Abstract
Activation T-DNA tagging can generate dominant gain-of-function mutants by overexpression of a particular endogenous gene. We identified an activation-tagged mutant, sturdy, exhibiting a stiff inflorescence stem, thicker leaves, shorter siliques, larger seeds, round-shaped flowers, and delayed growth. It is most important that unlike its wild-type counterpart, this mutant is less prone to lodging. Cloning of STURDY revealed that in sturdy, there is an open reading frame containing a single intron encoding a patatin-like homolog. The T-DNA is inserted into the 3′ region of the second exon. The mutant phenotype was shown to be the result of overexpression of STURDY by mRNA analysis and transgenic studies. Preliminary histological studies have revealed an increase in cell number in the inflorescence stem of mutant plants; however, additional studies are needed to better understand the overexpression phenotype.
Lodging occurs when crop plants fail to withstand environmental challenges, resulting in extensive financial losses in agriculture each year throughout the world. Crop plants fall to the ground prematurely, making them recalcitrant to mechanical harvest. Even in areas where manual harvesting is practiced, lodged plants can absorb moisture from the ground surface, interfering with the desiccation process and compromising the quality of the grain. In 1992, it was reported that lodging in winter wheat cost ≤£130 million in loss of yield alone (Berry et al., 1998) and reduced the grain quality standard (Hagberg falling number) from a U.K. 5-year average of 287 to 254 (Home Grown Cereal Authority, 1993). Through conventional breeding methods, dwarf traits have been identified and introgressed into various crop species, resulting in shorter plants with stiffer stems less prone to lodging. For example, the gibberellin-insensitive semidwarf Rht genes in wheat and the dw genes in sorghum have been used to reduce plant height (Quinby and Karper, 1954; Gale and Youssefian, 1985). However, these genetic alterations in plant stature are limited to certain crops or varieties and are often accompanied with negative pleiotropic effects including reduced yield and lower grain protein levels. The progress in recombinant DNA technologies recently has provided alternative approaches for improvement of agronomic crops. We are interested in studying plant stem structure at the molecular level and identifying target genes to genetically modify crops with improved lodging resistance.
In principle, there are at least three aspects in which genetic engineering can be applied to improve lodging resistance. Many genes implicated in cell wall biosynthesis, composition, and lignification have been cloned (for review, see Boudet, 1998; Reiter, 1998). By altering the composition of the stem cell walls, it may be possible to increase stem strength. A change in stem strength alternatively could come from altering the organization of the stem structure (e.g. rearrangement of vascular bundles). Several homeobox genes were shown to preferentially express in the vascular tissues of Arabidopsis (Baima et al., 1995; Tornero et al., 1996). An Arabidopsis mutant displaying the transformation of collateral vascular bundles into amphivasal vascular bundles recently was identified (Zhong et al., 1999). An increase in mass could also result in a stronger stem structure. Studies of transgenic plants demonstrated the possibility of modifying the stem mass by varying the level of auxins or cytokinins (Medford et al., 1989; Romano et al., 1991). However, no mutant or transgenic plant showing a strengthened stem phenotype with the normal plant height has been reported.
We initiated an activation T-DNA screen to isolate mutants displaying unusual stem phenotypes. First described by Hayashi et al. (1992), activation T-DNA tagging produces dominant mutations caused by increased or ectopic expression of endogenous genes. This approach allows discovery of novel genes in which recessive, loss-of-function mutations have no obvious phenotype. In this study, we report the recovery of a dominant gain-of-function mutation affecting stem, leaf surface, silique length, seed size, flower shape, and growth rate. The mutant allele, named sturdy, was cloned and shown to be related to the patatin family of proteins. All phenotypes observed in sturdy mutants are due to overexpression of the gene. A preliminary microscopic examination of sturdy inflorescence stems revealing their unusual structure is presented.
RESULTS
Identification of a Dominant, Stiff-Stem Mutant, Sturdy, in an Activation T-DNA-Tagging Population
We transformed Arabidopsis plants (Columbia ecotype) with a binary vector, pMON29963, to generate an activation T-DNA-tagging population. The T-DNA region of pMON29963 contains four enhancers of the cauliflower mosaic virus (CaMV)-35S promoter stacked near the right border, and the selection marker neomycin phosphotransferase II gene (NPTII) driven by the nopaline synthase promoter close to the left border. The transformants were first germinated on medium containing kanamycin and resistant seedlings were transferred to soil for phenotypic evaluation. Under our growth conditions, Arabidopsis plants of the Columbia ecotype can reach an average height of 40 cm (Huang et al., 1998) at the global proliferative arrest stage (Hensel et al., 1994). The stems typically cannot physically sustain growth and the plants bend toward the soil surface or lodge. In screening 4,200 transformants, we identified a single lodge-resistant plant. It appeared to have stiffer and thicker inflorescence stems and branches than wild-type plants, so it was named sturdy. This phenotype was observed in 83 of 114 T2 progeny, suggesting that sturdy is a dominant mutation caused by a T-DNA insertion at a single locus (χ2 = 0.3).
