4967–4985. doi: 10.3390/molecules19044967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Huang H, et al. Non-biased and efficient global amplification of a single-cell cDNA library. Nucleic Acids Res. 2014;42(2):e12. doi: 10.1093/nar/gkt965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27(6):863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sun J, Nishiyama T, Shimizu K, Kadota K. TCC: an R package for comparing tag count data with robust normalization strategies. BMC Bioinformatics. 2013;14:219. doi: 10.1186/1471-2105-14-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tang M, Sun J, Shimizu K, Kadota K. Evaluation of methods for differential expression analysis on multi-group RNA-seq count data. BMC Bioinformatics. 2015;16:361. doi: 10.1186/s12859-015-0794-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11(3):R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Saito R, et al. A travel guide to Cytoscape plugins. Nat Methods. 2012;9(11):1069–1076. doi: 10.1038/nmeth.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Subach OM, Cranfill PJ, Davidson MW, Verkhusha VV. An enhanced monomeric blue fluorescent protein with the high chemical stability of the chromophore. PLoS ONE. 2011;6(12):e28674. doi: 10.1371/journal.pone.0028674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Igarashi T, et al. Lentivirus-mediated expression of angiostatin efficiently inhibits neovascularization in a murine proliferative retinopathy model. Gene Ther. 2003;10(3):219–226. doi: 10.1038/sj.gt.3301878. [DOI] [PubMed] [Google Scholar]
- 98.Faul F, Erdfelder E, Lang AG, Buchner A. G*Power 3: a flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav Res Methods. 2007;39(2):175–191. doi: 10.3758/BF03193146. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.