Abstract
More than 40 diseases, most of which primarily affect the nervous system, are caused by expansions of simple sequence repeats dispersed throughout the human genome. Expanded trinucleotide repeat diseases were discovered first and remain the most frequent. More recently tetra-, penta-, hexa- and even dodeca-nucleotide repeat expansions have been identified as the cause of human disease, including some of the most common genetic disorders seen by neurologists. Repeat expansion diseases include both causes of myotonic dystrophy (DM1 and DM2), the most common genetic cause of amyotrophic lateral sclerosis/frontotemporal dementia (C9ORF72), Huntington disease and eight other polyglutamine disorders including the most common forms of dominantly inherited ataxia, the most common recessive ataxia (Friedreich ataxia), and the most common heritable mental retardation (Fragile X Syndrome). Here I review distinctive features of this group of diseases that stem from the unusual, dynamic nature of the underlying mutations. These features include marked clinical heterogeneity and the phenomenon of clinical anticipation. I then discuss the diverse molecular mechanisms driving disease pathogenesis, which vary depending on the repeat sequence, size and location within the disease gene, and whether the repeat is translated into protein. I conclude with a brief clinical and genetic description of individual repeat expansion diseases that are most relevant to neurologists.
Keywords: Trinucleotide, polyglutamine diseases, repeat instability, C9ORF72, anticipation, expanded repeats, myotonic dystrophy, Spinocerebellar ataxia
Introduction
Microsatellites, also known as simple sequence repeats or tandem repeats, are normally polymorphic nucleotide sequences scattered throughout the human genome. Although such repeats have long been used in linkage studies to define the genetic basis of human diseases, it only became clear over the past several decades that expansions of simple sequence repeats directly cause many human diseases. The vast majority of so-called repeat expansion diseases primarily affect the nervous system. Here I discuss general features of the repeat expansion diseases most relevant to neurologists, emphasizing their distinctive features that stem from the dynamic nature of the underlying mutations. Because there are more than two dozen such diseases, they cannot all be discussed in detail. For more information on specific repeat expansion diseases, I refer the reader to definitive chapters in this volume on Fragile X Syndrome and Fragile X Tremor Ataxia Syndrome (FXS/FXTAS; chapter 25), the CAG/polyglutamine diseases (chapter 11), spinal and bulbar muscular atrophy (SBMA; chapter 38), Huntington disease (HD; chapter 17), the dominantly inherited ataxias known as the spinocerebellar ataxias (SCAs; chapter 12), the recessive disorder Friedreich ataxia (chapter 13 on recessive ataxias), C9ORF72 frontotemporal dementia/amyotrophic lateral sclerosis (chapter 27 on ALS), and Unverricht-Lundborg myoclonic epilepsy (EPM1; chapter 30 on genetic epilepsies). This review does not cover the class of congenital neurocognitive disorders caused by polyalanine-encoding expansions (125), but does discuss oculopharyngeal muscular dystrophy (OPMD), an adult onset neuromuscular disorder caused by polyalanine expansion.
The discovery of dynamic repeat expansions shattered the conventional rules of Mendelian inheritance. These rules posit fixed (static) mutations that cause disease through an autosomal dominant, autosomal recessive, or X-linked mechanism, yielding a similar phenotype within families and across generations. In contrast, expanded repeats are unstable (dynamic) mutations that often change size in successive generations. Moreover, depending on the size of the mutation, repeat expansion diseases can manifest with markedly varied phenotypes. Nearly all are primarily neurological diseases and some are among the most common genetic disorders seen by a neurologist. Thus, this category of diseases, unheard of 30 years ago, is now well known to neurologists.
The first nine repeat expansions discovered were all trinucleotide repeats, suggesting that this repeat unit might be a constant feature of this new class of diseases (107). As Table 1 shows, however, new genetic discoveries revealed that the size of repeats ranges from trinucleotides (the vast majority) to tetranucleotides (DM2), pentanucleotides (SCA10, SCA31), hexanucleotides (C9ORF72 FTD/ALS, SCA36) and even dodecanucleotides (EPM1). Additional repeat expansion diseases likely will be discovered that further widen the range of sequence repeats underlying human disease.
Table 1.
Repeat expansions causing neurologic disease
| • CAG – at least 10 diseases (Huntington disease, spinal and bulbar muscular atrophy, dentatorubral-pallidoluysian atrophy and seven SCAs) |
| • CGG – fragile X, fragile X tremor ataxia syndrome, other fragile sites (GCC, CCG) |
| • CTG – myotonic dystrophy type 1, Huntington disease-like 2, spinocerebellar ataxia type 8, Fuchs corneal dystrophy |
| • GAA – Friedreich ataxia |
| • GCC – FRAXE mental retardation |
| • GCG – oculopharyngeal muscular dystrophy |
| • CCTG – myotonic dystrophy type 1 |
| • ATTCT – spinocerebellar ataxia type 10 |
| • TGGAA – spinocerebellar ataxia type 31 |
| • GGCCTG – spinocerebellar ataxia type 36 |
| • GGGGCC – C9ORF72 frontotemporal dementia/amyotrophic lateral sclerosis |
| • CCCCGCCCCGCG – EPM1 (myoclonic epilepsy) |
General Features
Despite their diversity, repeat expansion diseases share numerous features that stem from the underlying genetics (Table 2). All repeat expansions, for example, arise from normally polymorphic repeats in the population. The degree of polymorphism among normal repeats ranges from disease to disease. But in general, repeats at the high end of the normal range (“mutable normal” repeats) have an increased propensity to further expand upon transmission, moving into the pathogenic range. This means that de novo mutations can occur in previously unaffected families. A small percentage of HD, for example, is sporadic (e.g. 98), and the same is true for numerous autosomal dominant repeat expansion diseases. An important implication of this fact is that a patient showing features consistent with a certain repeat expansion disease, yet lacking a family history of similar disease, may still harbor the mutation. Thus, in the right clinical setting the absence of a documented family history should not dissuade the physician from performing a genetic test to confirm or exclude the suspected diagnosis.
Table 2.
General features of repeat expansion diseases
| • Arise from normally existing polymorphic repeats |
| • Expansions are unstable (dynamic), often changing size when transmitted to next generation |
| • Longer repeats tend to cause more severe, earlier onset disease |
| • Clinical anticipation is common: earlier onset, more severe disease in successive generations |
| • Highly variable phenotype, primarily reflecting differences in repeat size |
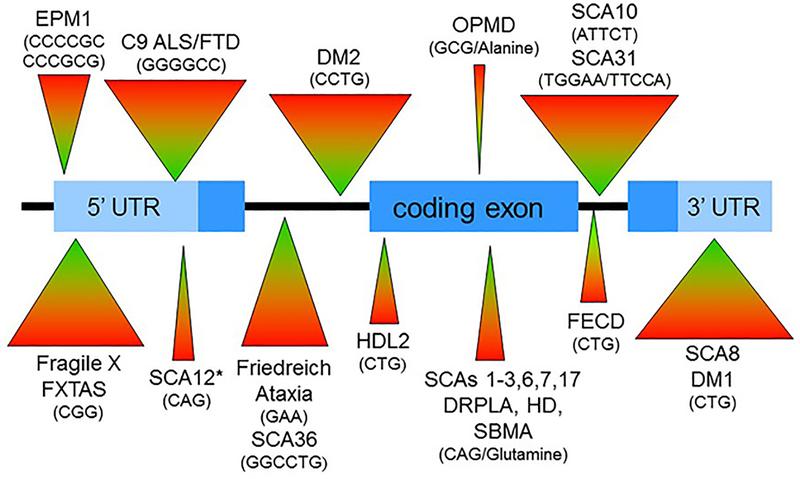
Repeat expansions are inherently dynamic, often changing size when transmitted to the next generation. As a result, even in the same family, the phenotypic range of disease in affected individuals can vary remarkably. The degree of within-generation and across-generation change varies among the disorders, in part because the size of expansions varies from less than 30 repeats (e.g. OPMD, SCA6) to upwards of one or several thousand repeats (e.g. DM1, DM2, FXS, SCA10, SCA36). Figure 1 shows a schematic of a gene into which various repeat sequences underlying repeat expansion diseases have been placed according to their known location in their respective disease genes. Some reside in the 5’ or 3’ untranslated regions (UTR), others in protein-coding exons, and still others in introns. In figure 1, the markedly different sizes of pathogenic expansions are suggested by the varying sized triangles associated with each identified disease.
Figure 1. Schematic of gene showing repeat expansions that cause neurological diseases.
Repeat sequences linked to the indicated diseases are placed schematically into the appropriate gene locations. The differing sizes of the associated triangles roughly reflect the range of repeat expansion sizes in each disease. *SCA12 repeat is actually in an intron that also has promoter elements.
The diseases tend to show a striking genotype-phenotype correlation between repeat length and disease severity. The longer the repeat, the more severe the disease and the earlier the onset of symptoms. This is particularly well illustrated for the CAG/polyglutamine diseases, nine disorders in which relatively modest CAG repeat expansions encode abnormally long stretches of glutamine in the respective disease proteins. In all nine CAG/polyglutamine diseases, there is a strong inverse correlation between repeat length and age of symptom onset: longer repeats cause earlier disease that usually has more profound signs and symptoms (3,36,40,59,62,66,70,75,108,115,122,126,129). The size of the expanded CAG repeat is itself the key determinant for age of onset, accounting for 45% to 70% of the effect, depending on the specific disease. The remaining contribution to age of onset comes from other, minor genetic modifiers and environmental factors that remain under investigation. The relationship between repeat size and age of onset/disease severity is also robust for DM1, Friedreich ataxia, FXS/FXTAS, and several of the non-polyQ SCAs caused by expanded repeats. In contrast, the correlation between repeat length and disease severity in SCA8 is much less robust, and in DM2 and C9ORF72-mediated ALS/FTD is weak if present at all. Intriguingly, all three of these latter diseases are caused by often quite large and complex repeats, suggesting that still unidentified molecular features at these loci influence the behavior of the pathogenic repeats.
Repeat expansion diseases typically show clinical anticipation – that is, the tendency for disease severity to increase in successive generations of a family. This phenomenon had been noted by astute physicians long before the mutational basis was discovered but had been dismissed as biologically implausible; perhaps, doubters argued, the apparent worsening of disease across generations was merely the byproduct of detection bias. The discovery of expanded repeat mutations, and the recognition that expanded repeats are inherently dynamic with longer repeats tending to be increasingly unstable, shed new light on the phenomenon. We now understand that anticipation is explained by the tendency for expanded repeats to further enlarge upon transmission to the next generation, coupled with the fact that longer expansions tend to cause earlier onset, more severe disease.
The extent of anticipation and the degree to which there are parent-of-origin effects influencing anticipation vary across the diseases. For example, several CAG/polyglutamine diseases show a marked paternal effect. Children born to fathers harboring a mutation in HD, SCA2 or SCA7, for example, may develop disease much earlier than their father, in some cases even before the father has developed symptoms (e.g. 138,140). In contrast, the most severe form of myotonic dystrophy type I (DM1), congenital myotonic dystrophy, is almost always transmitted from the mother (6,141). Not all repeat expansions diseases show significant anticipation or a clear parent of origin effect. OPMD, for example, does not show anticipation, which may reflect the exceptionally small expansion size in this disease.
The mode of inheritance varies among the repeat expansion diseases. While the majority are inherited in an autosomal dominant manner, at least two are autosomal recessive (Friedreich’s ataxia and EPM1), two are X-linked recessive (FXS, FRAXE mental retardation) and two are X-linked yet have a dominant toxic mechanism of action (FXTAS and SBMA). The differing modes of inheritance reflect different molecular mechanisms of disease, discussed in the next section.