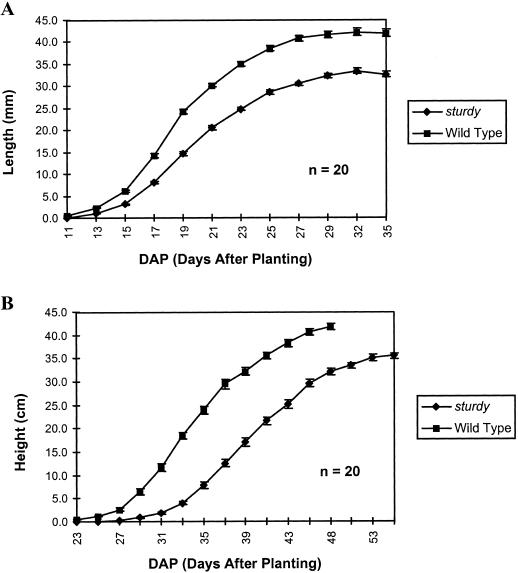
As shown in Figure 1, B and C, 2-week-old sturdy plants are small and dark green in color, similar to gibberellin-deficient mutants. However, sturdy did not respond significantly different than wild-type plants when active gibberellin analogs were applied. Moreover, unlike gibberellin-insensitive mutants, sturdy plants grow to a height comparable with wild-type plants with only a slight (15%) reduction in height. However, it takes about 55 d for sturdy plants to reach global proliferative arrest stage compared with 47 d for wild-type plants grown under the same conditions. The growth rate was monitored through the plants' life cycle by measuring leaf expansion and plant height. The most significant difference is that sturdy mutants' stems are an average of 40% thicker than those of the wild-type plants. These results are summarized in Table I and Figure 2. It appears that sturdy develops greater stiffness in its inflorescence stems and branches through an increase in diameter and at the expense of plant height and growth rate. The sturdy mutation affects other parts of the plant as well. Its leaves have a rougher surface, and its flowers and siliques are shorter (Fig. 1, D–G). The shape of mutant flowers develops as round instead of oval. Also, the mutant flower clusters appear to be more compact with the stigmas protruding out of the flower buds (Fig. 1H). When the length of the siliques and number of seeds in each silique were measured (Table II), we found that mutant siliques also produced fewer seeds; however, total seed weight remained about the same. On average, individual mutant seeds weighed 30% more than wild type. We noticed that leaves and stems of the sturdy mutant are brittle and prone to fracture.
Figure 1.
The phenotype of the sturdy mutant. A, Mature mutant plant. B and C, Two-week-old mutant and wild-type plants, respectively. D and E, Mutant and wild-type flowers, respectively. F and G, Mutant siliques and wild-type siliques, respectively. H, Topical view of a mutant flower cluster.
Table I.
Physical characterization of the sturdy plants
| Phenotype | Flowering Timea | Terminal Heightb | Time-to-Terminal Height | Stem Diameter |
|---|---|---|---|---|
| d | cm | d | mm | |
| sturdy | 30 ± 0.3 | 35.4 ± 0.63 | 55 ± 0.2 | 2.10 ± 0.04 |
| Wild type | 25 ± 0.2 | 41.8 ± 0.64 | 47 ± 0.3 | 1.50 ± 0.03 |
Twenty sturdy and wild-type plants for the measurement; each value represents the means ± se.
Number of days for the primary bolt to reach a height of 1 cm.
Height at which main stem produced at global proliferative arrest (Hensel et al., 1994).
Figure 2.
Growth rate comparison between sturdy and wild-type plants. A, Chart of length (cm) of the sixth leaf by days. Under our growth conditions, the fifth and sixth leaf are the initial adult leaves in most Arabidopsis plants and they continue to expand throughout their life cycle. We recorded the length of the sixth leaf every other day and used these data as one of the growth rate indicators. B, Chart of plant height (cm) by days. The height of the plants was recorded every other day and was also used as a growth rate indicator.
Table II.
Physical characterization of the silique and seed produced by sturdy plants
| Phenotype | Silique Length | No. of Seeds within a Silique | Total Seed Wt | Total No. of Seeds | Average Seed Wt |
|---|---|---|---|---|---|
| mm | mg | mg | |||
| sturdy | 8.0 ± 0.15 | 47.1 ± 1.35 | 25.3 | 941 | 0.027 |
| Wild type | 15.0 ± 0.33 | 60.9 ± 2.78 | 24.5 | 1,217 | 0.020 |
Twenty mature siliques were randomly picked from sturdy and wild-type plants for the measurement; each value represents the means ± se.
Cloning of the Genomic DNA of Sturdy
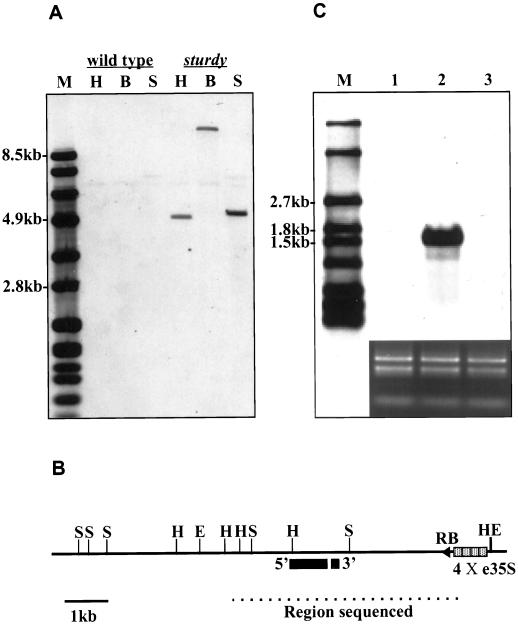
To clone sturdy, we first analyzed the mutant with a genomic DNA gel blot. Three different restriction enzymes (HindIII, BglII, and SpeI) were used to digest the genomic DNA of wild-type and mutant plants and the CaMV-35S enhancer was used as the probe for hybridization (Fig. 3A). DNA gel-blot analysis with three enzyme digests displayed a single band only on the lanes loaded with the mutant genomic DNA. This result, along with the T2 segregation ratio, confirmed that a single T-DNA insertion produced the dominant sturdy mutation. We then used the same probe to screen a library constructed from the genomic DNA of sturdy. Three positive clones were isolated independently and all of them contained the same 12-kb genomic DNA fragment. Figure 3B shows the restriction map and the sequenced region of the fragment.
Figure 3.
Genomic characterization of the sturdy mutant and identification of the STURDY gene. A, Genomic DNA gel-blot analysis of the sturdy mutant. A DNA gel blot containing 3 μg of wild-type and mutant genomic DNA digested with HindIII (H), BglII (B), and SpeI (S) hybridized with a CaMV-35S enhancer probe. The digoxigenin-labeled DNA fragments (Roche Molecular Biochemicals, Indianapolis) were used as markers (M). B, Restriction map of the sturdy mutant allele. The map shows HindIII (H), EcoRI (E), and SpeI (S) restriction sites of a 12-kb DNA fragment flanking the right border (RB) of the T-DNA isolated from the mutant genomic library. Indicated by the dashed line, approximately 5.5 kb of the fragment near the right border was sequenced, and an open reading frame (ORF) separated by one intron (black boxes) was identified. C, RNA gel-blot analysis of the sturdy mutant. An RNA gel blot loaded with 2 μg of total leaf RNA isolated from a wild type (1), sturdy mutant (2), and a second activation tagged line (3) were hybridized with a probe containing the sequence of the identified ORF. The DIG-labeled RNA fragments (Roche Molecular Biochemicals) were used as markers (M). On the bottom is an ethidium bromide stain of an agarose gel loaded with the RNA (1 μg per lane) used in the gel blot.
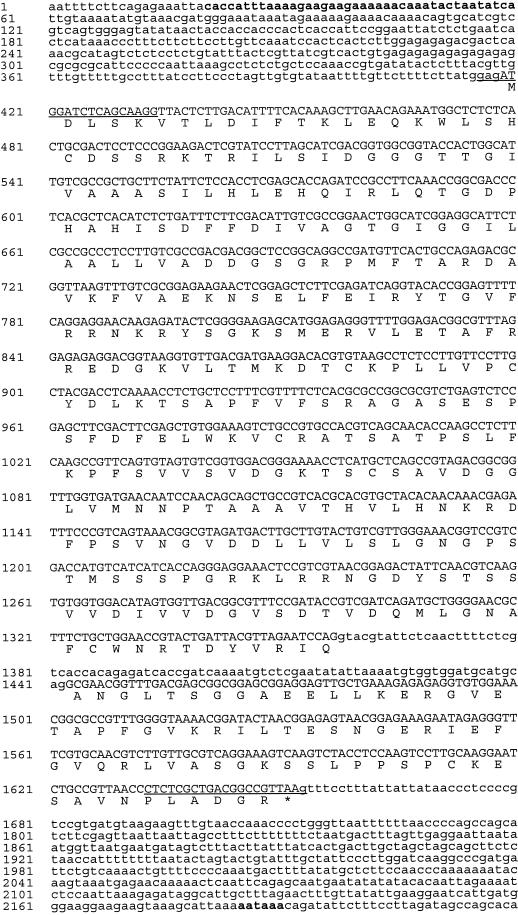
Based on previous reports, T-DNA activation tagging is likely to overexpress the nearest gene from the CaMV-35S enhancers to induce the mutant phenotypes (Hayashi et al., 1992; Kakimoto, 1996; Kardailsky et al., 1999; Weigel et al., 2000). An approximately 5.5-kb region of the genomic DNA near the T-DNA was sequenced to identify ORFs. One putative gene 2.7 kb from the CaMV-35S enhancers (Fig. 3B) was identified with two exons separated by a single intron. This putative gene is 1,234 bp and encodes 382 amino acids (Fig. 4). The CAAT and TATA boxes as well as a poly-A signal were also identified. To test if this ORF was overexpressed in the sturdy mutant, an RNA gel blot was prepared using equal amounts of RNA (2 μg) isolated from leaves of wild-type and mutant plants and probed with the ORF sequence. The RNA blot shown in Figure 3C clearly indicated that the ORF was highly expressed in the mutant leaves but was undetectable in the leaves of wild-type plants and in a second activation-tagging line grown under the same condition. This suggests that the putative gene identified by the screen is the STURDY gene.
Figure 4.
The nucleotide sequence of the STURDY gene and its flanking regions. The STURDY ORF is shown in uppercase letters and lowercase letters refer to the flanking and intron sequences of the STURDY gene. The CAAT and TATA sequences (positions 21–60) and the putative polyadenylation signal (positions 2,185–2,190) are in boldfaced font. The single-letter abbreviations for amino acids were used and an asterisk denotes a stop codon. The sequences used as primers for reverse transcriptase (RT)-PCR to isolate the STURDY cDNA are underlined.
Overexpression of the Putative STURDY cDNA in Wild-Type Plants Conferring the Sturdy Phenotype
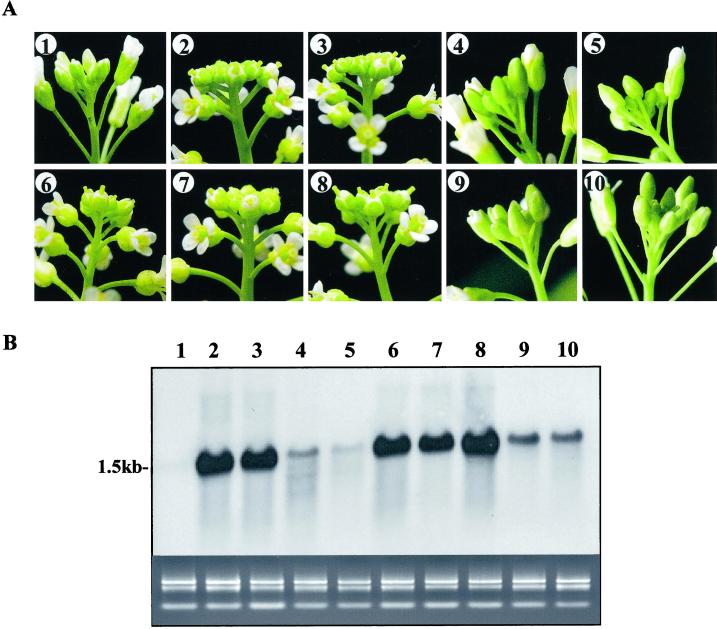
To demonstrate that overexpression of STURDY is solely responsible for the mutant phenotypes, we isolated the sturdy cDNA by RT-PCR and constructed a transformation vector, pMON42154, containing the cDNA driven by the enhanced CaMV-35S promoter. The sequence of the cDNA fragment amplified by the primer designed according to the genomic DNA also confirmed the location of the intron. Among 49 transgenic plants transformed with pMON42154, 26 of them clearly exhibited a sturdy-like phenotype. We selected five transgenic plants from each group for further analysis. As shown in Figure 5, plants numbered 2, 3, 6, 7, and 8 had the typical sturdy floral cluster, round and compact, whereas 1, 4, 5, 9, and 10 appeared to be normal. When the RNA samples prepared from leaves were analyzed on an RNA gel blot, only those sturdy-like transgenic plants showed strong expression of STURDY. These results confirm that the sturdy phenotype is caused by the overexpression of the STURDY gene identified in the T-DNA activation-tagging screen alone.