Arguably, the most striking feature about repeat expansion diseases is the markedly diverse phenotypes manifested for a given disease. This varied phenotype principally reflects differences in repeat size. In HD, for example, whereas most affected individuals develop midlife chorea with cognitive and psychiatric symptoms, individuals with the longest repeats can manifest in their teens or even earlier with bradykinesia and dystonia rather than chorea (so called Westphal or juvenile onset variant of HD). Likewise, another CAG/polyglutamine disorder, dentatorubral-pallidoluysian atrophy (DRPLA), can manifest early in life with dystonia, epilepsy and cognitive impairment or later with chorea and ataxia, again primarily reflecting differences in the size of the expansion. Originally described in Japan, the DRPLA mutation was soon also discovered to be the cause of Haw-River syndrome in the United States, which had been thought to be a distinct disorder because it is characterized by seizures, brain calcifications and ataxia (18,75,90). In DM1, the shortest pathogenic repeats can manifest simply with cataracts and late-life myotonia whereas the longest repeats cause congenital myotonic dystrophy with profound weakness and cognitive impairment. In Friedreich ataxia, the “classic” form of disease typically begins before age 25 with the hallmark features of progressive ataxia, sensory loss and areflexia, but individuals with the smallest expansions may not develop symptoms until quite late in life and can even manifest with spasticity and hyperreflexia rather than areflexia (37a).
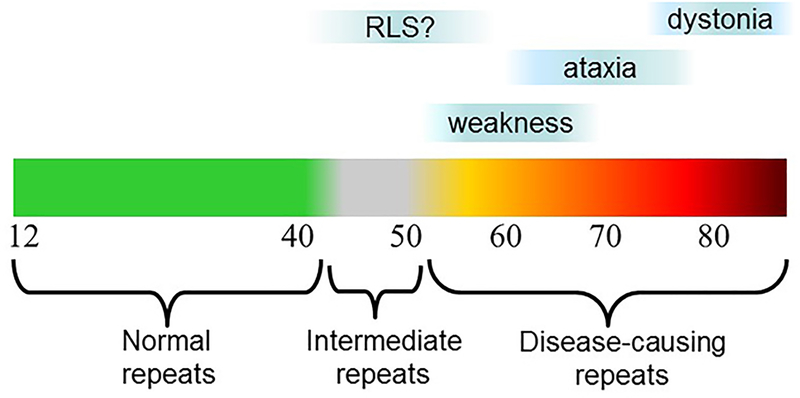
In SCA3, also known as Machado Joseph disease, disease features vary so much that clinicians have even categorized the disease into subtypes: Type I, early onset with dystonia; Type II, midlife onset with progressive ataxia and brainstem signs; and Type III, late life onset with distal leg weakness caused by motor neuronopathy and milder ataxia (37b). Figure 2 shows the range of disease repeats in SCA3, mapped onto these distinct subtypes, as well as a possible fourth phenotype: restless leg syndrome in individuals with intermediate expansions (a rare occurrence). The key determinant driving these distinct subtypes in SCA3 is disease repeat length, with the aging process itself likely being an additional superimposed factor in this, and other, CAG/polyglutamine diseases (e.g. 115). Similar graphs that map disease features and severity across the disease repeat range can be drawn for all of the CAG/polyglutamine diseases, as well as for most other repeat expansion diseases.
Figure 2. Relationship of repeat length to disease features in spinocerebellar ataxia type 3 (SCA3).
Normal CAG repeat length in SCA3 is 12 to ~44, with disease repeats ranging from ~60–87. Clinical subtypes 1 (dystonia), 2 (ataxia), and 3 (weakness) tend to be associated with the range of indicated repeat lengths. Rare intermediate repeat lengths may be associated with restless legs syndrome.
Genetic Testing
The fact that these diseases are due to a single, specific expanded repeat simplifies genetic testing. For many repeat expansion diseases, directed PCR-based screening of repeat length is straightforward, sensitive, specific, and inexpensive. In the right clinical setting, specific candidate gene testing is readily available and cost effective. For some repeat expansion diseases, however, particularly those diseases with very large and complicated expansions or with the possible interruption of pure repeats in some individuals, testing may require other screening methods (e.g. Southern blot hybridization) and the interpretation of results can be more complicated. For nearly all repeat expansion diseases, GeneReviews at the NCBI bookshelf provides expert reports with detailed, up-to-date descriptions of molecular genetic screening and analysis for a given disease (https://www.ncbi.nlm.nih.gov/books/NBK1116/). This author considers GeneReviews a go-to place to understand the nuances of genetic screening and the interpretation of test results for individual repeat expansion diseases.
Directed screening for specific repeat expansions has greatly aided neurologists in diagnosing patients with unexplained disease. But expanded repeats are not well captured by current next generation sequencing (NGS) techniques. Thus, as medical practice moves increasingly to exomic or full genomic sequencing of patients with undefined (presumably genetic) diseases, we must keep in mind that current NGS may miss repeat expansion diseases. Additional directed screening for specific expansions may still be required.
Diverse Molecular Mechanisms
The pathogenic mechanism for a given repeat expansion disease is determined by a variety of factors. Simplistically, the primary mechanism can be divided into toxic gain of function or deleterious loss of gene function, as outlined in Table 3. Predictably, the relatively few diseases with a loss of function mechanism are inherited in an autosomal recessive manner (Friedreich ataxia, EPM1) or an X-linked recessive manner (FXS). By contrast, those due to a toxic gain of function mechanism are inherited in an autosomal dominant manner (the overwhelming majority of diseases) or display X-linked dominant effects (SBMA, FXTAS). OPMD is a special case that normally shows autosomal dominant inheritance, but can, in individuals homozygous for the largest normal-sized repeat, follow an autosomal recessive pattern.
Table 3.
Primary mechanism of disease
| Toxic gain of function |
| • CAG/polyQ diseases |
| • Spinocerebellar ataxia types 8, 10, 12, 31, 36 |
| • C9ORF72 frontotemporal dementia/ amyotrophic lateral sclerosis |
| • Huntington disease-like 2 |
| • Myotonic dystrophy types 1 and 2 |
| • Oculopharyngeal muscular dystrophy |
| • Fragile X tremor ataxia syndrome |
| • Fuchs endothelia corneal dystrophy |
| Loss of function |
| • Friedreich Ataxia |
| • Fragile X syndrome |
| • Myoclonic epilepsy (Unverricht-Lundborg) |
Table 4 lists six factors that contribute to the pathogenic mechanisms underlying repeat expansion diseases. For example, the sequence of the repeated unit (e.g. CAG vs. CTG vs. GAA, etc.) influences the intramolecular structures that can be adopted by the repeat (e.g. 22), which in turn can influence transcription, translation, and binding to various RNA binding proteins. GC-rich sequences are more prone to adopt highly stable intramolecular folds that bind RNA-binding proteins (eg. DM1, FXTAS, C9ORF72, SCA36) in ways that disrupt splicing of essential genes. GC-rich sequences also are prone to hyper-methylation that can silence the gene (e.g. FXS, FRAXE). In contrast, the GAA repeat of Friedreich ataxia is prone to form DNA-RNA triplex structures that impede transcription, leading to marked reduction in expression of the encoded protein, frataxin. For repeats within protein-coding regions, the precise nucleotide sequence specifies the amino acid produced in the protein, with profound implications for the cause of disease. For CAG, it is glutamine, hence the name polyglutamine diseases, and for GCG, it is alanine which affects the behavior of the encoded protein in OPMD.
Table 4.
Factors contributing to pathogenic mechanisms of repeat expansion diseases
| • Sequence of repeat |
| • Size of repeat |
| • Location of repeat within gene |
| • Whether repeat encodes RNA or protein |
| • Function of repeat-containing gene |
| • Extent of meiotic and somatic instability |
As already discussed, in nearly all cases the size of the expansion in a given disease is the major driver of disease features and severity. But across the various diseases, the size of disease-causing expansions ranges from as small as 8–18 repeats in OPMD and only ~35–80 in most CAG/polyglutamine diseases, to as large as more than a thousand repeats in DM1, DM2, SCA10 and SCA36. The smallest disease-causing repeats are probably constrained because they reside in protein-coding regions of the gene. Even rather small changes to the amino acid repeat (e.g. polyglutamine, polyalanine) in the encoded disease protein will greatly affect the way it behaves (e.g. 124,147). The largest disease-causing repeats, in contrast, are found in introns or untranslated regions; in these regions there may be less constraint on the permissable size and a less clear-cut correlation between repeat length and disease correlation.
A fascinating example of the importance of repeat size is provided by FXS and FXTAS -- two distinct diseases that occur at the same repeat, either with a very large expansion (FXS) or a much smaller expansion (FXTAS). The CGG repeat that underlies both diseases resides in the promoter of the FMR1 gene. Males with FXS, the most common inherited form of mental retardation, have very large CGG expansions that become hypermethylated, effectively silencing FMR1 expression (12,103,130). Because the FMR1 protein is important for brain development and regulates synaptic activity, its absence leads to mental retardation, among other features of FXS. The full-blown CGG expansion in a FXS male is inherited from the mother, who in turn inherited a smaller repeat from her father (i.e., the maternal grandfather of FXS males). The grandfather’s much smaller CGG repeat is not hypermethylated and does not silence expression of the gene; in fact, it drives even stronger transcription at the FMR1 locus. The resultant CGG-containing RNA transcript is neurotoxic in two ways: 1) it forms RNA foci that sequester critical RNA splicing factors; and 2) it drives the aberrant production of aggregation-prone proteins across the CGG repeat through the recently discovered process of Repeat Associated Non-methionine translation, or RAN translation (26,50,137). As a result, maternal grandfathers in families with FXS frequently develop a late-life neurodegenerative syndrome characterized by tremor, ataxia and cognitive impairment, known as FXTAS (53). In short, differing sizes of the very same repeat fundamentally alter the way the sequence behaves and affects the brain in FXS versus FXTAS. The story of FXTAS and FXS is also a compelling illustration of clinical anticipation: A repeat that further enlarges across three generations of a family, going from causing a late-life neurodegenerative disease in the first generation to an early life neurodevelopmental syndrome in the third generation.
Precisely where an expanded repeat lies in a gene also influences how it affects the gene and thereby the molecular mechanism of disease. Large repeats in or near a promoter can profoundly influence expression of the gene as illustrated with FXS above. Repeats within introns can influence alternative splicing of the disease gene, globally perturb splicing by sequestering splicing factors, or perturb transcription (13,148,151). Repeats within the 3’UTR likewise can have a toxic RNA effect by disrupting RNA homeostasis (151). Finally, repeats within the protein-coding region of the gene are directly translated into protein, leading to altered properties of the encoded disease protein, with deleterious consequences for the nervous system.
The formally clean separation (107) of expanded repeats into those that are translated into protein and are exclusively neurodegenerative (“type I” repeat expansion diseases) versus those that are not translated to protein and are multisystem disorders (“type II” repeat expansion diseases), has broken down recently with the discovery of RAN translation. We now recognize that repeats in traditionally non-translated regions of the transcript (5’UTR, introns) can be subjected to RAN translation, resulting in the production of toxic protein species (26,152). Adding to this complexity is the discovery that many genes are transcribed in both the sense and antisense directions. As a result, a CAG repeat in one direction becomes CTG in the other (and vice versa), and GGGGCC becomes GGCCCC. These are not hypothetical examples, as there is emerging evidence for bidirectional expression in CAG/polyglutamine repeat diseases, the CTG repeat disease SCA8, and C9ORF72-mediated FTD/ALS (e.g. 51,52). In general, less antisense transcript is produced than sense transcript, but even a small amount of the alternative transcript could have profound effects. Some evidence suggests antisense transcript production may even increase in the disease state, aggravating the problem (e.g. 51).