Figure 5.
The floral phenotype of the transgenic plants overexpressing STURDY. A, The floral phenotype of 10 independent transgenic lines. B, RNA gel-blot analysis. RNA (6 μg) isolated from the leaves of the 10 transgenic lines was hybridized to the STURDY cDNA probe. On the bottom is an ethidium bromide stain of an agarose gel loaded with the RNA (1 μg per lane) used in the gel blot.
Molecular Characterization of the STURDY Gene
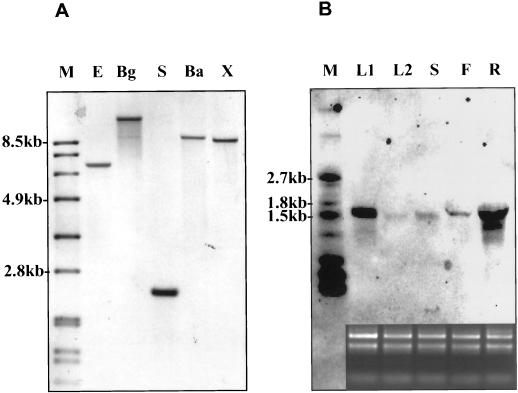
When the STURDY gene was used to search the GenBank sequence database by BLAST programs (Altschul et al., 1997), it matched a putative gene in an Arabidopsis chromosome 3 bacteria artificial chromosome clone, F16M2 (accession no. AL138648). No other significant homology was identified at the nucleotide level. The predicted protein sequence showed similarity to a group of patatin or patatin-like proteins. STURDY was compared with three other predicted patatin-like proteins from the Arabidopsis genome sequences along with patatin and patatin homologs from potato, tobacco, and rubber by protein sequence alignment using the MegAlign program (DNASTAR Inc., Madison, WI). As illustrated in Figure 6, striking resemblance exists across the entire sequence between STURDY and these patatin proteins. Patatin, a 40-kD glycoprotein, has been found in all Solanum species tested (Prat et al., 1990) and accounts for up to 40% of the total water soluble protein present in the tubers (Racusen and Foote, 1980). In potato, patatin is a multigene family with 10 to 18 members per haploid genome (Mignery et al., 1988; Twell and Ooms, 1988), and its expression often associates with tuber formation (Perl et al., 1991). However, patatin-like proteins have been found in other plant organs (Paiva et al., 1983; Vancanneyt et al., 1989) and other species as well (Vancanneyt et al., 1989; Ganal et al., 1991). We examined the genomic composition and expression pattern of the STURDY gene. Even under less stringent wash conditions (0.5× SSC, 50°C for the high stringent washing), no other homologous sequence of the STURDY gene was identified (Fig. 7A). This is consistent with sequence search results that failed to identify homologous Arabidopsis sequences. Also, the STURDY gene was found to be expressed in all tissues tested with highest expression levels in roots (Fig. 7B).
Figure 6.
Homology between STURDY amino acid sequence and those of patain and patatin-like proteins from Arabidopsis and other plant species. A consensus sequence between homologous proteins was identified and is shown at the top of the alignment. The amino acids in black boxes are those identical to the consensus sequence, and the amino acids in gray boxes are those similar to the consensus sequence. The positions of each sequence used in the alignment are listed on the right. The accession numbers of these sequences are STURDY, AL138648; Arabidopsis patatin-like protein 1 (AtPLP1), AC004697; Arabidopsis patatin-like protein 2 (AtPLP2), AL049655; Arabidopsis patatin-like protein 3 (AtPLP3), Z99707; tobacco, U68484; rubber, AJ223038; and potato, A24142.
Figure 7.
Genomic composition and expression pattern of the STURDY gene. A, DNA gel-blot analysis. A DNA gel blot containing 3 μg of EcoRI (E), BglII (Bg), SpeI (S), BamHI (Ba), and XbaI (X) digests of wild-type genomic DNAs were hybridized to the STURDY cDNA probe. B, RNA gel-blot analysis. RNA (2 μg) isolated from the leaves of sturdy mutant and RNA (20 μg) isolated from leaves (L2), stems (S), flowers (F), and roots (R) of wild type were hybridized to the STURDY cDNA probe. On the bottom is an ethidium bromide stain of an agarose gel loaded with the RNA (1 μg per lane) used in the gel blot.
Microscopic Analysis of Sturdy Mutant Stems
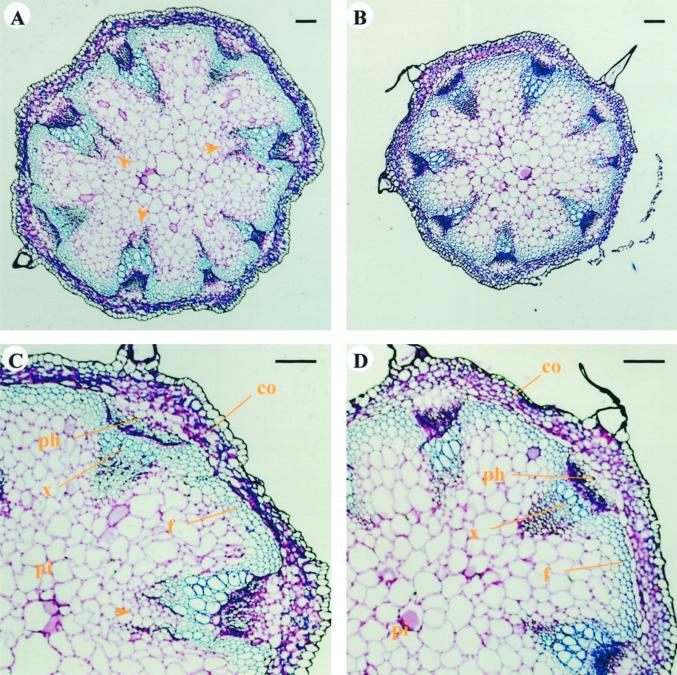
To investigate the stem structure of the sturdy mutant, we sectioned and analyzed the inflorescence stems from several individual mutant and wild-type plants. Due to the differences in their growth rate, the plant material was collected at a similar developmental stage based on height (14–15 cm) rather than days after planting. As shown in Figure 8, which represents the stem transverse sections of typical sturdy (A and C) and wild-type (B and D) plants, the diameter is significantly greater in sturdy plants compared with wild type at the same magnifications. In general, sturdy appears to increase in cell number rather than cell size. At this developmental stage, there are eight equally spaced vascular bundles in the wild-type plants. In sturdy plants, the number of vascular bundles remains the same, but many of them appear to be expanding and branching out at the tip toward the pith (indicated by the arrows in Fig. 8, A and C) as if it is undergoing the formation of extra vascular bundles. When stained with toluidine blue, xylem and fiber cells become blue due to the presence of lignin in cell walls. The sturdy stem has at least one more layer of blue-staining cells than wild type (Fig. 8). Therefore, the stiffness of sturdy stems could be explained by the presence of additional layers of lignin-containing cells.
Figure 8.
Cross sections of inflorescence stems of sturdy and wild-type plants. A and B, sturdy and wild type, respectively. C and D, Close-up sturdy and wild type. Arrows point to the branching vascular bundles. Bars = 100 μm. co, Cortex; f, interfascicular fibers; ph, phloem; pi, pith; x, xylem.
DISCUSSION
We described the isolation and characterization of a novel dominant Arabidopsis mutant, sturdy, generated by activation T-DNA tagging. The sturdy mutant was identified by its stiff inflorescence stem that supported its vertical growth and prevented lodging. The phenotype also includes thicker leaves, shorter siliques, larger seeds, round flowers, and delayed growth. By screening the genomic library prepared from sturdy genomic DNA using the CaMV-35S enhancer sequence as the probe, we discovered a putative gene of 1,234 bp linked to the CaMV-35S enhancer tetramer. This putative gene, containing a single intron, encodes a patatin-like protein that is highly expressed in the sturdy mutant and therefore is likely to be the STURDY gene. Transgenic studies confirmed that the sturdy phenotype resulted from the overexpression of the STURDY gene.
Patatin is a major soluble glycoprotein found in potato tubers (Racusen and Foote, 1980) and is encoded by a multigene family (Mignery et al., 1988; Twell and Ooms, 1988). Because of the diversity and complexity of the gene family, most of the molecular characterization of patatin has focused on cloning and expression of different classes of patatin genes (Bevan, et al., 1986; Rosahl et al., 1986; Pikaard et al., 1987; Köster-Töpfer et al., 1989; Rocha-Sosa, et al., 1989). The expression of patatin and patatin-like genes subsequently were detected in other plant organs (Paiva et al., 1983; Vancanneyt et al., 1989) and other species as well (Vancanneyt et al., 1989; Ganal et al., 1991). Although regarded as a storage protein, patatin has enzymatic activity. It exhibits esterase activity on a broad range of lipid substrates (Rosahl et al., 1987; Andrews et al., 1988; Högen and Willmitzer, 1990). These findings suggest that patatin or patatin-like proteins may possess other biological functions. We currently are investigating the enzymatic activities of the predicted STURDY gene product. Studying STURDY should enhance our understanding of the function(s) of patatin. This stiff inflorescence stem is the first reported phenotype caused by overexpression of a patatin-like protein. With the progression of the Arabidopsis genome sequencing project, many more patatin-like proteins are being discovered in Arabidopsis. Although they may not always share a high degree of homology at the nucleotide level, their protein sequences clearly indicate that they belong to the class of patatin-like proteins. It will be interesting to see whether overexpression of other patatin-like proteins will result in a similar phenotype.
The cross sections show that the inflorescence stem structure organization of the sturdy mutant retains a similar pattern to wild type, whereas the stem mass is significantly increased. It has been reported that the stem cell mass could be altered by varying the endogenous levels of auxins and cytokinins in transgenic plants (Medford et al., 1989; Romano et al., 1991). In addition, a phospholipase A2 involved in signal transduction of a disease-resistant reaction appeared to be a patatin (Senda et al., 1996). Therefore, it could be hypothesized that STURDY plays a role in the signal transduction pathway of hormone responses. It is interesting that mutations in a receptor protein kinase, ERECTA, cause a similar phenotype to sturdy (Torii et al., 1996). It is suggested that ERECTA participates in the coordination of cell growth pattern. We currently are investigating the relationship between ERECTA and STURDY. Modifications in the biochemical composition of the sturdy mutant alternatively may be responsible for the strengthened inflorescence stem phenotype. Slightly brittle leaves and an increase in the number of lignin-containing cells in stems of the sturdy mutant indicate biochemical alternations in cellular components by overexpressing of STURDY. Further detailed analysis on the physiology and biochemistry of the sturdy mutant is required to determine the cause of the mutant phenotype.
Although most genetic screens are based on mutants originating from loss-of-function, the activation T-DNA-tagging screen provides an alternative approach to identify mutants that are caused by gain-of-function. A related approach, with a complete CaMV-35S promoter oriented outward from a transposable Ds element, has also been used to isolate gain-of-function mutations (Wilson et al., 1996; Schaffer et al., 1998; Fridborg et al., 1999). These approaches enhance the possibility of discovering new genes in pathways that have been exhaustively studied by conventional genetic screens, or genes that have either no or lethal mutant phenotypes when inactivated. A particular useful feature of these methods of gene discovery is the potential for direct application of isolated genes. For example, in a genetic screen for an agronomically desirable phenotype, conventional loss-of-function approaches are likely to isolate genes that require inactivation for their application. In contrast, activation T-DNA and transposon tagging identify genes that can be applied by overexpression. Because gene knockout technologies such as antisense, chimeric gene targeting (Hohn and Puchta, 1999) and homologous recombination (Puchta and Hohn, 1996) are still commercially problematic, overexpression remains the most useful technique for genetic engineering.