For a given repeat expansion disease, the function of the gene product and how repeat expansion influences it expression are critically important in disease pathogenesis. In the case of Friedreich ataxia (20), the GAA expansion reduces expression of frataxin, a protein implicated in mitochondrial function. Individuals homozygous for the expansion have markedly reduced levels of frataxin, resulting in impaired mitochondrial function, oxidative stress in the nervous system and heart, and the symptoms of ataxia, sensory loss and cardiomyopathy that are so characteristic of the disorder. A small fraction of Friedreich ataxia patients are compound heterozygotes, having a single GAA expansion with an inactivating mutation or deletion in the second allele (4,45,55). This point underscores the fact that Friedreich ataxia is, without question, a loss of function disorder due to deficiency of frataxin, a protein critically important for mitochondrial function in neurons and cardiomyocytes. In contrast, in DM1 the expanded repeat does not greatly alter expression of the encoded protein known as DM protein kinase. It does, however, profoundly perturb normal RNA splicing in skeletal and cardiac muscle and, to a lesser extent, in the brain. The expanded repeat-containing DM1 transcript sequesters vital splicing factors in muscle, including muscleblind (69,110), and the resultant failure to generate appropriately spliced muscle gene products causes the myotonia so characteristic of the disease. A third illustration of the importance of the affected gene product is the entire class of CAG/polyglutamine disorders; as discussed further below, the clinical and pathological differences across this class of disorders reflect the specific protein context in which the expanded polyglutamine tract occurs in each disease.
The extent of meiotic instability for a given repeat determines the extent of anticipation for a repeat expansion disorder. The degree of meiotic instability depends on numerous factors including the size of the repeat (38,44), whether the repeat is pure or interrupted (24), the sequence of the repeat, and the positioning of the repeat relative to nucleosomes and other chromosomal factors. Compared to meiotic instability, it has been more difficult to gauge the role of somatic instability in repeat expansion diseases, though it clearly occurs in numerous disorders including HD and DM1 (e.g. 5,134). Emerging evidence in the CAG/polyglutamine diseases supports the view that somatic instability within the nervous system contributes significantly to disease severity (131). Moreover, the longer the repeat, the greater the likelihood of somatic instability (145). DNA repair factors have been implicated as potential genetic modifiers of age of onset in Huntington’s disease (49) and possibly other CAG/polyglutamine diseases (67). Perhaps polymorphisms in specific DNA repair factors modulate the degree of somatic instability and thereby influence disease severity (91,120). In SCA2, individuals in whom the CAG repeat contains CAA interruptions can manifest with dominantly inherited parkinsonism rather than disabling ataxia; this clinical difference may reflect a differing degree of somatic instability with pure versus interrupted CAG repeats.
The Diseases
Here I briefly describe individual diseases or groups of related repeat expansion diseases, presented roughly in the chronological order of their discovery. I remind the reader that this neurogenetics volume includes definitive chapters that provide much more information on specific diseases. I also refer the reader to GeneReviews for up-to-date reports on nearly all repeat expansion diseases.
Fragile X syndrome (FXS), fragile X tremor ataxia syndrome (FXTAS), and other fragile X sites
FXS is named after the folate-sensitive fragile site at the FRAXA locus on the X chromosome. The most common cause of inherited mental retardation, FXS typically affects males, varies greatly in severity, and is associated with dysmorphic features including enlarged head, ears and testicles (see chapter 25 in this volume for a more thorough description). Scientists were puzzled for years that the risk of FXS increased from one generation to the next. Indeed, this particular example of anticipation carried its own name, the Sherman paradox. The discovery in 1991 that FXS and its underlying fragile site are caused by an expanded CGG repeat that changes size over generations explained the paradox. Normal-sized repeats are polymorphic, ranging from 6 to 52, with repeats at the high end of this range being increasingly prone to further expand (“mutable normal”). In FXS families the repeat sizes span a wide range, from “premutations” of ~60–200 repeats (typically found in maternal grandfathers) to full mutations of several thousand repeats (found in affected FXS males). The mothers of affected FXS males have variably sized expansions and are prone to premature ovarian failure.
As described earlier, the molecular mechanism of FXS is a loss of expression of the developmentally important nervous system protein, FMRP. Full expansions promote hypermethylation of the FMR1 promoter and reduce translation of the transcript, effectively silencing expression the gene (12,39,103,130). Further supporting a loss of function as the basis of disease is the fact that some FXS is caused by inactivating mutations rather than a CGG expansion (33,47).
Maternal grandfathers of affected FXS males, who typically harbor a premutation allele, are at risk for developing FXTAS (53). This late-life neurodegenerative disorder is characterized by progressive ataxia, essential-type tremor, cognitive impairment and occasionally parkinsonism. Brain imaging often shows T2-weighted signal abnormalities in the middle cerebellar peduncles as well as more generalized brain atrophy. In males with late-life, progressive cognitive impairment, imbalance and incoordination who have a grandson with mental retardation, screening for the FXS/FXTAS repeat expansion should be considered.
In FXTAS, the premutation does not silence the gene but instead permits continued expression of the FMRP transcript. The transcript may even accumulate to higher levels than normal, and is thought to cause disease through a combination of toxic RNA and toxic protein effects linked to RAN translation (137), as discussed earlier.
Numerous other fragile sites are caused by GC-rich trinucleotide expansions, in some cases associated with mental retardation. These include FRAXE mental retardation (GCC), as well as the FRAXE (GCC) and FRA16A (CCG) fragile sites (e.g. 72, 106).
Myotonic dystrophy types 1 and 2 (DM1, DM2)
Myotonic dystrophy is an autosomal dominant multi-system disease characterized principally by myotonic myopathy. There are two major forms of myotonic dystrophy, both caused by repeat expansions. DM1, also known as Steinert disease, is caused by a CTG expansion in the 3’UTR of the DMPK gene (7,16,43,54). DM2, which is much less common than DM1 and was previously known as proximal myotonic myopathy, is caused by a CCTG repeat in intron 1 of the CNBP gene (formerly named ZNF9)(84). Despite their similarities, DM1 and DM2 differ in important molecular and clinical respects. Most importantly, DM1 shows robust repeat length/disease severity correlation as well as significant anticipation, whereas DM2 does not.
DM1 is characterized by progressive weakness and myotonia, often associated with cataracts, cardiac arrhythmias, endocrinopathy and cognitive impairment (32). The range of severity is broad, with differences in repeat length being the key driver of disease severity. Table 5 depicts the relationship between CTG repeat length, age of onset and disease severity, including mild, classic and congenital forms of disease. “Mild” disease may manifest simply with premature cataracts and baldness, with electromyographically detectable myotonia. “Classic” disease typically manifests in young adulthood and includes distal weakness, symptomatically and often disabling myotonia, as well as significant cardiac conduction defects in addition to cataracts and baldness. Classic disease, when presenting in teen years, is also known as “juvenile” disease. “Congenital” DM1, in which the affected parent is nearly always the mother, is present at birth. The infant is floppy, facial and jaw muscles are weak resulting in failure to thrive, and mental retardation and development delay are common. Rather than displaying myotonia, congenital DM muscles display features of arrested fiber development. Table 5 highlights that that some unaffected individuals have repeats in the “mutable normal” range of 35–49 repeats. Such metastable alleles are prone to expand when transmitted to the next generation; new mutations in families arise through this process. Table 5 also illustrates overlap in the range of repeat lengths across these various classes. An important, life-threatening feature of DM1 is cardiac involvement which can lead to sudden cardiac death. Repeat length and cardiac abnormalities also are correlated in DM1 (95). Genetic testing for the CTG expansion in DM1 is relatively straightforward. In less than 5% of DM1 patients, the CTG repeat is interrupted by other trinucleotides but the clinical and genetic significance since of these interruptions is unknown (68).
Table 5.
CTG repeat length/phenotype correlation in myotonic dystrophy type 1
| Phenotype | Clinical Features | Repeat Size | Age of Onset |
|---|---|---|---|
| Mutable normal | None | 35–49 | NA |
| Mild | Cataracts Mild myotonia |
50-~150 | 20–70 years |
| Classic | Weakness Myotonia Cataracts Balding Cardiac arrhythmia |
~100-~1000 | 10–30 years |
| Congenital | Infantile hypotonia Respiratory deficits Intellectual disability Classic signs develop later |
~700- >1000 | Birth to 10 years |
DM2 commonly presents as proximal muscle weakness with variable myotonia, hence its former name proximal myotonic myopathy (121). Like DM1, it too shows marked clinical heterogeneity ranging from mild forms of disease that may be difficult to detect, to profound and disabling proximal muscle weakness. There is no congenital form of disease nor is there apparent anticipation. Cardiac involvement is less common in DM2 than in DM1 (118), but still requires careful monitoring. Whereas in DM1 cognitive impairment is well described (19,46), DM2 shows much less cognitive involvement (109). The CCTG repeat expansion in DM2 is complex, including repeat elements in addition to the CCTG repeat, and is prone to an extreme range of pathogenic expansions, from 75 units to as many as 11,000 units (mean of roughly 5000 repeats) (68). Despite this broad range, there is little evidence for correlation between repeat length and disease severity; reasons for this lack of relationship are currently unknown.
The molecular mechanism of disease may be as well worked out for DM1 as it is for any repeat expansion disease. The CTG expansion resides in the 3’UTR of the DMPK transcript, where it does not alter expression of the disease protein, but does form RNA foci and bind to and sequester essential splicing factors (69,110). This toxic RNA effect leads to a failure to generate appropriately spliced isoforms of key muscle genes, leading to myotonia and other symptoms of disease. Loss of DMPK function is well tolerated in mouse models (21) and thus nucleotide-based gene-silencing approaches as potential disease-modifying therapy represent an attractive strategy for this disabling disorder (136). The pathogenic basis of DM2 is less clear, but leading candidates include a toxic RNA effect (e.g. 89).
CAG/polyglutamine diseases
At least nine diseases belong to the CAG/polyglutamine group (11,58,70,74,77,99,105,114,122). These diseases are thoroughly described in four chapters in this volume: the polyglutamine diseases as a whole (chapter 11); SBMA (chapter 38); HD (chapter 17); and the SCAs (chapter 12). Here, I focus on shared features across the class as well as distinctive findings in specific disorders that shed light on disease mechanisms.
All nine are dominantly inherited disorders except for SBMA which is an X-linked disorder with dominant toxic features. All are classified as rare diseases. HD, the best known among them, is also the most common, with SCA3 next in line. Six are dominantly inherited ataxias (also known as SCAs) including the four most common SCAs among the 40 discovered thus far (SCAs 1,2,3,6). A seventh disorder, DRPLA, can be thought of as a hybrid between SCA and HD. In all nine, the primary pathogenic mechanism is believed to be proteotoxicity emanating from the encoded disease protein (see Table 6). Other than sharing a common glutamine repeat, the various disease proteins are entirely unrelated and serve very different cellular functions. The distinctive clinical and pathological features of individual CAG/polyglutamine diseases are believed to stem primarily from this differing protein context. At least two other repeat expansion diseases may share elements with the polyglutamine diseases: In SCA8, the antisense transcript can encode a polyglutamine protein through RAN translation, and the CAG repeat in SCA12 theoretically could encode polyglutamine though evidence supporting this is less clear.
Table 6.
Polyglutamine diseases and their encoded proteins
| CAG/Polyglutamine Diseases | Encoded protein(s) | Function |
|---|---|---|
| Huntington disease | Huntingtin | Scaffold protein associated with autophagy |
| Spinal and bulbar muscular atrophy | Aandrogen receptor | Hormone-dependent transcription factor |
| Dentatorubral-pallidoluysian atrophy | Atrophin-1 | Transcription cofactor |
| Spinocerebellar ataxia type I | ATXN1 | Transcription cofactor |
| Spinocerebellar ataxia type 2 | ATXN2 | RNA binding protein implicated in RNA homeostasis |
| Spinocerebellar ataxia type 3 | ATXN3 | Deubiquitinase |
| Spinocerebellar ataxia type 6 | CACNA1A subunit (Cav2.1)/α1ACT | Calcium channel subunit/transcription factor |
| Spinocerebellar ataxia type 7 | ATXN7 | Component of SAGA acetyltransferase complex |
| Spinocerebellar ataxia type 17 | TATA Binding Protein | General transcription factor |
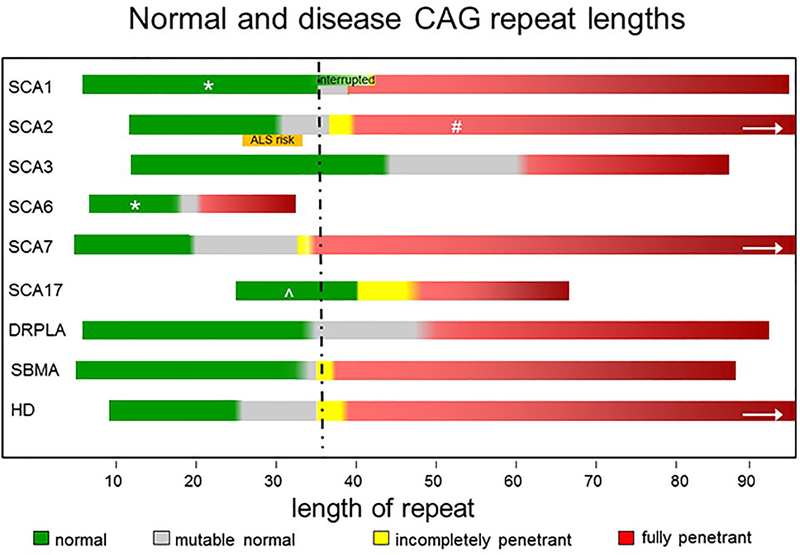
Figure 3 compares the normal and disease repeat lengths for the CAG/polyglutamine diseases. Several important features can be gleaned from the comparison. First, a repeat length of roughly 35–40 is a common disease threshold across polyglutamine disorders. This repeat size conforms to in vitro biochemical studies showing that polyglutamine proteins begin to aggregate at roughly the same threshold. This is a relevant point because aggregation of the disease protein, particularly in neurons, is a common pathological hallmark of the polyglutamine disorders. There are, however, important exceptions to this repeat length threshold. For example, SCA6 is caused by an expansion well below this threshold; it is also, however, the only CAG/polyglutamine disease in which the encoded protein is a trans-membrane protein, a calcium channel subunit. Tethered close to the membrane, the glutamine repeat in SCA6 may be particularly sensitive to small changes in length. Recently, a second polyglutamine-containing protein was discovered to be expressed from the SCA6 locus, a transcription factor known α1ACT, raising another possibility for the basis of a shorter disease threshold: potentially profound effects of modest repeat expansions on transcription function.In contrast, for SCA3 and SCA17, normal repeats exist that are even larger than the putative threshold of 35. The respective disease proteins in SCA3 and SCA17 are a deubiquitinase implicated in protein quality control (ATXN3) and an essential general transcription factor (TATA binding protein). Whether these proteins possess special properties that would permit a larger normal polyglutamine tract remains to be determined.
Figure 3. Normal and disease repeat lengths for the CAG/polyglutamine diseases.
Normal (green), incompletely penetrant (yellow) and fully penetrant (red) repeat lengths are shown for each disease. Deepening color of red illustrates increased disease severity and earlier age of onset with longer repeat lengths. Arrows illustrate that the longest disease repeats in spinocerebellar ataxia type 2 (SCA2), SCA7 and Huntington disease are >100 repeats. Gray regions represent size range of intermediate length repeats that may be prone to further expansion. High normal SCA2 alleles (orange) are a risk factor for ALS. At SCA1 locus, normal repeats can be longer when they are interrupted by CAT residues. DRPLA, dentatorubral-palliodoluysian atrophy; SBMA, spinobulbar muscular atrophy
* Normal repeat length in SCA1 and SCA6 are modifiers of age of onset, and SCA6 normal repeat length is also a modifier of age of onset in SCA2.
# In SCA2 disease repeats interrupted by CAA can be associated with dominantly inherited Parkinson disease rather than ataxia.
^ Normal repeat in SCA17 is an imperfect CAG repeat interrupted with CAA residues.
A second feature for many CAG/polyglutamine diseases is a narrow range of modestly expanded repeats for which disease is incompletely penetrant (colored yellow in figure 3). Clearly, factors other than the repeat length itself influence whether disease will become manifest. A particularly telling example is SCA1, where perfect CAG repeats begin to cause disease at approximately 40 repeats, whereas CAG repeats interrupted by a histidine-coding CAT can extend several repeats further before causing disease. The simplest hypothesis (not proven) is that the disease protein is stabilized when histidine interrupts the polyglutamine tract. For all polyglutamine diseases, in the fully penetrant repeat range, the smaller the expansion the wider the predicted potential age of symptom onset. In HD, for example, individuals with 40 repeats may experience their first disease symptoms in the 30s or 70s, whereas individuals with repeat length of 80 would always show signs of disease before age 30. Careful genetic analysis of HD individuals whose repeat lengths are near the lower end of the pathogenic range has identified additional genetic modifiers that influences age of onset (49).
Understandably, the study of polyglutamine disorders has focused on the expanded repeat, but insights have also been gained from studying the normal repeat. In both SCA6 and SCA1, the size of the normal repeat contributes modestly to age of onset (135a and b), and the size of the normal SCA6 repeat may be a modifier of disease onset in SCA2 (111). Moreover, at the SCA2 locus, repeats at the high end of the normal range are an important genetic risk factor for ALS. Intriguingly, some SCA2 expansions can manifest with ALS (116,143) and SCA2 itself is characterized by motor neuropathy in addition to progressive ataxia. The SCA2 protein, ATXN2, is an RNA binding protein implicated in stress granule dynamics and RNA/protein homeostasis, which are critically important pathways in ALS and presumably also in SCA2. This discovery underscores the point that, even in the normal range, repeat length variation influences the behavior of polyglutamine disease proteins.
Other repeat expansion SCAs
At least four other SCAs are caused by repeat expansions. One of these, SCA12, is caused by a CAG expansion that may or may not encode polyglutamine, and at least one other, SCA8, can generate polyglutamine from the antisense strand.
Spinocerebellar ataxia type 8 (SCA8)
Accounting for approximately 2–5% of all dominant ataxia, SCA8 typically manifests as a relatively mild cerebellar ataxia that is compatible with a normal life span, though the phenotype varies considerably (31,60). The disease-associated CTG repeat (76) resides in a highly polymorphic locus transcribed in both directions, and the complexity at this locus may help explain why the SCA8 repeat expansion is not fully penetrant. The CTG repeat is flanked by a polymorphic CTA repeat and the combined CTG/CTA length is usually reported in genetic test results. The normal repeat length is 15–50 triplets, with pathogenic expansions typically in the range of 80–250 triplets, though affected individuals have been described with repeats spanning from 71 to over 1300. Because repeat expansions across this broad range are incompletely penetrant, physicians should exercise caution in concluding that an expansion at the SCA locus is necessarily the cause of a patient’s ataxia. This incomplete penetrance also means that most ataxic individuals identified to have the SCA8 expansion are sporadic cases without a family history of dominant ataxia. Although the basis of the incomplete penetrance remains uncertain, scientific investigations of the SCA8 locus have shed important light on mechanisms of repeat expansion disease. Evidence exists at this locus for both toxic RNA and toxic protein effects as disease mechanisms (30, 152). The locus is transcribed in both directions (97), and RAN translation can generate multiple homopolymeric proteins, including an expanded polyglutamine protein from the antisense (CAG) strand (152).
Spinocerebellar ataxia type 10 (SCA10)
SCA 10 is a slowly progressive, relatively pure cerebellar ataxia that is often accompanied by generalized seizures (132, 133). The mutation is a pentanucleotide ATTCT expansion (93) residing in an intron of the ATXN10 gene. SCA10 is particularly prevalent among individuals of Amerindian origin in Mexico and Brazil. Intriguingly, the frequency of seizures, which usually manifest after ataxia has begun, is much greater among Mexicans (~60% with seizures) than Brazilians (less than 5% with seizures). This phenotypic difference may reflect population differences in the extent of interruptions in the highly complex disease repeat (94). The ATTCT repeat normally is 10–32 units in length, whereas fully penetrant alleles are between ~ 800 and 4,500 repeats in length. Incompletely penetrant repeats of 282 to ~ 800 have also been reported. The massive size of the repeat requires southern blot analysis to determine repeat length. There is a modest inverse correlation of repeat length to age of onset, accounting for approximately one third of the variation in age of onset, and anticipation has been described with paternal transmission (92). The primary disease mechanism is still under investigation but may be a toxic RNA effect (151).
Spinocerebellar ataxia type 12 (SCA12)
SCA12 is a rare, dominantly inherited form of ataxia that often begins with an action tremor of the arms resembling essential tremor (56,104). Disease typically begins in adulthood and slowly progresses to involve mild ataxia and spasticity. SCA12 is most common in persons of Indian descent but has also been described in persons of European descent (10,17,128). The CAG repeat expansion occurs within or just upstream of an exon in the PPP2R2B gene which encodes a phosphatase important in the nervous system. Normal repeats are between 4 and 32 triplets, whereas fully penetrant disease-causing expansions are 66 triplets or longer. Several individuals have been reported with expansions between ~40 and 66 suggesting that alleles in this range have reduced penetrance and may cause disease. Whether the CAG repeat encodes polyglutamine or instead effects expression of specific splice variants of the encoded phosphatase, remains uncertain.
Spinocerebellar ataxia type 31 (SCA31)
SCA 31 is a relatively pure cerebellar ataxia with a mean onset at approximately 60 years of age and compatible with a normal life span (100). Originally described in Japan, where it is the third most common form of SCA, SCA31 has since been discovered in other ethnic populations (64,119). It is associated with a complex, large insertion that contains multiple repeated nucleotide sequences, most notably a pentanucleotide TGGAA. A 1.5–2 kilobase insertion lacking the TGGAA pentanucleotide repeat is occasionally seen in control individuals, but insertions that include the pentanucleotide repeat are found in individuals with disease. The insertion lies within an intron found in two genes encoded in opposite directions, the TK2 (thymidine kinase 2) and BEAN (brain expressed, associated with NEDD4) genes. Thus, the encoded pentanucleotide can be either TGGAA or UUCCA. Emerging evidence supports a dominant toxic gain of function mechanism that may include both RNA toxicity and the production of RAN peptides (63,101). Cerebellar Purkinje cells are selectively vulnerable and show distinctive pathological features including changes in nuclear morphology and the accumulation of halo-like amorphous material in Purkinje cells (150).