The patatin gene family provided a model system for studying developmental and metabolic gene expression in the late 1980s and early 1990s. Since then, research interests in patatin have declined due to the lack of physiological relevance. By using an activation T-DNA-tagging approach, we found that a patatin-like protein, STURDY, is implicated in stem structure. Although many questions are yet to be answered, our findings shed new light on the biological function of patatin and, hopefully, provoke renewed interest in research related to patatin. Meanwhile, we continue to study the strengthened stem phenotype caused by overexpression of STURDY and apply the technology to crop species to enhance lodging resistance.
MATERIALS AND METHODS
Plasmid Construction
The binary vector, pMON29963, used to generate the activation T-DNA-tagging population was constructed similar to methods described by Hayashi et al. (1992). It contains four stacked enhancers each corresponding to the −90 to −342 region of the original CaMV-35S promoter (Fang et al., 1989). This enhancer repeat was inserted into the T-DNA region 15 bp from the right border. Near the left border was the neomycin phosphotransferase II gene (NPTII) driven by the nopaline synthase promoter conferring kanamycin resistance. All other genetic elements used in the binary vector were identical to those described by Ye et al. (1999).
PCR primers, 5′-CTTCTAGAGATGGATCTCAGCAAGG-3′ and 5′-CGGGATCCTTAACGGCCGTCAGCGAG-3′, were designed according to the sequences at the beginning and end of the STURDY reading frame (Fig. 4) with the addition of restriction sites XbaI and BamHI for convenient cloning. These primers were used in a RT-PCR reaction to amplify a 1.1-kb cDNA fragment from RNA extracted from the leaves of sturdy mutants. The PCR product was inserted into the TA cloning vector (Invitrogen, San Diego) for further manipulation. To detect any mutations generated by PCR, the RT-PCR amplified cDNAs were sequenced and compared with genomic DNA containing the STURDY gene. The STURDY cDNA was then inserted into the binary transformation vector for Agrobacterium tumefaciens-mediated transformation into Arabidopsis. The resulting plasmid, pMON42154, contains the STURDY cDNA under transcriptional control of an enhanced 35S promoter. The −90 to −342 region of the original CaMV-35S promoter (Fang et al., 1989) was quadrupled to produce the enhanced 35S promoter.
Plant Material, Growth Conditions, and Transformation
Seeds of Arabidopsis ecotype Columbia (Col-0) were obtained from Lehle Seeds (Tucson, AZ). Plants were grown in potting soil (Scotts, Marysville, OH) in a growth chamber at 24°C with a 16-hr photoperiod (120 μmol m−2 sec−1). The binary Ti plasmids pMON29963 and pMON42154 were introduced into Arabidopsis via A. tumefaciens-mediated vacuum infiltration (Bechtold et al., 1993). To select the transgenic plants, seeds collected from vacuum-infiltrated plants were surface sterilized and germinated on kanamycin (40 mg L−1) containing Murashige and Skoog medium (M0404, Sigma, St. Louis) supplemented with Suc (1% [w/v]) and MES [2-(N-morpholino)ethanesulfonic acid; 0.5 g L−1] at pH 5.8.
Isolation of Genomic DNA and DNA Gel-Blot Analysis
Genomic DNA was isolated from leaves of Arabidopsis plants as previously described (Coleman and Kao, 1992). Genomic DNA (3 μg) was digested with restriction enzymes, separated on a 0.7% (w/v) agarose gel, and transferred to nylon membranes (Roche Molecular Biochemicals). Prehybridization, hybridization, washing, and detection of the membrane were conducted using the nonradioactive DIG system (Roche Molecular Biochemicals) following the manufacturer's protocols.
Isolation of Total RNA and RNA-Blot Analysis
Total RNA was isolated from leaves, inflorescence stems, roots, and flowers of wild-type and sturdy mutants by TRIzol Reagent from Life Technologies (Gaithersburg, MD) following the manufacturer's protocols. Total RNA samples (2, 6, or 30 μg) were electrophoresed on 1% (w/v) agarose/formaldehyde gels and transferred to nylon membranes (Roche Molecular Biochemicals). The blots were analyzed by the same DIG system described above following the manufacturer's protocols for RNA gel-blot analysis.
Genomic Library Construction and Screening
Approximately 30 μg of genomic DNA was isolated from sturdy using the same method described in the DNA gel-blot analysis. The genomic library was constructed by partial digestion with BglII and then using the Lambda FIX II/XhoI Partial Fill-In Vector Kit (Stratagene, La Jolla, CA) following the manufacturer's instructions. The same nonradioactive DIG system was also used for screening the library. Using the CaMV-35S enhancer as a probe, three independent clones were isolated from about 15,000 plaques screened. All three clones were identical in sequence and contained a 12-kb BglII cut of the genomic fragment.
Sequence Analysis
The genomic and cDNA clones were sequenced using in-house facilities (ABI Prism 377, Perkin Elmer, Foster, CA). The STURDY gene, including its promoter region containing CAAT and TATA boxes, coding region, and poly-A signal, was identified by GENSCAN1.0, a software written by Chris Burge (Stanford University, CA) modeling Arabidopsis. The STURDY coding region and its translated protein sequence were used to search the GenBank database by applying BLASTN and BLASTP 2.0.8 (Altschul et al., 1997). Homology alignment of various patatin and patatin-like protein sequences was done by the MegAlign software (DNASTAR Inc.) using the clustal method with PAM250 residue weight table. Conserved amino acid residues were defined by no larger than three distance units.
Microscopy
Tissue samples were collected from first and second internodes of wild-type and sturdy plants (14–15 cm) in the growth chamber. Tissue (approximately 5-mm slices) was vacuum infiltrated in a fixative solution containing 4% (w/v) paraformaldehyde and 0.5% (v/v) glutaraldehyde in 100 mm phosphate buffer (pH 7.0). Tissue samples were then placed in 30% (v/v) ethanol. Further processing was done on a Tissue-Tek VIP automated processor (Sakura Finetek, Torrance, CA) through a series of graded ethanol wash steps: 50%, 75%, and 85% (v/v) ethanol at 37°C for an hour each; 95% and 100% (v/v) ethanol at 40°C and 45°C for 2 h each; and finally with paraffin at 58°C for 10 h with four solution changes. Sections 6 μm thin were cut from embedded tissue using a microtome (Reichert Jung 2030 model, Leica Co, Deerfield, IL) and mounted on Probeon Plus slides (Fisher Scientific, Pittsburgh), which were then dried overnight at 37°C.
For toluidine blue O stain, slides were deparaffinized using a series of xylene and ethanol washes (5 min each) and then rehydrated. Slides were stained for 5 min in 1% (w/v) toluidine blue O (Fisher Scientific) in 1% (w/v) Borax (Sigma) and then rinsed in water for 2 min and dehydrated quickly through a series of ethanol washes (95%, 95%, 100%, and 100% [v/v] for 1 min each). These were subsequently placed into two changes of xylene and mounted with Permount (Fisher Scientific). Slides were observed under a microscope (BH-2; Olympus, Melville, NY).
ACKNOWLEDGMENTS
We thank Debbie Stone, Wendi Zumalt, and Stacy Minor for plant transformation; Mark Woerner and Richard Ornber for their expertise and suggestions in microscopy; and Claire CaJacob, Charles Romano, and Tedd Elich for their valuable comments on the manuscript.
LITERATURE CITED
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein data base search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews DL, Beames B, Summers MD, Park WD. Characterization of the lipid acyl hydrolase activity of the major potato (Solanum tuberosum) tyber protein, patatin, by cloning and abundant expression in a baculovirus vector. Biochem J. 1988;252:199–206. doi: 10.1042/bj2520199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baima S, Nobili F, Sessa G, Luchetti S, Ruberti I, Morelli G. The expression of the Athb-8 homeobox gene is restricted to provascular cells in Arabidopsis thaliana. Development. 1995;121:4171–4182. doi: 10.1242/dev.121.12.4171. [DOI] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Paris Life Sci. 1993;316:1194–1199. [Google Scholar]
- Berry P, Sylvester-Bradley R, Scott RK, Clare RW, Spink JH, Baker CJ. Proceedings of the Sixth Home Grown Cereal Authority Research and Development Conference on Cereals and Oilseeds. London: Home Grown Cereal Authority; 1998. Factors affecting lodging; pp. 11.1–11.11. [Google Scholar]
- Bevan M, Barker R, Goldsbrough A, Jarvis M, Kavanagh T, Iturriaga G. The structure and transcription start site of a major potato tuber protein gene. Nucleic Acids Res. 1986;14:4625–4638. doi: 10.1093/nar/14.11.4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudet A-M. A new view of lignification. Trends Plant Sci. 1998;3:67–71. [Google Scholar]
- Coleman CE, Kao T-H. The flanking regions of two Petunia inflata S alleles are heterogeneous and contain repetitive sequences. Plant Mol Biol. 1992;18:725–737. doi: 10.1007/BF00020014. [DOI] [PubMed] [Google Scholar]
- Fang RX, Nagy F, Sivasubramaniam S, Chua NH. Multiple cis regulatory elements for maximal expression of cauliflower mosaic virus 35S promoter in transgenic plants. Plant Cell. 1989;1:141–150. doi: 10.1105/tpc.1.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridborg I, Kuusk S, Moritz T, Sundberg E. The Arabidopsis dwarf mutant shi exhibits reduced gibberellin responses conferred by overexpression of a new putative zinc finger protein. Plant Cell. 1999;11:1019–1031. doi: 10.1105/tpc.11.6.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale MD, Youssefian S. Dwarfing genes in wheat. In: Russel GE, editor. Progress in Plant Breeding. London: Don Butterworth and Co; 1985. pp. 1–35. [Google Scholar]
- Ganal MW, Bonierbale MW, Roeder MS, Park WD, Tanksley SD. Genetic and physical mapping of the patatin genes in potato and tomato. Mol Gen Genet. 1991;225:501–509. doi: 10.1007/BF00261693. [DOI] [PubMed] [Google Scholar]
- Hayashi H, Czaja I, Lubenow H, Schell J, Walden R. Activation of a plant gene by T-DNA tagging: auxin-independent growth in vitro. Science. 1992;258:1350–1353. doi: 10.1126/science.1455228. [DOI] [PubMed] [Google Scholar]
- Hensel LL, Nelson MA, Richmond TA, Bleecker AB. The fate of inflorescence meristem is controlled by developing fruits in Arabidopsis. Plant Physiol. 1994;106:863–876. doi: 10.1104/pp.106.3.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Högen R, Willmitzer L. Biochemical and genetic analysis of different patatin isoforms expressed in various organ of potato (Solanum tuberosum) Plant Sci. 1990;66:221–230. [Google Scholar]
- Hohn B, Puchta H. Gene therapy in plants. Proc Natl Acad Sci USA. 1999;96:8321–8323. doi: 10.1073/pnas.96.15.8321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Home Grown Cereal Authority. Cereal Statistics 1993. London: Home Grown Cereal Authority; 1993. [Google Scholar]
- Huang S, Raman AS, Ream JE, Fujiwara H, Cerny RE, Brown SM. Overexpression of 20-oxidase confers a gibberellin-overproduction phenotype in Arabidopsis. Plant Physiol. 1998;118:773–781. doi: 10.1104/pp.118.3.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakimoto T. CKI1, a histidine kinase homolog implicated in cytokinin signal transduction. Science. 1996;274:982–985. doi: 10.1126/science.274.5289.982. [DOI] [PubMed] [Google Scholar]