Spinocerebellar ataxia type 36 (SCA36)
This slowly progressive cerebellar ataxia typically begins in mid-life, involving balance problems followed later by dysarthria and limb ataxia (61). Sensorineural hearing loss usually occurs and less often motor neuropathy can be a disabling feature. Cerebellar symptoms are initially midline, affecting gait and balance. Consistent with this, initial brain atrophy occurs in the superior vermis of the cerebellum, but as disease progresses atrophy extends throughout the cerebellum and into the brainstem. The disease is caused by a large hexanucleotide (GGCCTG) repeat expansion in the first intron of the NOP56 gene (73). Normal repeats are usually between 3 and 14, whereas in individuals with disease the expansion is at least 650 repeats extending to more than 2000 repeats. A few individuals have also been described with small expansions (only 20–35 units), though the molecular basis for this is uncertain (102). The pathogenic mechanism is also uncertain but RNA foci containing the repeat have been described, suggesting a toxic RNA effect (85).
Huntington disease-like 2 (HDL2)
Families with a dominantly inherited disease resembling HD (chorea, cognitive impairment and psychiatric disturbance) may instead have HDL2 (2). This rare phenocopy of HD is caused by a CTG repeat expansion in the Junctophilin-3 (JPH3) gene (57). Normal repeats are between 6 and 28, whereas expanded repeats are between ~41 and 58 repeats. Disease typically occurs in midlife and recapitulates many features of HD, with weight loss being a frequent finding. Similar to HD, some individuals with HDL2 can present with juvenile onset disease resembling the Westphal variant of HD (rigidity, parkinsonism, dystonia). The brain MRI often resembles that of HD, showing selective atrophy of the basal ganglia and cortex with relative sparing of the brainstem and cerebellum. The diagnosis of HDL2 cannot be established without molecular genetic testing for the repeat expansion. The pathogenic mechanism remains uncertain and, as with other repeat expansion diseases, may have multiple components. Located in an alternatively spliced exon of the JPH3 gene, the repeat can be transcribed in both directions, leading to CUG (more common) or CAG (less common) repeat-containing transcripts. While a dominant RNA toxic effect may occur, the repeat expansion also reduces levels of the Junctophilin-3 protein (123), which could prove deleterious to neurons.
Friedreich ataxia
Friedreich ataxia is the most common autosomal recessive ataxia, present primarily in Indian and European populations. Before the disease mutation was discovered, Friedreich ataxia was defined as a young onset progressive ataxia with sensory loss, scoliosis, areflexia and cardiomyopathy occurring before age 25. Other disabling features of disease include hearing loss, motor weakness, and diabetes. The discovery of a GAA repeat expansion in the FRDA gene (20) soon led to the recognition that the classic definition of disease, requiring onset before age 25, was incorrect. While most Friedreich ataxia meets this classic definition, roughly a quarter of individuals develop signs of disease after age 25 (e.g. 37a). Moreover, late onset Friedreich ataxia may not show the classically described areflexia and is less likely to have significant cardiac involvement. Most affected individuals are homozygous for the expansion but a small percentage are compound heterozygotes who have an inactivating or deletion mutation in one allele and an expansion in the other allele (4,45,55). There is robust repeat length-disease correlation, with the size of the smaller of the two expansions showing inverse correlation with age of symptom onset and a direct correlation with the probability of significant cardiac dysfunction (37a,41,96). Occasionally, however, disease features can vary widely within a family despite similar sized expansions (9) indicating that other factors influence disease severity beyond the mutation.
The basis of disease is impairment in mitochondrial function due to loss of frataxin, a protein required for the assembly of iron-sulfur cluster enzymes in the mitochondria. Frataxin fails to be expressed principally because the GAA expansion directly impedes transcription, although a contributing factor is expansion-induced epigenetic silencing of the upstream promotor (13,25,82). Aggressive, ongoing efforts to develop preventive therapies include gene replacement strategies, compounds to boost frataxin expression and anti-oxidant approaches, although no compound has yet been proven to slow the disease (71). Friedreich ataxia can be viewed as a mitochondrial disorder with all the attendant neurologic problems seen in mitochondrionopathies. Aggressive clinical management (28) should include close attention to cardiac status, as cardiac abnormalities are the most common cause of death. Annual evaluations should include an EKG and echocardiogram, with referral to cardiology for patients showing abnormalities (87).
Unverricht-Lundborg myoclonic epilepsy (EPM1)
EPM1 is the most common cause of myoclonic epilepsy in North America, typically beginning between 6 and 15 years of age and progressing over time (29). The initial symptom can be either action- or stimulus-induced myoclonus or generalized tonic-clonic seizures, but eventually both are present in affected persons. Ataxia also is a common feature. The EEG shows photosensitive spike and wave abnormalities and the background can be slowed. While cognition is generally normal, mild intellectual deficits may develop over time. The myoclonus is progressive and can be very disabling, leading to wheelchair use for approximately one third of affected individuals.
This autosomal recessive disease is caused by expansion of a dodecamer repeat in the CSTB gene which encodes the lysosomal protein cystatin B (8,65,78, 146). Normal repeats are 2–3 units in length and expansios srange from 30 to ~125 repeats. Most persons with EPM1 will be homozygous for expansions though a small percentage will have an activating mutation on one allele and an expansion on the other. While some evidence supports a correlation between repeat length and disease severity, this correlation is less robust than in many other repeat expansion diseases (e.g. 79).
Oculopharyngeal muscular dystrophy (OPMD)
OPMD is a dominantly inherited neuromuscular disorder characterized by adult onset progressive weakness, ptosis, ophthalmoparesis and dysphagia (86,139). The cause is a small GC(N) expansion in the polyadenylate binding protein 2 (PABP2) gene that modestly enlarges a polyalanine tract in the protein (15). OPMD is one of at least nine polyalanine diseases (125), the remainder of which are congenital neurocognitive disorders in which the expansions occur in transcription factors. In contrast, OPMD is an adult onset, progressive, degenerative disease. Reminiscent of the CAG/polyglutamine diseases, OPMD is a proteinopathy: the enlarged alanine tract promotes aggregation of the disease protein, resulting in the formation of intranuclear inclusions in skeletal muscle (124).
The normal GC(N) repeat length is typically 6 units and expansions are between 8 and 18 in disease. The GC(N) expansions can be either GCG or a mixture of GCG and GCA, both of which encode alanine. A distinctive feature of OPMD is that while most individuals possess a single expanded allele, some affected persons are compound heterozygotes with one allele containing 7 repeats and other 9 repeats. Remarkably, a small percentage of OPMD presents in an autosomal recessive manner wherein affected individuals are homozygous for alleles of 7 repeats. Evidence does not support anticipation in OPMD, but there is some support for a correlation between repeat length and disease severity (113).
As in the CAG/polyglutamine diseases, subtle changes in the size of the repeated amino acid tract can have major consequences for the polyalanine-containing disease protein (e.g. 81). PABP2 is a multifunctional protein implicated in transcriptional regulation, polyadenylation of mRNA, and nucleocytoplasmic transport. Its sequestration into inclusions in skeletal muscle likely both depletes the cell of an important nuclear factor and sequesters other vital proteins. As in other repeat expansion diseases, several molecular mechanisms could simultaneously contribute to the disease process.
Fuchs endothelial corneal dystrophy (FECD)
FECD is included here among the neurological repeat expansion diseases because it affects vision, is relatively common, and is one of the most recently described repeat expansion diseases. FECD is a degenerative condition characterized by progressive loss of corneal endothelium, thickening of the Descemet’s membrane and deposition of extracellular matrix in the cornea. This process results in progressive corneal edema and visual loss, typically after age 60. At least five other genes or genetic loci are associated with FECD (1), but the most common form - a late onset form – is associated with modest expansion of an intronic CTG repeat in the transcription factor four (TCF4) gene (83,127,149). Normal CTG repeats are between 10 and 37, and pathogenic repeats are greater than 50. Little is known about how the expansion contributes to disease, but the current leading hypothesis is a toxic RNA effect (148).
C90RF72 FTD/ALS
In the 6 years since a GGGGCC (or G4C2) hexanucleotide expansion in the C9ORF72 gene was discovered to cause familial ALS and FTD (112), our understanding of the ALS/FTD disease spectrum has advanced tremendously (27,42,48). The “C9 story” is one of rapid discoveries and unexpected biological convergences. Advances that have been made possible by the discovery of the G4C2 expansion have led to emerging view that perturbation of RNA homeostasis in neurons is a major cause of neurodegenerative disease, whether ALS or FTD.
An autosomal dominant disease, C9ORF72-mediated neurodegeneration can present as ALS or FTD even in the same family (23, 88,142,144). It is by far the most common form of familial ALS, accounting for about a third of ALS families and 5–10% of sporadic cases in an ALS clinic. It is also a common cause of FTD, explaining about one fourth of familial FTD. Age of symptom onset ranges from 30 to 70 years of age with a mean onset in the late 50s. C9-mediated ALS most often resembles typical ALS, can be bulbar or limb onset, can progress rapidly (though not always) and can be associated with later cognitive symptoms. Thus, C9-mediated ALS should be evaluated and treated just as in any ALS patient. The pattern of C9-mediated FTD most commonly is behavioral variant FTD, with the full range of behavioral and cognitive symptoms including disinhibition, apathy and executive dysfunction. Less commonly, C9-mediated FTD presents semantic variant primary progressive aphasia (PPA) or nonfluent variant PPA, and, very rarely, can resemble corticobasal syndrome, progressive supranuclear palsy or an HD-like syndrome. Occasionally parkinsonian features are seen in C9-mediated ALS or FTD.
Normal G4C2 repeats are ~25 units or less, and high penetrance disease alleles are typically greater than ~60 repeat units, ranging up to more than 4,000 units; rarely, repeats between 47 and 60 segregate with disease in families. This pure GC repeat poses technical challenges for genetic screening especially as a larger expansion (144). A repeat-primed PCR assay is typically used to detect smaller expansions (<80), but accurately sizing larger repeats requires other techniques (e.g. Southern blot hybridization) that only provides a rough estimate of length. Even then, extensive somatic mosaicism at the locus means that repeat lengths estimated from blood may not accurately reflect repeats in vulnerable brain tissue.
Perhaps in part because of difficulties in accurately defining disease repeat length, C9-mediated disease differs from most other repeat expansion diseases in that scientists have not established a clear correlation between repeat length and disease severity (e.g. 142, 144). There are, however, potential biological explanations for this lack of correlation, including the fact that the expansion is prone to variable hypermethylation which will affect expression of the gene and presumably disease expression (117). Epigenetic effects also may explain why some individuals with C9 expansions remain asymptomatic until very late in life.
The RNA binding protein TDP43 accumulates in neuronal aggregates in C9-mediated FTD and ALS, placing C9-mediated neurodegeneration into the class of TDP43 proteinopathies. A wealth of genetic, animal modeling and biochemical studies have rapidly shed light on the molecular mechanism of this disease. Though initial studies revealed modest reduction in expression of the C9ORF72 gene product (a protein of still uncertain function), it now seems clear that a partial loss of gene function, or haploinsufficiency, is not a major component of disease pathogenesis. Instead, compelling cases have been made both for RNA toxicity (the formation of RNA foci, sequestration of critical RNA-binding proteins and aberrant splicing) and proteotoxicity (RAN translation from both the sense and antisense transcripts, generating five distinct dipeptide proteins that vary in toxicity and propensity to aggregate) (14,34,35,42,48,80). Very likely, both pathways contribute to disease though many questions remain unanswered. For example, if dipeptide species are an important toxic element, why do they tend to accumulate in certain brain regions that are not particularly vulnerable to disease? The remarkably fast progress in this repeat expansion disease gives one confidence that answers will be found. In any case, enough is already known to justify aggressive ongoing efforts to use nucleotide-based or pharmacological strategies to reduce levels of the expanded repeat-containing transcripts as potential disease-modifying therapy.