- Kardaisky I, Shukla V, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D. Activation tagging of the floral inducer FT. Science. 1999;286:1962–1965. doi: 10.1126/science.286.5446.1962. [DOI] [PubMed] [Google Scholar]
- Medford JL, Horgan R, El-Sawi Z, Klee HJ. Alternations of endogenous cytokinins in transgenic plants using a chimeric isopentenyl transferase gene. Plant Cell. 1989;1:403–413. doi: 10.1105/tpc.1.4.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignery GA, Pikaard CS, Park WD. Molecular characterization of the patatin multigene family. Gene. 1988;62:27–44. doi: 10.1016/0378-1119(88)90577-x. [DOI] [PubMed] [Google Scholar]
- Paiva E, Lister RM, Park WD. Induction and accumulation of major tuber proteins of potato in stems and petioles. Plant Physiol. 1983;71:161–168. doi: 10.1104/pp.71.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perl A, Aviv D, Willmitzer L, Galun E. In vitro tuberization in transgenic potatoes harboring β-glucuronidase linked to a patatin promoter: effects and sucrose levels and photoperoids. Plant Sci. 1991;73:87–95. [Google Scholar]
- Pikaard CS, Brusca JS, Hannapel DJ, Park WD. The two classes of genes for the major potato tuber protein, patatin, are differentially expressed in the tubers and roots. Nucleic Acid Res. 1987;15:1979–1994. doi: 10.1093/nar/15.5.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prat S, Frommer WB, Höfgen R, Keil M, Kosmann J, Köster-Töpfer M, Liu X-Y, Müller B, Peña-Cortés H, Rocha-Sosa M, Sánchez-Serrano JJ, Sonnewald U, Willmitzer L. Gene expression during tuber development in potato plants. FEBS Lett. 1990;286:334–338. doi: 10.1016/0014-5793(90)81281-r. [DOI] [PubMed] [Google Scholar]
- Puchta H, Hohn B. From centiMorgans to base pairs: homologous recombination in plants. Trends Plant Sci. 1996;1:340–348. [Google Scholar]
- Quinby JR, Karper RE. Inheritance of height in sorghum. Agron J. 1954;46:211–216. [Google Scholar]
- Racusen D, Foote M. A major soluble glycoprotein of potato tuber. J Food Biochem. 1980;4:43–52. [Google Scholar]
- Reiter W-D. The molecular analysis of cell wall components. Trends Plant Sci. 1998;3:27–32. [Google Scholar]
- Rocha-Sosa M, Sonnewaid U, Frommer W, Stratmann M, Schell J, Willmitzer L. Both developmental and metabolic signals activate the promoter of a class I patatin. EMBO J. 1989;8:23–29. doi: 10.1002/j.1460-2075.1989.tb03344.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano CP, Hein MB, Klee HJ. Inactivation of auxin in tobacco transformed with the indoleacetic acid-lysine synthetase gene of Pseudomonas savastanoi. Gene Dev. 1991;5:438–446. doi: 10.1101/gad.5.3.438. [DOI] [PubMed] [Google Scholar]
- Rosahl S, Schell J, Willmitzer L. Expression of a tuber-specific storage protein in transgenic tobaco plants: demonstration of an esterase activity. EMBO J. 1987;6:1155–1159. doi: 10.1002/j.1460-2075.1987.tb02348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosahl S, Schmit R, Schell J, Willmitzer L. Isolation and characterization of a gene from Solanum tuberosum encoding patatin, the major storage protein of potato tubers. Mol Gen Genet. 1986;203:214–220. [Google Scholar]
- Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carré JA, Coupland G. The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell. 1998;93:1219–1229. doi: 10.1016/s0092-8674(00)81465-8. [DOI] [PubMed] [Google Scholar]
- Senda K, Yoshioka H, Doke H, Kawakita K. A cytosolic phospholipase A2 from potato tissues appears to be patatin. Plant Cell Physiol. 1996;37:347–353. doi: 10.1093/oxfordjournals.pcp.a028952. [DOI] [PubMed] [Google Scholar]
- Torii KU, Mitsukawa N, Oosumi T, Matsuura Y, Yokoyama R, Whittier RF, Komeda Y. The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell. 1996;8:735–746. doi: 10.1105/tpc.8.4.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornero P, Conejero V, Vera P. Phloem-specific expression of a plant homeobox gene during secondary phases of vascular development. Plant J. 1996;9:639–648. doi: 10.1046/j.1365-313x.1996.9050639.x. [DOI] [PubMed] [Google Scholar]
- Twell D, Ooms G. Structural diversity of the patatin gene family in potato cv. Désirée Mol Gen Genet. 1988;212:325–336. doi: 10.1007/BF00334703. [DOI] [PubMed] [Google Scholar]
- Vancanneyt G, Sonnewald U, Höfgen R, Willmitzer L. Expression of a patatin-like protein in the anthers of potato (Solanum tuberosum) and pepper (Capsicum annuum) flowers. Plant Cell. 1989;1:533–540. doi: 10.1105/tpc.1.5.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Ahn JH, Blázquez MA, Borevitz JO, Christensen SK, Fankhauser C, Ferrándiz C, Kardailsky I, Malancharuvil EJ, Neff MM, Nguyen JT, Sato S, Wang ZY, Xia Y, Dixon RA, Harrison MJ, Lamb CJ, Yanofsky MF, Chory J. Activation tagging in Arabidopsis. Plant Physiol. 2000;122:1003–1013. doi: 10.1104/pp.122.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson K, Long D, Swinburne J, Coupland G. A dissociation insertion causes a semidominant mutation that increases expression of TINY, an Arabidopsis gene related to APETALA2. Plant Cell. 1996;8:659–671. doi: 10.1105/tpc.8.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye GN, Stone D, Pang SZ, Creely W, Gonzalez K, Hinchee M. Arabidopsis ovule is the target for Agrobacterium in planta vacuum infiltration transformation. Plant J. 1999;19:249–257. doi: 10.1046/j.1365-313x.1999.00520.x. [DOI] [PubMed] [Google Scholar]
- Zhong R, Taylor JJ, Ye Z-H. Transformation of the collateral vascular bundles into amphivascular bundles in an Arabidopsis mutant. Plant Physiol. 1999;120:53–64. doi: 10.1104/pp.120.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]