References
- 1.Afshari NA, Igo RP Jr, Morris NJ, et al. (2017). Genome-wide association study identifies three novel loci in Fuchs endothelial corneal dystrophy. Nat Commun 8:14898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson DG, Walker RH, Connor M, et al. (2017). A systematic review of the Huntington disease-like 2 phenotype. J Huntingtons Dis 6:37–46. [DOI] [PubMed] [Google Scholar]
- 3.Andrew SE, Goldberg YP, Kremer B, et al. (1993). The relationship between trinucleotide (CAG) repeat length and clinical features of Huntington’s disease. Nat Genet 4:398–403. [DOI] [PubMed] [Google Scholar]
- 4.Anheim M, Mariani LL, Calvas P, et al. (2012). Exonic deletions of FXN and early-onset Friedreich ataxia. Arch Neurol 69:912–6. [DOI] [PubMed] [Google Scholar]
- 5.Anvret M, Ahlberg G, Grandell U, et al. (1993). Larger expansions of the CTG repeat in muscle compared to lymphocytes from patients with myotonic dystrophy. Hum Mol Genet 2:1397–400. [DOI] [PubMed] [Google Scholar]
- 6.Ashizawa T, Dubel JR, Dunne PW, et al. (1992). Anticipation in myotonic dystrophy. II. Complex relationships between clinical findings and structure of the GCT repeat. Neurology 42:1877–83. [DOI] [PubMed] [Google Scholar]
- 7.Aslandis C, Jansen G, Amemiya C, et al. (1992). Cloning of the essential myotonic dystrophy region and mapping of the putative defect. Nature 355:548–51. [DOI] [PubMed] [Google Scholar]
- 8.Assenza G, Benvenga A, Gennaro E, et al. (2017). A novel c132–134 del mutation in Unverricht-Lundborg disease and the review of literature of heterozygous compound patients. Epilepsia 58:e31–e35. [DOI] [PubMed] [Google Scholar]
- 9.Badhwar A, Jansen A, Andermann F, et al. (2004). Striking intrafamilial phenotypic variability and spastic paraplegia in the presence of similar homozygous expansions of the FRDA1 gene. Mov Disord 19:1424–31. [DOI] [PubMed] [Google Scholar]
- 10.Bahl S, Virdi K, Mittal U, et al. (2005). Evidence of a common founder for SCA12 in the Indian population. Ann Hum Genet 69:528–34. [DOI] [PubMed] [Google Scholar]
- 11.Banfi S, Servadio A, Chung MY, et al. (1994). Identification and characterization of the gene causing type 1 spinocerebellar ataxia. Nat Genet 7:513–20. [DOI] [PubMed] [Google Scholar]
- 12.Bell MV, Hirst MC, Nakahori Y, et al. (1991). Physical mapping across the fragile X: hypermethylation and clinical expression of the fragile X syndrome. Cell 64:861–6. [DOI] [PubMed] [Google Scholar]
- 13.Bidichandani SI, Ashizawa T, Patel PI (1998). The GAA triplet-repeat expansion in Friedreich ataxia interferes with transcription and may be associated with an unusual DNA structure. Am J Hum Genet 62:111–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boeynaems S, Bogaert E, Kovacs D, et al. (2017). Phase separation of C9orf72 dipeptide repeats perturbs stress granule dynamics. Mol Cell 65:1044–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brais B, Bouchard JP, Xie YG, et al. (1998). Short GCG expansions in the PABP2 gene cause oculopharyngeal muscular dystrophy. Nat Genet 18:164–7. [DOI] [PubMed] [Google Scholar]
- 16.Brook JD, McCurrach ME, Harley HG, et al. (1992). Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3’ end of a transcript encoding a protein kinase family member. Cell 68:799–808. [DOI] [PubMed] [Google Scholar]
- 17.Brussino A, Graziano C, Giobbe D, et al. (2010). Spinocerebellar ataxia type 12 identified in two Italian families may mimic sporadic ataxia. Mov Disord 25:1269–73. [DOI] [PubMed] [Google Scholar]
- 18.Burke JR, Wingfield MS, Lewis KE, et al. (1994). The Haw River syndrome: dentatorubropallidoluysian atrophy (DRPLA) in an African-American family. Nat Genet 7:521–4. [DOI] [PubMed] [Google Scholar]
- 19.Cabada T, Iridoy M, Jericó I, et al. (2017). Brain involvement in myotonic dystrophy Type 1: A morphometric and diffusion tensor imaging study with neuropsychological correlation. Arch Clin Neuropsychol 32:401–412. [DOI] [PubMed] [Google Scholar]
- 20.Campuzano V, Montermini L, Moltò MD, et al. (1996). Friedreich’s ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science 271:1423–7. [DOI] [PubMed] [Google Scholar]
- 21.Carrell ST, Carrell EM, Auerbach D, et al. (2016). Dmpk gene deletion or antisense knockdown does not compromise cardiac or skeletal muscle function in mice. Hum Mol Genet 25:4328–4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen JL, VanEtten DM, Fountain MA, et al. (2017). Structure and dynamics of RNA repeat expansions that cause Huntington’s Disease and myotonic dystrophy type 1. Biochemistry [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chio A, Borghero G, Restagno G, et al. (2012). Clinical characteristics of patients with familial amyotrophic lateral sclerosis carrying the pathogenic GGGGCC hexanucleotide repeat expansion of C9ORF72. Brain 135:784–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chung MY, Ranum LP, Duvick LA, et al. (1993). Evidence for a mechanism predisposing to intergenerational CAG repeat instability in spinocerebellar ataxia type I. Nat Genet 5:254–8. [DOI] [PubMed] [Google Scholar]
- 25.Chutake YK, Lam C, Costello WN, et al. (2014). Epigenetic promoter silencing in Friedreich ataxia is dependent on repeat length. Ann Neurol 76:522–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cleary JD, Ranum LP (2017). New developments in RAN translation: insights from multiple diseases. Curr Opin Genet Dev 44:125–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cooper-Knock J, Shaw PJ, Kirby J (2014). The widening spectrum of C9ORF72-related disease; genotype/phenotype correlations and potential modifiers of clinical phenotype. Acta Neuropathol 127:333–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Corben LA, Lynch D, Pandolfo M, et al. (2014). Consensus clinical management guidelines for Friedreich ataxia. Orphanet J Rare Dis 9:184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crespel A, Ferlazzo E, Franceschetti S, et al. (2016). Unverricht-Lundborg disease. Epileptic Disord 18:28–37. [DOI] [PubMed] [Google Scholar]
- 30.Daughters RS, Tuttle DL, Gao W, et al. (2009). RNA gain-of-function in spinocerebellar ataxia type 8. PLoS Genet 5:e1000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Day JW, Schut LJ, Moseley ML, et al. (2000). Spinocerebellar ataxia type 8: clinical features in a large family. Neurology 55:649–57. [DOI] [PubMed] [Google Scholar]
- 32.De Antonio M, Dogan C, Hamroun D, et al. (2016). Unravelling the myotonic dystrophy type 1 clinical spectrum: A systematic registry-based study with implications for disease classification. Rev Neurol (Paris) 172:572–580. [DOI] [PubMed] [Google Scholar]
- 33.De Boulle K, Verkerk AJ, Reyniers E, et al. (1993). A point mutation in the FMR-1 gene associated with fragile X mental retardation. Nat Genet 3:31–5. [DOI] [PubMed] [Google Scholar]
- 34.DeJesus-Hernandez M, Finch NA, Wang X, et al. (2017). In-depth clinico-pathological examination of RNA foci in a large cohort of C9ORF72 expansion carriers. Acta Neuropathol [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, et al. (2011). Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72:245–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Doyu M, Sobue G, Mukai E, et al. (1992). Severity of X-linked recessive bulbospinal neuronopathy correlates with size of the tandem CAG repeat in androgen receptor gene. Ann Neurol. 32:707–10. [DOI] [PubMed] [Google Scholar]
- 37a. Dürr A, Cossee M, Agid Y, et al. (1996). Clinical and genetic abnormalities in patients with Friedreich’s ataxia. N Engl J Med 335:1169–75. [DOI] [PubMed] [Google Scholar]
- 37b. Durr A, Stevanin G, Cancel G, et al. (1996). Spinocerebellar ataxia 3 and Machado-Joseph disease: clinical, molecular, and neuropathological features. Ann Neurol 39:490–9. [DOI] [PubMed] [Google Scholar]
- 38.Eichler EE, Holden JJ, Popovich BW, et al. (1993). Length of uninterrupted CGG repeats determines instability in the FMR1 gene. Nat Genet 5:168–73. [DOI] [PubMed] [Google Scholar]
- 39.Feng Y, Zhang F, Lokey LK, et al. (1994). Translational suppression by trinucleotide repeat expansion at FMR1. Nat Genet 8:88–94. [DOI] [PubMed] [Google Scholar]
- 40.Figueroa KP, Coon H, Santos N, et al. (2017). Genetic analysis of age at onset variation in spinocerebellar ataxia type 2. Neurol Genet 3:e155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Filla A, De Michele G, Cavalcanti F, et al. (1996). The relationship between trinucleotide (GAA) repeat length and clinical features in Friedreich ataxia. Am J Hum Genet 59:554–60. [PMC free article] [PubMed] [Google Scholar]
- 42.Freibaum BD, Taylor JP (2017). The role of dipeptide repeats in C9ORF72-related ALS-FTD. Front Mol Neurosci 10:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fu YH, Pizzuti A, Fenwick RG Jr, et al. (1991). An unstable triplet repeat in a gene related to myotonic muscular dystrophy. Cell 67:1047–58. [DOI] [PubMed] [Google Scholar]
- 44.Fu YH, Kuhl DP, Pizzuti A, et al. (1995). Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Science 268:731–4. [DOI] [PubMed] [Google Scholar]
- 45.Galea CA, Huq A, Lockhart PJ, et al. (2016). Compound heterozygous FXN mutations and clinical outcome in friedreich ataxia. Ann Neurol 79:485–95. [DOI] [PubMed] [Google Scholar]
- 46.Gallais B, Gagnon C, Mathieu J, et al. (2017). Cognitive decline over time in adults with myotonic dystrophy type 1: A 9-year longitudinal study. Neuromuscul Disord 2:61–72. [DOI] [PubMed] [Google Scholar]
- 47.Gedeon AK, Baker E, Robinson H, et al. (1992). Fragile X syndrome without CCG amplification has an FMR1 deletion. Nat Genet 19:341–4. [DOI] [PubMed] [Google Scholar]
- 48.Gendron TF, Petrucelli L (2017). Disease mechanisms of C9ORF72 repeat expansions. Cold Spring Harb Perspect Med pii: a024224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Genetic Modifiers of Huntington’s Disease (GeM-HD) Consortium (2015). Identification of genetic factors that modify clinical onset of Huntington’s disease. Cell 162:516–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Green KM, Linsalata AE, Todd PK (2016). RAN translation-What makes it run? Brain Res 1647:30–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gudde AE, van Heeringen SJ, de Oude AI, et al. (2017). Antisense transcription of the myotonic dystrophy locus yields low-abundant RNAs with and without (CAG)n repeat. RNA Biol 19:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haeusler AR, Donnelly CJ, Rothstein JD (2016). The expanding biology of the C9orf72 nucleotide repeat expansion in neurodegenerative disease. Nat Rev Neurosci 17:383–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hagerman PJ, Hagerman RJ (2015). Fragile X-associated tremor/ataxia syndrome. Ann N Y Acad Sci 1338:58–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Harley HG, Brook JD, Rundle SA, et al. (1992). Expansion of an unstable DNA region and phenotypic variation in myotonic dystrophy. Nature 355:545–6. [DOI] [PubMed] [Google Scholar]
- 55.Hoffman-Zacharska D, Mazurczak T, Zajkowski T, et al. (2016). Friedreich ataxia is not only a GAA repeats expansion disorder: implications for molecular testing and counselling. J Appl Genet 57:349–55. [DOI] [PubMed] [Google Scholar]
- 56.Holmes SE, O’Hearn EE, McInnis MG, et al. (1999). Expansion of a novel CAG trinucleotide repeat in the 5’ region of PPP2R2B is associated with SCA12. Nat Genet 23:391–2. [DOI] [PubMed] [Google Scholar]
- 57.Holmes SE, O’Hearn E, Rosenblatt A, et al. (2001). A repeat expansion in the gene encoding junctophilin-3 is associated with Huntington disease-like 2. Nat Genet 29:377–8. [DOI] [PubMed] [Google Scholar]
- 58.Huntington’s Disease Collaborative Research Group (1993). A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell 72:971–83. [DOI] [PubMed] [Google Scholar]
- 59.Igarashi S, Tanno Y, Onodera O, et al. (1992). Strong correlation between the number of CAG repeats in androgen receptor genes and the clinical onset of features of spinal and bulbar muscular atrophy. Neurology 42:2300–2. [DOI] [PubMed] [Google Scholar]
- 60.Ikeda Y, Dalton JC, Moseley ML, et al. (2004). Spinocerebellar ataxia type 8: molecular genetic comparisons and haplotype analysis of 37 families with ataxia. Am J Hum Genet 75:3–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ikeda Y, Ohta Y, Kobayashi H, et al. (2012). Clinical features of SCA36: a novel spinocerebellar ataxia with motor neuron involvement (Asidan). Neurology 79:333–41. [DOI] [PubMed] [Google Scholar]
- 62.Illarioshkin SN, Igarashi S, Onodera O (1994).Trinucleotide repeat length and rate of progression of Huntington’s disease. Ann Neurol 36:630–5. [DOI] [PubMed] [Google Scholar]
- 63.Ishiguro T, Sato N, Ueyama M, et al. (2017). Regulatory role of RNA chaperone TDP-43 for RNA misfolding and repeat-associated translation in SCA31. Neuron 94:108–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ishikawa K, Dürr A, Klopstock T, et al. (2011). Pentanucleotide repeats at the spinocerebellar ataxia type 31 (SCA31) locus in Caucasians. Neurology 77:1853–5. [DOI] [PubMed] [Google Scholar]
- 65.Joensuu T, Kuronen M, Alakurtti K, et al. (2007). Cystatin B: mutation detection, alternative splicing and expression in progressive myclonus epilepsy of Unverricht-Lundborg type (EPMI) patients. Eur J Hum Genet 15:185–93. [DOI] [PubMed] [Google Scholar]
- 66.Johansson J, Forsgren L, Sandgren O (1998). Expanded CAG repeats in Swedish spinocerebellar ataxia type 7 (SCA7) patients: effect of CAG repeat length on the clinical manifestation. Hum Mol Genet 7:171–6. [DOI] [PubMed] [Google Scholar]
- 67.Jones L, Houlden H, Tabrizi SJ (2017). DNA repair in the trinucleotide repeat disorders. Lancet Neurol 16:88–96. [DOI] [PubMed] [Google Scholar]
- 68.Kamsteeg E-J, Kress W, Catalli C, et al. (2012). Best practice guidelines and recommendations on the molecular diagnosis of myotonic dystrophy types 1 and 2. Eur J Hum Genet 20:1203–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kanadia RN, Johnstone KA, Mankodi A, et al. (2003). A muscleblind knockout model for myotonic dystrophy. Science 302:1978–80. [DOI] [PubMed] [Google Scholar]
- 70.Kawaguchi Y, Okamoto T, Taniwaki M, et al. (1994). CAG expansions in a novel gene for Machado-Joseph disease at chromosome 14q32.1. Nat Genet 8:221–8. [DOI] [PubMed] [Google Scholar]
- 71.Kearney M, Orrell RW, Fahey M, et al. (2016). Pharmacological treatments for Friedreich ataxia. Cochrane Database Syst Rev 30:CD007791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Knight SJL, Flannery AV, Hirst MC, et al. (1993). Trinucleotide repeat expansion and hypermethylation of a CpG island in FRAXE mental retardation. Cell 74:127–34. [DOI] [PubMed] [Google Scholar]
- 73.Kobayashi H, Abe K, Matsuura T, et al. (2011). Expansion of intronic GGCCTG hexanucleotide repeat in NOP56 causes SCA36, a type of spinocerebellar ataxia accompanied by motor neuron involvement. Am J Hum Genet 89:121–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Koide R, Ikeuchi T, Onodera O, et al. (1994). Unstable expansion of CAG repeat in hereditary dentatorubral-pallidoluysian atrophy (DRPLA). Nat Genet 6:9–13. [DOI] [PubMed] [Google Scholar]
- 75.Komure O, Sano A, Nishino N (1995). DNA analysis in hereditary dentatorubral-pallidoluysian atrophy: correlation between CAG repeat length and phenotypic variation and the molecular basis of anticipation. Neurology 45:143–9. [DOI] [PubMed] [Google Scholar]
- 76.Koob MD, Moseley ML, Schut LJ, et al. (1999). An untranslated CTG expansion causes a novel form of spinocerebellar ataxia (SCA8). Nat Genet 21:379–84. [DOI] [PubMed] [Google Scholar]
- 77.La Spada AR, Wilson EM, Lubahn DB, et al. (1991). Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature 352:77–9. [DOI] [PubMed] [Google Scholar]
- 78.Lalioti MD, Scott HS, Buresi C, et al. (1997). Dodecamer repeat expansion in cystatin B gene in progressive myoclonus epilepsy. Nature 386:847–51. [DOI] [PubMed] [Google Scholar]
- 79.Lalioti MD, Scott HS, Genton P, et al. (1998). A PCR amplification method reveals instability of the dodecamer repeat in progressive myoclonus epilepsy (EPM1) and no correlation between the size of the repeat and age at onset. Am J Hum Genet 62:842–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lehmer C, Oeckl P, Weishaupt JH, et al. (2017). Poly-GP in cerebrospinal fluid links C9orf72-associated dipeptide repeat expression to the asymptomatic phase of ALS/FTD. EMBO Mol Med [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li L, Ng NK, Koon AC, et al. (2017). Expanded polyalanine tracts function as nuclear export signals and promote protein mislocalization via eEF1A1 factor. J Biol Chem 292:5784–5800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li Y, Lu Y, Polak U, et al. (2015). Expanded GAA repeats impede transcription elongation through the FXN gene and induce transcriptional silencing that is restricted to the FXN locus. Hum Mol Genet 24:6932–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li YJ, Minear MA, Rimmler J, et al. (2011). Replication of TCF4 through association and linkage studies in late-onset Fuchs endothelial corneal dystrophy. PLoS One 6:e18044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liquori CL, Ricker K, Moseley ML, et al. (2001). Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science 293:864–7. [DOI] [PubMed] [Google Scholar]
- 85.Liu W, Ikeda Y, Hishikawa N, et al. (2014). Characteristic RNA foci of the abnormal hexanucleotide GGCCUG repeat expansion in spinocerebellar ataxia type 36 (Asidan). Eur J Neurol 21:1377–86. [DOI] [PubMed] [Google Scholar]
- 86.Luigetti M, Lo Monaco M, Mirabella M, et al. (2015). Oculopharyngeal muscular dystrophy: Clinical and neurophysiological features. Clin Neurophysiol 126:2406–8. [DOI] [PubMed] [Google Scholar]
- 87.Lynch DR, Regner SR, Schadt KA, et al. (2012). Management and therapy for cardiomyopathy in Friedreich’s ataxia. Expert Rev Cardiovasc Ther 10:767–77. [DOI] [PubMed] [Google Scholar]
- 88.Majounie E, Reoton EA, Mok K, et al. (2012). Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. The Lancet Neurology 11:323–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Margolis JM, Schoser BG, Moseley ML, et al. (2006). DM2 intronic expansions: evidence for CCUG accumulation without flanking sequence or effects on ZNF9 mRNA processing or protein expression. Hum Mol Genet 15:1808–15. [DOI] [PubMed] [Google Scholar]
- 90.Maruyama S, Saito Y, Nakagawa E (2012). Importance of CAG repeat length in childhood-onset dentatorubral-pallidoluysian atrophy. J Neurol 259:232934. [DOI] [PubMed] [Google Scholar]
- 91.Mason AG, Tomé S, Simard JP, et al. (2013). Expression levels of DNA replication and repair genes predict regional somatic repeat instability in the brain but are not altered by polyglutamine disease protein expression or age. Hum Mol Genet 23:1606–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Matsuura T, Fang P, Lin X, et al. (2004). Somatic and germline instability of the ATTCT repeat in spinocerebellar ataxia type 10. Am J Hum Genet 74:1216–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Matsuura T, Yamagata T, Burgess DL, et al. (2000). Large expansion of the ATTCT pentanucleotide repeat in spinocerebellar ataxia type 10. Nat Genet 26:191–4. [DOI] [PubMed] [Google Scholar]
- 94.McFarland KN, Liu J, Landrian I, et al. (2014). Repeat interruptions in spinocerebellar ataxia type 10 expansions are strongly associated with epileptic seizures. Neurogenetics 15:59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Melacini P, Villanova C, Menegazzo E (1995). Correlation between cardiac involvement and CTG trinucleotide repeat length in myotonic dystrophy. J Am Coll Cardiol 25:239–45. [DOI] [PubMed] [Google Scholar]
- 96.Montermini L, Richter A, Morgan K, et al. (1997). Phenotypic variability in Friedreich ataxia: role of the associated GAA triplet repeat expansion. Ann Neurol 41:675–82. [DOI] [PubMed] [Google Scholar]
- 97.Moseley ML, Zu T, Ikeda Y, et al. (2006). Bidirectional expression of CUG and CAG expansion transcripts and intranuclear polyglutamine inclusions in spinocerebellar ataxia type 8. Nat Genet 38:758–69. [DOI] [PubMed] [Google Scholar]
- 98.Myers RH, MacDonald ME, Koroshetz WJ, et al. (1992). De novo expansion of a (CAG)n repeat in sporadic Huntington’s disease. Ann Neurol 32:707–10. [DOI] [PubMed] [Google Scholar]
- 99.Nagafuchi S, Yanagisawa H, Sato K, et al. (1994). Dentatorubral and pallidoluysian atrophy expansion of an unstable CAG trinucleotide on chromosome 12p. Nat Genet 6:14–8. [DOI] [PubMed] [Google Scholar]
- 100.Nakamura K, Yoshida K, Matsushima A, et al. (2017). Natural history of Spinocerebellar Ataxia Type 31: a 4-year prospective study. Cerebellum 16:518–524. [DOI] [PubMed] [Google Scholar]
- 101.Niimi Y, Takahashi M, Sugawara E, et al. (2013). Abnormal RNA structures (RNA foci) containing a penta-nucleotide repeat (UGGAA)n in the Purkinje cell nucleus is associated with spinocerebellar ataxia type 31 pathogenesis. Neuropathology 33:600–11. [DOI] [PubMed] [Google Scholar]
- 102.Obayashi M, Stevanin G, Synofzik M, et al. (2015). Spinocerebellar ataxia type 36 exists in diverse populations and can be caused by a short hexanucleotide GGCCTG repeat expansion. J Neurol Neurosurg Psychiatry 86:986–95. [DOI] [PubMed] [Google Scholar]
- 103.Oberlé I, Rousseau F, Heitz D, et al. (1992). Instability of a 550-base pair DNA segment and abnormal methylation in fragile X syndrome. Science 252:1097–102. [DOI] [PubMed] [Google Scholar]
- 104.O’Hearn E, Holmes SE, Calvert PC, et al. (2001). SCA-12: Tremor with cerebellar and cortical atrophy is associated with a CAG repeat expansion. Neurology 56:299–303. [DOI] [PubMed] [Google Scholar]
- 105.Orr HT, Chung MY, Banfi S, et al. (1993). Expansion of an unstable trinucleotide CAG repeat in spinocerebellar ataxia type 1. Nat Genet 4:221–6. [DOI] [PubMed] [Google Scholar]
- 106.Parrish JE, Oostra BA, Verkerk AJ, et al. (1994). Isolation of a GCC repeat showing expansion in FRAXF, a fragile site distal to FRAXA and FRAXE. Nat Genet 8:229–35. [DOI] [PubMed] [Google Scholar]
- 107.Paulson HL, Fischbeck KH (1996). Trinucleotide repeats in neurogenetic disorders. Ann Rev Neurosc. 119:79–107. [DOI] [PubMed] [Google Scholar]
- 108.Penney JB Jr, Vonsattel JP, MacDonald ME (1997). CAG repeat number governs the development rate of pathology in Huntington’s disease. Ann Neurol 41:689–92. [DOI] [PubMed] [Google Scholar]
- 109.Peric S, Rakocevic Stojanovic V, Mandic Stojmenovic G, et al. (2017). Clusters of cognitive impairment among different phenotypes of myotonic dystrophy type 1 and type 2. Neurol Sci 38:415–423. [DOI] [PubMed] [Google Scholar]
- 110.Philips AV, Timchenko LT, Cooper TA (1998). Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science 280:737–41. [DOI] [PubMed] [Google Scholar]
- 111.Pulst SM, Santos N, Wang D, et al. (2005). Spinocerebellar ataxia type 2: polyQ repeat variation in the CACNA1A calcium channel modifies age of onset. Brain 128:2297–303. [DOI] [PubMed] [Google Scholar]
- 112.Renton AE, Majounie E, Waite A, et al. (2011). A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72:257–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Richard P, Trollet C, Stojkovic T, et al. (2017). Correlation between PABPN1 genotype and disease severity in oculopharyngeal muscular dystrophy. Neurology 88:359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Riess O, Schöls L, Bottger H, et al. (1997). SCA6 is caused by moderate CAG expansion in the alpha1A-voltage-dependent calcium channel gene. Hum Mol Genet. 6:1289–93. [DOI] [PubMed] [Google Scholar]
- 115.Rosenblatt A, Kumar BV, Mo A (2012). Age, CAG repeat length, and clinical progression in Huntington’s disease. Mov Disord 27:272–6. [DOI] [PubMed] [Google Scholar]
- 116.Ross OA, Rutherford NJ, Baker M, et al. (2011). Ataxin-2 repeat-length variation and neurodegeneration. Hum Mol Genet 20:3207–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Russ J, Liu EY, Wu K (2015). Hypermethylation of repeat expanded c9orf72 is a clinical and molecular disease modifier. Acta Neuropathol 129:39–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sansone VA, Brigonzi E, Schoser B, et al. (2013). The frequency and severity of cardiac involvement in myotonic dystrophy type 2 (DM2): Long-term outcomes. Int J Cardiol 168:1147–53. [DOI] [PubMed] [Google Scholar]
- 119.Sato N, Amino T, Kobayashi K, et al. (2009). Spinocerebellar ataxia type 31 is associated with “inserted” penta-nucleotide repeats containing (TGGAA)n. Am J Hum Genet 85:544–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Schmidt MH, Pearson CE (2016). Disease-associated repeat instability and mismatch repair. DNA Repair (Amst) 38:117–26. [DOI] [PubMed] [Google Scholar]
- 121.Schneider C, Ziegler A, Ricker K, et al. (2000). Proximal myotonic myopathy: evidence for anticipation in families with linkage to chromosome 3q. Neurology 55:383–8. [DOI] [PubMed] [Google Scholar]
- 122.Schöls L, Vieira-Saecker AM, Schöls S, et al. (1995). Trinucleotide expansion within the MJD1 gene presents clinically as spinocerebellar ataxia and occurs most frequently in German SCA patients. Hum Mol Genet 4:1001–5. [DOI] [PubMed] [Google Scholar]
- 123.Seixas AI, Holmes SE, Takeshima H, et al. (2012). Loss of junctophiin-3 contributes to Huntington disease-like 2 pathogenesis. Ann Neurol 71:245–57. [DOI] [PubMed] [Google Scholar]
- 124.Shanmugam V, Dion P, Rochefort D, et al. (2000). PABP2 polyalanine tract expansion causes intranuclear inclusions in oculopharyngeal muscular dystrophy. Ann Neurol 48:798–802. [PubMed] [Google Scholar]
- 125.Shoubridge C, Gecz J (2012). Polyalanine tract disorders and neurocognitive phenotypes. Adv Exp Med Biol 769:185–203. [DOI] [PubMed] [Google Scholar]
- 126.Snell RG, MacMillan JC, Cheadle JP, et al. (1993). Relationship between trinucleotide repeat expansion and phenotypic variation in Huntington’s disease. Nat Genet 4:393–7. [DOI] [PubMed] [Google Scholar]
- 127.Soliman AZ, Xing C, Radwan SH, et al. (2015). Correlation of severity of Fuchs endothelial corneal dystrophy with triplet repeat expansion in TCF4. JAMA Ophthalmol 133:1386–91. [DOI] [PubMed] [Google Scholar]
- 128.Srivastava AK, Choudhry S, Gopinath MS, et al. (2001). Molecular and clinical correlation in five Indian families with spinocerebellar ataxia 12. Ann Neurol 50:796–800. [DOI] [PubMed] [Google Scholar]
- 129.Stine OC, Pleasant N, Franz ML (1993). Correlation between the onset age of Huntington’s disease and length of the trinucleotide repeat in IT-15. Hum Mol Genet 2:1547–9. [DOI] [PubMed] [Google Scholar]
- 130.Sutcliffe JS, Nelson DL, Zhang F, et al. (1992). DNA methylation represses FMR-1 transcription in fragile X syndrome. Hum Mol Genet 1:397–400. [DOI] [PubMed] [Google Scholar]
- 131.Swami M, Hendricks AE, Gillis T, et al. (2009). Somatic expansion of the Huntington’s disease CAG repeat in the brain is associated with an earlier age of disease onset. Hum Mol Genet 18:3039–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Teive HA, Ashizawa T (2013). Spinocerebellar ataxia type 10: from Amerindians to Latin Americans. Curr Neurol Neurosci Rep 13:393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Teive HA, Munhoz RP, Arruda WO, et al. (2012). Spinocerebellar ataxias: genotype-phenotype correlations in 104 Brazilian families. Clinics (Sao Paulo) 67:443–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Telenius H, Kremer B, Goldberg YP, et al. (1994). Somatic and gonadal mosaicism of the Huntington disease gene CAG repeat in brain and sperm. Nat Genet 6:409–14. [DOI] [PubMed] [Google Scholar]
- 135a.Tezenas du Montcel S, Durr A, Bauer P, et al. (2014). Modulation of the age at onset in spinocerebellar ataxia by CAG tracts in various genes. Brain 137:2444–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135b.Tezenas du Montcel S, Durr A, Rakowicz M, et al. (2014). Predication of the age at onset in spinocerebellar ataxia type 1, 2, 3, and 6. J Med Genet 51:479–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Thornton CA, Wang E, Carrell EM (2017). Myotonic dystrophy: approach to therapy. Curr Opin Genet Dev 44:135–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Todd PK, Oh SY, Krans A, et al. (2013). CGG repeat-associated translation mediates neurodegeneration in fragile X tremor ataxia syndrome. Neuron 78:440–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Trang H, Stanley SY, Thorner P, et al. (2015). Massive CAG repeat expansion and somatic instability in maternally transmitted infantile spinocerebellar ataxia type 7. JAMA Neurol 72:219–23. [DOI] [PubMed] [Google Scholar]
- 139.Trollet C, Gidaro T, Klein P et al. (2001). Oculopharyngeal Muscular Dystrophy In: Pagon RA, Adam MP, Ardinger HH et al. Gene Reviews (Internet). University of Washington, Seattle. [PubMed] [Google Scholar]
- 140.Trottier Y, Briancalana V, Mandel JL (1994). Instability of CAG repeats in Huntington’s disease: relation to parental transmission and age of onset. J Med Genet 31:377–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Tsilfidis C, MacKenzie AE, Mettler G, et al. (1994). Correlation between CTG trinucleotide repeat length and frequency of severe congenital myotonic dystrophy. Nat Genet 1:192–5. [DOI] [PubMed] [Google Scholar]
- 142.van Blitterswijk M, DeJesus-Hernandez M, Niemantsverdriet E, et al. (2013). Association between repeat sizes and clinical and pathological characteristics in carriers of C9ORF72 repeat expansions (Xpansize-72): a cross-sectional cohort study. Lancet Neurol 12:978–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Van Damme P, Veldink JH, van Blitterswijk M, et al. (2011). Expanded ATXN2 CAG repeat size in ALS identifies genetic overlap between ALS and SCA2. Neurology 76:2066–72. [DOI] [PubMed] [Google Scholar]
- 144.Van Mossevelde S, van der Zee J, Cruts M, et al. (2017). Relationship between C9orf72 repeat size and clinical phenotype. Curr Opin Genet Dev 44:117–124. [DOI] [PubMed] [Google Scholar]
- 145.Veitch NJ, Ennis M, McAbney JP, et al. (2007). Inherited CAG.CTG allele length is a major modifier of somatic mutation length variability in Huntington disease. DNA Repair (Amst) 6:789–96. [DOI] [PubMed] [Google Scholar]
- 146.Virtaneva K, D’amato E, Miao J et al. (1997). Unstable minisatellite expansion causing recessively inherited myoclonus epilspsy, EPM1. Nat Genet 15:393–6. [DOI] [PubMed] [Google Scholar]
- 147.Walters WH, Murphy RM (2009). Examining polyglutamine peptide length: a connection between collapsed conformations and increased aggregation. J Mol Biol 393:978–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wieben ED, Aleff RA, Tang X, et al. (2017). Trinucleotide repeat expansion in the transcription factor 4 (TCF4) gene leads to widespread mRNA splicing changes in Fuchs’ endothelial corneal dystrophy. Invest Ophthalmol Vis Sci 58:343–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wieben ED, Aleff RA, Tosakulwong N, et al. (2012). A common trinucleotide repeat expansion within the transcription factor 4 (TCF4, E2–2) gene predicts Fuchs corneal dystrophy. PLoS One 7:e49083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Yoshida K, Asakawa M, Suzuki-Kouyama E, et al. (2014). Distinctive features of degenerating Purkinje cells in spinocerebellar ataxia type 31. Neuropathology 34:261–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Zhang N, Ashizawa T (2017). RNA toxicity and foci formation in microsatellite expansion diseases. Curr Opin Genet Dev 44:17–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zu T, Gibbens B, Doty NS, et al. (2011). Non-ATG-initiated translation directed by microsatellite expansions. Proc Natl Acad Sci USA 108:260–5. [DOI] [PMC free article] [PubMed] [Google Scholar]